Abstract
Aims
The major cardiac cell types composing the adult heart arise from common multipotent precursor cells. Cardiac lineage decisions are guided by extrinsic and cell-autonomous factors, including recently discovered long noncoding RNAs (lncRNAs). The human lncRNA CARMEN, which is known to dictate specification toward the cardiomyocyte (CM) and the smooth muscle cell (SMC) fates, generates a diversity of alternatively spliced isoforms.
Methods and results
The CARMEN locus can be manipulated to direct human primary cardiac precursor cells (CPCs) into specific cardiovascular fates. Investigating CARMEN isoform usage in differentiating CPCs represents therefore a unique opportunity to uncover isoform-specific functions in lncRNAs. Here, we identify one CARMEN isoform, CARMEN-201, to be crucial for SMC commitment. CARMEN-201 activity is encoded within an alternatively spliced exon containing a MIRc short interspersed nuclear element. This element binds the transcriptional repressor REST (RE1 Silencing Transcription Factor), targets it to cardiogenic loci, including ISL1, IRX1, IRX5, and SFRP1, and thereby blocks the CM gene program. In turn, genes regulating SMC differentiation are induced.
Conclusions
These data show how a critical physiological switch is wired by alternative splicing and functional transposable elements in a long noncoding RNA. They further demonstrated the crucial importance of the lncRNA isoform CARMEN-201 in SMC specification during heart development.
Keywords: Cardiac precursor cells, Smooth muscle cells, Long noncoding RNAs, Splicing, Transposable elements
Graphical Abstract
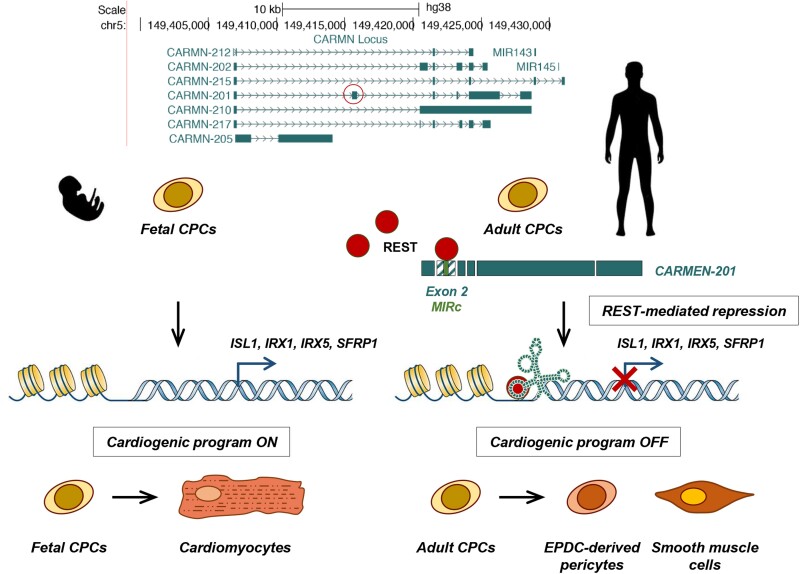
Graphical Abstract.
1. Introduction
The mammalian heart is composed of several cell types that derive from mesodermal progenitor cells of the first and second heart fields.1 The distinct lineages arise from multipotent cardiovascular precursors.2–4 In this framework, very few studies have evaluated transcriptional regulation in primary human cardiac precursor cells (CPCs). We have previously derived clonal populations of CPCs from the fetal and the adult human heart.5,6 Their comparison allows for the dissection of the molecular mechanisms controlling cardiac specification and differentiation. Understanding the processes regulating cardiac cell programing and reprograming provides also novel insights for the treatment of cardiovascular disease.
Next-generation sequencing coupled to the assessment of the epigenetic landscape has shown that mammalian genomes produce thousands of noncoding transcripts. Of these, long noncoding (lnc)RNAs represent the most heterogeneous and diverse class of RNA molecules. lncRNAs can be multiexonic, spliced, capped, and polyadenylated.7 Current estimates predict the existence of approximately 200 000 lncRNAs in human, of which very few have been fully characterized. lncRNAs are implicated in a variety of functions that define cell identity and behavior. They exert Cis- and Trans-regulatory functions, controlling chromatin remodeling and transcription. Cis-acting lncRNAs operate in close vicinity to their site of transcription.8 On the other hand, Trans-acting lncRNAs leave their site of transcription and exert functions at remote locations in the genome.9 In this case, lncRNAs partner with proteins such as chromatin remodelers to modify the local chromatin environment at target locations. In the cardiovascular system, several lncRNAs have been identified as key players in cell differentiation and homeostasis. Braveheart, Fendrr, and Meteor have been involved in mesoderm and cardiac differentiation.10–12Myheart controls CM hypertrophy, and Wisper is a regulator of cardiac fibroblasts, critical for the development of fibrosis.13,14 Then, SMILR and SENCR have been associated with SMC proliferation and differentiation.15,16
The way in which functions are encoded in lncRNAs’ sequences remains enigmatic. It is thought that lncRNAs comprise modular assemblages of functional elements, composed of structural motifs that interact with proteins or other nucleic acids.17 Recent studies have implicated repetitive transposable elements as a source of functional lncRNA domains.18,19 In this context, lncRNA loci can produce a variety of transcripts through alternative splicing.7,20 It has been speculated that these alternative isoforms, by containing different combinations of exon sequences, thereby exert diverse functions. Some years ago, we identified CARMEN, a conserved lncRNA essential for cardiogenesis.21CARMEN has been previously referred to as MIR143HG because the locus hosts MIR-143 and MIR-145, two microRNAs (miRNAs) important for SMC differentiation.22 Nevertheless, the miRNA precursor represents only one CARMEN isoform among several others. All other isoforms are lncRNA splice variants, with no apparent coding potential, for which functions remain to be fully defined. We showed previously that three isoforms were involved in the cardiac specification in human fetal CPCs.21 Moreover, we reported that one isoform referred earlier as to CARMEN-7 (formally CARMEN-201 or ENST00000505254), was differentially expressed in CPCs committing to the CM vs. the SMC lineage.6 Interestingly, the CARMEN locus can be manipulated to direct CPCs into specific cardiovascular fates. Investigating CARMEN isoform usage in differentiating CPCs represents therefore a unique opportunity to uncover isoform-specific function in lncRNAs. Here, we demonstrate that CARMEN-201 controls specification into the SMC lineage in human CPCs and that this function is encoded within an alternatively spliced exon containing a MIRc short interspersed nuclear element (SINE).
2. Methods
Methods are described in detail in the Supplementary Information available online.
2.1. Human CPCs
Fetal heart biopsies were collected at 5 weeks of gestation following abortion, and adult atrial appendages were obtained from patients undergoing cardiac surgery. CPCs were isolated by enzymatic digestion as previously described.5,6
2.2. Human plasma samples
Plasma samples were collected from patients admitted to the Lausanne University Hospital with a diagnosis of myocardial infarction.
2.3. Study approval
The study was approved by the Lausanne University Hospital Ethics Committee and the Swiss Ethics Committee (Human cardiac precursor cells: Protocols 22/03 and 178/09; Human plasma from patients with myocardial infarction: Protocol PB_2018-00231 and 94/15) and was conducted according to the Declaration of Helsinki. Written informed consent was obtained from all patients included in the study.
2.4. GapmeR-mediated knockdown
CARMEN-201 silencing was obtained by adding CARMEN-201-specific GapmeRs targeting Exon 2 to CPCs at a final concentration of 20 nM.
2.5. siRNA-mediated knockdown
siRNA transfection was performed using RNAiMax (ThermoFisher). CPCs were transfected with the indicated siRNA at a final concentration of 10 nM.
2.6. RNA extraction, RT-PCR, and real-time PCR
Total RNA from plasma and from cultured cells was extracted using the miRNeasy Serum/Plasma Advanced kit and the miRNeasy kit (Qiagen).
2.7. Absolute quantification of CARMEN isoform expression
pBluescript SK + plasmids containing CARMEN-201, CARMEN-205 or CARMEN-217 cDNA were synthesized (GenScript, USA). Data were converted into transcript copy per cell, assuming 100% efficiency in the conversion of RNA into cDNA.
2.8. Subcellular fractionation
Cells were harvested and lysed. The lysate was centrifuged at 3800 g for 2 min. The supernatant represented the cytoplasmic fraction. The pellet was used to produce the nuclear fraction.
2.9. CRISPR/Cas9-mediated Exon 2 deletion
A CRISPR/Cas9-D10A Nickase-based strategy was used to delete Exon 2 in the CARMEN gene.
2.10. CRISPR-On activation
We used the CRISPR/dCas9-based Synergistic Activator Mediator (SAM) gain of function system to activate CARMEN isoform expression in fetal CPCs.23
2.11. RNA sequencing
Sequencing libraries were prepared according to Illumina RNA Seq library kit instructions. Libraries were sequenced with the Illumina HiSeq2000 (100 bp; PE).
2.12. TRIAGE analysis
The Transcriptional Regulatory Inference Analysis from Gene Expression (TRIAGE) analysis, providing a means to identify cell type-specific regulatory genes has been described.24
2.13. Uniform Manifold Approximation and Projection density plots
We used the data provided by Asp et al., 2019 (accession number is European Genome-phenome Archive (EGA): EGAS00001003996). The Uniform Manifold Approximation and Projection (UMAP) was generated using Nebulosa package.
2.14. Lentiviral vectors
The SIN-cppt-CMV-EGFP-WHV plasmid was a kind gift from Dr Nicole Deglon (University of Lausanne, Lausanne, Switzerland). EGFP sequences were replaced by either the wild-type CARMEN-201 Exon 2 or a mutated Exon 2 containing scrambled MIRc sequences.
2.15. RNA pulldown and identification of CARMEN-201 protein partners
The pBluescript SK + plasmids containing C-201 Ex2 or C-201 mutEx2 were used to synthesize biotinylated probes. The precleared lysate was incubated with either no probe, biotinylated C-201 Ex2 or biotinylated mutC-201 Ex2. Proteins were loaded on 12% SDS-PAGE.
2.16. Tandem mass spectrometry
For identifying CARMEN-201 protein partners, tryptic peptide mixtures were injected on an Ultimate RSLC 3000 nanoHPLC system (Dionex, Sunnyvale, CA, USA).
2.17. Western blotting
Proteins associated with biotinylated C-201 transcript were resolved by SDS-PAGE and electroblotted onto PVDF membranes (GE Healthcare).
