Abstract
Cleavage and polyadenylation of pre-mRNAs is a necessary step for gene expression and function. Majority of human genes exhibit multiple polyadenylation sites, which can be alternatively used to generate different mRNA isoforms from a single gene. Alternative polyadenylation (APA) of pre-mRNAs is important for the proteome and transcriptome landscape. APA is tightly regulated during development and contributes to tissue-specific gene regulation. Mis-regulation of APA is linked to a wide range of pathological conditions. APA-mediated gene regulation in the heart is emerging as a new area of research. Here, we will discuss the impact of APA on gene regulation during heart development and in cardiovascular diseases. First, we will briefly review how APA impacts gene regulation and discuss molecular mechanisms that control APA. Then, we will address APA regulation during heart development and its dysregulation in cardiovascular diseases. Finally, we will discuss pre-mRNA targeting strategies to correct aberrant APA patterns of essential genes for the treatment or prevention of cardiovascular diseases. The RNA field is blooming due to advancements in RNA-based technologies. RNA-based vaccines and therapies are becoming the new line of effective and safe approaches for the treatment and prevention of human diseases. Overall, this review will be influential for understanding gene regulation at the RNA level via APA in the heart and will help design RNA-based tools for the treatment of cardiovascular diseases in the future.
Keywords: Alternative polyadenylation, Heart development, Cardiovascular disease, 3′UTR length and gene regulation
1. Introduction
Cleavage and polyadenylation is a process where the 3′ end of eukaryotic pre-mRNAs are cleaved at a specific ‘CA’ dinucleotide site known as poly(A) site (PAS) followed by the addition of a poly (A) tail of ∼100–200 adenosine nucleotides at the 3′ end of pre-mRNAs.1 3′ cleavage and polyadenylation of pre-mRNAs are critical for nuclear export, mRNA stability, localization, and translation.2–9 Cleavage and polyadenylation are tightly coupled with transcription and splicing.10,11 Genome-wide studies revealed that 60–70% of human genes have multiple PASs.12,13
Alternative polyadenylation (APA) is defined as the differential use of multiple PASs within a given pre-mRNA. Depending on the location of these PAS, APA can directly affect gene expression or generate diverse mRNA isoforms, providing an additional layer of regulation.14 APA regulates gene expression and function in a cell and tissue-specific manner, and its dysregulation is frequently observed under diseased conditions. Therefore, deciphering APA regulation in health vs. disease will help understand organism development and provide new therapeutic options for diseases in which APA regulation goes awry.
2. Impact of APA on gene regulation
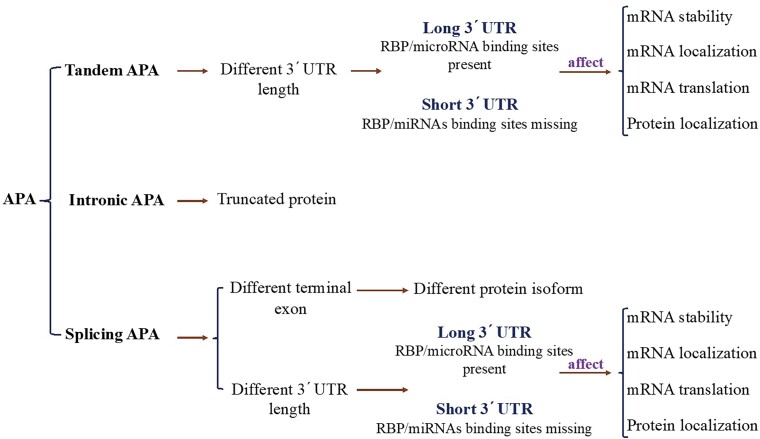
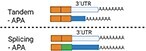
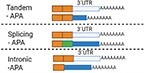
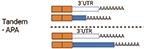
There are different types of APA: tandem-APA, splicing-APA (Coding Region-APA),15,16 and intronic-APA (IPA).2,17,18 Tandem-APA is the most common type, in which PASs reside in different locations within the 3′UTR, resulting in transcripts with different lengths of 3′UTR. In this type of APA, the protein-coding region is not affected (Figure 1). In splicing-APA, both coding region and 3′UTR length are affected (Figure 1). In intronic-APA, the PAS is in the intron, leading to the generation of truncated proteins (Figure 1). In this review, we will mostly focus on tandem-APA regulation.
Figure 1.
Different types of APA.
2.1. Tandem-APA in gene expression and protein output
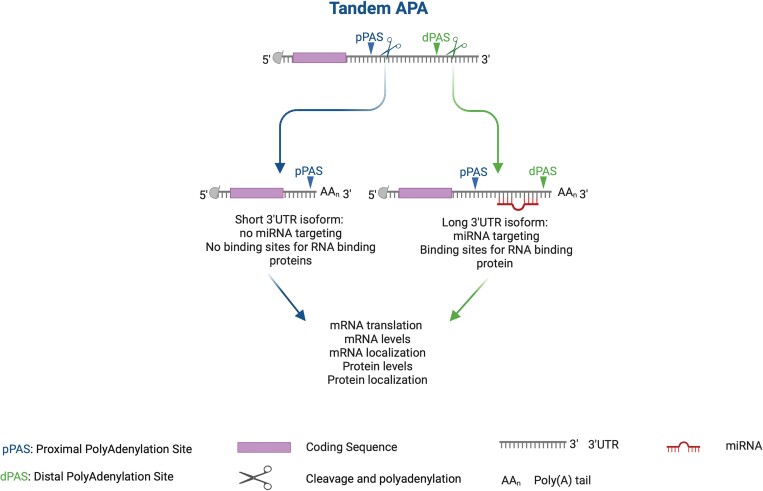
In tandem-APA, proximal PAS (pPAS) or distal PAS (dPAS) is used preferentially to generate transcripts with short or long 3′UTRs, respectively (Figure 2). mRNAs with long 3′UTRs often harbour cis-acting elements for microRNA (miRNA) binding sites or/and RNA-binding protein (RBP) sites that are missing in transcripts with short 3′UTR isoforms, so they are regulated differently19 (Figure 2). miRNAs bind to 3′UTRs and affect mRNA translation or stability20,21 (Figure 2). RBPs can also bind to the cis-acting elements within the 3′UTR to regulate mRNA stability, mRNA translation, or mRNA localization.22,23 Moreover, RBPs and miRNAs could interact with each other to potentiate or antagonize their functions through binding to 3′UTRs of a given mRNA.24–26 Tandem-APA regulates mRNA stability3,27–31 and translation3,6,30,32–34 by modulating positive or negative cis/trans-acting factors that bind 3′UTRs (Figures 1 and 2).
Figure 2.
Tandem-APA. A gene that contains >1 PAS in its 3′UTR is susceptible to APA regulation. In this example, this gene has 2 PASs: proximal poly(A) site (pPAS) and distal poly(A) site (dPAS) in the same 3′UTR. The usage of pPAS or dPAS generates short or long 3′UTR isoforms, respectively. Image created using Biorender.
In general, pPAS usage shortens 3′UTR length enabling an escape from miRNAs/RBPs mediated regulation (Figure 2).2,3 A study showed that mRNA transcripts with >1 kb 3′UTR had shorter mRNA half-lives.27 Other studies also showed a negative correlation between 3′UTR length and mRNA abundance.3,28–30 However, there is also evidence that longer 3′UTR isoforms are more stable.6,31 miRNAs21,35 and some RBPs, such as TIA1,36 TTP,26,37 and AUF-1,38 bind to the 3′UTR and destabilize mRNAs, whereas RBP HuR can stabilize transcripts.39 The different actions of these RBPs can explain the differences in these studies.
3′UTR also can act as competing endogenous RNAs (ceRNAs) to regulate gene expression.40–42 When 3′UTRs shorten, they lose miRNA target sites allowing these miRNAs to bind to other genes.40–42 For instance, PTEN and EPS15 share the same miRNA binding sites and thus are ceRNA partners. Shortening of 3′UTR of EPS15 frees up miRNAs, making them available to bind to the 3′UTR of PTEN mRNA in turn repressing PTEN expression.42
mRNA isoforms with shorter 3′UTRs are associated with polysomes, leading to increased protein production.43 All these studies demonstrate that tandem-APA-mediated changes in 3′UTR length can affect both mRNA levels and mRNA translation.
2.2. Tandem 3′UTR APA in mRNA and protein localization
Tandem-APA can regulate mRNA localization. Unique sequences within the 3′UTRs can function as ‘barcodes’ for RBP recognition and transport to specific subcellular compartments.44,45 Genome-wide analysis revealed that mRNAs with shorter 3′UTRs are more enriched in the cytoplasm than the nucleus.4 There is also evidence that mRNAs with long 3′UTRs are more likely to be localized to the endoplasmic reticulum (ER), whereas shorter 3′UTR isoforms are often localized to the mitochondrial envelope and to ribosomal subunits.5
It has been recently discovered that 3′UTR can drive protein localization independent of mRNA localization. CD47, which encodes for integrin-associated protein, has two isoforms with short and long 3′UTR. Long 3′UTR of CD47 can direct GFP protein localization to the plasma membrane, whereas the short 3′UTR to the ER, independent of their mRNA localization.9 Subsequent studies identified a meshwork assembled by RBP TIS11B intertwined with ER facilitating 3′UTR’s interactions with RBPs HuR and SET, controlling protein localization.46,47 Altogether, these results indicate that tandem-APA-mediated 3′UTR length changes can affect mRNA localization as well as protein localization independent of mRNA localization within the cell.
2.3. Splicing-APA in gene regulation
Splicing-APA is mediated by alternative splicing (AS) of exons that contain PASs,2 which usually involves the last exon definition.48 PASs can also be present in introns.49 If PASs are within introns, a truncated protein is generated. In some cases, genes with intronic PASs are expressed at low levels.50
Preferential inclusion or exclusion of introns and exons with PASs via AS can modulate protein output as well as mRNA levels. Based on the different locations of PASs in exons/introns, splicing-APA can impact gene expression, mRNA isoform generation, and protein output (Figure 1).14,51
A well-known example of splicing-APA is the immunoglobulin (Ig) M heavy-chain (µ) gene. IgM has two different isoforms generated via splicing-APA, one is a surface antigen receptor (µm) and the other is a secreted isoform (µs).16,52 When B cells become plasma cells, differential IgM splicing leads to predominantly pPAS usage, generating soluble IgM (µs)16,52 In sum, splicing-APA increases protein diversity and influences gene expression.
