Abstract
Purpose:
Patients with small-cell lung cancer (SCLC) have an exceptionally poor prognosis, calling for improved real-time noninvasive biomarkers of therapeutic response.
Experimental Design:
We performed targeted error-correction sequencing on 171 serial plasmas and matched white blood cell (WBC) DNA from 33 patients with metastatic SCLC who received treatment with chemotherapy (n = 16) or immunotherapy-containing (n = 17) regimens. Tumor-derived sequence alterations and plasma aneuploidy were evaluated serially and combined to assess changes in total cell-free tumor load (cfTL). Longitudinal dynamic changes in cfTL were monitored to determine circulating cell-free tumor DNA (ctDNA) molecular response during therapy.
Results:
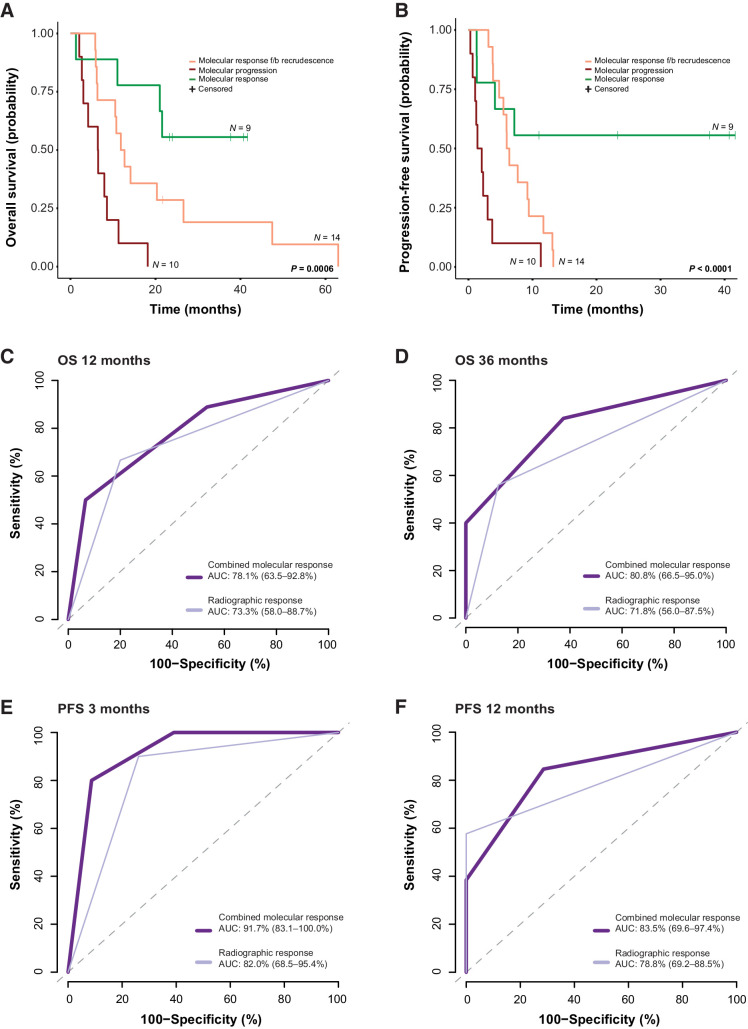
Combined tiered analyses of tumor-derived sequence alterations and plasma aneuploidy allowed for the assessment of ctDNA molecular response in all patients. Patients classified as molecular responders (n = 9) displayed sustained elimination of cfTL to undetectable levels. For 14 patients, we observed initial molecular responses, followed by ctDNA recrudescence. A subset of patients (n = 10) displayed a clear pattern of molecular progression, with persistence of cfTL across all time points. Molecular responses captured the therapeutic effect and long-term clinical outcomes in a more accurate and rapid manner compared with radiographic imaging. Patients with sustained molecular responses had longer overall (log-rank P = 0.0006) and progression-free (log-rank P < 0.0001) survival, with molecular responses detected on average 4 weeks earlier than imaging.
Conclusions:
ctDNA analyses provide a precise approach for the assessment of early on-therapy molecular responses and have important implications for the management of patients with SCLC, including the development of improved strategies for real-time tumor burden monitoring.
Translational Relevance.
Longitudinal analyses of sequence and structural ctDNA landscapes reflect clinical outcomes and can accelerate rapid noninvasive detection of therapeutic response for patients with small-cell lung cancer.
Introduction
Small-cell lung cancer (SCLC) accounts for approximately 15% of all lung cancer cases worldwide and is characterized by an aggressive clinical course leading to a 5-year survival rate of approximately 5% (1, 2). Most patients with SCLC display early tumor spread, requiring platinum-based chemotherapy as the backbone of treatment regimens in all stages of the disease (3). Despite the incorporation of immune-checkpoint inhibitors in first-line treatments for patients with extensive-stage SCLC (4, 5), the vast majority of patients develop therapeutic resistance prior to the detection of measurable lesions on conventional imaging (6). Patients with relapsed disease infrequently undergo tumor biopsies for molecular testing, which presents a major barrier for translational research efforts in this setting (6). To this end, minimally invasive approaches can be particularly informative and accurately characterize early responses to systemic therapy in patients with SCLC, ultimately enabling timely interventions and improving patient outcomes.
Circulating tumor DNA (ctDNA), which is released from tumor cells into the bloodstream, can be used to identify and track relapsed disease and monitor tumor burden changes during therapy (7–9). Although ctDNA dynamics during therapy have been studied in several solid tumors; there is a limited number of studies involving longitudinal ctDNA analyses for patients with SCLC (7, 8, 10–12). These studies provide preliminary evidence to support the value of residual ctDNA to predict disease relapse following curative-intent therapy in patients with limited-stage SCLC (7, 8) and to delineate the genomic profile and subclonal architecture of extensive-stage SCLC (9–11). Importantly, sequence mutation-based ultrasensitive liquid biopsy approaches do not fully capture the evolving landscape of cancer during therapy; to this end, evaluation of ctDNA copy-number profiles and plasma aneuploidy may complement ctDNA response assessments (13–15). However, integrative multifeature approaches for longitudinal molecular response evaluation leveraging sequence and structural alterations in ctDNA have not yet been explored for patients with SCLC.
We thus hypothesized that combined analyses of somatic sequence and chromosomal arm-level structural alterations (the latter termed plasma aneuploidy) in ctDNA would enhance the sensitivity of ctDNA molecular responses to dynamically predict clinical outcomes in patients with SCLC undergoing systemic therapies. Here, we used an ultrasensitive targeted next-generation sequencing approach of plasma and matched white blood cell (WBC) DNA (14, 16–21) to detect and deconvolute the origin of variants identified in ctDNA and performed comprehensive longitudinal analyses of the landscape of tumor-derived sequence alterations combined with plasma aneuploidy. Using this combined tiered approach, we precisely tracked dynamic changes in circulating tumor burden and evaluated molecular responses as a real-time biomarker of clinical outcomes compared with conventional radiographic imaging.
Materials and Methods
Cohort description
Tumor biopsies and serial plasma samples were collected from 33 patients with SCLC who received systemic therapies, including immunotherapy, for metastatic disease across two medical centers (Johns Hopkins Hospital, n = 17; Vanderbilt University Medical Center, n = 16). Patients received treatment with either platinum-based combination or immunotherapy-containing regimens; platinum-based combination chemotherapy was the most common among patients who received first-line treatment during analysis for the study (14/20 patients who received first-line treatment) and the remaining 6 patients received chemoimmunotherapy regimens, while immunotherapy-containing regimens were the most common among patients who received second- or third-line treatment (11/13 patients). No significant differences in survival outcomes were observed for the first-line cohort between treatment groups. Detailed clinical characteristics including treatment and outcomes are summarized in Supplementary Table S1. Therapeutic response was assessed radiographically and radiographic best overall response (BOR) was used for subsequent statistical analyses [mean time to first imaging assessment = 8 weeks (median 6 weeks); mean time to BOR assessment = 11 weeks (median 7 weeks)]. Radiographic treatment response assessment was retrospectively abstracted from clinical records based on the treating provider's assessment of disease change as indicated by imaging at each point of radiographic disease evaluation (e.g., partial response, stable disease, progressive disease). To compare the dynamics of radiographic response and molecular response, time to BOR was determined as the time from treatment start to best-recorded response (most often at the first radiographic timepoint). For patients who received immunotherapy-containing regimens, durable clinical benefit (DCB) was defined as lack of disease progression or death within 6 months of treatment initiation, and nondurable clinical benefit (NDB) was defined as disease progression or death within 6 months of treatment initiation. Platinum sensitivity was defined as BOR of partial or complete response at first restaging after first-line platinum chemotherapy with progression >60 days after completion of chemotherapy. Platinum resistance was defined as BOR of stable disease, mixed response or progressive disease at initial restaging after first-line platinum chemotherapy, or best radiographic response of partial response at first restaging followed by progression <60 days within the completion of chemotherapy. Patients who received chemoimmunotherapy combination regimens as first-line treatment (n = 7) were not evaluable for platinum sensitivity (Supplementary Table S1). Progression-free survival (PFS) and overall survival (OS) were defined as the time elapsed between the date of treatment initiation and the date of disease progression, or the date of death, respectively. The study was conducted in accordance with the Declaration of Helsinki, approved by the Institutional Review Boards of each medical center, and patients provided written informed consent for sample acquisition for research purposes.
Samples and next-generation sequencing
Serial blood samples were collected in K2EDTA tubes (BD Vacutainer) prior to treatment, during the course of treatment (average time to first blood sample collection after initiation of treatment = 9 weeks; median = 3 weeks), and at the time of clinical progression. The volume of whole blood used for the isolation of plasma and buffy-coat samples was 10 mL, corresponding to one K2EDTA collection tube per time point. Cell-free DNA (cfDNA), isolated from 1.9 to 5 mL plasma (n = 139), and matched WBC DNA (n = 32) were processed for targeted error-correction sequencing (TEC-seq), as previously described (ref. 18; Supplementary Tables S2–S3). In addition, WBC DNA samples from an independent group of 56 patients with lung cancer were processed for TEC-seq using identical methods (18) and also used for downstream analyses of copy-number profiles (Supplementary Table S3). Briefly, cfDNA was extracted from plasma using the Qiagen Circulating Nucleic Acids kit (Qiagen GmbH). TEC-seq libraries were prepared from 3 to 125 ng cfDNA and 75 ng of matched WBC DNA, the latter sheared to a size of 200 bp, followed by targeted capture using a custom set of hybridization probes (Personal Genome Diagnostics) and sequenced using 100-bp paired-end reads on Illumina HiSeq 2500 instruments. TEC-seq characteristics are shown in Supplementary Tables S2–S3. Regions captured by TEC-seq across 58 genes encompassing 99,359 bases were compared with the genomic locations of mutations previously reported from whole-genome sequencing of tumor samples from 110 patients with SCLC in George and colleagues (22) to evaluate the suitability of the targeted panel for the capture of SCLC somatic mutations (Supplementary Table S4). Specific to capture of the RB1 locus, which is frequently mutated in SCLC, these comparisons revealed that TEC-seq would capture 106 of 110 tumors (96%) and 31% (29/35 mutations) of sequence mutations detected within RB1 coding regions for these tumors, supporting the utility of the panel for the reliable characterization of SCLC genomic profiles through ctDNA analyses.
