Abstract
Purpose:
Tumor antigens are central to antitumor immunity. Recent evidence suggests that peptides from noncanonical (nonC) aberrantly translated proteins can be presented on HLA-I by tumor cells. Here, we investigated the immunogenicity of nonC tumor HLA-I ligands (nonC-TL) to better understand their contribution to cancer immunosurveillance and their therapeutic applicability.
Experimental Design:
Peptides presented on HLA-I were identified in 9 patient-derived tumor cell lines from melanoma, gynecologic, and head and neck cancer through proteogenomics. A total of 507 candidate tumor antigens, including nonC-TL, neoantigens, cancer-germline, or melanocyte differentiation antigens, were tested for T-cell recognition of preexisting responses in patients with cancer. Donor peripheral blood lymphocytes (PBL) were in vitro sensitized against 170 selected nonC-TL to isolate antigen-specific T-cell receptors (TCR) and evaluate their therapeutic potential.
Results:
We found no recognition of the 507 nonC-TL tested by autologous ex vivo expanded tumor-reactive T-cell cultures while the same cultures demonstrated reactivity to mutated, cancer-germline, or melanocyte differentiation antigens. However, in vitro sensitization of donor PBL against 170 selected nonC-TL, led to the identification of TCRs specific to three nonC-TL, two of which mapped to the 5′ UTR regions of HOXC13 and ZKSCAN1, and one mapping to a noncoding spliced variant of C5orf22C. T cells targeting these nonC-TL recognized cancer cell lines naturally presenting their corresponding antigens. Expression of the three immunogenic nonC-TL was shared across tumor types and barely or not detected in normal cells.
Conclusions:
Our findings predict a limited contribution of nonC-TL to cancer immunosurveillance but demonstrate they may be attractive novel targets for widely applicable immunotherapies.
Translational Relevance.
Recent evidence suggests that peptides derived from noncanonical (nonC) aberrantly translated proteins can be presented on HLA-I by tumor cells, but detailed studies of their immunogenicity are lacking. Our findings provide key insights for the clinical exploitation of nonC HLA-I ligands as targets for vaccines or T-cell therapies. We found that peptides derived from nonC proteins were frequently presented on HLA-I of patient-derived tumor cell lines (TCL) across different tumor types. Unlike neoantigens, cancer-germline, or melanocyte differentiation antigens, nonC HLA-I ligands did not frequently elicit antitumor T-cell responses in patients with cancer, suggesting they play a limited role in immune surveillance and immune-editing. However, in vitro raised T-cell responses and T-cell receptors targeting 3 nonC peptides recognized their specific antigens naturally presented by tumor cells, targeted multiple TCL, and did not or barely target normal cells tested. These findings support that specific nonC HLA-I peptides may represent valuable targets for widely applicable immunotherapies.
Introduction
Tumor antigens are central to antitumor immunity. Peptides derived from tumor antigens presented on HLA molecules (pHLA) on the surface of cancer cells can elicit protective and therapeutic T-cell responses (1). The existence of T cells targeting non-mutated tumor-associated antigens (TAA) and cancer-germline antigens (CGA) in patients with cancer is well established (2, 3). Their shared expression in a substantial fraction of tumors has led to the development of widely applicable vaccines or T cell–based therapies (4, 5). However, off-tumor toxicities have been reported (6–8). Technological advances in next-generation sequencing and tandem mass spectrometry coupled with high-throughput immunologic and HLA multimer screens have expedited the systematic discovery of the personalized landscape of antigens contributing to tumor immunogenicity. Accumulating evidence demonstrates that neoantigens arising from nonsynonymous somatic mutations (NSM) greatly contribute to the immunogenicity of human tumors. For instance, neoantigen-specific T cells are frequently detected in patients with cancer (9–13) and mutational load correlates with the clinical benefit of immune checkpoint blockade (14). Their foreign nature and high tumor specificity together with the antitumor responses observed following transfer of neoantigen-specific T cells in selected patients (15–18) render these attractive targets. Yet, existing techniques still fail to capture most antigens targeted by tumor-reactive T cells and this constitutes a major obstacle for the development of immunotherapy.
Tumor antigen discovery efforts thus far have largely investigated the immunogenicity of selected genomically annotated proteins or NSM in coding regions, limited to only 2% of the genome. However, up to 75% of the genome can be transcribed and, potentially, translated (19). Emerging data demonstrate that peptides derived from alternative open reading frames (ORF) or from allegedly noncoding regions referred to as noncanonical (nonC) or cryptic antigens are frequently presented on HLA-I molecules (20–23). A fraction of such aberrant translation events has been postulated to be specifically presented on tumor cells, thus substantially expanding the repertoire of targetable tumor antigens (24–27). Their non-mutated nature and occasional shared presentation across different tumors, has further attracted attention to nonC proteins as targets for immunotherapy.
Despite the potential of tumor-specific nonC HLA-I ligands as a source of tumor antigens, their systematic identification in humans remains challenging. Spontaneous T-cell responses against peptides derived from nonC proteins have been rarely identified using cumbersome and time-consuming immunologic screens of tumor cDNA libraries (28–30). A growing number of recent studies have exploited immunopeptidomics to identify these antigens (22–27, 31), but their immunogenicity and their selective expression in cancer remains largely unexplored. Here, we investigated the presentation and immunogenicity of nonC antigens across different cancer types to better understand their contribution to cancer immunosurveillance and to address their therapeutic potential. We employed a proteogenomics pipeline (20) to identify nonC HLA-I ligands derived from off-frame translation of coding sequences and noncoding regions [UTR, noncoding RNA (ncRNA), intronic and intergenic] in patient-derived tumor cell lines (TCL) of different histological types. We further modified the pipeline to select peptides preferentially presented by cancer cells and evaluated their natural or induced immunogenicity by assessing preexisting and in vitro sensitized T-cell responses.
Materials and Methods
Patient characteristics
Patients Gyn-1, Gyn-2, Gyn-3, Gyn-4, H&N-1, H&N-3, Mel-1, Mel-2, and Mel-3 were chosen for this study on the basis of availability of autologous TCL and matched lymphocytes. Patient characteristics are summarized in Supplementary Table S1.
Establishment of patient-derived TCL
A small fragment (2–4 mm3) of tumor biopsies or surgically resected tumor was cultured in RPMI1640 plus (Lonza) containing 10% FBS Hyclone (GE Healthcare), 100 U/mL penicillin (Lonza), 100 μg/mL streptomycin (Lonza), and 25 mmol/L HEPES (Thermo Fisher Scientific) at 37°C in 5% CO2. The medium was replaced once every month until the TCL was established and then further expanded in T2 media containing RPMI1640 plus (Lonza), 10% to 20% FBS (Gibco), depending on the TCL, 100 U/mL penicillin (Lonza), 100 μg/mL streptomycin (Lonza), and 25 mmol/L HEPES (Thermo Fisher Scientific) or cryopreserved until used. TCL were regularly tested for Mycoplasma by PCR of cell culture supernatants and were authenticated on the basis of the identification of patient-specific somatic mutations and HLA molecules.
Tumor-infiltrating lymphocyte expansion
Small tumor fragments (2–4 mm3) were cultured in individual wells of a 24-well plate in T-cell media consisting of RPMI1640 plus (Lonza) supplemented with 10% human AB serum (BST), 100 U/mL penicillin (Lonza), 100 μg/mL streptomycin (Lonza), 2 mmol/L l-Glutamine (Lonza), 25 mmol/L HEPES (Thermo Fisher Scientific) and 6e6 IU IL2 (Proleukin) at 37°C and 5% CO2. Fresh media containing IL2 was added on day 5 and media was changed, or tumor-infiltrating lymphocyte (TIL) were split when confluent every other day thereafter. T cells were expanded independently for 15 to 30 days and cryopreserved until use. In some cases, T cells underwent a rapid expansion protocol (REP), as explained below.
Rapid expansion protocol
T cells were expanded for 14 days using 30 ng/mL anti-CD3 (BioLegend, catalog no. 317303, RRID:AB_571924), 3e3 IU/mL of IL2 (Proleukin) and irradiated allogeneic peripheral blood mononuclear cell (PBMC; 50 Gy) pooled from three donors as feeder cells in T-cell medium RPMI1640 plus (Lonza):AIM-V (Gibco) containing 5% human AB serum (BST), 100 U/mL penicillin (Lonza), 100 μg/mL streptomycin (Lonza), 2 mmol/L l-Glutamine (Lonza), 12.5 mmol/L HEPES (Thermo Fisher Scientific). After day 6, half of the medium was replaced with fresh T-cell medium containing IL2 every other day. Cells were split when confluent, harvested on day 14, and cryopreserved until use.
PBMC isolation
PBMCs were obtained using a Ficol density gradient (Lymphoprep, Stem Cell) from pheresis or whole blood and cryopreserved for cell sorting, DNA extraction for whole-exome sequencing (WES) and to expand B cells ex vivo.
T-cell sorting from peripheral blood lymphocyte
Peripheral blood lymphocytes (PBL) were sorted on the basis of the expression of surface markers previously described to enrich for tumor-reactive T cells in peripheral blood such as PD-1 (32). Briefly, PBMCs were thawed and rested overnight without cytokines. Following CD8+ enrichment using CD8 microbeads (Miltenyi Biotec, catalog no. 130–045–201, RRID:AB_2889920), the Fc receptor was blocked (Miltenyi Biotec, catalog no. 130–059–901, RRID:AB_2892112) and cells were stained with the following antibodies for 30 minutes at 4°C: CD3-PECy7 (BD Biosciences, catalog no. 557851, RRID:AB_396896, 0.5:50), CD8-APCH7 (BD Biosciences, catalog no. 560179, RRID:AB_1645481, 1:50), PD1-PE (BioLegend, catalog no. 329906, RRID:AB_940483, 0.75:50), CD38-APC (BioLegend, catalog no. 303510, RRID:AB_314362, 0.5:50), and HLA-DR BV605 (BioLegend, catalog no. 307639, RRID:AB_11219187, 0.75:50). CD3+CD8+ cells expressing PD1hi alone or in combination with HLA-DR and CD38 were sorted in BD FACS AriaTM and expanded using a REP as previously specified.
Generation of autologous antigen-presenting cell
B cells were isolated from cryopreserved PBMCs by positive selection using CD19+ microbeads (Miltenyi Biotec, catalog no. 130–050–301, RRID:AB_2848166) and expanded through CD40-CD40 L stimulation by culturing cells for 4 to 5 days with irradiated NIH3T3 feeder cells constitutively expressing CD40 L at 37°C in 5% CO2 in B cell medium. Iscove's IMDM media (Gibco) containing 10% human AB serum (Biowest), 100 U/mL penicillin and 100 μg/mL streptomycin (Lonza), 2 mmol/L l-Glutamine (Lonza), and supplemented with 200 U/mL IL4 (Peprotech). Up to three rounds of stimulation and expansion were performed consecutively. B cells were cryopreserved from day 5 to 6 until use. When used after cryopreservation, B cells were thawed in B cell medium containing DNAse (Pulmozyme, Roche) 20 hours before use in coculture assays. Alternatively, CD4+ T cells were isolated from PBMCs by positive selection using CD4+ microbeads (Miltenyi Biotec, catalog no. 130–045–101, RRID:AB_2889919) or FACS sorting and subsequently expanded through a REP.
Peptides
The amino acid sequences of the identified tumor antigen HLA-I ligands were purchased from JPT Peptide Technologies (Berlin, Germany) as crude and used for screening, in vitro sensitization (IVS), and mass spectrometry (MS) validation with synthetic peptides. HPLC peptides were supplied by JPT Peptide Technologies (Berlin, Germany) and used in coculture experiments to confirm the reactivities. Selected endogenous HLA-I ligands were ordered from Thermo Fisher Scientific as crude (PePotec grade 3) with one stable isotope-labeled amino acid and used for Parallel Reaction Monitoring (PRM) validation.