2.18. RNA immunoprecipitation
The RNA immunoprecipitation (RIP) experiment was conducted as described.14
2.19. RIP following by sequencing
RIP was performed using anti-REST IgG. REST-associated transcripts were purified using the RNeasy isolation kit (Qiagen). Sequencing libraries were prepared according to Illumina RNA Seq library kit instructions. Libraries were sequenced with the Illumina HiSeq2000 (100 bp; PE).
2.20. Chromatin immunoprecipitation followed by real-time quantitative PCR
Chromatin immunoprecipitation followed by real-time quantitative PCR (ChIP-qPCR) was performed using the Pierce magnetic ChIP Kit (Thermo Scientific) and the ChIPAb + REST Kit (Millipore) according to the manufacturers’ instructions.
2.21. Immunohistochemistry
Cells were fixed in 2% paraformaldehyde, permeabilized in 0.3% Triton-X100 and processed for immunostaining using appropriate antibodies. Coverslips were mounted with VECTASHIELD Antifade Mounting medium with DAPI (VECTOR LABORATORIES).
2.22. RNA BaseScope in situ hybridization
RNA BaseScope in situ hybridization was used to detect CARMEN-201 expression in adult CPCs in culture or in sections of human hearts. In situ detection was performed with the BaseScope kit (ACD biotech, 323900).
2.23. Masson’s trichome staining
Paraffin tissue sections were also processed for Masson’s trichrome staining and analyzed with a Zeiss Axioscan Z1 (Carl Zeiss).
2.24. Statistics
All data were collected from at least three independent experiments, performed at least in triplicates. Data throughout the paper are expressed as mean ± SEM. Statistical analysis: ANOVA with post hoc Tukey.
3. Results
3.1. CARMEN isoforms are differentially expressed in CPCs committing to the CM vs. the SMC fate
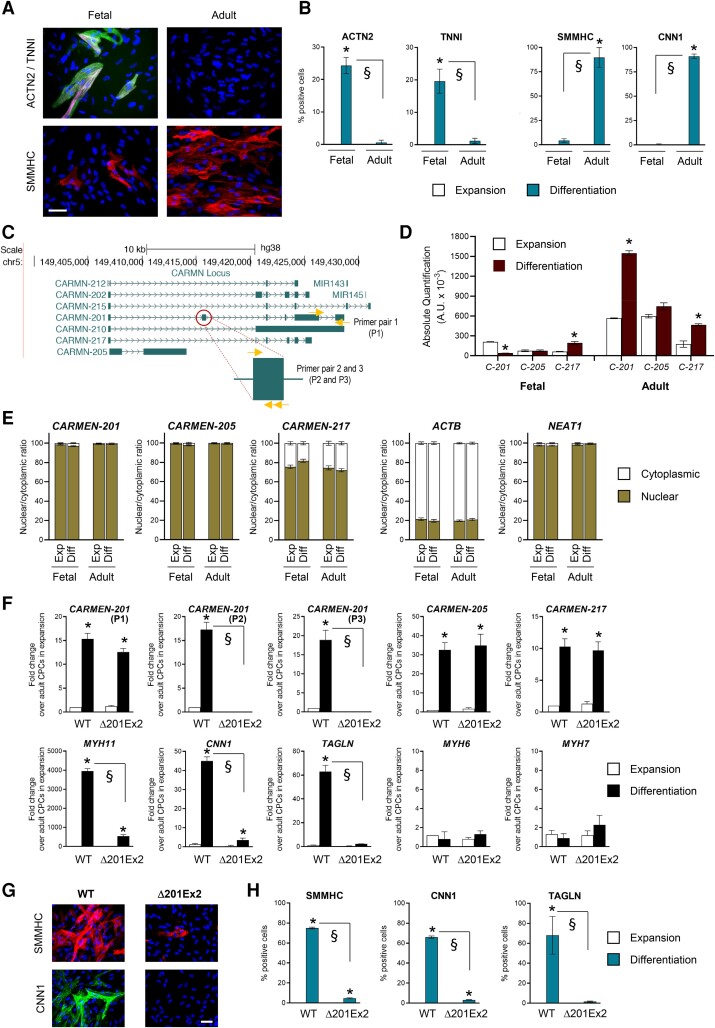
To study the importance of CARMEN isoforms in cardiac differentiation, we took advantage of primary CPCs isolated from the human heart. We isolated CPCs from the fetal heart at 5 weeks of gestation, hereafter referred to as fetal CPCs.5 Adult CPCs were isolated from atrial appendages of cardiac patients.6 Fetal CPCs have a high propensity to differentiate into CMs whereas adult CPCs preferentially produce SMCs. Indeed, 7 days after inducing differentiation, CMs, expressing ACTN2, ACTC1, MYH6, MYH7, and TNN1, were readily detected in fetal CPC cultures. In contrast, adult CPCs differentiated into SMCs expressing ACTA2, CALD1, CNN1, and MYH11 (Figure 1A and B; see Supplementary material online, Figure S1A). We next analyzed CARMEN expression under these two experimental conditions. Using capture long-read sequencing,25 we detected seven annotated isoforms, i.e. CARMEN (C)-201, -202, -205, -210, -212, -215, and -217 (GENCODE v33; Figure 1C). C-215 is the precursor of MIR-143 and MIR-145 whereas all other isoforms represent lncRNA transcripts whose expression terminates upstream of the miRNAs. All isoforms were significantly expressed in adult CPCs during SMC differentiation. In contrast, one isoform, C-201, was downregulated during the differentiation of fetal CPCs into CMs (see Supplementary material online, Figure S1B). Absolute quantification confirmed that C-201 represented the main isoform in adult CPCs but the least abundant in fetal CPCs (Figure 1D), suggesting its involvement in SMC differentiation. All CARMEN isoforms but the miRNA precursor (C-215) were more abundant in the nucleus than the cytoplasm, a feature compatible with the postulated function of lncRNAs as regulators of gene expression (Figure 1E; see Supplementary material online, Figure S1C).
Figure 1.
CARMEN-201 controls SMC specification via its second exon. (A and B) Representative images and quantification of ACTN2-positive TNNI-positive CMs and SMMHC-positive SMCs in differentiating fetal and adult CPC cultures. Scale bar: 50 µm. (C) Annotated CARMEN isoforms. The CARMEN-201second exon is highlighted. (D) Absolute quantification of CARMEN-201 (C-201), CARMEN-205 (C-205) and CARMEN-217 (C-217) in differentiating fetal and adult CPCs. (E) Nuclear and cytoplasmic levels of CARMEN-201, CARMEN-205, CARMEN-217, ACTB, and NEAT. (F) Expression of CARMEN-201, CARMEN-205, CARMEN-217, SMC markers (MYH11; CNN1; TAGLN), and CM markers (MYH6; MYH7) in adult WT and Δ201Ex2 CPC clones lacking CARMEN-201 Exon 2. (G and H) Representative images and quantification of SMMHC-positive CNN1-positive TAGLN-positive SMCs in cultures of differentiating adult WT or Δ201Ex2 CPC clones. Scale bar: 50 µm. Data represent mean values ± SEM; *P < 0.05 as compared with fetal CPCs in expansion; §P < 0.05 compared with the indicated conditions (n = 3–6). ANOVA with post hoc Tukey. See also see Supplementary material online, Figure S1–S3.
3.2. CARMEN-201 controls SMC specification via its second exon
We next evaluated the involvement of C-201 in SMC differentiation using a knockdown approach. Antisense oligonucleotides (GapmeRs) were designed to target the C-201 second exon, uniquely present in this isoform (Figure 1C). C-201 was downregulated in adult CPCs following GapmeR transfection (see Supplementary material online, Figure S2A). The anti-C-201 GapmeRs affected no other isoforms, demonstrating the specificity of the approach. C-201 silencing had no effects on the differentiation of fetal CPCs into CMs but completely blocked the capacity of adult CPCs to produce SMCs (see Supplementary material online, Figure. S2B and C). To explore the importance of the C-201second exon in SMC specification, we produce adult CPCs lacking the exon using CRISPR/Cas9 deletion. Guide (g)RNAs were designed to remove the second exon without affecting any other exons (see Supplementary material online, Figure S3A). C-201 Exon 2-deleted adult CPC clones were derived. Importantly, the C-201 isoform was still expressed in adult CPCs lacking C-201 Exon 2. However, the transcript was reduced by the size of the second exon (see Supplementary material online, Figure S3B). Endogenous C-201 expression was similarly induced in both wild-type and deleted adult CPC (Δ201Ex2) clones during differentiation as detected by using a primer pair amplifying the 3′ end of the transcript (Figure 1F; primer pair P1). The deletion of the second exon was verified using two primer pairs spanning the exon (i.e. P2; P3). Next, we evaluated the capacity of deleted adult CPCs to produce SMCs. Measurement of marker gene expression as well as immunostaining demonstrated that adult CPCs lacking C-201 Exon 2 lost their ability to differentiate into SMCs (Figure 1F–H).
3.3. CARMEN-201 induction is sufficient to trigger an SMC gene program in undifferentiated fetal CPCs
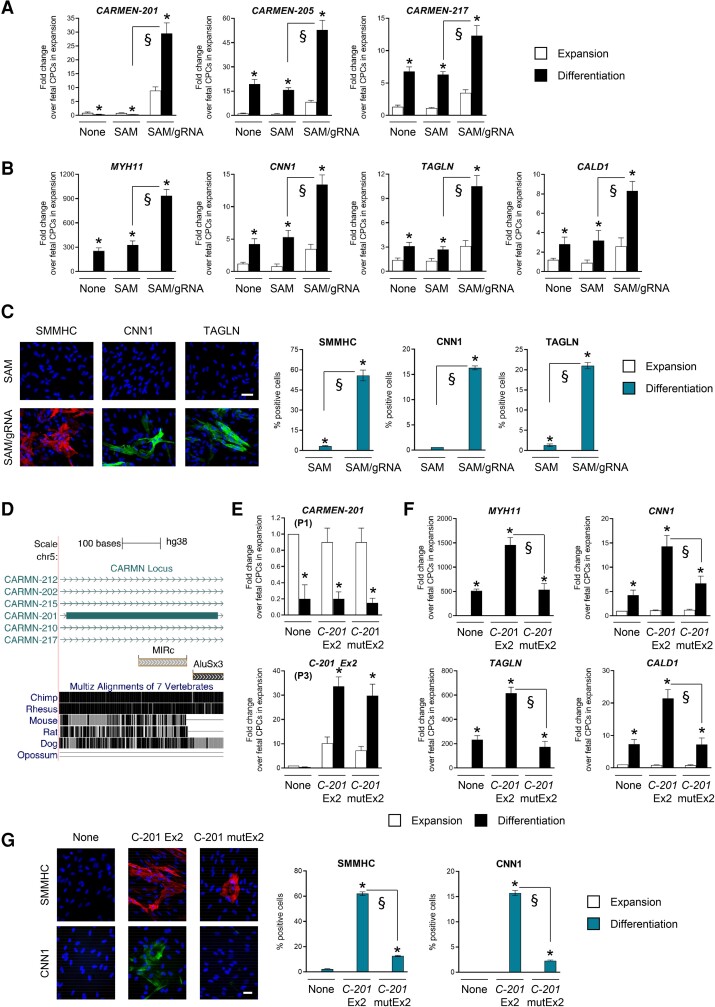
To evaluate the capacity of C-201 to redirect fetal CPCs into the SMC lineage, we used a CRISPR-On approach. We targeted transcriptional activators (dCas9-VP64; MS2-p65-HSF1), 200 bp upstream of the C-201 transcriptional start site (TSS) via expression of a modified gRNA containing MS2 aptamers (SAM system;23). Endogenous C-201 expression was downregulated in fetal CPCs in the absence of gRNA but markedly increased when the gRNA was expressed (Figure 2A). Compared to the large C-201 induction, the other CARMEN isoforms, as well as the two hosted miRNAs, were marginally activated (Figure 2A; see Supplementary material online, Figure S3C). We tested therefore the effects of C-201 manipulation on the fate of normally cardiogenic fetal CPCs. Strikingly, cells with forced C-201 transcription produced a large amount of SMCs, indicating that C-201 expression was sufficient to adopt an SMC fate (Figure 2B and C). Production of CMs was minimally affected, likely reflecting the differentiation of untransfected fetal CPCs (see Supplementary material online, Figure S3D and E). Globally, the expression of C-201 was found to be necessary and sufficient for inducing SMC specification in CPCs.