3. Regulation of APA
3.1. Cleavage and polyadenylation factors
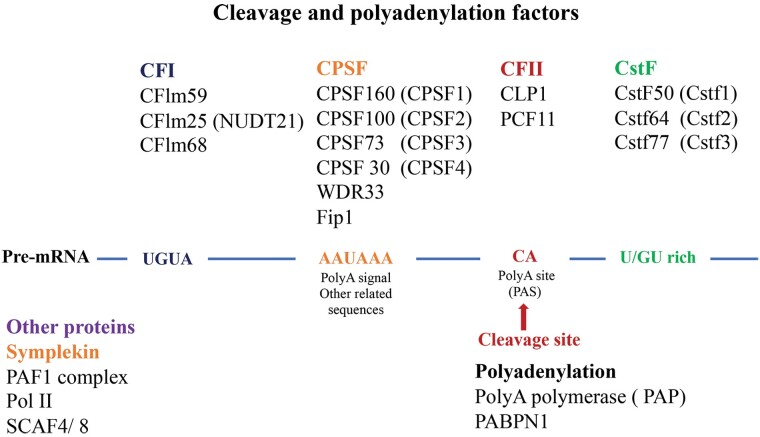
There are several cis-acting sequences in the 3′UTR required for cleavage and polyadenylation of pre-mRNAs in mammals. The essential elements include a hexameric poly(A) signal (typically AAUAAA) and a cleavage site which usually is a ‘CA’ dinucleotide located 10–30 bases downstream of the poly(A) signal (Figure 3).2 In mammals, there is a variable U/G-U-rich region (downstream sequence element) located at 20–40 bases downstream of the PAS (Figure 3). A conserved UGUA or UAUA sequence and a U-rich element (upstream sequence element) are usually located upstream of the poly(A) signal2 (Figure 3).
Figure 3.
Cis-acting RNA sequences and proteins necessary for cleavage and polyadenylation of pre-mRNAs. Cis-acting elements include upstream ‘UGUA’ and downstream ‘U/GU-rich’ auxiliary elements as well as ‘AAUAAA’ poly(A) signal and ‘CA’ cleavage site where polyA is added. Proteins include CFIm, CPSF, CstF, CFII complexes, Symplekin, PAP, and others.
These sequences are recognized by 3′ end processing proteins. The metazoan 3′ end processing machinery includes a large protein complex composed of cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CFIm), cleavage factor II (CFIIm), poly(A) polymerase (PAP), poly(A)-binding protein (PABP), and Symplekin53,54 (Figure 3). At the initiation step, components of CPSF and CstF bind to the polyA signal and the U/G-U-rich region, respectively, determining the region for cleavage.
CPSF has six subunits: CPSF-160 (CPSF1), CPSF-100 (CPSF2), CPSF-73 (CPSF3), CPSF-30 (CPSF4), WDR33, and hFIP1.55 CPSF-30 and WDR33 proteins recognize and bind to the poly(A) signal.56 CPSF-73 is the endonuclease, cleave the mRNA at the 3′ end, 20–40 nt downstream of the poly(A) signal57,58 (Figure 3). CPSF-73 is associated with CPSF-100 and Symplekin proteins as a stable complex on 3′ end of pre-mRNA to guide polyadenylation.59
CstF has three subunits: CstF-50 (CSTF1), CstF-64 (CSTF2), and CstF-77 (CSTF3).60 CstF-64 binds directly to the GU-rich region, whereas Cstf-50 and Cstf77 associate with both CstF-64 and Symplekin, reinforcing the binding of CPSFs and CstFs to the pre-mRNAs.2,61,62
CFIm complex has three proteins CFIm25 (CPSF5/NUDT21), CFIm59 (CPSF7), and CFIm68 (CPSF6), which bind to the UGUA element in the pre-mRNA.63,64 CFIIm is a heterodimer complex that consists of polyadenylation factor hCLP1 and PCF11.17,65 They target the ‘CA’ cleavage site, and weakly or transiently regulate 3′ end processing.66 The mechanism by which these proteins regulate APA has been extensively reviewed.16,67,68
3.2. Co-transcriptional regulation of APA
Cleavage and polyadenylation occur co-transcriptionally. RNA polymerase II (Pol II) is required for efficient 3′ end processing. Factors that influence transcriptional elongation can affect APA. In general, slow transcription elongation is associated with pPAS usage, generating shorter 3′UTR isoforms.69,70 Pol II ChIP-seq analysis revealed that Pol II binding was enriched in PASs that are being utilized for 3′ end processing, coincident with where CstF77 is recruited to the PASs.71 This indicates that Pol II deposition may influence PAS usage and APA.
In yeast, mutated Rpb1(Pol II) slowed transcription and resulted in global mRNA 3′UTR shortening.72 Moreover, both global and reporter-based nuclear run-on assays showed that highly expressed genes tend to display Pol II pausing at pPASs in cells, and human and mouse tissues.13
Transcription factors and Pol II-associated proteins affect APA via interactions with cleavage and polyadenylation factors. Transcription factor TFIID interacts with CPSF complex.16 CDC73, a component of the Pol II and chromatin-associated human Paf1 complex, interacts with both CPSF and CstF complexes and affects mRNA 3′ end processing.73 These results, and others not discussed here, demonstrate a strong association between APA regulation and transcription.
3.3. The role of splicing factors and regulators in APA regulation
Spliceosome components U1 snRNP,74 U1A,75,76 U2 snRNP, and its auxiliary factor U2AF6577,78 are involved in the recruitment of 3′ end processing factors and stimulation of mRNA cleavage and polyadenylation. Excess levels of U1A inhibit PAP activity and 3′ end processing.79 U1 snRNP recruits CPSF160 to the 3′UTR and stabilizes the interaction and enhances cleavage and polyadenylation of pre-mRNAs.74 U1 snRNP is important in protecting pre-mRNAs from premature cleavage and polyadenylation at cryptic intronic PASs. Depletion of U1 snRNA at 5′ splice site disrupted U1 snRNP function leading to widespread premature 3′ end cleavage and polyadenylation.80 Consistently, antisense morpholino oligonucleotide (AMO) against U1 snRNA promoted cryptic intronic PAS usage leading to premature cleavage and polyadenylation.81 Reducing the expression of U1 by AMO without affecting its splicing function led to widespread 3′UTR shortening.82 These studies reveal that U1A and U1 snRNP play multiple roles in regulating APA.
U2 snRNP directly interacts with CPSF and regulates 3′ processing.77,78 The subunit of U2 snRNP auxiliary factor U2AF65 binds to CFIm and stimulates 3′ end cleavage and polyadenylation when it is tethered to the 3′ end of adenovirus L3 pre-mRNA.83 RBPs with roles in pre-mRNA splicing and AS,84,85 such as SRp20, SRSF7,55 hnRNPA2/B1,86 hnRNP1,87 RBFOX2,88 CELF2,89 MBNL,90 NOVA,91 bind 3′UTRs and/or affect APA patterns.18 These results collectively indicate that splicing factors/regulators can regulate APA.
3.4. Epigenetic regulation of APA
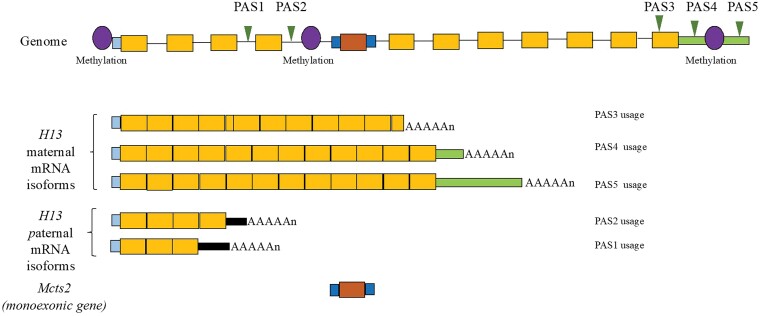
Epigenetic regulation can influence APA.92 Allele-specific DNA methylation affects APA of imprinted gene H13.93H13 has five PASs93 (Figure 4). DNA methylation downstream of PASs inhibits specific PAS usage, determining allele-dependent H13 and MCTS2 expression93 (Figure 4), linking methylation to APA and gene expression.
Figure 4.
Gene structure and mRNA isoforms generated from H13/Mcts locus. Monoexonic gene Mcts2 resides in the fourth intron of H13 gene. Two PASs (PAS1 and PAS2) of H13 gene are located before Mcts2, whereas three (PAS3, PAS4, and PAS5) are located on the last exon of H13. There are three CpG methylation sites: H13 promoter, Mcts2 promoter, and one between PAS4 and PAS5. Among these three CpG islands, H13 promoter is not methylated in both maternal and paternal alleles in neonatal mouse brain, while Mcts2 promoter and H13 3′ end were methylated in the maternal allele but not in the paternal allele. Correspondingly, PAS3, PAS4, and PAS5 were primarily utilized in maternal, and PAS1 and PAS2 were utilized in paternal alleles.
Genetic ablation of DNA methyltransferases DNMT1 and DNMT3b led to APA changes in 546 genes in HCT116 cells. Majority (75%) of these genes displayed 3′UTR shortening.94 DNA methylation between pPAS and dPAS within the 3′UTRs interrupted CTCF-mediated chromatin remodelling.94 CTCF regulates transcription by recruiting cohesion, forming a chromatin loop.95,96 CTCF-binding sites located between two PASs are often associated with cohesin complex component RAD21 and Pol II binding.94 Notably, depletion of RAD21 or CTCF-binding site restored dPAS usage. A model was proposed in which DNA methylation between pPAS and dPAS can prevent CTCF binding to that region supporting transcription elongation to reach to the dPAS.94 These studies support a strong link between epigenetic regulation and APA.
