Abstract
Background
Unplanned hospital readmissions are serious medical adverse events, stressful to patients, and expensive for hospitals. This study aims to develop a probability calculator to predict unplanned readmissions (PURE) within 30-days after discharge from the department of Urology, and evaluate the respective diagnostic performance characteristics of the PURE probability calculator developed with machine learning (ML) algorithms comparing regression versus classification algorithms.
Methods
Eight ML models (i.e. logistic regression, LASSO regression, RIDGE regression, decision tree, bagged trees, boosted trees, XGBoost trees, RandomForest) were trained on 5.323 unique patients with 52 different features, and evaluated on diagnostic performance of PURE within 30 days of discharge from the department of Urology.
Results
Our main findings were that performances from classification to regression algorithms had good AUC scores (0.62–0.82), and classification algorithms showed a stronger overall performance as compared to models trained with regression algorithms. Tuning the best model, XGBoost, resulted in an accuracy of 0.83, sensitivity of 0.86, specificity of 0.57, AUC of 0.81, PPV of 0.95, and a NPV of 0.31.
Conclusions
Classification models showed stronger performance than regression models with reliable prediction for patients with high probability of readmission, and should be considered as first choice. The tuned XGBoost model shows performance that indicates safe clinical appliance for discharge management in order to prevent an unplanned readmission at the department of Urology.
Keywords: Unplanned readmissions, Urology, Machine Learning, Algorithms, XGBoost
Plain Language Summary
Study need and importance
Unplanned readmissions form a consistent problem for many hospitals. Unplanned readmission rates can go up as high as to 35%, and may differ significantly between respective hospital departments. In addition, in the field of Urology readmission rates can be greatly influenced by type of surgery performed and unplanned readmissions in patients can go up as high as 26%. Although predicting unplanned readmissions for individual patients is often complex, due to multiple factors that need to be taken into account (e.g. functional disability, poor overall condition), there is evidence that these can be prevented when discharge management is evaluated with an objective measuring tool that facilitate such risk stratification between high and low risk patients. However, to the best of our knowledge, the latter risk stratification using ML driven probability calculators in the field of Urology have not been evaluated to date. Using ML, calculated risk scores based on analysing complex data patterns on patient level can support safe discharge and inform concerning the risk of having an unplanned readmission.
What we found
Eight ML models were trained on 5.323 unique patients with 52 different features, and evaluated on diagnostic performance. Classification models showed stronger performance than regression models with reliable prediction for patients with high probability of readmission, and should be considered as first choice. The tuned XGBoost model shows performance that indicates safe clinical appliance for discharge management in order to prevent an unplanned readmission at the department of Urology. Limitations of our study were the quality and presence of patient data on features, and how to implement these findings in clinical setting to transition from predicting to preventing unplanned readmissions.
Interpretation for clinicians
ML models based on classification should be first choice to predict unplanned readmissions, and the XGBoost model showed the strongest results.
Introduction
Unplanned readmissions form a consistent problem for many hospitals, rates can go up as high as to 35%, and differ significantly between hospital departments [1]. Departments with a heterogenous patient population often experience high unplanned readmission rates (e.g. Intensive Care Unit (ICU), Internal medicine, Geriatric medicine) due to the complexity of care, heterogenous patient population, and suboptimal discharge management on individual patient level [2]. In addition, in the field of Urology readmission rates can be greatly influenced by type of surgery performed and readmissions in patients can go up as high as 26% [3]. Although predicting unplanned readmissions for individual patients is often complex, due to multiple features that need to be taken into account (e.g. functional disability, poor overall condition), there is evidence that these can be prevented when discharge management is evaluated with an objective measuring tool that facilitate such risk stratification between high and low risk patients [4, 5]. The latter risk stratification using Machine Learning (ML) driven probability calculators in the field of Urology have not been evaluated to date.
Using ML, calculated risk scores based on analysing complex data patterns can support safe discharge on patient level, and can be used with capacity management on a department level. The physician team can assess high-risk scores by evaluation of the responsible modifiable (i.e. can act on) risk factors on patient level. With this information, the physician team may evaluate if the patient is safe for discharge, needs to stay admitted in order to optimize specific modifiable features, and if discharged whether bed capacity needs to be taken into account for possible unplanned readmission. The use of such ML driven algorithms in clinical setting has shown to be feasible application in predicting unplanned readmissions [6]. Moreover, shared decision-making based on individualised risk stratification reduces the risk of unplanned readmission up to 13%. This includes informing the patient about the current situation, optimizing specific features before discharge, and discussing what factors (i.e. features) carry risk and could lead to an unplanned readmission [7].
From a ML methodological point of view algorithms are commonly trained with limited set of features (i.e. variables), such as length of stay, acuity of admission, comorbidity, and emergency department utilization in the 6 months before admission (LACE). While larger sets of features are available in the patient chart during clinical admission which can be applied to train algorithms with [8, 9]. Also, there are few comparisons between regression and classification based algorithms in context of unplanned readmissions [10].
Our primary aim was to develop a ML-driven probability calculator to predict unplanned readmissions (PURE) within 30-days after discharge for patients that had a clinical admission at the department of Urology. Our second aim was to evaluate the difference performance of the PURE probability calculator developed using ML algorithms, comparing regression versus classification algorithms. We hypothesized it is feasible to develop a strong performing PURE probability calculator, and there is no difference in performance when developed with ML algorithms using classification versus regression algorithms.
Methods
Guidelines
This study followed the guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research, and the guidelines for Transparent Reporting of Multivariable Prediction Models for Individual Prognosis or Diagnosis (TRIPOD) [11, 12].
Data safety
To ensure proper handling of privacy-sensitive patient data, the independent Scientific Research Advisory Committee (Adviescommissie Wetenschappelijk Onderzoek—ACWO) within the OLVG was consulted and agreed (study number WO 21.099 – PURE) with the use of these data from the hospital population.
Data source
A retrospective cohort study design was used, and data of 7.570 unique patients with documentation present in the database (Clarity) of the Electronical Medical Records (EMR) (EPIC, Wisconsin, United States) were extracted using a SQL query. Patients with a clinical admission at the department of Urology of a community hospital in Amsterdam between January 2015 and October 2021 were included. Patients that deceased during clinical admission were excluded. To prevent repeated measures and data leakage, one admission or readmission per patient was included in the dataset.
Unplanned readmission
The primary outcome was a 30 day unplanned hospital readmission at the department of Urology, and readmissions were defined as clinical admissions within 30 days of discharge from previous clinical admission at the department of Urology.
