Abstract
Metabolic networks are interconnected and influence diverse cellular processes. The protein-metabolite interactions that mediate these networks are frequently low affinity and challenging to systematically discover. We developed mass spectrometry integrated with equilibrium dialysis for the discovery of allostery systematically (MIDAS) to identify such interactions. Analysis of 33 enzymes from human carbohydrate metabolism identified 830 protein-metabolite interactions, including known regulators, substrates, and products as well as previously unreported interactions. We functionally validated a subset of interactions, including the isoform-specific inhibition of lactate dehydrogenase by long-chain acyl–coenzyme A. Cell treatment with fatty acids caused a loss of pyruvate-lactate interconversion dependent on lactate dehydrogenase isoform expression. These protein-metabolite interactions may contribute to the dynamic, tissue-specific metabolic flexibility that enables growth and survival in an ever-changing nutrient environment.
Metabolites are the small-molecule substrates, intermediates, and end products of metabolic pathways, and their interactions with proteins also communicate metabolic status to diverse cellular processes (Fig. 1A). Such regulatory interactions—both covalent and noncovalent—adapt cell behavior to dynamic nutrient availability and metabolic demand. The identification of protein-metabolite interactions (PMIs) has been sporadic, and strategies to discover such interactions are limited. Some progress has been made (1, 2), but the nature of many PMIs complicates their identification. For example, to maximize dynamic regulatory potential, metabolites frequently interact with their target proteins with an affinity close to their cellular concentrations—often low micromolar to low millimolar. Therefore, we developed the highly sensitive mass spectrometry integrated with equilibrium dialysis for the discovery of allostery systematically (MIDAS) platform to enable the systematic discovery of PMIs, including both low- and high-affinity interactions (3).
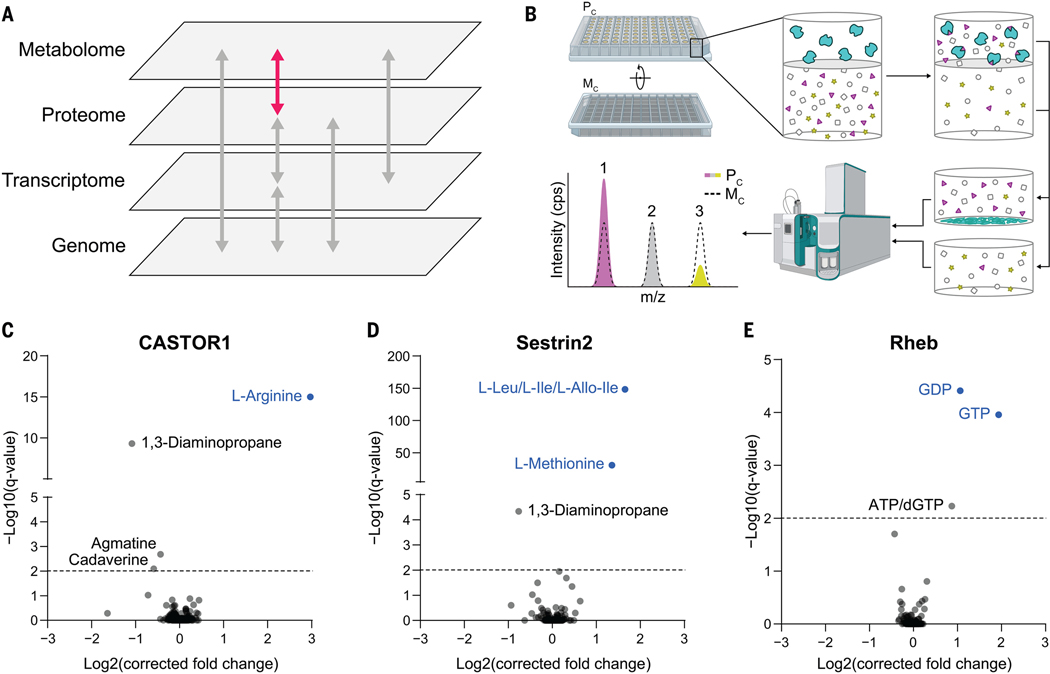
Fig. 1. MIDAS is a platform for the systematic discovery of PMIs.
(A) Biological systems are organized into domains of information (labeled gray panes). Flow of information within and between these domains is transmitted through direct interactions and underlies biological function (arrows). The MIDAS platform provides PMI discovery (pink arrow). (B) The MIDAS platform is an equilibrium dialysis tandem FIA-MS approach. (Top left and top center) Purified proteins (cyan) are loaded into the protein chamber (Pc) and defined pools of metabolites into the metabolite chamber (Mc), separated by a protein-impermeable dialysis membrane. (Top right) The system is incubated to relative equilibrium. (Bottom right and bottom center) Proteins are removed by precipitation, metabolites in the Pc and Mc are sampled, and the relative abundance of metabolites from both chambers are quantified using FIA-MS. (Bottom left) PMIs are observed as an increase (1) or decrease (3) in metabolite abundance in the Pc relative to the Mc (dotted peak). Metabolites that have equal abundance in the Pc relative to the Mc (2) are defined as noninteracting with the target protein. cps, counts per second; m/z, mass/charge ratio. (C to E) Volcano plots of MIDAS analyses of the mTORC1 regulators CASTOR1, Sestrin2, and Rheb. Significant PMIs are labeled; previously known interactions are blue. All proteins were screened by triplicate equilibrium dialysis and technical triplicate FIA-MS injections. Significant PMIs identified by MIDAS are labeled and have a Q < 0.01 (dotted line).
The MIDAS platform detects PMIs
MIDAS leverages the biophysical principle of equilibrium dialysis (Fig. 1B). Briefly, a purified protein is separated from a defined library of metabolites by a semipermeable dialysis membrane that allows diffusion of metabolites but not protein. After incubation, the system achieves relative equilibrium, such that the concentration of free (i.e., noninteracting) metabolites is similar in the protein and metabolite chambers (Fig. 1B, gray outlined symbols). However, the total concentration of those metabolites that interact with the protein is higher or lower in the protein chamber relative to the metabolite chamber dependent on binding affinity and mode of interaction (Fig. 1B, magenta triangles and yellow stars). The protein is then denatured and removed from the protein chamber, and the relative abundances of all metabolites from both chambers is quantified by high-throughput flow injection analysis–mass spectrometry (FIA-MS). The fold change between the chambers is determined and then normalized and corrected to remove nonspecific interactions (see materials and methods). A positive fold change indicates a direct PMI and is dependent on the binding affinity of the interaction. A negative fold change can result from the enzymatic conversion of the metabolite at a reaction rate faster than the diffusion rate across the membrane. PMIs that are not disrupted during protein denaturation—both covalent and noncovalent—also produce negative fold changes as the metabolite is removed with the protein.