3.5. Role of RNA modifications on APA regulation
Methylation and pseudouridylation of RNAs can influence APA.97 m6A modification mediated by VIRMA and METTL3 can affect APA, likely through the association of VIRMA with CFIm25 and CFIm68.98 Depletion of YTHDC1, m6A reader protein, resulted in genome-wide APA changes in mouse oocytes,99 likely through interactions with CFIm68. m6A methyltransferase subunit WTAP promoted intron retention, polyadenylation, and exon skipping at potential G-quadruplex-forming sequences, which altered splicing-APA.100
Thirty per cent 3′UTRs101,102 in humans are pseudouridylated, suggesting a potential role for pseudouridylation in APA regulation. Depletion of pseudouridine synthases PUS1, PUS7, RPUSD4, and TRUB1, which bind to 3′UTR, led to 3′UTR lengthening via APA in HepG2 cells.103 These studies suggest that RNA modifications may play an important role in regulating APA through mechanisms that remain to be elucidated.
3.6. Influence of ubiquitination on APA
Ubiquitination of cleavage and polyadenylation factors can impact APA. Depletion of ubiquitin hydrolase USP22 disrupted recruitment of CPSF-73 to the 3′ end of IRF1 pre-mRNA and resulted in defective cleavage and polyadenylation of IRF1 pre-mRNA.104
Another study unveiled a role for ubiquitin ligase MAGE-A11 in APA regulation.105 MAGE-A11 is primarily expressed in male germline but is aberrantly activated in cancer cells, promoting tumour progression.105 Activation of MAGE-A11 led to ubiquitination of PCF11, which disassociated CFIm25 and resulted in 3′UTR shortening of mRNAs.105 3′UTR shortening mediated by MAGE-A11 led to elevated oncogene expression.105 About 50% of 3′UTR shortening events regulated by MAGE-A11 overlapped with CFIm25 depletion, showing that PCF11 ubiquitination but not degradation led to CFIm25 dissociation from 3′ end of pre-mRNAs. These studies indicate that ubiquitination can act as an additional regulatory layer for APA control.
4. APA in heart development and cardiovascular disease
The role of APA in cancer and other human diseases was extensively reviewed.17,68,106 APA-mediated gene regulation in the heart is being recognized as an important contributor to cardiac gene expression and function. In this section, we summarized APA regulation during heart development and in diseased hearts.
4.1. APA regulation during heart development
APA is highly regulated during cell proliferation28,32,107 and differentiation,108,109 processes essential for heart development. APA is regulated in a tissue-specific manner depending on the availability of cleavage and specificity factors as well as APA regulators.110–113
A study used mouse embryos to profile global poly(A) usage and found that genes underwent 3′UTR lengthening as the embryos developed.114 Consistently, single-cell RNA (scRNA) sequencing revealed global 3′UTR lengthening in all cell types between embryonic day 9.5 (E9.5) and E13.5 stages during mouse embryo development.115 This result is not surprising, as 3′UTR lengthening occurs during cell differentiation.114,116 However, it is not well understood how (i) APA affects cardiac gene expression during development, (ii) APA is regulated during heart development, and (iii) cleavage and polyadenylation machinery or RBPs involved in APA are regulated during heart development.
Using long-read cDNA sequencing and poly(A)click-seq (PAC-Seq), we examined splicing-APA-mediated regulation of Tropomyosin (Tpm1) by RBPs during rat heart development.88Tpm1 is an actin-binding protein required for cytoskeletal functions and for contraction in muscle cells. TPM1 is necessary for heart development and mutations or changes in TPM1 is linked to human heart diseases. Tpm1 has four terminal exons which are differentially spliced in a tissue-specific and development-dependent manner.117–121 Muscle-specific Tpm1 isoforms generated via splicing-APA were increased during rat heart development,88 correlating with the high demand for contraction of adult hearts. We showed that developmentally regulated RBPs RBFOX2 and PTBP1 antagonistically regulate muscle-specific Tpm1 isoforms via splicing-APA in rat myoblasts.88,122
Transcription factor NKX2-5 is implicated in APA regulation in the embryonic heart. Nkx2-5 is necessary for heart development123 and establishing transcriptional networks for cardiac differentiation.124NKX2-5 is mutated in patients with congenital heart defects.125–128 A recent study linked Nkx2-5 to APA regulation.129 Essential cardiac genes cardiac troponin T (Tnnt2) and ATPase sarcoplasmic/endoplasmic reticulum Ca2+ Transporting 2 (Atp2a2) mRNAs displayed longer 3′UTRs in Nkx2-5 mutant hearts and in Nkx2-5 depleted embryonic cardiomyocytes.129 Genomic analysis showed Nkx2-5 occupancy in transcription start sites as well as downstream genomic regions near PAS, suggesting a possible role for Nkx2-5 in co-transcriptional APA regulation.129 Supporting this, Nkx2-5 depletion promoted serine 2 phosphorylation of Pol II and its consequent binding to downstream regions of Tnnt2 and Atp2a2, which facilitated the recruitment of cleavage and polyadenylation machinery to the dPAS, resulting in 3′UTR lengthening.129 Phosphorylation of Pol II-serine 2 mediates transcription-coupled 3′ end processing through promoting binding of cleavage and polyadenylation factors to PASs.130
A recent study successfully identified non-polyadenylated and polyadenylated transcripts in single cells of mouse embryos during heart morphogenesis using VASA-seq, contributing to better understanding APA regulation during heart development.131 These results suggest that transcription factors and RBPs can influence APA-mediated cardiac gene expression during heart development.
4.2. Dysregulation of APA in cardiac hypertrophy
Cardiac hypertrophy is the enlargement of the heart in response to physiological stimuli or pathological stress. Pathological cardiac hypertrophy is an early adaptive process to increase heart function, but eventually can lead to heart failure.
Genome-wide APA changes were identified in mouse and rat hypertrophic hearts132–134 and in human failing hearts;135 315 tandem-APA events were identified in transverse aortic constriction (TAC)-induced hypertrophic hearts, and 60% of which resulted in 3′UTR shortening.132 The hallmark of cardiac hypertrophy is the reactivation of foetal gene expression.136,137 Importantly, hypertrophic mouse hearts displayed embryonic-like APA patterns.132
Another study also showed global 3′UTR shortening in TAC-induced hypertrophic mouse hearts,134 which mimicked embryonic heart APA. Moreover, 3′UTR length changes resulted in differential association of these mRNAs with translating polysomes.134 These results suggest that during cardiac hypertrophy APA changes can alter both mRNA levels and translation, affecting protein output.
Calcium homeostasis and cell cycle are important for cardiac development and hypertrophy.138,139 Genes involved in calcium homeostasis and cell cycle, i.e. Asph, Egfr, Pard3, and Arpp21, used different terminal exons at embryonic vs. adult stages.134,140–142 Thus, APA may contribute to foetal gene reactivation during hypertrophy.
APA regulators are altered during cardiac hypertrophy. Star-PAP is a nuclear polyA polymerase, which regulates 3′ end processing of selective mRNAs under cellular stress.143 Star-PAP depletion resulted in the loss of the longest 3′UTR isoform of NQO1,133 which is a critical gene for heart function. Both Star-PAP and NQO1 proteins were reduced in hypertrophic rat hearts. Ectopic expression of Star-PAP and longest 3′UTR isoform of NQO1 reversed hypertrophic gene response in cardiomyocytes.133 These suggested that APA regulation of NQO1 via Star-PAP is a contributor to cardiac hypertrophy. Another study also linked Star-PAP to 3′ end processing in hypertrophic hearts. Star-PAP interacted with AS regulator RBM10 and regulated APA of anti-hypertrophic genes including HAND2, TGFBR3, ERBB3, RHEBL1, COL5A1, CACNA1G, and HO-1, affecting their expression144 (Table 1). Mechanisms by which RBM10 and Star-PAP co-ordinately mediate APA need further investigation.
Table 1.
Genes regulated via APA and APA regulators relevant to heart development and cardiovascular diseases
| Heart disease | APA types | Genes regulated via APA | APA factors/regulators or RBPs | References |
|---|---|---|---|---|

|
|
Arfgef2, Lrrc58, Ppm1k, Klf4, NQ0, CLOCK, Gsk3b, Camkk2, Ccnd2, Faf2, Mtus1, Etf1, Utp6, Pdzrn3, Ctc1, Asph, Egfr, Pard3, Arpp2, CACNA1G, Rheb11, HAND2, TGFBR3, ERBB3, RHEBL1, COL5A1, and HO-1 | Star-PAP, RBM10 | 132–134,143,144 |
|
|
CDC42EP3, PIGK, RCAN1, WEE1, FBRSL1,SERF, COL1A, FN1, and TGFβR1 | PABPN1, CPSF30, PCF11, CSTF64 | 135,145 |

|

|
KCNH2 | HuR, PABPN1 | 69 |

|
|
Irf2bp2, Ddx5, Timp2, Ub32n and Flt1 | CPSF6, CFIm68 | 146,147 |

|
|
ATP1B1 and SLC7A1 | Undefined | 148–150 |
Recent advancements in sequencing technology and computational algorithms allow precise determination of APA patterns151 (Table 2). scAPAatlas of human and mouse tissues is a good source to assess APA at the single-cell level.151 According to the scAPAatlas, CLOCK, a transcription factor that regulates circadian rhythm, has four different mRNA transcripts with varying 3′UTR lengths generated via APA151 (Table 1). In atrial cardiomyocytes, predominantly long 3′UTR-containing transcripts of CLOCK were identified; whereas in ventricular cardiomyocytes, predominantly short 3′UTR isoforms were detected.151 Single nucleotide polymorphisms in CLOCK are associated with obesity161 and ischaemia/reperfusion.162 Cardiomyocyte-specific expression of CLOCK mutant under chronic desynchronic light cycle increased expression of cardiac hypertrophy markers,163 linking APA regulation of CLOCK to cardiac hypertrophy.
Table 2.