Features
Based on findings of several studies and clinical impact, 53 features were included, and some features, such as vitals or laboratory (lab) results, contained over time data within each admission.
These features are split into the following six categories:
Patient characteristics
Lab results
Medication
Health care logistics
Medical history
Type of surgery
(For a detailed overview, see Appendix.)
Bias
Possible bias could originate from arbitrarily choosing a set of features by the researchers, incomplete documentation of data on features, and unknown lab results from external parties that were not included.
Missing data
Missing data, was checked for the Missing At Random (MAR) assumption, and platelet count (82.6% missing) was dropped as feature. All remaining continuous features with missing data (serum creatinine, hemoglobin, BMI, alcohol use, systolic and diastolic blood pressure, and smoking history), were imputed using multiple imputation by chained equations [13] (MICE) with a default number of multiple imputations (5), 100 iterations (maxit), and the Predictive Mean Matching (PMM) settings for imputing numerical data. Non-continuous features with missing data were coded to ‘No’ or ‘Absent’, and therefore showed no missing data. More information considering imputed features can be found in Table 1.
Table 1.
Missing values per feature: count and percentage
| Variable | Count | Percentage |
|---|---|---|
| Platelet count | 6254 | 82.6 |
| Serum creatinine | 4050 | 53.5 |
| Hemoglobin | 3742 | 49.4 |
| BMI | 2187 | 28.9 |
| Alcohol use | 1645 | 21.7 |
| Systolic blood pressure | 902 | 11.9 |
| Diastolic blood pressure | 902 | 11.9 |
| Smoking history | 507 | 6.7 |
Study size
Specific information about patient characteristics can be found in Table 3 in Appendix.
Imbalanced outcome
Of all observations, 10% of all patients had an unplanned readmission. This indicates a class imbalance and poses a potential problem when performing classification, as classification leans towards the class with the most observations and can skew the performance of an algorithm [14]. Observations on outcome were rebalanced using Synthetic Minority Oversampling Technique (SMOTE) and synthetized observations (i.e. oversampling) based on existing observations, combined with removing existing observations (i.e. undersampling) to create a specified balance. To prevent data leakage, data was split into a train and test set and resampling was only performed on the training set. Patients with an unplanned readmission were oversampled to 36% and patients without an unplanned readmission were set to 64% using undersampling.
Model development
For modelling and evaluating, only supervised ML was applied. To achieve the first aim of this study, developing a PURE probability calculator, the following regression algorithms were used: 1) Logistic Regression, Penalized Logistic Regression 2) LASSO, and 3) RIDGE. The following classification algorithms were used: 4) Normal Decision Tree, 5) Bagged Trees, 6) Boosted Trees, 7) XG Boosted Trees, and 8) Random Forest. The available data was split to a ratio of 70:30 to create a training, and test set respectively. More information concerning patient characteristics between the train- and the test data can be found in the Appendix in Table 4. To ensure a fitting sampling strategy, 5-fold cross validation on the training set was applied. Before using the data for training and evaluating the models, all data were corrected for outliers and examined for confounding using correlation analysis and Principal Component Analysis (PCA) [15]. Centering and scaling was configured as extra setting in the regression algorithms to apply during training. Feature engineering (variable selection) was evaluated using the RandomForest algorithm to identify the predictive value for each feature, with importance measured in mean decrease of accuracy per feature [16].
Model evaluation
To achieve the second aim of this study, evaluate differences in diagnostic performance characteristics of the regression and classification algorithms, the following metrics were used: accuracy, sensitivity, specificity, Area Under the Curve (AUC), Positive Predictive Value (PPV), and Negative Predictive Value (NPV).
Software
Data pre-processing and analysis were performed using R Version 4.0.2, and R-studio Version 1.3.1073 (R-Studio, Boston, MA, USA). All code is made available via https://github.com/koenwelvaars/PURE_study.
Results
In total, 7.570 unique patients were included with 52 different features.
Study size
Starting with 7.570 observations, the process of over and undersampling using SMOTE changed the original number observations. SMOTE was only applied to the train set to prevent leakage of information into the test set. In the training of models, 5.323 observations were included. More information on selection of observations and each taken step in this process is shown in Fig. 1.
Fig. 1.
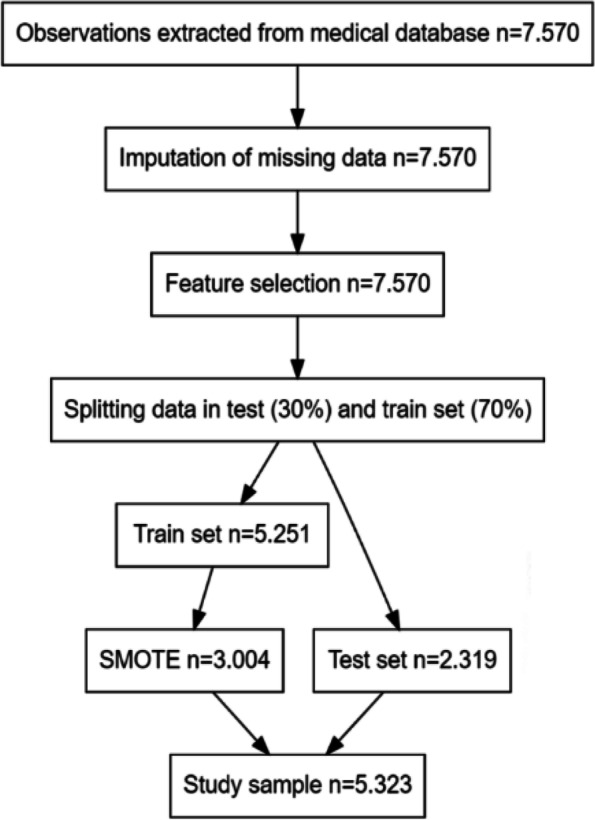
Flow diagram observations
Feature selection
The feature importance of the 52 features were evaluated with a RandomForest algorithm training 2500 trees and features were included based on two criteria:
the feature had a good predictive value (> = 10% importance);
the feature was expected to have clinical importance.
