Abstract
Diterpenes are the major defensive small molecules enabling soft corals to survive without a tough exterior skeleton, and until now their biosynthetic origin has remained intractable. Furthermore, biomedical application of these molecules has been hampered by lack of supply. Here, we identify and characterize coral-encoded terpene cyclase genes that produce the eunicellane precursor of eleutherobin and cembrene, representative precursors for the >2,500 terpenes found in octocorals. Related genes are found in all sequenced octocorals and form their own clade, indicating a potential ancient origin concomitant with the split between the hard and soft corals. Eleutherobin biosynthetic genes are colocalized in a single chromosomal region. This demonstrates that, like plants and microbes, animals also harbor defensive biosynthetic gene clusters, supporting a recombinational model to explain why specialized or defensive metabolites are adjacently encoded in the genome.
Graphical Abstract:
Introduction
In the Ordovician, the ancestors of corals split into two groups: the hexacorals (hard corals and anemones) and octocorals (soft corals, sea pens, and relatives)1. Like other sedentary invertebrates, corals are exposed to predation and have adapted both physical and chemical defenses2. Today, hexacorals appear to specialize in peptide venoms, injected by stinging nematocysts, while the octocorals almost universally contain small defensive isoprenoids, primarily diterpenes. As of early 2021, 2,347 structurally distinct diterpene natural products have been described from diverse octocorals, including soft corals, gorgonians, sea fans, sea pens, and others3. Because of their potent biological activity, coral compounds are considered to be drug leads for cancers and other diseases, including a few commercial products4. A limitation is that the coral diterpenes must be supplied by harvesting coral reef animals or by impractical chemical syntheses5.
Here, we sought to understand how corals synthesize defensive terpenoids. Many marine invertebrates rely on defensive symbiotic bacteria, which provide their hosts with defensive toxins. While octocorals are not known to contain abundant bacterial symbionts, like other corals they often partner with photosynthetic dinoflagellates6. However, many terpene-producing octocorals (such as sea pens) lack symbiotic dinoflagellates7. For these reasons, we hypothesized that the corals themselves synthesize defensive diterpenes. There are no terpene or other defensive small molecule biosynthetic pathways so far characterized from any marine animal genome, and indeed animals are often thought to be chemically depauperate in comparison to microbes. Discovery of coral defensive biosynthetic genes would demonstrate that marine animals and not just symbionts produce important defensive small molecules, shed light on a key biochemical event that may have enabled the rise of the soft-bodied corals, and provide a strategy for supplying important marine drug leads. This work describes the discovery of terpene biosynthetic gene clusters encoded in the animal genome. Genomic and transcriptomic sequencing of the eleutherobin producer Erythropodium caribaeorum lead to the heterologous expression and in vitro characterization of two diterpene producing enzymes. These genes are flanked by sequences predicted to encode for tailoring enzymes also involved in the biosynthetic pathway, and phylogenetic analysis implies that these terpene cyclases play an ancient evolutionary role in coral defense.
Results
Identification of TPS genes in Erythropodium caribaeorum
We focused on the biosynthesis of the coral diterpene eleutherobin (1), a defensive metabolite and potent anti-tubulin cancer lead whose further development has been stymied by the challenge of supply (Fig. 1a)5. Erythropodium caribaeorum was collected by SCUBA in Lauderdale by the Sea, Florida, USA. The specimens contained desmethyleleutherobin, derived from the eunicellane skeleton, as well as abundant other diterpenes, whose structures were mostly derived from the briarane skeleton (Fig. 1b). In turn, the eunicellane and briarane skeletons were proposed to be linked via a common cembrene intermediate, as cembranes are ubiquitous throughout octocorals but the instability of the unmodified cembrene skeleton makes their isolation rare8. The E. caribaeorum metatranscriptome was sequenced, assembled, and analyzed. Initial analysis by BLAST revealed no putative TPS genes. TPSs are famously sequence divergent, and BLAST depends upon overall sequence similarity. Thus, we instead applied a hidden Markov model (HMM) based on multiple sequence alignment of known diterpene cyclases (Supplementary Figure. 1 and Supplementary Table 1). HMM searches are more sensitive because they reflect highly conserved, essential motifs, such as active sites in an enzyme, an advantage which has been previously employed with excellent effect to discover new bacterial terpene synthases9. Our HMM search revealed eight hits that had the right protein sequence length (approximately 400 amino acid residues) and the correct distance between active site motifs to be putative TPSs (Fig. 1c). Moreover, structure prediction of these cyclases revealed a likely structural similarity to characterized TPSs from bacteria (Extended Data Figure 1 and Supplementary Table 2). BLAST searching using these sequences revealed at least nine related TPSs in E. caribaeorum (Figure 1, Supplementary Table 3). Metagenomic analysis provided a 1.4 Gb DNA assembly (42.5 % GC, N50 ≥1,924 bp). The assembled contigs were binned, revealing that >98% of assembled reads originated in the coral genome, with only ~0.8% and 0.3% originating in dinoflagellates and other microbes, respectively. No sequences similar to bacterial TPSs were present in the metagenome. Of the TPSs identified in the transcriptome, one originated in a dinoflagellate contig (Fig. 1b and Supplementary Table 3). Further analysis showed that similar proteins are encoded in all available Symbiodinium spp. dinoflagellate genomes/transcriptomes, and in dinoflagellate-derived sequences from hard and soft corals, sponges, and a few other cases (Fig. 4), indicating that such genes are unlikely to be involved in defensive terpene synthesis. The remaining TPSs were embedded in genomic contexts that were clearly coral in origin, flanked by genes from coral central metabolism, such as 40S ribosomal protein-like and transient receptor potential channel-like genes (Extended Data Fig. 2 and Supplementary Table 4). Strikingly, multiple relatives of the octocoral TPS sequences were found in all octocoral genomes and transcriptomes in available databases, including those from all major octocoral groups, while they were absent from hexacorals (Extended Data Fig. 3). This implied that TPSs may be ubiquitous in octocorals.
