Abstract
Due to the surge in interest in using single nucleotide polymorphisms (SNPs) for genotyping a facile and affordable method for this is an absolute necessity. Here we introduce a procedure that combines an easily automatable single tube sample preparation with an efficient high throughput mass spectrometric analysis technique. Known point mutations or single nucleotide polymorphisms are easily analysed by this procedure. It starts with PCR amplification of a short stretch of genomic DNA, for example an exon of a gene containing a SNP. By shrimp alkaline phosphatase digest residual dNTPs are destroyed. Allele-specific products are generated using a special primer, a conditioned set of α-S-dNTPs and α-S-ddNTPs and a fresh DNA polymerase in a primer extension reaction. Unmodified DNA is removed by 5′-phosphodiesterase digestion and the modified products are alkylated to increase the detection sensitivity in the mass spectrometric analysis. All steps of the preparation are simple additions of solutions and incubations. The procedure operates at the lowest practical sample volumes and in contrast to other genotyping protocols with mass spectrometric detection requires no purification. This reduces the cost and makes it easy to implement. Here it is demonstrated in a version using positive ion detection on described mutations in exon 17 of the amyloid precursor protein gene and in a version using negative ion detection on three SNPs of the granulocyte-macrophage colony stimulating factor gene. Preparation and analysis of SNPs is shown separately and simultaneously, thus demonstrating the multiplexibility of this genotyping procedure. The preparation protocol for genotyping is adapted to the conditions used for the SNP discovery method by denaturing HPLC, thus demonstrating a facile link between protocols for SNP discovery and SNP genotyping. Results corresponded unanimously with the control sequencing. The procedure is useful for high throughput genotyping as it is required for gene identification and pharmacogenomics where large numbers of DNA samples have to be analysed. We have named this procedure the ‘GOOD Assay’ for SNP analysis.
INTRODUCTION
Single nucleotide polymorphisms (SNPs) are thought to be ideally suited as genetic markers for establishing genetic linkage and as indicators of genetic diseases (1). In some cases a single SNP is responsible for a genetic disease. According to estimates the human genome may contain >3 million SNPs. Due to their propensity they lend themselves to very high resolution genotyping (2). The SNP consortium, a joint effort of 10 major pharmaceutical companies, has announced the development of 300 000 SNP markers and their placement in the public domain by mid 2001 (3). These facts increase the need for high throughput genotyping technologies. In order to make large-scale genotyping feasible, facile and affordable methods are required that can be operated at very high throughput and at very low cost. Recently, the application of oligonucleotide arrays (DNA chips) to large-scale SNP genotyping was demonstrated (4). A drawback of DNA chips is that although a large number of SNPs can be genotyped simultaneously, genomic DNA does not suffice as substrate due to low signal/noise and the lack of sensitivity. PCR has to be applied prior to DNA chip analysis to generate a sufficient amount of template thus dramatically limiting the vast genotyping capacity. As SNPs occur at an average rate of one every several hundred bases highly multiplex PCR or whole genome amplification strategies are required, which are very difficult to establish. For this reason DNA chips currently are predominantly used for re-sequencing of genes with high variability or for expression profiling.
In many cases genotyping, especially as in the elucidation of polygenic diseases, where large DNA collections have to be scanned, requires a method of analysis that can provide high throughput analysis of a limited number of markers on a large number of individuals (5). The presented method is ideally suited for this purpose.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI) has revolutionised the mass spectrometric analysis of biomolecules (6). MALDI has been applied to the analysis of DNA in variations that range from the analysis of PCR products (7,8), to approaches using sequencing (9), allele specific termination (10), single nucleotide primer extension reactions (11–14) and hybridisation with PNAs (15,16). The major drawback of these approaches is that they heavily rely on stringent purification procedures prior to MALDI analysis that do not lend themselves to easy automation and contribute to the cost of a reaction. Spin column purification and/or magnetic bead technology and reversed-phase purification are frequently applied.
We recently showed that the analysis of nucleic acids by MALDI is strongly dependent on the charge state, and a 100-fold increase in analysis sensitivity can be achieved, when the DNA is conditioned to carry one positive charge or one negative charge (17–19). These key breakthroughs set the stage for the implementation of modification chemistry into a molecular biological procedure, as such modified DNA products are also significantly less susceptible to adduct formation. The issue of using a MALDI mass spectrometer for genotyping SNPs is thereby shifted to the efficiency of producing allele-specific products.
