Abstract
Purpose:
Encorafenib + cetuximab (E+C) is an effective therapeutic option in chemorefractory BRAFV600E metastatic colorectal cancer (mCRC). However, there is a need to improve the efficacy of this molecular-targeted therapy and evaluate regimens suitable for untreated BRAFV600E in patients with mCRC.
Experimental Design:
We performed a series of in vivo studies using BRAFV600E mCRC tumor xenografts. Mice were randomized to receive 5-fluoruracil (5-FU), irinotecan, or oxaliplatin regimens (FOLFIRI or FOLFOX), (E+C) or the combination. Patients received long-term treatment until disease progression, with deescalation strategies used to mimic maintenance therapy. Transcriptomic changes after progression on cytotoxic chemotherapy or targeted therapy were assessed.
Results:
Antitumor activity of either FOLFIRI or E+C was better as first-line treatment as compared with second-line, with partial cross-resistance seen between a cytotoxic regimen and targeted therapy with an average 62% loss of efficacy for FOLFIRI after E+C and a 45% loss of efficacy of E+C after FOLFIRI (P < 0.001 for both). FOLFIRI-treated models had upregulation of epithelial–mesenchymal transition (EMT) and MAPK pathway activation, where E+C treated models had suppressed MAPK signaling. In contrast, with chemotherapy with E+C, EMT and MAPK signaling remained suppressed. FOLFOX or FOLFIRI, each in combination with E+C, were the most active first-line treatments as compared with E+C or to chemotherapy alone. Furthermore, FOLFOX in combination with E+C as first-line induction therapy, followed by E+C ± 5-FU as maintenance therapy, was the most effective strategy for long-term disease control.
Conclusions:
These results support the combination of cytotoxic chemotherapy and molecular-targeted therapy as a promising therapeutic approach in the first-line treatment of BRAFV600E mCRC.
Translational Relevance.
The dual blockade with encorafenib + cetuximab is an effective therapeutic option in chemorefractory BRAFV600E-mutant metastatic colorectal cancer (mCRC). A relevant clinical issue is how to improve the efficacy of this molecular targeted therapy in this poor-prognosis subtype. Here, we report the results of an in vivo preclinical study with the aim of defining treatment strategies that could help to improve the clinical management of patients with BRAFV600E-mutant mCRC, by using relevant preclinical models for BRAFV600E-mutant mCRC, such as four patient-derived tumor xenografts as well as HT29-derived tumor xenografts. In this study, we have compared different potential therapeutic strategies of combining and sequencing chemotherapy regimens with encorafenib and cetuximab and explored the potential mechanism associated with adaptive resistance to the treatments. In conclusion, these in vivo preclinical results provide a rationale for the combination of chemotherapy and molecular targeted drugs as potential treatment of choice for the first-line setting, as it is currently being evaluated in the ongoing BREAKWATER randomized phase III study in patients with BRAFV600E-mutant mCRC.
Introduction
Colorectal cancer is the third most commonly diagnosed cancer in males and the second in females, with 1.9 million new cases and 0.9 million deaths in 2020 according to the World Health Organization (WHO; ref. 1). There have been significant developments in colorectal cancer research over the last few years, enabling us to better characterize individual tumors and to classify them according to certain molecular or genetic features (2–5). Mutations of BRAF are found in 8% to 15% of patients with colorectal cancer. The most frequent mutation is BRAFV600E (6). Detection of the BRAFV600E mutation has relevant clinical, prognostic, and therapeutic implications. Previous studies have demonstrated higher rates of peritoneal and metastatic lymph node involvement, less benefit from standard chemotherapy treatments, and shorter survival in patients with the BRAFV600E mutation (6). A subgroup of BRAFV600E-mutant colorectal cancer could be also associated with high levels of microsatellite instability (MSI-H) and may arise from sessile serrated adenomas (6, 7).
As BRAFV600E-mutant colorectal cancer has emerged as a distinct clinical entity, multiple clinical trials have tried to replicate the successes of targeted therapies that have been developed for BRAFV600E-mutant melanoma (8). However, less than 10% patients with BRAFV600E-mutant metastatic CRC (mCRC) responded to BRAF inhibitor monotherapy in early-phase clinical trials (8). A key finding was the identification of a specific molecular adaptive feedback after BRAF inhibition, which results in increased signaling through activation of the EGFR pathway (9, 10). This discovery led to clinical studies that have evaluated the combination of BRAF and EGFR inhibitors, with promising antitumor activity (11–14). The results of the BEACON CRC study provide first support for encorafenib + cetuximab (E+C) to be a new standard of care for patients with BRAFV600E mCRC who have received prior systemic therapy (15). However, a major challenge is to improve the efficacy of this molecular targeted combination and to define its role in earlier phases of the disease, such as in the first-line setting. Finally, the ANCHOR CRC study is the first prospective study using a BRAF inhibitor–based therapy in first-line BRAFV600E mCRC, although the durability of response did not appear superior to cytotoxic regimens (16). The optimal approach to combining targeted therapy with cytotoxic chemotherapy is needed, to guide subsequent clinical trials.
Here, we report the results of preclinical in vivo studies, in which we have evaluated and compared different potential therapeutic strategies of combining and sequencing chemotherapy regimens with encorafenib and cetuximab. We have investigated which types of combination treatments and corresponding sequences of administration could optimize the therapeutic efficacy of blocking BRAFV600E-mutant signaling with E+C. Furthermore, we explored the potential mechanism associated with adaptive resistance to the treatments.
Materials and Methods
Tumor xenografts
Four different patient-derived tumor xenografts (PDXs) were established from tissue biopsies collected from patients with mCRC under a research laboratory protocol approved by UT MD Anderson Cancer Center Institutional Review Board (IRB, Houston, TX), and all patients provided written informed consent for specimens to be used for research purposes including implantation in xenografts. All in vivo experiments and procedures were approved by Institutional Animal Care and Use Committee. All in vivo experiments utilizing PDXs were performed according to NIH NCI recommendations summarized in SOP50102: PDX Implantation, Expansion, and Cryopreservation (subcutaneous).