Figure 2.
The CARMEN-201 Exon 2 contains a functional transposable element implicated in SMC specification. (A) Expression of CARMEN isoforms and (B) SMC markers (MYH11; CNN1; TAGLN; CALD1) in differentiating fetal cells either untransfected (None), transfected with the SAM system in the absence of gRNA (SAM) or with the SAM system with a gRNA targeting sequences upstream the CARMEN TSS (SAM/gRNA). (C) Representative images and quantification of SMCs in cultures of differentiating fetal CPCs transfected as in (A). Scale bar: 50 µm. (D) Position of the MIRc transposable element in the CARMEN-201second exon, and sequence conservation. (E) Expression of CARMEN-201 using either a primer pair specific for the endogenous transcript (P1) or the exogenous Exon 2 (P3), and (F) SMC markers (MYH11; CNN1; TAGLN; CALD1) in differentiating fetal CPCs either not transduced (None), transduced with a lentiviral vector encoding CARMEN(C)-201 Ex2 or transduced with a lentiviral vector encoding a mutated C-201 Ex2 (C-201 mutEx2). (G) Representative images and quantification of SMMHC-positive CNN1-positive SMCs in cultures of differentiating fetal CPCs transfected as in (F). Scale bar: 50 µm. Data represent mean values ± SEM; *P < 0.05 as compared with fetal CPCs in expansion; §P < 0.05 compared with the indicated conditions (n = 3–6). ANOVA with post hoc Tukey. See also Supplementary material online, Figure S3 and S4.
3.4. The CARMEN-201second exon contains a functional transposable element that drives SMC commitment
The results above prompted us to evaluate the role of the C-201 second exon in SMC specification. We looked at its primary structure (397 nucleotides) and detected a SINE, which was identified as a 126 nucleotide-long Mammalian-wide Interspersed Repeat (MIR)c element. Of note, the exon is highly conserved in primates but not found in other species (Figure 2D). Intriguingly, MIRc is part of a catalog of predicted Repeat Insertion Domains of LncRNAs (RIDLs) promoting nuclear localization,18 a feature consistent with the pronounced nuclear enrichment of C-201 (Figure 1E). The exon also contains a partial ALU sequence.
To study the possible role of the MIRc element in determining SMC specification, we produced two lentiviral vectors for overexpressing the entire Exon 2 in fetal CPCs (see Supplementary material online, Figure S4A). The first version contained wild-type MIRc sequences (C-201 Ex2) whereas, in the second vector, the whole MIRc sequence was scrambled (C-201 mutEx2), thus maintaining length and sequence composition (see Supplementary material online, Figure S4A). Endogenous C-201 expression was downregulated in differentiating fetal CPCs as expected (Figure 2E; P1). However, significant wild-type and mutated C-201 Exon 2 expression was measured in the respective transduced groups as judged by using Exon 2-specific primers (P3). We next evaluated SMC and CM differentiation (Figure 2F and G; see Supplementary material online, Figure S4B–D). Non-transduced fetal CPCs differentiated into CMs. In sharp contrast, overexpression of wild-type C-201 Exon 2 forced fetal CPCs to adopt an SMC fate, as evidenced by the expression of SMC markers and immunostaining. Importantly, when the MIRc element was mutated (C-201 mutEx2), SMC differentiation was not observed, supporting a critical role for this transposable element in the capacity of C-201 to direct CPCs into the SMC lineage.
3.5. Identification of upstream regulators of CM and SMC specification in human CPCs
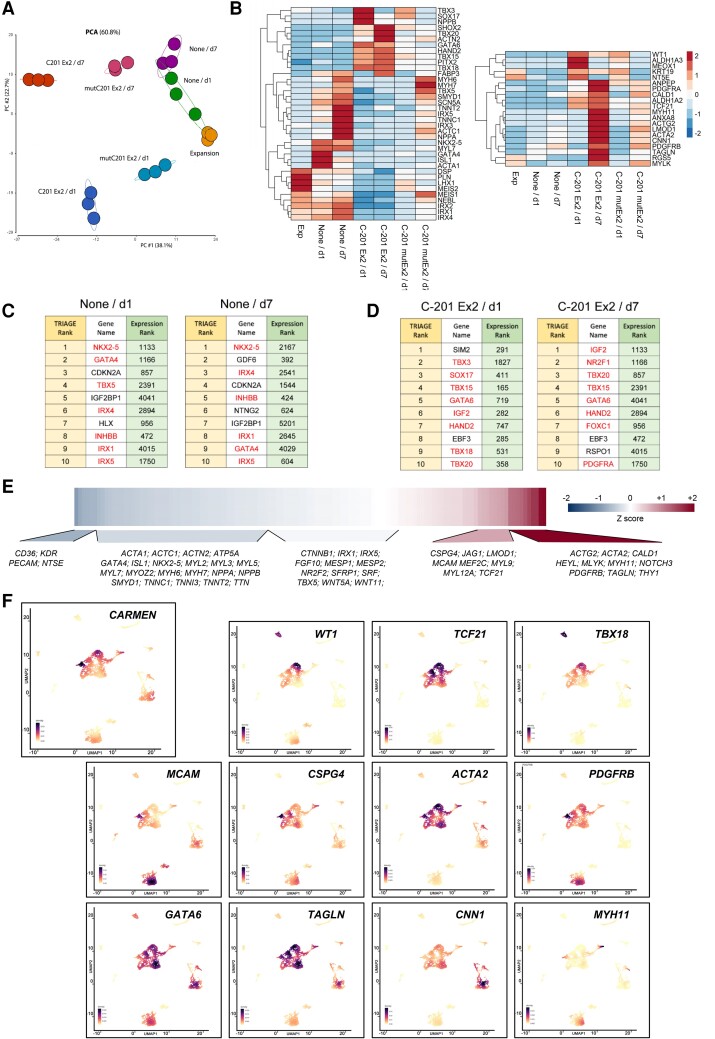
To better understand the processes leading to a switch in cell identity, we profiled the CPC transcriptomes under different experimental conditions. Principal component analysis (PCA) was conducted to evaluate differentiation when C-201 Exon 2 was expressed and not expressed (Figure 3A). Samples of differentiating fetal CPCs (None) revealed temporal changes (d0/Expansion; d1; d7) characterizing CM specification. In contrast, fetal CPCs overexpressing wild-type C-201 Exon 2 deviated significantly from the original differentiation track. Importantly, CPC samples with overexpression of the MIRc-mutated C-201 Exon 2 were transcriptionally similar to untransfected samples, indicating again that the transposable element was necessary for SMC commitment.
Figure 3.
Transcriptomic analysis and identification of upstream regulators of CM and SMC specification in fetal CPCs with or without C-201 Exon 2 overexpression. (A) Principal Component Analysis (PCA) visualizing transcriptomic data in a two-dimensional space. (B) Expression heatmap of regulators and markers of the CM and the SMC fate. The heatmaps show scaled TPM values. (C and D) TRIAGE transformation of input RNA Seq data predicts regulatory genes controlling cell differentiation based on TRIAGE rank order (left) compared with ranking observed using simple gene expression (right). Control differentiation (None) vs. differentiation following C-201 Exon 2 overexpression (C-201 Ex2) 1 (d1) or 7 days (d7) after transduction. Regulators implicated in cardiovascular differentiation are highlighted in red. (E) Human heart single-cell data analysis reveals genes positively and negatively correlated with CARMEN expression during development. (F) UMAP plots showing epicardium-, pericyte-, and SMC-specific expression of CARMEN in the human heart 6.5 weeks post-conception. See also Supplementary material online, Figure S4 and S5.
We next analyzed the transcriptomic data in detail (Figure 3B; see Supplementary material online, Figure S4E). Although fetal CPC differentiation was associated with early expression of cardiac TFs (e.g. ISL1; TBX5; GATA4; NKX2-5) and late expression of genes associated with CM excitation-contraction coupling (e.g. MYH6; MYH7; SCN5A; TNNT2), C-201 Exon 2-overexpressing cells induced genes characterizing epicardial (e.g. WT1; TCF21; TBX18), pericyte (e.g. ACTA2; PDGFRB; NTS5E) and SMC lineages (e.g. TAGLN; CNN1; MYH11; MYLK). The two distinct transcriptomes were enriched with relevant terms in a gene ontology analysis (see Supplementary material online, Figure S4F). Several factors characterizing the SHF and the outflow tract (OFT) were differentially expressed under the two experimental conditions (e.g. GATA6; HAND2; ISL1; MEIS1; MEIS2; PITX2), suggesting a developmental origin for the fetal and adult CPCs. Interestingly, undifferentiated fetal CPCs (Expansion) expressed factors from the Iroquois family of TFs (IRX1; IRX3; IRX4; IRX5), which are known to be activated in mesodermal tissues, in particular the dorsal mesoderm from which the heart derives.26
To identify regulators of specification during either CM or SMC differentiation, we took advantage of TRIAGE (Transcriptional Regulatory Inference Analysis of Gene Expression), a novel metric for inferring genes orchestrating cell identity.24 TRIAGE is based on the observation that broad H3K27me3 occupancy at promoters enriches for genes driving cell fates. TRIAGE calculates a repressive score for each gene, which can be combined with any type of genome-wide sequencing data to predict genes governing cell differentiation. We therefore identified upstream regulators of fetal CPC specification with and without C-201 Exon 2 overexpression, 1 and 7 days after induction of differentiation. The results of this inference analysis are presented in a way to compare rank orders based on input gene expression (right column) vs. TRIAGE ranking (left column) (Figure 3C and D). The top-ranked TRIAGE candidates revealed that key mesodermal and cardiac TFs were involved in the commitment of untransfected fetal CPCs into the CM fate whereas determinants of pericyte and SMC identity were activated following C-201 Exon 2 expression. The complete lists of TRIAGE regulators identified under the two different experimental conditions are presented in Supplementary material online, Table S1. Analysis of cell identity (ARCHS4;27) and associated biological pathways (GO BP) using all identified TRIAGE candidates validated their functional roles in CM and SMC commitment (see Supplementary material online, Figure S5A).