Cloning, in vitro transcription of RNA, and electroporation
The HLA sequences of interest or predicted ORF of the immunogenic nonC tumor ligands (nonC-TL) peptides were cloned into pcDNA3.1 using BamH1 and EcoR1 containing a Kozak motif upstream of the start codon. HLA-I sequences were obtained from IPD-IMGT/HLA and codon-optimized. The predicted ORF were constructed from the second nearest upstream in-frame start codon (ATG, CTG, or GTG) to the first in-frame stop codon downstream; the sequence was not codon optimized nor additional start codons were added. All the plasmids were synthesized by Genscript.
For in vitro transcription (IVT) of RNA the plasmids were linearized with Not-I followed by phenol-chloroform extraction and precipitation with sodium acetate and ethanol. Next, 1 μg of DNA was used as a template to generate RNA by IVT using HiScribe# T7 ARCA mRNA Kit with tailing (New England) following the manufacturer's instructions. RNA was precipitated using LiCl₂, resuspended at 1 μg/μL in molecular grade H₂O, and stored at −80° until use.
From 0.5–1e6 TCL, healthy human cells, and B cells were harvested and resuspended in 100 μL of Opti-MEM media (Gibco) and transferred into a sterile 0.2-cm cuvette (VWR electroporation cuvettes). From 4 to 8 μg of RNA encoding for the sequence of interest were added for electroporation. Cells were electroporated at 150 V, 20 ms, and 1 pulse using an ECM 830 BTX-Electroporator. After electroporation, cells were resuspended in pre-warmed specific media containing DNAse (Pulmozyme, Roche). After 20 hours cells at 37°C and 5% CO₂, cells were harvested, washed with PBS, and used in coculture assays. A GFP RNA electroporation control was included for each cell line and assessed by flow cytometry as a transfection control.
Coculture assays: IFNγ enzyme-linked immunospot assays and detection of activation marker 4–1BB using flow cytometry
T cells were thawed into T-cell medium supplemented with 3,000 IU IL2 (Proleukin) and DNAse (Pulmozyme, Roche) 3 to 4 days before coincubation with target cells. All cocultures were performed in the absence of exogenously added cytokines. Cells were stained with CD3-APCH7 (BD Biosciences, catalog no. 560176, RRID:AB_1645475, 0.3:40), CD8-PECy7 (BD Biosciences, catalog no. 557746, RRID:AB_396852, 0.1:40), CD4-PE (BD Biosciences, catalog no. 555347, RRID:AB_395752, 0.3:40), and CD137-APC (BD Biosciences, catalog no. 550890, RRID:AB_398477, 0.5:40), and in some cases mTRB-FITC (Thermo Fisher Scientific, catalog no. 11–5961–82, RRID:AB_465323, 0.2:40) for 30 minutes at 4°C, washed with staining buffer containing propidium iodide (1:2,000) and acquired in BD FACSLyric, BD FACSCanto (RRID:SCR_018055) or BD FACSCelesta (RRID:SCR_019597). In parallel, IFNy secretion was detected using IFNγ capture (MABTECH, catalog no. 3420–3-1000, RRID:AB_907282) and detection antibodies (MABTECH, catalog no. 3420–6-1000, RRID:AB_907272), and assessed by enzyme-linked immunospot (ELISPOT) assay following the manufacturer instructions. ELISPOT plates were analyzed and counted in ELISPOT reader. For all the assays, plate-bound anti-CD3 (BioLegend, catalog no. 317303, RRID:AB_571924, 1 μg/mL) was used as a positive control. Media, and/or autologous antigen-presenting cell (APC) pulsed with irrelevant peptides were used as negative controls.
All T-cell populations expanded from patients (TILs or sorted PBLs) were tested for autologous tumor recognition and only tumor-reactive T-cell populations were interrogated for recall T-cell responses to specific antigens. From 2e4 to 5e4 ex vivo expanded TIL, sorted PBL or enriched populations of tumor-reactive lymphocytes were cocultured with 1e5 to 2e5 peptide-pulsed autologous APC (either B cells, or CD4+ T cells). T-cell reactivities were considered positive if the number of IFNγ spots were greater than double the amount of the irrelevant control condition and greater than 40 spots. In addition, reactivities had to be observed in at least two independent experiments. Crude peptides preparations were used for screening, and the reactivities were further confirmed with HPLC grade peptides. Experiments were performed at least twice.
Enrichment of tumor-reactive and antigen-specific T cells
Either expanded TIL, sorted PBL, or IVS T cells were cocultured with tumor cells or peptide-pulsed autologous APC for 20 hours. CD3+CD8+ cells expressing 4–1BB were sorted in BD FACS AriaTM or BD Influx and expanded using a REP as previously specified. The same antibodies and dilutions used for cocultures described above were scaled up for staining 4–1BB+ T cells.
HLA restriction element determination
COS-7 cells were transfected with plasmids encoding the individual HLA molecules using Lipofectamine 2000 (Life Technologies). After resting overnight, cells were harvested and pulsed with the corresponding peptides for 2 hours, washed, and used as targets in coculture assays.
In vitro sensitization of PBL
HLA-A*11:01 donor PBMCs were stimulated with 5 independent peptide pools (PP) each containing up to 35 nonC-TL selected by the prediction score to bind to HLA-A*11:01 according to NetMHCpan4.0. Cells underwent three consecutive rounds of stimulation every 7 days with 0.25 μg/mL per peptide and a combination of IL21, IL7, and IL2. More specifically, at day 0, 5e6 donor PBMC were cultured in 24-well plates with OpTmizer media (Gibco) containing IL21 (Peprotech 25 ng/mL) and the corresponding PP at 0.25 μg/mL per peptide. On day 6, IL2 (Proleukin 18 IU/mL) and IL7 (Peprotech 10 ng/mL) were added. For STIM2 (day 7) and STIM3 (day 14), T cells from the previous STIM were harvested, counted, and restimulated with autologous irradiated PBMC (50 Gy) pulsed with the corresponding PP at 1:10 ratio. Thereafter, fresh OpTmizer media (Gibco) containing IL2 (18 IU/mL) and IL7 (10 ng/mL) was replaced when medium looked acidified, or cells required splitting.
De novo T-cell responses were evaluated after three stims by coculturing IVS T cells with autologous B cells pulsed with the corresponding PP and analyzing 4–1BB upregulation by flow cytometry as described above. T cells recognizing the corresponding PP were sorted on the basis of 4–1BB expression and expanded for 14 days in a REP (enrichment of antigen-specific T cells). To identify the specific peptide recognized within the PP, sorted populations were cocultured with B cells pulsed with individual peptides. The recognition was confirmed using HPLC purified peptides.
T-cell receptor sequencing and PBL transduction
The T-cell receptor (TCR) locus was sequenced by multiplex single-cell RNA sequencing (RNA-seq) of enriched antigen-specific T-cell populations. The samples were multiplexed using TotalSeq barcodes. Sequencing was done on an Illumina NS6000 with an S1 flowcell and v1 chemistry. Mapping, quantification, and clonotype definitions were done using cell ranger multi software (version 6.1.1 using the reference vdj_GRCh38_alts_ensembl-5.0.0). Demultiplexing and subsequent analysis was using the packages Seurat (version 4.0.3) and scRepertoire (version 1.3.5); Seurat::HTODemux was run using default parameters to obtain singlets.
TRA V-J-encoding sequences and TRB V-D-J-encoding sequences were combined to sequences encoding the mouse constant TRA and TRB chains (33), respectively. Mouse constant regions were modified, as previously described (34, 35). The full-length TRB and TRA chains were cloned separated by a furin SGSG P2A linker into pMSGV1 retroviral vector (GenScript). Transient retroviral supernatants were generated by transfecting the vector encoding the TCR of interest (MSGV1) and envelope (RD114) into 293GP cells using Lipofectamine 2000 (Life Technologies). PBLs were activated in T-cell medium supplemented with 50 ng/mL anti-CD3 and 300 IU/mL IL2 for 3 days before retroviral transduction. Retroviral supernatants were harvested at 24 and 48 hours, centrifuged to discard cell debris, and diluted 1:1 with medium and used to transduce the activated lymphocytes using the spinoculation method, as previously described (15).
Normal human cell lines
Normal human cell lines were purchased from Promocell, thawed and cultured following the manufacturer's instructions in the recommended media without antibiotics. Cells were split when confluent with Dettaching kit (Promocell), cultured at the recommended concentration and expanded no more than 4 passages until use. HCM-c (Cat: C-128810) were cultured in myocyte growth medium (Catalog no.: C-39275). HREpC-c (Catalog no.: C-12665) were cultured in Renal Epithelial Cell GM media (Catalog no.: C-39606). HSAEpC-c (Catalog no.: C-12642) were cultured in Small Airway Epithelial cell GM (Catalog no.: C-39175). NHEM.f-c (Catalog no.: C-12400) were cultured in Melanocyte growth medium (Catalog no.: C-39415).
Whole-exome sequencing
To identify the tumor-specific NSM, genomic DNA was purified from a cell pellet of patient-derived TCL and matched PBMC. WES libraries were generated by exome capture of approximately 20,000 coding genes using SureSelect human All exon V6 kit (Agilent Technologies) and paired-end sequencing was performed on a HiSeq sequencer (Illumina) at Macrogen. The average sequencing depth ranged from 100 to 150 for each of the individual libraries generated. Alignments of WES to the reference human genome build hg19 were performed using novoalign MPI from novocraft. Duplicates were marked using Picard's MarkDuplicates tool. Insertion and deletion (indel) realignment and base recalibration were performed according to GATK best-practices. Samtools was used to create tumor and normal pileup files. Four independent mutation callers (Varscan, SomaticSniper, Mutect and Strelka) were used to call somatic NSM. The genomic coordinates from VCF files containing tumor-specific mutations were converted from hg19 to hg38 assemblies.
Genotype-Tissue Expression and The Cancer Genome Atlas RNA analyses
The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEX) data from paired tumor and healthy data was obtained and analyzed using GEPIA (02/05/2022) and plotted using R. RNA levels are expressed as Log2 (TPM+1), the density of color in each block represents the median expression value of a gene in a given tissue, normalized by the maximum median expression value across all blocks. Abbreviation of tumor types: ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangio carcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Purification of HLA-I peptides
Purified anti-HLA-I clone W6/32 (ATCC, catalog no. HB-95, RRID:CVCL_7872) antibodies were cross-linked to protein-A Sepharose 4B conjugate beads (Invitrogen) with dimethyl pimelimidate dihydrochloride (Sigma-Aldrich) in 0.2 mol/L Sodium Borate buffer pH 9 (Applichem). From 5e7 to 3e8 tumor cells (See Supplementary Table S1) were snap-frozen, thawed, and lysed with PBS containing 0.6% CHAPS (Applichem) and Protease inhibitor Cocktail Complete (Roche). The cell lysates were sonicated (Misonix 3000) and cleared by centrifugation for 1 hour at max speed to obtain the soluble fraction containing the pHLA complexes. The HLA-I affinity chromatography was performed using a 96-well single-use micro-plate with 3 μm glass fiber and 10-μm polypropylene membranes (Agilent). Sep-Pak tC18 100 mg Sorbent 96-well plates (Waters) were used for peptide purification and concentration as previously described (36). Peptides were eluted with 500 μL of 32,5% ACN in 0.1% TFA, lyophilized, and further cleaned and desalted with TopTips (PolyLC Inc.)
LC/MS-MS acquisition
Acclaim Pep-Map nanoViper, C18 (Thermo Scientific) at a flow rate of 15 μL/min using a Thermo Scientific Dionex Ultimate 3000 chromatographic system (Thermo Scientific). Peptides were separated using a C18 analytical column of 75 μm × 250 mm, 1.8 μm, 100Å (Waters) or 25 μm × 250 mm, 1.8 μm, 100Å (Waters). Orbitrap Fusion Lumos Tribrid (Thermo Scientific) mass spectrometer was operated in data-dependent acquisition (DDA) mode. Survey MS scans were acquired in the orbitrap with the resolution (defined at 200 m/z) set to 120,000. The top speed (most intense) ions per scan were fragmented in the linear ion trap (CID) and detected in the Orbitrap with the resolution set to 30,000. Quadrupole isolation was employed to selectively isolate peptides of 400 to 600 m/z. Included charged states were 2 and 3. Target ions already selected for MS/MS were dynamically excluded for 10 seconds.