Low-passage tumors were implanted subcutaneously (s.c.) into 4- to 6-week-old female immunodeficient nu/nu mice from Envigo Laboratories for expansion. Once tumors reached 2,000 mm3, tumors were collected, cut into 2 mm3 pieces, and implanted into cohorts of 20 to 40 nu/nu mice for consequent experiments. Four PDXs (B1003b, BB8140, C5002, and C5003) were used. Each PDX harbored a BRAFV600E mutation and were derived from patients naïve to BRAF- or EGFR-targeted therapy. BB8140, C5002, and C5003 are microsatellite-stable tumors, whereas B1003b is a MSI-H tumor. In addition, the HT29 colorectal cancer cell line (which possesses the BRAFV600E mutation) was purchased from the ATCC. The cell line was passaged in vitro in McCoy 5A Medium (Sigma-Aldrich), which was supplemented with 10% FBS (Sigma-Aldrich),100 U/mL penicillin and 100 μg/mL streptomycin. The HT29 cell line was routinely screened for the presence of Mycoplasma (Mycoplasma Detection Kit, Roche Diagnostics). To establish an in vivo model, 106 HT29 cells were suspended in 200 mL of Matrigel (BD Biosciences) and injected subcutaneously into female nude mice.
Evaluation of efficacy in PDX models across lines of therapy
Four- to 6-week-old female balb/c athymic (nuÞ/nuÞ) mice were purchased from Charles River Laboratories. Mice were maintained in accordance with the institutional guidelines of the MD Anderson and University of Campania “L. Vanvitelli” Animal Care and Use Committee. Two PDX models were implanted subcutaneously in the lateral flanks. Once tumors reached 200 mm3, mice were randomized into either vehicle, FOLFIRI, or E+C treatments. Encorafenib was administered by oral gavage (5 mg/kg) twice daily. For FOLFIRI treatment, mice were injected intraperitoneally with irinotecan (40 mg/kg) and levofolinate calcium (30 mg/kg) first, followed by 5-fluorouracil (5-FU; 55 mg/kg), once a week. Cetuximab at the dose of 1 mg was injected intraperitoneally twice a week. The mice were treated until their tumors reached a size that was 100% larger than the starting size. Tumor size and body weight were measured twice a week. At 100% progression, tumors were collected and passaged for second-line treatments. Tumor fragments at a size of 2 mm3 were implanted into the flanks of new mice. Once tumors reached 200 mm3, they were randomized to the alternate therapy they had received in the first-line (i.e., FOLFIRI → E+C; or E+C → FOLFIRI) or to the same therapy they had received in the first-line (i.e., FOLFIRI → FOLFIRI; E+C → E+C). Mice were treated until tumors reached a size of 100% larger than the starting size, mice were sacrificed, and the tumors were harvested. Tumor size and body weights were measured twice a week.
Evaluation of combination treatments
To assess the utility of combination therapy, mice from PDX models were randomized to one of four arms: vehicle; FOLFIRI; E+C; or FOLFIRI + encorafenib and cetuximab. Mice were treated for 21 days, with tumor size and body weight measured bi weekly. Separately, HT29-bearing mice were distributed into 10 groups consisting of 10 animals per group: vehicle; cetuximab; encorafenib; cetuximab + encorafenib; FOLFIRI; FOLFIRI + encorafenib; FOLFIRI + encorafenib and cetuximab; FOLFOX; FOLFOX + encorafenib; FOLFOX + encorafenib, and cetuximab. After 2 weeks, when tumors reached a mean volume of 200 to 400 mm3, treatment was initiated. Cetuximab, encorafenib, and FOLFIRI-based regimen were dosed as previously described. For modeling of the FOLFOX regimen, mice were injected intraperitoneally with oxaliplatin (12 mg/kg) and levofolinate calcium (30 mg/kg) first, followed by 5-FU (55 mg/kg), once a week. Treatment was continued for 6 weeks. Subsequently, for the maintenance regimen experiment, 10 mice/group were treated with vehicle, FOLFOX + encorafenib, or FOLFOX + encorafenib and cetuximab. At the completion of 21 days of treatment, that was defined as “induction treatment,” mice in the FOLFOX + encorafenib arm switched to encorafenib alone; otherwise, mice in the FOLFOX + encorafenib and cetuximab arm switched to E+C. This “maintenance treatment” was continued for 8 weeks, and afterward, animals were followed for an additional 17-week follow-up period. For a third experiment, 40 mice were treated for 21 days with vehicle or with FOLFOX + encorafenib and cetuximab. At the end of this induction treatment, treated mice were randomized into three groups (10 mice/group) and treated for 8 weeks with 5-FU alone, with E+C or with the combination of 5-FU + encorafenib and cetuximab. After this maintenance treatment, mice were followed for an additional 17 weeks. The mice were euthanized after tumors grew to 2,000 mm3 or after they became moribund. Duration from treatment initiation to sacrifice was used as a surrogate of overall survival (OS).
RNA sequencing and analysis
RNA was collected using Qiagen RNEasy Micro Kit (catalog No. 74004, Qiagen), according to the manufacturer's protocol. RNA was sent to Admera for sequencing. STAR (version 2.7.2b) aligner (17) was used for mapping raw BAM/FASTQ files to the human reference genome (GRCh38), and Biobambam (version 0.0.191; ref. 18) was used to mark duplicate reads. GENCODE v22 (19) was used for annotation. Reads were counted using HTSeq (version 0.11.0; ref. 20). Raw reads from PDX model samples were first processed with Xenome (version 1.0.1; ref. 21) for the classification of human and mouse reads. Classified human reads were processed as described above. Mouse reads were mapped and annotated using a mouse reference genome (GRCm39), and GENCODE vM26. DESeq2 (version 1.36.0; ref. 22) was used for normalization and differential gene expression analysis. Genes that are not expressed were filtered prior to normalization. Genes with an absolute log2 fold-change over 1.5 and FDR < 0.05 were considered differentially expressed. The fgsea (v1.22.0; 23) package was used for Gene Set Enrichment Analysis (GSEA) with genes preranked by fold-change values and visualized using the ggplot2 (24) package. MAPK-activation score was calculated as previously described (25) and relative gene expression was visualized using the pheatmap (26) package.
Histology procedures, whole slide imaging, and quantitative image analysis
Tissues were collected in 10% neutral buffered formalin and processed routinely after 24 to 48 hours of fixation. After processing, tissues were paraffin-embedded and sectioned 3 to 4 μm thick with a microtome and placed on positively charged glass slides. Sections were deparaffinized and stained with hematoxylin and eosin (H&E). IHC staining was completed using a Leica Bond RX autostainer with Leica detection kit. Brightfield whole slide imaging was performed at 20× magnification using an Aperio AT2 scanner. Image analysis was performed using tuned Leica algorithms specific for each marker and evaluated in Imagescope/eslide manager. Images for figures were captured using Imagescope v12.4.3.5008. PhosphoErk1/2 (No.4370) antibody used for IHC staining was from Cell Signaling Technology. Pancytokeratin (No.10403R) antibody was from Bioss Antibodies Inc.