Here we implement the principle of sensitivity increasing chemical modifications into a facile molecular biological procedure for generating binary, allele-specific products and sample conditioning with detection by MALDI mass spectrometry. We have termed this genotyping procedure the ‘GOOD Assay’. This assay requires no purification and is achieved by simple addition of reagent solutions into a single reaction tube and incubations.
We have applied the GOOD Assay to a mutation in codon 717 in the amyloid precursor protein gene which is associated with autosomal dominant Alzheimer’s disease (20,21) and to three single nucleotide polymorphisms of the human gene for granulocyte-macrophage colony stimulating factor (GM-CSF). GM-CSF is a growth factor with a proliferative stimulus for bone marrow neutrophil stem cell precursors. It plays an important role as a component of the regulation of the neutrophil-mediated inflammatory response (22,23).
MATERIALS AND METHODS
Oligonucleotides were synthesized by an in-house synthesis service and were all HPLC purified. For the amplification of exon 17 of APP primers 5′-CTTAATTTGTTTTCAAGGTGTTC-3′ (forward) and 5′-TGCAGTCAAGTTTACCTAC-3′ (reverse) were used to give a 183 bp PCR product. 5′-TGT-CATAGCGACAGTGAPTTNH2PTC-3′ was used as primer for the primer extension reaction. This primer contains phosphorothioate bridges (PT) 5′ and 3′ of TNH2. TNH2 is an amino modifier C6 dT (Glen Research, Sterling, VA). Its amino functionality was used for attaching a positive charge tag according to Gut et al. (18) and Bartlett-Jones et al. (24). For the amplification of part of GM-CSF, primers 5′-TGTGCACATGGTGGTCAT-3′ (forward) and 5′-GAATCTCCTGGCCCTTATC-3′ (reverse) were used to give a 500 bp PCR product. 5′-CTTACTGGACTGAGGTTPTGC-3′ (a C/A SNP is located at position 99 of the PCR product), 5′-CCAGGAAGTCCAAACTGPTTG-3′ (a C/T SNP is located at position 345 of the PCR product) and 5′-GAGAGCCCTCAGGAAPTGG-3′ (a C/T SNP is located at position 417 of the PCR product) were used as primers for the primer extension reaction. Phosphorothioate groups inhibit the 5′-phosphodiesterase digest. The phosphate between nucleoside 1 and 2 from the 3′-end remains as the negative charge tag on the products of 5′-phosphodiesterase digest and alkylation.
dNTPs, Taq DNA polymerase and 5′-phosphodiesterase (calf spleen) were purchased from Boehringer Mannheim (Mannheim, Germany), α-S-dNTPs were purchased from Amersham (Little Chalfont, UK). α-S-ddNTPs were provided by Amersham (Little Chalfont, UK). Thermosequenase and shrimp alkaline phosphatase were purchased from Amersham Buchler (Braunschweig, Germany). PlatinumTM Taq DNA polymerase high fidelity (TPHF) was purchased from Gibco BRL (Karlsruhe, Germany). Standard reagents were purchased from Aldrich (Steinheim, Germany).
The GOOD Assay for positive ion mode MALDI genotyping
PCR. One microlitre of genomic DNA (10 ng), extracted from full blood according to the procedure described in Sambrook et al. (25), was used as template for PCR. Five picomoles of the forward and 5 pmol of the reverse primers were used in 40 mM Trisbase, 32 mM (NH4)2SO4, 50 mM KCl and 2 mM MgCl at pH 8.8 with 200 µM dNTPs and 0.2 U Taq DNA polymerase in a 10 µl vol. The reaction was denatured for 2 min at 95°C, then thermocycled for 20 s at 95°C, 30 s at 58°C and 30 s at 72°C 30 times.
SAP digest. Shrimp alkaline phosphatase (SAP) (0.5 µl, 1 U/µl) was added to the PCR reaction and incubated at 37°C for 1 h. The SAP was denatured for 10 min at 90°C.