Mass spectrometry data analysis of HLA-I peptides with Peptide-PRISM
Peptide-PRISM was used as previously described (20) without including random substitutions nor proteasome-spliced peptides. Briefly, for each identified fragment ion mass spectrum the Top 10 candidates were first identified by de novo sequencing with PEAKS X Studio (RRID:SCR_022841) and later aligned to a database containing a 3-frame translated transcriptome (Ensembl90) and 6-frame translated genome (hg38). In addition, vcf files from somatic mutation calling were used to interrogate NSM in a personalized fashion. All identified string matches were categorized into CDS (in-frame with annotated protein), 5′-UTR (contained in annotated mRNA, overlapping with 5′-UTR), Off-frame (off-frame contained in the coding sequence), 3′-UTR (all others that are contained in an mRNA), ncRNA (contained in annotated ncRNA), Intronic (intersecting any annotated intron) or Intergenic. Then, for each fragment ion mass spectrum, the category with the highest priority (CDS>5′UTR>Off-frame>3′UTR>ncRNA>Intronic>Intergenic) was identified, and all other hits among the 10 de novo candidates were discarded. The FDR was calculated for each category in a stratified mixture model considering the peptide length and database size. The same pipeline was applied to immunopeptidomics data obtained from HLA ligand atlas (37) including various tissues and HLA alleles. The predicted ORF from the nonC HLA-I ligands identified in the healthy immunopeptidome were retrieved and used to filter out the nonC HLA-I ligands from our tumor samples derived from the same ORF. All identified peptides were filtered to FDR 0.01. In addition, peptides with a de novo score (ALC) smaller than 30 and the sequences that could not be unequivocally assigned to a single category (Top location count = 1) were filtered out (Supplementary Data 1). For peptides derived from NSM, the FDR was set at 0.02 (Supplementary Data 2).
HLA-I typing and prediction of binding to patient-specific HLA molecules
HLA typing was determined from the WES data using the PHLAT algorithm (Supplementary Data 3). Eluted ligand likelihood percentile rank scores for binding to the patient's HLA molecules were obtained for all unique peptides ≥ 8 Aa eluted from TCL using NetMHCpan 4.0. The threshold for binding was set to < 2%-tile rank.
Validation of HLA-I peptides with synthetic peptides
Spectrum validation of the experimentally eluted HLA-I ligand tumor antigen candidates was performed by computing the similarity of the spectra acquired in the sample with the corresponding non-labeled synthetic peptide from the library. Briefly, synthetic crude peptides obtained from JPT were acquired in a pool using LC/MS-MS in conditions similar to those previously used to analyze samples to generate a spectral library. Peptide sequences were identified by database search with PEAKS-X Pro (PEAKS Studio RRID:SCR_022841) using a database containing Swiss-Prot as well as all the tumor antigen candidates interrogated. The search was exported as “for third party” format and imported into Skyline software (RRID:SCR_014080) to generate the library. The experimentally acquired HLA-I tumor samples were uploaded into Skyline and the similarity of the fragments (b/y ions) from the library (synthetic) versus endogenous (sample) were analyzed considering library dot product (dotp) values, which range from 0 to 1 and, being dotp = 1 the closest match.
Validation of HLA-I peptides with isotope-labeled peptides
For each selected peptide, a synthetic isotope-labeled peptide at one chosen amino acid was spiked into the samples and used as an internal standard for PRM detection. The amount of internal standard peptide to be spiked in each sample was evaluated using dilution curves and the final concentration was chosen on the basis of a good chromatographic signal and no trace detectable of potential unlabeled traces from the synthetic internal standard. For Mel-3 TCL 30% of the sample (total of 5e7 cells) and for Mel-1 50% of the sample (total 1e8) was analyzed by PRM using different MS machines. Orbitrap Eclipse (Thermo Fisher Scientific) coupled to an EASY-nanoLC 1000 UPLC system (Thermo Fisher Scientific) with a 50 cm C18 chromatographic column. A PRM method was used for data acquisition with a quadrupole isolation window set to 1.4 m/z and MS2 scans over a mass range of m/z 250 to 1,800, with detection in the Orbitrap mass analyzer at a 240 K resolution. MS2 fragmentation was performed using HCD fragmentation at a normalized collision energy of 30%, the AGC was set at 100,000, and the maximum injection time at 502 ms. All data were acquired with Thermo Xcalibur (RRID:SCR_014593).
For data analysis, fragment ion chromatographic traces corresponding to the targeted precursor peptides were evaluated with Skyline software v.21.2. Verification of the endogenous peptides was based on: (i) the number of detected traces, (ii) co-elution of endogenous traces, (iii) co-elution of endogenous and internal standard peptides, (iv) correlation of the fragment ions relative intensities between endogenous and internal standard peptides and, (v) expected retention time.
CRISPR/cas9 knock-out
Single-guide RNA (sgRNA) targeting the predicted ORF for 5′U-HOXC13 were designed with CRISPOR tool (http://crispor.tefor.net/). sgRNA specifically binding the genomic peptide location or upstream with the highest predicted knock out (KO) efficiency were selected. Next, sgRNA were cloned into lenti-Cas9-v2 (RRID:Addgene_52961) with BsmBI.
Lentiviral supernatants were generated by co-transfecting HEK293 cells with lenti-Cas9-v2 encoding the sgRNA of interest, psPAX2 and pMD2G plasmids with PEI (Sigma) and non-supplemented DMEM media (Gibco). Media was replaced after 12 hours with Opti-MEM (Gibco) containing 2% FBS (Gibco). Supernatants were collected at 42 to 48 hours after transfection and filtered through a 0.45-μm low-protein binding filter (Millipore). Patient-derived TCL were infected with lentiviral supernatants containing polybrene (Sigma) final concentration of 8 μg/mL followed by spinoculation 900 g, 32°C, 50 minutes. Five days post-infection, T2 media (RPMI containing 10% FBS supplemented with Pen/Strep and l-Glut) was replaced with T2 media containing puromycin 1 μg/mL to select the cells efficiently transduced. Thereafter, media was replaced with fresh media containing puromycin when acidified or cells split when confluent. To evaluate the KO efficiency, puromycin-resistant cells were used as targets in coculture assays with 5′U-HOXC13 specific T cells.
Statistics
Experiments were performed without duplicates, unless otherwise specified, data were reported as mean ± SEM. All experiments were repeated at least twice. For HLA-I ligand identification with Peptide-PRISM, the FDR was calculated for each category in a stratified mixture model as previously described (20) considering the peptide length and database size.
Study approval
This work was conducted in accordance with the Declaration of Helsinki. Samples were obtained through a study approved by the Vall d'Hebron Hospital ethical committee (PR(AG)482/2017). All patients provided a written, informed consent.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD036856. The source data underlying Fig. 5A was downloaded from GEPIA (02/05/2022), data for Supplementary Fig. S5 was downloaded from PRIDE (identifiers PXD022150 and PXD004894). All other data are available from the corresponding author on reasonable request.
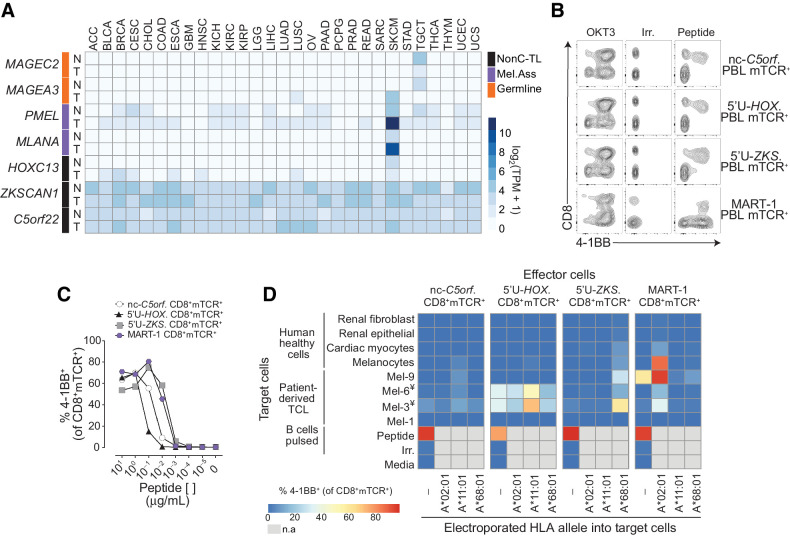
Figure 5.
Evaluation of the tumor specificity of the three immunogenic nonC-TL. A, RNA expression analysis in tumors (T) and matched healthy tissues (N) of the canonical genes encoding immunogenic nonC-TL compared with TAA and CGA. TCGA and GTEX data were obtained from GEPIA. B, CD8 coreceptor activation-dependence of PBL transduced with antigen-specific TCRs. FACS plots show the expression of 4–1BB by CD8 (gated on CD3+mTCR+) after coculture with peptide pulsed B cells. B cells pulsed with an irrelevant (Irrel.) peptide were used as negative control. C, TCR-transduced T cells purified by FACS sorting (CD8+mTCR+) were cocultured with B cells pulsed with serial dilutions of the corresponding peptide. SD mean is plotted. D, Expression and translation of nonC-TL in healthy human cells and selected TCLs was indirectly evaluated by coculturing control and electroporated target cells with RNA encoding the specified HLA alleles with sort purified TCR-transduced T cells. T-cell activation was assessed by measuring 4–1BB expression on CD8+mTCR+. ¥TCL naturally expressing HLA-A*11:01. n.a, non-assessed.
Results
NonC tumor HLA-I ligands are frequently identified in patient-derived TCLs
We first sought to determine whether tumor-specific nonC HLA-I ligands could be identified in 9 short-term cultured TCL derived from 4 patients with gynecologic cancer (Gyn), 3 patients with melanoma (Mel), and 2 patients with head and neck (H&N) cancer (Supplementary Table S1). These samples were selected irrespective of the tumor histology, solely based on the availability of matched ex vivo expanded TIL and/or peripheral blood tumor-reactive lymphocyte populations.
To this end, peptides bound to HLA-I were isolated and analyzed by LC/MS-MS using state-of-the-art procedures. Amino acid (Aa) sequences were identified through a previously described pipeline, Peptide-PRISM (20), with some modifications (Fig. 1A). Briefly, for each MS spectrum, the top 10 candidates were first identified by de novo sequencing and later mapped to a database including the 3-frame transcriptome and 6-frame genome. In addition, WES information of each TCL was included to interrogate the presentation of mutated peptides derived from cancer-specific NSM. The FDR was calculated independently for each genomic category considering the search space and peptide length in a stratified mixture model as previously described (20). Following this strategy and selecting a 0.01 FDR, we identified 839 nonC peptides presented on HLA-I in all the TCLs studied, ranging from 0.5% to 5.4% of the total eluted peptides (Fig. 1B).
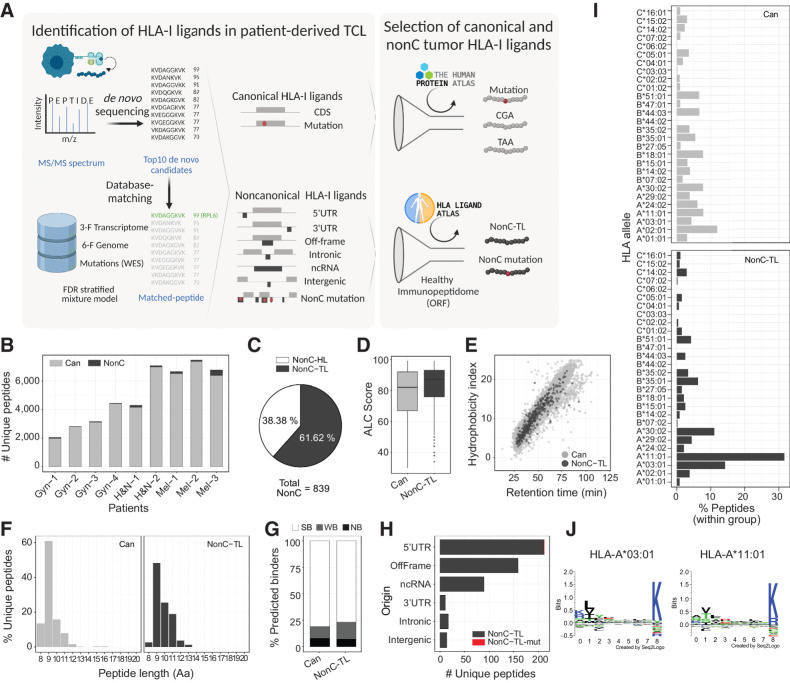
Figure 1.