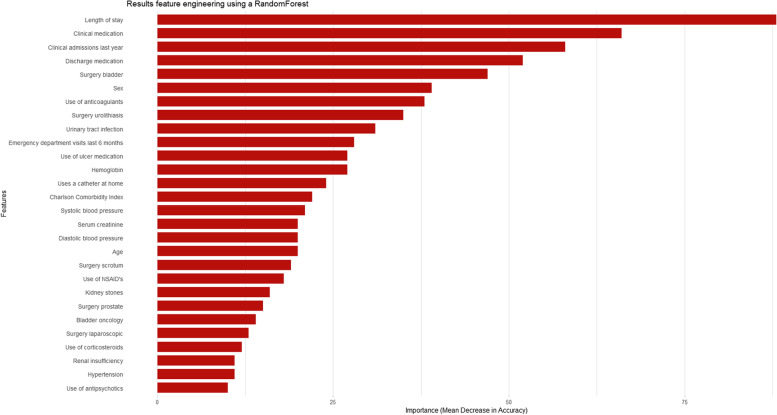
In the final model, 28 features were included ranging from length of stay to use of antipsychotics. Feature importance was calculated and the importance per feature can be found in Fig. 2. This figure indicates an overall performance per feature and does not indicate a negative or positive effect on outcome. Consult Fig. 5 in the Appendix for information on all features, where red features were included and blue features were not.
Fig. 2.
Feature importance
Evaluate performance differences between regression and classification algorithms
To assess the baseline performance, models were trained on selected features and without hyperparameter tweaking. The only non-default setting was the number of trees (default is 500) as trained by the RandomForest algorithm, which was set to 2000.
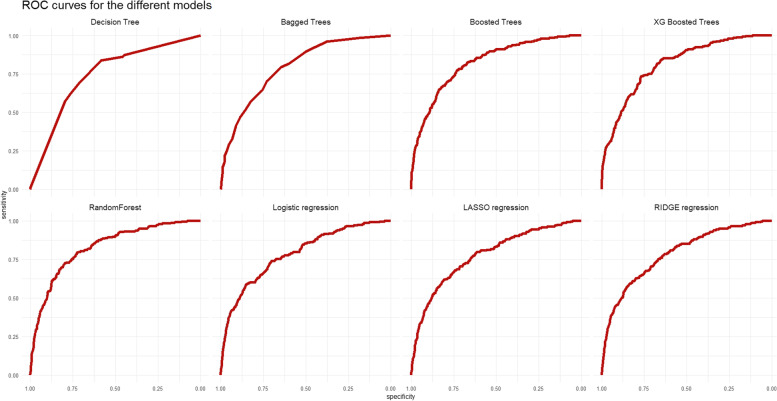
Evaluated on the test set, most models had good AUC scores ranging from 0.62 to 0.82. For AUC, a score above 0.80 indicates a strong discriminative ability. The models showed a better performance in predicting positives in comparison to negatives based on the balance between sensitivity and specificity. The Positive Predictive Value (PPV) scores for all models did not drop below 0.92, indicating that 92% of patients predicted positive were truly readmitted to the hospital. Information of other metrics are shown in Table 2. As seen in the ROC curve plot in Fig. 3, models trained based on classification algorithms (straight lines) show a stronger performance and outperform models trained on regression algorithms (dotted lines). A Wilcoxon test was used to test for statistically significant difference between metrics of the classification algorithms as a group (Decision tree, bagged trees, boosted trees, XG boosted trees, RandomForest), and regression algorithms as a group (Logistic regression, LASSO, and RIDGE regression). Only specificity showed a statistically significant difference with a p-value of 0.0358, whereas sensitivity, AUC, PPV, and NPV did not (p-values of 0.1314, 0.0512, 0.1745, 0.0583, 0.0714 respectively).
Table 2.
Evaluation of performance of regression and classification algorithms
| Algorithm | Accuracy | Sensitivity | Specificity | AUC | PPV | NPV |
|---|---|---|---|---|---|---|
| Decision Tree | 0.68 | 0.80 | 0.57 | 0.75 | 0.94 | 0.24 |
| Bagged Trees | 0.70 | 0.83 | 0.57 | 0.79 | 0.94 | 0.27 |
| Boosted Trees | 0.74 | 0.85 | 0.63 | 0.82 | 0.95 | 0.31 |
| XG Boosted Trees | 0.72 | 0.84 | 0.60 | 0.81 | 0.95 | 0.30 |
| RandomForest | 0.72 | 0.88 | 0.55 | 0.71 | 0.95 | 0.34 |
| Logistic regression | 0.71 | 0.88 | 0.53 | 0.79 | 0.94 | 0.32 |
| LASSO regression | 0.62 | 0.96 | 0.28 | 0.62 | 0.92 | 0.45 |
| RIDGE regression | 0.62 | 0.97 | 0.27 | 0.62 | 0.92 | 0.50 |
Fig. 3.
ROC curves of models
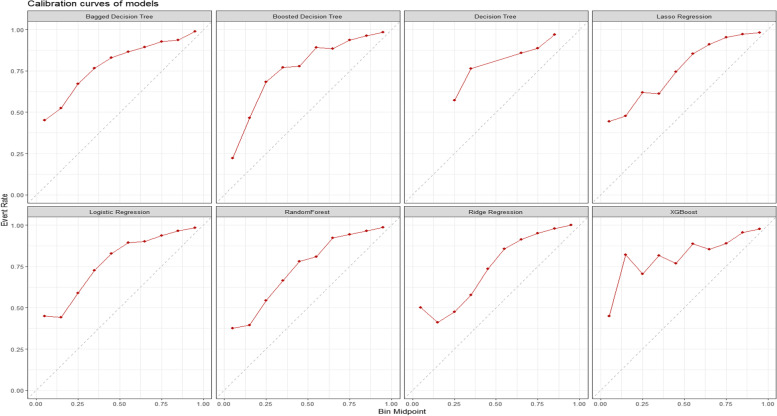
The calibration curves of all trained models show that resampling with SMOTE mainly created an underestimation of predicting positives for our case of 30-day unplanned readmissions. If left without additional calibration, this would lead to a scenario where there would be few patients with a prediction of high risk of having a 30-day unplanned readmission. More information can be found in Fig. 4.
Fig. 4.
Calibration curves of models
Evaluation of the final model used as probability calculator for unplanned readmissions withing 30 days
An XGBoost model, a serial tree-based ensemble learner, showed the strongest overall performance and was chosen as the final model. The model using a boosted trees algorithm also shows a strong performance, but was not chosen due to three reasons being 1) less robust to overfitting, 2) cannot apply cross validation on each iteration, and 3) performs less accurate as compared to XGBoost on smaller datasets.