The MIDAS metabolite library comprises 401 compounds that represent a sizable fraction of the water-soluble, chemically stable, FIA-MS–detectable, and commercially available components of the human metabolome (fig. S1A and data S1). Because of the intrinsic differences in chemical structure and ionization properties, not all metabolites could be analyzed with the same FIA-MS parameters. We profiled each metabolite individually for its optimal FIA-MS ionization and detection conditions (data S2) and, guided by these criteria, divided the library into four pools for multiplexed analysis (fig. S1B and data S1). We developed rapid FIA-MS methods, optimized for each pool, that enabled quantification of the constituent metabolites.
We performed a pilot validation study using proteins with well-characterized metabolite interactors. We analyzed three human proteins that regulate mechanistic target of rapamycin complex 1 (mTORC1): cytosolic arginine sensor for mTORC1 subunit 1 (CASTOR1), which binds arginine (4); Sestrin2, which binds leucine, isoleucine, and methionine (5); and Rheb, which hydrolyzes guanosine triphosphate (GTP) to guanosine diphosphate (GDP) (fig. S1C) (6). In each case, the known metabolite ligands of these proteins were the most enriched interactors detected (Fig. 1, C to E; see table S1 for metabolite abbreviations). In addition to known interactions, polyamine derivatives (1,3-diaminopropane, agmatine, and cadaverine) were found to bind CASTOR1 and Sestrin2, which suggests potential feedback regulation given that the mTORC1 pathway promotes polyamine synthesis in some cancers (7). Thus, MIDAS effectively identified known PMIs—regulators, substrates, and products.
MIDAS reveals inter- and intrapathway interactions across carbohydrate metabolism
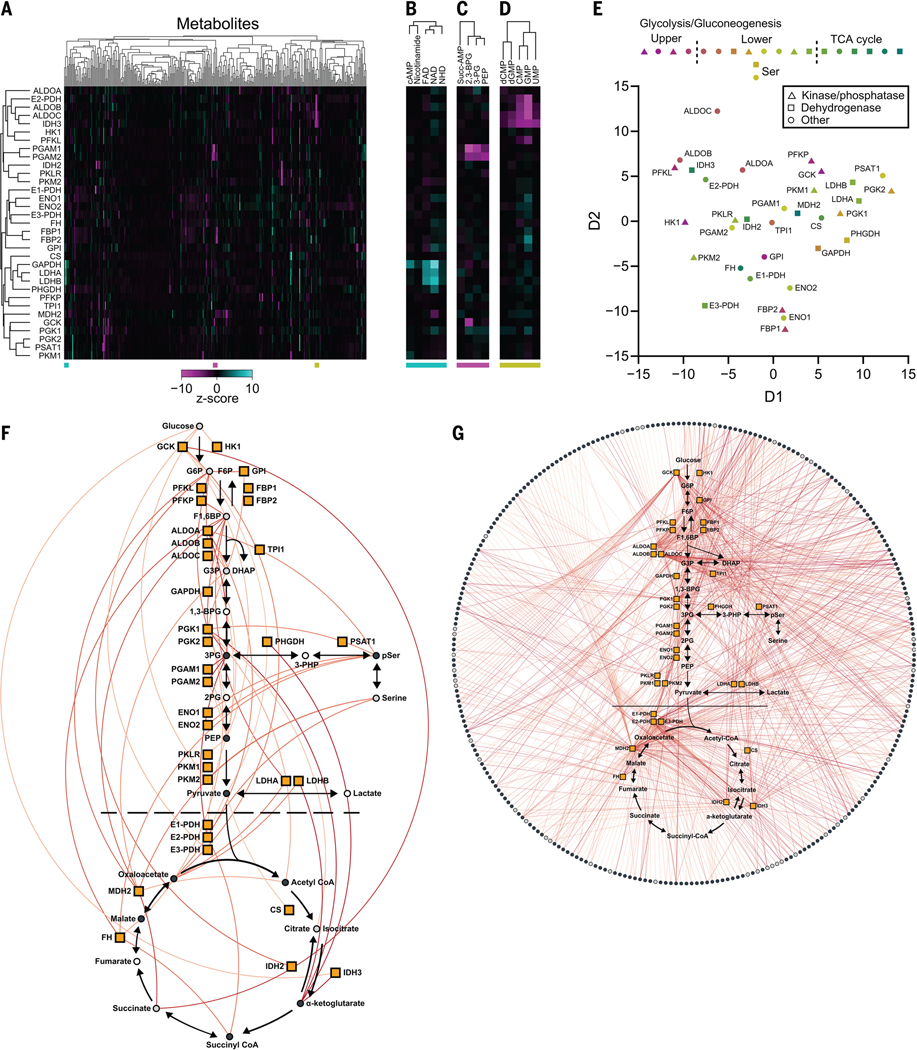
The enzymes of carbohydrate metabolism drive most cellular energy production and biosynthetic precursor generation and are known to be regulated by metabolite interactions. Therefore, we used MIDAS to profile 33 human enzymes spanning glycolysis, gluconeogenesis, the tricarboxylic acid (TCA) cycle, and the serine biosynthetic pathway that branches from glycolysis (fig. S1C). In total, we identified 830 putative PMIs, many of which were previously unknown (data S4). Unsupervised hierarchical clustering (Fig. 2, A to D) and multidimensional scaling (Fig. 2E) of the PMI dataset demonstrated that structurally and functionally related proteins frequently had similar metabolite interactions. For example, phosphoglycerate mutase 1 and 2 (PGAM1 and PGAM2), enolase 1 and 2 (ENO1 and ENO2), fructose-1,6-bisphosphatase 1 and 2 (FBP1 and FBP2), and lactate dehydrogenase A and B (LDHA and LDHB) all clustered closely with their isoform counterparts. However, this was not observed across all enzyme isoforms and isozymes, nor would it be expected given that divergent evolution enables distinct metabolic function and regulation, particularly when reflected in cell type-specific isoform expression. For example, pyruvate kinase muscle isoform 1 (PKM1) is primarily expressed in adult tissues, whereas pyruvate kinase muscle isoform 2 (PKM2) is expressed in fetal tissues and many cancer cells (8). The difference between PKM1- and PKM2-metabolite interactomes may reflect their specific, context-dependent function and regulation. Additionally, isocitrate dehydrogenase isozymes (IDH2 and IDH3), which catalyze similar chemistry but are evolutionarily and structurally unrelated (9), exhibited distinct metabolite interactomes. We observed clustering of multiple nicotinamide adenine dinucleotide (NAD)–dependent dehydrogenases: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), LDHA, LDHB, mitochondrial malate dehydrogenase (MDH2), and 3-phosphoglycerate dehydrogenase (PHGDH), which suggests that enzyme reaction class can drive the protein-metabolite interactome (Fig. 2E). An analogous clustering of structurally and functionally related metabolites was also apparent, including nicotinamide-containing metabolites and flavin-adenine dinucleotide (Fig. 2B), phosphate-containing organic acids (Fig. 2C), and several nucleotide monophosphates (Fig. 2D).