Primer extension reaction. Twenty-five picomoles of the charge tagged primer were added together with 100 µM α-S-dNTPs (reduced set, at least one of the four bases is omitted and in some cases replaced with the corresponding α-S-ddNTP) and 0.5 U Thermosequenase. The reaction volume was increased to 20 µl by the addition of water. An initial denaturing step of 3 min at 95°C was used followed by 40 cycles of 20 s at 95°C, 1 min at 58°C and 1 min at 62°C.
Primer removal. One microlitre of a 0.5 M acetic acid solution was added to the processed extension reaction resulting in a reaction pH <7.0. Then 2 µl of 5′-phosphodiesterase, that was previously dialysed against ammonium citrate (0.1 M, pH 6.0), was added and the reaction incubated for 45 min at 37°C.
Alkylation reaction. Forty-five microlitres of acetonitrile, 15 µl of triethylammonium bicarbonate solution (pH 8.5) and 14 µl of CH3I were added. The reaction was incubated at 40°C for 25 min. Upon cooling a biphasic system was obtained. Twenty microlitres of the upper layer was sampled and diluted in 45 µl of 40% acetonitrile. This solution was directly used to transfer the samples onto the matrix.
Sample preparation for MALDI analysis. The α-cyano-4-hydroxy-cinnamic acid methyl ester (CNME) matrix was prepared by spotting 0.5 µl of a 1% solution in acetone onto the target and spotting 0.3 µl of a solution of the sample in 40% acetonitrile on top of the dried matrix. Forty percent acetonitrile dissolves the surface layer of the matrix allowing a concentrated incorporation of the analytes into the matrix surface. By this preparation, a very thin and fine crystalline matrix layer was achieved. CNME was synthesized according to the method of Gut et al. (18).
Mass spectrometric analysis. Spectra were recorded on a Bruker Reflex II time-of-flight mass spectrometer. This mass spectrometer is equipped with a reflector and upgraded with delayed extraction. Spectra were recorded in positive ion mode and detected linearly. Acceleration potentials typically chosen were 20 kV. For delayed extraction the acceleration potential was switched with a delay of 200 ns.
The GOOD Assay for negative ion mode MALDI genotyping
PCR. One microlitre of genomic DNA (5 ng) extracted from full blood according to the procedure described in Sambrook et al. (25) was used as template for PCR. Five picomoles of the forward and 5 pmol of the reverse primer were used in 1 µl of TPHF (10×) buffer (pH 8.9), 0.5 µl MgSO4 (50 mM), 200 µM dNTPs and 0.075 U TPHF DNA polymerase in a 10 µl vol. The reaction was denatured for 1 min at 94°C, then thermocycled for 15 s at 94°C, 30 s at 56°C and 1 min at 68°C 30 times (the identical PCR protocol as for the WAVE Denaturing HPLC detection of the SNPs is used in one-fifth of the volume).
SAP digest. One and a half microlitres MgSO4 (50 mM) was added, otherwise the procedure was done as above.
Primer extension. This was the same as above, except that in the case of multiplexing the amount of each primer was reduced to 8 pmol.
Primer digest, the alkylation reaction and sample preparation for MALDI analysis. These were done the same as described above.
Mass spectrometric analysis. Spectra were recorded in negative ion mode, detected linearly with an acceleration potential of typically 18 kV. For delayed extraction the acceleration potential was switched with a delay of 200 ns.