Identification and characteristics of nonC HLA-I ligands presented by patient-derived TCLs. A, Diagram depicting the pipeline used to identify HLA-I ligands derived from canonical (Can) and nonC proteins presented by patient-derived TCL. The top 10 candidates for each MS spectrum were identified by de novo sequencing and aligned to a database containing the 3-frame transcriptome, 6-frame genome, and the NSM identified by WES. The FDR was calculated for each category shown using a stratified mixture model (left). All canonical peptides containing mutations as well as peptides derived from CGA or TAA were further studied. For nonC HLA-I ligands, healthy immunopeptidomics data from HLA ligand atlas was used to filter out peptides presented in healthy tissues at the ORF level to obtain the nonC-TL (right). B, Number of canonical and nonC HLA-I peptides identified per patient. C, Percentage of nonC peptides derived from predicted ORF present or absent in a healthy tissue immunopeptidome data set (nonC-HL and nonC-TL, respectively). D, ALC identification score of canonical and nonC-TL. E, Predicted hydrophobicity index (y axis) and retention time (x axis). Each dot represents a unique peptide sequence. F, Length distribution of unique HLA-I peptide sequences. Only peptides < 20 Aa are depicted. G, Percentage of peptides predicted to bind to the patient-specific HLA alleles according to NetMHCPan4.0. Peptides were categorized into strong binders (SB; %-tile ran ≤ 0.5), weak binders (WB; %-tile rank = 0.5–2), or nonbinders (NB; %-tile rank > 2). H, Number of nonC-TL originated from each of the ORF categories noted. I, HLA allele binding preference of Can and nonC-TL. For each peptide, only the min rank predicted by NetMHCpan4.0 was considered. J, Consensus peptide binding motif of the two HLAs predicted to present the majority of the nonC peptides identified. Image downloaded from NetMHCpan4.0 motif viewer. In all the analyses shown, the FDR threshold was set at 0.01 and ALC score at 30. (A, Created with BioRender.com.)
To select nonC peptides preferentially presented by tumor cells, immunopeptidomics data from samples available from the HLA ligand atlas (38) was used to filter out peptides presented in healthy tissues (Fig. 1A). Although LC/MS-MS is less sensitive than RNA-seq, this technique is capable of capturing canonical as well as aberrant translation. Given that we leveraged an immunopeptidomics database derived from donors presenting a fraction of all potential HLA alleles, nonC peptides were excluded at the ORF level rather than the Aa sequence to overcome a potential bias toward frequent alleles. As a result, we found that from a total of 839 unique nonC peptides detected in our tumor samples, 322 (38.38%) were predicted to derive from ORFs also present in healthy tissue (nonC-HL). Hence, 517 (61.6%) were considered preferentially presented on tumor HLA-I and referred to as nonC-TL (Fig. 1C). NonC-TL displayed similar characteristics as peptides derived from canonical proteins such as the MS identification score (ALC) or the correlation of the retention time with the hydrophobicity index (Fig. 1D and E). In addition, nonC-TL exhibited expected HLA-I ligand features as shown by the length distribution ranging from 8 to 12 Aa and the high percentage of peptides predicted to bind to the patient's HLA alleles according to NetMHCpan4.0 (Fig. 1F and G). Moreover, as with the HLA-I peptides derived from canonical proteins, most of the nonC-TL were validated by MS using synthetic or isotope labeled peptides, (Supplementary Figs. S1–S2). Altogether, these analyses indicate that our approach accurately identified the HLA-I ligand repertoire including nonC-TL presented by patient-derived TCL.
Next, we evaluated the genomic origin of the identified nonC-TL. Consistent with previous studies (20, 27, 39), we found that translation of 5′UTR was the main origin followed by off-frame and ncRNA (Fig. 1H). Peptides derived from 3′UTR, intronic and intergenic regions were less frequently detected. In addition, one nonC peptide derived from a 5′UTR containing a tumor-specific mutation was detected in patient Gyn-3 (Fig. 1H). We noticed a clear bias in the HLA-I binding preference distribution of nonC ligands towards HLA-A*11:01 and HLA-A*03:01 alleles according to NetMHCpan4.0 (Fig. 1I; Supplementary Fig. S3). Both HLA alleles bind peptides with a similar motif containing basic residues at p9 (Fig. 1J), a unique feature among all the alleles studied (Supplementary Fig. S4). Indeed, the binding preference of nonC peptides to both HLA-A*11:01 and HLA-A*03:01 alleles has been previously reported in other immunopeptidomics studies (20, 21), however the exact mechanism underpinning this bias is still unknown. Overall, our results showed that nonC-TL are frequently detected in patient-derived TCLs. Importantly, 76 of our nonC-TL were found in published HLA-I immunopeptidomics datasets from melanoma resection samples (refs. 40, 41; Supplementary Fig. S5), supporting that HLA-I presentation of nonC-TL is not an artifact of in vitro cultured cells and that these peptides can be naturally presented in vivo.
NonC-TL constitute an abundant source of candidate tumor antigens
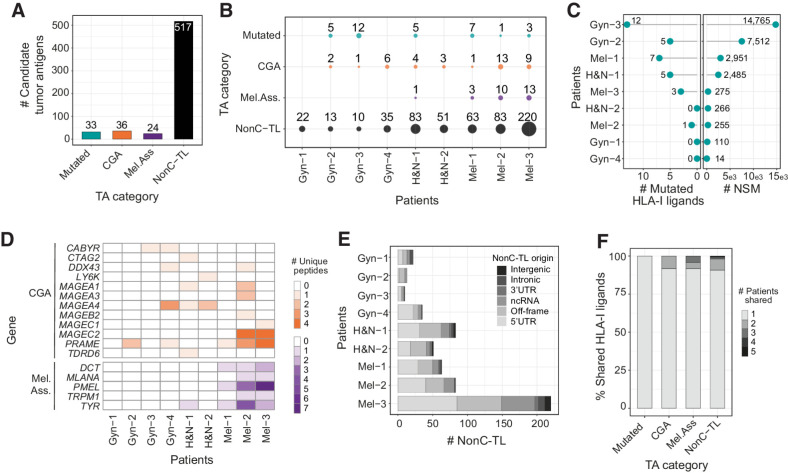
To examine whether nonC-TL represent an attractive source of tumor antigens that could be exploited therapeutically we first compared the number of nonC-TL eluted from HLA-I with those derived from conventional tumor antigen sources, including peptides encoded by canonical coding regions derived from somatically mutated gene products, and from TAA such as CGA or melanoma-associated antigens arising from melanocyte differentiation proteins. While the number of HLA-I ligands derived from canonical tumor antigens ranged from 24 to 36 peptides, nonC-TL outnumbered the other categories, with 517 unique tumor antigen candidates in all the TCL studied (Fig. 2A). Of note, this observation was consistent across most of the patients studied (Fig. 2B). In more detail, a total of 33 mutated peptides were detected in 6 of 9 patients. Despite the number of eluted HLA-I ligands containing mutations was low compared with the total NSM identified by WES, these results are in line with previous immunopeptidomics studies where few mutations are typically detected (refs. 41–43; Fig. 2B and C). Moreover, 36 peptides derived from 12 genes encoding for CGA and 24 peptides derived from 5 melanoma-associated antigens were identified in 8 of 9 patients (Fig. 2D). Furthermore, nonC-TL mainly originated from 5′UTR and off-frame translation in most patients, while peptides derived from intergenic and intronic regions were not or barely detected (Fig. 2E).
Figure 2.
Candidate tumor antigens presented on HLA-I from patient-derived TCL identified through proteogenomics. A, Total number of unique peptide sequences derived from candidate tumor antigens (TA) by category. Data from all patients were pooled together. Only unique peptide sequences were considered. B, The number and category of TA are displayed for each TCL. C, The number of mutated peptides eluted from HLA-I (left) and number of NSM identified by WES (right) are displayed for each patient. D, Heat map displaying the number of epitopes derived from specific CGA or melanoma-associated antigens per patient. E, Number of nonC-TL originated from each ORF category per patient. F, Percentage of candidate TA uniquely identified in one patient or shared by TA category. The FDR threshold was set at 0.02 for mutations and 0.01 for all other categories.
One advantage of exploiting non-mutated antigens over private mutations as targetable tumor antigens is the fact that they can be shared across patients, which could facilitate the development of off-the-shelf vaccines or T cell–based therapies. Similar to CGA or melanoma-associated antigens, we observed that ∼10% of nonC-TL were shared among at least 2 patients (Fig. 2F). In contrast, all mutated HLA-I ligands detected were derived from private mutations and thus, exclusively identified in a single patient. Of note, nonC-TL showed the highest number of shared peptides, including one and three sequences identified in 5 and 4 patients, respectively (Fig. 2F). Altogether, this data highlights nonC-TL as promising targets for the development of therapeutic interventions because they constitute a broader spectrum of candidate tumor antigens compared with peptides derived from mutations, CGA, or melanoma-associated antigens and can be shared across patients.
Tumor-reactive T cells in patients with cancer preferentially recognize neoantigens and TAA rather than nonC-TL
To further evaluate the role of nonC-TL in cancer immunosurveillance we assessed the presence of spontaneous T-cell responses targeting personalized candidate tumor antigens including nonC-TL, mutated, CGA and melanoma-associated antigens in the 9 patients with cancer. To this end, ex vivo expanded tumor-infiltrating lymphocyte (TIL) or PBL populations reactive to their corresponding autologous TCL were cocultured with autologous APCs pulsed with the synthetic peptides identified through proteogenomics. T-cell reactivity was tested by IFNγ release and the upregulation of the activation cell-surface marker 4–1BB assessed by ELISPOT and flow cytometry, respectively (Fig. 3A and B; Supplementary Fig. S6).
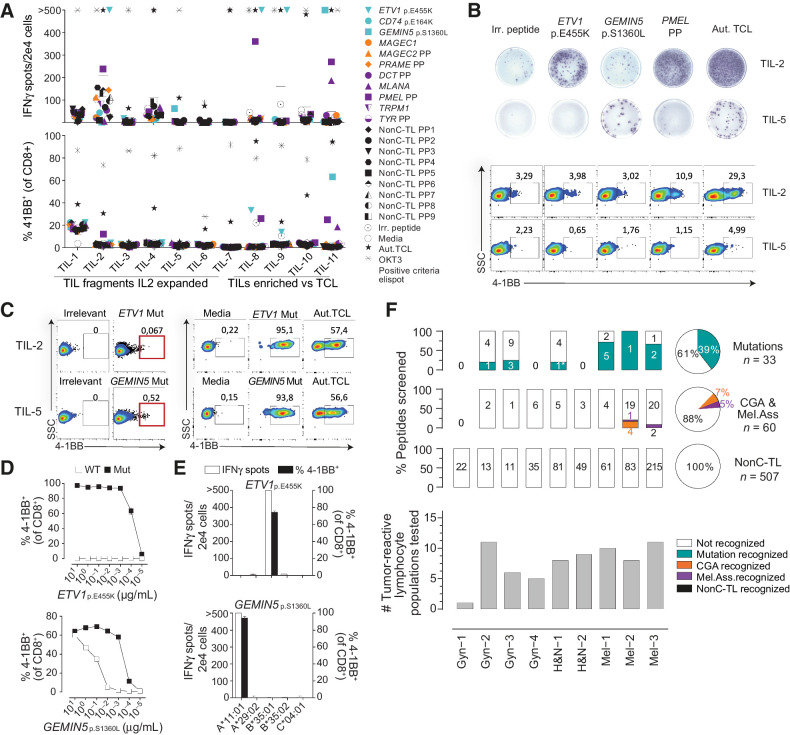
Figure 3.