Fig. 2. The protein-metabolite interactome of human carbohydrate metabolism.
(A) Heatmap representation of MIDAS PMIs of 33 enzymes in human carbohydrate metabolism. Heatmap values are the z-score log2(corrected fold change) for all metabolites in the MIDAS metabolite library on a per-protein basis. Clustering was performed by one minus the Pearson correlation. Positive (cyan) and negative (magenta) metabolite z-score log2(corrected fold change) have a maximum and minimum cutoff of 10 and −10, respectively. MIDAS analysis of all proteins was performed by triplicate equilibrium dialysis and technical triplicate FIA-MS injections. (B to D) Excerpt examples of metabolite clustering from (A). Colored bars (bottom) indicate the location of the extracted heatmaps from (A). (E) Multidimensional scaling (MDS) of 33 human enzymes in carbohydrate metabolism based on their MIDAS PMIs. MDS distance values were generated from the z-score log2(corrected fold change) for all metabolites in the MIDAS metabolite library on a per-protein basis. (F and G) Significant intrapathway (F) and interpathway (G) interactions (colored lines) between metabolites (circles) and 33 enzymes in human carbohydrate metabolism (orange boxes) (plots generated in Electrum). Metabolites with (light gray circles) and without (dark gray circles) isomers in the same screening pool are shown. Metabolites not present in the library (open circles) are also indicated. Significant PMIs identified by MIDAS have a Q < 0.01 and are colored by increasing significance, from light orange to red.
Analysis of the 830 putative PMIs identified by the MIDAS platform showed that carbohydrates exhibited the largest number of interactions with enzymes from carbohydrate metabolism (Fig. 2F and fig. S2). This likely reflects both substrate-product relationships as well as the allosteric or orthosteric regulation of these enzymes by upstream or downstream metabolites (i.e., feedforward and feedback regulation). Most noncarbohydrate PMIs involved amino acids, nucleotides, and fatty acid derivatives. Such PMIs not only represent substrates and products of enzymes in these pathways but may reveal both intra- and interpathway regulation of carbohydrate metabolism (Fig. 2G). Because MIDAS is an in vitro platform that lacks the intracellular compartmentalization found in vivo, some of the putative PMIs are not predicted to occur in intact cells (10); however, given the physiological plasticity of protein and metabolite intracellular localization, such PMIs should not necessarily be ignored. We compared MIDAS data with previously reported PMIs in the BRENDA and Recon3D databases (11, 12) using Fisher’s exact test and found that MIDAS significantly identified known substrates and products (P < 2.0 × 10−12) and activators and inhibitors (P < 4.7 × 10−8). We propose that these MIDAS data provide a detailed view of the integration of local and distal metabolic information in carbohydrate metabolism.
Structural analysis of metabolite interactions with enolase and fumarase
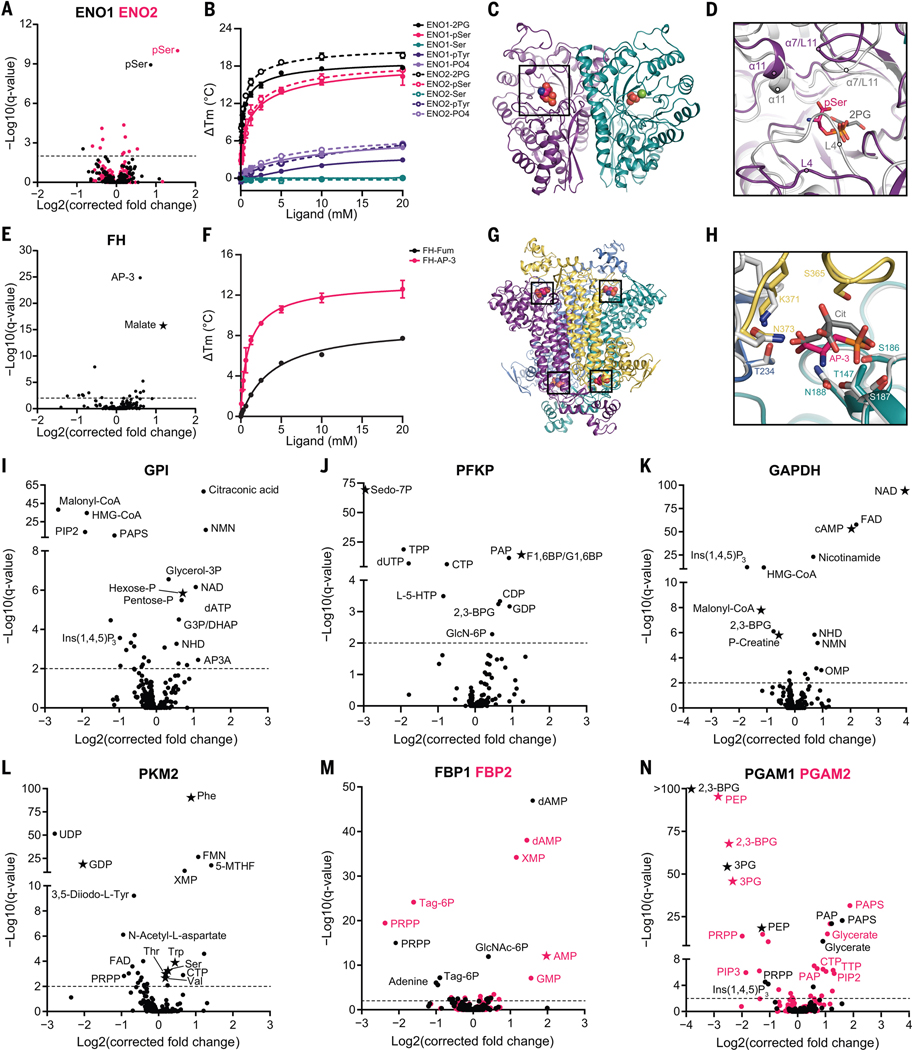
We selected a subset of PMIs for deeper bioinformatic, biochemical, and structural analysis. Enolase catalyzes the penultimate step in glycolysis, and the most enriched metabolite for both isoforms (ENO1 and ENO2) was phosphoserine (pSer) (Fig. 3A). pSer is the immediate precursor for serine biosynthesis, which diverges from glycolysis upstream of enolase (Fig. 2F). Serine allosterically activates PKM2 (13), the enzyme immediately downstream of enolase in glycolysis. Differential scanning fluorimetry (DSF) (14), which measures the changing thermal stability of a protein upon ligand binding, showed that pSer (but not serine, phosphotyrosine, or phosphate) stabilized both ENO1 [apparent dissociation constant (Kd app) = 1.38 mM] and ENO2 (KD app = 1.15 mM) (Fig. 3B) with low affinity similar to their substrate 2-phosphoglycerate (2PG) (Kd app = 0.298 mM and 0.289 mM, respectively). X-ray crystallography of the pSer-ENO2 complex showed that pSer was asymmetrically bound to the ENO2 dimer at one of the two active sites and partially overlapped with the 2PG phosphate binding site (Fig. 3, C and D, and fig. S3, A and B). Furthermore, pSer promoted an open active site conformation relative to the substrate-bound complex, observed as repositioning of loops 4 and 11 and a helices 7 and 11 (Fig. 3D). pSer only weakly inhibited in vitro enolase activity (fig. S3C). Thus, this binding event might modulate other enolase activities, such as one of its reported moonlighting functions (15, 16).