RESULTS
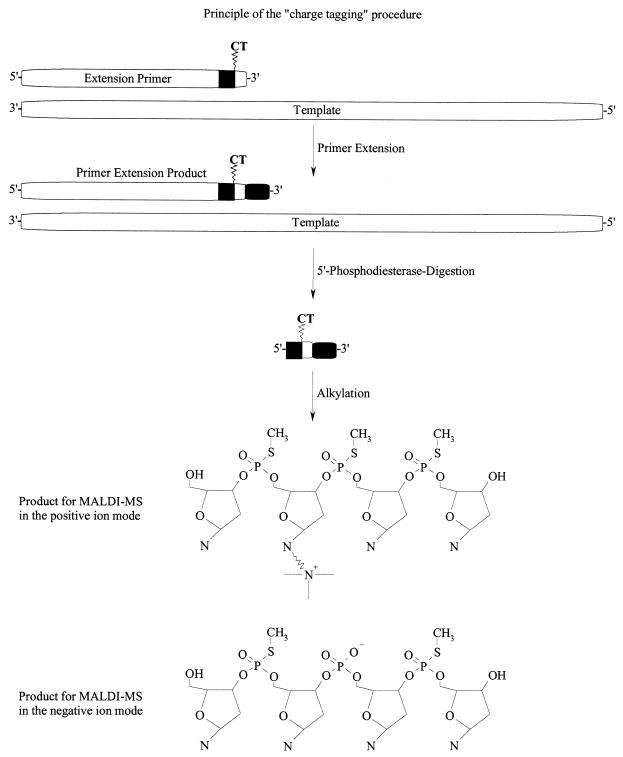
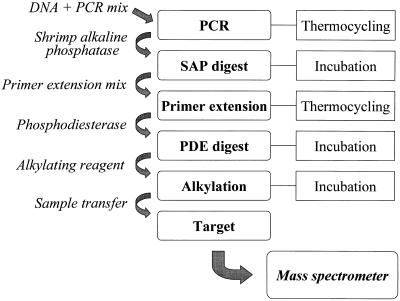
A procedure was established for the facile analysis of known SNPs. The procedure was comprised of amplifying a specific stretch of DNA containing a mutation of interest. Residual dNTPs that disturb an allele-specific product generation are removed with shrimp alkaline phosphatase. A new, tailored set of α-S-dNTPs and α-S-ddNTPs, a primer conditioned for sensitive detection by MALDI mass spectrometry and a DNA polymerase are added for thermocyclic primer extension (Fig. 1). Templates, primers of the PCR and parts of the primers used for the primer extension are digested with 5′-phosphodiesterase. Phosphorothioate bridges incorporated into the conditioned primers inhibit the full digestion of the allele-specific products. By quantitatively alkylating the phosphorothioate bridges the products are conditioned for sensitive detection by MALDI. The procedure is executed by the simple addition of reagents for the next reaction step and further incubation (Fig. 2).
Figure 1.
Principle of the method. A primer containing the charge tag near the 3′-end is used to make an allele-specific product with α-S-dNTP building blocks. The charge tag function is neighboured by phosphorothioate bridges and extension with α-S-dNTPs results in a product with a phosphorothioate backbone. The method can be expanded by the use of α-S-ddNTPs. After extension, the part of the primer containing regular phosphate bonds is digested off with 5′-phosphodiesterase. The first phosphorothioate group from the 5′-end inhibits the digestion of the product. The phosphorothioate functions are quantitatively alkylated and the products are transferred to MALDI analysis without purification.
Figure 2.
Pipetting scheme for the GOOD Assay starting from genomic DNA to the assignment of an allele after MALDI analysis. The reaction sequence is executed in a single tube.
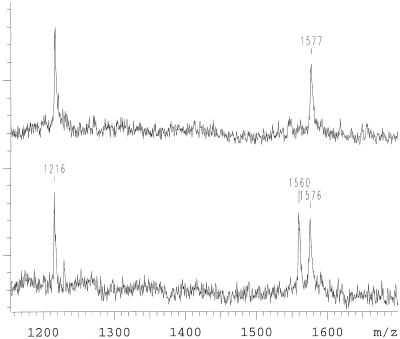
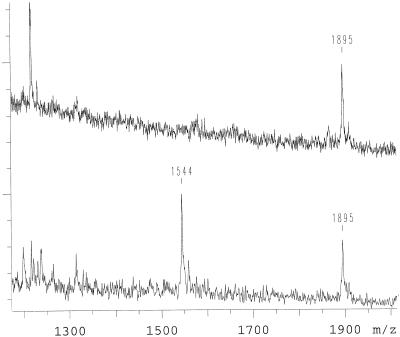
For positive ion detection a G→A mutation in position 1 of codon 717 of the amyloid precursor protein gene (APP) is shown. This mutation results in a valine to isoleucine amino acid transition. The PCR product of exon 17 encompasses nine known mutations. For the generation of an allele-specific product a primer modified as described in Materials and Methods was positioned directly next to position 1 of codon 717. Priming of the APP exon 17 PCR product with the modified oligonucleotide is efficient. By using different combinations of α-S-dNTPs and α-S-ddNTPs different allele-specific products were generated. Using α-S-dGTP, α-S-dATP and α-S-ddTTP a wild-type template results in a product of the sequence 5′-ATCTCGT-3′ with a measured mass of 1894 Da, while the mutation results in 5′-ATCTCAT-3′ with a mass of 1878 Da (Fig. 3). Equivalent results were obtained by omitting the terminating base and only using α-S-dGTP and α-S-dATP (Fig. 4). The products are 5′-ATCTCG-3′ with a measured mass of 1576 Da for the wild-type and 5′-ATCTCA-3′, with a mass of 1560 Da for the mutant. We found that incorporation of α-S-dNTPs opposite complementary bases is efficient, fidelity is very high, and the absence of the next complementary base results in termination. There is also the possibility to separate the product bands more by using two terminating bases (Fig. 5). In this approach α-S-dGTP, α-S-ddATP and α-S-ddTTP were added and resulted in a product of the sequence 5′-ATCTCGT-3′ with the measured mass of 1895 Da for the wild type and 5′-ATCTCA-3′, with a mass of 1544 