Statistical analysis
The statistical analyses were carried out using the GraphPad Prism (version 9 for Windows) and SPSS package (version 21.0 for Windows, SPSS Inc.). The Student t test was used to evaluate the statistical significance of differences between treatments. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. All the tests were two-sided, with P < 0.05 to indicate statistical significance.
Data availability
The raw data generated in this study are available upon request from the corresponding author.
Results
Cancer cell resistance to first-line therapy impairs efficacy of second-line therapy in BRAFV600E PDXs
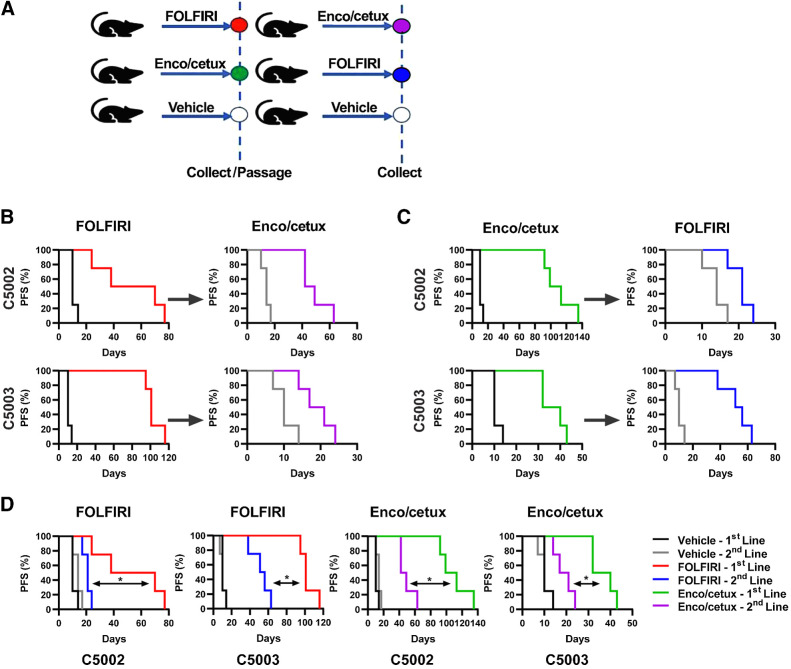
In order to better understand the optimal sequence of administering molecular targeted therapies and chemotherapies in BRAFV600E-mutant mCRC, we first designed an in vivo experiment with PDX (Fig. 1A). Mice were randomized to receive either FOLFIRI (27) or E+C. Mice were treated until tumor progression. Tumors were then collected and directly reimplanted into mice for the second-line treatment, that consisted in the alternate therapy (Fig. 1A).
Figure 1.
Resistance to first-line therapy impairs the efficacy of second-line therapy. BRAFV600E-mutated colorectal cancer PDX models C5002 and C5003 were treated with FOLFIRI or encorafenib + cetuximab (Enco/cetux) until progression. Cross-over to alternate therapy until progression. A,In vivo sequencing of therapy model. B, Progression-free survival (PFS) of C5002 and C5003 for FOLFIRI first-line treatment and E+C second-line treatment (n = 4). C, PFS for E+C first-line treatment and FOLFIRI second-line treatment (n = 4). D, PFS for first and second-line FOLFIRI or E+C treatment. (*, P < 0.05)
In particular, in C5002 and C5003 PDX models, we compared the efficacy of FOLFIRI as first-line treatment, E+C as second-line treatment (Fig. 1B), E+C as first-line treatment, and FOLFIRI as second-line (Fig. 1C). In the C5002 PDX model, E+C had greater antitumor activity than FOLFIRI in first-line treatment, whereas both regimens performed similarly in second-line treatment (Fig. 1B and C). In addition, both E+C and FOLFIRI were significantly more effective as first-line as compared with second-line treatment (P = 0.0067 and P = 0.0062, respectively; Fig. 1D). Importantly, prior treatment with FOLFIRI reduced the duration of disease control with E+C, with a similar effect of prior E+C on FOLFIRI efficacy. In the C5003 PDX model, E+C and FOLFIRI had better antitumor activity in first-line than in second-line treatment (P = 0.0062 and P = 0.0180, respectively; Fig. 1B–D). In contrast to C5002 PDX, FOLFIRI had greater efficacy than E+C as first-line treatment in C5003 PDX (Fig. 1B–D). Similar to C5002, prior treatment with the alternate regimen reduced subsequent efficacy. Finally, to confirm that these tumors were resistant to first-line therapies, we also re-treated resistant C5002 and C5003 PDX with the same first-line therapy with little or no response.
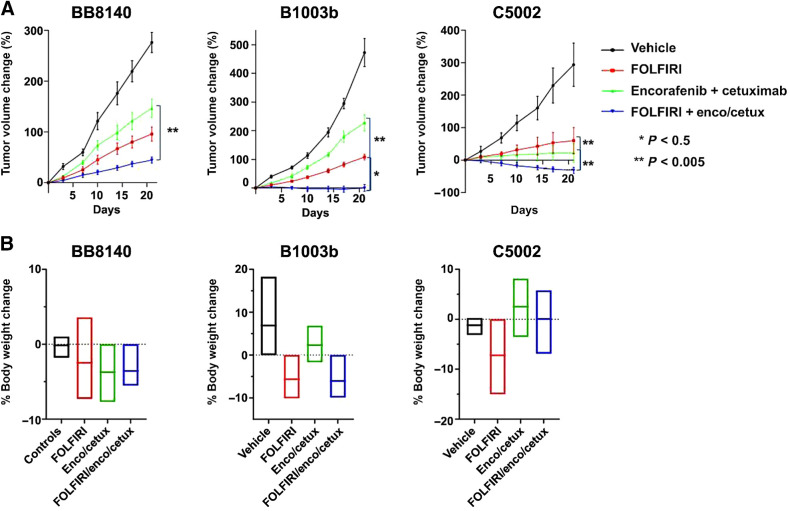
Figure 2.
Additive benefit of cytotoxic chemotherapy to BRAF + EGFR–targeted therapy. Mice were randomized into one of four arms and dosed for 21 days: vehicle, FOLFIRI, E+C, or FOLFIRI + E+C. A, Percent change in tumor volume from day 0, ± SD of n = 8. B, Mean percent change in body weights from day 0, ± SD of n = 8. *, P < 0.05; **, P < 0.005.