Fig. 1. Terpene biosynthesis in the eleutherobin producer, Erythropodium caribaeorum.
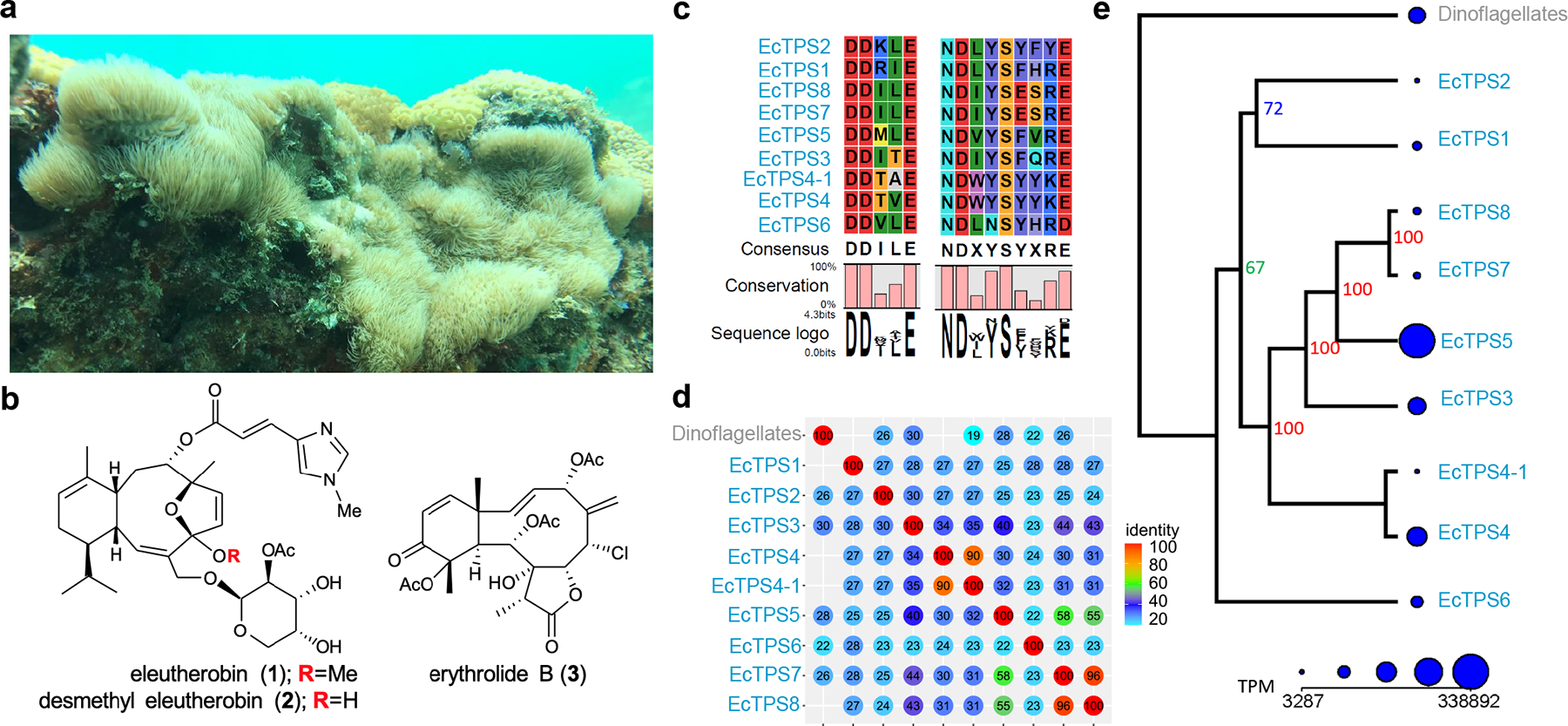
(a) The gorgonian octocoral E. caribaeorum in its native habitat (b) structures of eunicellane (1 and 2) and briarane (3) diterpenoids from octocorals. (c) Alignment of the TPS divalent metal binding motifs for EcTPSs (d) the pairwise identities of nine TPSs found in E. caribaeorum. An additional TPS. An additional TPS was attributed to Symbiodinium sp. dinoflagellates living within the coral. (e) A phylogenetic tree is shown with the TPSs and their relative expression levels, in transcripts per million (TPM).