To further investigate the association of CARMEN expression with blood vessel development in vivo, we used data reporting the comprehensive transcriptional analysis of the embryonic human heart at the single-cell level at different stages of gestation.28 To understand how CARMEN expression was related to gene network changes, we performed a correlation analysis of CARMEN against all genes in all cells across all developmental time points (Figure 3E). CARMEN abundance was associated preferentially with pericyte and SMC gene programs, and not with the expression of CM or endothelial cell markers. We next reanalyzed data generated at 6.5 weeks post-conception,28 an important point in development corresponding to the formation of the cardiac vasculature. Newly generated UMAP density plots revealed lineage-specific markers, reflecting the cellular diversity of the embryonic human heart including epicardial cells (WT1; TCF21; TBX18), pericytes (MCAM; CSPG4; ACTA2; PDGFRB), and SMCs (GATA6; TAGLN; CNN1; MYH11) (Figure 3F). CARMEN was found expressed preferentially in these cells, substantiating an important role for this locus in pericyte and SMC specification from the epicardium during the development of the human heart. In contrast, CMs and endothelial cells, marked by TBX5; GATA4; NKX2-5; MYH6; and PECAM; KDR, did not expressed CARMEN at this developmental stage (see Supplementary material online, Figure S5B). Interestingly, ALDH1A2 and the COUP transcription factor NR2F2, critical for atrial identity, were also expressed in CARMEN-expressing cells.
3.6. The MIRc element is a binding module for the RE1 silencing transcription factor
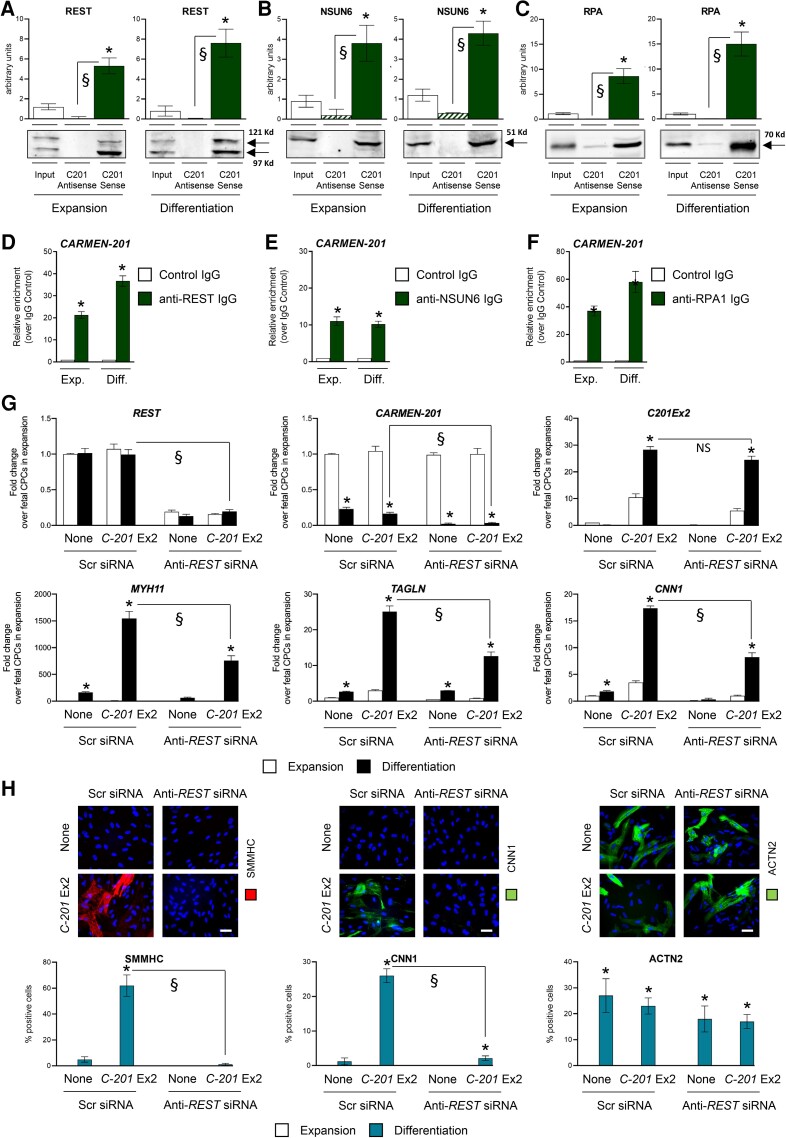
Many lncRNAs function by interacting with proteins. To identify C-201 protein partners, we performed a RNA pulldown assay. Biotinylated C-201 Exon 2 was used as a bait to purify C-201-associated proteins from adult CPC lysates. As control, we used a mutated C-201 Exon 2 lacking the MIRc element. Proteins were identified by mass spectrometry. Three proteins were detected as significantly associated to C-201 Exon 2, namely the transcriptional repressor RE1 Silencing Transcription Factor (REST; aka Neuron Restrictive Silencer Factor), the RNA methyltransferase NOP2/Sun RNA Methyltransferase 6 (NSUN6), and the Replication Protein A1 (RPA1), a protein implicated in stabilization of single-stranded DNA (see Supplementary material online, Figure S6A; see Supplementary material online, Table S2). We first confirmed the association of each protein with C-201 by pulling down the full C-201 transcript and quantifying the amount of bound proteins by Western blotting (Figure 4A–C; see full unedited gels in Supplementary Information). An antisense C-201 transcript was used as a control. The results demonstrated the specific interaction of C-201 with REST, NSUN6, and RPA1 in proliferating and differentiating adult CPCs. We next performed a RIP assay using antibodies directed against REST, NSUN6, and RPA1, respectively (Figure 4D–F). Quantitative measurement of bound CARMEN isoforms confirmed the interaction of C-201 with REST, NSUN6, and RPA1 during CPC expansion and differentiation. No other CARMEN isoforms were found associated with REST (see Supplementary material online, Figure S6B). In contrast, small amounts of C-205 were detected as bound to NSUN6, and C-217 appeared to interact with both NSUN6 and RPA1 (see Supplementary material online, Figure S6C–D). Finally, to determine whether REST possessed intrinsic propensity to bind MIRc-containing transcripts, we performed a REST RIP coupled to RNA profiling. REST-bound transcripts were found significantly enriched in sequences containing a MIRc element as compared with transcripts not bound by REST with similar length distribution and orientation. Nevertheless, the enrichment is also higher when exploring repeat-containing genes in general, suggesting that global REST binding to RNA molecules could require additional transposable elements (see Supplementary material online, Figure S6E; see Supplementary material online, Table S3).
Figure 4.
Identification of CARMEN-201 protein partners. (A) Quantification of REST, (B) NSUN6 and (C) RPA1 by Western blotting in a protein pulldown assay using biotinylated sense or antisense CARMEN-201 transcript in adult CPC lysates. Graphs show mean values ± SEM *P < 0.05 as compared with input; §P < 0.05 comparing sense and antisense probe (n = 3). (D–F) Quantification of CARMEN-201 enrichment after RNA immunoprecipitation using a control immunoglobulin G (IgG) or IgG directed against REST (anti-REST IgG), NSUN6 (anti-NSUN6 IgG) and RPA1 (anti-RPA1 IgG). Graphs show mean values ± SEM; *P < 0.05 as compared with control (n = 3). (G) Expression of REST, CARMEN-201 and SMC markers (MYH11; CNN1; TAGLN) in differentiating fetal CPCs either not transduced (None), transduced with a lentiviral vector encoding C-201 Exon 2 (C-201 Ex2), and treated with either a scrambled siRNA (Scr siRNA) or a siRNA directed against REST (Anti-REST siRNA). (H) Representative images and quantification of SMMHC-positive CNN1-positive SMCs and ACTN2-psoitive CMs in cultures of differentiating fetal CPCs transfected as in (G). Scale bars: 50 µm. Data represent mean values ± SEM; *P < 0.05 as compared with fetal CPCs in expansion; §P < 0.05 compared with indicated conditions (n = 3–6). ANOVA with post hoc Tukey. See also Supplementary material online, Figure S6.
To evaluate the role of REST in C-201-mediated CPC specification, we first used a knockdown approach. Fetal CPCs were transfected with control or REST siRNA, and induced to differentiate into SMC following C-201 Exon 2 overexpression (Figure 4G and H). Under control conditions, C-201 Exon 2 expression forced CPCs to adopt an SMC specification. In sharp contrast, REST knockdown abolished the capacity of C-201-expressing CPCs to commit to the SMC lineage. To confirm these results, we tested the effects of REST silencing in adult CPCs spontaneously differentiating into SMC (see Supplementary material online, Figure S6F). Again, in the absence of REST, adult CPCs were unable to produce an SMC progeny. Interestingly, C-201 appeared downregulated in differentiating REST-deficient CPCs. This was also confirmed using RNA BaseScope in situ hybridization in adult CPCs (see Supplementary material online, Figure S6G). The resetting of C-201 expression following REST knockdown mimicked therefore what observed in differentiating fetal CPCs (see Supplementary material online, Figure S1A and B). Moreover, the cellular distribution of the C-201 isoform was modified following REST silencing (see Supplementary material online, Figure S6H). Significant cytoplasmic enrichment was evident in the absence of REST, in contrast to what was measured under basal conditions. This observation suggested therefore that REST, which carries a nuclear localization signal, might contribute to retaining C-201 in the nucleus via its capacity to bind the MIRc element in the second exon.