Methods to identify APA patterns
| Method | Fragmentation | Analysis | Ref. | Limitations | Advantage |
|---|---|---|---|---|---|
| Microarray | Restriction endonucleases | Affymetrix Power Tools, etc. | 32,114,152–154 | Low resolution and sensitivity | Simple |
| 3′-Seq | Heat shearing | Map reads to ref. seq. by Bowtie and uses (CS_J)=∑j = 1πwj*j to determine differential poly(A) usage | 107 | May cause bias during fragmentation and ligation | Simple, better sensitivity |
| 3P-Seq | Restriction endonucleases | Reads mapping, tailing, etc. | 155 | May cause bias during fragmentation and ligation, complex experimental process | Simple, better sensitivity |
| PAS-Seq | RNA Fragmentation Buffer | Reads mapping, poly(A) site clustering, 3′UTR database establishing, etc. | 109 | May cause bias during fragmentation and ligation | Simple, better sensitivity |
| 3′READS | RNA Fragmentation Buffer | Reads mapping, poly(A) site clustering, 3′UTR database establishing, etc. | 116 | May cause bias during fragmentation and ligation | Simple, better sensitivity |
| PolyA-seq | No fragmentation | Reads mapping and tailing, determine poly(A) site with | 12 | May cause bias during ligation and size selection | Simple, no fragmentation, better sensitivity |
| 2P-seq | Partial digestion with RNase T1 | Reads mapping, poly(A) site clustering, etc. | 6 | May cause bias during fragmentation, and ligation; complex procedure | Better sensitivity |
| MAPS | No fragmentation | Reads mapping, etc. | 156 | May cause bias during capturing and ligation | No fragmentation, better sensitivity |
| PAT-Seq | Partial digestion with RNase T1 | Reads trimming, tail length and poly(A) site determination, etc. | 157 | May cause bias during fragmentation, size selection, and ligation; complex | Better sensitivity |
| SAPAS | Heat shearing | Reads mapping, poly(A) site clustering, etc. | 158 | May cause bias during fragmentation | Better sensitivity |
| IVT-SAPAS | Heat shearing | Reads mapping, poly(A) site clustering, etc. | 159 | May cause bias during fragmentation, and ligation; complex | Better sensitivity |
| PAC-Seq | No fragmentation | Reads mapping, poly(A) site clustering, etc. | 160 | May cause bias during click it process | Simple, no fragmentation, better sensitivity |
| Long-read sequencing | No fragmentation | Reads mapping, etc. | 88 | Coverage issues | Simple, no fragmentation |
4.3. APA regulation and APA regulators in heart failure
In heart disease, APA is aberrantly regulated. In a study, genome-wide APA changes were identified in human failing hearts.135 About half of these APA changes led to 3′UTR shortening.135CDC42EP3, PIGK, RCAN1, and Wee1 displayed significant 3′UTR shortening, whereas FBRSL1 and SERF2 showed 3′UTR lengthening in human failing hearts135 (Table 1). Importantly, changes in their 3′UTR lengths correlated with changes in their protein levels, suggesting that 3′UTR length changes are reflected in protein levels. Consistently, dysregulation of 3′ end processing proteins PABPN1, CPSF30, and PCF11 were identified in human failing hearts135 (Table 1). Mechanisms responsible for altered expression of PABPN1, CPSF30, and PCF11 in failing hearts and the consequences of these changes on APA need to be further investigated.
CSTF64 expression was increased in left ventricles and cardiac fibroblasts of human failing hearts.145 Profibrotic genes including Col1A, Fn1, and TGFβR1 exhibited a shift to a pPAS usage rather than dPAS, resulting in increased protein levels of fibrotic genes, contributing to the fibrosis.145 Loss of CSTF64 in cardiac fibroblasts isolated from heart failure patients led to increased dPAS usage of profibrotic genes reducing their expression145 (Table 1).
These findings suggest that APA factors are susceptible to dysregulation in diseased hearts. The contribution of APA factors/regulators to heart diseases is largely unknown. It would be important to determine whether changes in APA and/or APA regulators are secondary to failing hearts or primary drivers of heart failure.
4.4. APA regulation of ion channels involved in cardiac arrhythmias
Mutations in genes encoding ion channels can lead to cardiac arrhythmias.164 The Kv11.1 voltage-gated potassium channel (KCNH2 gene) mediates the rapid activation of delayed rectifier current (IKr) and is responsible for action potential repolarization in the heart.165 Changes in Kv11.1 levels or KCNH2 mutations are associated with cardiac arrhythmia.166KCNH2 isoforms with different N- and C-terminal termini are identified.69 Among these isoforms, PASs in intron 9 lead to the generation of a C-terminal domain (CTD) truncated isoform, which is non-functional.69 Consistently, mutations in KCNH2 gene that affects the CTD were identified in long QT syndrome (LQTS) patients.167,168 RBPs HuR69 and PABPN1169 were identified as inhibitors of PAS usage in intron 9 and promoters of full-length KCNH2 expression and proper IKr. Further studies are needed to determine whether modulating KCNH2 APA in LQTS could prevent arrhythmias (Table 1).
4.5. APA in atherosclerosis
Atherosclerosis is characterized by vascular plaque formation due to inflammation-mediated accumulation of fats, cholesterol, and other substances in the arterial walls. Plaques narrow and clog arteries, limit blood flow, and can potentially burst.170 mTORC1 pathway is activated during atherosclerosis and has been identified as a critical contributor to atherosclerosis progression. Activation of mTORC-1 resulted in dramatic shortening of 3′UTRs of 846 genes, which in turn increased their polysome association and translation.146 Of these APA-regulated genes, Irf2bp2,171Ddx5,172Timp2,173 and Ub32n174 important for the development of atherosclerosis were up-regulated. mTOR activation regulates phosphorylation and subcellular localization of CPSF618 (Table 1), likely contributing to mTORC-mediated APA changes.
Another example of atherosclerosis-associated gene regulated by APA is Flt1 (Table 1). Flt1 encodes for the vascular endothelial growth factor receptor 1 (VEGFR1).175 Inhibition of VEGFR1 reduces angiogenesis and atherosclerosis.176Flt1 has a PAS located in intron 13, which generates the soluble form of FLT1.147 Soluble FLT1 functions as a decoy sequestering VEGF and inhibiting angiogenesis and atherosclerosis.175,177
4.6. APA in hypertension
Hypertension or high blood pressure is a major risk factor for heart failure, atrial fibrillation, heart valve disease, atherosclerosis, aortic syndromes, and ischaemia.178,179 Several genes related to hypertension harbour polymorphisms in their 3′UTRs that affect their APA regulation.148–150,180ATP1B1 encodes for ATPase Na+/K+ transporting subunit beta 1, which is important for maintaining Na+/K+ ions across the cell membrane. Increased ATP1B1 expression is linked to hypertension in rats.181 A polymorphic T-rich sequence is found downstream of the pPAS of ATP1B1.148 This polymorphic T-rich sequence promotes pPAS usage, generating an ATP1B1 short 3′UTR isoform that increased ATP1B1 protein levels, promoting hypertension148 (Table 1).
Another hypertension-related gene regulated via APA is SLC7A1 (Table 1). SLC7A1 encodes for high affinity cationic amino acid transporter 1.182 Decreased SLC7A1 protein is associated with the onset of hypertension. There is a polymorphic SNP ss52051869 upstream of the human SLC7A1 pPAS.149,150 Minor T allele of the SNP promotes the generation of the SLC7A1 long 3′UTR isoform, whereas the C allele often generates the short 3′UTR isoform.149,150 The SLC7A1 long 3′UTR isoform results in decreased SLC7A1 protein, highly associated with the onset of hypertension.149,150
5. Identification of global APA patterns
APA was originally detected by microarray, which had limited resolution.32,152,153 Currently, RNA-sequencing methods are used to determine PAS usage and APA.6,12,107,109,116,155–160 These methods may include fragmentation and ligation processes, which could generate bias. Correspondingly, several bioinformatic tools were designed to extract PAS/clusters from RNA-sequencing reads.183Table 2 summarizes the sequencing methods and analyses used for APA detection.
6. APA in cardiovascular disease prevention and therapy
6.1. APA as a biomarker for predicting disease severity
One of the difficulties in treating cardiovascular diseases is that the disease progresses without obvious phenotypes and can irreversibly damage the heart. For example, adults can start accumulating lipids in vessels without symptoms, which can quietly result in atherosclerosis or even thrombosis. These conditions are not easy to treat and might be fatal.184 Heart failure is typically preceded by cardiac hypertrophy, which initially is an adaptive response to increased cardiac function and shows no significant cardiac dysfunction. However, the development of heart failure leads to high rates of morbidity even with the current treatments.185
Since APA patterns are different in normal vs. diseased hearts (Table 1), APA profiles in blood cells might be used as biomarkers. APA dysregulation correlates with different grades of tumour and a variety of tumour types, making it a good biomarker for cancer types.186 Remarkably, three histologically indistinguishable mouse lymphoma subtypes were distinguished with >74% accuracy based on APA signatures.187 Limited work has been done to profile APA patterns in blood from patients with cardiovascular diseases. APA profiling of blood cells and heart tissues might help better understand molecular mechanisms and gene signatures of cardiovascular diseases. Advancements in scRNA-sequencing provide gene regulatory profiles in a single cell.188 Future studies are needed to develop scAPAatlas of human heart diseases. A new study generated tools to determine the potential consequences of pathogenic variants of APA in a wide range of pathological conditions.189 APA profiles and predictions on gene expression in blood might help with the prediction and prevention of cardiovascular diseases at early stages.
6.2. Modulating APA for treatment and prevention of cardiovascular diseases
Correcting APA patterns of disease-causing genes could help restore essential gene expression and function in diseased hearts. For instance, CD47 is increased in human atherosclerotic arteries, and antibody against CD47 ameliorates atherosclerosis.190CD47 has short and long 3′UTR isoforms. The long 3′UTR isoform of CD47 is the cell surface ligand responsible for immune checkpoint function while the short 3′UTR isoform is not. Preventing the generation of CD47 long 3′UTR isoform without affecting the short isoform could eliminate pro-atherosclerotic lesions. Thus, APA-targeted gene therapy may provide a specific way to regulate different gene isoforms with different functions.