Da for the mutant. Five further combinations were tried. All combinations gave allele-specific, distinguishable products. For the examples shown in Figures 3 and 4 the allele product masses are separated by only 16 Da. Wrong incorporations were never observed. In some cases, where more than one base was added for an allele specific, early termination of the primer extension was detected, but full-length primer extension products were predominant. Residual primer (1216 Da) was observed and can be used as the mean for quality control. For confirmation, 10 patient DNAs that were genotyped blind in this study were sequenced. The mass spectrometric genotyping corresponded with the DNA sequencing result.
Figure 3.
Mutation analysis of an individual affected by a G→A mutation in position 1 of the APP 717 codon. α-S-dGTP, α-S-dATP and α-S-ddTTP are used in the primer extension reaction. At 1216 Da residual primer is observed. The mutant allele is observed at 1878 Da and wild-type allele at 1894 Da. The top is the spectrum of an individual who is wild type, the bottom an individual carrying the mutation on one allele.
Figure 4.
Another variant of a mutation analysis as it is shown in Figure 3. α-S-dGTP and α-S-dATP are used in this primer extension reaction. The mutant allele is observed at 1560 Da and the wild-type allele at 1576 Da. The top is the spectrum of an individual who is wild type, the bottom an individual carrying the mutation on one chromosome. There is a 1 Da difference between the wild-type alleles of the two analyses. This is due to rounding of the raw data. It represents the largest difference detected in this study.
Figure 5.
Another variant of a mutation analysis as it is shown in Figure 3. α-S-dGTP, α-S-ddATP and α-S-ddTTP are used in the primer extension reaction. The mutant allele is observed at 1544 Da and wild-type allele at 1895 Da. The top is the spectrum of an individual who is wild type, the bottom an individual carrying the mutation on one chromosome.
Assays for published point mutations in codons 665, 670/671, which are in exon 16 and 692/693, 713, 716, which are in exon 17 were also established and gave appropriate mass signals with wild-type DNA. Experiments with DNA carrying mutations were not done, as we are not in possession of DNA from individuals harbouring these mutations.
For negative ion detection we show three different polymorphisms of the human gene for GM-CSF: a C→A transition at position 99, a C→T transition at position 345 and a C→ T transition at position 417 of the generated PCR product. Extension primers were positioned immediately next to the position of the SNPs. The products after primer extension, 5′-phosphodiesterase digestion and alkylation were 4mers containing a single negative charge derived from the phosphate bridge between nucleoside 1 and 2 from the 3′-end of the extension primer.
The different polymorphisms were investigated individually (Fig. 6) and in multiplex (Fig. 7). The products at position 99 with the sequences TGCC or TGCA have corresponding masses 1191 and 1215 Da, the products at position 345 with the sequences GTGC or GTGT have corresponding masses 1231 and 1246 Da, and the products at position 417 with the sequences AGGC or AGGT have corresponding masses 1240 and 1255 Da. The alleles of all polymorphisms can be easily called both in individual and multiplex analysis. As a control the patient DNAs used in this study were sequenced for confirmation. The mass spectrometric analysis corresponded with the DNA sequences in all cases. Fifteen patient DNAs were used for this study.