3.7. CARMEN-201 inhibits the CM fate via REST-mediated repression of cardiogenic transcription factor expression
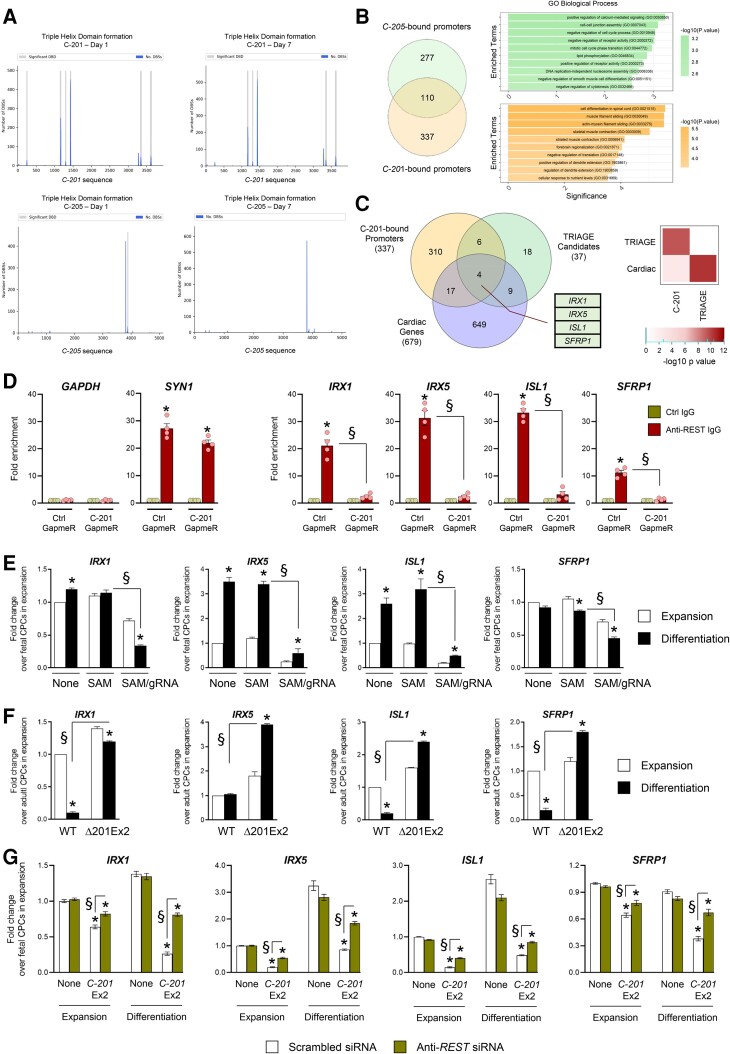
The association of C-201 with REST suggested a mechanism involving the targeting of the repressor to important regulatory loci to control cell fate in differentiating CPCs. In addition, RPA1, a C-201 protein partner, has been implicated in RNA:DNA triple helix stabilization,29 indicating that C-201 could interact with DNA sequences at target promoters. Thus, we took advantage of Triplex Domain Finder (TDF), an application developed to detect DNA-binding domains (DBDs) in lncRNAs.30 TDF identifies also the DNA regions bound by the selected lncRNAs, i.e. gene promoters containing binding sites for the lncRNA DBDs. Because REST had been associated with repression, we sought to identify C-201 target promoters within the list of downregulated genes following C-201 Exon 2 overexpression. Several DBDs were predicted in C-201, in particular in the sequences spanning the second exon (Figure 5A). As a control, we performed a similar analysis for C-205 and C-217. The C-205 transcript was found to contain distinct DBDs (Figure 5A) whereas C-217 was not predicted to contain significant DBDs (not shown). In total, 447 gene promoters were identified as potentially bound by C-201, and 387 by C-205, respectively. Among those, 337 were uniquely associated with C-201. These genes were related to GO Biological Processes defining striated muscle contraction (Figure 5B). In order to identify relevant targets of C-201/REST action, we crossed the list of C-201-bound genes as predicted by TDF with the list of TRIAGE candidates and the list of validated cardiac genes (Human Protein Atlas—ENSEMBL) (Figure 5C; see Supplementary material online, Table S4). Hypergeometric tests explored the significance of the overlaps and revealed four primary candidates: IRX1; IRX5; ISL1; and SFRP1. ISL1 is a member of the LIM homeodomain family of a transcription factor, crucial for the development of the SHF. The two Iroquois homeobox transcription factors IRX1 and IRX5 have been involved in developmental patterning in the embryonic heart. Finally, SFRP1 is a modulator of the WNT pathway that plays important roles in cardiac specification and differentiation. Importantly, the single-cell analysis demonstrated that these factors were not expressed in CARMEN-expressing cells in the embryonic human heart at 6.5 weeks of gestation (see Supplementary material online, Figure S5B). Importantly, all four genes contained REST binding sites as determined by chromatin immunoprecipitation followed by sequencing (ChIP-Seq) in a study interrogating REST binding in various cell types (see Supplementary material online, Figure S7A; Gene Expression Omnibus: GSM803369; GSM1010735; GSM1010804).31 We evaluated therefore REST occupancy at the promoters of IRX1, IRX5, ISL1, and SFRP1 in differentiating fetal and adult CPCs by ChIP-quantitative real-time (q)PCR. As expected, REST occupied the promoter of the different candidate genes solely in adult CPCs expressing C-201, and not in fetal CPCs (see Supplementary material online, Figure S7B). In these experiments, we used GAPDH as a negative control (REST occupancy in neither fetal nor adult CPCs) and SYN1 as a positive control (REST occupancy in both fetal and adult CPCs). Then, to formally demonstrate the dependence of REST targeting to IRX1; IRX5; ISL1; and SFRP1 on C-201 action, we performed an additional experiment in adult CPCs with or without C-201 silencing. Consistently, REST occupancy at candidate promoters in adult CPCs was blunted following GapmeR-mediated C-201 knockdown (Figure 5D).
Figure 5.
Identification of IRX1, IRX5, SFRP1 and ISL1 as target genes of C-201 action. (A) Significant DNA-binding domains (DBDs) identified in the mature sequence of C-201 and C-205 when analyzed against the differentially downregulated genes on Day 1 and Day 7 following induction of differentiation. Graph shows the number of DNA-binding sites (DBS) for each DBD. (b) Venn diagram illustrating the overlap of promoters predicted to form triple helices with C-201 and C-205. Functional enrichment analysis of the isoform-specific bound promoters. Graph shows the negative logarithm of the P value. (C) Venn diagram illustrating the identification of IRX1, IRX5, ISL1, and SFRP1 as common to the indicated lists of genes. Hypergeometric tests were performed to explore the significance of the overlap (D) REST occupancy at the promoters of IRX1, IRX5, SFRP1, and ISL1 in adult CPCs with or without GapmeR-mediated C-201 silencing as determined by ChIP-qPCR. Occupancy at the GAPDH and the SYN1 promoters was used as negative and positive controls, respectively. (E) Expression of IRX1, IRX5, ISL1, and SFRP1 in differentiating fetal cells either untransfected (None), transfected with the SAM system in the absence of gRNA (SAM) or with the SAM system with a gRNA targeting sequences upstream the CARMEN TSS (SAM/gRNA). (F) Expression of IRX1, IRX5, ISL1, and SFRP1 in differentiating adult CPCs treated with Scrambled siRNA (Scr SiRNA) or Anti-REST siRNA. (G) Expression of IRX1, IRX5, ISL1, and SFRP1 in differentiating fetal CPCs either untransfected, transfected with a lentiviral vector encoding C-201 Ex2 or a mutated C-201 Ex2 (C-201 mutEx2), treated with either a scrambled siRNA (Scr siRNA) or a siRNA directed against REST (Anti-REST siRNA). Data represent mean values ± SEM; *P < 0.05 as compared with CPCs in expansion; §P < 0.05 compared with indicated conditions (n = 3–6). ANOVA with post hoc Tukey. See also Supplementary material online, Figure S7.
To validate the relevance of ISL1, IRX1, IRX5, and SFRP1 in CPC specification, we first determined expression in fetal CPCs induced to differentiate into SMC following forced C-201 expression using CRISPR-On (Figure 5E). Each candidate was downregulated after induction of C-201 expression. We then measured expression in adult CPC clones lacking C-201 Exon 2 (Δ201Ex2), i.e. not able to activate an SMC gene program. ISL1, IRX1, IRX5, and SFRP1 expression was restored in these cells during differentiation as compared with what observed in wild-type cells (Figure 5F). Furthermore, we evaluated expression after manipulating REST. We observed the re-expression of the four factors in differentiating adult CPCs after REST knockdown (see Supplementary material online, Figure S7C). Moreover, REST silencing allowed also the re-expression of ISL1, IRX1, IRX5, and SFRP1 in fetal CPCs overexpressing C-201 Exon 2 (Figure 5G). Altogether, these findings supported that the four candidates were under control by C-201 via REST-mediated repression.