Preexisting T-cell responses to candidate tumor antigens in patients with cancer. For each patient, reactivity was evaluated by co-incubating 2e4 T cells (TIL or PBL sorted on the basis of specific markers, e.g., PD1hi), with 2e5 autologous APC pulsed with 1 μg/mL of selected peptides either alone or in pools (PP). IFNγ ELISPOT and 4–1BB upregulation by FACS were used to measure T-cell responses after 20 hours. A, Reactivity to tumor antigen candidates for Mel-3. The number of IFNγ spots per well (top) and the percentage of cells expressing 4–1BB (bottom) are shown. Mutated peptides are plotted in turquoise, CGA in orange, melanoma-associated in purple, and nonC-TL in black. B, Representative ELISPOT results (top) and flow cytometry plots (bottom) for TIL-2 and TIL-5 from Mel-3 with the targets specified. C, TIL populations recognizing the mutated HLA-I peptides indicated were enriched by flow cytometry sorting of 4–1BB+ lymphocytes and expanded for 14 days. Plot showing gates used for sorting (left) and recognition of the targets specified after expansion (right). D, T-cell reactivity of neoantigen-enriched T-cell populations to serial dilutions of the WT or mutant (Mut) ETV1p.E455K and GEMIN5p.S1360L peptides. E, Neoantigen-enriched population was cocultured with COS-7 cells transfected with the indicated individual HLA-I alleles and pulsed with the corresponding peptides to determine the restriction element. F, Summary of the reactivity against candidate tumor antigens in all patients studied. The percentage and the absolute number of recognized and non-recognized peptides within each category are shown per patient (bar plot) and for all the patients studied (pie chart). The number of tumor-reactive lymphocytes tested for each patient is shown on the bottom. Plotted cells were gated on live CD3+CD8+lymphocytes. ‘>’ denotes greater than 500 spots/2e4 cells. *Mutation recognized previously identified. Experiments were performed twice. Aut.TCL: autologous tumor cell line.
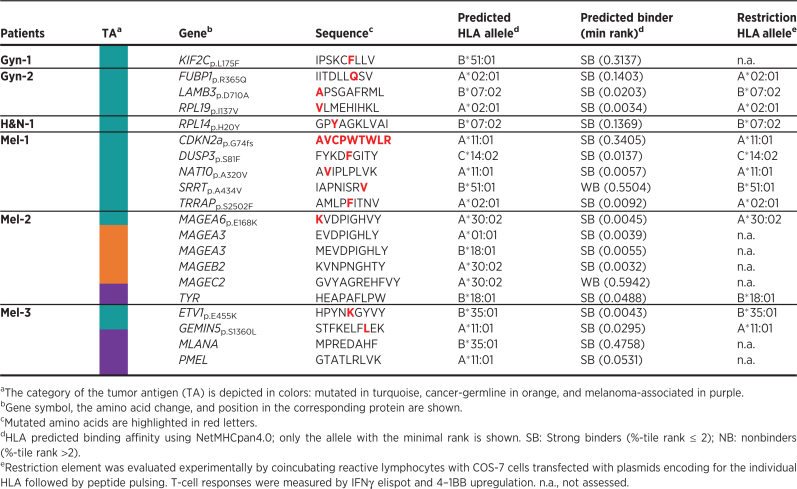
In patient Mel-3, we detected T-cell reactivity to at least one candidate peptide in 5 of 11 different tumor-reactive TIL populations interrogated. TILs recognized two neoantigens (ETV1p.E455K and GEMIN5p.S1360L) and two immunogenic peptides derived from melanoma-associated antigens (PMEL and MLANA). However, no reactivity was detected to any of the 9 peptides derived from CGA nor the 215 nonC-TL tested (Fig. 3A). To further test and characterize the T-cell responses observed, antigen-specific T cells were enriched by flow cytometry based-sorting of 4–1BB+ cells followed by ex vivo expansion (Fig. 3C). The neoantigen-specific enriched populations showed a higher response to the mutated peptide than to the wild-type (WT) counterparts as shown by peptide titration experiments, although T cells exhibited a variable functional avidity to their cognate antigen (Fig. 3D). ETV1p.E455K and GEMIN5p.S1360L were restricted to HLA-B*35:01 and HLA-A*11:01, respectively (Fig. 3E). Importantly, coculture experiments showed that both neoantigen-specific T cells isolated recognized the autologous TCL, ultimately demonstrating that these peptides are naturally processed and presented by the tumor (Fig. 3C).
We used the same strategy to identify and characterize preexisting T-cell responses targeting candidate tumor antigens from all patients included in the study (Fig. 3F). In total, screening of preexisting tumor-reactive T cells for recognition of 600 tumor antigen candidates led to the detection of 20 immunogenic peptides in the 9 patients with cancer studied (Fig. 3F; Table 1). Of note, we could detect T-cell responses to at least one neoantigen in the six patients in which neoantigen candidates were detected through immunopeptidomics. Overall, 13 of the 33 mutated HLA-I ligands tested were immunogenic, representing 39% of the total neoantigen candidates. Furthermore, four immunogenic peptides derived from CGA and three derived from melanoma-associated antigens were also recognized by naturally occurring tumor-reactive T cells. A detailed characterization of the antigen-specific T cells isolated including autologous tumor recognition, HLA restriction, functional avidity and WT counterpart recognition for neoantigens is shown in Supplementary Fig. S7–S10. In contrast, none of the 507 unique nonC-TL candidates interrogated were able to elicit a recall immune response in any of the studied patients. Altogether, these results reveal that although nonC-TL were frequently detected in TCL, antigens derived from canonical regions were preferentially recognized by tumor-reactive lymphocytes.
Table 1.
Immunogenic tumor antigens identified in patients with cancer.
NonC-TL recognized by in vitro sensitized T cells are shared across patient-derived TCL
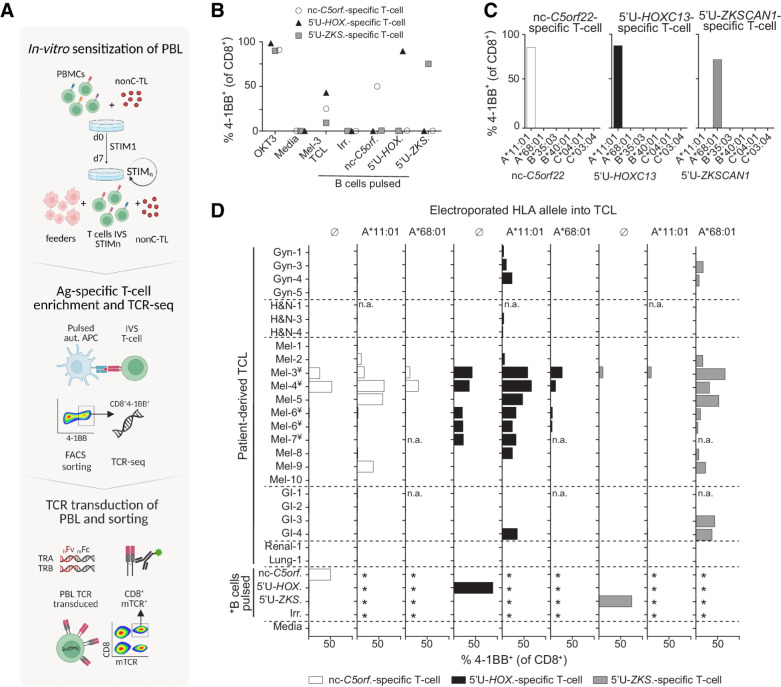
Although we did not detect preexisting T-cell responses targeting nonC-TL, we reasoned that these antigens could still be immunogenic, and thus could represent attractive targets for T-cell therapies or vaccines. To address this question, we investigated the presence of naïve T cells specific to nonC-TL- in the repertoire of healthy individuals carrying the corresponding HLA restriction alleles. As such peptide specific TCR are in very low frequency we sought to enrich nonC-TL–specific T cells through IVS in a non-autologous HLA-matched setting (Fig. 4A).
Figure 4.
IVS of donor PBLs identified three nonC-TL shared across patient-derived TCLs. A, Donor PBL were IVS via three consecutive rounds of stimulation with 170 selected nonC-TL predicted to bind to HLA-A*11:01. Reactive T cells were enriched through FACS sorting based on CD8+ 4–1BB+ expression after 20 hours coculture with autologous B cells pulsed with the specific peptides and expanded for 14 days. The top 1 αβ pairs were cloned into a retroviral vector to transduce PBL and CD8+ mTCRB+ cells were FACS sorted to obtain a pure transduced population. B, Reactivity of antigen-specific T cells generated by IVS following FACS sorting enrichment. Frequency of 4–1BB+ on CD8+ cells after 20 hours coculture with B cells pulsed with the HPLC peptides specified is depicted. C, Restriction element was evaluated by coculturing enriched T-cell populations with COS-7 cells expressing the donor HLA alleles and pulsed with the corresponding peptides. D, Expression and translation of the immunogenic nonC-TL in multiple patient-derived TCL indirectly evaluated through the detection of 4–1BB expression of nonC antigen-specific T cells cocultured with TCL left untreated or electroporated with RNA encoding the specified HLA-I alleles. *B cells were not electroporated. ¥TCL naturally expressing HLA-A*11:01. n.a., not assessed. (A, Created with BioRender.com.)
Of the total 507 nonC-TL tested for recognition in all patients, we selected 170 peptides predicted to bind to HLA-A*11:01 according to NetMHCpan4.0, an allele expressed in 14% of the Caucasian population (http://allelefrequencies.net). Through IVS of PBL from an HLA-A*11:01 donor, we detected, isolated, and expanded T cells specifically recognizing three nonC-TL (Fig. 4B). Two peptides mapped to 5′-UTR regions of the canonical genes HOXC13 (5′U-HOXC13) and ZKSCAN1 (5′U-ZKSCAN1), and one peptide mapped to a noncoding spliced variant of C5orf22 gene (nc-C5orf22, Supplementary Fig. S11). The ORF encoding these nonC-TL were considerably short, with up to 21, 46, or 49 amino acids respectively, as confirmed by the recognition of APC electroporated with RNA encoding the predicted ORF (Supplementary Fig. S12). In addition, the loss of recognition of TCL transduced with Cas9-sgRNA targeting the 5′UTR HOXC13 genomic locus unequivocally confirmed the specificity of the T cells for this antigen and the exact genomic location from which it is transcribed and translated (Supplementary Fig. S13).
Although these three immunogenic nonC-TL were originally detected in Mel-3 TCL through immunopeptidomics, we investigated whether these antigens could also be expressed in other TCLs. We exploited the high sensitivity of the nonC antigen-specific T cells identified to evaluate the expression and translation of these nonC antigens in a panel of 24 patient-derived TCL. T cells were cocultured with TCL artificially expressing the restriction element of interest (i.e., HLA-A*11:01 for 5′U-HOXC13 and nc-C5orf22, and HLA-A*68:01 for 5′U-ZKSCAN1; Fig. 4C), in addition to the endogenous HLA alleles. Strikingly, we found that the three nonC-TL evaluated were frequently expressed and detected by nonC-TL–specific T cells in patient-derived TCL, as observed by 4–1BB upregulation when the relevant HLA was expressed (Fig. 4D). Of note, 5′U-HOXC13 and 5′U-ZKSCAN1 were detected in melanoma, but also in other less immunogenic tumor types such as gastrointestinal cancers (GI) or gynecologic malignancies. Furthermore, the recognition of several HLA-A*11:01+ TCL by nonC-TL–specific T cells without transfecting any additional HLA, showed that nc-C5orf22 and 5′U-HOXC13 can be naturally processed and presented on HLA-I (Fig. 4D). Altogether, these results show that nonC-TL are shared across tumor types and can be naturally presented and recognized by T cells.