For a subset of 5 patients, we performed tumor/normal whole-exome sequencing (WES) in order to evaluate the concordance between copy-number profiles computed from tumor WES compared with plasma aneuploidy. Tumor DNA was extracted following macrodissection of formalin-fixed paraffin-embedded (FFPE) tumor tissue using the Qiagen DNA FFPE kit (Qiagen GmbH) and matched WBC DNA was extracted using the Qiagen DNA Blood Mini kit (Qiagen GmbH). Tumor and normal DNA were sheared to a target fragment size of 200 bp and processed for WES as previously described (23). Target enrichment was performed using Agilent SureSelect in-solution capture reagents and SureSelect XT Human All Exon V4 probes (Agilent). Prepared libraries were paired-end (100 bp) sequenced using Illumina HiSeq 2500 instruments (Personal Genome Diagnostics). WES technical metrics are shown in Supplementary Table S5.
Sequence alteration calling and characterization of variant origin
Primary processing of plasma next-generation sequencing data was performed as previously described (17). Briefly, Illumina CASAVA software (v1.8) was used for demultiplexing and masking of dual index adaptor sequences. Sequence reads were subsequently aligned to the hg19 human reference genome using NovoAlign with additional realignment of select regions using the Needleman–Wunsch method (24). Comparison between the genomic coordinates of sequence changes in hg19 and the most recent hg38 genome assembly revealed that the TEC-seq gene panel does not overlap any annotated regions of change between the two assemblies.
Somatic mutations, consisting of point mutations, small insertions, or deletions, were identified across targeted regions using VariantDx (24) as previously described (17). Following variant calling, the origin of plasma variants (tumor-derived, clonal hematopoiesis (CH)-derived, and germline) was assessed using a tumor-agnostic WBC-informed approach, where plasma variants were filtered by matched WBC DNA sequencing to remove germline variants and variants related to clonal hematopoiesis. To eliminate the very small chance of buffy-coat contamination by circulating tumor cells, which would result in the classification of tumor-specific mutations as CH-derived, information related to variant recurrence in COSMIC was used. Variants detected in WBC DNA TEC-Seq that were also found to be lung cancer hotspots were characterized as tumor-derived and not CH-derived, despite being detected in the matched normal sample. Variants detected in plasma samples were cross-referenced against the COSMIC v95 database (cancer.sanger.ac.uk; ref. 25) for the annotation of hotspot alterations using OpenCRAVAT (26). A conservative COSMIC frequency threshold of 25 occurrences was used to define a lung cancer hotspot (17). Of the variants that were not classified as cancer hotspots, those with a variant allele fraction (VAF) ≥25% in all WBC and plasma samples from the same individual were classified as germline. All cancer hotspots and nongermline variants were further compared against matched WBC sequence data to confirm variant origin. Alterations that were detected in matched WBC sequencing or in the canonical clonal hematopoiesis gene DNMT3A were classified as CH-derived. For all variants detected in plasma but not in matched WBC DNA samples (based on a threshold of ≥3 supermutants), we performed an additional evaluation of their supermutant counts in the WBC DNA sequence data to eliminate CH variants with levels below the TEC-seq level of detection (17, 18, 27). To this end, plasma variants with a supermutant count of ≥1 in WBC were classified as CH-derived and alterations with a WBC supermutant count of 0 were assigned a tumor-derived origin. Plasma variants that were assigned a tumor-derived origin were visually inspected using the Integrative Genomic Viewer and considered for downstream analyses. For patient (PT) 16, for whom matched WBC sequencing data were not available, all variants were considered tumor-derived with the exception of a DNMT3A mutation that was classified as CH-derived and as such excluded from downstream analyses. A summary of the pipeline used for variant origin classification is shown in Supplementary Fig. S1.
Evaluation of plasma aneuploidy
The genome-wide copy-number profiles of plasma samples were analyzed by modeling the variability of sequence coverage across the genome using the CNVkit method (28). Briefly, sequencing reads mapped to target regions of interest (Supplementary Table S2), as well as background reads that were not selectively captured, were both considered to construct a genome-wide map of coverage depth. The depth of coverage was corrected by subtracting out the variability explained by GC content, size of targeted region, and sequence mappability. Sequencing reads from matched WBC DNA samples were used to construct a reference panel for the evaluation of somatic copy-number profiles from plasma. The coverage profile of each plasma sample was corrected with respect to the reference panel to derive the relative copy-number profile for each genomic region (bin). Circular binary segmentation (CBS; ref. 29) was applied to copy-ratio profiles to identify genomic segments at constant copy numbers. To derive plasma aneuploidy (PA) scores, the following steps were applied: first, in each plasma or matched WBC DNA sample, the log copy ratio of segments was adjusted to have a mean value of 0 and transformed into a linear scale. For each chromosomal arm, a weighted average of the copy-ratio values of overlapping segments was calculated, where the weights represented the fraction of the arm overlapping the segment. For plasma samples, arm-level copy-ratio values were transformed into z-scores using the mean and standard deviation of the copy ratio of each arm in the reference panel. Similarly, arm-level copy-ratio values for each WBC DNA sample were calculated by applying a leave-one-out approach. Arm-level z-scores for each plasma and matched WBC sample were combined to calculate a per-sample PA score (13). PA scores from WBC DNA samples ranged from −1.54 to 4.90; a conservative PA threshold of 4.90 was thus selected for the distinction between normal ploidy and tumor aneuploidy in plasma (PA scores > 4.90 were considered representative of tumor aneuploidy; Supplementary Fig. S2A). To further support the use of a PA score of 4.90 as a threshold, we profiled copy-number alterations and resultant aneuploidy scores derived from the most aberrant chromosomal arms from an independent set of 56 WBC DNA samples (processed and captured using the same targeted sequencing panel used for this study) and compared the distribution of aneuploidy scores with that of the 32 matched WBC DNA samples used as the normal reference for the aneuploidy analyses. These comparisons showed that the distribution of aneuploidy scores was comparable across the new set of unmatched WBC DNA samples (representing “healthy” state) and the 32 matched WBC DNA samples previously analyzed, with no significant differences observed in the median scores for each set (Supplementary Fig. S2B). All samples included in the set of 56 WBC DNA samples had an aneuploidy score below 4.90, further supporting the use of this conservative threshold for the determination of tumor-derived aneuploidy in plasma samples. Plasma copy-number profiles were further compared with copy-number profiles generated using an orthogonal method, ichorCNA (30), to confirm regions of aneuploidy (Supplementary Fig. S2C). These comparisons revealed that nonzero ichorCNA tumor fraction scores were assigned to almost all matched WBC DNA samples, resulting in a large overlap in ichorCNA scores between plasma and matched normal DNA samples. In contrast, PA scores more clearly distinguished plasma from normal DNA samples (MW U test P value = 2.1e−15), demonstrating increased specificity for the detection of cell-free tumor load (cfTL) compared with ichorCNA (Supplementary Fig. S2C). Genome-wide copy-number profiles for tumor samples were derived from WES data using FACETS (31).
Analysis of cfTL and molecular response classification
Sequence alterations and PA were used to determine the cfTL dynamics for each patient using a combined tiered approach, based on combined mutation and aneuploidy dynamics, or PA for patients without detectable mutations. Mutation-based ctDNA responses (n = 27) were computed using the mutant allele fraction (MAF) of the most abundant tumor-derived mutation at each timepoint as a measure of cfTL, where a reduction to 0% MAF indicated complete elimination of cfTL (Supplementary Fig. S2D–S2I). To establish that a maximal MAF reduction to 0% was sufficient to indicate complete elimination of cfTL, statistical modeling of MAF uncertainty was performed using data from a mutant pool dilution series previously characterized in Phallen and colleagues (17) based on tumor cell lines spiked into unrelated wild-type DNA (Supplementary Fig. S2D–S2I). MAF estimates for 14 distinct mutations (12 SNVs, 2 Indels) across seven genes (BRAF, EGFR, ERBB2, KRAS, NRAS, PIK3CA, and TP53) were used to calculate coefficient of variation (CV) % estimates per unique variant at each dilution level. A simulated dilution series with a wider range of MAF levels and replicates (n = 30 variants; 0.1%–80% MAF sampled at 2,000× coverage; n = 10 replicates per level) was developed to supplement the experimental data. Experimental and simulated data were separately analyzed in a nonlinear decay model to evaluate the relationship between reference MAF and %CV. Subsequently, 95% uncertainty limits were calculated across the range of reference MAF values. Based on these analyses, 2 samples prepared from <3 ng cfDNA (Supplementary Table S3) were excluded from molecular response analyses, given the uncertainty in determining ctDNA dynamics for these samples. PA-based ctDNA responses (n = 6) were determined using PA scores as a binary measure of cfTL, where a PA score above 4.90 indicated plasma cancer aneuploidy, while a PA score below or equal to 4.90 indicated normal ploidy.
Patients who displayed sustained complete elimination of cfTL from baseline to the final sampling time point were classified in the molecular response category. For patients with both mutation-based and PA-based responses evaluable, we required both mutation and PA elimination to classify patients in the ctDNA molecular response category. Patients who displayed elimination of cfTL between baseline and intermediate time points followed by an increase in cfTL at the final time point analyzed for the study were assigned a classification of molecular response followed by recrudescence. Finally, patients who displayed persistence of cfTL across all sampled plasma time points were classified in the molecular progression category. Of 33 patients included in this study, we anticipated that approximately 1/3 would exhibit molecular progression and ∼1/3 would have a molecular response. Assuming 10 molecular responders and 10 molecular progressors, we would have 80% power to detect a difference in the log hazards for OS of −2.12 with a type 1 error of 0.05, assuming a probability of death of 1 and 0.25 within the study interval among nonresponders and responders, respectively.
For 3 patients (PT 2, PT 6, and PT 24), tumor-derived sequence alterations were identified only at the final timepoint analyzed and were undetectable across the initial time points evaluated. For PT 6, PA scores indicated the presence of aneuploidy across all sampled time points and were used as a readout of cfTL to assign a classification of molecular progression. The remaining 2 patients were assigned a classification of molecular response followed by recrudescence as both patients displayed undetectable cfTL across the first 3 timepoints analyzed following treatment initiation. This pattern was suggestive of an initial molecular response, with cfTL only detected at the final time point at the time of disease progression. For a single patient (PT25), partial concordance was observed between sequence and structural dynamics, and classification of molecular progression was assigned, based on the persistence of tumor-specific sequence mutations across all time points analyzed (Supplementary Table S6).