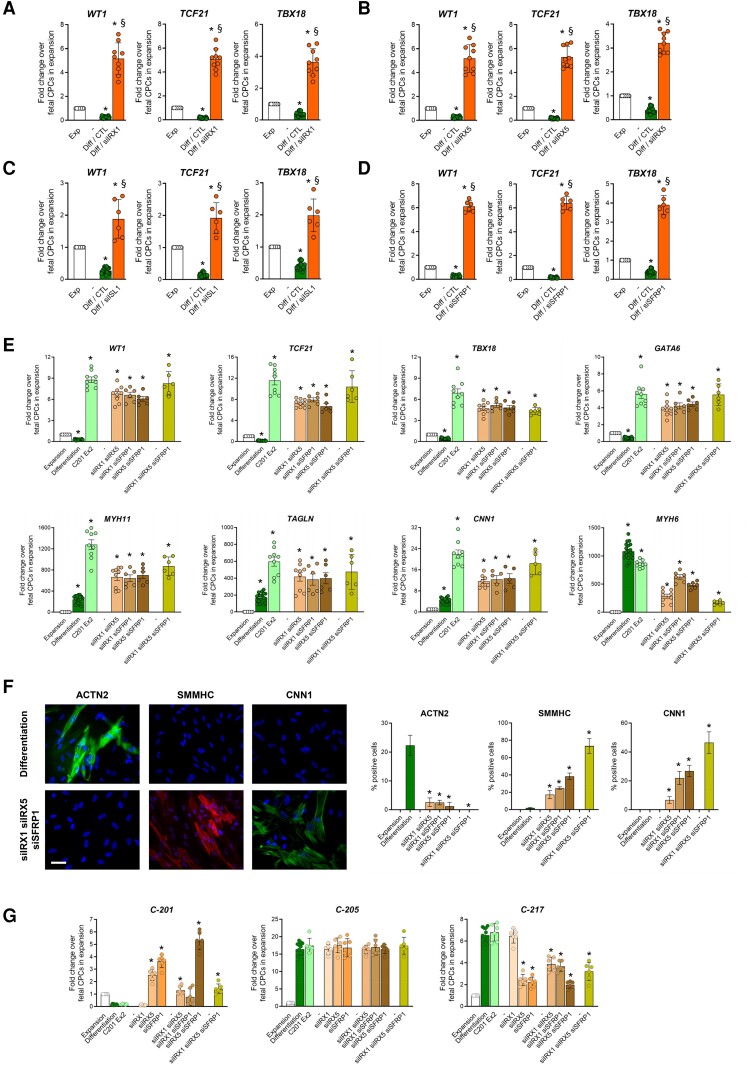
3.8. ISL1, IRX1, IRX5, and SFRP1 silencing promotes the SMC fate
We next proceeded to validate the importance of ISL1, IRX1, IRX5, and SFRP1 in controlling specification into the CM vs. the SMC lineage. To mimic REST-mediated repression, we used a siRNA approach to knockdown each candidate in fetal CPCs normally committing to the CM lineage. We first evaluated the effects of individual factor silencing on the capacity of fetal CPCs to produce functional progeny. Knocking down either IRX1, IRX5 or SFRP1 did not affect other candidate gene expression but ISL1 knockdown slightly decreased IRX1, IRX5, and SFRP1 levels (see Supplementary material online, Figure S8A–D), suggesting ISL1 lies upstream of these factors in the cardiac regulatory network, in accordance with its role as a pioneer transcription factor in the developing heart.32 We then investigated the re-specification of fetal CPCs into the SMC fate following siRNA-mediated silencing of each factor individually or in combination. In differentiating fetal CPCs, knocking down either IRX1, IRX5, ISL1, or SFRP1 restored epicardial gene expression, i.e. WT1, TCF21, and TBX18, confirming that downregulation of these critical cardiogenic factors was a mandatory step in reprograming CPCs into the smooth muscle fate (Figure 6A–D). In addition, individual factor silencing also resulted in the re-expression of GATA6, HAND2, PDGFRA, and TBX20 (see Supplementary material online, Figure S8E–H), which are known to mark bipotential cardiac precursors giving rise to CMs and SMCs.33 Nevertheless, manipulating each candidate separately has little impact on SMC gene expression (see Supplementary material online, Figure S8I–L). In fact, ISL1 knockdown had even a negative effect on late SMC maker expression. This suggested that ISL1 operated at the onset of CPC specification but was also necessary for late-stage differentiation.
Figure 6.
Validation of candidate cardiogenic factor downregulation. (A–D) Expression of epicardial genes (WT1, TCF21, TBX18) in differentiating fetal CPCs following IRX1, IRX5, ISL1, and SFRP1 silencing. (E) Epicardial (WT1, TCF21, TBX18) SMC (MYH11, TAGLN, CNN1) and CM marker (MYH6) expression in differentiating fetal CPCs overexpressing C-201 Ex2, and in fetal CPCs treated with siRNAs directed against the indicated factors in combination. (F) Representative images and quantification of ACTN2-positive CMs and SMMHC-positive CNN1-positive SMCs in cultures of differentiating fetal CPCs transfected as in (E). Scale bars: 50 µm. Expression of CARMEN-201 (C-201), CARMEN-205 (C-205) and CARMEN-217 (C-217) in differentiating fetal CPCs treated as in (E). Data represent mean values ± SEM; *P < 0.05 as compared with fetal CPCs in expansion; §P < 0.05 as compared with fetal CPCs in differentiation (n = 6–12). ANOVA with post hoc Tukey. See also see Supplementary material online, Figure S8.
We tested therefore combinations of siRNAs targeting IRX1, IRX5, and SFRP1 (Figure 6E). As a positive control for the activation of the SMC gene program, C-201 Exon 2-overexpressing fetal CPCs were included in the experiment. Each combination was associated with a large induction of epicardial and SMC gene expression as compared with individual knockdown, with maximal impact achieved when all three factors were downregulated simultaneously. This manipulation was as potent as C-201 Exon 2 overexpression in inducing SMC genes in normally cardiogenic CPCs. Accordingly, massive SMC differentiation was observed by immunostaining following IRX1, IRX5 and SFRP1 knockdown (Figure 6F). Interestingly, commitment occurred at the expense of the cardiogenic lineage (Figure 6E and F) but had no impact on endothelial cell production (not shown). Our data indicated therefore that IRX1, IRX5, and SFRP1 downregulation was sufficient to redirect fetal CPCs into the epicardial and the SMC lineages. Nevertheless, as mentioned above, ISL1 expression appeared necessary during SMC differentiation. To formally demonstrate this point, we performed an additional experiment in which the four factors were silenced together (see Supplementary material online, Figure S8M). In this case, ISL1 silencing produced a slight negative effect on SMC marker expression induced by combined IRX1, IRX5 and SFRP1 knockdown, sustaining a role for ISL1 in the late stage of SMC differentiation.
Interestingly, manipulating IRX1, IRX5, or SFRP1 had a striking effect on CARMEN isoform expression (Figure 6G). Indeed, while endogenous C-201 was downregulated during specification in differentiating fetal CPCs, its expression was reactivated after IRX5 and SFRP1 knockdown, and even more so when IRX5 and SFRP1 were silenced together, suggesting C-201 was negatively regulated by the two cardiogenic factors. Remarkably, C-217 expression demonstrated a mirror image, consistent with coordinated regulation of the two isoforms and suggesting a switch might operate during SMC specification. C-205 was not modulated under these different conditions.
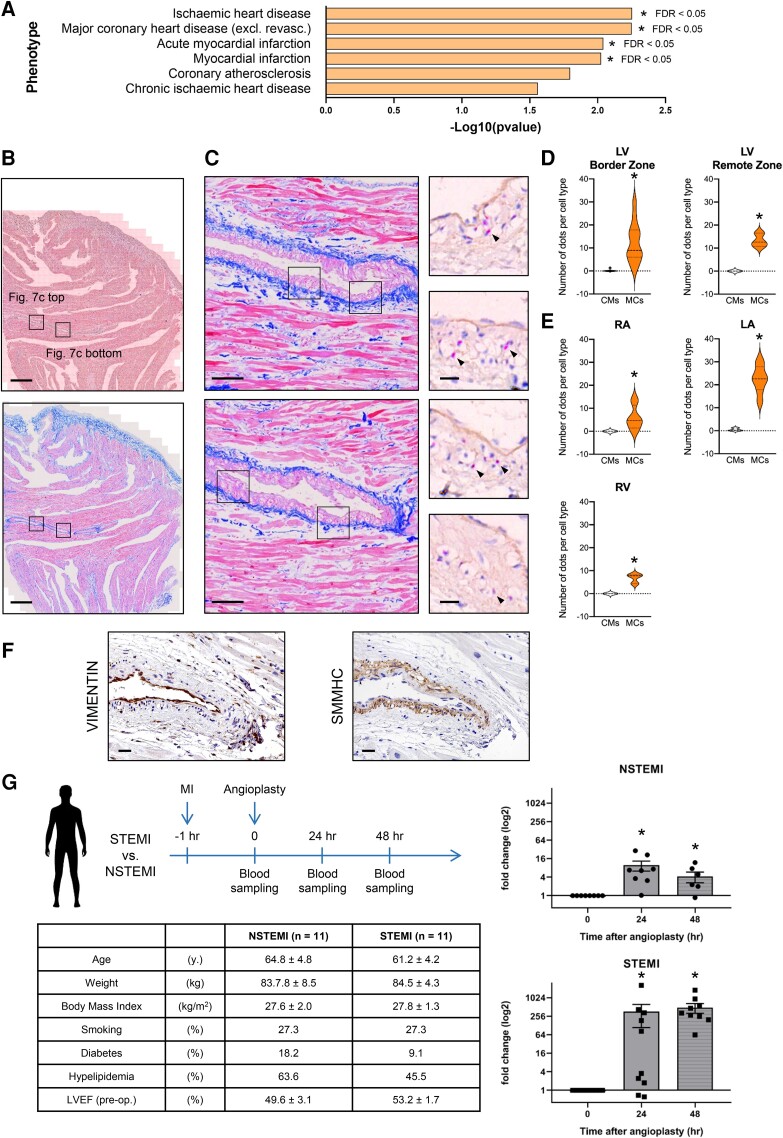
3.9. CARMEN-201 expression is increased in response to myocardial infarction in humans
In an attempt to evaluate the relevance of our findings in disease, we queried the association of CARMEN with cardiovascular traits using CTG-VIEW (https://view.genoma.io). We identified important phenotypes related to cardiovascular conditions as strongly associated with CARMEN (Figure 7A). This prompted us to investigate whether C-201 was differentially expressed in the damaged myocardium. We first used RNA BaseScope in situ hybridization to localize C-201 expression in the human heart. Samples were collected from explanted hearts of transplant patients, and expression was compared in CMs vs. mural cells (Figure 7B–E). C-201 was found uniquely expressed in mural cells of large coronary vessels. Immunostaining for VIMENTIN (marking endothelial cells and fibroblasts) and smooth muscle myosin heavy chain (SMMHC; marking SMCs) supported C-201 expression being associated primarily with SMCs (Figure 7F; see Supplementary material online, Figure S8N). C-201 expression seemed equally distributed in the ventricular and atrial vasculature. Next, we measured C-201 expression in the blood of patients experiencing acute coronary syndrome, with no prior history of cardiac disease. Plasma samples were obtained during angioplasty that took place less than 1 h after myocardial infarction, and at 24 and 48 h thereafter (Figure 7G). Individuals were classified based on the presence or absence of ST elevation, namely STEMI and non-STEMI (NSTEMI). C-201 was not expressed immediately after infarction, suggesting that the transcript was not induced under basal conditions. However, the amounts of transcripts dramatically increased after 1 and 2 days. Importantly, circulating C-201 concentrations were more elevated in STEMI vs. NSTEMI patients. Altogether, it suggested that C-201 was expressed in large vessels of the heart and responded acutely to hemodynamic stress with an expression being proportional to the severity of the disease.
Figure 7.