Fig. 3. MIDAS identifies known and previously undescribed metabolite interactions with enzymes from human carbohydrate metabolism.
(A) Volcano plot of MIDAS metabolite interactions with ENO1 (black) and ENO2 (pink). (B) Ligand-induced DSF melting point analysis of ENO1 (solid lines, solid circles) and ENO2 (dotted lines, open circles) with 2PG (black), pSer (pink), serine (Ser; teal), phosphotyrosine (pTyr; purple), and phosphate (PO4; light purple). (C) X-ray crystal structure of the pSer-ENO2 complex [Protein Data Bank (PDB) ID: 7MBH]. pSer (black box), phosphate ions (orange and red spheres), magnesium ion (green sphere), and monomers within the ENO2 dimer (purple and teal) are displayed. (D) Magnified view of the ENO2 active site with pSer (pink) or 2PG (gray) bound (2PG-ENO2; PDB: 3UCC) (53). Secondary structure is labeled in the pSer-ENO2 (purple) and 2PG-ENO2 (light gray) costructures. (E) Volcano plot of MIDAS metabolite interactions with fumarase (FH). (F) Ligand-induced DSF melting point analysis of FH with fumarate (Fum; black) and AP-3 (pink). [(B) and (F)] Line of best fit was determined from triplicate experiments, each with sextuplicate technical replicates using the specific binding and Hill slope equation from GraphPad Prism 9. Means ± SDs are plotted from triplicate experiments. (G) X-ray crystal structure of the AP-3–FH complex (PDB: 7LUB). AP-3 (black boxes) and monomers within the FH tetramer (purple, yellow, teal, and light blue) are shown. (H) Magnified view of the FH active site with AP-3 (pink) or citrate (Cit; gray) bound (Escherichia coli Cit-FH structure, light gray; PDB: 1FUO) (18). Side chains that coordinate the AP-3 interaction with FH are labeled and colored according to FH monomers from (G). (I to N) Volcano plots of MIDAS metabolite interactions with GPI; 6-phosphofructokinase, platelet type (PFKP); GAPDH; PKM2; FBP1 (black) and FBP2 (pink); and PGAM1 (black) and PGAM2 (pink). [(A), (E), and (I) to (N)] Stars indicate a previously known human PMI primarily sourced from BRENDA (https://www.brenda-enzymes.org/index.php). MIDAS analysis of all proteins was performed by triplicate equilibrium dialysis and technical triplicate FIA-MS injections. Specific, significant PMIs identified by MIDAS are labeled (see table S1 for metabolite abbreviations). Significant PMIs have a Q < 0.01 (dotted line). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
We identified 2-amino-3-phosphonopropionic acid (AP-3), a component of phosphonate metabolism and the transamination product of 3-phosphonopyruvate (17), as a putative interactor with fumarase, an enzyme in the TCA cycle that catalyzes the reversible hydration of fumarate to malate (Fig. 3E). AP-3 induced the thermal stabilization of fumarase (Kd app = 0.98 mM) similar to its substrate, fumarate (Kd app = 3.87 mM) (Fig. 3F). Kinetic assays demonstrated that AP-3 competitively inhibited fumarase (fig. S3D), and consistent with this, the crystal structure of the complex revealed that AP-3 binds in the active site of fumarase similarly to the known inhibitor citrate (Fig. 3, G and H, and fig. S3E) (18). Although detected in human tissues and ubiquitous in microbial metabolism (19–21), little is known about AP-3 metabolism in humans and the consequences of fumarase modulation by AP-3. These findings demonstrate that without a priori information, MIDAS can identify previously unreported, low-affinity, and functionally impactful PMIs.
MIDAS identified known and previously unknown interactions
MIDAS identified PMIs with previously known substrates, products, and regulators (Fig. 3, I to N, and fig. S3, F to L, stars). For example, glucose-6-phosphate isomerase (GPI) interacted with its substrates glucose-6-phosphate and fructose-6-phosphate (hexose-P) (Fig. 3I); phosphofructokinase (PFKP) interacted with its product, fructose 1,6-bisphosphate (F1,6BP/G1,6BP), and alternative substrate, sedoheptulose-7-phosphate (Sedo-7P) (Fig. 3J) (22); GAPDH interacted with its substrate, NAD, and regulators, cyclic adenosine monophosphate (cAMP), creatine-phosphate (P-creatine), and malonyl-coenzyme A (CoA) (Fig. 3K) (23–26); PKM2 interacted with GDP and multiple amino acid regulators (Fig. 3L) (27); and PGAM1 and PGAM2 interacted with their substrates 3-phosphoglycerate (3PG), 2,3-bisphosphoglycerate (2,3-BPG), and phosphoenolpyruvate (PEP) (Fig. 3N).