Statistical analyses
The Fisher exact test was used to assess concordance between sequence and structural response classifications and to evaluate categorical variables associated with the best overall radiographic response. Survival curves were generated using the Kaplan–Meier method (survminer package) and compared using log-rank tests. Univariate and multivariate Cox proportional hazards regression analyses were used to assess the impact of ctDNA molecular responses on overall and PFS. ROC analyses for molecular response classifications were performed using the pROC package (32). A logistic regression model incorporating sequence response and PA response classifications was generated to evaluate the accuracy of combined molecular responses for the prediction of radiographic response. All analyses were performed using R Statistical Software (v 4.0.2; R Core Team 2020; ref. 33).
Data availability
Next-generation sequence data can be retrieved from the European Genome-Phenome Archive (EGA; accession number EGAS00001006831).
Results
Cohort and overall approach
We evaluated ctDNA from serial plasma samples (n = 139) obtained from 33 patients with SCLC over a median follow-up duration of 11 months (range, 1–63 months; Fig. 1A). Patients received treatment with either chemotherapy (n = 16; 14 first-line, 1 second-line, 1 third-line) or immunotherapy-containing regimens (n = 17; 6 first-line chemoimmunotherapy, 7 second-line immunotherapy, 4 third-line immunotherapy; Fig. 1B). Clinical characteristics for patients in the study cohort are summarized in Supplementary Table S1. Matched WBC DNA deep sequencing was performed for all evaluable cases (n = 32) to identify germline alterations and variants attributed to CH, and enable accurate characterization of plasma variants based on origin (Materials and Methods).
Figure 1.
Study methodology and cohort analyzed. Overall, 33 patients with SCLC who received systemic treatment with either chemotherapy or immunotherapy-containing regimens were analyzed for this study. A, High-depth targeted error-correction sequencing (TEC-seq) was performed on cfDNA extracted from serial plasma samples collected at baseline and longitudinally throughout the course of treatment, alongside matched WBC DNA. Sequence and structural alterations were directly detected in cfDNA for each patient at each time point analyzed and used to evaluate longitudinal changes in cfTL using a combined tiered approach. WES was further performed on pretreatment tumor and matched WBC DNA derived from buffy coat from a subset of 5 patients and used to evaluate the concordance between copy-number profiles computed from tumor next-generation sequence data and PA. Finally, molecular response classifications were assigned to each patient based on dynamic changes in cfTL and used to predict clinical outcomes. B, Swimmer plot showing disease course in each patient, treatment, and samples collected. The mean time to first blood sample collection after initiation of treatment for patients included in the study was 9 weeks (median 3 weeks). The mean time to first imaging assessment in this cohort was 8 weeks (median 6 weeks), and the mean time to BOR assessment was 11 weeks (median 7 weeks).
To track longitudinal dynamic changes in cfTL and capture the evolving ctDNA landscape during therapy, we developed a combined tiered approach for molecular response determination based on mutation and PA dynamics in ctDNA. For mutation-based ctDNA analyses, we performed qualitative assessment of tumor-derived mutations detected in serial plasma samples from each patient and evaluated their kinetics in variant allele frequencies during therapy across sampled time points (Materials and Methods). To investigate aneuploidy changes in ctDNA, chromosomal arm-level copy-number profiles were assessed and the most aberrant chromosomal arm representations in each sample were used to construct a PA score (Materials and Methods). In selected patients, PA profiles were cross-referenced to segmentation-based copy-number profiles of the matched tumor to assess concordance. We combined sequence mutations and aneuploidy in ctDNA to estimate circulating cfTL (Materials and Methods), and cfTL was tracked longitudinally to characterize ctDNA molecular responses and evaluate concordance with clinical outcomes.
Landscape of ctDNA sequence alterations
Sequence alterations were analyzed in plasma at baseline (pretreatment), serially after the initiation of systemic therapy, and at the time of disease progression. Plasma variants detected were classified as germline, tumor- or CH-derived (Materials and Methods; Supplementary Fig. S1; Supplementary Table S6). Of the 137 unique cfDNA plasma variants, 8 variants were of germline origin, while 47 variants (35%) were CH-derived. CH variants predominantly affected genes known to be involved in clonal hematopoiesis (34, 35), including DNMT3A, TP53, and ATM (Fig. 2). Importantly, CH variants were also detected in genes not canonically associated with clonal hematopoiesis, including KIT, EGFR, BRCA1, and BRCA2; these were accurately characterized as CH-derived using our tumor-agnostic WBC DNA-informed approach and were subsequently excluded from the assessment of ctDNA molecular responses. These findings highlight the importance of correctly classifying variants by origin, prior to longitudinal tracking of tumor-derived variants and cfTL in plasma (Fig. 2).
Figure 2.
Landscape of sequence alterations in plasma. The type and origin of variants detected across plasma samples analyzed for this study are shown. Alteration frequencies for each gene across plasma samples analyzed are shown on the right and per-sample mutation counts are displayed in the bar plot on top. Panels indicating sampling time point, treatment type, PA scores, combined molecular response, OS, and PFS are shown below. Germline variants (filled circles) were observed in a total of 8 genes (FLT3, FGFR1, CDK6, ALK, POLE, BRAF, PTCH1, and BRCA2) on a patient-specific basis. Variants attributed to CH (crosses) were identified in canonical CH genes (DNMT3A, TP53, and ATM) in >50% (18/33) of patients analyzed for the study and across 14 additional genes at a lower prevalence. Overall, tumor-derived sequence alterations (solid border) were detected in plasma from 30 of the 33 patients analyzed and were most frequently detected in the TP53 gene, which was mutated in 26 patients. Of these 26 patients, 3 shared a mutation in RB1.
Tumor-derived sequence alterations were identified across plasma samples for 30 of 33 patients. At baseline, 27 patients had ≥1 (median 2; range, 1–9) tumor-derived alteration detected, at a median ctDNA-MAF of 21% (range, 0.1%–73.1%; Supplementary Fig. S3). In 3 patients, tumor-derived somatic mutations were detected at the final timepoint, but were either undetectable at baseline (n = 1) or could not be evaluated at this time point due to the unavailability of baseline samples (n = 2). The genomic landscape of ctDNA mutations revealed that nonsynonymous somatic alterations in TP53 were the most frequent among patients with detectable tumor-derived mutations in this cohort (26/30 cases; 87%), consistent with the nearly universal inactivation of TP53 in SCLC tumors (22, 36, 37). Three of these 26 cases (12%) shared a comutation in RB1, consisting of nonsynonymous, splice-site alterations and indels (Fig. 2). The overall mutation frequency in RB1 was lower than expected as homozygous RB1 deletions were not accounted for by our analyses. Tumor-derived mutations were also detected at <10% prevalence across genes involved in PI3K signaling, DNA damage repair, and RAS/RTK signaling, including PIK3CA (10%), PALB2 (10%), EGFR (10%), PTEN (7%), BRAF (7%), BRCA1 (6%), BRCA2 (6%), and KIT (3%; Fig. 2). Tumor-derived alterations in KRAS and NRAS were identified in 3% (1/30) and 7% (2/30) of patients, respectively (Fig. 2). Mutations in NOTCH family genes were not identified in this cohort (Fig. 2; Supplementary Table S6). Comparison between tumor and plasma mutation profiles in 5 patients with matched tumor WES revealed that tumor-specific mutations in TP53, IDH1, and KIT were detectable in plasma from 3 patients (patients 5, 11, and 14) and were correctly classified using our tumor-agnostic WBC-informed approach (Supplementary Table S6).
Plasma aneuploidy
In addition to tumor-derived sequence alterations, genome-wide chromosomal arm-level somatic copy-number aberrations (SCNA) were evaluated at each timepoint and used to construct a PA score to quantify the extent of deviation from a copy-neutral state (normal ploidy; Materials and Methods). Recurrent copy-number losses were observed across chromosome 10q and 17p, regions known to harbor genomic losses in SCLC tumors, and across chromosomal arm 3p, consistent with the frequent loss of this region across lung cancers (refs. 22, 36, 38, 39; Fig. 3A and B; Supplementary Fig. S4A). Similarly, frequent copy-number gains were identified across chromosomal arms 1p, 3q, 5p, and 8q that are recurring in SCLC (refs. 36, 39; Fig. 3A and B; Supplementary Fig. S4A). To investigate the concordance between tumor tissue and plasma-derived aneuploidy, we computed copy-number profiles from WES in 5 patients and evaluated their concordance with our plasma aneuploidy approach as well as an orthogonal approach (ichorCNA). Using the exome-derived copy-number profiles as the ground truth, we found that copy-number profiles from both our PA approach and ichorCNA closely resembled the tumor-derived profiles in patients with detectable aneuploidy in plasma (Fig. 3B; Supplementary Fig. S4B–S4F). Importantly, assessment of PA scores enabled subsequent evaluation of cfTL and ctDNA dynamics during therapy for all 3 patients with undetectable tumor-derived sequence alterations (Fig. 3C–E; Supplementary Table S7).
Figure 3.
Chromosomal arm-level somatic copy-number profiles and PA. Targeted sequencing data were used to assess genome-wide arm-level SCNAs across serial plasma samples. Representative examples of genome-wide SCNA profiles are shown for 2 patients. A, Patient 10 displayed multiple copy-number gains (chromosomes 1, 3q, 5p, 7, 12p, 15, 17, and 20) and losses (chromosomes 2, 3p, 4, 5q, 8, 9q, 10, 13, and 16) in plasma at baseline sampling. Analysis of these regions across follow-up (week 3–73) plasma samples collected after cisplatin/etoposide treatment indicated a return to normal ploidy, which was detected prior to the radiographic assessment of partial or complete (PR/CR) response at week 11. B, Patient 5 displayed widespread genome-wide copy-number aberrations across tumor (WES; top) and matched serial plasma samples (bottom). This included gains across chromosomes 1, 8, 12, 13, 17, 18, 19, and 20 and losses across chromosomes 2, 3p, 4, 5q, 7, 10q, 11, 14, 16, 21, and 22. All SCNAs detected in the tumor were captured in cfDNA at baseline and persistently across all follow-up time points analyzed in plasma after initiation of second-line ipilimumab/nivolumab combination therapy. This was consistent with a radiographic assessment of PD 5 weeks after initiation of ipilimumab/nivolumab combination therapy. C–E, The most aberrant alterations in individual chromosome arm-level copy number were used to calculate a genome-wide composite measure of PA (termed PA score) for each sample. Examples of longitudinal trends in PA scores are shown for 3 patients who had no detectable tumor-derived sequence alterations across plasma time points analyzed. C, For patient 3, aneuploidy was detected in plasma 4 weeks after baseline sampling, followed by a reduction to normal ploidy by the week 8 timepoint, consistent with an OS time exceeding 21 months from baseline sampling. In contrast, radiographic assessment at week 6 indicated PD for this patient. D–E, Similarly, in patients 19 and 29, aneuploidy was detected at baseline followed by a reduction to normal ploidy by week 3–4 and week 5–7 time points, respectively. This preceded radiographic assessments of PR at weeks 6 and 15, respectively.