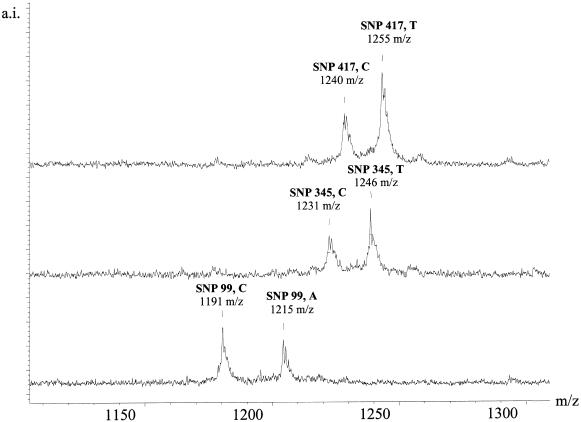
Figure 6.
The GOOD Assay in negative ion mode applied to polymorphisms in position 99, 345 and 417 of GM-CSF with individuals that are heterozygous. The signals corresponding to the two alleles are easily assigned and distinguished.
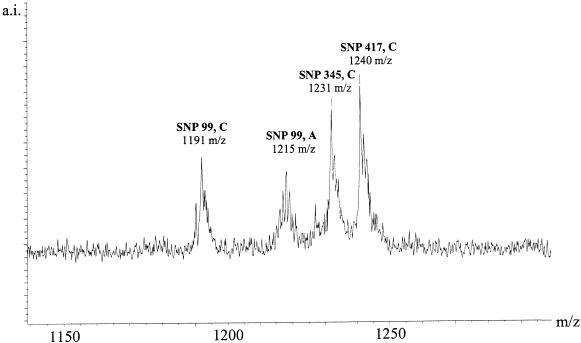
Figure 7.
Multiplexed genotyping by the GOOD Assay in negative ion mode of an individual that is heterozygous in position 99 and homozygous allele C in position 345 and homozygous allele C in position 417 of GM-CSF.
DISCUSSION
A novel procedure for genotyping SNPs termed the GOOD Assay is demonstrated. The GOOD Assay implements sensitivity increasing DNA modifications into a primer extension procedure for generating allele-specific products and subsequent analysis by MALDI mass spectrometry without requiring purification. It is applied to genotyping mutations in the APP gene in healthy and affected individuals and to three SNPs in GM-CSF in single and multiplex. Two variants of the GOOD Assay are demonstrated, one a variant with positive ion detection, which requires a specially modified oligonucleotide, but has the advantage that it has a greater scope for multiplexing and a more accessible variant with negative ion detection.
Here we demonstrate how to generate allele-specific products that are exclusively detectable under the chosen MALDI conditions. The entire process requires only one tube or microtitre plate into which in successive reaction steps appropriate reagents are dispensed. The product is transferred onto the MALDI target without purification. This is in stark contrast to any other SNP genotyping method using MALDI as the detection system (7–16). Of the starting PCR <0.5% are used for the MALDI sample preparation and in the MALDI analysis only a fraction of this is used. With a PCR machine that is capable of handling smaller sample volumes reliably, reagent consumption could be further reduced dramatically.
As was recently shown, charge tagging and backbone neutralisation of DNA result in a dramatic increase in sensitivity and decrease in susceptibility to form adducts with constituents (17,18). The GOOD Assay provides a streamlined method for the integration of these modifications into allele-specific product generation. Due to the increased detectability of modified products the reaction mixture can be diluted. Through the dilution in the GOOD Assay reaction constituents are diluted without the allele-specific products falling beneath the detection threshold. The biphasic system of the alkylation reaction also helps to remove some disturbing reagents (i.e. detergents from enzymes). The modified allele-specific products are observable without requiring purification prior to MALDI analysis. The bandwidth of reagents that are tolerated by the process is quite large, so that different PCR conditions (DNA polymerases and buffers) can be coupled into the procedure. In the example shown with GM-CSF, the PCR is done according to the suppliers specification with the provided buffer.