Fig. 4. A distinct TPS clade is found in all sequenced octocoral genomes and transcriptomes.
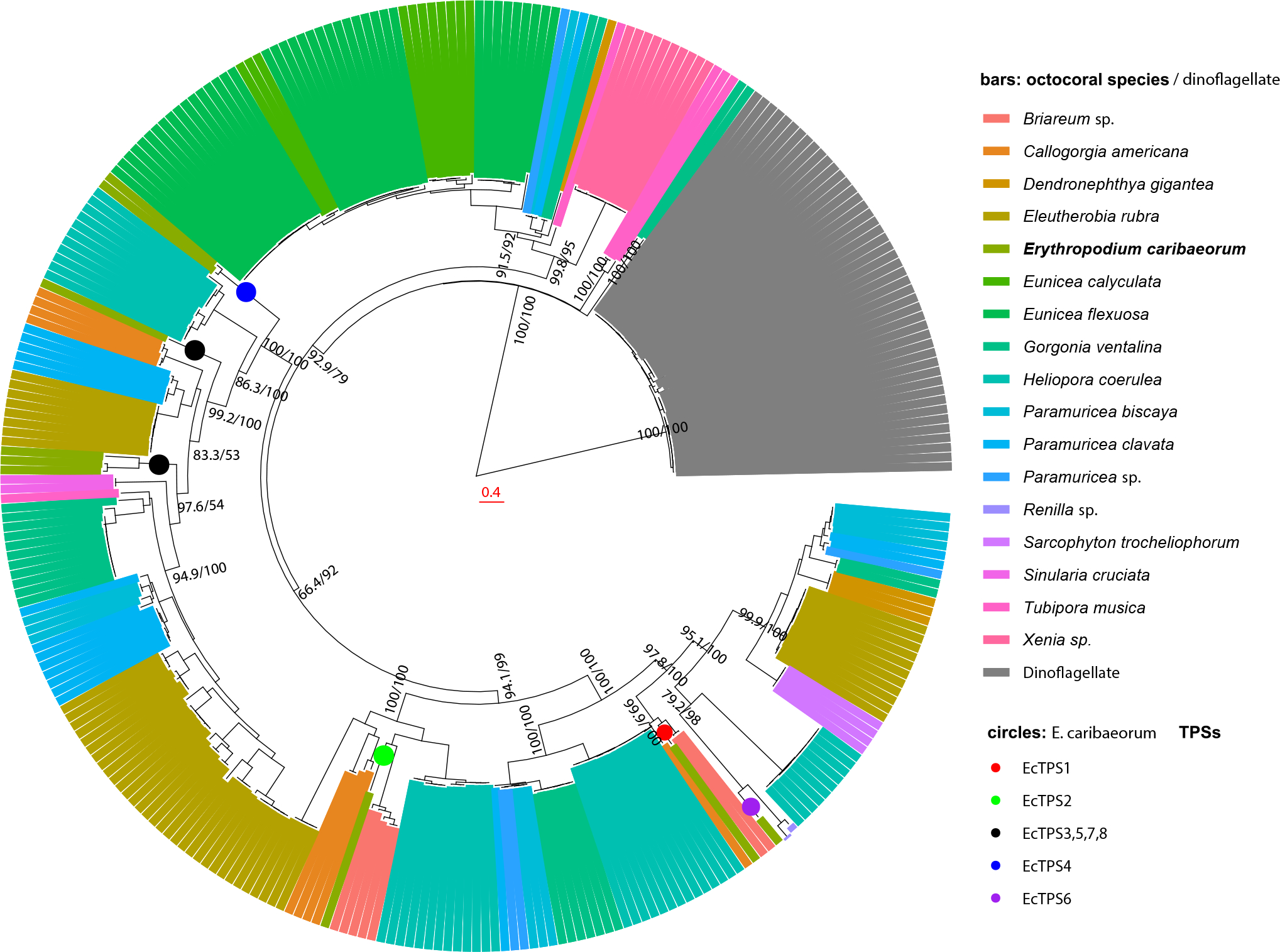
This maximum likelihood tree generated in IQ-tree includes publicly available TPS genes related to the nine E. caribaeorum TPSs. The numbers at branch points are the SH-aLRT support / ultrafast bootstrap support, in percent. The scale bar is expected number of substitutions per site. Octocorals show a highly diverse array of TPS genes, spanning nine clades that are widely distributed amongst octocoral orders and families. An additional single clade comprises related TPS genes from sponges, hexacorals, octocorals, and Symbiodinium spp. dinoflagellates, and therefore it is attributed to dinoflagellate symbionts in these animals. The dinoflagellate clade is very distantly related to the coral clade (see Extended Data Fig. 7).