Dynamics of cfTL predict therapeutic response
To evaluate changes in the clonal stoichiometry of ctDNA under the distinct selective pressure of immunotherapy and chemotherapy, patients were grouped into chemotherapy-only and immunotherapy-containing treatment groups for downstream analyses of cfTL changes. Combined analyses of ctDNA mutations and PA were performed to evaluate longitudinal dynamic changes in cfTL for each patient. We defined cfTL as the contribution of the most abundant tumor-derived sequence and/or PA score > 4.9 (Materials and Methods), and tracked cfTL during the course of therapy (14). Statistical analyses of the uncertainty in cfTL measurement at <1% maximal MAF revealed that complete elimination of cfTL according to mutation and PA dynamics was required to precisely determine the absence of detectable ctDNA during therapy (Supplementary Fig. S2). Through this approach, we were able to assess cfTL in all 33 patients and assign a molecular response classification based on distinct patterns of cfTL dynamics during therapy (Materials and Methods). A classification of molecular response was assigned to patients who displayed sustained complete elimination of cfTL from baseline to the final sampling time point, and patients who displayed persistence of cfTL across all time points analyzed were classified in the molecular progression category. Patients who displayed complete elimination of cfTL from baseline to intermediate time points followed by an increase at final sampling were assigned a classification of molecular response followed by recrudescence.
We found significant concordance between sequence mutation and longitudinal PA dynamics across patients analyzed, with 95% (19/20) of cases with both evaluable metrics showing concordant responses (P < 0.001, Fisher exact test; Supplementary Fig. S5). Overall, 9 patients were classified as molecular responders from our cohort and 10 as molecular progressors, resulting in a difference in the log hazards of OS of −2.32 (95% CI: −3.9 to −0.74), which exceeds the effect size that we were powered to detect. In patients who were classified as molecular responders, we observed complete elimination of cfTL after a median time of 4 weeks following the initiation of therapy (range, 2–8 weeks; Fig. 4A). Six molecular responders had a best radiographic response assessment of either partial or complete response (PR/CR). In one molecular responder, cfTL was detected in plasma 4 weeks after initiation of immunotherapy, followed by elimination across all subsequent follow-up time points. These trends were consistent with the prolonged OS time of this patient exceeding 21 months from the time of baseline sampling. In contrast, radiographic assessment at week 6 indicated progressive disease (PD), which did not capture long-term clinical outcomes in this patient. For 14 patients, we observed an initial molecular response, followed by a recrudescence of cfTL (Fig. 4B) or emergence of new clones for 3 patients (Fig. 4C). Among molecular progressors, we found persistence of cfTL across all time points analyzed (Fig. 4D).
Figure 4.
Dynamic changes in cfTL during therapy. Longitudinal changes in cfTL across plasma time points analyzed for each patient were used to assign a combined molecular response classification. Representative examples are shown of patients who were assigned to each of the 3 classifications. A, Patient 10 was classified as a molecular responder based on the complete elimination of cfTL, assessed using tumor-derived sequence alterations, between baseline and week 5 sampling during cisplatin/etoposide chemotherapy treatment (indicated by the green shaded area). A reduction in PA scores to undetectable levels from baseline was also observed in this patient. B and C, Patients 26 and 21 were assigned a classification of molecular response followed by recrudescence based on the elimination of cfTL between baseline and intermediate time points [during atezolizumab/etoposide/carboplatin (purple) and carboplatin/etoposide (green) treatment, respectively], after which an increase in cfTL was observed at the final time points analyzed. C, In patient 21, a shift in mutation profiles defined by the presence of tumor-derived RET (p.Y314F) and TP53 (p.V173E) mutations at recrudescence, which were not present at the baseline timepoint, was observed. D, Patient 1 was classified as a molecular progressor based on the persistence of cfTL, defined by a tumor-derived TP53 (p.C135S) sequence alteration, across all time points analyzed during treatment with nivolumab (blue). E, Combined molecular responses were significantly associated with clinical evaluations of best radiographic response (P = 0.003, Fisher exact test). F, A broader comparison between the elimination of cfTL at any timepoint analyzed for the study and radiographic assessments further revealed concordance (P = 0.001, Fisher exact test) between each variable. G, Molecular responses were determined on average 4 weeks prior to best radiographic response assessments in 28 patients with comparable ctDNA and imaging assessments in this cohort (mean 5.61 weeks vs. 10.21 weeks; P = 0.01 Mann–Whitney U test). Patients without baseline plasma samples available (n = 2) and cases with discordant molecular and radiographic responses (n = 3) were excluded from analyses. Mean times to response assessment are shown alongside standard error for each modality.
Overall, molecular response classifications assigned based on longitudinal cfTL changes were consistent with radiographic response assessments (Fig. 4E, P = 0.003, Fisher exact test) and were associated with DCB (Supplementary Fig. S6A; P = 0.019, Fisher exact test) for patients treated with immunotherapy-containing regimens. These results demonstrate the predictive value of early on-therapy cfTL elimination for determining therapeutic responses to both systemic chemotherapy and immunotherapy in SCLC. Concordance with radiographic responses was also observed for the elimination of cfTL at any time point following the initiation of therapy (Fig. 4F; P = 0.001, Fisher exact test), suggesting the presence of cfTL elimination, in addition to the time to elimination, may be relevant for the determination of therapeutic outcomes in patients. A logistic regression model incorporating structural and sequence response predictions in the subset of 20 patients with both metrics evaluable confirmed that combined molecular responses outperformed individual features (sequence mutation and PA dynamics) for the prediction of therapeutic response (Supplementary Fig. S5C). Molecular responses derived from ctDNA were identified on average 4 weeks prior to best radiographic assessments in this cohort (Fig. 4G; P = 0.01, Mann–Whitney U test).
ctDNA molecular responses predict clinical outcomes
Finally, we assessed whether cfTL dynamics were associated with PFS and OS (Fig. 5; Supplementary Fig. S6B–S6E). Patients classified as molecular responders attained longer OS and PFS compared with patients who displayed initial molecular responses followed by recrudescence or molecular progression (median OS–OS not reached vs. 12.35 vs. 6.48 months, respectively, P = 0.0006 and median PFS–PFS not reached vs. 6.18 vs. 1.74 months, respectively, P < 0.0001; Fig. 5A and B). These trends were consistent across patients who received first-line treatment (median OS 21.01 vs. 12.25 vs. 6.44 months, respectively, P = 0.036 and median PFS not reached vs. 6.18 vs. 2.99 months, respectively, P < 0.001) and among patients who received either second- or third-line treatments (median OS not reached vs. 34.78 vs. 6.51 months, respectively, P = 0.015 and median PFS 7.17 vs. 7.41 vs. 1.22 months, respectively, P = 0.051). We repeated these analyses after the removal of patients without baseline plasma samples (n = 2); however, we did not identify any changes in the associations between ctDNA molecular responses and OS and PFS (Supplementary Fig. S7). Importantly, ctDNA molecular responses remained a significant predictor of OS (molecular response vs. molecular progression HR = 0.09, 95% CI = 0.02–0.42, P = 0.002; molecular response f/b recrudescence vs. molecular progression HR = 0.14, 95% CI = 0.04–0.48, P = 0.002) and PFS (molecular response vs. molecular progression HR = 0.02, 95% CI = 0.00–0.16, P < 0.001; molecular response f/b recrudescence vs. molecular progression HR = 0.05, 95% CI = 0.01–0.26, P < 0.001) after accounting for clinical covariates in a multivariate Cox proportional hazards regression model (Supplementary Tables S8–S9). To evaluate the importance of matched WBC filtering for the accurate assessment of circulating tumor burden, longitudinal changes in cfTL were reanalyzed for each patient, considering all plasma variants, without filtering out CH-derived alterations. CH-unfiltered ctDNA responses were not significantly associated with either OS or PFS (Supplementary Fig. S8), demonstrating the importance of matched WBC analyses for the accurate tracking of cfTL.
Figure 5.
Combined tiered ctDNA molecular responses and the prediction of survival outcomes. Of the 33 patients included in the study cohort, patients who were assigned a classification of molecular response (green) based on longitudinal dynamic changes in cfTL displayed superior (A) overall survival (OS; median survival not reached vs. 12.35 vs. 6.48 months; log-rank P 0.0006) and (B) progression-free survival (PFS; median survival not reached vs. 6.18 vs. 1.74 months; log-rank P < 0.0001) compared with patients with an initial molecular response followed by recrudescence (orange) or molecular progression (red). C and D, Molecular responses derived from ctDNA (dark purple) were a stronger predictor of OS at 12 (AUC 78.1 vs. 73.3) and 36 (AUC 80.0 vs. 71.8) months, and (E and F) early post-therapy PFS at 3 (AUC 91.7 vs. 82.0) and 12 (AUC 83.5 vs. 78.8) months, compared with radiographic assessment (PR/CR vs. SD/PD/MR; light purple). f/b, followed by.
The predictive ability of ctDNA molecular responses for the determination of landmark OS (12, 36, and 64 months) and PFS at 3, 6, 9, 12, and 14 months was assessed comparatively to best radiographic assessments for each patient. These analyses demonstrated that ctDNA molecular responses more accurately predicted OS at 12 months (AUC 78.1% vs. 73.3%), 36 months (AUC 80.8% vs. 71.8%), and importantly at 64 months (AUC 87.3% vs. 67.6%) compared with imaging (Fig. 5C and D; Supplementary Fig. S6B). Similarly, ctDNA molecular responses displayed a higher predictive performance for PFS at 3 months (AUC 91.7% vs. 82.0%), 12 months (AUC 83.5% vs. 78.8%), and 14 months (AUC 92.9% vs. 76.8%), compared with radiographic assessments (Fig. 5E and F), and were similarly predictive of PFS at 6 (AUC 74.1% vs. 77.1%) and 9 months (AUC 71.1% vs. 77.3%; Supplementary Fig. S6C–S6E). Taken together, these findings suggest that ctDNA molecular responses may be particularly informative in predicting durable therapeutic responses and long-term clinical outcomes.
Discussion
Previous efforts to characterize ctDNA alterations in SCLC have largely focused on tracking mutations during the course of treatment, rather than a combined evaluation of the evolving mutation and PA landscapes (7, 8, 10–12). We developed an approach for multifeature assessment of circulating tumor burden by incorporating mutation and PA dynamics, allowing for more sensitive detection of circulating tumor load during therapy. This tumor-agnostic WBC DNA-informed strategy may provide a comprehensive assessment of tumor-derived sequence and large-scale chromosomal aberrations that are representative of longitudinal changes in tumor burden under the selective pressure of treatment. Our findings highlight the value of longitudinal ctDNA molecular responses to capture and interpret clinical outcomes for patients with SCLC.