In vivo efficacy of FOLFIRI in combination with BRAF + EGFR inhibitors
Next, we evaluated the therapeutic potential of combining FOLFIRI with E+C. Three PDX models were used. Mice were randomized to single treatments or to the combination of chemotherapy plus molecular targeted agents and were treated for 21 days. FOLFIRI + encorafenib and cetuximab was the most active among all the treatment arms in the three PDXs (Fig. 2A). Tumor growth inhibition varied among the three PDXs. Significant tumor regression was observed in C5002 PDX. Significant tumor growth inhibition occurred as best response in B1003b PDX, whereas delayed tumor progression was reported in BB8140 PDX. The mice tolerated the treatment well and were able to maintain their body weight. (Fig. 2B).
Pathway analysis following chemotherapy and/or molecular targeted therapy in BRAFV600E PDX
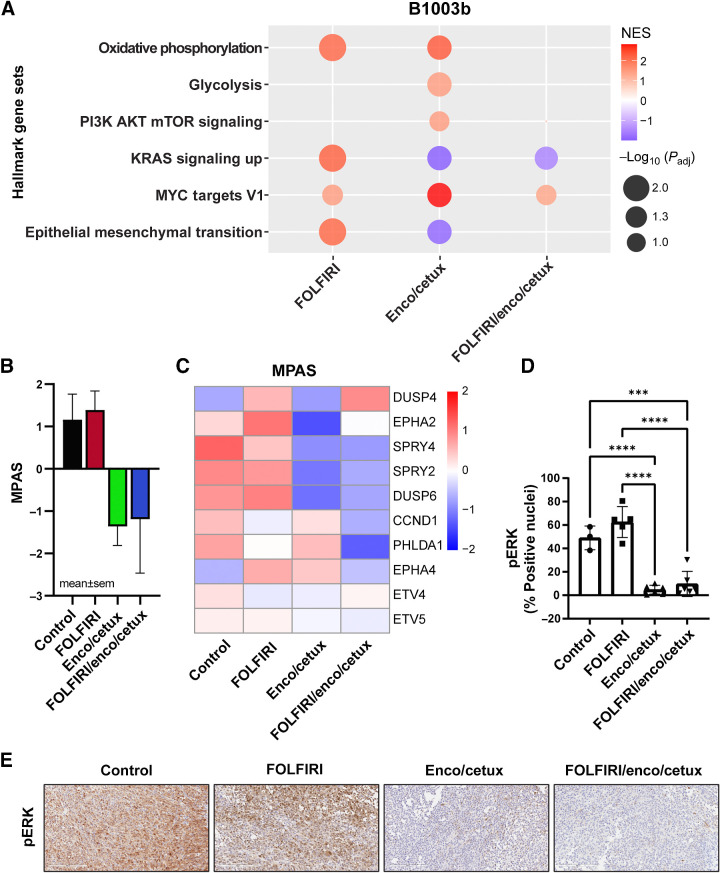
In order to gain knowledge on the differential transcriptomic changes induced by treatment, RNA was extracted and sequenced from B1003 PDX samples after 21 days of treatment. Pathway analysis demonstrated several differentially expressed gene signatures (Supplementary Fig. S1). Of interest, the oxidative phosphorylation (OXPHOS) pathway was upregulated in both tumors in the FOLFIRI group and in the E+C group, whereas it was downregulated in tumors from mice that were treated with the combination of FOLFIRI and E+C (Fig. 3A). Similarly, glycolysis and PI3K/AKT/mTOR-signaling pathways were upregulated in the E+C groups and significantly downregulated in the combo group. The KRAS-signaling pathway was significantly upregulated in FOLFIRI-treated tumors (Fig. 3A), but it was downregulated in tumors treated with either E+C or with the combination of chemotherapy and molecular-targeted agents (Fig. 3A). It was interesting that MYC target V1 genes were upregulated in the E+C treatment group and downregulated in chemo and in the chemo plus encorafenib and cetuximab treatment group. Moreover, the epithelial-to-mesenchymal transition (EMT) pathway was upregulated in the FOLFIRI treatment group as compared with vehicle, whereas it was downregulated in the E+C treatment group as well as in FOLFIRI + encorafenib and cetuximab–treated tumors (Fig. 3A). Consistent with these observations, MAPK-associated genes using an MAPK activation score (MPAS) have been evaluated (25). DUSP6, that acts downstream to KRAS and is a biomarker of MAPK-signaling, was less expressed in tumors from mice treated with E+C or in those treated with the full combination as compared with tumors from mice treated with FOLFIRI or with vehicle (Fig. 3B and C). DUSP4, which negatively regulates members of the MAPK superfamily is overexpressed in the combination-treated tumors compared with FOLFIRI or tumors treated with targeted agents (Fig. 3B and C). We further validated our findings on the transcriptomic changes under cytotoxic chemotherapy or targeted therapy by investigating changes in key markers of MAPK-signaling pathways. We performed IHC staining using B1003b PDX tumor tissues (Fig. 3D and E). Phospho-ERK expression as a measure of MAPK pathway activity was found relatively upregulated in FOLFIRI treatment; that was successfully inhibited in E+C and in the combination of FOLFIRI + encorafenib and cetuximab treatment (Fig. 3D and E).
Figure 3.
Transcriptional changes after cytotoxic chemotherapy and targeted therapy of B1003b 21-day treated tumors. A, GSEA analysis of hallmark gene sets between treatment samples (FOLFIRI, E+C, or FOLFIRI + E+C) relative to controls. A bubble plot was colored by normalized enrichment score (NES) and the size of the bubbles represent adjusted P values. Red bubbles indicate gene sets that are more enriched after treatment (NES > 0), whereas blue bubbles represent gene sets enriched in control samples (NES < 0). B, MPAS and C, Heatmap of 10 MAPK target genes average expression after cytotoxic chemotherapy and targeted therapy. D, Percentage of positive nuclei/cells in were compared between treatment groups (n = 4–6). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. E, IHC staining (20×) identifying the pERK in B1003b tissue among four treatments.