The nature of the matrix chosen for the MALDI analysis in the GOOD Assay does not correspond to traditionally used matrices. It is a non-acidic matrix. As such, it might act as a discriminator for the modified products against other components of the procedure.
In the mass range of 1000 to 3000 Da, into which the products of the GOOD Assay come to lie, isotopic resolution can be achieved, and the resolution Δm/m then is better than 3000 (Fig. 6). Mass spectrometric detection ideally lends itself to the detection of multiple SNPs in a single preparation, as the resolution power easily allows the distinction of peaks separated by as little as 4 Da in this mass range. The smallest mass difference found between two natural bases (T and A) is 9 Da. The smallest mass difference of two alleles of the interleaved SNPs in the multiplex of GM-CSF is 6 Da. This was resolved without problems. For the positive charge tag version the bandwidth for multiplexing can be increased by attaching charge tag reagents of varying masses to primers of different SNPs.
Overalkylation of the 3′ and 5′ ends of the DNA products is not a problem if reaction conditions are followed strictly.
The current limitations of the GOOD Assay in positive ion mode detection are the commercial availability of only TNH2 for the introduction of a charge tag near the 3′-end of the extension primer. It limits the positioning of the primer for the extension reaction. We have synthesized CCT, GCT and ACT accordingly and used them successfully. The synthesis and integration of these compounds will be shown elsewhere. This limitation does not exist for the procedure using negative charge tag technology. In return the degree of freedom for multiplexing is more limited.
It was the aim of this work to provide an efficient and affordable method for the analysis of SNPs. Another of the main criteria was that the procedure can be directly transferred to commercially available liquid handling robots. The GOOD Assay has been implemented on a liquid handling robot (RoboAmp 2000, MWG, Ebersberg, Germany).
With commercially available MALDI mass spectrometers being capable of recording 20 000 spectra per day a single system could be used to generate 100 000 genotypes from up to 20 000 different individuals. Reagent consumption of the GOOD Assay is already very low and has further scope for optimisation. This moves the cost of a single genotype to a fraction of cost of genotyping with current technology. It has the added advantage over gel-based technologies that each found allele gives an absolute, measured mass. This significantly contributes to the ease of allele calling.
Therapies that produce symptomatic benefit are now available for Alzheimer’s disease and trials are underway examining whether early treatments modify disease progression. If and when such disease modifying therapies become available there may be increased need to screen large numbers of samples for the presence of pathogenic point mutations. It is in the rapid and accurate screening of very large samples for mutations (in the APP or other genes) that the GOOD Assay may be particularly valuable.
The smallest fraction of genetic disease are monogenetic and a large proportion of genes responsible for monogenic diseases have been identified. In comparison the identification of genes involved in polygenic diseases is very difficult (5). Many diseases that have a major impact on the quality of life, such as cancer, cardiovascular disease, obesity or diabetes, are polygenic and therefore very difficult to identify. Their identification requires the analysis of statistically relevant numbers of samples (>1000 individuals). The presented method for SNP genotyping provides a practical solution for tackling high throughput genotyping, which is a prerequisite for the identification of the genes involved in these ailments. The effort of the SNPs consortium will provide ample numbers of markers to choose from (3).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Roman Pawlik for the synthesis of oligonucleotides. α-S-ddNTPs were kindly provided by Dr Alan Hamilton (Amersham, UK). This work was supported by Bruker-Daltonik GmbH, Bremen, in the project Genwaage of the Bundesministerium für Bildung und Forschung (BMBF). The DNA samples were from a collection of British families provided with the assistance of Dr Martin Rossor and Dr John Janssen and supported by grants from the medical research council (MRC).