The sensitive detection of ultra-low levels of ctDNA in a background of nontumor cfDNA in peripheral blood has presented a major challenge for liquid biopsies. This is exacerbated by the possible misclassification of variants related to clonal hematopoiesis as tumor-derived alterations, particularly as CH variants can occur within known oncogenic driver genes for solid tumors (9, 27, 34, 35). To this end, optimized next-generation sequencing (NGS) approaches that enable the specific detection and filtering of CH variants from candidate ctDNA alterations can help to significantly reduce the biological noise inherent to tumor-agnostic NGS approaches (17). Using ultrasensitive targeted error-correction sequencing of plasma and matched WBC DNA samples, we showed that the landscape of alterations detected in ctDNA is representative of the SCLC genomic landscape, including mutations in TP53 and copy-number aberrations in chromosomes 1p, 3, 5p, 8q, 10q, and 17p, which are regions known to harbor genomic gains and losses in SCLC tumors (22). The high mutant allele fractions (average 18%) of tumor-derived sequence alterations identified in ctDNA in this study were consistent with previous reports in SCLC (8, 12, 15), suggesting an increased cfTL for SCLC tumors. Importantly, these characterizations were enabled by the precise detection and filtering out of alterations attributed to clonal hematopoiesis, which were detected in 35% of cases in our cohort, demonstrating the importance of correctly classifying CH variants for accurate quantification of circulating tumor load in patients. Filtering out biological noise related to CH variants allowed for accurate determination of ctDNA molecular response in a tumor-agnostic manner. Moreover, as this approach to mutational analysis was based on tracking the maximal MAF across all tumor-derived ctDNA alterations detected at each time point, rather than the numeric value of individual mutations, our method was not dependent upon the detection of low-frequency subclonal alterations that are likely to be missed by most hybrid capture NGS approaches with a limit of detection in the 0.01% to 0.1% range (40).
In addition to evaluating ctDNA molecular responses by mutation trends (14, 16–21), we implemented a combined tiered approach that utilizes both on-target and off-target reads from TEC-seq NGS outputs to determine copy-number profiles and PA (13) and combined sequence alteration dynamics with PA to determine ctDNA molecular responses. Such an approach may maximally leverage on-target and off-target NSG data for the simultaneous assessment of both somatic mutations and PA. Analyses of the landscape of large-scale structural alterations in plasma have previously demonstrated that aneuploidy and its plasticity during the therapeutic intervention can provide real-time insight into tumor responses and identify early resistance (41–43). Nevertheless, previously reported methods for aneuploidy detection in plasma may not be compatible with targeted NGS data and do not utilize a normal DNA reference set for either the correction of plasma coverage profiles or the computation of thresholds for the determination of PA (44). As a proof of concept, we evaluated the concordance between tumor tissue-derived and plasma-derived copy-number profiles for a subset of patients and found a high degree of concordance. This combined tiered approach to ctDNA response evaluation allowed for the detection of molecular responses in all patients included in our study cohort earlier than radiographic responses.
We identified 3 distinct patterns of ctDNA molecular response that were reflective of clinical outcomes; such that sustained early ctDNA elimination differentiated patients with ctDNA elimination followed by recrudescence and patients in whom ctDNA burden persisted during therapy. Overall, patients with sustained ctDNA elimination attained longer PFS and OS, supporting the importance of ctDNA complete elimination as a reliable indicator of durable tumor response to systemic therapy in patients with SCLC (7, 45, 46). In contrast, we observed a clear pattern of ctDNA molecular progression among patients with disease progression. The longitudinal nature of our analyses allowed for the detection of additional ctDNA kinetics patterns including ctDNA recrudescence that is associated with distinct clinical courses. ctDNA recrudescence denotes an increase in levels of mutations that were detected prior to therapy but were completely eliminated during therapy and recrudesce as therapeutic resistance emerges. Our analyses supported that ctDNA recrudescence was distinct from ctDNA progression, the latter denoting persistent or increasing ctDNA levels across longitudinally sampled time points. Accordingly, we found significant differences in clinical outcomes between patients with ctDNA recrudescence compared with those with ctDNA progression. Although patients with ctDNA recrudescence had inferior outcomes compared with patients with sustained ctDNA elimination, they attained significantly longer PFS and OS compared with patients with ctDNA persistence.
Our study has several limitations, predominantly related to the small sample size of the cohort analyzed. Due to its retrospective nature, best radiographic responses were retrospectively abstracted from the clinical records, as RECIST evaluations were not available. Although blood sampling was overall harmonized across patients included in the study, blood collection time points were not fully standardized with respect to imaging evaluations. This variation in blood sampling and imaging time points may have introduced a bias in the comparison between time to molecular and best radiographic responses in this study. The uncertainty in cfTL measurement below 1% maximal MAF and incomplete coverage of the RB1 gene may represent additional limitations.
Future prospective studies will be essential to validate our findings in the clinical setting. Our analyses supported the predictive nature of ctDNA response with respect to therapeutic response, independent of the treatment administered. Therefore, prospective validation of these findings in clinical trials can be pursued in settings where patient selection is needed to better prognosticate clinical outcomes. As an example, in the CASPIAN study of first-line durvalumab with platinum-etoposide followed by durvalumab, 17% of patients with extensive-stage (ES) SCLC were alive at 3 years (4), highlighting a patient population with differential clinical outcomes that may be captured by ctDNA molecular response approaches. The predictive value of ctDNA kinetics can also be prospectively validated in the limited-stage setting after chemoradiation, especially as immunotherapy approaches are tested in this setting (ADRIATIC; NCT03703297, ACHILES; NCT03540420, NRG-LU005; NCT03811002). By implementing ctDNA kinetics as an early endpoint of clinical outcome, immune-checkpoint blockade may thus be reserved for the patients most likely to benefit. In addition to enabling the standardization of blood collection and imaging time points relative to each assessment and treatment time points and RECIST response assessments, prospective analyses will facilitate the further assessment of the optimal timing for ctDNA molecular response evaluation after initiation of treatment with either systemic platinum-based chemotherapy or immunotherapy-containing regimens. Such efforts will also be critical to further evaluate the durability of ctDNA molecular responses and their value in predicting long-term PFS and OS.
In summary, we showed that dynamic molecular responses following systemic chemotherapy or immunotherapy can be reliably captured through a comprehensive assessment of ctDNA sequence and structural landscapes prior to radiologic progression. These results suggest that therapeutic response monitoring based on combined molecular response criteria may be used to provide guidance on clinical endpoints in SCLC, including both PFS and OS. Given the rapid clinical progression typically observed in patients with SCLC, our combined tiered approach to ctDNA-based tumor burden assessment may provide a foundation for the early identification of durable molecular responses or resistance, when the decision to continue or switch to an alternative more effective therapy will have the most clinical benefit. Larger prospective studies should be pursued to determine whether therapeutic decisions based on longitudinal ctDNA dynamics can significantly improve outcomes for patients with SCLC.
Supplementary Material
Supplementary Tables S1-S9
Schematic summarizing the branched logic used to determine the origin of sequence alterations identified in plasma NGS. Variants representing lung cancer hotspots were classified as tumor-derived, independent of their presence in matched WBC DNA NGS. Variants with a variant allele fraction (VAF) >25% in all WBC DNA and plasma samples from a patient were classified as germline. Alterations that were detected in matched WBC DNA sequencing or in the canonical clonal hematopoiesis (CH) gene DNMT3A were classified as CH-derived. For all variants detected in plasma using TEC-Seq but not in matched WBC DNA samples (based on a threshold of >3 supermutants), we performed an additional evaluation of their supermutant counts in the WBC DNA sequence data to exclude potential CH origin. Plasma variants with a supermutant count of >1 in matched WBC DNA TEC-Seq data were classified as CH-derived and alterations with a matched WBC DNA supermutant count of 0 were assigned a tumor-derived origin.
(A) PA scores were assessed across sequenced plasma (n=139) and matched white blood cell (n=32) samples from patients analyzed in the study cohort to determine a threshold for determination of aneuploidy in plasma. A PA score of 4.90, which was the maximum score observed in WBC DNA samples, was selected as the threshold for determination of aneuploidy in plasma. (B) Distribution of PA scores in the panel of 32 matched normal WBC samples analyzed for this study and an independent held out set of 56 WBC normal samples that were processed and analyzed using the same targeted panel and sequencing approach (Methods). The maximum PA score observed for both normal panels was 4.90, supporting the use of this value as the threshold for determination of aneuploidy in plasma. No significant differences were observed in the median PA score (indicated by dotted lines) for each normal panel. (C) Comparison between plasma aneuploidy scores and tumor fraction scores determined using an orthogonal method (ichorCNA) in matched normal DNA (blue) and plasma samples from all timepoints (yellow), plasma samples with detectable tumor-derived sequence mutations (orange) and plasma timepoints with undetectable sequence alterations (grey). Non-zero ichorCNA tumor fraction values were assigned to most samples resulting in significant overlap between the distributions of tumor fractions in plasma and matched normal DNA samples. In contrast, comparisons performed using plasma aneuploidy scores revealed significant differences in the distribution of assigned values across matched normal DNA samples (reflecting normal ploidy) and plasma samples (reflecting tumor aneuploidy). (D-I) Statistical modelling of MAF dynamics was performed using a set of known tumor-specific mutations previously characterized in Phallen et al, Science Transl Med, 2017 from a mixture of DNA from tumor cell lines spiked into unrelated wild-type DNA at dilutions ranging from 0.1-100%. Mutations in this set with replicate data were evaluated for (D, G) variability in MAF estimate followed by (E, H) calculation of the coefficient of variation (CV), and (F, I) finally projection of the relative percentage uncertainty in the MAF estimate. Analyses of mutation data from Phallen et al (2017) are shown in (D-F) and results from a simulation study of 30 mutations ranging in MAF from 0.1% to 80% using a simple statistical model of variability are shown in (G-I). Both analyses of mutation data from Phallen et al (2017) and simulation data showed a significant increase in CV as MAF decreases. For example, given a maximal MAF of 1%, the real 95% uncertainty in MAF estimate was shown to range from -50% to 50% (0.5% MAF to 1.5% MAF). Thus, a greater relative reduction is required to determine elimination of ctDNA for lower maximal MAF values. Based on these results, complete elimination of cell-free tumor load (to either 0% max MAF or undetectable PA) was deemed to be the most appropriate threshold for precise determination of molecular response.
Sequence alterations that were classified as germline (median 44.9%, range 27.3% - 73.1%) had the highest overall MAFs, compared to both tumor-derived (median 10.2%, range 0.1% - 75.7%) and clonal-hematopoiesis-derived (CH-derived) variants (median 0.8%, range 0.1 – 86.7%). Tumor-derived variants had significantly higher MAFs compared to CH-derived variants. P values were calculated using the Mann-Whitney U test. **** p < 0.0001
(A) Genome-wide log2 copy ratios derived from targeted error-correction sequencing (TEC-Seq) data using CNVkit (Methods) are shown for each plasma sample, with copy number gains indicated in red and losses indicated in blue. Each row corresponds to a single plasma sample and rows are ordered with respect to consecutive sampling timepoints analyzed for each patient. (B-F) Genome-wide copy number profiles are shown for patients with matched tumor (WES) and plasma samples analyzed. We observed a high concordance between copy number profiles derived by whole exome sequencing, our approach (CNVkit) and ichorCNA for patients with detectable aneuploidy in plasma. (B) Patient 3 had undetectable ctDNA according to tumor-specific sequence alterations. However, this patient was classified as a molecular responder based on decrease of plasma aneuploidy to normal ploidy by the week 8 timepoint. (C) Patient 5 was classified as a molecular progressor based on the presence of tumor-specific sequence alterations and plasma aneuploidy across all timepoints analyzed. (D) Similarly, patient 9 was classified as a molecular progressor based on the persistence of mutations and plasma aneuploidy, which were detectable across all sampled timepoints from this patient. (E, F) Plasma copy number profiles for patients 11 and 14 indicated ploidy close to normal ploidy across all timepoints analyzed. These patients were classified as molecular responders based on tumor-derived sequence alterations.