APA patterns can be modulated using genome editing (CRISPR or Talon), RNA editing, antisense oligonucleotides (ASOs), or small molecules. The polyA signal, PAS or PASs, or the length of 3′UTR can be edited to modulate APA.
CRISPR-mediated genome editing was used to modulate PAS usage of CCND1 and in turn its expression.191 The whole region between short and long mRNA 3′UTR was removed to maintain only the short Calm1 isoform. Removal of Calm1 long isoform disorganized dorsal root ganglion and hippocampal neuron activation in mice.192
Facioscapulohumeral muscular dystrophy (FSHD) patients have increased DUX4 expression in skeletal muscle. Using CRISPR-genome editing DUX4 APA was modulated to reduce DUX4 expression in FSHD patient-derived myoblasts.193 Genome editing is an efficient method of manipulating APA. However, potential off-target effects should be considered during design.194
ASO-based therapy is another way to alter APA. Modified ASOs that do not cause mRNA degradation is successfully used in clinics to modulate AS of SMA gene to treat children with spinal muscular dystrophy.195,196 ASOs were also used to block the PAS of DUX4, which successfully reduced DUX4 expression in the FSHD patient xenografts model without inducing cell toxicity.197 ASOs blocking intronic PAS in Kv11.1 mRNA successfully repressed generation of truncated Kv11.1 isoform and up-regulated Kv11.1 full-length isoform.198 One of the limitations of ASOs is tissue-specific targeting and delivery.199 ASOs also might have off-target effects and special consideration need to be taken while performing ASO experiments and two or more ASOs should be used for these studies.200
Small molecules can also be used to modulate APA. They can bind specific RNA structures and alter RNA splicing, cleavage of RNA, pre-RNA translation, and deactivate non-coding RNAs.201 Small molecule derived from N,N’-bis(2-quinolinyl)pyridine-2,6-dicarboxamide was able to preferentially stabilize and isolate G-quadruplex DNA and RNA motifs.202 Small molecules T4 and T5 were identified to promote pPAS usage in many transcripts with longer introns.7
7. Concluding remarks
APA has been recognized as an important RNA processing step that can directly affect gene expression and function. It is implicated in a variety of biological processes. It is tightly regulated, but its regulation goes awry in diseased conditions. A large protein complex controls mRNA cleavage and polyadenylation efficiency. Transcription regulation, epigenetic regulation, splicing factors/regulators, RNA modifications, and ubiquitination also closely intertwine with APA.
We touched upon the importance of APA-mediated gene regulation during cardiac development and in heart diseases. There are many unanswered questions that need deeper investigation. Are APA patterns altered similarly in different cardiovascular diseases? Which APA regulators or RBPs are involved in APA dysregulation in cardiovascular diseases of different aetiologies? How can we effectively and specifically target APA regulation of genes in the heart for therapy? How APA-mediated gene regulation contributes to heart development? Thus, understanding the molecular mechanisms responsible for APA dysregulation in cardiovascular diseases will provide new RNA-based therapies with nucleotide-level precision. The translational research using advanced molecular tools to manipulate APA in disease models as well as investigations on APA regulators and APA-related biological pathways in cardiovascular diseases require more attention.
Acknowledgements
We thank Kuyumcu-Martinez lab members for their critical insights.
Contributor Information
Jun Cao, Faculty of Environment and Life, Beijing University of Technology, Xueyuan Road, Haidian District, Beijing 100124, PR China.
Muge N Kuyumcu-Martinez, Department of Biochemistry and Molecular Biology, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77573, USA; Department of Neurobiology, University of Texas Medical Branch, Galveston, TX 77555, USA; Institute for Translational Sciences, University of Texas Medical Branch, 301 University Blvd, Galveston, TX 77573, USA.
Funding
This work was supported in part by grants from American Heart Association [20TPA35490206], National Institutes of Health/NHLBI [1R01HL157780-01A1], Additional Ventures Single Ventricle Research Fund and UTMB JSMEF Pilot to M.N.K-M. J.C. was funded by the National Natural Science Foundation of China [82200328]. The contents of the manuscript are solely the responsibility of the authors and do not necessarily represent the official views of NIH and other grant agencies/foundations.
References
- 1. Proudfoot NJ, Brownlee GG. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature 1976;263:211–214. [DOI] [PubMed] [Google Scholar]
- 2. Masamha CP, Wagner EJ. The contribution of alternative polyadenylation to the cancer phenotype. Carcinogenesis 2018;39:2–10. [DOI] [PubMed] [Google Scholar]
- 3. Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009;138:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neve J, Burger K, Li W, Hoque M, Patel R, Tian B, Gullerova M, Furger A. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res 2016;26:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng LC, Zheng D, Zhang Q, Guvenek A, Cheng H, Tian B. Alternative 3′ UTRs play a widespread role in translation-independent mRNA association with the endoplasmic reticulum. Cell Rep 2021;36:109407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spies N, Burge CB, Bartel DP. 3′ UTR-isoform choice has limited influence on the stability and translational efficiency of most mRNAs in mouse fibroblasts. Genome Res 2013;23:2078–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Araki S, Nakayama Y, Sano O, Nakao S, Shimizu-Ogasawara M, Toyoshiba H, Nakanishi A, Aparicio S. Decoding transcriptome dynamics of genome-encoded polyadenylation and autoregulation with small-molecule modulators of alternative polyadenylation. Cell Chem Biol 2018;25:1470–1484.e75. [DOI] [PubMed] [Google Scholar]
- 8. Mayr C. Regulation by 3′-untranslated regions. Annu Rev Genet 2017;51:171–194. [DOI] [PubMed] [Google Scholar]
- 9. Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015;522:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 2014;15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell 2002;108:501–512. [DOI] [PubMed] [Google Scholar]
- 12. Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res 2012;22:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, Tian B. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol 2011;7:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi Y. Alternative polyadenylation: new insights from global analyses. RNA 2012;18:2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rehfeld A, Plass M, Krogh A, Friis-Hansen L. Alterations in polyadenylation and its implications for endocrine disease. Front Endocrinol (Lausanne) 2013;4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell 2011;43:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan F, Hankey W, Wagner EJ, Li W, Wang Q. Alternative polyadenylation of mRNA and its role in cancer. Genes Dis 2021;8:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitschka S, Mayr C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat Rev Mol Cell Biol 2022;23:779–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mayr C. What are 3′ UTRs doing? Cold Spring Harb Perspect Biol 2019;11:a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- 21. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szostak E, Gebauer F. Translational control by 3′-UTR-binding proteins. Brief Funct Genomics 2013;12:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia-Maurino SM, Rivero-Rodriguez F, Velazquez-Cruz A, Hernandez-Vellisca M, Diaz-Quintana A, De la Rosa MA, Diaz-Moreno I. RNA binding protein regulation and cross-talk in the control of AU-rich mRNA fate. Front Mol Biosci 2017;4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ 2011;18:1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci 2012;13:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Iqbal MB, Johns M, Cao J, Liu Y, Yu SC, Hyde GD, Laffan MA, Marchese FP, Cho SH, Clark AR, Gavins FN, Woollard KJ, Blackshear PJ, Mackman N, Dean JL, Boothby M, Haskard DO. PARP-14 combines with tristetraprolin in the selective posttranscriptional control of macrophage tissue factor expression. Blood 2014;124:3646–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE Jr. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 2003;13:1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 2014;510:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng D, Wang R, Ding Q, Wang T, Xie B, Wei L, Zhong Z, Tian B. Cellular stress alters 3′UTR landscape through alternative polyadenylation and isoform-specific degradation. Nat Commun 2018;9:2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen M, Lyu G, Han M, Nie H, Shen T, Chen W, Niu Y, Song Y, Li X, Li H, Chen X, Wang Z, Xia Z, Li W, Tian XL, Ding C, Gu J, Zheng Y, Liu X, Hu J, Wei G, Tao W, Ni T. 3′ UTR lengthening as a novel mechanism in regulating cellular senescence. Genome Res 2018;28:285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mitra M, Johnson EL, Swamy VS, Nersesian LE, Corney DC, Robinson DG, Taylor DG, Ambrus AM, Jelinek D, Wang W, Batista SL, Coller HA. Alternative polyadenylation factors link cell cycle to migration. Genome Biol 2018;19:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008;320:1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thivierge C, Tseng HW, Mayya VK, Lussier C, Gravel SP, Duchaine TF. Alternative polyadenylation confers Pten mRNAs stability and resistance to microRNAs. Nucleic Acids Res 2018;46:10340–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rhinn H, Qiang L, Yamashita T, Rhee D, Zolin A, Vanti W, Abeliovich A. Alternative alpha-synuclein transcript usage as a convergent mechanism in Parkinson’s disease pathology. Nat Commun 2012;3:1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 2015;25:651–665. [DOI] [PubMed] [Google Scholar]