Putative eleutherobin TPS is embedded in a BGC
Unlike the situation found in microbes and plants, animal-derived biosynthetic genes are not thought to be grouped into biosynthetic gene clusters (BGCs)10. When biosynthetic genes are colocalized, bioinformatics analysis often enables the genes and their products to be correlated with high certainty, but absent a BGC it is very challenging to determine function from sequence11. Since we did not expect animal genes to be clustered, we worried that each coral TPS would require experimental validation. Unexpectedly, however, we found that putative eleutherobin (elu) genes were clustered together in the animal chromosome. elu contained genes for terpene cyclization (EcTPS1), a cytochrome P450 for the needed oxidative steps, and an acyltransferase that might transfer either acetate or N-methylurocanate to hydroxyl groups that decorate the eleutherobin skeleton (Fig. 2 and Supplementary Table 4). Additionally, the elu cluster contained a number of other genes that might be involved in either resistance or biosynthesis.
Fig. 2. Terpene biosynthetic pathways in octocorals are often found in animal-encoded gene clusters.
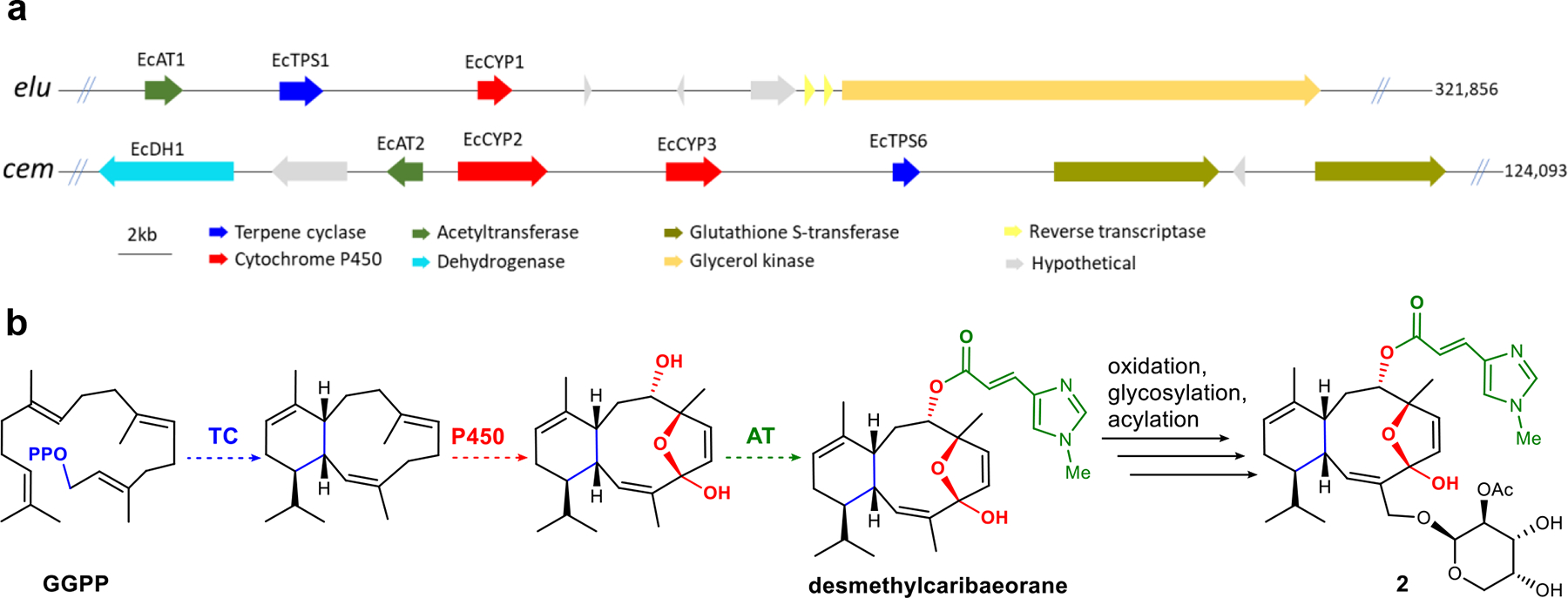
(a) Putative E. caribaeorum biosynthetic gene clusters containing terpene cyclases that were characterized in this study (see Fig. 3). (b) Proposed biogenesis of desmethyleleutherobin (2) starting from GGPP.