(A) Overall, 27 patients had cell-free tumor load (cfTL) detected using tumor-derived sequence alterations, of which 20 patients also had evaluable plasma aneuploidy (PA)-based responses. Comparison between ctDNA responses determined using mutation-based and plasma aneuploidy-based responses in these 20 patients revealed significant concordance (p < 0.001, Fisher’s exact test). (B) Concordance was also observed between complete elimination of cfTL based on tumor-derived sequence alterations and plasma aneuploidy dynamics in these patients (p < 0.001, Fisher’s exact test). For these comparisons, plasma aneuploidy dynamics were assessed using a binarized classification of PA scores (PA score > 4.90 indicative of tumor aneuploidy, PA score <4.90 indicative of normal ploidy): PA undetectable (normal ploidy across all timepoints analyzed), PA response (aneuploidy detected at baseline or T1 sampling followed by a reduction to normal ploidy by final sampling), PA response followed by recrudescence (aneuploidy detected at baseline and final sampling with a reduction to normal ploidy at intermediate timepoints), PA progression (aneuploidy detected across all timepoints analyzed). (C) A logistic regression model combining both mutation-based and aneuploidy-based ctDNA responses in these 20 patients demonstrated that the combination of both sequence and structural dynamics outperformed either metric for the prediction of overall radiographic response (Combined model AUC = 0.90, Sequence alterations AUC = 0.85, Structural Alterations AUC = 0.74). (D-E) Further analyses of molecular responses derived from mutation-based analyses of cfTL changes in 27 patients with evaluable tumor-derived mutation trends demonstrated significant associations with both overall (OS) and progression-free (PFS) survival (log-rank p = 0.02 and p < 0.0001, respectively), independent of treatment modality (Chemotherapy vs immunotherapy-containing regimens). (F-G) cfTL elimination according to mutation-based analyses was also predictive of both OS and PFS (log-rank p = 0.01 and P < 0.0001, respectively). (H-I) Similarly, PA-based ctDNA responses showed significant associations with both OS (p =0.003) and PFS (log-rank p=0.001).
(A) Comparison between ctDNA molecular responses and clinical benefit determined through radiographic imaging for patients treated with immunotherapy-containing regimens revealed concordance between each assessment (p = 0.019, Fisher’s exact test). For these comparisons, durable clinical benefit (DCB) was defined as a lack of progression or death within 6 months of treatment initiation; non-durable benefit (NDB) was defined as progression or death within 6 months of treatment initiation. Comparison between ctDNA molecular responses and radiographic assessments (PR/CR vs SD/PD/MR) for the determination of (B) overall survival (OS) at 64 months (maximum OS observed in patients with date of death recorded) and progression-free survival (PFS) at (C) 6 months, (D) 9 months and (E) 14 months (maximum PFS observed in patients with clinically documented progression following systemic treatment), is shown for all 33 patients included in the study cohort.
Survival analyses of 31 patients with evaluable baseline plasma samples revealed that ctDNA molecular responses were significantly associated with both (A) overall survival-OS and (B) progression-free survival-PFS. Patients classified as molecular responders had a significantly longer OS and PFS compared to patients who were assigned a classification of molecular response f/b recrudescence or molecular progression. Comparison between molecular responses and radiographic assessments (PR/CR vs SD/PD/MR) in these patients revealed that ctDNA molecular responses were a stronger predictor of (C) OS at 12 and (D) 36 months and PFS at (E) 3 and (F) 12 months, compared to best overall radiographic responses.
Longitudinal changes in cfTL were re-analyzed for each patient, considering all plasma variants independent of origin. Survival analyses revealed that CH-unfiltered ctDNA responses were not associated with either (A) overall survival-OS or (B) progression-free survival-PFS. Comparison between CH-unfiltered ctDNA responses and radiographic assessments (PR/CR vs SD/PD/MR) in these patients revealed that CH-unfiltered responses we not predictive of (C) OS at 12 and (D) 36 months, and PFS at (E) 3 and (F) 12 months, compared to best overall radiographic responses.
Acknowledgments
This work was supported in part by the NIH grant CA121113 (V. Anagnostou and V.E. Velculescu), the Bloomberg–Kimmel Institute for Cancer Immunotherapy (V. Anagnostou), the Johns Hopkins SKCCC core grant NCI CCSG P30 CA006973 (V. Anagnostou and V.E. Velculescu), the ECOG–ACRIN Thoracic Malignancies Integrated Translational Science Center grant UG1CA233259 (V. Anagnostou and V.E. Velculescu), the V Foundation (V. Anagnostou and V.E. Velculescu), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (V.E. Velculescu), the Commonwealth Foundation (V. Anagnostou and V.E. Velculescu), the International Lung Cancer Foundation (L. Sivapalan), the LUNGevity Foundation (V. Anagnostou), and Stand Up To Cancer–LUNGevity–American Lung Association Lung Cancer Interception Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT23-17). Work in the Lovly lab was supported by the NIH (grant numbers U54CA217450-01, U01CA224276-01, P30-CA086485, and UG1CA233259), a Vanderbilt–Ingram Cancer Center Young Ambassadors Award, and a Lung Cancer Foundation of America/International Association for the Study of Lung Cancer Lori Monroe Scholarship. W.T. Iams was supported by an NCCN Young Investigator Award. Stand Up To Cancer is a division of the Entertainment Industry Foundation. The indicated Stand Up To Cancer grant is administered by the American Association for Cancer Research, the scientific partner of SU2C.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
W.T. Iams reports other support from Genentech, Jazz Pharma, G1 Therapeutics, Mirati, Bristol Myers Squibb, Takeda, Janssen, EMD Serono, Elevation Oncology, Catalyst, OncLive, Chardan, Clinical Care Options, Cello Health, and Curio Science outside the submitted work. S.C. Scott reports personal fees from Genentech, Foundation Medicine, and Regeneron and grants from Mirati Therapeutics outside the submitted work. J.R. White reports other support from Resphera Biosciences LLC outside the submitted work. V.E. Velculescu reports grants, personal fees, and other support from Delfi Diagnostics during the conduct of the study as well as other support from Viron Therapeutics and Epitope outside the submitted work; in addition, V.E. Velculescu has various patents pending, issued, licensed, and with royalties paid from Delfi Diagnostics, LabCorp, Qiagen, Sysmex, Agios, Genzyme, Esoterix, Ventana, and ManaT Bio; is a founder of Delfi Diagnostics and serves on the board of directors and as a consultant for this organization; and owns Delfi Diagnostics stock, which is subject to certain restrictions under university policy. Additionally, Johns Hopkins University owns equity in Delfi Diagnostics. V.E. Velculescu divested his equity in Personal Genome Diagnostics (PGDx) to LabCorp in February 2022. V.E. Velculescu is an inventor on patent applications submitted by Johns Hopkins University related to cancer genomic analyses and cell-free DNA for cancer detection that have been licensed to one or more entities, including Delfi Diagnostics, LabCorp, Qiagen, Sysmex, Agios, Genzyme, Esoterix, Ventana, and ManaT Bio. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. V.E. Velculescu is an advisor to Viron Therapeutics and Epitope. These arrangements have been reviewed and approved by Johns Hopkins University in accordance with its conflict-of-interest policies. C.L. Hann reports grants from AbbVie and Genentech; grants and personal fees from Amgen, AstraZeneca, and BMS; and personal fees from Janssen during the conduct of the study. C.M. Lovly reports grants from U54CA217450-01, U01CA224276-01, P30-CA086485, UG1CA233259, Vanderbilt-Ingram Cancer Center Young Ambassadors Award, and Lung Cancer Foundation of America/International Association for the Study of Lung Cancer Lori Monroe Scholarship during the conduct of the study as well as personal fees from Amgen, AstraZeneca, Blueprints Medicine, Cepheid, D2G Oncology, Daiichi Sankyo, Eli Lilly, EMD Serono, Foundation Medicine, Genentech, Janssen, Medscape, Pfizer, Puma, Roche, and Takeda outside the submitted work; in addition, C.M. Lovly chairs a data and safety monitoring board for a Janssen study of amivantamab plus lazertinib, a role that is uncompensated and not directly related to this study. V. Anagnostou reports grants from AstraZeneca, Personal Genome Diagnostics, Delfi Diagnostics, and Bristol Myers Squibb during the conduct of the study as well as grants from AstraZeneca, Bristol Myers Squibb, Delfi Diagnostics, and Personal Genome Diagnostics outside the submitted work; in addition, V. Anagnostou has a patent for 17/779,936 issued and a patent for 17/047,006 issued and is an inventor on patent applications (63/276,525, 17/779,936, 16/312,152, 16/341,862, 17/047,006, and 17/598,690), submitted by Johns Hopkins University, related to cancer genomic analyses, ctDNA therapeutic response monitoring, and immunogenomic features of response to immunotherapy that have been licensed to one or more entities. Under the terms of these license agreements, the University and inventors are entitled to fees and royalty distributions. No disclosures were reported by the other authors.
Authors' Contributions
L. Sivapalan: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. W.T. Iams: Resources, data curation, investigation, methodology, writing–review and editing. Z. Belcaid: Conceptualization, investigation, methodology, writing–review and editing. S.C. Scott: Data curation, investigation, methodology, writing–review and editing. N. Niknafs: Data curation, formal analysis, visualization, methodology, writing–review and editing. A. Balan: Data curation, formal analysis, visualization, methodology, writing–review and editing. J.R. White: Formal analysis, visualization, methodology. P. Kopparapu: Data curation, investigation, methodology. C. Cann: Data curation, writing–review and editing. B.V. Landon: Data curation, investigation, methodology, writing–review and editing. G. Pereira: Resources, data curation. V.E. Velculescu: Resources, data curation, supervision, methodology, writing–review and editing. C.L. Hann: Resources, data curation, supervision, funding acquisition, methodology, writing–review and editing. C.M. Lovly: Resources, data curation, supervision, funding acquisition, methodology, writing–review and editing. V. Anagnostou: Conceptualization, resources, formal analysis, supervision, funding acquisition, visualization, methodology, writing–original draft, writing–review and editing.