- 36. Meyer C, Garzia A, Mazzola M, Gerstberger S, Molina H, Tuschl T. The TIA1 RNA-binding protein family regulates EIF2AK2-mediated stress response and cell cycle progression. Mol Cell 2018;69:622–635 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 2005;19:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA 2010;1:457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brennan CM, Steitz JA. Hur and mRNA stability. Cell Mol Life Sci 2001;58:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fan Z, Kim S, Bai Y, Diergaarde B, Park HJ. 3′-UTR shortening contributes to subtype-specific cancer growth by breaking stable ceRNA crosstalk of housekeeping genes. Front Bioeng Biotechnol 2020;8:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li L, Wang D, Xue M, Mi X, Liang Y, Wang P. 3′UTR Shortening identifies high-risk cancers with targeted dysregulation of the ceRNA network. Sci Rep 2014;4:5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park HJ, Ji P, Kim S, Xia Z, Rodriguez B, Li L, Su J, Chen K, Masamha CP, Baillat D, Fontes-Garfias CR, Shyu AB, Neilson JR, Wagner EJ, Li W. 3′ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet 2018;50:783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Passacantilli I, Panzeri V, Bielli P, Farini D, Pilozzi E, Fave GD, Capurso G, Sette C. Alternative polyadenylation of ZEB1 promotes its translation during genotoxic stress in pancreatic cancer cells. Cell Death Dis 2017;8:e3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eliscovich C, Singer RH. RNP Transport in cell biology: the long and winding road. Curr Opin Cell Biol 2017;45:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayya VK, Duchaine TF. Ciphers and executioners: how 3′-untranslated regions determine the fate of messenger RNAs. Front Genet 2019;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma W, Mayr C. A membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 2018;175:1492–1506 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma W, Zheng G, Xie W, Mayr C. In vivo reconstitution finds multivalent RNA-RNA interactions as drivers of mesh-like condensates. Elife 2021;10:e64252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol 2008;28:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dubbury SJ, Boutz PL, Sharp PA. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 2018;564:141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Devany E, Park JY, Murphy MR, Zakusilo G, Baquero J, Zhang X, Hoque M, Tian B, Kleiman FE. Intronic cleavage and polyadenylation regulates gene expression during DNA damage response through U1 snRNA. Cell Discov 2016;2:16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lutz CS. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol 2008;3:609–617. [DOI] [PubMed] [Google Scholar]
- 52. Chang JW, Yeh HS, Yong J. Alternative polyadenylation in human diseases. Endocrinol Metab (Seoul) 2017;32:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 2010;38:2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xiang K, Tong L, Manley JL. Delineating the structural blueprint of the pre-mRNA 3′-end processing machinery. Mol Cell Biol 2014;34:1894–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwich OD, Blumel N, Keller M, Wegener M, Setty ST, Brunstein ME, Poser I, Mozos IRL, Suess B, Munch C, McNicoll F, Zarnack K, Muller-McNicoll M. SRSF3 and SRSF7 modulate 3′UTR length through suppression or activation of proximal polyadenylation sites and regulation of CFIm levels. Genome Biol 2021;22:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 2014;28:2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mandel CR, Kaneko S, Zhang HL, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 2006;444:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 2006;444:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sullivan KD, Steiniger M, Marzluff WF. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 2009;34:322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Darmon SK, Lutz CS. mRNA 3′ end processing factors: a phylogenetic comparison. Comp Funct Genomics 2012;2012:876893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev 1997;11:2755–2766. [DOI] [PubMed] [Google Scholar]
- 62. Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 1999;63:405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brown KM, Gilmartin GM. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 2003;12:1467–1476. [DOI] [PubMed] [Google Scholar]
- 64. Yang Q, Gilmartin GM, Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci U S A 2010;107:10062–10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schafer P, Tuting C, Schonemann L, Kuhn U, Treiber T, Treiber N, Ihling C, Graber A, Keller W, Meister G, Sinz A, Wahle E. Reconstitution of mammalian cleavage factor II involved in 3′ processing of mRNA precursors. RNA 2018;24:1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Y, Liu L, Qiu Q, Zhou Q, Ding J, Lu Y, Liu P. Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res 2021;40:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 2017;18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mohanan NK, Shaji F, Koshre GR, Laishram RS. Alternative polyadenylation: an enigma of transcript length variation in health and disease. Wiley Interdiscip Rev RNA 2022;13:e1692. [DOI] [PubMed] [Google Scholar]
- 69. Gong Q, Stump MR, Zhou Z. Regulation of Kv11.1 potassium channel C-terminal isoform expression by the RNA-binding proteins HuR and HuD. J Biol Chem 2018;293:19624–19632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dharmalingam P, Mahalingam R, Yalamanchili HK, Weng T, Karmouty-Quintana H, Guha A, Thandavarayan RA. Emerging roles of alternative cleavage and polyadenylation (APA) in human disease. J Cell Physiol 2022;237:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fusby B, Kim S, Erickson B, Kim H, Peterson ML, Bentley DL. Coordination of RNA polymerase II pausing and 3′ end processing factor recruitment with alternative polyadenylation. Mol Cell Biol 2016;36:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yague-Sanz C, Vanrobaeys Y, Fernandez R, Duval M, Larochelle M, Beaudoin J, Berro J, Labbe S, Jacques PE, Bachand F. Nutrient-dependent control of RNA polymerase II elongation rate regulates specific gene expression programs by alternative polyadenylation. Genes Dev 2020;34:883–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci U S A 2009;106:755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lutz CS, Murthy KG, Schek N, O’Connor JP, Manley JL, Alwine JC. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 1996;10:325–337. [DOI] [PubMed] [Google Scholar]
- 75. Kaida D. The reciprocal regulation between splicing and 3′-end processing. Wiley Interdiscip Rev RNA 2016;7:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schek N, Cooke C, Alwine JC. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol 1992;12:5386–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Koga M, Satoh T, Takasaki I, Kawamura Y, Yoshida M, Kaida D. U2 snRNP is required for expression of the 3′ end of genes. PLoS One 2014;9:e98015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 2006;23:195–205. [DOI] [PubMed] [Google Scholar]
- 79. Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 1994;76:531–541. [DOI] [PubMed] [Google Scholar]
- 80. Venters CC, Oh JM, Di C, So BR, Dreyfuss G. U1 snRNP telescripting: suppression of premature transcription termination in introns as a new layer of gene regulation. Cold Spring Harb Perspect Biol 2019;11:a032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010;468:664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, Dreyfuss G. U1 snRNP determines mRNA length and regulates isoform expression. Cell 2012;150:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J 2006;25:4854–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 1992;6:837–847. [DOI] [PubMed] [Google Scholar]
- 85. Bradley T, Cook ME, Blanchette M. SR proteins control a complex network of RNA-processing events. RNA 2015;21:75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Movassat M, Crabb TL, Busch A, Yao C, Reynolds DJ, Shi Y, Hertel KJ. Coupling between alternative polyadenylation and alternative splicing is limited to terminal introns. RNA Biol 2016;13:646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J 2007;26:2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cao J, Routh AL, Kuyumcu-Martinez MN. Nanopore sequencing reveals full-length tropomyosin 1 isoforms and their regulation by RNA-binding proteins during rat heart development. J Cell Mol Med 2021;25:8352–8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chatrikhi R, Mallory MJ, Gazzara MR, Agosto LM, Zhu WS, Litterman AJ, Ansel KM, Lynch KW. RNA binding protein CELF2 regulates signal-induced alternative polyadenylation by competing with enhancers of the polyadenylation machinery. Cell Rep 2019;28:2795–2806 e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Batra R, Charizanis K, Manchanda M, Mohan A, Li M, Finn DJ, Goodwin M, Zhang C, Sobczak K, Thornton CA, Swanson MS. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell 2014;56:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008;456:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Soles LV, Shi Y. Crosstalk between mRNA 3′-end processing and epigenetics. Front Genet 2021;12:637705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc’his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev 2008;22:1141–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nanavaty V, Abrash EW, Hong C, Park S, Fink EE, Li Z, Sweet TJ, Bhasin JM, Singuri S, Lee BH, Hwang TH, Ting AH. DNA methylation regulates alternative polyadenylation via CTCF and the cohesin complex. Mol Cell 2020;78:752–764 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kim S, Yu NK, Kaang BK. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med 2015;47:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 2017;6:e25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Frye M, Blanco S. Post-transcriptional modifications in development and stem cells. Development 2016;143:3871–3881. [DOI] [PubMed] [Google Scholar]
- 98. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, Wang F, Wang X, Shen B, Wang Y, Feng X, He C, Liu J. VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 2018;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 2018;14:e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Horiuchi K, Kawamura T, Hamakubo T. Wilms’ tumor 1-associating protein complex regulates alternative splicing and polyadenylation at potential G-quadruplex-forming splice site sequences. J Biol Chem 2021;297:101248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014;515:143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015;11:592–597. [DOI] [PubMed] [Google Scholar]
- 103. Martinez NM, Su A, Burns MC, Nussbacher JK, Schaening C, Sathe S, Yeo GW, Gilbert WV. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol Cell 2022;82:645–659.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chipumuro E, Henriksen MA. The ubiquitin hydrolase USP22 contributes to 3′-end processing of JAK-STAT-inducible genes. FASEB J 2012;26:842–854. [DOI] [PubMed] [Google Scholar]