To assess whether performance of the XGBoost model can be improved, an automated grid search was executed on the train set to tune hyperparameters. The final model with optimized hyperparameters was evaluated on the test set and resulted in an improvement of 11% on accuracy (0.83) while other metrics showed similar performances, indicating that the original XGBoost model already had a strong overall performance. Additional information of the hyperparameters can be found in the Appendix. To assess performance bias in the final model, additional subgroup analysis were performed on sex, age groups, and surgery (yes/no). Statistical differences between the original dataset and subgroups were measured using DeLong’s test to compare two ROC curves. Within the subgroup sex, both male and female showed no significant difference with p-values of 0.4084 and 0.1428 respectively. Age was categorized into groups 18 – 45, 45 – 65, and 65 + , and showed no significant differences with p-values 0.0951, 0.8226, and 0.3019 respectively. Participants with surgery were compared to participants with no surgery and with p-values of 0.8182, and 0.5023 no significant differences were found. No subgroup analysis was performed on COVID-19 since inclusion of patients was limited to the department of Urology and did not suffer in patient care as compared to the department of Pulmonary Diseases for example.
Discussion
Predictive models based on classification algorithms have a stronger performance compared to regression algorithms. The best performing model, the XGBoost model, had good diagnostic performance characteristics that can safely be applied as a risk calculator in clinical setting.
For the clinical department of Urology, evidence on applied ML in predicting unplanned readmissions is scarce. This is the first ML driven probability calculator with accurate prediction of unplanned readmission for Urology patients. Our study shows similar results (AUC 0.62 – 0.82) as compared to earlier studies on performance of predicting 30-day unplanned readmissions (AUC 0.21 – 0.88) [1]. Also, results on features having a high importance on outcome (e.g. length of stay, previous admission and medication) were comparable. We found that using a broader set of features led to a stronger performance as compared to only using LACE, and provides a more detailed risk stratification [9].
Limitations
The results of this study should be interpreted in light of strengths and weaknesses. Strengths being an elaborate comparison using a multitude of features and ML techniques to develop models with. Weaknesses being the quality and presence of patient data on features, and no implementation of PURE in clinical practice to investigate transitioning from predicting to preventing unplanned readmissions.
Features with high importance do not show causal relationship and do not compare to features investigation in a randomized controlled trial. Therefore, feature importance should be evaluated thoroughly on model performance and clinical utility. The selection of features was partly arbitrarily chosen based on earlier scientific findings, and if expected to have a relevant clinical impact based on experiences from the clinical staff of Urology. Missing values of non-continuous features were coded to ‘No’ or ‘Absent’, and could show an incorrect importance as a consequence of incomplete discrete documentation of data in the patient chart. Based on clinical experience and discharge management in the hospital, a period was applied to extract mean values of the last 24 h before discharge in order to make use of features with over time data (e.g. blood pressure). This poses a problem for generalizing our findings, since other hospitals could apply a different period and a set of discharge management choices.
Most ML applications are specific and opt to improve patient care concerning patients suffering from urolithiasis, renal cell carcinoma, bladder cancer, and prostate cancer. As a more generic problem, prevention of unplanned readmissions by applying ML should be further studied in order to evaluate the efficacy on functional outcomes, reduce avoidable stress for patients and improve patient satisfaction [17]. In addition, shared decision making using risk-stratifying predictions of a ML model can decrease the risk up to 13%. Physicians are able to optimize specific outcomes (e.g. complications, infections) more easily by using a calculated risk stratification individual patient level, and discuss these findings with the patient in order to create awareness of potential risks [7, 18–20].
Aside from developing a best performing model, more investigation is necessary in order to determine what features lead to an improved performance. Also, the positive or negative impact of features on outcome need to be elucidated for a better understanding of the clinical value. Follow up studies should focus on varying such dependencies with a more in depth analysis of feature selection, and evaluate if a similar performance as compared to the PURE model is still achieved. In order to transition from predicting to preventing unplanned readmissions, this in depth analysis should also include a comparison of impact of non-modifiable (i.e. static, cannot act on) versus modifiable (i.e. dynamic, can act on) features on model performance and clinical utility.
In order to assess generalizability of the findings in our study, external validation by deploying the model using the same parameter settings and features, is a step that needs to be taken using a specific data sampling method. Other studies show similarities in improved results by applying resampling, but not much drift in calibration, suggesting that the impact of resampling effects on calibration are more case-sensitive as compared to other evaluation metrics. Although distorting calibration, our models trained on resampled data can still have clinical utility whereas the model can have poor calibration yet a strong discriminating performance [21, 22]. Hospitals have differences in patient population, discharge management, and even clinical workflows, which could affect performance of the model. Using transfer learning (i.e. the application of knowledge gained from completing one task to help solve a related problem), our model can be deployed in other hospitals and should be compared an evaluated if the same performance is acquired.
Overall conclusion
It is feasible to develop a risk calculator with a strong performance in predicting unplanned readmissions for the department of Urology. In addition, regression based models are outperformed by classification based models and the latter should be a first pick for use of ML in order to predict unplanned readmissions.
Acknowledgments
Role of the funder/sponsor
The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Statement of human and animal rights
This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent applying the hospital patient data for this study was obtained from the independent Scientific Research Advisory Committee (ACWO), and individual informed consent was deemed unnecessary due to the size of the population as long as applied for this study.
Conflict of interest disclosures
None reported.
Abbreviations
- PURE
Predicting unplanned readmissions
- ML
Machine learning
Appendix
Table 3.