MIDAS also uncovered many previously unknown PMIs from diverse metabolic pathways (Fig. 3, I to N, and fig. S3, F to L, circles). For example, acyl-CoA, inositol phosphates, nicotinamides, adenine nucleotides, and downstream glycolytic intermediates interacted with GPI (Fig. 3I); inositol-1,4,5-trisphosphate [Ins(1,4,5)P3], 2,3-BPG, and 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) interacted with GAPDH (Fig. 3K); and PKM2 interacted with flavins, 5-methyltetrahydrofolate (5-MTHF), and a thyroid hormone intermediate 3,5-diiodo-L-tyrosine (Fig. 3L). PKM2 is known to be allosterically regulated in vitro by thyroid hormone T3 (28). Interpathway metabolite interactions were also detected with the enzymes glucokinase (GCK), liver 6-phosphofructokinase (PFKL), aldolase B (ALDOB), triosephosphate isomerase 1 (TPI1), phosphoglycerate kinase 1 (PGK1), phosphoserine aminotransferase 1 (PSAT1), and isocitrate dehydrogenase 2 (IDH2) (fig. S3, F to L). Together, these results suggest that MIDAS detects extensive protein-metabolite interplay across the metabolic network.
MIDAS analysis of multiple isoforms of metabolic enzymes demonstrated both shared and distinct metabolite interactions. Fructose bisphosphatase catalyzes the conversion of fructose-1,6-bisphosphate to fructose-6-phosphate. Both isoforms (FBP1 and FBP2) interacted with various nucleotide monophosphates and 5-phospho-D-ribose 1-diphosphate (PRPP), the end product of the pentose phosphate pathway and substrate for purine and pyrimidine metabolisms (Fig. 3M). However, only FBP1 showed an interaction with glucosamine-6-phosphate, an often-rate-limiting intermediate in the hexosamine pathway, which is derived from fructose-6-phosphate. These findings may reflect the expression differences between FBP1 (gluconeogenic tissues) and FBP2 (nongluconeogenic tissues) (https://www.gtexportal.org/home/). Similarly, isoforms of phosphoglycerate mutase (PGAM1 and PGAM2) interacted with a large set of metabolites, almost all of which were identical between them, except for Ins(1,4,5)P3 with PGAM1 and phosphatidylinositol-4,5-bisphosphate C-6 (PIP2) and phosphatidylinositol-3,4,5-trisphosphate C-6 (PIP3) with PGAM2 (Fig. 3N). This might reflect differential membrane recruitment and/or regulation of phosphoglycerate mutase isoforms by phosphoinositide kinases, which are activated by growth factor signaling (29). PMI differences between isoforms or isozymes may inform their specific function and regulation.
LDHA is inhibited by adenosine triphosphate (ATP)
Lactate dehydrogenase (LDH) catalyzes the reduction of pyruvate to lactate coincident with the oxidation of NADH to NAD. Consumption of pyruvate, the end product of glycolysis, by LDH competes with its mitochondrial uptake and oxidation by the TCA cycle to maximize ATP production. When mitochondrial pyruvate oxidation is limited, such as in hypoxia or aerobic glycolysis, LDH is required to regenerate NAD to enable continued glycolytic flux. The LDH reaction is reversible and is required to use lactate, a major circulating carbohydrate in mammals (30), as a fuel to support cellular functions. LDH is thus a key node in carbohydrate metabolism.
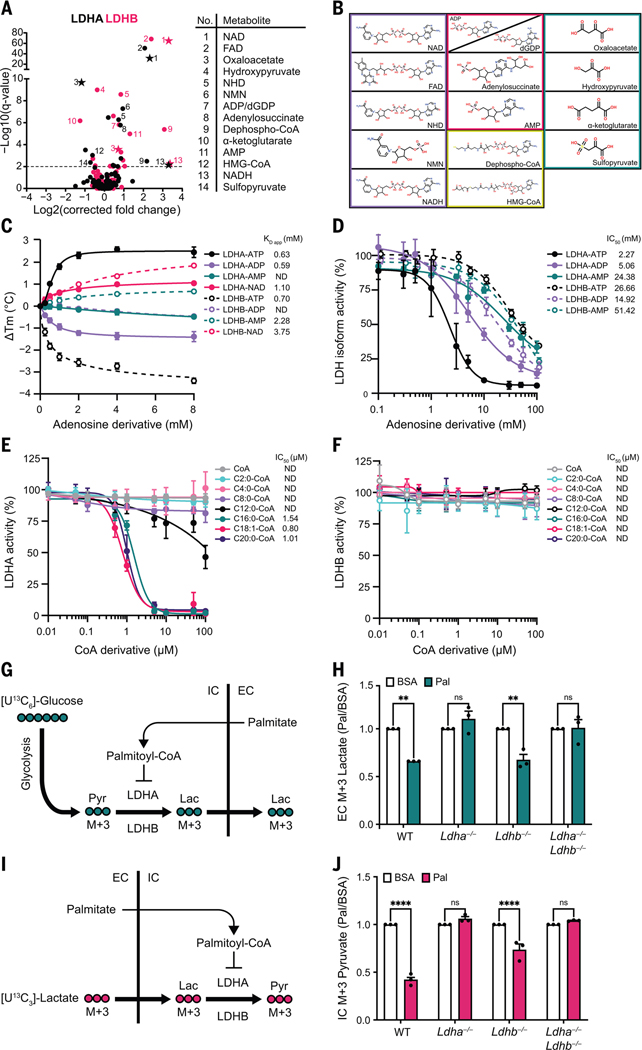
The two major isoforms, LDHA and LDHB, have distinct substrate reaction kinetics and tissue expression (31). MIDAS analysis of LDHA and LDHB revealed interactions with several metabolites, most of which were common to both proteins (Fig. 4A). These included the substrates NADH and NAD and the structurally related nucleotides, nicotinamide mononucleotide and flavin adenine dinucleotide, as well as the competitive inhibitor, oxaloacetate (32), and other keto acids related to the LDH substrates lactate and pyruvate (Fig. 4, A and B). We also observed two other classes of interacting metabolites, adenosine nucleotides and free and acylated CoA. Using DSF, we found that ATP interacted with LDHA and LDHB with a KD app = 0.636 mM and 0.697 mM, respectively (Fig. 4C), which is a low and biologically relevant affinity given that the intracellular steady-state ATP concentration range is 1 to 8 mM (33). The observed interactions of either LDH isoform with adenosine diphosphate (ADP) and adenosine monophosphate (AMP) may not be physiologically relevant given the disparity between the KD app values and the cellular concentrations of ADP and AMP [~0.4 and ~0.04 mM, respectively (34)] (Fig. 4C). Enzymatic activity assays of the two LDH isoforms further supported this conclusion because both AMP and ADP inhibited LDHA and LDHB only at supraphysiological concentrations (Fig. 4D). Despite similar binding affinities to both LDHA and LDHB (Fig. 4C), ATP inhibited only the LDHA isoform, with a half maximal inhibitory concentration (IC50) of 2.3 mM, and this inhibition appeared to be competitive with NAD and lactate (Fig. 4D and fig. S4A). This isoform-specific inhibition could relate to the opposing effects of ATP binding on the thermal stability of the two proteins (Fig. 4C).