Characterization of eunicellane and cembrene TPSs
To determine whether EcTPS1 is the eunicellane synthase involved in eleutherobin biosynthesis, we expressed the recombinant gene in Escherichia coli and purified the soluble protein. When incubated with synthetic geranylgeranyl pyrophosphate (GGPP), EcTPS1 produced a single compound, as gauged by gas chromatography-mass spectrometry (GCMS) (Fig. 3). The reaction was scaled up, and isolation, purification, and structure elucidation using spectroscopy (Supplementary Note 1). led to the identification of the EcTPS1 product as the known eunicellane, klysimplexin R (4)12. This result suggested that elu was correctly identified as the eleutherobin cluster. To further validate this result, four of the other most highly expressed TPSs in E. caribaeorum were treated in the same manner as EcTPS1. All four were expressed in soluble fashion, but only one other protein (EcTPS6) produced a product (Fig. 3). Control experiments including no enzyme and all 5 enzyme reactions are shown in Extended Data Fig. 4. The remaining three proteins likely utilize substrates not present in the reaction mixture and are thus possibly not diterpene cyclases. The single EcTPS6 product was scaled up, purified, and analyzed spectroscopically to reveal that EcTPS6 is a cembrene synthase13. Since cembrene (5) is the precursor to >1,000 defensive coral terpenes, the expression of cembrene and eunicellane cyclases in these experiments provided a biochemical basis for 2 of the >50 major carbon skeletons that explain the thousands of known coral defensive terpenes8,14,15.
Fig. 3. Synthesis of coral terpenes from synthetic GGPP analogues in vitro.
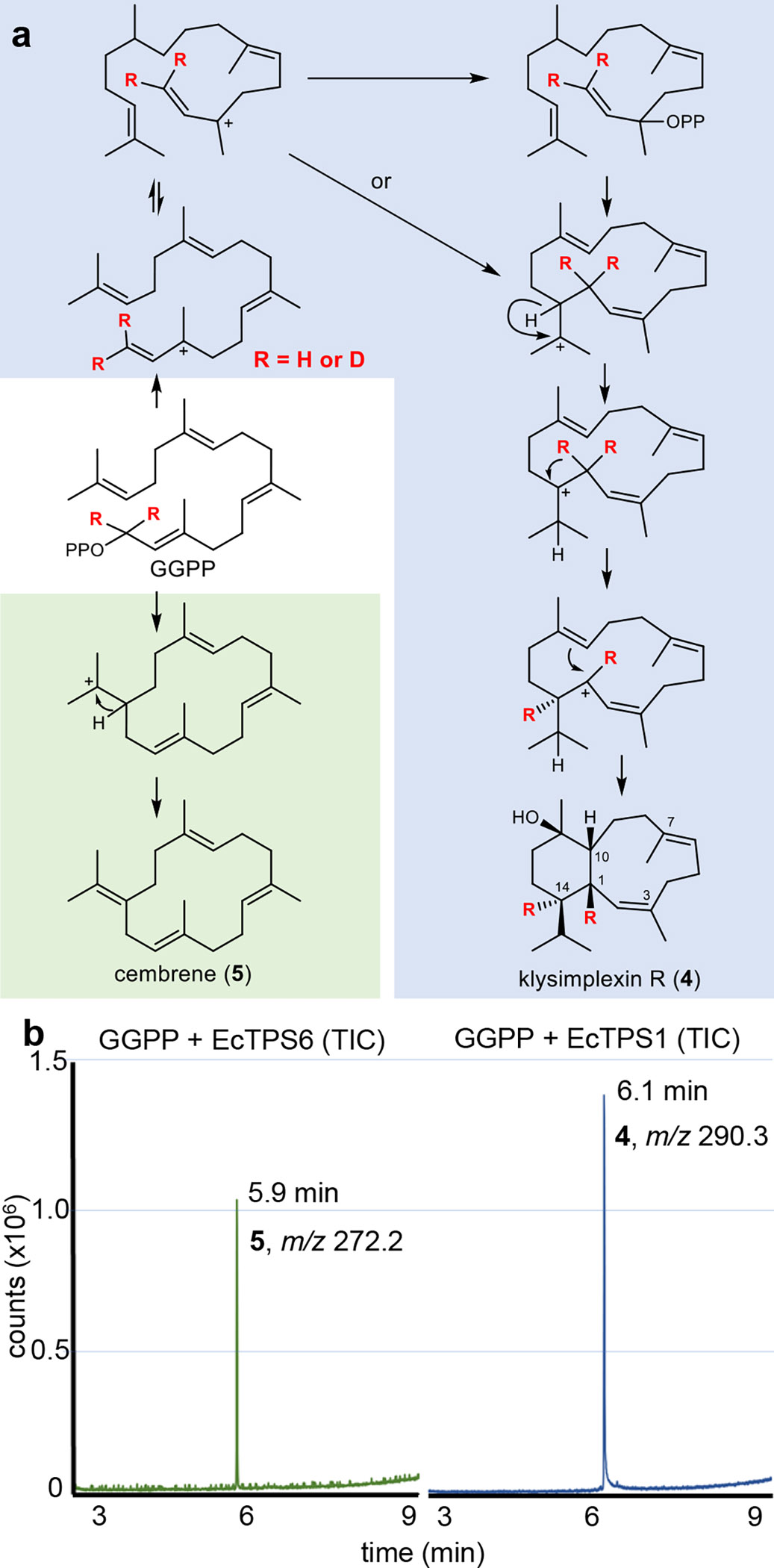
(a) The coral terpene precursors klysimplexin R (4) and cembrene (5) were synthesized by incubating GGPP with purified coral enzymes EcTPS1 and EcTPS6, respectively. EcTPS1 was also treated with deuterated GGPP, resulting in deuterium migration supporting an unprecedented cyclization mechanism (see text for details). (b) GCMS traces (total ion current) from incubation of EcTPS1 (right) and EcTPS6 (left). Analytical scale assays indicated that 4 (tr=6.11 min, m/z = 290.3) and 5 (tr=5.88 min, m/z = 272.2) were the sole cyclized products. The no-enzyme control and other 3 enzyme reactions are shown in Extended Data Fig. 5. The resulting electron impact mass spectra, as well as a full 2D NMR data set, support the structures of the compounds as shown (Supplementary Note 1).