Patient characteristics
| Unplanned readmission within 30 days | ||||
|---|---|---|---|---|
| Yes ( N = 774) | No ( N = 6796) | P -value | Total ( N = 7570) | |
| Charlson Comorbidity Index | ||||
| Mean (SD) | 1.48 (2.03) | 0.998 (1.74) | < 0.001 | 1.05 (1.77) |
| Median [Min, Max] | 0 [0, 10.0] | 0 [0, 11.0] | 0 [0, 11.0] | |
| Age | ||||
| Mean (SD) | 70.3 (15.7) | 64.4 (17.5) | < 0.001 | 65.0 (17.4) |
| Median [Min, Max] | 74.0 [20.0, 103] | 68.0 [13.0, 109] | 69.0 [13.0, 109] | |
| BMI | ||||
| Mean (SD) | 26.4 (5.48) | 25.9 (4.92) | 0.0181 | 26.0 (4.99) |
| Median [Min, Max] | 25.6 [13.3, 66.5] | 25.3 [13.3, 53.1] | 25.3 [13.3, 66.5] | |
| Systolic blood pressure | ||||
| Mean (SD) | 134 (17.2) | 131 (18.8) | < 0.001 | 131 (18.7) |
| Median [Min, Max] | 133 [93.0, 182] | 129 [85.0, 210] | 129 [85.0, 210] | |
| Diastolic blood pressure | ||||
| Mean (SD) | 74.5 (8.18) | 74.4 (9.04) | 0.644 | 74.4 (8.96) |
| Median [Min, Max] | 74.0 [53.0, 105] | 74.0 [44.0, 126] | 74.0 [44.0, 126] | |
| Creatinine blood | ||||
| Mean (SD) | 115 (90.9) | 95.5 (64.2) | < 0.001 | 97.5 (67.7) |
| Median [Min, Max] | 91.0 [37.0, 1260] | 83.0 [21.0, 1480] | 84.0 [21.0, 1480] | |
| Hemoglobin | ||||
| Mean (SD) | 7.67 (1.12) | 7.72 (1.22) | 0.246 | 7.71 (1.21) |
| Median [Min, Max] | 7.70 [4.10, 11.6] | 7.80 [4.00, 11.6] | 7.80 [4.00, 11.6] | |
| Clinical medication | ||||
| Mean (SD) | 51.7 (34.0) | 30.4 (25.7) | < 0.001 | 32.6 (27.4) |
| Median [Min, Max] | 44.0 [7.00, 227] | 22.0 [0, 267] | 24.0 [0, 267] | |
| Home medication | ||||
| Mean (SD) | 12.8 (8.47) | 8.07 (7.33) | < 0.001 | 8.55 (7.59) |
| Median [Min, Max] | 11.0 [0, 48.0] | 6.00 [0, 60.0] | 6.00 [0, 60.0] | |
| Clinical admissions last year | ||||
| Mean (SD) | 0.860 (1.52) | 0.311 (0.732) | < 0.001 | 0.367 (0.862) |
| Median [Min, Max] | 0 [0, 11.0] | 0 [0, 9.00] | 0 [0, 11.0] | |
| ED visits last 6 months | ||||
| Mean (SD) | 0.382 (0.890) | 0.144 (0.503) | < 0.001 | 0.169 (0.560) |
| Median [Min, Max] | 0 [0, 8.00] | 0 [0, 8.00] | 0 [0, 8.00] | |
| Length of Stay | ||||
| Mean (SD) | 3.98 (5.44) | 2.21 (3.35) | < 0.001 | 2.39 (3.66) |
| Median [Min, Max] | 3.00 [0, 97.0] | 1.00 [0, 65.0] | 1.00 [0, 97.0] | |
| Sex | ||||
| Female | 153 (19.8%) | 2439 (35.9%) | < 0.001 | 2592 (34.2%) |
| Male | 621 (80.2%) | 4357 (64.1%) | 4978 (65.8%) | |
| History of smoking | ||||
| No | 656 (84.8%) | 5577 (82.1%) | 0.0702 | 6233 (82.3%) |
| Yes | 118 (15.2%) | 1219 (17.9%) | 1337 (17.7%) | |
| Use of alcohol | ||||
| No | 420 (54.3%) | 3308 (48.7%) | 0.00363 | 3728 (49.2%) |
| Yes | 354 (45.7%) | 3488 (51.3%) | 3842 (50.8%) | |
| Interpreter needed | ||||
| No | 738 (95.3%) | 6618 (97.4%) | 0.00183 | 7356 (97.2%) |
| Yes | 36 (4.7%) | 178 (2.6%) | 214 (2.8%) | |
| Fluency in Dutch | ||||
| No | 78 (10.1%) | 783 (11.5%) | 0.255 | 861 (11.4%) |
| Yes | 696 (89.9%) | 6013 (88.5%) | 6709 (88.6%) | |
| Uses a catheter at home | ||||
| No | 716 (92.5%) | 6563 (96.6%) | < 0.001 | 7279 (96.2%) |
| Yes | 58 (7.5%) | 233 (3.4%) | 291 (3.8%) | |
| Use of anticoagulants | ||||
| No | 116 (15.0%) | 2392 (35.2%) | < 0.001 | 2508 (33.1%) |
| Yes | 658 (85.0%) | 4404 (64.8%) | 5062 (66.9%) | |
| Use of NSAID's | ||||
| No | 529 (68.3%) | 4697 (69.1%) | 0.692 | 5226 (69.0%) |
| Yes | 245 (31.7%) | 2099 (30.9%) | 2344 (31.0%) | |
| Use of corticosteroids | ||||
| No | 686 (88.6%) | 6533 (96.1%) | < 0.001 | 7219 (95.4%) |
| Yes | 88 (11.4%) | 263 (3.9%) | 351 (4.6%) | |
| Use of antipsychotics | ||||
| No | 715 (92.4%) | 6578 (96.8%) | < 0.001 | 7293 (96.3%) |
| Yes | 59 (7.6%) | 218 (3.2%) | 277 (3.7%) | |
| Use of ulcer medication | ||||
| No | 380 (49.1%) | 4086 (60.1%) | < 0.001 | 4466 (59.0%) |
| Yes | 394 (50.9%) | 2710 (39.9%) | 3104 (41.0%) | |
| Oncology | ||||
| Absent | 700 (90.4%) | 6358 (93.6%) | 0.0014 | 7058 (93.2%) |
| Present | 74 (9.6%) | 438 (6.4%) | 512 (6.8%) | |
| Medication | ||||
| No | 83 (10.7%) | 1793 (26.4%) | < 0.001 | 1876 (24.8%) |
| Yes | 691 (89.3%) | 5003 (73.6%) | 5694 (75.2%) | |
| Comorbidity | ||||
| Absent | 607 (78.4%) | 5995 (88.2%) | < 0.001 | 6602 (87.2%) |
| Present | 167 (21.6%) | 801 (11.8%) | 968 (12.8%) | |
| Surgery | ||||
| No | 354 (45.7%) | 4325 (63.6%) | < 0.001 | 4679 (61.8%) |
| Yes | 420 (54.3%) | 2471 (36.4%) | 2891 (38.2%) | |
P-values calculated with Student’s T-test for numeric variables and Chi-squared test for categorical variables
Detailed overview of explanatory variables.