REFERENCES
- 1.Landegren U., Kaiser,R., Caskey,C.T. and Hood,L. (1988) Science, 242, 229–237. [DOI] [PubMed] [Google Scholar]
- 2.Cooper D.N., Smith,B.A., Cooke,H.J., Niemann,S. and Schmidke,D. (1985) Hum. Genet., 69, 201–205. [DOI] [PubMed] [Google Scholar]
- 3.Marshall E. (1999) Science, 284, 406–407. [DOI] [PubMed] [Google Scholar]
- 4.Wang D.G., Fan,J.-B., Siao,C.-J., Berno,A., Young,P., Sapolsky,R., Ghandour,G., Perkins,N., Winchester,E., Spencer,J., Kruglyak,L., Stein,L., Hsie,L., Topaloglou,T., Hubbell,E., Robinson,E., Mittmann,M., Morris,M.S., Shen,N., Kilburn,D., Rioux,J., Nusbaum,C., Rozen,S., Hudson,T.J., Lipshutz,R., Chee,M. and Lander,E.S. (1998) Science, 280, 1077–1082. [DOI] [PubMed] [Google Scholar]
- 5.Hager J., Dina,C., Francke,S., Dubois,S., Houari,M., Vatin,V., Vaillant,E., Lorentz,N., Basdevant,A., Clement,K., Guy-Grand,B. and Froguel,P. (1998) Nature Genet., 20, 304–308. [DOI] [PubMed] [Google Scholar]
- 6.Karas M. and Hillenkamp,F. (1988) Anal. Chem., 60, 2299–2301. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y.-H., Bai,J., Zhu,Y., Liang,X., Siemieniak,D., Venta,P.J. and Lubman,D.M. (1995) Rapid Commun. Mass Spectrom., 9, 735–743. [DOI] [PubMed] [Google Scholar]
- 8.Ch’ang L.-Y., Tang,K., Schell,M., Ringelberg,C., Matteson,K.J., Allman,S.L. and Chen,C.H. (1995) Rapid Commun. Mass Spectrom., 9, 772–774. [DOI] [PubMed] [Google Scholar]
- 9.Taranenko N.I., Allman,S.L., Golovlev,V.V., Taranenko,N.V., Isola,N.R. and Chen,C.H. (1998) Nucleic Acids Res., 26, 2488–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Little D.P., Braun,A., Darnhofer-Demar,B., Frilling,A., Li,Y., McIver,R.T. and Köster,H. (1997) J. Mol. Med., 75, 745–750. [DOI] [PubMed] [Google Scholar]
- 11.Haff L. and Smirnov,I.P. (1997) Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haff L.A. and Smirnov,I.P. (1997) Nucleic Acids Res., 25, 3749–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei Z., Ono,T. and Smith,L.M. (1998) Nucleic Acids Res., 26, 2827–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross P., Hall,L., Smirnov,I. and Haff,L. (1998) Nature Biotech., 16, 1347–1351. [DOI] [PubMed] [Google Scholar]
- 15.Ross P.L., Lee,K. and Belgrader,P. (1997) Anal. Chem., 69, 4197–4202. [DOI] [PubMed] [Google Scholar]
- 16.Griffin T.J., Tang,W. and Smith,L.M. (1997) Nature Biotech., 15, 1368–1372. [DOI] [PubMed] [Google Scholar]
- 17.Gut I.G. and Beck,S. (1995) Nucleic Acids Res., 23, 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gut I.G., Jeffery,W.A., Pappin,D.J.C. and Beck,S. (1997) Rapid Commun. Mass Spectrom., 11, 43–50. [Google Scholar]
- 19.Berlin K. and Gut,I.G. (1999) Rapid Commun. Mass Spectrom., 13, 1739–1743. [DOI] [PubMed] [Google Scholar]
- 20.The Alzheimer’s Disease Collaborative Group (1995) Nature Genet., 11, 219–222.7550356 [Google Scholar]
- 21.Hardy J. (1997) Trends Neurosci., 4, 154–159. [DOI] [PubMed] [Google Scholar]
- 22.Adachi T. and Alam,R. (1998) Am. J. Physiol., 275, 623–633. [DOI] [PubMed] [Google Scholar]
- 23.Fossati G., Mazzucchelli,I., Gritti,D., Ricevuti,G., Edwards,S.W., Moulding,D.A. and Rossi,M.L. (1998) J. Mol. Med., 6, 943–951. [DOI] [PubMed] [Google Scholar]
- 24.Bartlet-Jones M., Jeffery,W.A., Hansen,H.F. and Pappin,D.J.C. (1994) Rapid Commun. Mass Spectrom., 8, 737–742. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.