Eunicellane TPS enzymatic mechanism
EcTPS1 is a potential biocatalyst to simplify eleuthrobin supply and analog generation, so we further characterized its biochemical properties. EcTPS1 kinetic parameters (Km 44 μM, kcat 0.02 s−1) are similar to those of other characterized TPSs (Extended Data Fig. 5). To investigate the mechanism of cyclization, 1,1-dideutero-GGPP was synthesized and incubated with EcTPS1. The resulting product was purified to homogeneity and characterized spectroscopically (Supplementary Note 1). Surprisingly, the two deuterium atoms were found on C-1, and C-14, implying a different biochemical mechanism than that found in bacterial eunicellane synthesis16. We propose a cyclization cascade mechanism involving initial bond formation between C-1 and C-14 with E to Z isomerization of the Δ2,3 bond to afford a cembrenyl cation (Fig. 3a). This double bond geometry might be the result of either C2-C3 bond rotation in the delocalized carbocation (as shown) or geranyl linalool pyrophosphate as a possible intermediate. A series of 1,2-hydride shifts, first from C-14 to C-18 then from C-1 to C-14, generate the allylic cembrenyl carbocation, which then undergoes bond formation between C-1 and C-10 to afford the eunicelyl carbocation. Carbocation quenching by water affords diterpene alcohol klysimplexin R (4). To further support this unprecedented mechanism, we performed density functional theory (DFT) calculations, revealing that the bicyclic carbocation generated after C-1 – C-10 bond formation is at the pathway global minimum, while the pyramidal hydride shift intermediates represent theoretical transition states consistent with the proposed mechanism (Extended Data Fig. 6 and Supplementary Note 2).
Terpene biosynthesis and BGCs are ubiquitous in octocorals
The octocoral fossil record goes back to the Ordovician period or possibly earlier, with several modern groups such as helioporids and sea pens arising in the Jurassic17. Transcriptomes have been obtained for all of these groups, including all major octocoral orders. We found all available transcriptomes encode one or more coral TPSs. A phylogenetic analysis revealed that the coral TPSs comprise a single clade, distinct from TPSs found in other organisms (Fig. 4) and suggesting a possible ancient origin. We used the 15 octocoral transcriptomes representing all of the available species with high-quality sequences and performed a phylogenetic analysis using OrthoFinder (Supplementary Table 5). The TPS and animal phylogenetic trees were compared using PACo18, revealing significant coevolutionary congruence (P = 0.00011). Further work is required with more coral species to fully elucidate this feature. In addition, both hard and soft corals, as well as other organisms, harbored the dinoflagellate TPS from symbiotic Symbiodinium spp. As found in elu, many coral TPSs that we could identify in E. caribaeorum and elsewhere were chromosomally adjacent to cytochromes P450 (Fig. 2 and Extended Data Fig. 7, 8). This includes TPS genes found in genomic assemblies four other species for which genome sequences are available: Xenia sp., Dendronephthya gigantea, Paramuricea clavata, and Renilla muelleri, which show sequences resembling tailoring enzymes such as P450, AT and DH nearby predicted TPS genes. Although not all predicted TPS sequences are found in such clusters, the octocoral genomes so far sequenced contain such clusters. Since most coral terpenes undergo oxidative additions and rearrangements, potentially these P450s tailor terpene hydrocarbons19. Like coral TPSs, the TPS-linked P450s are clearly coral in origin, being most closely related to similar genes from corals and bearing only distant similarity to proteins from other organisms. These findings potentially explain in part the observation that coral terpene pathways are found within BGCs. Moreover, although only a few octocoral TPS-linked P450s are currently available because of the small number of octocoral genomes, these genes appear to have evolved in a branch of terpene-specific P450 enzymes (Extended Data Fig. 9). Along with the presence of mobile elements (ie. reverse transcriptases, Fig. 2), the phylogenetic similarity between these terpene-specific P450 enzymes supports the hypothesis that paralogous genes within clusters arise via gene duplication and divergence events. This has been proposed for plant BGCs and can be extended to other enzymes within these clusters10. Cytochromes P450 are commonly found clustered with TPSs in plants, fungi, and other organisms10. One possible explanation for why defensive compound genes cluster is that the loss of a single gene from the pathway could lead to overproduction of toxic intermediates, leading to an addiction-like property. For