References
- 1. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol 2016;893:1–19. [DOI] [PubMed] [Google Scholar]
- 2. van Meerbeeck JP, Fennell DA, De Ruysscher DKM. Small-cell lung cancer. Lancet 2011;378:1741–55. [DOI] [PubMed] [Google Scholar]
- 3. Rudin CM, Ismaila N, Hann CL, Malhotra N, Movsas B, Norris K, et al. Treatment of small-cell lung cancer: American Society of Clinical Oncology endorsement of The American College of Chest Physicians guideline. J Clin Oncol 2015;33:4106–11. [DOI] [PubMed] [Google Scholar]
- 4. Goldman JW, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide versus platinum-etoposide alone in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): updated results from a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2021;22:51–65. [DOI] [PubMed] [Google Scholar]
- 5. Liu SV, Reck M, Mansfield AS, Mok T, Scherpereel A, Reinmuth N, et al. Updated overall survival and PD-L1 subgroup analysis of patients with extensive-stage small-cell lung cancer treated with atezolizumab, carboplatin, and etoposide (IMpower133). J Clin Oncol 2021;39:619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:725–37. [DOI] [PubMed] [Google Scholar]
- 7. Iams WT, Kopparapu PR, Yan Y, Muterspaugh A, Zhao Z, Chen H, et al. Blood-based surveillance monitoring of circulating tumor DNA from patients with SCLC detects disease relapse and predicts death in patients with limited-stage disease. JTO Clin Res Reports 2020;1:100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith JT, Balar A, Lakhani DA, Kluwe C, Zhao Z, Kopparapu P, et al. Circulating tumor DNA as a biomarker of radiographic tumor burden in SCLC. JTO Clin Res Reports 2021;2:100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sivapalan L, Murray JC, Canzoniero JV, Landon B, Jackson J, Scott S, et al. Liquid biopsy approaches to capture tumor evolution and clinical outcomes during cancer immunotherapy. J Immunother Cancer 2023;11:e005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devarakonda S, Sankararaman S, Herzog BH, Gold KA, Waqar SN, Ward JP, et al. Circulating tumor DNA profiling in small-cell lung cancer identifies potentially targetable alterations. Clin Cancer Res 2019;25:6119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernandez-Cuesta L, Perdomo S, Avogbe PH, Leblay N, Delhomme TM, Gaborieau V, et al. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine 2016;10:117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang L, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun 2018;9:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4:162ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phallen J, Leal A, Woodward BD, Forde PM, Naidoo J, Marrone KA, et al. Early noninvasive detection of response to targeted therapy in non–small cell lung cancer. Cancer Res 2019;79:1204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohan S, Foy V, Ayub M, Leong HS, Schofield P, Sahoo S, et al. Profiling of circulating free DNA using targeted and genome-wide sequencing in patients with SCLC. J Thorac Oncol 2020;15:216–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015;6:7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017;9:eaan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, et al. Dynamics of tumor and immune responses during immune checkpoint blockade in non-small cell lung cancer. Cancer Res 2019;79:1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hwang M, Canzoniero JV, Rosner S, Zhang G, White JR, Belcaid Z, et al. Peripheral blood immune cell dynamics reflect antitumor immune responses and predict clinical response to immunotherapy. J Immunother Cancer 2022;10:e004688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Georgiadis A, Durham JN, Keefer LA, Bartlett BR, Zielonka M, Murphy D, et al. Noninvasive detection of microsatellite instability and high tumor mutation burden in cancer patients treated with PD-1 blockade. Clin Cancer Res 2019;25:7024–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vega DM, Nishimura KK, Zariffa N, Thompson JC, Hoering A, Cilento V, et al. Changes in circulating tumor DNA reflect clinical benefit across multiple studies of patients with non–small-cell lung cancer treated with immune checkpoint inhibitors. JCO Precis Oncol 2022;6:e2100372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anagnostou V, Niknafs N, Marrone K, Bruhm DC, White JR, Naidoo J, et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat cancer 2020;1:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 2015;7:283ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2019;47:D941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pagel KA, Kim R, Moad K, Busby B, Zheng L, Hynes-Grace M, et al. OpenCRAVAT, an open source collaborative platform for the annotation of human genetic variation. Biorxiv 2019;794297. Available from: http://biorxiv.org/content/early/2019/10/06/794297.abstract [Google Scholar]
- 27. Leal A, van Grieken NCT, Palsgrove DN, Phallen J, Medina JE, Hruban C, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun 2020;11:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Talevich E, Shain AH, Botton T, BBC CNVkit.: Genome-wide copy number detection and visualization from targeted DNA sequencing. PLOS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557–72. [DOI] [PubMed] [Google Scholar]
- 30. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2020. Available from: https://www.r-project.org/. [Google Scholar]
- 34. Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25:1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Naylor SL, Johnson BE, Minna JD, Sakaguchi AY. Loss of heterozygosity of chromosome 3p markers in small-cell lung cancer. Nature 1987;329:451–4. [DOI] [PubMed] [Google Scholar]
- 39. The Clinical Lung Cancer Genome Project (CLCGP) and Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med 2013;5:209ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet 2018;19:269–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012;4:162ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chan KCA, Jiang P, Chan CWM, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Douville C, Springer S, Kinde I, Cohen JD, Hruban RH, Lennon AM, et al. Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc Natl Acad Sci U S A 2018;115:1871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Normanno N, Rossi A, Morabito A, Signoriello S, Bevilacqua S, Di Maio M, et al. Prognostic value of circulating tumor cells’ reduction in patients with extensive small-cell lung cancer. Lung Cancer 2014;85:314–9. [DOI] [PubMed] [Google Scholar]
- 46. Hiltermann TJN, Pore MM, van den Berg A, Timens W, Boezen HM, Liesker JJW, et al. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol 2012;23:2937–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1-S9
Schematic summarizing the branched logic used to determine the origin of sequence alterations identified in plasma NGS. Variants representing lung cancer hotspots were classified as tumor-derived, independent of their presence in matched WBC DNA NGS. Variants with a variant allele fraction (VAF) >25% in all WBC DNA and plasma samples from a patient were classified as germline. Alterations that were detected in matched WBC DNA sequencing or in the canonical clonal hematopoiesis (CH) gene DNMT3A were classified as CH-derived. For all variants detected in plasma using TEC-Seq but not in matched WBC DNA samples (based on a threshold of >3 supermutants), we performed an additional evaluation of their supermutant counts in the WBC DNA sequence data to exclude potential CH origin. Plasma variants with a supermutant count of >1 in matched WBC DNA TEC-Seq data were classified as CH-derived and alterations with a matched WBC DNA supermutant count of 0 were assigned a tumor-derived origin.
(A) PA scores were assessed across sequenced plasma (n=139) and matched white blood cell (n=32) samples from patients analyzed in the study cohort to determine a threshold for determination of aneuploidy in plasma. A PA score of 4.90, which was the maximum score observed in WBC DNA samples, was selected as the threshold for determination of aneuploidy in plasma. (B) Distribution of PA scores in the panel of 32 matched normal WBC samples analyzed for this study and an independent held out set of 56 WBC normal samples that were processed and analyzed using the same targeted panel and sequencing approach (Methods). The maximum PA score observed for both normal panels was 4.90, supporting the use of this value as the threshold for determination of aneuploidy in plasma. No significant differences were observed in the median PA score (indicated by dotted lines) for each normal panel. (C) Comparison between plasma aneuploidy scores and tumor fraction scores determined using an orthogonal method (ichorCNA) in matched normal DNA (blue) and plasma samples from all timepoints (yellow), plasma samples with detectable tumor-derived sequence mutations (orange) and plasma timepoints with undetectable sequence alterations (grey). Non-zero ichorCNA tumor fraction values were assigned to most samples resulting in significant overlap between the distributions of tumor fractions in plasma and matched normal DNA samples. In contrast, comparisons performed using plasma aneuploidy scores revealed significant differences in the distribution of assigned values across matched normal DNA samples (reflecting normal ploidy) and plasma samples (reflecting tumor aneuploidy). (D-I) Statistical modelling of MAF dynamics was performed using a set of known tumor-specific mutations previously characterized in Phallen et al, Science Transl Med, 2017 from a mixture of DNA from tumor cell lines spiked into unrelated wild-type DNA at dilutions ranging from 0.1-100%. Mutations in this set with replicate data were evaluated for (D, G) variability in MAF estimate followed by (E, H) calculation of the coefficient of variation (CV), and (F, I) finally projection of the relative percentage uncertainty in the MAF estimate. Analyses of mutation data from Phallen et al (2017) are shown in (D-F) and results from a simulation study of 30 mutations ranging in MAF from 0.1% to 80% using a simple statistical model of variability are shown in (G-I). Both analyses of mutation data from Phallen et al (2017) and simulation data showed a significant increase in CV as MAF decreases. For example, given a maximal MAF of 1%, the real 95% uncertainty in MAF estimate was shown to range from -50% to 50% (0.5% MAF to 1.5% MAF). Thus, a greater relative reduction is required to determine elimination of ctDNA for lower maximal MAF values. Based on these results, complete elimination of cell-free tumor load (to either 0% max MAF or undetectable PA) was deemed to be the most appropriate threshold for precise determination of molecular response.
Sequence alterations that were classified as germline (median 44.9%, range 27.3% - 73.1%) had the highest overall MAFs, compared to both tumor-derived (median 10.2%, range 0.1% - 75.7%) and clonal-hematopoiesis-derived (CH-derived) variants (median 0.8%, range 0.1 – 86.7%). Tumor-derived variants had significantly higher MAFs compared to CH-derived variants. P values were calculated using the Mann-Whitney U test. **** p < 0.0001
(A) Genome-wide log2 copy ratios derived from targeted error-correction sequencing (TEC-Seq) data using CNVkit (Methods) are shown for each plasma sample, with copy number gains indicated in red and losses indicated in blue. Each row corresponds to a single plasma sample and rows are ordered with respect to consecutive sampling timepoints analyzed for each patient. (B-F) Genome-wide copy number profiles are shown for patients with matched tumor (WES) and plasma samples analyzed. We observed a high concordance between copy number profiles derived by whole exome sequencing, our approach (CNVkit) and ichorCNA for patients with detectable aneuploidy in plasma. (B) Patient 3 had undetectable ctDNA according to tumor-specific sequence alterations. However, this patient was classified as a molecular responder based on decrease of plasma aneuploidy to normal ploidy by the week 8 timepoint. (C) Patient 5 was classified as a molecular progressor based on the presence of tumor-specific sequence alterations and plasma aneuploidy across all timepoints analyzed. (D) Similarly, patient 9 was classified as a molecular progressor based on the persistence of mutations and plasma aneuploidy, which were detectable across all sampled timepoints from this patient. (E, F) Plasma copy number profiles for patients 11 and 14 indicated ploidy close to normal ploidy across all timepoints analyzed. These patients were classified as molecular responders based on tumor-derived sequence alterations.