Fig. 4. ATP and long-chain acyl-CoAs inhibit LDH in an isoform-specific manner.
(A) Volcano plots of MIDAS metabolite interactions with LDHA (black) and LDHB (pink). Specific, significant metabolites are numbered and labeled. Stars indicate a previously known human PMI, primarily sourced from BRENDA (https://www.brenda-enzymes.org/index.php). MIDAS analysis of LDHA and LDHB was performed by triplicate equilibrium dialysis and technical triplicate FIA-MS injections. Significant PMIs identified have a Q < 0.01 (dotted line). (B) Metabolite classes that interact with LDHA and LDHB from (A) (nicotinamides and dinucleotides, purple; adenosine nucleotide derivatives, pink; CoA derivatives, yellow; keto acids, teal). (C) Ligand-induced DSF melting point analysis of LDHA (solid lines, filled circles) and LDHB (dotted lines, open circles) with ATP (black), ADP (light purple), AMP (teal), and NAD (pink). KD app was determined from triplicate experiments, each with sextuplicate technical replicates, by fitting the specific binding and Hill slope equation from GraphPad Prism 9. Means ± SDs are plotted from triplicate experiments. (D) Enzyme activity of LDHA (solid lines, filled circles) and LDHB (dotted lines, open circles) treated with ATP (black), ADP (light purple), or AMP (teal). (E and F) Enzyme activity of LDHA or LDHB treated with CoA (gray), acetyl-CoA (C2:0-CoA; cyan), butyryl-CoA (C4:0-CoA; light pink), octanoyl-CoA (C8:0-CoA; light purple), lauroyl-CoA (C12:0-CoA; black), palmitoyl-CoA (C16:0-CoA; teal), oleoyl-CoA (C18:1-CoA; pink), and saturated arachidoyl-CoA (C20:0-CoA; purple). [(D) to (F)] IC50 was determined from triplicate experiments, each with triplicate technical replicates using GraphPad Prism 9. ND, not determined. Means ± SDs are plotted from triplicate experiments. (G) Schematic of [U13C6]-glucose metabolism in cells treated with palmitate-conjugated BSA after inhibition of the mitochondrial pyruvate carrier with UK5099. Pyr, pyruvate; Lac, lactate; IC, intracellular; EC, extracellular. (H) Fold change of extracellular [U13C3]-lactate collected from the growth media of the indicated H9c2 cell lines in response to treatment with palmitate-conjugated BSA (Pal) relative to BSA-vehicle control (BSA). Absolute abundance is displayed in fig. S4H. (I) Schematic of [U13C3]-lactate metabolism in cells treated with palmitate-conjugated BSA after inhibition of the mitochondrial pyruvate carrier with UK5099. (J) Fold change of intracellular [U13C3]-pyruvate in indicated H9c2 cell lines in response to treatment with palmitate-conjugated BSA (Pal) relative to BSA-vehicle control (BSA). Absolute abundance is displayed in fig. S4J. [(H) and (J)] Experiments were performed in triplicate, and means ± SDs are displayed. A two-way analysis of variance (ANOVA) and Sidak’s multiple comparison test (GraphPad Prism 9) was performed between Pal and BSA samples (ns, not significant; **P < 0.005; ****P < 0.0001).
LDHA, but not LDHB, is inhibited by fatty acyl-CoAs in vitro and in cells
We investigated the putative interaction between the LDH isoforms and CoA or CoA conjugated to short-, medium-, or long-chain fatty acids (i.e., acyl-CoAs). Esterification of long-chain (>12 carbons) fatty acids to CoA is required for their intracellular diffusion and transport into the mitochondrial matrix, where they undergo β oxidation to fuel ATP production (35). The accumulation of these long-chain acyl-CoA species is a signal of carbon fuel excess (36). We observed that acyl-CoAs inhibited LDHA as a function of fatty acid chain length. Neither CoA alone nor any acyl-CoA with a fatty acid chain length of up to eight carbons affected enzyme activity, and C12:0-CoA (lauroyl-CoA) only inhibited LDHA with an IC50 >100 μM (Fig. 4E). However, long-chain acyl-CoAs, such as C16:0-CoA (palmitoyl-CoA), C18:1-CoA (oleoyl-CoA), and C20:0-CoA (arachidoyl-CoA), all inhibited LDHA with IC50 values of ~1 μM (Fig. 4E). The inhibition of LDHA by palmitoyl-CoA was noncompetitive with respect to both NAD and lactate, which suggests that it likely binds to LDHA outside of the active site (fig. S4B). Notably, LDHB, which shares 75% amino acid sequence identity with LDHA, was completely insensitive to all tested acyl-CoAs, even at concentrations up to 100 μM (Fig. 4F).
Having observed that palmitoyl-CoA inhibited LDHA but not LDHB, we used two orthogonal approaches to test for a physical interaction. In a DSF assay, low-micromolar concentrations of palmitoyl-CoA (similar to the IC50) induced the formation of a distinct thermolabile species of LDHA and a thermostable species of LDHB (fig. S4C). LDHA and LDHB also bound to palmitoyl-CoA immobilized on agarose beads, and the binding of either protein was disrupted by free palmitoyl-CoA but not by buffer or C2:0-CoA (acetyl-CoA) (fig. S4D). These data indicate that LDHA and LDHB directly interact with palmitoyl-CoA with low-micromolar affinity.
Given that palmitoyl-CoA inhibited LDHA at physiological concentrations, we tested whether this inhibition occurs in cells. We performed metabolic tracing experiments using H9c2 rat cardiomyoblasts, which were chosen because of their native expression of both isoforms, wherein we deleted the Ldha gene, the Ldhb gene, or both (fig. S4E). We treated cells with 13C-labeled glucose in the presence or absence of bovine serum albumin (BSA)–conjugated palmitate, which allows for efficient delivery of the fatty acid into the cell, where it is esterified to palmitoyl-CoA (Fig. 4G). We used mass spectrometry to measure the uptake and assimilation of 13C into lactate. All four cell lines [wild-type (WT), Ldha−/−, Ldhb−/−, and Ldha−/−Ldhb−/−] showed a similar (~80%) increase in intracellular palmitate after incubation with its BSA conjugate (fig. S4F). Palmitate decreased the labeling of lactate from 13C-glucose in WT and Ldhb−/− cells but not in cells lacking LDHA (Fig. 4H and fig. S4, G and H), which demonstrates that palmitate inhibition of glucose-to-lactate conversion is dependent on LDHA in these cells. Multiple enzymes in carbohydrate metabolism are sensitive to acyl-CoA abundance (37–41), so to more specifically interrogate the conversion of lactate to pyruvate by LDH, we performed experiments wherein we followed the conversion of 13C-lactate to 13C-pyruvate (Fig. 4I). Again, treatment with palmitate blunted the generation of m+3 pyruvate in WT and Ldhb−/− cells, but pyruvate labeling in Ldha−/− or Ldha−/−Ldhb−/− cells was unaffected (Fig. 4J and fig. S4, I and J).