TCRs targeting nonC antigens can display cancer-specific recognition
A potential concern that has not yet been addressed regarding the therapeutic targeting of nonC-TL is whether they are tumor specific. To investigate this, we analyzed the RNA expression of the canonical genes encoding for the three immunogenic nonC-TL in several solid tumors and matched healthy tissues from repository data (GEPIA). We compared their expression pattern to PMEL and MLANA as examples of melanoma-associated antigens, and MAGEA3 and MAGEC2 representing CGA (Fig. 5A). Whereas C5orf22 and ZKSCAN canonical genes displayed a variable but ubiquitous expression among tissues, the expression of HOXC13 canonical gene in healthy tissues appeared to be restricted to melanocytes, resembling the expression pattern of MLANA. This raised the possibility that the identified immunogenic nonC-TL might not be tumor specific. However, RNA transcript level cannot distinguish canonical from aberrant translation. In addition, contrary to the RNA-seq data analysis, the healthy immunopeptidome data that was used to select for nonC HLA-I ligands derived from ORFs absent in nonmalignant cells suggested that the nonC translation of these peptides was tumor specific.
To gain further insights into the selective expression of these nonC-TL in tumor cells and their applicability as targets for cancer immunotherapy, we empirically evaluated the expression and translation of the selected immunogenic nonC-TL in several human healthy cell types by exploiting the ability of antigen-specific T cells to detect their cognate peptides with high sensitivity. To this end, we first sequenced the TCR locus of the nonC-TL–specific T cells identified by IVS and the most frequent TCR-α/β pairs of each of the populations were cloned into a retroviral vector and used to transduce PBL. In addition, given that the canonical HOXC13 gene expression pattern resembled MLANA, we generated DMF5 TCR-transduced cells as an example of a TCR tested in clinical trials, which recognizes the melanoma-associated antigen MART-127–35 (encoded by MLANA and restricted to HLA-A*0201) that is expressed both in melanoma cells and healthy melanocytes (8). Given that the TCRs recognizing nonC-TL were CD8-dependent, as opposed to the MART-1–specific TCR (Fig. 5B), all PBL TCR transduced cells were sorted on the basis of CD8 and mTCR expression (CD8+mTCR+). Peptide titration experiments to measure functional avidity of TCR transduced cells evidenced that nc-C5orf22 and 5′U-HOXC13–specific TCRs required higher concentrations of minimal peptide to become activated, while 5′U-ZKSCAN1 and MART-1 TCR transduced cells were more sensitive at detecting their cognate antigen (Fig. 5C).
Next, the four antigen-specific CD8+ mTCR+ cells were cocultured with human cells of different origin, including normal melanocytes, cardiac myocytes, renal epithelial, and fibroblast, as well as a few of the previously tested melanoma cell lines. As in the previous experiment used to evaluate the expression of nonC-TL antigens in patient-derived TCL (Fig. 4D), expression of the specific HLA alleles restricting antigen recognition of the TCRs evaluated was exogenously enforced in the target cells and T-cell activation was evaluated by measuring 4–1BB upregulation in mTCR+ cells by flow cytometry following coculture (Fig. 5D; Supplementary Fig. S14). As expected, MART-1 TCR transduced cells strongly recognized human melanocytes electroporated with the HLA-A*02:01, as well as 3 of the 4 patient-derived melanoma cell lines tested. Surprisingly, cardiac myocytes were also recognized by MART-1 TCR transduced cells, albeit to a limited extent. 5′U-ZKSCAN1 TCR transduced cells displayed preferential recognition of melanoma cells compared with normal melanocytes and cardiac myocytes when target cells were electroporated with the relevant allele HLA-A*68:01. Importantly, 5′U-HOXC13 displayed recognition of two of the four melanoma cell lines included but did not recognize cardiac myocytes and barely recognized normal melanocytes when electroporated with the corresponding allele HLA-A*11:01. The lower sensitivity of the nonC C5orf22 TCR and/or limited expression of the antigen precluded us from reaching a conclusion regarding the tumor specific expression of this antigen. Overall, these results indicate that the 5′U-HOXC13 peptide is the nonC antigen with the highest tumor specificity followed by 5′U-ZKSCAN1. Importantly, both nonC antigens displayed a superior tumor specific profile compared with MART-1. Although we cannot rule out the possibility that these antigens could be expressed and translated in additional normal cell types, our findings suggest that the aberrant translation giving rise to the nonC peptides studied occurs preferentially in tumor cells rather than in normal cells. Overall, our results demonstrate that nonC-TL are a promising alternative source of tumor antigens to neoantigens, CGA and TAA not only because they can be naturally presented but also because they can be immunogenic and expressed across diverse tumor types but not, or at very low levels, in healthy cells.
Discussion
Tumor antigens play an integral role driving protective or therapeutic antitumor immunity. Recent evidence suggests that peptides derived from nonC proteins can be systematically detected through proteogenomics and are specifically presented on HLA-I by tumor cells. However, their contribution to tumor immune surveillance and their immunogenicity has not been explored in detail.
The proteogenomics pipeline we used, Peptide-PRISM, is independent of RNA-seq and Ribo-seq but enables the detection of HLA-I ligands potentially originating from any region of the genome, including CDS, UTR, off-frame, ncRNA, intronic, and intergenic regions (Fig. 1A and H). Like previous immunopeptidomics studies (20–24, 31, 27, 39, 44), we observed that nonC HLA-I ligands were frequently detected across different cancer types (Fig. 1B). Because we were interested in tumor-specific candidates we used healthy immunopeptidome data to exclude peptides derived from ORFs present in nonmalignant cells. As a result, 61.5% of the nonC HLA-I–ligands detected were preferentially presented by tumors, referred to as nonC-TL (Fig. 1C). Importantly, we studied the repertoire of presented tumor antigen candidates in 9 patient-derived TCL. Our work shows that nonC-TL (n = 517) outnumber the HLA-I ligands derived from conventional tumor antigens such as mutations (n = 33), CGA antigens (n = 36), and melanoma-associated antigens (n = 24; Fig. 2A). In line with previous reports (41–43), the number of HLA-I ligands derived from NSM was relatively low compared with the NSM identified by WES (Fig. 2C). Notably, our data adds a considerable number of mutated HLA-I peptides to those identified through immunopeptidomics so far, underscoring the performance of Peptide-PRISM at detecting neoantigen candidates.
NonC proteins were frequently presented in patient-derived TCLs, being the main source of candidate tumor antigens, but preexisting T-cell responses targeting nonC-TL were not detected in any of the patients studied (Fig. 3F; Supplementary Fig. S15). In contrast, nearly 65% of the antigens recognized by T cells were derived from mutations (n = 13), 20% from CGA (n = 4) and 15% from melanoma-associated antigens (n = 3; Table 1; Supplementary Fig. S15). Moreover, the isolated antigen-specific T cells recognized the autologous TCL (Supplementary Fig. S7; Fig. 10), demonstrating that the peptides identified were bona fide tumor antigens. To our knowledge, the existence and frequency of naturally occurring T cells targeting nonC-TL compared with conventional tumor antigens has not previously been investigated in such detail. The majority of studies identifying nonC peptides presented on HLA-I did not investigate their immunogenicity in patients (20, 22–24, 31, 39, 45). A few explored their immunogenicity through IVS of PBL from healthy donors which detects naïve rather than antigen-experienced T cells (21, 46–48), or immunized mouse models (25). Only one proteogenomics report evaluated T-cell responses against nonC HLA-I ligands as well as other relevant tumor antigens derived from melanoma-associated antigens and CGA identified from patient-derived TCL and tumor samples (27). Although they reported some degree of reactivity to one of the 571 nonC HLA-I peptides evaluated, our results are in accordance with their findings because the percentage of reactive peptides clearly favored melanocyte differentiation antigens, supporting that nonC HLA-I ligands are not as immunogenic.
One limitation of our study lies in the nature of the proteogenomics approach used combined with the stringent and uniform 0.01 FDR threshold set to select the tumor antigen candidates. Although this pipeline potentially detects peptides originating from any region of the genome, the FDR calculation using a stratified mixture model could result in an underestimation of nonC ligands with an increased search space, for example intergenic regions. Consequently, some of the previously described nonC sources were not properly interrogated such as endogenous retroviral elements, or not considered in this study such as RNA editing or peptide splicing (49, 50). Overall, our findings predict a limited contribution of nonC-TL to tumor immune surveillance.
Despite we could not detect recall T-cell responses to nonC-TL, we were able to isolate and expand T cells specifically targeting three nonC-TL through IVS of non-autologous HLA-A*11:01+ PBL. We found that two immunogenic nonC-TL derived from the aberrant translation of 5′UTR of HOXC13 and ZKSCAN1 genes were frequently detected in patient-derived melanoma TCL as well as other less immunogenic tumor types such as GI or Gyn. In addition, an immunogenic nonC-TL derived from a noncoding spliced variant of C5orf22 gene was also detected in several melanoma TCL. These results revealed that nonC-TL can be immunogenic and shared across tumor types, thus representing attractive targets for off-the-shelf vaccines or T-cell therapies.
The paradoxical lack of recognition of nonC-TL in patients with cancer in light of the fact that at least a fraction of these were proven to be immunogenic could be explained through different mechanisms. For instance, we screened ex vivo expanded lymphocytes which frequently present a skewed oligoclonal TCR repertoire, which could lead to the depletion of some tumor-reactive clones (51). Although this could be the case, theoretically, this would negatively impact on the T-cell recognition of all antigen categories equally. Another possible explanation is that the level of expression of nonC-TL may be sufficient for presentation on tumor HLA-I, but inadequate for efficient cross-presentation in vivo, leading to defective priming of T cells. In line with this hypothesis, nonC peptides are thought to be less abundant and largely originated from disordered or unstable proteins with shorter half-lives compared with functionally annotated proteins (22, 52). In addition, some nonC ORF are so small that they do not require processing (39). These characteristics can facilitate the accessibility of nonC peptides into the HLA-I antigen presentation pathway (52, 53). Moreover, some studies have demonstrated that rapidly degraded proteins and minimal epitopes are unable to provoke cross-priming by APC as opposed to stable, full-length antigens (54). Altogether, this data suggests that the native characteristics of the aberrant translation events giving rise to nonC-TL (i.e., low abundance and instability) could account, at least in part, for the lack of recognition in patients with cancer. An alternative explanation for our findings is that nonC-TL could also be presented by mTEC cells in the thymus, leading to partial central tolerance and, consequently, limiting the abundance of T cells targeting nonC-TL in periphery. This, combined with the low level of expression and/or poor priming could hamper the detection of antigen-experienced T cells targeting nonC-TL in patients with cancer. Furthermore, one could speculate that a fraction of these aberrantly translated proteins may be more efficiently eliminated via nonsense-mediated mRNA decay (NMD) due to the presence of premature translation termination codons, thus reducing the abundance of these type of antigens (55). In this case, the inhibition of NMD may be an effective strategy to increase their protein expression level and, eventually, triggering a T-cell response. Further studies should be performed to address this possibility.