example, in the case of terpene biosynthesis if a plant loses an important TPS, it will be subject to predation, while if it loses the clustered P450, it will accumulate a toxic, unmodified terpene hydrocarbon that damages the plant10. Here, we extend these ideas to animals, showing that this property of BGCs is universal in all forms of life so far studied. Indeed, when the crude hexane extracts of E. caribaeorum were analyzed by GCMS, the presence of klysimplexin R (4) was not detectable. This is in agreement with previous feeding experiments using radiolabelled GGPP which were only able to detect trace amounts of unoxidized diterpenes with no klysimplexin R (4) observed20. While 4 was previously found to not be particularly toxic toward a panel of human cancer cell-lines, 5 was previously found to be toxic toward both a human cancer cell line as well as brine shrimp12,21. The toxicity of 4 and 5 toward the gorgonian host is still unknown, and the role of tailoring enzymes coupled to additional resistance mechanisms will be of future interest.
Like elu, several coral TPS BGCs also contain acyltransferases, and phylogenetic analysis shows that these belong to a larger group of coral alcohol acylases. They are also more distantly related to acyltransferases from plants and fungi. Acyltransferase EcAT1 from the elu cluster is predicted to be a membrane bound protein, with a single transmembrane helix located at its N-terminus. The remainder of the protein resembles trichothecene 15-O-acetyltransferase (TRI3), which has an available co-crystal structure with a sesquiterpene substrate22. Threading of EcAT1 with the terpene-TRI3 crystal structure revealed that EcAT1 has the correct residues positioned adjacent to the AATase active site motifs, which would allow it to acylate a diterpene substrate (Extended Data Fig. 10 and Supplementary Table 6). In a similar vein to EcCYP1, previous work has shown that while the diterpene backbone lacking urocanylation is stable, it is not detectable in crude extracts of E. caribaeorum23.
Discussion
Evolution and roles of BGCs
This work provides the experimental evidence that animals contain BGCs. While a few animals have uncharacterized BGCs derived from recent horizontal gene transfer from bacteria, prior to this report no animal BGC has been identified or characterized24. Certain kinds of genes were previously known to be clustered in animals, often reflecting the demands of recombination such as found in sex-specific25 or human immune system genes26. However, the rationale for why animals would need to cluster metabolic genes is not a priori obvious.
Several ideas have been advanced to explain the presence of BGCs in plants and microbes, among which the most commonly invoked include the need to: 1) transfer whole BGCs between organisms horizontally; 2) coordinate expression of genes that might otherwise lead to toxic or useless intermediates; and 3) prevent recombinational deletion of key pathway genes, as found in TPS-P450 pairs in plants and fungi27. The presence of ancient, animal-derived BGCs in corals suggests that the first two ideas are not relevant to animal biology and thus may be less important in maintaining BGCs. Therefore, the addiction model may be more important in explaining the universality of biosynthetic gene clustering in the diverse forms of life on earth. In turn, this indicates that other defensive compounds in animals may also originate in BGCs, greatly easing biotechnological access to chemical diversity needed to synthesize potential drugs such as eleutherobin.
The presumption that sessile marine organisms have evolved elaborate and potent chemical defenses remains a tenet in marine natural products drug discovery. The fact that the evolution of stony skeletons as well as terpenoids in corals coincides with the divergence of soft corals and hard corals supports this underlying claim. Thus, the characterization of the EcTPSs and elu constitute not only a biotechnological advancement in the field of marine natural products, but also provides molecular evidence in support of the fundamental principle driving this field.
Methods
Animal material
Specimens of E. caribaeorum were collected by hand using SCUBA off the coast of Ft. Lauderdale, FL at a depth of approximately −5 m, preserved by both direct freezing and by incubation in RNAlater followed by flash freezing, and shipped to the University of Utah on dry ice. Field identification of E. caribaeorum was performed based on gross morphological features, including light pink polyps and a cream-colored rind with red underside. The tentative assignment was confirmed using chemical and sequencing methods (described below) identifying typical E. caribaeorum terpenes and housekeeping genes.