To test the possibility that upstream or downstream intermediates in fatty acid metabolism inhibit LDHA, we performed 13C-glucose and 13C-lactate tracing experiments in the presence of triacsin C, an inhibitor of acyl-CoA synthase, which catalyzes fatty acid conjugation to CoA (fig. S5, A and C) (42). In both experiments, triacsin C prevented palmitate-mediated inhibition of lactate and pyruvate labeling (fig. S5, B and D), thus demonstrating that conjugation to CoA is required for palmitate to inhibit LDHA activity. To determine whether catabolism of acyl-CoAs is required for their inhibition of LDHA, we performed experiments using 2,2-dimethyl-palmitate (DiMePal) or 2,2-dimethyl-stearate (DiMeSte) (fig. S6A). DiMePal and DiMeSte are dimethylated fatty acid analogs that can be conjugated to CoA by acyl-CoA synthase but cannot be further metabolized through β oxidation (fig. S6, A and B). Similar to palmitoyl-CoA, DiMePal-CoA inhibited LDHA but not LDHB in vitro (fig. S6, C and D). Tracing with either 13C-glucose or 13C-lactate was inhibited by DiMePal or DiMeSte (fig. S6, E to I). These results suggest that the inhibition of LDHA by palmitate is mediated by long-chain acyl-CoAs and not by upstream or downstream fatty acid intermediates.
Discussion
Both ATP and long-chain acyl-CoAs preferentially inhibited LDHA but not LDHB. LDHA and LDHB, the two dominant isoforms of LDH, are expressed in a tissue-specific pattern such that the liver almost exclusively expresses LDHA, whereas the heart has high expression of LDHB (fig. S7, A and B). The IC50 for inhibition by ATP is well within the range of normal intracellular ATP concentrations, so LDHA may be partially inhibited in all cells with normal energy status. Given that the liver, the most LDHA-dominant tissue, catabolizes multiple substrates, inhibition by ATP might be a mechanism to spare carbohydrates, like lactate, for other tissues. The liver and heart have very different metabolic demands that mirror their LDHA and LDHB expression differences, especially in the context of fatty acids. The heart is a metabolic omnivore (43), acquiring energy from multiple nutrient sources. Expression of LDHB enables carbohydrate metabolism, particularly lactate uptake and catabolism, even in the context of active fatty acid metabolism (and potentially high acyl-CoA concentration). The liver plays a distinct and critically important role in organismal metabolic homeostasis. LDHA inhibition by acyl-CoAs could be a mechanism for the unexpected interplay of lactate, fatty acids, and gluconeogenesis observed in animal studies (44, 45). Analysis of 928 cancer cell lines from DepMap (46, 47) has revealed a stronger negative correlation between lactate and long-chain acyl-carnitines (intermediates in fatty acid metabolism) in the 70 cell lines that primarily express LDHA (LDHAHi LDHBLo) relative to 858 cell lines that express both LDHA and LDHB (LDHAHi LDHBHi) (fig. S7, C to F). LDHA-specific inhibitors have been proposed to block aerobic glycolysis in cancers (48, 49), where perhaps the isoform-specific regulatory mechanism or mechanisms of ATP and acyl-CoAs could be exploited therapeutically.
This interpathway regulation between fatty acid and carbohydrate metabolisms is just one potential example of the myriad metabolite-driven regulatory events that enforce organismal homeostasis, which is vital to appropriately respond to stressors such as the feed-fast cycle, exercise, and infection. Interactions between proteins and metabolites may mediate much of this control. We validated MIDAS as a platform for the discovery of these critical mechanisms, particularly for the detection of low-affinity interactions. In complement to recent discoveries of functionally important PMIs (50–52), MIDAS identified hundreds of putative interactions with the enzymes of carbohydrate metabolism. Therefore, MIDAS serves as a conduit to identify, understand, and exploit previously unknown modes of metabolic regulation across the protein-metabolite interactome.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Rutter laboratory for helpful discussions and comments on the manuscript. We thank D. E. Gottschling (Calico Life Sciences LLC) for helpful discussions throughout the project. We thank Roche and Navitor for providing proteins for MIDAS analyses. We thank director C. Davey of the University of Utah Mutation Generation and Detection Core for providing CRISPR reagents, cell genotyping services, and construction of bacterial expression vectors. We thank the director B. Luo of the University of Utah Drug Discovery Core Facility for generating mutant cell lines. Metabolomics analysis was performed in the Metabolomics Core Facility at the University of Utah. Mass spectrometry equipment was obtained through NCRR Shared Instrumentation grants 1S10OD016232-01,1S10OD018210-01A1, and 1S10OD021505-01. The support and resources from the Center for High Performance Computing at the University of Utah are gratefully acknowledged. J.R. is an investigator of the Howard Hughes Medical Institute.
Funding:
This study received support from National Institutes of Health grants T32DK091317 (K.G.H.), T32DK007115 (K.G.H.), U54DK110858 (J.R.), R35GM131854 (J.R.), R01DK108859 (D.R.T.), R01GM103369 (A.P.V.), R35CA242379 (M.G.V.H.), R01CA201276 (M.G.V.H.), T32-GM007287 (S.B.), U54DK110858 (J.E.C.), R01HD04346 (M.J.P.), R01HD105311 (M.J.P.), R01 GM125944 (F.J.S.), R01 DK112854 (F.J.S.), F99CA253744 (J.A.B.), 5K00CA212445 (A.J.B.), T32DK091317 (A.L.), and F30CA243440 (J.M.W.); National Science Foundation DBI-1661375 and IIS-1513616 (B.W.); National Research Foundation of Korea grant 2021R1F1A1063558 (H.-C.A.); National Natural Science Foundation of China grant 32071207 (C.-H.Y.); German Research Foundation SFB860 (K.T.); Conselho Nacional de Desenvolvimento Científico e Tecnológico 308769/2019-8 (M.C.N.) and 154690/2018-0 (M.A.A.A.); Fundação de Amparo á Pesquisa do Estado de São Paulo 2008/08262-6 (M.C.N.); Coordenação de Aperfeipçoamento de Pessoal de Nível Superior 88882.328384/2010-01 (M.A.A.A.); Calico LLC (J.R.); the Nora Eccles Treadwell Foundation (J.R. and C.P.H.); the Howard Hughes Medical Institute (J.R.); a Howard Hughes Faculty Scholars grant (M.G.V.H.); the Emerald Foundation (M.G.V.H.); the Lustgarten Foundation (M.G.V.H.); the Ludwig Center at MIT (M.G.V.H.); the MIT Center for Precision Cancer Medicine (M.G.V.H.); the University of Nottingham (I.D.); and the University of West Virginia (B.A.W.).