Evaluation of induction and maintenance regimens in vivo
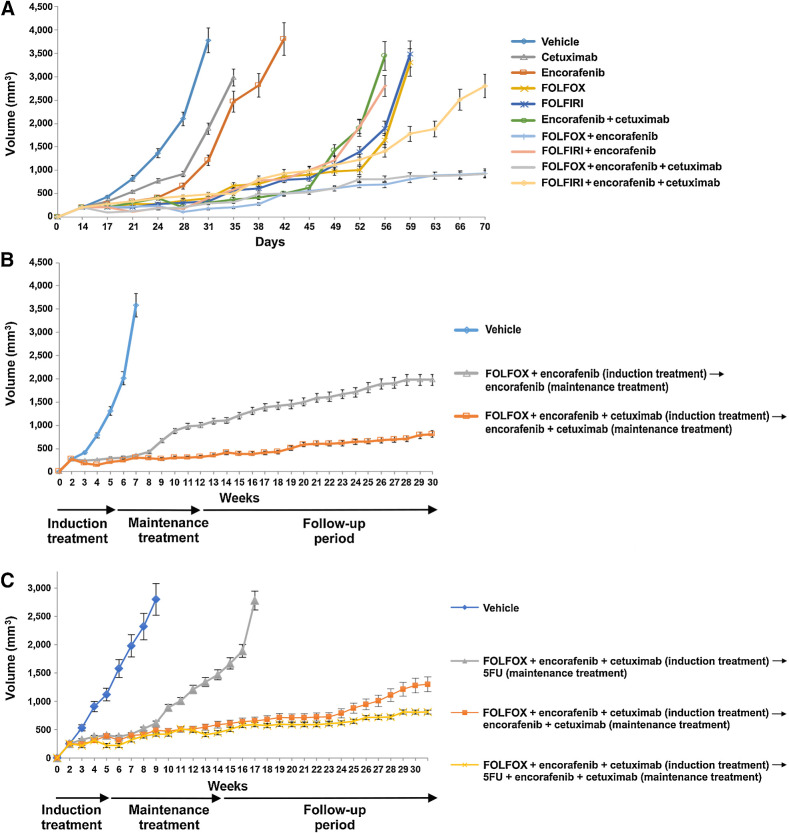
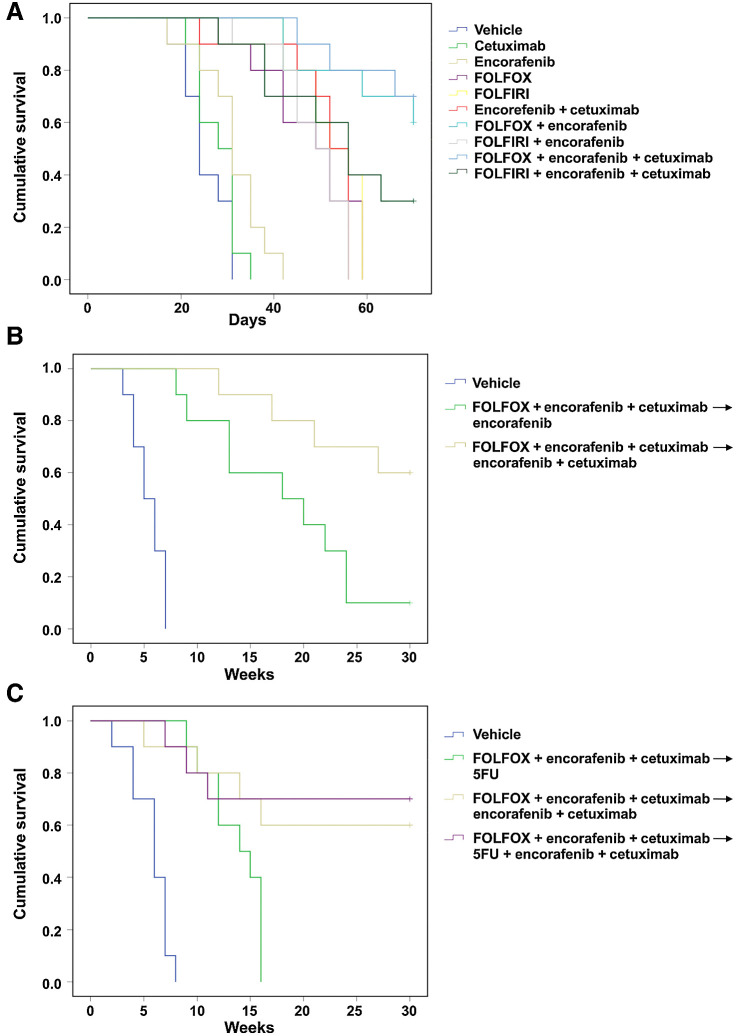
To evaluate alternate induction and maintenance regimens, we compared FOLFOX or FOLFIRI chemotherapies alone and in combination with molecular-targeted therapies in HT29 tumor xenografts models (Fig. 4A). In our preliminary data, we chose FOLFIRI as our chemotherapy of choice because of its clinical relevance and limited toxicity in extended treatment in the in vivo setting; however, the evaluation of other clinically relevant options, such as FOLFOX, is crucial. For these experiments, we used colorectal cancer cell lines because these models have been recognized as being more suitable for long-term experiments and were readily available at the site conducting the experiment. In particular, we examined the optimal sequence of administering molecular targeted therapies and chemotherapies in HT29 colorectal cancer-derived cell lines bearing the BRAFV600E mutation and microsatellite-stable profile. In the HT29 tumor xenograft model, the combination of encorafenib and cetuximab was superior to either encorafenib (HR, 0.045; confidence interval; 95% CI, 0.005–0.38; P = 0.004) and cetuximab (HR, 0.052; CI, 0.006–0.43; P = 0.006). Both FOLFOX and FOLFIRI were active and equivalent in duration of tumor control to E+C. The combination of E+C and either FOLFOX or FOLFIRI was superior to the cytotoxic chemotherapy alone (HR, 0.115; 95% CI, 0.025-–0.53 and HR, 0.63; 95% CI, 0.22–1.73; P = 0.006 and P = 0.36, respectively) or E+C alone (HR, 0.15; 95% CI, 0.033–0.68 and HR, 0.58; 95% CI, 0.21–1.60; P = 0.014 and P = 0.29, respectively). There was a numerically longer duration of control with the FOLFOX regimen than FOLFIRI (70% vs. 30% survival at end of experiment; Figs. 4A and 5A). It was notable that FOLFOX + encorafenib appeared equivalent to FOLFOX + E+C in this model, despite the superiority of E+C over encorafenib alone. The positive effect seen with FOLFOX+ encorafenib regimen could be explained by the continuous treatment with oxaliplatin-based chemotherapy.
Figure 4.