In our work, we addressed the tumor specific expression of nonC antigens, an essential aspect for the development of immunotherapeutic interventions. Previous studies have used healthy RNA-seq data to exclude nonC HLA-I ligands presented in healthy tissues. Alternatively, Ribo-seq could potentially be used to select tumor-specific nonC ORF. However, this technique is relatively new, the sensitivity is still limited and little data from healthy tissue is currently available. Instead, we leveraged a healthy immunopeptidome dataset to select nonC HLA-I ligands absent in nonmalignant cells. Although immunopeptidomics is less sensitive, we believe it is more relevant, because it can detect peptides derived from both canonical and aberrant translation. To gain additional insights into the tumor specificity of the three immunogenic nonC identified, we indirectly evaluated their expression and translation in healthy human cells derived from several vital organs as well as melanocytes by using lymphocytes transduced with antigen specific TCRs. Our results revealed that the 5′U-HOXC13 peptide was the nonC antigen with the highest tumor-specificity, because TCR-transduced T cells targeting this antigen recognized several TCL, but barely melanocytes or any other healthy human cell tested. In fact, both 5′U-HOXC13 and 5′U-ZKSCAN1 nonC antigens displayed a far superior tumor specific profile compared with MART-1, as TCR DMF5 displayed strong recognition of melanocytes (>90%). Unexpectedly, DMF5 TCR transduced T cells recognized cardiac myocytes at a similar level as TCR transduced T cells targeting the nonC-TL 5′U-ZKSCAN1 (10% to 20%). Although DMF5 TCR has shown objective tumor responses in patients it also evidenced severe on-target toxicities in skin, eyes, and ears due to the high expression of MART-1 in melanocytes (8), but toxicities related to cardiac myocyte recognition has never been reported. These findings suggest that the low level of expression of these nonC-TL in healthy tissues that we observed (Fig. 5D) are insufficient to trigger undesired off-tumor toxicities. Overall, our data supports that nonC-TL can display a tumor-specific profile that is compatible with their use as therapeutic targets. Nonetheless, these data must be interpreted with caution, because we cannot rule out the possibility that these immunogenic nonC-TL are expressed and translated in other healthy cells. Interestingly, two of the immunogenic nonC-TL identified derived from 5′UTRs. Previous work has shown that alternative initiation factor eIF2A is essential for cancer progression, during which it mediates initiation of translation at unconventional 5′ sites (56), potentially explaining some level of tumor specificity observed, particularly, for 5′U-HOXC13 nonC-TL. Gaining insights into the exact mechanism driving the aberrant translation of each of the targetable nonC-TL could further elucidate their cancer specificity and prove critical to select patients that could be treated with TCR gene engineered T cells targeting nonC-TL.
Overall, our results demonstrate that nonC-TL constitute an abundant source of candidate tumor antigens compared with peptides derived from mutations, CGA, or tissue differentiation antigens. NonC-TL are shared across tumor types and can be naturally presented by cancer cells and recognized by T cells. More importantly, they are not detected or at very low levels in healthy cells. Therapeutic interventions such as vaccines or TCR gene engineered T cells targeting nonC-TL could overcome the defective endogenous T-cell responses observed in patients with cancer by enhancing the de novo priming of T cells or administering large numbers of effector cells that can directly attack tumor cells. In fact, it is tempting to speculate that the lack or defective T-cell response to nonC tumor antigens in patients with cancer could be advantageous, because these antigens and the HLA presenting them are less exposed to immune selective pressure and, consequently, tumors would less likely present antigen escape variants selected through immunoediting. Our findings predict a limited contribution of nonC-TL to cancer immunosurveillance and, at the same time, underscore nonC-TL as a promising source of antigens for the development for therapeutic interventions.
Supplementary Material
Tumor cell line HLA-I typing and tumor type
Includes a large list of all the peptides and details of the HLA-I ligands identified at <=0.01 FDR and >=30 ALC score
Includes the list of peptides derived from NSM identified at 0.02 FDR and ALC>=30
Show patient characteristics and relevant experimental information
Supplementary Figure 1. Validation of HLA-I ligand tumor antigen candidates with synthetic peptidesSupplementary Figure 2. Validation of selected HLA-I ligand tumor antigen candidates identified in Mel-1 and Mel-3 with isotope-labeled peptidesSupplementary Figure 3. Frequency of nonC peptides detected in HLA-I.Supplementary Fig. 4. HLA-I allele binding motif of all patients included in the study.Supplementary Figure 5. A fraction of nonC-TL can be detected in publicly available immunopeptidomics datasets from human tumor biopsiesSupplementary Figure 6. Evaluation of pre-existing T-cell responses to candidate tumor antigens in cancer patientsSupplementary Figure 7. Enrichment of antigen-specific T cells from cancer patients by flow cytometry-based sorting.Supplementary Figure 8. Deconvolution of peptide pools containing recognized CGA or melanoma-associated antigens.Supplementary Figure 9. Functional avidity and specificity of neoantigen-specific T cells isolated from cancer patients.Supplementary Figure 10. HLA restriction element and recognition of autologous tumor cell lines by antigen-specific T cells isolated from cancer patients.SupplementaryFigure 11. Genomic location of the three immunogenic nonC-TL identified through IVS.Supplementary Figure 12. Recognition of the predicted ORFs encoding the immunogenic nonC-TL by TCR transduced cells.Supplementary Figure 13. Loss of recognition of TCL transfected with Cas9-sgRNA targeting the predicted 5’U-HOXC13 ORF.Supplementary Figure 14. Assessment of expression and translation of the immunogenic nonC-TL in human healthy cells.Supplementary Figure 15. NonC-TL are the main source of candidate tumor antigens identified by proteogenomics but tumor-reactive T cells preferentially recognize neoantigens derived from NSM.
Acknowledgments
We thank the patients for their participation in this study, Steven A. Rosenberg for providing valuable reagents and support for NGS studies, R. Pujol for helpful scientific discussion, J. Gonzalez for bioinformatics support, CRG/UPF Flow Cytometry Unit for assistance with cell sorting, and CRG/UPF and IRB Proteomics Units for technical support. A. Gros and this work were funded by the Comprehensive Program of Cancer Immunotherapy & Immunology II (CAIMI-II) supported by the BBVA Foundation (53/2021), Institute Carlos III (MS15/00058 and PI17/01085), AECC (IDEAS197PORT), and La Fundació La Marató de TV3 (201919–30). We thank CERCA Programme / Generalitat de Catalunya for institutional support. M. Lozano-Rabella was supported by the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) (2018FI_B 00946). A. Garcia-Garijo was supported by Generalitat PERIS award (SLT017/20/000131). A. Yuste-Estevanez was supported by the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) (2021 FI_B 00365). J. Palomero was supported by the Beatriu de Pinós programme (BP 2018), cofounded by the Agency for Management of University and Research Grants (AGAUR) and European Union's Horizon 2020.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
M. Lozano-Rabella reports grants from Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) during the conduct of the study. A. Garcia-Garijo reports grants from Generalitat de Catalunya during the conduct of the study. A. Yuste-Estevanez reports grants from Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) during the conduct of the study. J. Martín-Liberal reports personal fees from Astellas, Bristol-Myers Squibb, MSD, Novartis, Pierre Fabre, Pfizer, Roche, Sanofi, and Highlight Therapeutics outside the submitted work. I. Matos reports grants from Roche and ESMO and personal fees from Daiichi Sankyo outside the submitted work. J.M. Piulats reports grants from BeiGene and Mirati outside the submitted work. I. Brana reports grants from AstraZeneca, Bicycle Therapeutics, Celgene, Dragonfly, GlaxoSmithKline, Gliknik, ISA Pharmaceuticals, Janssen Pharmaceuticals, Kura, Nanobiotics, Novartis, Northern Biologics, Orion Pharma, Odonate Therapeutics, Regeneron, Pfizer, Sanofi, Pharmamar, Seattle Genetics, Shattuck Labs, and VCN Biosciences; grants and personal fees from Boehringer Ingelheim, Bristol-Myers Squibb, Immutep, Merck Serono, and Merck Sharp & Dohme; and personal fees from Achilles Therapeutics, Cancer Expert Now, eTheRNA Immunotherapies, PCI Biotech, and Pierre Fabre outside the submitted work. In addition, I. Brana is a member of the CAIMI-II program funded by BBVA Foundation. E. Muñoz-Couselo reports personal fees from BMS, MSD, Novartis, Pierre Fabre, and Sanofi outside the submitted work. E. Garralda reports grants from Novartis, Roche, Thermo Fisher, AstraZeneca, Taiho, and BeiGene and other support from Roche/Genentech, F. Hoffmann-La Roche, Ellipses Pharma, Neomed Therapeutics1 Inc., Boehringer Ingelheim, Janssen Global Services, Seattle Genetics, TFS, Alkermes, Thermo Fisher, Bristol-Myers Squibb, MabDiscovery, Anaveon, F-Star Therapeutics, Hengrui, Sanofi, Merck Sharp & Dohme, Roche, Lilly, Novartis, Agios Pharmaceuticals, Amgen, Bayer, BeiGene USA, Blueprint Medicines, BMS, Cellestia Biotech, Debiopharm, Forma Therapeutics, Genentech Inc., Genmab B.V., GSK, Glycotope Gmbh, Incyte Biosciences, Incyte Corporation, ICO, Kura Oncology Inc., Lilly, S.A, Loxo Oncology Inc., Macrogenics Inc., Menarini Ricerche Spa, Merck, Sharp & Dohme de España, S.A, Nanobiotix, S.A, Novartis Farmacéutica, S.A., Pfizer, SLU, PharmaMar, S.A.U., Pierre Fabre Medicament, Principia Biopharma Inc., Psioxus Therapeutics Ltd., Sanofi, Sierra Oncology, Sotio A.S., and Symphogen A.S. outside the submitted work. A. Gros reports grants from Comprehensive Program of Cancer Immunotherapy and Immunology II (CAIMI-II) (53/2021) supported by the BBVA Foundation, Instituto de Salud Carlos III (MS15/00058, PI17/01085), Asociación Española Contra el Cáncer (AECC) (IDEAS197PORT), La Fundació la Marató de TV3 (201919-30), and Ministerio de Ciencia, Innovación y Universidades (PID2020-118529RB-I00) during the conduct of the study as well as personal fees from Achilles Therapeutics, Singula Bio, BioNTech, Instil Bio, Pact Pharma, and Vall d'Hebron Institute of Oncology; grants and personal fees from Roche; and grants from Merck KGaA outside the submitted work. In addition, A. Gros has a patent for E-059-2013/0 licensed and with royalties paid from Intima Bioscience Inc., Intellia Therapeutics, Inc., Tailored Therapeutics, LLC, Cellular Biomedicine Group, Inc., and Geneius Biotechnology, Inc.; a patent for E-085-2013/0 licensed and with royalties paid from Intima Bioscience Inc., Intellia Therapeutics, Inc., and Geneius Biotechnology, Inc.; and a patent for E-149-2015/0 licensed and with royalties paid from Intima Bioscience Inc., Intellia Therapeutics, Inc., and Tailored Therapeutics, LLC. No disclosures were reported by the other authors.
Authors' Contributions
M. Lozano-Rabella: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft. A. Garcia-Garijo: Investigation, methodology, writing–review and editing. J. Palomero: Investigation, methodology, writing–review and editing. A. Yuste-Estevanez: Investigation, methodology. F. Erhard: Software, methodology. R. Farriol-Duran: Conceptualization. J. Martin-Liberal: Resources. M. Ochoa de Olza: Resources. I. Matos: Resources. J.J. Gartner: Formal analysis, methodology. M. Ghosh: Methodology. F. Canals: Methodology. A. Vidal: Resources. J.M. Piulats: Resources. X. Matías-Guiu: Resources. I. Brana: Resources. E. Muñoz-Couselo: Resources. E. Garralda: Resources. A. Schlosser: Software, methodology. A. Gros: Conceptualization, resources, supervision, funding acquisition, investigation, project administration, writing–review and editing.