(A) Overall, 27 patients had cell-free tumor load (cfTL) detected using tumor-derived sequence alterations, of which 20 patients also had evaluable plasma aneuploidy (PA)-based responses. Comparison between ctDNA responses determined using mutation-based and plasma aneuploidy-based responses in these 20 patients revealed significant concordance (p < 0.001, Fisher’s exact test). (B) Concordance was also observed between complete elimination of cfTL based on tumor-derived sequence alterations and plasma aneuploidy dynamics in these patients (p < 0.001, Fisher’s exact test). For these comparisons, plasma aneuploidy dynamics were assessed using a binarized classification of PA scores (PA score > 4.90 indicative of tumor aneuploidy, PA score <4.90 indicative of normal ploidy): PA undetectable (normal ploidy across all timepoints analyzed), PA response (aneuploidy detected at baseline or T1 sampling followed by a reduction to normal ploidy by final sampling), PA response followed by recrudescence (aneuploidy detected at baseline and final sampling with a reduction to normal ploidy at intermediate timepoints), PA progression (aneuploidy detected across all timepoints analyzed). (C) A logistic regression model combining both mutation-based and aneuploidy-based ctDNA responses in these 20 patients demonstrated that the combination of both sequence and structural dynamics outperformed either metric for the prediction of overall radiographic response (Combined model AUC = 0.90, Sequence alterations AUC = 0.85, Structural Alterations AUC = 0.74). (D-E) Further analyses of molecular responses derived from mutation-based analyses of cfTL changes in 27 patients with evaluable tumor-derived mutation trends demonstrated significant associations with both overall (OS) and progression-free (PFS) survival (log-rank p = 0.02 and p < 0.0001, respectively), independent of treatment modality (Chemotherapy vs immunotherapy-containing regimens). (F-G) cfTL elimination according to mutation-based analyses was also predictive of both OS and PFS (log-rank p = 0.01 and P < 0.0001, respectively). (H-I) Similarly, PA-based ctDNA responses showed significant associations with both OS (p =0.003) and PFS (log-rank p=0.001).
(A) Comparison between ctDNA molecular responses and clinical benefit determined through radiographic imaging for patients treated with immunotherapy-containing regimens revealed concordance between each assessment (p = 0.019, Fisher’s exact test). For these comparisons, durable clinical benefit (DCB) was defined as a lack of progression or death within 6 months of treatment initiation; non-durable benefit (NDB) was defined as progression or death within 6 months of treatment initiation. Comparison between ctDNA molecular responses and radiographic assessments (PR/CR vs SD/PD/MR) for the determination of (B) overall survival (OS) at 64 months (maximum OS observed in patients with date of death recorded) and progression-free survival (PFS) at (C) 6 months, (D) 9 months and (E) 14 months (maximum PFS observed in patients with clinically documented progression following systemic treatment), is shown for all 33 patients included in the study cohort.
Survival analyses of 31 patients with evaluable baseline plasma samples revealed that ctDNA molecular responses were significantly associated with both (A) overall survival-OS and (B) progression-free survival-PFS. Patients classified as molecular responders had a significantly longer OS and PFS compared to patients who were assigned a classification of molecular response f/b recrudescence or molecular progression. Comparison between molecular responses and radiographic assessments (PR/CR vs SD/PD/MR) in these patients revealed that ctDNA molecular responses were a stronger predictor of (C) OS at 12 and (D) 36 months and PFS at (E) 3 and (F) 12 months, compared to best overall radiographic responses.
Longitudinal changes in cfTL were re-analyzed for each patient, considering all plasma variants independent of origin. Survival analyses revealed that CH-unfiltered ctDNA responses were not associated with either (A) overall survival-OS or (B) progression-free survival-PFS. Comparison between CH-unfiltered ctDNA responses and radiographic assessments (PR/CR vs SD/PD/MR) in these patients revealed that CH-unfiltered responses we not predictive of (C) OS at 12 and (D) 36 months, and PFS at (E) 3 and (F) 12 months, compared to best overall radiographic responses.
Data Availability Statement
Next-generation sequence data can be retrieved from the European Genome-Phenome Archive (EGA; accession number EGAS00001006831).



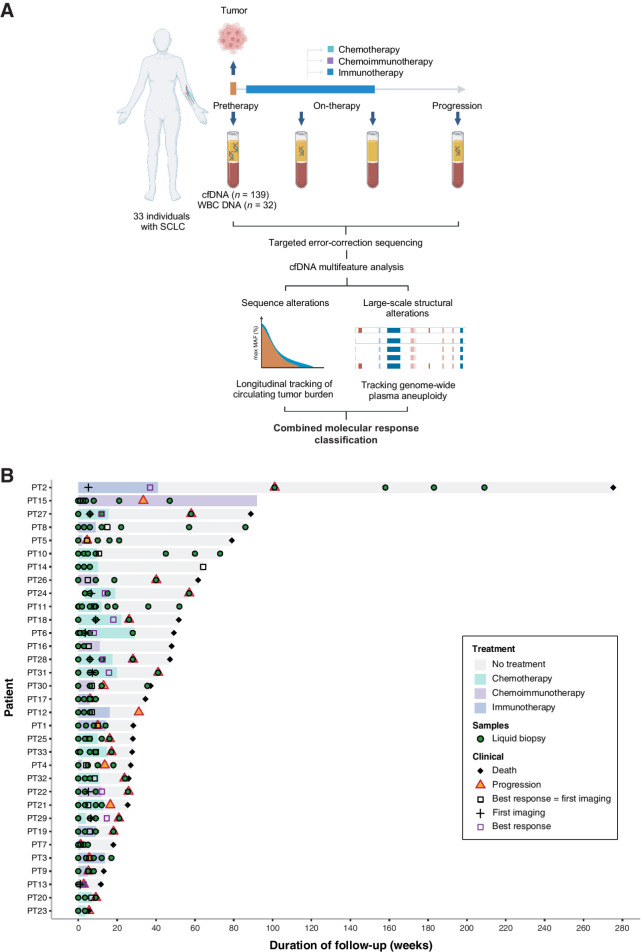
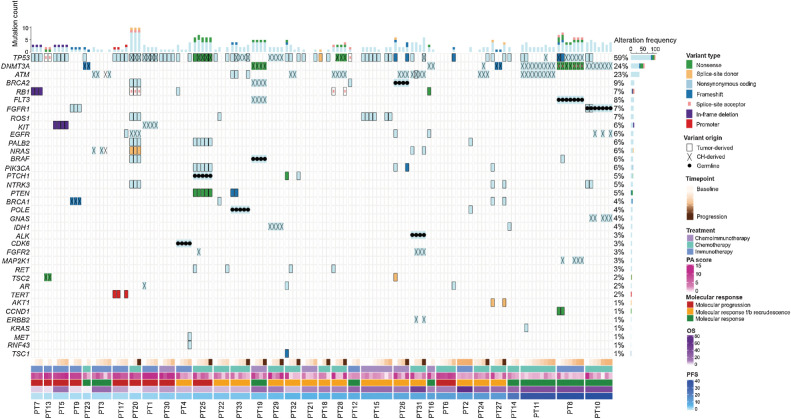
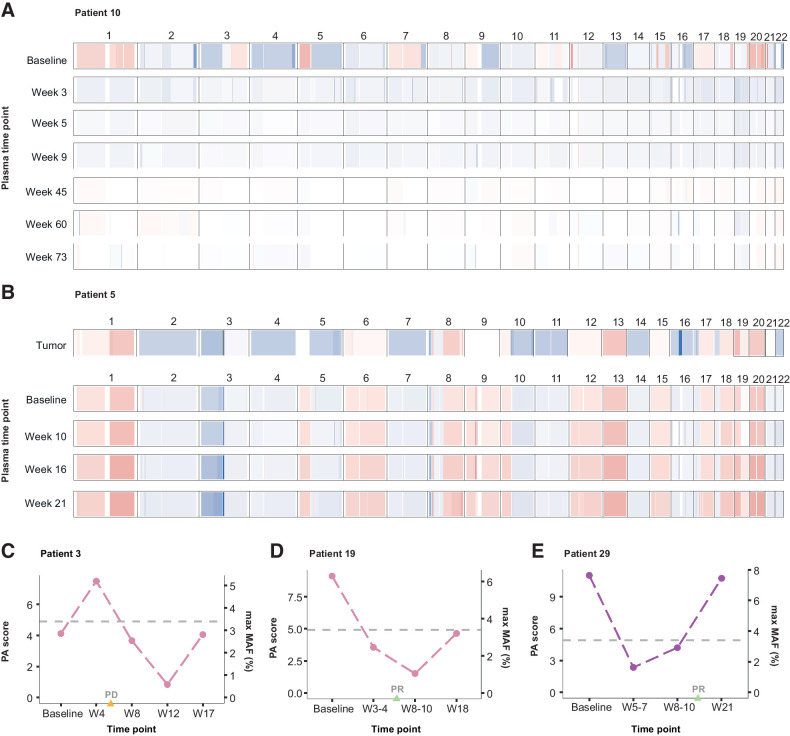
![Figure 4. Dynamic changes in cfTL during therapy. Longitudinal changes in cfTL across plasma time points analyzed for each patient were used to assign a combined molecular response classification. Representative examples are shown of patients who were assigned to each of the 3 classifications. A, Patient 10 was classified as a molecular responder based on the complete elimination of cfTL, assessed using tumor-derived sequence alterations, between baseline and week 5 sampling during cisplatin/etoposide chemotherapy treatment (indicated by the green shaded area). A reduction in PA scores to undetectable levels from baseline was also observed in this patient. B and C, Patients 26 and 21 were assigned a classification of molecular response followed by recrudescence based on the elimination of cfTL between baseline and intermediate time points [during atezolizumab/etoposide/carboplatin (purple) and carboplatin/etoposide (green) treatment, respectively], after which an increase in cfTL was observed at the final time points analyzed. C, In patient 21, a shift in mutation profiles defined by the presence of tumor-derived RET (p.Y314F) and TP53 (p.V173E) mutations at recrudescence, which were not present at the baseline timepoint, was observed. D, Patient 1 was classified as a molecular progressor based on the persistence of cfTL, defined by a tumor-derived TP53 (p.C135S) sequence alteration, across all time points analyzed during treatment with nivolumab (blue). E, Combined molecular responses were significantly associated with clinical evaluations of best radiographic response (P = 0.003, Fisher exact test). F, A broader comparison between the elimination of cfTL at any timepoint analyzed for the study and radiographic assessments further revealed concordance (P = 0.001, Fisher exact test) between each variable. G, Molecular responses were determined on average 4 weeks prior to best radiographic response assessments in 28 patients with comparable ctDNA and imaging assessments in this cohort (mean 5.61 weeks vs. 10.21 weeks; P = 0.01 Mann–Whitney U test). Patients without baseline plasma samples available (n = 2) and cases with discordant molecular and radiographic responses (n = 3) were excluded from analyses. Mean times to response assessment are shown alongside standard error for each modality.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/0fd3/10261918/b4ec9a226dc4/2310fig4.jpg)