Antitumor efficacy with different treatment combination and sequence in BRAFV600E HT29 tumor xenograft. HT29 cells were injected subcutaneously into the left flanks of nude mice and treated with different combination regimen and in different sequence. A, After 2 weeks (average tumor size 200–400 mm3) mice were treated for 6 weeks with: vehicle, encorafenib, cetuximab, alone and in the combination, FOLFOX or FOLFIRI alone, with encorafenib and in combination with E+C. B, After 2 weeks of s.c. injection, mice were treated with vehicle, FOLFOX + encorafenib (10 mice) and FOLFOX + E+C (10 mice). At the end of 3 weeks of therapy, defined as induction treatment, treated mice were randomized into encorafenib alone or E+C, respectively (maintenance treatment). C, After 2 weeks of s.c. injection, mice were treated with vehicle (10 mice), FOLFOX + E+C (30 mice) for 3 weeks, defined as induction treatment. Subsequently treated mice were randomized into 5-FU alone (10 mice), E+C (10 mice), and 5-FU + E+C (10 mice; maintenance treatment).
Figure 5.
Effects of different treatment combinations and sequences on the survival in BRAFV600E HT29 tumor xenograft. A, Mice were monitored for survival until 70 days following tumor cell injection. Differences in animal survival among groups were evaluated by use of the Mantel–Cox log-rank test. B–C, Mice were monitored for survival until 30 weeks following tumor cell injection. Differences in animal survival among groups were evaluated by use of the Mantel–Cox log-rank test.
On the basis of these findings, we performed a second experiment to evaluate if a maintenance treatment (either encorafenib alone or encorafenib in combination with cetuximab) could be able to prevent and/or delay the onset of cancer cell resistance after the induction treatment and also if FOLFOX + encorafenib may be sufficient in this setting (Fig. 4B). Mice were first treated with an induction of FOLFOX + encorafenib or with FOLFOX + encorafenib and cetuximab. At the end of 3 weeks of therapy, mice were transitioned to receive encorafenib alone in combination with cetuximab, respectively. The maintenance treatment was continued for 8 weeks, and afterward, mice were monitored for an additional 17 weeks (follow-up period; Fig. 4B). Although the combination of FOLFOX and encorafenib was similar to FOLFOX + E+C after the induction period, the best disease control rate was observed for the group of mice that were treated with FOLFOX + encorafenib and cetuximab followed by encorafenib and cetuximab maintenance, resulting in an improved OS (HR, 0.26; 95% CI, 0.077–0.87; P = 0.029; Figs. 4B and 5B).
Finally, a third experiment was performed to evaluate if the addition of 5-FU as maintenance treatment could improve the clinical efficacy of this therapeutic combination and sequence. Mice were treated with FOLFOX + encorafenib and cetuximab for 3 weeks (induction treatment). Then, they were randomized to the following treatment arms: 5-FU alone, 5-FU + encorafenib and cetuximab or encorafenib and cetuximab (maintenance treatment; Fig. 4C). Maintenance treatment was continued for 8 weeks, and afterward, mice were monitored for an additional 17 weeks (follow-up period). In the 5-FU monotherapy maintenance arm, tumors started to regrow shortly after the cessation of the induction treatment. On the contrary, both maintenance arms that contained encorafenib and cetuximab (with or without 5-FU) were effective in controlling disease progression (Fig. 4C). Furthermore, a long-term additional clinical benefit by adding 5-FU to encorafenib and cetuximab as maintenance treatment was observed (Fig. 4C). The antitumor activity resulted in a significantly prolonged survival of mice for 5-FU + E+C versus 5-FU alone (HR, 0.25; 95% CI, 0.068–0.91; P = 0.036) or for 5-FU + E+C versus 5-FU alone (HR, 0.76; 95% CI, 0.17–3.42; P = 0.73; Fig. 5C).
Discussion
In recent years, significant progress has been made in characterizing mCRC through the identification of distinct molecular subgroups driven by genomic factors. One such subgroup is defined by the presence of the BRAFV600E mutation. There is an urgent unmet need for implementing a better treatment strategy for this poor-prognosis disease group (2, 8, 28). Here, we report the results of an in vivo preclinical study with the aim of defining treatment strategies that could help to improve the clinical management of patients with BRAFV600E-mutant mCRC. Because a high proportion of these patients will have limited chances of receiving a second-line therapy after first-line treatment for the rapid deterioration of their clinical conditions, a key issue is to maximize the treatment efficacy within the first-line setting. A first clinically relevant finding from this study is that either cytotoxic chemotherapy or molecular-targeted therapy (E+C) are more effective in first-line as compared with second-line treatment. This is consistent with clinical data on the triplet of encorafenib, cetuximab, and binimetinib, where cross-trial comparison of ANCHOR and BEACON results show response rates of 48%, 34%, and 22% for first-, second-, and third-line therapy, respectively. Furthermore, acquired resistance to FOLFIRI reduces the antitumor activity of subsequent E+C. Similarly, initial therapy with E+C impairs the efficacy of FOLFIRI upon progression.
We have evaluated the changes of key pathways that occurred following different therapies by tumor RNA sequencing (RNA-seq). The results of this study suggest the potential involvement of two mechanisms of acquired resistance, such as EMT and KRAS/MAPK signaling pathways (29, 30). Both of these pathways were quite upregulated in FOLFIRI-treated tumors, suggesting that EMT and KRAS activation could be mechanisms of adaptation to chemotherapy, as has been previously identified (29, 30). In contrast, both pathways were downregulated after treatment with E+C, and were not overexpressed with the combination, suggesting an ability of the combination of cytotoxic and targeted therapies to intercept these critical adaptation mechanisms. In addition, Myc, downstream target of RAS/RAF/ERK pathway, responsible of c-Myc stability (31), was found downregulated in the combination group. Collectively, these findings support the concept that the addition of molecular targeted therapy to chemotherapy could be a promising strategy to overcome cancer cell resistance that occurs following chemotherapy. However, further data and validation in broader models and settings are necessary to confirm our results. An additional interesting observation was the impact of the therapies on the OXPHOS and glycolysis pathway. Mitochondrial OXPHOS is one of the major sources of energy for colorectal cancer, and high levels have been associated with reduced patient survival in patients with colorectal cancer (32, 33). In addition, the pathway is emerging as a mechanism of resistance to targeted therapies including BRAF, as suggested by its upregulation after targeted therapy alone in our model (34). Similarly, cytotoxic drug treatment, including irinotecan, could enhance the OXPHOS pathway (35). Furthermore, increasing evidence suggests that glycolysis in cancer is associated with drug (36) resistance. Interestingly, the results of the present study show that OXPHOS and glycolysis pathways were downregulated following FOLFIRI + encorafenib and cetuximab treatment. Although further research is needed, these results highlight the potential for combination therapy to change the transcriptional adaptation leading to improved response prolonging disease control in patients.