Footnotes
Competing interests: K.G.H. and J.R. are inventors of MIDAS technology that has been licensed to Atavistik Bio, for which K.G.H. is a consultant and J.R. is a founder. F.C. and F.J.S. have financial interest in Furanica, Inc. F.J.S. has financial interest in Creegh Pharma, Inc. S.R.H. is an employee of Calico Life Sciences. All other authors declare that they have no competing interests.
Data and materials availability:
All data are available in the main text, supplementary materials, or online, as indicated. Protein expression plasmids generated for this study are available from Addgene. The MIDAS R analysis tool is available at https://github.com/KevinGHicks/MIDAS. The Electrum vizualization tool is available at https://github.com/Electrum-app/Electrum/releases/tag/v0.0.0. The coordinates for atomic models have been deposited in the Protein Data Bank under accession codes 7MBH (pSer-ENO2) and 7LUB (AP-3-FH). MIDAS FIA-MS spectra for proteins analyzed in this study are available at www.ebi.ac.uk/metabolights/MTBLS7237.
REFERENCES AND NOTES
- 1.Diether M, Sauer U, Curr. Opin. Microbiol. 39, 16–23 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Diether M, Nikolaev Y, Allain FH, Sauer U, Mol. Syst. Biol. 15, e9008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orsak T et al. , Biochemistry 51, 225–232 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Chantranupong L et al. , Cell 165, 153–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfson RL et al. , Science 351, 43–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoki K, Li Y, Xu T, Guan KL, Genes Dev. 17,1829–1834 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zabala-Letona A et al. , Nature 547, 109–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christofk HR et al. , Nature 452, 230–233 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Sun P, Liu Y, Ma T, Ding J, Cell Discov. 6, 94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alam MT et al. , Nat. Commun. 8, 16018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunk E et al. , Nat. Biotechnol. 36, 272–281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang A et al. , Nucleic Acids Res. 49, D498–D508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaneton B et al. , Nature 491, 458–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niesen FH, Berglund H, Vedadi M, Nat. Protoc. 2, 2212–2221 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Alemany R et al. , Am. J. Hematol. 72, 234–242 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Feo S, Arcuri D, Piddini E, Passantino R, Giallongo A, FEBS Lett. 473, 47–52 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver T, Banaszak L, Biochemistry 35, 13955–13965 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Tan SA, Tan LG, Clin. Physiol. Biochem. 7, 303–309 (1989). [PubMed] [Google Scholar]
- 20.Newman RH, Tate KR, Commun. Soil Sci. Plant Anal. 11, 835–842 (1980). [Google Scholar]
- 21.Clark LL, Ingall ED, Benner R, Nature 393, 426 (1998). [Google Scholar]
- 22.Nakahigashi K et al. , Mol. Syst Biol. 5, 306 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galvàn-Pena S et al. , Nat. Commun. 10, 338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano M, Funayama S, de Oliveira MB, Bruel SL, Gomes EM, Comp. Biochem. Physiol. B 102, 873–877 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Oguchi M, Gerth E, Fitzgerald B, Park JH, J. Biol. Chem. 248, 5571–5576 (1973). [PubMed] [Google Scholar]
- 26.Kulkarni RA et al. , Cell Chem. Biol. 24, 231–242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan M et al. , Biochem. J. 475, 1821–1837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashizawa K, McPhie P, Lin KH, Cheng SY, Biochemistry 30, 7105–7111 (1991). [DOI] [PubMed] [Google Scholar]
- 29.Fruman DA et al. , Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui S et al. , Nature 551, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pesce A, McKay RH, Stolzenbach F, Cahn RD, Kaplan NO, J. Biol. Chem. 239, 1753–1761 (1964). [PubMed] [Google Scholar]
- 32.Wiese EK et al. , Nat. Metab. 3, 954–968 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traut TW, Mol. Cell. Biochem. 140, 1–22 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Gowans GJ, Hardie DG, Biochem. Soc. Trans. 42, 71–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houten SM, Violante S, Ventura FV, Wanders RJ, Annu. Rev. Physiol. 78, 23–44 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Koves TR et al. , Cell Metab. 7, 45–56 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y et al. , Cell Biochem. Funct. 26, 233–237 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Thompson AL, Cooney GJ, Diabetes 49, 1761–1765 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Jenkins CM, Yang J, Sims HF, Gross RW, J. Biol. Chem. 286, 11937–11950 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerbey AL et al. , Biochem. J. 154, 327–348 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishida Y et al. , Mol. Cell 59, 321–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomoda H, Igarashi K, Omura S, Biochim. Biophys. Acta 921, 595–598 (1987). [PubMed] [Google Scholar]
- 43.Taegtmeyer H, Circulation 72, IV1–IV8 (1985). [PubMed] [Google Scholar]
- 44.Sindelar DK, Balcom JH, Chu CA, Neal DW, Cherrington AD, Diabetes 45, 1594–1604 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Sindelar DK et al. , Diabetes 46, 187–196 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Ghandi M et al. , Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H et al. , Nat. Med. 25, 850–860 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Billiard J et al. , Cancer Metab. 1, 19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rai G et al. , J. Med. Chem. 60, 9184–9204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bezerra GA et al. , lUCrJ 7, 693–706 (2020). [Google Scholar]
- 51.Bezerra GA et al. , Biochimie 183, 100–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao Q et al. , Nat. Commun. 12, 3440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin J, Chai G, Brewer JM, Lovelace LL, Lebioda L, J. Inorg. Biochem. 111, 187–194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text, supplementary materials, or online, as indicated. Protein expression plasmids generated for this study are available from Addgene. The MIDAS R analysis tool is available at https://github.com/KevinGHicks/MIDAS. The Electrum vizualization tool is available at https://github.com/Electrum-app/Electrum/releases/tag/v0.0.0. The coordinates for atomic models have been deposited in the Protein Data Bank under accession codes 7MBH (pSer-ENO2) and 7LUB (AP-3-FH). MIDAS FIA-MS spectra for proteins analyzed in this study are available at www.ebi.ac.uk/metabolights/MTBLS7237.