- 105. Yang SW, Li L, Connelly JP, Porter SN, Kodali K, Gan H, Park JM, Tacer KF, Tillman H, Peng J, Pruett-Miller SM, Li W, Potts PR. A cancer-specific ubiquitin ligase drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3′ end processing complex. Mol Cell 2020;77:1206–1221.e07. [DOI] [PubMed] [Google Scholar]
- 106. Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet 2019;20:599–614. [DOI] [PubMed] [Google Scholar]
- 107. Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, Agami R. E2f mediates enhanced alternative polyadenylation in proliferation. Genome Biol 2012;13:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Grassi E, Santoro R, Umbach A, Grosso A, Oliviero S, Neri F, Conti L, Ala U, Provero P, DiCunto F, Merlo GR. Choice of alternative polyadenylation sites, mediated by the RNA-binding protein Elavl3, plays a role in differentiation of inhibitory neuronal progenitors. Front Cell Neurosci 2018;12:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 2011;17:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. MacDonald CC. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond (2018 update). Wiley Interdiscip Rev RNA 2019;10:e1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, Eisman RC, Andrews J, Kaufman T, Cherbas P, Celniker SE, Graveley BR, Lai EC. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 2012;1:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol 2005;6:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ni T, Yang Y, Hafez D, Yang W, Kiesewetter K, Wakabayashi Y, Ohler U, Peng W, Zhu J. Distinct polyadenylation landscapes of diverse human tissues revealed by a modified PA-seq strategy. BMC Genomics 2013;14:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A 2009;106:7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Agarwal V, Lopez-Darwin S, Kelley DR, Shendure J. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat Commun 2021;12:5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 2013;10:133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil 2015;36:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lin JJ, Eppinga RD, Warren KS, McCrae KR. Human tropomyosin isoforms in the regulation of cytoskeleton functions. Adv Exp Med Biol 2008;644:201–222. [DOI] [PubMed] [Google Scholar]
- 119. Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil 2001;22:5–49. [DOI] [PubMed] [Google Scholar]
- 120. Wieczorek DF, Jagatheesan G, Rajan S. The role of tropomyosin in heart disease. Adv Exp Med Biol 2008;644:132–142. [DOI] [PubMed] [Google Scholar]
- 121. Schevzov G, Whittaker SP, Fath T, Lin JJ, Gunning PW. Tropomyosin isoforms and reagents. Bioarchitecture 2011;1:135–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cao J, Verma SK, Jaworski E, Mohan S, Nagasawa CK, Rayavara K, Sooter A, Miller SN, Holcomb RJ, Powell MJ, Ji P, Elrod ND, Yildirim E, Wagner EJ, Popov V, Garg NJ, Routh AL, Kuyumcu-Martinez MN. RBFOX2 is critical for maintaining alternative polyadenylation patterns and mitochondrial health in rat myoblasts. Cell Rep 2021;37:109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Akazawa H, Komuro I. Cardiac transcription factor Csx/Nkx2-5: its role in cardiac development and diseases. Pharmacol Ther 2005;107:252–268. [DOI] [PubMed] [Google Scholar]
- 124. Arminan A, Gandia C, Bartual M, Garcia-Verdugo JM, Lledo E, Mirabet V, Llop M, Barea J, Montero JA, Sepulveda P. Cardiac differentiation is driven by NKX2.5 and GATA4 nuclear translocation in tissue-specific mesenchymal stem cells. Stem Cells Dev 2009;18:907–918. [DOI] [PubMed] [Google Scholar]
- 125. McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 2003;42:1650–1655. [DOI] [PubMed] [Google Scholar]
- 126. Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 1998;281:108–111. [DOI] [PubMed] [Google Scholar]
- 127. Abou Hassan OK, Fahed AC, Batrawi M, Arabi M, Refaat MM, DePalma SR, Seidman JG, Seidman CE, Bitar FF, Nemer GM. NKX2-5 mutations in an inbred consanguineous population: genetic and phenotypic diversity. Sci Rep 2015;5:8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Costa MW, Guo G, Wolstein O, Vale M, Castro ML, Wang L, Otway R, Riek P, Cochrane N, Furtado M, Semsarian C, Weintraub RG, Yeoh T, Hayward C, Keogh A, Macdonald P, Feneley M, Graham RM, Seidman JG, Seidman CE, Rosenthal N, Fatkin D, Harvey RP. Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ Cardiovasc Genet 2013;6:238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nimura K, Yamamoto M, Takeichi M, Saga K, Takaoka K, Kawamura N, Nitta H, Nagano H, Ishino S, Tanaka T, Schwartz RJ, Aburatani H, Kaneda Y. Regulation of alternative polyadenylation by Nkx2-5 and Xrn2 during mouse heart development. Elife 2016;5:e16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 2004;13:67–76. [DOI] [PubMed] [Google Scholar]
- 131. Salmen F, De Jonghe J, Kaminski TS, Alemany A, Parada GE, Verity-Legg J, Yanagida A, Kohler TN, Battich N, van den Brekel F, Ellermann AL, Arias AM, Nichols J, Hemberg M, Hollfelder F, van Oudenaarden A. High-throughput total RNA sequencing in single cells using VASA-seq. Nat Biotechnol 2022;40:1780–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Park JY, Li W, Zheng D, Zhai P, Zhao Y, Matsuda T, Vatner SF, Sadoshima J, Tian B. Comparative analysis of mRNA isoform expression in cardiac hypertrophy and development reveals multiple post-transcriptional regulatory modules. PLoS One 2011;6:e22391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sudheesh AP, Mohan N, Francis N, Laishram RS, Anderson RA. Star-PAP controlled alternative polyadenylation coupled poly(A) tail length regulates protein expression in hypertrophic heart. Nucleic Acids Res 2019;47:10771–10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Soetanto R, Hynes CJ, Patel HR, Humphreys DT, Evers M, Duan G, Parker BJ, Archer SK, Clancy JL, Graham RM, Beilharz TH, Smith NJ, Preiss T. Role of miRNAs and alternative mRNA 3′-end cleavage and polyadenylation of their mRNA targets in cardiomyocyte hypertrophy. Biochim Biophys Acta 2016;1859:744–756. [DOI] [PubMed] [Google Scholar]
- 135. Creemers EE, Bawazeer A, Ugalde AP, van Deutekom HW, van der Made I, de Groot NE, Adriaens ME, Cook SA, Bezzina CR, Hubner N, van der Velden J, Elkon R, Agami R, Pinto YM. Genome-wide polyadenylation maps reveal dynamic mRNA 3′-end formation in the failing human heart. Circ Res 2016;118:433–438. [DOI] [PubMed] [Google Scholar]
- 136. Reddy S, Zhao M, Hu DQ, Fajardo G, Hu S, Ghosh Z, Rajagopalan V, Wu JC, Bernstein D. Dynamic microRNA expression during the transition from right ventricular hypertrophy to failure. Physiol Genomics 2012;44:562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Cox EJ, Marsh SA. A systematic review of fetal genes as biomarkers of cardiac hypertrophy in rodent models of diabetes. PLoS One 2014;9:e92903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev 2007;87:521–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tyser RC, Miranda AM, Chen CM, Davidson SM, Srinivas S, Riley PR. Calcium handling precedes cardiac differentiation to initiate the first heartbeat. Elife 2016;5:e17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Wang L, Zhang H, Hasim A, Tuerhong A, Hou Z, Abdurahmam A, Sheyhidin I. Partition-defective 3 (PARD3) regulates proliferation, apoptosis, migration, and invasion in esophageal squamous cell carcinoma cells. Med Sci Monit 2017;23:2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Fukumoto S, Koyama H, Hosoi M, Yamakawa K, Tanaka S, Morii H, Nishizawa Y. Distinct role of cAMP and cGMP in the cell cycle control of vascular smooth muscle cells: cGMP delays cell cycle transition through suppression of cyclin D1 and cyclin-dependent kinase 4 activation. Circ Res 1999;85:985–991. [DOI] [PubMed] [Google Scholar]
- 142. Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel) 2017;9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Laishram RS. Poly(A) polymerase (PAP) diversity in gene expression—star-PAP vs canonical PAP. FEBS Lett 2014;588:2185–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mohan N, Kumar V, Kandala DT, Kartha CC, Laishram RS. A splicing-independent function of RBM10 controls specific 3′ UTR processing to regulate cardiac hypertrophy. Cell Rep 2018;24:3539–3553. [DOI] [PubMed] [Google Scholar]
- 145. Neupane R, Youker K, Yalamanchili HK, Cieslik KA, Karmouty-Quintana H, Guha A, Thandavarayan RA. Cleavage stimulating factor 64 depletion mitigates cardiac fibrosis through alternative polyadenylation. Biochem Biophys Res Commun 2022;597:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Chang JW, Zhang W, Yeh HS, de Jong EP, Jun S, Kim KH, Bae SS, Beckman K, Hwang TH, Kim KS, Kim DH, Griffin TJ, Kuang R, Yong J. mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat Commun 2015;6:7218. [DOI] [PubMed] [Google Scholar]
- 147. Thomas CP, Raikwar NS, Kelley EA, Liu KZ. Alternate processing of Flt1 transcripts is directed by conserved cis-elements within an intronic region of FLT1 that reciprocally regulates splicing and polyadenylation. Nucleic Acids Res 2010;38:5130–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Prasad MK, Bhalla K, Pan ZH, O’Connell JR, Weder AB, Chakravarti A, Tian B, Chang YP. A polymorphic 3′UTR element in ATP1B1 regulates alternative polyadenylation and is associated with blood pressure. PLoS One 2013;8:e76290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yang Z, Venardos K, Jones E, Morris BJ, Chin-Dusting J, Kaye DM. Identification of a novel polymorphism in the 3′UTR of the L-arginine transporter gene SLC7A1: contribution to hypertension and endothelial dysfunction. Circulation 2007;115:1269–1274. [DOI] [PubMed] [Google Scholar]
- 150. Yang Z, Kaye DM. Mechanistic insights into the link between a polymorphism of the 3′UTR of the SLC7A1 gene and hypertension. Hum Mutat 2009;30:328–333. [DOI] [PubMed] [Google Scholar]
- 151. Yang X, Tong Y, Liu G, Yuan J, Yang Y. scAPAatlas: an atlas of alternative polyadenylation across cell types in human and mouse. Nucleic Acids Res 2022;50:D356–D364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 2005;33:201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. D’Mello V, Lee JY, MacDonald CC, Tian B. Alternative mRNA polyadenylation can potentially affect detection of gene expression by affymetrix genechip arrays. Appl Bioinformatics 2006;5:249–253. [DOI] [PubMed] [Google Scholar]