As a practical matter, first-line chemotherapy and combination therapies can result in higher toxicities and prolonged treatment durations. As such, optimal maintenance regimens are needed. This traditionally has been evaluated on the basis of clinical experience and is rarely modeled preclinically. Here, we provide evaluation of multiple maintenance regimens to help guide further treatment strategies for first-line combination BRAF therapies. As FOLFIRI has less cumulative toxicity mandating maintenance approaches, we focused on FOLFOX modeling. The results of the present study suggest that, after an induction course of FOLFOX + E+C, maintenance therapy of 5-FU with E+C provides improved duration of disease control compared with E+C or 5-FU alone. This finding is consistent with broader current practice to drop the oxaliplatin but maintain the other components of the regimen, and this management approach is recommended in the BREAKWATER study.
There are limitations to this work. Although PDX models have been well-validated for prediction of response in patients, the data on correlation between duration of disease control in vivo and in patients are less well established. The subcutaneous models do not fully replicate the microenvironment of mCRC, and the xenografts models are conducted in immunosuppressed models, thereby failing to account for any immune impact of the therapies. The models utilized do not readily develop acquired resistance through genomic mechanisms such as acquisition of KRAS, NRAS, or MAPK21 mutations, and therefore may not reflect altered cross-resistance to genomic mechanisms of resistance. As reported by Corcoran and colleagues, in 48% of patients that were treated with selective inhibitors of BRAF and EGFR (±a MEK inhibitor), KRAS or NRAS mutations were found at disease progression, with approximately one out of five patients having tumors with multiple subclonal RAS mutations (37). Additional questions are not able to fully addressed, such as whether FOLFOX or FOLFIRI is a better combination partner in the population, and we caution that the results of the single murine model to compare the two cytotoxic backbones may not reflect clinical results in patients. Recent data reported in abstract form from the safety lead of the BREAKWATER demonstrate high levels of response and disease control with both cytotoxic backbones (38).
In conclusion, these in vivo preclinical results provide a rationale for the combination of chemotherapy and molecular targeted drugs as a potential treatment of choice for the first-line setting and also provides a potential preclinical foundation to the ongoing BREAKWATER randomized phase III study in patients with BRAFV600E-mutant mCRC (17, 38).
Supplementary Material
Differentially enriched hallmark gene sets after cytotoxic chemotherapy and targeted therapy of B1003b PDX tumor samples.
Acknowledgments
A research grant that partially covered the costs of the study was provided by Regione Campania (I-Cure Research Project, grant number: Cup 21C17000030007, to F. Ciardiello and L. Altucci). This work was also supported by Cancer Center Support Grant – Gastrointestinal Program (Project Number: 5P30 CA016672–46). O.E. Villarreal was supported by the CPRIT Training Program (RP210028).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
S. Napolitano reports grants from Regione Campania (I-Cure Research Project) and Cancer Center Support Grant – Gastrointestinal Program during the conduct of the study. G. Martini reports personal fees from Servier and Incyte outside the submitted work. C. Della Corte reports personal fees from Novartis, AstraZeneca, and Roche as well as grants from Pfizer, personal fees from MSD, and other support from Amgen outside the submitted work. D. Ciardiello reports personal fees from Sanofi, BMS, and Merck-Serono during the conduct of the study. O.E. Villarreal reports grants from Cancer Prevention & Research Institute of Texas (CPRIT) during the conduct of the study. V.K. Morris reports other support from BioNTech, Pfizer, Bristol Myers Squibb, and Novartis as well as other support from RedX Pharma outside the submitted work. J. Tabernero reports personal fees from Array Biopharma, AstraZeneca, Bayer, Boehringer Ingelheim, Cardiff Oncology, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche Ltd, Genentech Inc, HalioDX SAS, Hutchison MediPharma International, Ikena Oncology, Inspirna Inc, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Scandion Oncology, Scorpion Therapeutics, Seattle Genetics, Servier, Sotio Biotech, Taiho, Tessa Therapeutics, TheraMyc and Tolremo Therapeutics, personal fees from Oniria Therapeutics and Imedex/HMP, Medscape Education, MJH Life Sciences, and PeerView Institute for Medical Education and Physicians Education Resource (PER) outside the submitted work. F. Ciardiello reports other support from Roche, Pfizer, Pierre Fabre, Merck KGaA, Servier, Amgen, and MSD during the conduct of the study as well as other support from Bayer and Eisai outside the submitted work. S. Kopetz reports other support from Pfizer during the conduct of the study; other support from Lutris, Iylon, Frontier Medicines, Xilis, Navire, Genentech, EMD Serono, Merck, Holy Stone Healthcare, Novartis, Lilly, Boehringer Ingelheim, AstraZeneca/MedImmune, Bayer Health, Redx Pharma, Ipsen, HalioDx, Lutris, Jacobio, Pfizer, Repare Therapeutics, Inivata, GlaxoSmithKline, Jazz Pharmaceuticals, Iylon, Xilis, AbbVie, Amal Therapeutics, Gilead Sciences, Mirati Therapeutics, Flame Biosciences, Servier, Carina Biotech, Bicara Therapeutics, Endeavor BioMedicines, Numab, Johnson & Johnson/Janssen, Genomic Health, Frontier Medicines, Replimune, Taiho Pharmaceutical, Cardiff Oncology, Ono Pharmaceutical, Bristol-Myers Squibb-Medarex, Amgen, Tempus, Foundation Medicine, Harbinger Oncology, Inc, Takeda, CureTeq, Zentalis, Black Stone Therapeutics, NeoGenomics Laboratories, Accademia Nazionale Di Medicina, Sanofi, Biocartis, Guardant Health, Array BioPharma, Genentech/Roche, EMD Serono, MedImmune, Novartis, Amgen, Lilly, and Daiichi Sankyo outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
S. Napolitano: Conceptualization, data curation, supervision, validation, investigation, methodology, writing–original draft, writing–review, and editing. M. Woods: Data curation, validation, investigation, visualization, methodology, writing–original draft, writing–review, and editing. H. Lee: Resources, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review, and editing. V. De Falco: Data curation, formal analysis, and validation. G. Martini: Data curation, supervision, investigation, writing–review, and editing. C. Della Corte: Data curation, supervision, validation, investigation, and visualization. E. Martinelli: Supervision, investigation, writing–review, and editing. V. Famiglietti: Data curation, software, formal analysis, and methodology. D. Ciardiello: Data curation, validation, investigation, visualization, and methodology. A. Anderson: Formal analysis, and methodology. N. Wall Fowlkes: Data curation and investigation. O.E. Villarreal: Data curation and formal analysis. A. Sorokin: Data curation, supervision, validation, investigation, and methodology. P. Kanikarla: Data curation, formal analysis, validation, investigation, and methodology. O. Coker: Data curation and validation. V. Morris: Data curation, investigation, visualization, writing–review, and editing. L. Altucci: Resources, funding acquisition, validation, and investigation. J. Tabernero: Supervision, writing–review, and editing. T. Troiani: Data curation, supervision, investigation, and visualization. F. Ciardiello: Conceptualization, resources, data curation, supervision, funding acquisition, project administration, writing–review, and editing. S. Kopetz: Conceptualization, resources, data curation, supervision, funding acquisition, project administration, writing–review, and editing.