- Patient characteristics
- ◦ Age
- ◦ Sex
- ◦ Charlson Comorbidity Index (CCI)
- ◦ BMI
- ◦ Smoking history
- ◦ Use of alcohol
- ◦ Fluency in Dutch
- Lab results during clinical admission
- ◦ Mean diastolic blood pressure within 24 hours before discharge
- ◦ Mean systolic blood pressure within 24 hours before discharge
- ◦ Mean platelet count within 24 hours before discharge
- ◦ Last serum creatinine before discharge
- ◦ Last hemoglobin before discharge
- Currently active medication during admission
- ◦ Total count of clinical medications
- ◦ Total count of discharge medications
- ◦ Use of anticoagulants
- ◦ Use of NSAID’s
- ◦ Use of corticosteroids
- ◦ Use of antipsychotics
- ◦ Use of ulcer medication
- Health care logistics at the time of admission
- ◦ Total count of clinical admissions in the last year
- ◦ Total count of emergency department visits last 6 months
- ◦ Total length of stay
- ◦ Interpreter needed
- ◦ Home use of catheter
- Medical history
- ◦ Hypercholesteremia
- ◦ Diabetes type I or type II
- ◦ Hypertension
- ◦ Rheumatoid arthritis
- ◦ Atrial fibrillation
- ◦ Renal insufficiency
- Cerebrovascular disorders
- ◦ Ischemic cardiovascular disease
- ◦ Peripheral vascular disease
- ◦ Heart failure
- ◦ Cardiovascular disease
- ◦ Kidney stones
- ◦ Urinary tract infection
- ◦ Testicular oncology
- ◦ Bladder oncology
- ◦ Ureteral oncology
- ◦ Urethra oncology
- ◦ Renal oncology
- ◦ Prostate oncology
- ◦ Renal pelvis oncology
- Type of surgery
- ◦ Open abdomen
- ◦ Laparoscopic
- ◦ Scrotum
- ◦ Penis
- ◦ Prostrate
- ◦ Urethral
- ◦ Ureterorenoscopy
- ◦ Urolithiasis
- ◦ Bladder
Patient characteristics between the train and the test dataset can be found in Table 4.
Table 4.
Patient characteristics between train and test data
| Train (N = 6008) | Test (N = 4638) | P-value | Total (N = 10,646) | |
|---|---|---|---|---|
| Use of antipsychotics | ||||
| No | 5710 (95.0%) | 4492 (96.9%) | < 0.001 | 10,202 (95.8%) |
| Yes | 298 (5.0%) | 146 (3.1%) | 444 (4.2%) | |
| Hypertension | ||||
| No | 5718 (95.2%) | 4420 (95.3%) | 0.796 | 10,138 (95.2%) |
| Yes | 290 (4.8%) | 218 (4.7%) | 508 (4.8%) | |
| Renal insufficiency | ||||
| No | 5830 (97.0%) | 4514 (97.3%) | 0.405 | 10,344 (97.2%) |
| Yes | 178 (3.0%) | 124 (2.7%) | 302 (2.8%) | |
| Use of corticosteroids | ||||
| No | 5632 (93.7%) | 4440 (95.7%) | < 0.001 | 10,072 (94.6%) |
| Yes | 376 (6.3%) | 198 (4.3%) | 574 (5.4%) | |
| Surgery laparoscopic | ||||
| No | 5862 (97.6%) | 4528 (97.6%) | 0.896 | 10,390 (97.6%) |
| Yes | 146 (2.4%) | 110 (2.4%) | 256 (2.4%) | |
| Bladder oncology | ||||
| No | 5734 (95.4%) | 4464 (96.2%) | 0.0441 | 10,198 (95.8%) |
| Yes | 274 (4.6%) | 174 (3.8%) | 448 (4.2%) | |
| Surgery prostate | ||||
| No | 5318 (88.5%) | 4220 (91.0%) | < 0.001 | 9538 (89.6%) |
| Yes | 690 (11.5%) | 418 (9.0%) | 1108 (10.4%) | |
| Kidney stones | ||||
| No | 5860 (97.5%) | 4484 (96.7%) | 0.00982 | 10,344 (97.2%) |
| Yes | 148 (2.5%) | 154 (3.3%) | 302 (2.8%) | |
| Use of NSAID's | ||||
| No | 4140 (68.9%) | 3214 (69.3%) | 0.682 | 7354 (69.1%) |
| Yes | 1868 (31.1%) | 1424 (30.7%) | 3292 (30.9%) | |
| Surgery scrotal | ||||
| No | 5874 (97.8%) | 4516 (97.4%) | 0.203 | 10,390 (97.6%) |
| Yes | 134 (2.2%) | 122 (2.6%) | 256 (2.4%) | |
| Age | ||||
| Mean (SD) | 66.7 (16.8) | 65.0 (17.7) | < 0.001 | 65.9 (17.2) |
| Median [Min, Max] | 71.0 [13.0, 103] | 69.0 [17.0, 106] | 70.0 [13.0, 106] | |
| Diastolic blood pressure | ||||
| Mean (SD) | 74.2 (8.85) | 74.7 (8.89) | 0.00368 | 74.5 (8.87) |
| Median [Min, Max] | 74.0 [44.0, 126] | 74.0 [44.0, 117] | 74.0 [44.0, 126] | |
| Creatinine blood | ||||
| Mean (SD) | 104 (80.5) | 97.7 (62.9) | < 0.001 | 101 (73.5) |
| Median [Min, Max] | 87.0 [21.0, 1270] | 84.0 [28.0, 1480] | 85.0 [21.0, 1480] | |
| Systolic blood pressure | ||||
| Mean (SD) | 132 (18.2) | 132 (18.7) | 0.956 | 132 (18.4) |
| Median [Min, Max] | 130 [87.0, 210] | 130 [85.0, 200] | 130 [85.0, 210] | |