- 154. Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 2009;4:e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 2011;469:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Fox-Walsh K, Davis-Turak J, Zhou Y, Li H, Fu XD. A multiplex RNA-seq strategy to profile poly(A+) RNA: application to analysis of transcription response and 3′ end formation. Genomics 2011;98:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Harrison PF, Powell DR, Clancy JL, Preiss T, Boag PR, Traven A, Seemann T, Beilharz TH. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA 2015;21:1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 2011;21:741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Fu Y, Ge Y, Sun Y, Liang J, Wan L, Wu X, Xu A. IVT-SAPAS: low-input and rapid method for sequencing alternative polyadenylation sites. PLoS One 2015;10:e0145477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Routh A, Ji P, Jaworski E, Xia Z, Li W, Wagner EJ. Poly(A)-clickseq: click-chemistry for next-generation 3-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res 2017;45:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Valladares M, Obregon AM, Chaput JP. Association between genetic variants of the clock gene and obesity and sleep duration. J Physiol Biochem 2015;71:855–860. [DOI] [PubMed] [Google Scholar]
- 162. Skrlec I, Milic J, Steiner R. The impact of the circadian genes CLOCK and ARNTL on myocardial infarction. J Clin Med 2020;9:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Durgan DJ, Tsai JY, Grenett MH, Pat BM, Ratcliffe WF, Villegas-Montoya C, Garvey ME, Nagendran J, Dyck JR, Bray MS, Gamble KL, Gimble JM, Young ME. Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int 2011;28:187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Grant AO. Cardiac ion channels. Circ Arrhythm Electrophysiol 2009;2:185–194. [DOI] [PubMed] [Google Scholar]
- 165. de la Pena P, Dominguez P, Barros F. Functional characterization of Kv11.1 (hERG) potassium channels split in the voltage-sensing domain. Pflugers Arch 2018;470:1069–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Anderson CL, Kuzmicki CE, Childs RR, Hintz CJ, Delisle BP, January CT. Large-scale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat Commun 2014;5:5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Fukumoto D, Ding WG, Wada Y, Fujii Y, Ichikawa M, Takayama K, Fukuyama M, Kato K, Itoh H, Makiyama T, Omatsu-Kanbe M, Matsuura H, Horie M, Ohno S. Novel intracellular transport-refractory mutations in KCNH2 identified in patients with symptomatic long QT syndrome. J Cardiol 2018;71:401–408. [DOI] [PubMed] [Google Scholar]
- 168. Biliczki P, Girmatsion Z, Harenkamp S, Anneken L, Brandes RP, Varro A, Marschall C, Herrera D, Hohnloser SH, Nattel S, Ehrlich JR. Cellular properties of C-terminal KCNH2 long QT syndrome mutations: description and divergence from clinical phenotypes. Heart Rhythm 2008;5:1159–1167. [DOI] [PubMed] [Google Scholar]
- 169. Stump MR, Nguyen RT, Drgastin RH, Search D, Gong Q, Zhou Z. Regulation of Kv11.1 isoform expression by polyadenylate binding protein nuclear 1. Int J Mol Sci 2021;22:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Ross R. Atherosclerosis—an inflammatory disease. N Engl J Med 1999;340:115–126. [DOI] [PubMed] [Google Scholar]
- 171. Chen HH, Keyhanian K, Zhou X, Vilmundarson RO, Almontashiri NA, Cruz SA, Pandey NR, Lerma Yap N, Ho T, Stewart CA, Huang H, Hari A, Geoffrion M, McPherson R, Rayner KJ, Stewart AF. IRF2BP2 reduces macrophage inflammation and susceptibility to atherosclerosis. Circ Res 2015;117:671–683. [DOI] [PubMed] [Google Scholar]
- 172. Fan Y, Chen Y, Zhang J, Yang F, Hu Y, Zhang L, Zeng C, Xu Q. Protective role of RNA helicase DEAD-box protein 5 in smooth muscle cell proliferation and vascular remodeling. Circ Res 2019;124:e84–e100. [DOI] [PubMed] [Google Scholar]
- 173. Di Gregoli K, George SJ, Jackson CL, Newby AC, Johnson JL. Differential effects of tissue inhibitor of metalloproteinase (TIMP)-1 and TIMP-2 on atherosclerosis and monocyte/macrophage invasion. Cardiovasc Res 2016;109:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Kumar SU, Kumar DT, Bithia R, Sankar S, Magesh R, Sidenna M, George Priya Doss C, Zayed H. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and bioinformatics approach. Front Genet 2020;11:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Luttun A, Tjwa M, Moons L, Wu Y, Angelillo-Scherrer A, Liao F, Nagy JA, Hooper A, Priller J, De Klerck B, Compernolle V, Daci E, Bohlen P, Dewerchin M, Herbert JM, Fava R, Matthys P, Carmeliet G, Collen D, Dvorak HF, Hicklin DJ, Carmeliet P. Revascularization of ischemic tissues by PlGF treatment, and inhibition of tumor angiogenesis, arthritis and atherosclerosis by anti-Flt1. Nat Med 2002;8:831–840. [DOI] [PubMed] [Google Scholar]
- 176. Camare C, Pucelle M, Negre-Salvayre A, Salvayre R. Angiogenesis in the atherosclerotic plaque. Redox Biol 2017;12:18–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Sasagawa T, Jinno-Oue A, Nagamatsu T, Morita K, Tsuruga T, Mori-Uchino M, Fujii T, Shibuya M. Production of an anti-angiogenic factor sFLT1 is suppressed via promoter hypermethylation of FLT1 gene in choriocarcinoma cells. BMC Cancer 2020;20:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Fuchs FD, Whelton PK. High blood pressure and cardiovascular disease. Hypertension 2020;75:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Rosendorff C, Black HR, Cannon CP, Gersh BJ, Gore J, Izzo JL Jr, Kaplan NM, O’Connor CM, O’Gara PT, Oparil S; American Heart Association Council for High Blood Pressure Research, American Heart Association Council on Clinical Cardiology, American Heart Association Council on Epidemiology and Prevention . Treatment of hypertension in the prevention and management of ischemic heart disease: a scientific statement from the American Heart Association Council for High Blood Pressure Research and the Councils on Clinical Cardiology and Epidemiology and Prevention. Circulation 2007;115:2761–2788. [DOI] [PubMed] [Google Scholar]
- 180. Faruque MU, Chen G, Doumatey A, Huang H, Zhou J, Dunston GM, Rotimi CN, Adeyemo AA. Association of ATP1B1, RGS5 and SELE polymorphisms with hypertension and blood pressure in African-Americans. J Hypertens 2011;29:1906–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Melzi ML, Syren ML, Assael BM, Sereni F, Aperia A. Increased renal tubular Na-K-ATPase activity in Milan hypertensive rats in the prehypertensive period. Pediatr Nephrol 1991;5:700–703. [DOI] [PubMed] [Google Scholar]
- 182. Bai X, Moraes TF, Reithmeier RAF. Structural biology of solute carrier (SLC) membrane transport proteins. Mol Membr Biol 2017;34:1–32. [DOI] [PubMed] [Google Scholar]
- 183. Chen W, Jia Q, Song Y, Fu H, Wei G, Ni T. Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics 2017;15:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Strong JP, Malcom GT, Newman WP III, Oalmann MC. Early lesions of atherosclerosis in childhood and youth: natural history and risk factors. J Am Coll Nutr 1992;11:51S–54S. [DOI] [PubMed] [Google Scholar]
- 185. Tham YK, Bernardo BC, Ooi JY, Weeks KL, McMullen JR. Pathophysiology of cardiac hypertrophy and heart failure: signaling pathways and novel therapeutic targets. Arch Toxicol 2015;89:1401–1438. [DOI] [PubMed] [Google Scholar]
- 186. Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 2014;5:5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res 2009;69:9422–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Gao Y, Li L, Amos CI, Li W. Analysis of alternative polyadenylation from single-cell RNA-seq using scDaPars reveals cell subpopulations invisible to gene expression. Genome Res 2021;31:1856–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Bogard N, Linder J, Rosenberg AB, Seelig G. A deep neural network for predicting and engineering alternative polyadenylation. Cell 2019;178:91–106 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kojima Y, Volkmer JP, McKenna K, Civelek M, Lusis AJ, Miller CL, Direnzo D, Nanda V, Ye J, Connolly AJ, Schadt EE, Quertermous T, Betancur P, Maegdefessel L, Matic LP, Hedin U, Weissman IL, Leeper NJ. CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 2016;536:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Wang Q, He G, Hou M, Chen L, Chen S, Xu A, Fu Y. Cell cycle regulation by alternative polyadenylation of CCND1. Sci Rep 2018;8:6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Bae B, Gruner HN, Lynch M, Feng T, So K, Oliver D, Mastick GS, Yan W, Pieraut S, Miura P. Elimination of Calm1 long 3′-UTR mRNA isoform by CRISPR-Cas9 gene editing impairs dorsal root ganglion development and hippocampal neuron activation in mice. RNA 2020;26:1414–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Joubert R, Mariot V, Charpentier M, Concordet JP, Dumonceaux J. Gene editing targeting the DUX4 polyadenylation signal: a therapy for FSHD? J Pers Med 2020;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids 2015;4:e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Corey DR. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat Neurosci 2017;20:497–499. [DOI] [PubMed] [Google Scholar]
- 196. Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov 2012;11:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Chen JC, King OD, Zhang Y, Clayton NP, Spencer C, Wentworth BM, Emerson CP Jr, Wagner KR. Morpholino-mediated knockdown of DUX4 toward facioscapulohumeral muscular dystrophy therapeutics. Mol Ther 2016;24:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Gong Q, Zhou Z. Regulation of isoform expression by blocking polyadenylation signal sequences with morpholinos. Methods Mol Biol 2017;1565:141–150. [DOI] [PubMed] [Google Scholar]
- 199. Chan JJ, Zhang B, Chew XH, Salhi A, Kwok ZH, Lim CY, Desi N, Subramaniam N, Siemens A, Kinanti T, Ong S, Sanchez-Mejias A, Ly PT, An O, Sundar R, Fan X, Wang S, Siew BE, Lee KC, Chong CS, Lieske B, Cheong WK, Goh Y, Fam WN, Ooi MG, Koh BTH, Iyer SG, Ling WH, Chen J, Yoong BK, Chanwat R, Bonney GK, Goh BKP, Zhai W, Fullwood MJ, Wang W, Tan KK, Chng WJ, Dan YY, Pitt JJ, Roca X, Guccione E, Vardy LA, Chen L, Gao X, Chow PKH, Yang H, Tay Y. Pan-cancer pervasive upregulation of 3′ UTR splicing drives tumourigenesis. Nat Cell Biol 2022;24:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Stainier DYR, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, Ingham PW, Schulte-Merker S, Yelon D, Weinstein BM, Mullins MC, Wilson SW, Ramakrishnan L, Amacher SL, Neuhauss SCF, Meng A, Mochizuki N, Panula P, Moens CB. Guidelines for morpholino use in zebrafish. PLoS Genet 2017;13:e1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Shao Y, Zhang QC. Targeting RNA structures in diseases with small molecules. Essays Biochem 2020;64:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Muller S, Kumari S, Rodriguez R, Balasubramanian S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat Chem 2010;2:1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]