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics 2022. CA Cancer J Clin 2022;72:7–33. [DOI] [PubMed] [Google Scholar]
- 2. Ciardiello F, Ciardiello D, Martini G, Napolitano S, Tabernero J, Cervantes A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin 2022;72:372–401. [DOI] [PubMed] [Google Scholar]
- 3. Parseghian CM, Napolitano S, Loree JM, Kopetz S. Mechanisms of innate and acquired resistance to anti-EGFR therapy: a review of current knowledge with a focus on rechallenge therapies. Clin Cancer Res 2019;25:6899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79–92. [DOI] [PubMed] [Google Scholar]
- 6. Ahn TS, Jeong D, Son MW, Jung H, Park S, Kim H, et al. The BRAF mutation is associated with the prognosis in colorectal cancer. J Cancer Res Clin Oncol 2014;140:1863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 2011;117:4623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee HM, Morris V, Napolitano S, Kopetz S. Evolving strategies for the management of BRAF-mutant metastatic colorectal cancer. Oncology (Williston Park) 2019;33:206–11. [PubMed] [Google Scholar]
- 9. Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100–3. [DOI] [PubMed] [Google Scholar]
- 11. Yang H, Higgins B, Kolinsky K, Packman K, Bradley WD, Lee RJ, et al. Antitumor activity of BRAF inhibitor vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res 2012;72:779–89. [DOI] [PubMed] [Google Scholar]
- 12. Hong DS, Morris VK, El Osta B, Sorokin AV, Janku F, Fu S, et al. Phase IB study of vemurafenib in combination with irinotecan and cetuximab in patients with metastatic colorectal cancer with BRAFV600E mutation. Cancer Discov 2016;6:1352–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kopetz S, Guthrie KA, Morris VK, Lenz HJ, Magliocco AM, Maru D, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG S1406). J Clin Oncol 2021;39:285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, et al. NCCN guidelines insights: colon cancer, version 2.2018. J Natl Compr Canc Netw 2018;16:359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 16. Grothey A, Yaeger R, Paez D, Tabernero J, j T, Yoshino T, et al. ANCHOR CRC: a phase 2, open-label, single arm, multicenter study of encorafenib (ENCO), binimetinib (BINI), plus cetuximab (CETUX) in patients with previously untreated BRAF V600E-mutant metastatic colorectal cancer (mCRC). Annals of Oncol 2019;30. [Google Scholar]
- 17. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2014;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tischler G, Leonard S. Biobambam: tools for read pair collation based algorithm on BAM files. Source Code Biol Med 2014;9:13. [Google Scholar]
- 19. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res 2012;22:1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Anders S, Pyl PT, Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, et al. Xenome–a tool for classifying reads from xenograft samples. Bioinformatics 2012;28:i172–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. Biorxiv 2021;060012. [Google Scholar]
- 24. Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York; 2016; ISBN 978–3-319–24277–4. [Google Scholar]
- 25. Wagle MC, Kirouac D, Klijn C, Liu B, Mahajan S, Junttila M, et al. A transcriptional MAPK pathway activity score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis Oncol 2018;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kolde R. pheatmap: Pretty Heatmaps 2019. Available from: https://CRAN.R-project.org/package=pheatmap/.
- 27. Kopetz S, Grothey A, Yaeger R, Ciardiello F, Desai J, Won Kim T, et al. BREAKWATER: randomized phase 3 study of encorafenib (enco) + cetuximab (cetux) ± chemotherapy for first-line (1L) treatment (tx) of BRAF V600E-mutant (BRAFV600E) metastatic colorectal cancer (mCRC). J Clin Oncol 2022;40(suppl 4):TPS211. [Google Scholar]
- 28. Johnson B, Kopetz S. Applying precision to the management of BRAF-mutant metastatic colorectal cancer. Target Oncol 2020;15:567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ni Q, Li M, Yu S. Research progress of epithelial-mesenchymal transition treatment and drug resistance in colorectal cancer. Technol Cancer Res Treat 2022;21:15330338221081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meng M, Zhong K, Jiang T, Liu Z, Kwan HY, Su T. The current understanding on the impact of KRAS on colorectal cancer. Biomed Pharmacother 2021;140:111717. [DOI] [PubMed] [Google Scholar]
- 31. Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000;14:2501–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rebane-Klemm E, Truu L, Reinsalu L, Puurand M, Shevchuk I, Chekulayev V, et al. Mitochondrial respiration in KRAS and BRAF mutated colorectal tumors and polyps. Cancers (Basel) 2020;12:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koit A, Shevchuk I, Ounpuu L, Klepinin A, Chekulayev V, Timohhina N, et al. Mitochondrial respiration in human colorectal and breast cancer clinical material is regulated differently. Oxid Med Cell Longev 2017;2017:1372640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell 2013;23:302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marx C, Sonnemann J, Maddocks ODK, Marx-Blümel L, Beyer M, Hoelzer D, et al. Global metabolic alterations in colorectal cancer cells during irinotecan-induced DNA replication stress. Cancer Metab 2022;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat 2018;38:1–11. [DOI] [PubMed] [Google Scholar]
- 37. Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov 2018;8:428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kopetz S, Yoshino Y, Kim TW, Desai J, Yaeger R, Cutsem EV, et al. BREAKWATER safety lead-in (SLI): encorafenib + cetuximab (EC) ± chemotherapy for first-line (1L) treatment (tx) of BRAF V600E-mutant (BRAFV600E) metastatic colorectal cancer (mCRC). J Clin Oncol 2022;40(suppl 4):134. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially enriched hallmark gene sets after cytotoxic chemotherapy and targeted therapy of B1003b PDX tumor samples.
Data Availability Statement
The raw data generated in this study are available upon request from the corresponding author.