| Charlson Comorbidity Index | ||||
| Mean (SD) | 1.15 (1.83) | 1.04 (1.80) | 0.00404 | 1.10 (1.82) |
| Median [Min, Max] | 0 [0, 11.0] | 0 [0, 10.0] | 0 [0, 11.0] | |
| Uses a catheter at home | ||||
| No | 5758 (95.8%) | 4432 (95.6%) | 0.509 | 10,190 (95.7%) |
| Yes | 250 (4.2%) | 206 (4.4%) | 456 (4.3%) | |
| Hemoglobin | ||||
| Mean (SD) | 7.71 (1.14) | 7.69 (1.19) | 0.629 | 7.70 (1.16) |
| Median [Min, Max] | 7.80 [4.00, 11.6] | 7.80 [4.00, 11.6] | 7.80 [4.00, 11.6] | |
| Use of ulcer medication | ||||
| No | 3324 (55.3%) | 2764 (59.6%) | < 0.001 | 6088 (57.2%) |
| Yes | 2684 (44.7%) | 1874 (40.4%) | 4558 (42.8%) | |
| ED visits last 6 months | ||||
| Mean (SD) | 0.207 (0.594) | 0.162 (0.558) | < 0.001 | 0.188 (0.579) |
| Median [Min, Max] | 0 [0, 8.00] | 0 [0, 7.00] | 0 [0, 8.00] | |
| Urinary tract infection | ||||
| No | 5818 (96.8%) | 4530 (97.7%) | 0.0115 | 10,348 (97.2%) |
| Yes | 190 (3.2%) | 108 (2.3%) | 298 (2.8%) | |
| Surgery urolithiasis | ||||
| No | 5462 (90.9%) | 4232 (91.2%) | 0.572 | 9694 (91.1%) |
| Yes | 546 (9.1%) | 406 (8.8%) | 952 (8.9%) | |
| Use of anticoagulants | ||||
| No | 1550 (25.8%) | 1608 (34.7%) | < 0.001 | 3158 (29.7%) |
| Yes | 4458 (74.2%) | 3030 (65.3%) | 7488 (70.3%) | |
| Sex | ||||
| Female | 1832 (30.5%) | 1556 (33.5%) | < 0.001 | 3388 (31.8%) |
| Male | 4176 (69.5%) | 3082 (66.5%) | 7258 (68.2%) | |
| Surgery bladder | ||||
| No | 4744 (79.0%) | 3878 (83.6%) | < 0.001 | 8622 (81.0%) |
| Yes | 1264 (21.0%) | 760 (16.4%) | 2024 (19.0%) | |
| Home medication | ||||
| Mean (SD) | 9.83 (7.80) | 8.55 (7.62) | < 0.001 | 9.27 (7.75) |
| Median [Min, Max] | 8.00 [0, 51.0] | 6.00 [0, 59.0] | 7.00 [0, 59.0] | |
| Clinical admissions last year | ||||
| Mean (SD) | 0.479 (1.03) | 0.361 (0.838) | < 0.001 | 0.428 (0.952) |
| Median [Min, Max] | 0 [0, 11.0] | 0 [0, 11.0] | 0 [0, 11.0] | |
| Clinical medication | ||||
| Mean (SD) | 37.6 (29.8) | 33.0 (27.9) | < 0.001 | 35.6 (29.1) |
| Median [Min, Max] | 28.0 [0, 253] | 25.0 [0, 209] | 26.0 [0, 253] | |
| Length of Stay | ||||
| Mean (SD) | 2.84 (3.72) | 2.39 (3.75) | < 0.001 | 2.64 (3.74) |
| Median [Min, Max] | 2.00 [0, 65.0] | 1.00 [0, 97.0] | 2.00 [0, 97.0] | |
| Oncology | ||||
| Absent | 5734 (95.4%) | 4464 (96.2%) | 0.0441 | 10,198 (95.8%) |
| Present | 274 (4.6%) | 174 (3.8%) | 448 (4.2%) | |
| Medication | ||||
| No | 1182 (19.7%) | 1200 (25.9%) | < 0.001 | 2382 (22.4%) |
| Yes | 4826 (80.3%) | 3438 (74.1%) | 8264 (77.6%) | |
| Comorbidity | ||||
| Absent | 5422 (90.2%) | 4158 (89.7%) | 0.326 | 9580 (90.0%) |
| Present | 586 (9.8%) | 480 (10.3%) | 1066 (10.0%) | |
| Surgery | ||||
| No | 3588 (59.7%) | 2988 (64.4%) | < 0.001 | 6576 (61.8%) |
| Yes | 2420 (40.3%) | 1650 (35.6%) | 4070 (38.2%) | |
P-values calculated with Student’s T-test for numeric variables and Chi-squared test for categorical variables
Detailed informed of hyperparameter optimization of the XGBoost model.
A grid-search was performed on the train set using 5-fold CV, to search for optimal parameter settings. Optimal parameter values found were: nrounds = 3000, eta = 0.015, max_depth = 5, gamma = 0.05, colsample_bytree = 1, min_child_weight = 1, and subsample = 0.5.
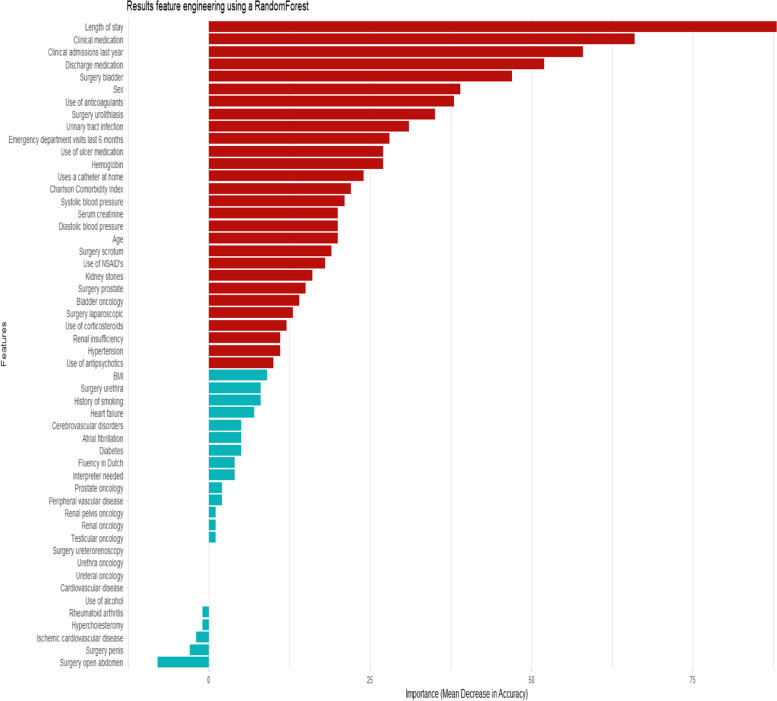
Importance of all features is shown in Fig. 5. This step was performed before feature selection for developing the models.
Fig. 5.
Importance of all features
Authors’ contributions
KW had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. EPH, MPJB and JND contributed equally as co-authors. Concept and design: All authors. Acquisition, analysis, modelling of data: KW. Interpretation of data: All authors. Drafting of the manuscript: All authors. Critical revision of the manuscript for important intellectual content: All authors. Statistical analysis: KW. Obtained funding: Not applicable. Administrative, technical, or material support: KW. Supervision: EPH, MPJB, JND. The author(s) read and approved the final manuscript.