References
- 1. Coulie PG, van den Eynde BJ, van der Bruggen P, Boon T. Tumor antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135–46. [DOI] [PubMed] [Google Scholar]
- 2. Brichard V, Pel Av, W6lfel T, W61fel C, de Plaen E, Leth6 B, et al. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. 1993. Available from: http://rupress.org/jem/article-pdf/178/2/489/1267835/489.pdf [DOI] [PMC free article] [PubMed]
- 3. van der Bruggen P, Traversari C, Chomez P, Lurquin C, de Plaen E, van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human. New Series 1991;254. [DOI] [PubMed] [Google Scholar]
- 4. Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 2011;29:917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006;314:126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother 2013;36:133–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Linette GP, Stadtmauer EA, v MM, Rapoport AP, Levine BL, Emery L, et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013;122:863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 2009;114:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kristensen NP, Heeke C, Tvingsholm SA, Borch A, Draghi A, Crowther MD, et al. Neoantigen-reactive CD8+ T cells affect clinical outcome of adoptive cell therapy with tumor-infiltrating lymphocytes in melanoma. J Clin Invest 2022;132:e150535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parkhurst MR, Robbins PF, Tran E, Prickett TD, Gartner JJ, Li J, et al. Unique neoantigens arise from somatic mutations in patients with gastrointestinal cancers. Cancer Discov 2019;9:1022–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stevanović S, Pasetto A, Helman SR, Gartner JJ, Prickett TD, Howie B, et al. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 2017;356:200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanada KI, Zhao C, Gil-Hoyos R, Gartner JJ, Chow-Parmer C, Lowery FJ, et al. A phenotypic signature that identifies neoantigen-reactive T cells in fresh human lung cancers. Cancer Cell 2022;40:479–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zacharakis N, Lutfi; HM, Seitter SJ, Kim SP, Gartner JJ, et al. Breast cancers are immunogenic: immunologic analyses and a phase II pilot clinical trial using mutation-reactive autologous lymphocytes. J Clin Oncol 2022;40:1741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51:202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344:641–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375:2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 2018;24:724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leidner R, Sanjuan Silva N, Huang H, Sprott D, Zheng C, Shih YP, et al. Neoantigen T-cell receptor gene therapy in pancreatic cancer. N Engl J Med 2022;386:2112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature 2012;489:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erhard F, Dölken L, Schilling B, Schlosser A. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol Res 2020;8:1018–26. [DOI] [PubMed] [Google Scholar]
- 21. Laumont CM, Daouda T, Laverdure JP, Bonneil É, Caron-Lizotte O, Hardy MP, et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from noncanonical reading frames. Nat Commun 2016;7:10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cuevas MVR, Hardy MP, Hollý J, Bonneil E, Durette C, Courcelles M, et al. Most noncanonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep 2021;34:108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scull KE, Pandey K, Ramarathinam SH, Purcell AW. Immunopeptidogenomics: Harnessing RNA-seq to illuminate the dark immunopeptidome. Mol Cell Proteomics 2021;20:100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao Q, Laverdure JP, Lanoix J, Durette C, Cote C, Bonneil E, et al. Proteogenomics uncovers a vast repertoire of shared tumor-specific antigens in ovarian cancer. Cancer Immunol Res 2020;8:544–55. [DOI] [PubMed] [Google Scholar]
- 25. Laumont CM, Vincent K, Hesnard L, Audemard É, Bonneil É, Laverdure JP, et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med 2018;10:eaau5516. [DOI] [PubMed] [Google Scholar]
- 26. Ouspenskaia T, Law T, Clauser KR, Klaeger S, Sarkizova S, Aguet F, et al. Thousands of novel unannotated proteins expand the MHC I immunopeptidome in cancer. Available from: 10.1101/2020.02.12.945840 [DOI] [PMC free article] [PubMed]
- 27. Chong C, Müller M, Pak HS, Harnett D, Huber F, Grun D, et al. Integrated proteogenomic deep sequencing and analytics accurately identify noncanonical peptides in tumor immunopeptidomes. Nat Commun 2020;11:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coulie PG, Lehmann F, Lethe B, Herman J, Lurquin C, Andrawiss M, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Immunology 1995;92:7976–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guilloux Y, Lucas S, Brichard VG, van Pel A, Viret C, de Plaen E, et al. A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J Exp Med 1996;183:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rong-Fu Wang B, Kawakami Y, Robbins PF, Rosenberg SA. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. 1996. Available from: https://rupress.org/jem/article-pdf/183/3/1131/499993/1131.pdf [DOI] [PMC free article] [PubMed]
- 31. Xiang H, Zhang L, Bu F, Guan X, Chen L, Zhang H, et al. A novel proteogenomic integration strategy expands the breadth of neo-epitope sources. Cancers 2022;14:3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 2016;22:433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cohen CJ, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res 2006;66:8878–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen CJ, Li YF, El-Gamil M, Robbins PF, Rosenberg SA, Morgan RA. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res 2007;67:3898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haga-Friedman A, Horovitz-Fried M, Cohen CJ. Incorporation of transmembrane hydrophobic mutations in the TCR enhance its surface expression and T-cell functional avidity. J Immunol 2012;188:5538–46. [DOI] [PubMed] [Google Scholar]
- 36. Chong C, Marino F, Pak H, Racle J, Daniel RT, Mü Ller M, et al. High-throughput and sensitive immunopeptidomics platform reveals profound interferon γ-mediated remodeling of the human leukocyte antigen (HLA) ligandome. Mol Cell Proteomics 2018;17:533–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marcu A, Bichmann L, Kuchenbecker L, Backert L, Kowalewski DJ, Freudenmann LK, et al. The HLA ligand atlas. A resource of natural HLA ligands presented on benign tissues. Available from: 10.1101/778944 [DOI]
- 38. Marcu A, Bichmann L, Kuchenbecker L, Kowalewski DJ, Freudenmann LK, Backert L, et al. HLA Ligand Atlas: a benign reference of HLA-presented peptides to improve T cell–based cancer immunotherapy. J Immunother Cancer 2021;9:e002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ouspenskaia T, Law T, Clauser KR, Klaeger S, Sarkizova S, Aguet F, et al. Unannotated proteins expand the MHC-I–restricted immunopeptidome in cancer. Nat Biotechnol 2022;40:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 2021;592:138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bassani-Sternberg M, Bräunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Newey A, Griffiths B, Michaux J, Pak HS, Stevenson BJ, Woolston A, et al. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J Immunother Cancer 2019;7:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kalaora S, Barnea E, Merhavi-Shoham E, Qutob N, Teer JK, Shimony N, et al. Use of HLA peptidomics and whole-exome sequencing to identify human immunogenic neo-antigens. Oncotarget 2016;7:5110–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cleyle J, Hardy MP, Minati R, Courcelles M, Durette C, Lanoix J, et al. Immunopeptidomic analyses of colorectal cancers with and without microsatellite instability. Mol Cell Proteomics 2022;21:100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang TY, Liu Q, Ren Y, Alam SK, Wang L, Zhu Z, et al. A pan-cancer transcriptome analysis of exitron splicing identifies novel cancer driver genes and neoepitopes. Mol Cell 2021;81:2246–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bartok O, Pataskar A, Nagel R, Laos M, Goldfarb E, Hayoun D, et al. Antitumor immunity induces aberrant peptide presentation in melanoma. Nature 2021;590:332–7. [DOI] [PubMed] [Google Scholar]
- 47. Bonté PE, Arribas YA, Merlotti A, Carrascal M, Zhang JV, Zueva E, et al. Single-cell RNA-seq–based proteogenomics identifies glioblastoma-specific transposable elements encoding HLA-I-presented peptides. Cell Rep 2022;39:110916. [DOI] [PubMed] [Google Scholar]
- 48. Nelde A, Flötotto L, Jürgens L, Szymik L, Hubert E, Bauer J, et al. Upstream open reading frames regulate translation of cancer-associated transcripts and encode HLA-presented immunogenic tumor antigens. Cell Mol Life Sci 2022;79:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mylonas R, Beer I, Iseli C, Chong C, Pak HS, Gfeller D, et al. Estimating the contribution of proteasomal spliced peptides to the HLA-I ligandome. Mol Cell Proteomics 2018;17:2347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verkerk T, Koomen SJI, Fuchs KJ, Griffioen M, Spaapen RM. An unexplored angle: T-cell antigen discoveries reveal a marginal contribution of proteasome splicing to the immunogenic MHC class I antigen pool. Proc Natl Acad Sci USA 2022;119:e2119736119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Poschke IC, Hassel JC, Rodriguez-Ehrenfried A, Lindner KAM, Heras-Murillo I, Appel LM, et al. The outcome of ex vivo TIL expansion is highly influenced by spatial heterogeneity of the tumor T-cell repertoire and differences in intrinsic in vitro growth capacity between T-cell clones. Clin Cancer Res 2020;26:4289–301. [DOI] [PubMed] [Google Scholar]
- 52. Dersh D, Hollý J, Yewdell JW. A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat Rev Immunol 2021;21:116–28. [DOI] [PubMed] [Google Scholar]
- 53. Yewdell JW. DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol 2011;32:548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, et al. CD8 + T cell cross-priming via transfer of proteasome substrates. Science 2004;304:1318–21. [DOI] [PubMed] [Google Scholar]
- 55. Supek F, Lehner B, Lindeboom RGH. To NMD or not to NMD: nonsense-mediated mRNA decay in cancer and other genetic diseases. Trends Genet 2021;37:657–68. [DOI] [PubMed] [Google Scholar]
- 56. Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, et al. Translation from unconventional 5′ start sites drives tumor initiation. Nature 2017;541:494–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tumor cell line HLA-I typing and tumor type
Includes a large list of all the peptides and details of the HLA-I ligands identified at <=0.01 FDR and >=30 ALC score
Includes the list of peptides derived from NSM identified at 0.02 FDR and ALC>=30
Show patient characteristics and relevant experimental information
Supplementary Figure 1. Validation of HLA-I ligand tumor antigen candidates with synthetic peptidesSupplementary Figure 2. Validation of selected HLA-I ligand tumor antigen candidates identified in Mel-1 and Mel-3 with isotope-labeled peptidesSupplementary Figure 3. Frequency of nonC peptides detected in HLA-I.Supplementary Fig. 4. HLA-I allele binding motif of all patients included in the study.Supplementary Figure 5. A fraction of nonC-TL can be detected in publicly available immunopeptidomics datasets from human tumor biopsiesSupplementary Figure 6. Evaluation of pre-existing T-cell responses to candidate tumor antigens in cancer patientsSupplementary Figure 7. Enrichment of antigen-specific T cells from cancer patients by flow cytometry-based sorting.Supplementary Figure 8. Deconvolution of peptide pools containing recognized CGA or melanoma-associated antigens.Supplementary Figure 9. Functional avidity and specificity of neoantigen-specific T cells isolated from cancer patients.Supplementary Figure 10. HLA restriction element and recognition of autologous tumor cell lines by antigen-specific T cells isolated from cancer patients.SupplementaryFigure 11. Genomic location of the three immunogenic nonC-TL identified through IVS.Supplementary Figure 12. Recognition of the predicted ORFs encoding the immunogenic nonC-TL by TCR transduced cells.Supplementary Figure 13. Loss of recognition of TCL transfected with Cas9-sgRNA targeting the predicted 5’U-HOXC13 ORF.Supplementary Figure 14. Assessment of expression and translation of the immunogenic nonC-TL in human healthy cells.Supplementary Figure 15. NonC-TL are the main source of candidate tumor antigens identified by proteogenomics but tumor-reactive T cells preferentially recognize neoantigens derived from NSM.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD036856. The source data underlying Fig. 5A was downloaded from GEPIA (02/05/2022), data for Supplementary Fig. S5 was downloaded from PRIDE (identifiers PXD022150 and PXD004894). All other data are available from the corresponding author on reasonable request.
Figure 5.
Evaluation of the tumor specificity of the three immunogenic nonC-TL. A, RNA expression analysis in tumors (T) and matched healthy tissues (N) of the canonical genes encoding immunogenic nonC-TL compared with TAA and CGA. TCGA and GTEX data were obtained from GEPIA. B, CD8 coreceptor activation-dependence of PBL transduced with antigen-specific TCRs. FACS plots show the expression of 4–1BB by CD8 (gated on CD3+mTCR+) after coculture with peptide pulsed B cells. B cells pulsed with an irrelevant (Irrel.) peptide were used as negative control. C, TCR-transduced T cells purified by FACS sorting (CD8+mTCR+) were cocultured with B cells pulsed with serial dilutions of the corresponding peptide. SD mean is plotted. D, Expression and translation of nonC-TL in healthy human cells and selected TCLs was indirectly evaluated by coculturing control and electroporated target cells with RNA encoding the specified HLA alleles with sort purified TCR-transduced T cells. T-cell activation was assessed by measuring 4–1BB expression on CD8+mTCR+. ¥TCL naturally expressing HLA-A*11:01. n.a, non-assessed.