CARMEN-201 expression is increased in response to myocardial infarction in humans. (A) Association of CARMEN with cardiovascular traits using CTG-VIEW. (B–E) Detection of C-201 in the failing human heart by RNA in situ hybridization assay (BaseScope). (B and C) Representative images of sections of an explanted heart obtained from a heart failure patient. (B). Top: Hematoxylin/Eosin; scale bar: 500 μm; Bottom: Masson Trichrome staining; scale bar: 500 μm. (C) Left: Masson Trichrome staining; scale bar: 100 µm; Right: Hematoxylin/Eosin; scale bar: 20 µm. Red dots: Positive C-201 BaseScope signals. (D and E) Quantification of C-201 expression in CMs and mural cells. Data represent mean values ± SEM; *P < 0.05; ANOVA with post hoc Tukey. Two patients; 5 sections per patient; 5 to 10 different areas per section. LV: Left ventricle; RV: Right ventricle; LA: Left atria; RA: Right atria. (F) Immunostaining detection of VIMENTIN-positive cells (endothelial cells and fibroblasts) and SMMHC-positive cells (SMCs) in adjacent sections of that used in b and c; scale bar: 25 µm. (G) Time course of blood sample collection, and expression of C-201 in plasma of STEMI and NSTEMI patients (Data represent mean values ± SEM; n = 11; *P < 0.01; ANOVA with post hoc Tukey), and Table presenting patient characteristics. See also Supplementary material online, Figure S8.
4. Discussion
In this study, we characterized for the first time the role of lncRNA isoforms in cell fate determination through a systematic examination of the human CARMEN locus. In primary human CPCs committing to the SMC lineage, the C-201 isoform associates with REST via its MIRc element targets the repressor to important cardiogenic loci, namely IRX1, IRX5, SFRP1 and ISL1, represses their expression and promotes SMC specification (see Graphical Abstract). Conversely, in CPCs adopting a CM fate, C-201 is not expressed, allowing IRX1, IRX5, ISL1 and SFRP1 expression and the subsequent activation of the cardiogenic program. Importantly, the MIRc-containing exon in C-201 is found in primates but not in other mammals, suggesting that MIRc-mediated functions controlling commitment into the SMC lineage has been integrated in the CARMEN locus only recently in evolution. The CARMEN locus has been involved in cardiogenesis, implicating however other isoforms than C-201.21 Our data suggest that coordinate regulation of C-201 and C-217 expression takes place during specification, providing a plausible mechanism for controlling specification. CARMEN isoforms appear to share a single promoter. Yet, additional transcription start sites have been recently detected in the C-201 isoform, suggesting transcriptional regulation might control C-201 expression in differentiating SMCs.34
Multipotent cardiovascular precursor cells expressing ISL1 give rise to both CMs and SMCs.2–4 In this context, our TRIAGE analysis identifies key cardiac TFs as regulators of fetal CPC differentiation. On the other hand, CPCs respecified into the SMC lineage after C-201 Exon 2 overexpression express a different gene program. Induction of GATA6, HAND2, PDGFRA and TBX20 in CPCs is a characteristic feature of cardiovascular intermediates capable of producing CMs and SMCs.33 We show also that commitment to the SMC lineage is characterized by the stepwise expression of markers of epicardium-derived cells (EPDCs) such as WT1, MEOX1, KRT19 and TBX18, and pericytes such as MCAM, CSPG4, ACTA2 and PDGFRB. During development, EPDCs establish the subepicardial mesenchyme, then migrate into the myocardium. These cells represent a known source of pericytes and SMCs for the forming coronary vasculature.35 In addition, genetic tracing experiments suggest that epicardial cells can also give rise to a myocardial progeny.36–38 Along the same line, a recent single-cell analysis identifies the juxta-cardiac field (JCF) contributing to both EPDCs and CMs.39 Trajectory analysis revealed a link between precursors from the JCF and the posterior SHF, supporting the postulated developmental origin of CARMEN-expressing CPCs. Of note, TRIAGE identifies IRX1 and IRX5 as important regulators of cardiogenesis. IRX1 is detected in the trabeculated and compact myocardium of the developing ventricular septum whereas IRX5 demonstrates a subendocardial to subepicardial gradient of expression.40 Consistently, our experiments show that knocking down IRX1 and IRX5 in fetal CPCs allows re-expression of epicardial and SMC markers, suggesting expression of the two factors is sufficient to maintain a cardiogenic identity in committed precursors. C-201 targets also SFRP1, a known WNT antagonist. Downregulation of the WNT pathway is an important step in establishing cardiac fates.41
In specifying CPCs, C-201 acts via REST-mediated repression. A role for C-201 in blocking REST activity during SMC determination, for instance via sequestering REST, is unlikely since REST silencing abolishes SMC commitment in C-201-expressing CPCs. Consistently, REST acts as a transcriptional repressor in the developing heart, where it is thought to repress adult cardiac gene expression.42–44 Accordingly, blockade of REST in the heart leads to cardiac dysfunction.45 In addition, our results suggest that temporal REST expression during the development of the heart also reflects the role of REST in cell fate determination. REST binds C-201 but not C-205 and C-217, further substantiating the importance of the C-201/REST complex for SMC differentiation. C-201 appears to be also indirectly under control by REST. C-201 relocalizes into the cytoplasm upon REST silencing. Therefore, the nuclear enrichment of C-201 could depend in part on its binding to REST. In this vein, REST binds C-201 via the MIRc repeat in the second exon, which was recently demonstrated to be associated with transcript nuclear localization.18
Then, C-201 associates with NSUN6 and RPA1. A recent study demonstrated a role for NSUN6 in methylating mRNAs and lncRNAs, such as MALAT1, NEAT1 AND XIST.46 Mechanistically, lncRNA m5C modification could be involved in transcript structure and stability.47C-201 associates also to RPA1. Importantly, RPA1 binds RNA with high affinity and promotes R-loop formation with homologous DNA.29 RNA-DNA hybrids initiate cellular processes regulating transcription and genome dynamics, two important determinants of cell specification.48,49 Thus, RPA1 might contribute to effective targeting of REST at regulatory loci via its capacity to stabilize C-201/promoter association, a feature consistent with the predicted DBDs in C-201.
CARMEN is expressed in adult tissues, particularly in the heart and the vasculature, reflecting expression in CMs and SMCs.50 An increasing body of evidence suggests CARMEN is associated with pathological states in the cardiovascular system.22 Relevant to the present work, CARMEN was recently demonstrated to regulate SMC differentiation and proliferation in atherosclerotic plaques.34 Unstable regions, in which high proliferation of dedifferentiated SMCs is observed, were characterized with decreased CARMEN levels. Consistently, SMCs adopting a synthetic phenotype characterized Carmen knockout mice. We have demonstrated previously that CARMEN is induced in the stressed mouse and human hearts.21 Interestingly, human CARMEN isoforms were found differentially expressed depending on the cardiac pathology, exemplifying again the complexity of the regulation of the CARMEN locus. Here, we show that C-201 levels increase in the blood during the acute phase of myocardial infarction. The likely source of circulating C-201 is the damaged heart. However, we cannot rule out the possibility that hemodynamic stress also stimulates release from the peripheral vasculature. Nevertheless, assuming a cardiac origin for C-201, its expression in CPCs could be part of the healing process initiated following injury. In this scenario, CPCs expressing C-201 would be specified preferentially into the SMC lineage. Increased myocardial tissue perfusion has been reported in cell-based regenerative therapies for heart disease. However, clinical trials failed to demonstrate functional improvement. This can be expected if precursors are diverted from the cardiogenic lineage secondary to C-201 expression. Our data propose therefore a mean to improve CM production via modulating C-201 expression in cardiac precursors. Finally, C-201 could represent an interesting biomarker for assessing the extent of cardiovascular damage in various pathological situations.
Altogether, this work demonstrates how a biological switch is encoded in lncRNA sequence to regulate cardiovascular specification. We have linked two key phenomena, namely alternative splicing and the presence of deeply-conserved transposable elements. lncRNAs display far greater levels of alternative splicing, although it has not been clear whether this reflects relaxed constraint or regulated production of isoforms with distinct functions.25 Here, we have shown an example where these two processes converge to produce functional transcript isoforms, and provided the first physiological role for a transposable element acting via a lncRNA during heart development.
Supplementary Material
Acknowledgements
We express our gratitude to Dr Gabriel Cuellar Partida, University of Queensland, Brisbane, Australia, for his help in analyzing the GWAS data. We are grateful to Dr Nicole Deglon and Dr Maria Del Rey, University of Lausanne Medical School, Switzerland, for the plasmids used to prepare lentiviral vectors. We thank the Genomic Technologies Facility, the Protein Analysis Facility, the Cellular Imaging Facility and the Mouse Pathology Facility at the University of Lausanne, Switzerland, for providing expertise in transcriptomics, proteomics, imaging and histology, respectively.
Contributor Information
Isabelle Plaisance, Experimental Cardiology Unit, Division of Cardiology, University of Lausanne Medical School, Lausanne, Switzerland.
Panagiotis Chouvardas, Department of Medical Oncology, Inselspital, University of Bern, Bern, Switzerland.
Yuliangzi Sun, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia.
Mohamed Nemir, Experimental Cardiology Unit, Division of Cardiology, University of Lausanne Medical School, Lausanne, Switzerland.
Parisa Aghagolzadeh, Experimental Cardiology Unit, Division of Cardiology, University of Lausanne Medical School, Lausanne, Switzerland.
Farhang Aminfar, Experimental Cardiology Unit, Division of Cardiology, University of Lausanne Medical School, Lausanne, Switzerland.
Sophie Shen, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia.
Woo Jun Shim, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia.
Francesca Rochais, Aix Marseille University, Marseille Medical Genetics, INSERM, U1251, Marseille, France.
Rory Johnson, Department of Medical Oncology, Inselspital, University of Bern, Bern, Switzerland; School of Biology and Environmental Science, University College Dublin, Dublin, Ireland.
Nathan Palpant, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia.
Thierry Pedrazzini, Experimental Cardiology Unit, Division of Cardiology, University of Lausanne Medical School, Lausanne, Switzerland.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors contribution
T.P. conceived the project, designed experiments, and wrote the paper. I.S. designed and performed all wet-lab experiments, with help from M.N., P.A. and F.A. P.C. and R.J. conducted the bioinformatic analyses. Y.S., S.S., W.J.S., and N.P. performed the TRIAGE, GWAS, and single-cell analyses. F.R. provided critical human material for RNA FISH experiment, and her expertise in heart development.
Funding
This work was supported by grants from the Swiss National Science Foundation (T.P.; Grant Nos CRSII5-1_173738 and 31003A_182322).
Data availability
All transcriptomic data has been deposited to GEO with the identifier GSE199930.