Funding
This work was supported by the OLVG Urology consortium.
Availability of data and materials
The data that support the findings of this study are available from OLVG but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the ACWO. The code is accessible via https://github.com/koenwelvaars/PURE_study.
Declarations
Ethics approval and consent to participate
This study was approved by the independent Scientific Research Advisory Committee. This study followed the guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research, and the guidelines for Transparent Reporting of Multivariable Prediction Models for Individual Prognosis or Diagnosis.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Koen Welvaars, Email: k.welvaars01@umcg.nl.
OLVG Urology Consortium:
J. A. van der Zee, G. A. van Andel, B. W. Lagerveld, M. C. Hovius, P. C. Kauer, and L. M. S. Boevé
References
- 1.Zhou H, Della PR, Roberts P, Goh L, Dhaliwal SS. Utility of models to predict 28-day or 30-day unplanned hospital readmissions: an updated systematic review. BMJ Open. 2016;6(6):e011060. doi: 10.1136/bmjopen-2016-011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaudeen N, Schnipper JL, Orav EJ, Wachter RM, Vidyarthi AR. Inability of providers to predict unplanned readmissions. J Gen Intern Med. 2011;26(7):771–6. doi: 10.1007/s11606-011-1663-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baack Kukreja J, Kamat AM. Strategies to minimize readmission rates following major urologic surgery. Ther Adv Urol. 2017;9(5):111–119. doi: 10.1177/1756287217701699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen MK, Meyer G, Uhrenfeldt L. Risk factors for acute care hospital readmission in older persons in Western countries: a systematic review. JBI Database System Rev Implement Rep. 2017;15(2):454–485. doi: 10.11124/JBISRIR-2016-003267. [DOI] [PubMed] [Google Scholar]
- 5.van der Does AMB, Kneepkens EL, Uitvlugt EB, Jansen SL, Schilder L, Tokmaji G, Wijers SC, Radersma M, Heijnen JNM, Teunissen PFA, Hulshof PBJE, Overvliet GM, Siegert CEH, Karapinar-Çarkit F. Preventability of unplanned readmissions within 30 days of discharge. A cross-sectional, single-center study. PLoS one. 2020;15(4):e0229940. doi: 10.1371/journal.pone.0229940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryu B, Yoo S, Kim S, Choi J. Development of prediction models for unplanned hospital readmission within 30 days based on common data model: a feasibility study. Methods Inf Med. 2021;60:e65–e74. doi: 10.1055/s-0041-1735166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker C, Zumbrunn S, Beck K, Vincent A, Loretz N, Müller J, Amacher SA, Schaefert R, Hunziker S. Interventions to improve communication at hospital discharge and rates of readmission: a systematic review and meta-analysis. JAMA Netw Open. 2021;4(8):e2119346. doi: 10.1001/jamanetworkopen.2021.19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baig M, Hua N, Zhang E, Robinson R, Armstrong D, Whittaker R, Robinson T, Mirza F, Ullah E. Predicting patients at risk of 30-day unplanned hospital readmission. Stud Health Technol Inform. 2019;8(266):20–24. doi: 10.3233/SHTI190767. [DOI] [PubMed] [Google Scholar]
- 9.Heppleston E, Fry CH, Kelly K, et al. LACE index predicts age-specific unplanned readmissions and mortality after hospital discharge. Aging Clin Exp Res. 2021;33:1041–1048. doi: 10.1007/s40520-020-01609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Futoma J, Morris J, Lucas J. A comparison of models for predicting early hospital readmissions. J Biomed Inform. 2015;56:229–238. doi: 10.1016/j.jbi.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 11.Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res. 2016;18(12):e323. doi: 10.2196/jmir.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–W73. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 13.Azur MJ, Stuart EA, Frangakis C, Leaf PJ. Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res. 2011;20(1):40–49. doi: 10.1002/mpr.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali A, Shamsuddin SM, Ralescu A. Classification with class imbalance problem: a review. Soft Computing Models in Industrial and Environmental Applications. 2015;7:176–204.
- 15.Lin Z, Yang C, Zhu Y, Duchi J, Fu Y, Wang Y, Jiang B, Zamanighomi M, Xu X, Li M, Sestan S, Zhao H, Wong WH. Simultaneous dimension reduction and adjustment for confounding variation. Proc Natl Acad Sci U S A. 2016;113(51):14662–7. doi: 10.1073/pnas.1617317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menze BH, Kelm BM, Masuch R, et al. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinformatics. 2009;10:213. doi: 10.1186/1471-2105-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suarez-Ibarrola R, Hein S, Reis G, Gratzke C, Miernik A. Current and future applications of machine and deep learning in urology: a review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J Urol. 2020;38(10):2329–2347. doi: 10.1007/s00345-019-03000-5. [DOI] [PubMed] [Google Scholar]
- 18.Jayakumar P, Moore MG, Furlough KA, Uhler LM, Andrawis JP, Koenig KM, Aksan N, Rathouz PJ, Bozic KJ. Comparison of an artificial intelligence-enabled patient decision aid vs educational material on decision quality, shared decision-making, patient experience, and functional outcomes in adults with knee osteoarthritis: a randomized clinical trial. JAMA Netw Open. 2021;4(2):e2037107. doi: 10.1001/jamanetworkopen.2020.37107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giordano C, Brennan M, Mohamed B, Rashidi P, Modave F, Tighe P. Accessing artificial intelligence for clinical decision-making. Front Digit Health. 2021;3:645232. doi: 10.3389/fdgth.2021.645232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henn J, Buness A, Schmid M, Kalff JC, Matthaei H. Machine learning to guide clinical decision-making in abdominal surgery-a systematic literature review. Langenbecks Arch Surg. 2021. 10.1007/s00423-021-02348-w. Epub ahead of print. PMID: 34716472. [DOI] [PMC free article] [PubMed]
- 21.Steyerberg EW, Harrell FE., Jr Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol. 2016;69:245–7. doi: 10.1016/j.jclinepi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Goorbergh R, van Smeden M, Timmerman D, Van Calster B. The harm of class imbalance corrections for risk prediction models: illustration and simulation using logistic regression. J Am Med Inform Assoc. 2022;29(9):1525–1534. doi: 10.1093/jamia/ocac093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from OLVG but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the ACWO. The code is accessible via https://github.com/koenwelvaars/PURE_study.