Translational perspective
LncRNAs regulate cell commitment and differentiation during development. Taking advantage of cardiac precursor cells isolated from the human fetal and adult heart, we identify a novel mechanism mediated by a specific isoform of the lncRNA CARMEN, which controls specification into the smooth muscle cell vs. the cardiomyocyte fate. These results have direct implications for cell therapy for heart disease. Moreover, CARMEN is associated with pathological states and represents an interesting biomarker for assessing the extent of damage in the cardiovascular system. Our data propose therefore a means to control cardiac cell identity and behavior during heart development and disease.
References
- 1. Meilhac SM, Buckingham ME. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol 2018;15:705–724. [DOI] [PubMed] [Google Scholar]
- 2. Kattman SJ, Huber TL, Keller GM. Multipotent flk-1 + cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 2006;11:723–732. [DOI] [PubMed] [Google Scholar]
- 3. Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1 + progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 2006;127:1151–1165. [DOI] [PubMed] [Google Scholar]
- 4. Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 2006;127:1137–1150. [DOI] [PubMed] [Google Scholar]
- 5. Gonzales C, Ullrich ND, Gerber S, Berthonneche C, Niggli E, Pedrazzini T. Isolation of cardiovascular precursor cells from the human fetal heart. Tissue Eng Part A 2012;18:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Plaisance I, Perruchoud S, Fernandez-Tenorio M, Gonzales C, Ounzain S, Ruchat P, Nemir M, Niggli E, Pedrazzini T. Cardiomyocyte lineage specification in adult human cardiac precursor cells via modulation of enhancer-associated long noncoding RNA expression. JACC BST 2016;1:472–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet 2018;19:535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 2020;21:102–117. [DOI] [PubMed] [Google Scholar]
- 9. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47–62. [DOI] [PubMed] [Google Scholar]
- 10. Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013;152:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 2013;24:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alexanian M, Maric D, Jenkinson SP, Mina M, Friedman CE, Ting CC, Micheletti R, Plaisance I, Nemir M, Maison D, Kernen J, Pezzuto I, Villeneuve D, Burdet F, Ibberson M, Leib SL, Palpant NJ, Hernandez N, Ounzain S, Pedrazzini T. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat Commun 2017;8:1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HS, Quertermous T, Chang CP. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014;514:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, Maric D, Maison D, Nemir M, Young RA, Schroen B, Gonzalez A, Ounzain S, Pedrazzini T. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med 2017;9:eaai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bell RD, Long X, Lin M, Bergmann JH, Nanda V, Cowan SL, Zhou Q, Han Y, Spector DL, Zheng D, Miano JM. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol 2014;34:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ballantyne MD, Pinel K, Dakin R, Vesey AT, Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, Joshi N, Dweck MR, Miano JM, McBride MW, Newby DE, McDonald RA, Baker AH. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation 2016;133:2050–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kirk JM, Kim SO, Inoue K, Smola MJ, Lee DM, Schertzer MD, Wooten JS, Baker AR, Sprague D, Collins DW, Horning CR, Wang S, Chen Q, Weeks KM, Mucha PJ, Calabrese JM. Functional classification of long non-coding RNAs by k-mer content. Nat Genet 2018;50:1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carlevaro-Fita J, Polidori T, Das M, Navarro C, Zoller TI, Johnson R. Ancient exapted transposable elements promote nuclear enrichment of human long noncoding RNAs. Genome Res 2019;29:208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 2013;9:e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ziegler C, Kretz M. The more the merrier-complexity in long non-coding RNA loci. Front Endocrinol (Lausanne) 2017;8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, Bussotti G, Pezzuto I, Notredame C, Heymans S, Guigo R, Johnson R, Pedrazzini T. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol 2015;89:98–112. [DOI] [PubMed] [Google Scholar]
- 22. Vacante F, Denby L, Sluimer JC, Baker AH. The function of miR-143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vascul Pharmacol 2019;112:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015;517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shim WJ, Sinniah E, Xu J, Vitrinel B, Alexanian M, Andreoletti G, Shen S, Sun Y, Balderson B, Boix C, Peng G, Jing N, Wang Y, Kellis M, Tam PPL, Smith A, Piper M, Christiaen L, Nguyen Q, Boden M, Palpant NJ. Conserved epigenetic regulatory logic infers genes governing cell identity. Cell Syst 2020;11:625–639.e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lagarde J, Johnson R. Capturing a long look at our genetic library. Cell Syst 2018;6:153–155. [DOI] [PubMed] [Google Scholar]
- 26. Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF. Patterning the embryonic heart: identification of five mouse iroquois homeobox genes in the developing heart. Dev Biol 2000;224:263–274. [DOI] [PubMed] [Google Scholar]
- 27. Lachmann A, Torre D, Keenan AB, Jagodnik KM, Lee HJ, Wang L, Silverstein MC, Ma'ayan A. Massive mining of publicly available RNA-seq data from human and mouse. Nat Commun 2018;9:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Asp M, Giacomello S, Larsson L, Wu C, Furth D, Qian X, Wardell E, Custodio J, Reimegard J, Salmen F, Osterholm C, Stahl PL, Sundstrom E, Akesson E, Bergmann O, Bienko M, Mansson-Broberg A, Nilsson M, Sylven C, Lundeberg J. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 2019;179:1647–1660.e1619. [DOI] [PubMed] [Google Scholar]
- 29. Mazina OM, Somarowthu S, Kadyrova LY, Baranovskiy AG, Tahirov TH, Kadyrov FA, Mazin AV. Replication protein A binds RNA and promotes R-loop formation. J Biol Chem 2020;295:14203–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuo CC, Hanzelmann S, Senturk Cetin N, Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt I, Kurian L, Costa IG. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res 2019;47:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rockowitz S, Lien WH, Pedrosa E, Wei G, Lin M, Zhao K, Lachman HM, Fuchs E, Zheng D. Comparison of REST cistromes across human cell types reveals common and context-specific functions. PLoS Comput Biol 2014;10:e1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao R, Liang X, Cheedipudi S, Cordero J, Jiang X, Zhang Q, Caputo L, Gunther S, Kuenne C, Ren Y, Bhattacharya S, Yuan X, Barreto G, Chen Y, Braun T, Evans SM, Sun Y, Dobreva G. Pioneering function of Isl1 in the epigenetic control of cardiomyocyte cell fate. Cell Res 2019;29:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noseda M, Harada M, McSweeney S, Leja T, Belian E, Stuckey DJ, Abreu Paiva MS, Habib J, Macaulay I, de Smith AJ, al-Beidh F, Sampson R, Lumbers RT, Rao P, Harding SE, Blakemore AI, Jacobsen SE, Barahona M, Schneider MD. PDGFRalpha demarcates the cardiogenic clonogenic Sca1 + stem/progenitor cell in adult murine myocardium. Nat Commun 2015;6:6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vacante F, Rodor J, Lalwani MK, Mahmoud AD, Bennett M, De Pace AL, Miller E, Van Kuijk K, de Bruijn J, Gijbels M, Williams TC, Clark MB, Scanlon JP, Doran AC, Montgomery R, Newby DE, Giacca M, O'Carroll D, Hadoke PWF, Denby L, Sluimer JC, Baker AH. CARMN Loss regulates smooth muscle cells and accelerates atherosclerosis in mice. Circ Res 2021;128:1258–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quijada P, Trembley MA, Small EM. The role of the epicardium during heart development and repair. Circ Res 2020;126:377–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008;454:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008;454:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A, Auer H, Achouri Y, Dubois C, Bondue A, Simons BD, Blanpain C. Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat Cell Biol 2014;16:829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tyser RCV, Ibarra-Soria X, McDole K, Arcot Jayaram S, Godwin J, van den Brand TAH, Miranda AMA, Scialdone A, Keller PJ, Marioni JC, Srinivas S. Characterization of a common progenitor pool of the epicardium and myocardium. Science 2021;371:eabb2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim KH, Rosen A, Bruneau BG, Hui CC, Backx PH. Iroquois homeodomain transcription factors in heart development and function. Circ Res 2012;110:1513–1524. [DOI] [PubMed] [Google Scholar]
- 41. Protze SI, Lee JH, Keller GM. Human pluripotent stem cell-derived cardiovascular cells: from developmental biology to therapeutic applications. Cell Stem Cell 2019;25:311–327. [DOI] [PubMed] [Google Scholar]
- 42. Kuwahara K, Saito Y, Takano M, Arai Y, Yasuno S, Nakagawa Y, Takahashi N, Adachi Y, Takemura G, Horie M, Miyamoto Y, Morisaki T, Kuratomi S, Noma A, Fujiwara H, Yoshimasa Y, Kinoshita H, Kawakami R, Kishimoto I, Nakanishi M, Usami S, Saito Y, Harada M, Nakao K. NRSF Regulates the fetal cardiac gene program and maintains Normal cardiac structure and function. EMBO J 2003;22:6310–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang D, Wu B, Wang P, Wang Y, Lu P, Nechiporuk T, Floss T, Greally JM, Zheng D, Zhou B. Non-CpG methylation by DNMT3B facilitates REST binding and gene silencing in developing mouse hearts. Nucleic Acids Res 2017;45:3102–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bingham AJ, Ooi L, Kozera L, White E, Wood IC. The repressor element 1-silencing transcription factor regulates heart-specific gene expression using multiple chromatin-modifying complexes. Mol Cell Biol 2007;27:4082–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuwahara K. Role of NRSF/REST in the regulation of cardiac gene expression and function. Circ J 2013;77:2682–2686. [DOI] [PubMed] [Google Scholar]
- 46. Selmi T, Hussain S, Dietmann S, Heiss M, Borland K, Flad S, Carter JM, Dennison R, Huang YL, Kellner S, Bornelov S, Frye M. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res 2021;49:1006–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol 2019;26:380–388. [DOI] [PubMed] [Google Scholar]
- 48. Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell 2012;46:115–124. [DOI] [PubMed] [Google Scholar]
- 49. Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet 2015;16:583–597. [DOI] [PubMed] [Google Scholar]
- 50. Papatheodorou I, Moreno P, Manning J, Fuentes AM, George N, Fexova S, Fonseca NA, Fullgrabe A, Green M, Huang N, Huerta L, Iqbal H, Jianu M, Mohammed S, Zhao L, Jarnuczak AF, Jupp S, Marioni J, Meyer K, Petryszak R, Prada Medina CA, Talavera-Lopez C, Teichmann S, Vizcaino JA, Brazma A. Expression atlas update: from tissues to single cells. Nucleic Acids Res 2020;48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All transcriptomic data has been deposited to GEO with the identifier GSE199930.