Chemical analysis of animal specimens
E. caribaeorum (40 g wet wt.) was extracted with MeOH:DCM 1:1 (100 mL) under ultrasound for 15 min. The solvent was evaporated, and the residue partitioned between MeOH and hexane (100 mL each). The MeOH layer was evaporated and the residue partitioned between H2O and DCM (100 mL), then the DCM layer evaporated. The residue was dissolved in MeOH and passed through a plug of HP20 then the solvent evaporated to provide a green oil (150 mg). This crude extract was fractionated by silica gel flash chromatography using a gradient of DCM/MeOH and fractions were analyzed by LCMS and 1H NMR. Fractions containing desmethyl eleutherobin were pooled and purified on semi-preparative HPLC using an RP C18 column and a gradient of ACN/H2O.
Metatranscriptome sequencing and analysis
Frozen animal tissue was pulverized under liquid nitrogen, and RNA was extracted using TRIzol (Invitrogen) and treated with DNA-free DNA removal kit (Invitrogen), following the manufacturer’s protocol. Yield and purity of total RNAs was assessed by spectrophotometry, and total RNA integrity determined by electrophoresis. Illumina library preparation and sequencing was performed at the Huntsman Cancer Institute’s High-Throughput Genomics (HCI-HTG) facility at the University of Utah. Poly(A) RNA from total RNA samples was purified using oligo(dT) magnetic beads, and mRNA sequencing libraries were prepared using the TruSeq Stranded mRNA Library Prep kit (Illumina). This approach enriches eukaryotic protein-coding RNA. Shot-gun sequencing was performed using an Illumina NovaSeq 6000 sequencer with a ~450 bp insert size and 2 ×150 bp paired-end runs to produce 100 M read-pairs. Raw reads were trimmed and adaptors removed by trimmomatic 0.39 and then assembled using rnaSPAdes v3.15.3 (189 Mb, 45 % GC, N50 ≥1,728 bp). Protein sequences were predicted from the transcriptome assembly using TransDecoder 5.5.0. (https://github.com/TransDecoder/TransDecoder/wiki).
Metagenome sequencing and analysis
Frozen tissue preserved in RNAlater (650 mg) was pulverized under liquid nitrogen then suspended in CTAB lysis buffer (11 mL containing hexadecyltrimethylammonium bromide 3% w/v, sodium chloride 1.4 M, sodium ethylenediaminetetraacetate 20 mM, polyvinylpyrrolidone 0.2 % w/v, β-mercaptoethanol 0.2 % v/v, Tris-HCL, 100 mM, pH=8.0). Proteinase K (0.3 mg/mL final concentration) was added followed by incubation for 1 hour at 55 °C and. The resulting suspension was extracted with chloroform/phenol solution (10 mL) then centrifuged (15 min, 4347 x g, room temperature). The clarified supernatant (top layer) was collected by pipette and washed with chloroform/isoamyl alcohol (10 mL) then centrifuged and the supernatant again recovered. To this solution was added sodium acetate solution (1 mL, NaOAc 3 M, pH=5.2) followed by cold isopropanol (10 mL). Upon cooling at −80 °C for 15 min, a precipitate formed which was collected by centrifugation (15 min, 4347 x g, 4 °C), washed with cold 75 % ethanol (4 mL) and dried on the lyophilizer. The pellet was reconstituted in pH 8.0 Tris buffer (1 mL) then treated with RNase (0.1 mg/mL final concentration) and incubated for 30 min at 37 °C followed by extraction with chloroform/phenol (1 mL), washing with chloroform isoamyl alcohol (1 mL) and precipitated by addition of sodium acetate solution (0.1 mL, NaOAc 3 M, pH=5.2) then 100 % ethanol (2 mL) and incubation on ice for 20 min. The suspension was centrifuged (15 min, 16,000 x g, 4 °C) and the pellet rinsed with cold, 75% ethanol (1 mL) to provide crude metagenomic DNA (41.8 μg; A260/A280=1.89 and A260/A230=1.24 by NanoDrop) which was dissolved in pH 8.0 TE buffer (100 μL ) and checked by gel electrophoresis (>48.5 kb).A portion of crude DNA (21 μg ) was further purified by spin-column clean-up using the Genomic DNA Clean & Concentrator-25 kit (Zymo Research) according to the manufacturers instructions. Purity and yield were determined by spectrophotometry and gel electrophoresis (4.3 μg; A260/A280=1.94 and A260/A230=2.30; ~10 kb). Illumina library preparation and sequencing was performed at the Huntsman Cancer Institute’s High-Throughput Genomics (HCI-HTG) facility at the University of Utah. Sequencing library preparation was performed using an Illumina TruSeq Nano DNA Sample Prep kit with a 450 bp mean insert size then sequenced on an Illumina NovaSeq 6000 sequencer using 2 × 150 bp runs. Adapter sequences were removed from raw reads and trimmed reads merged. Reads were assembled using metaSPADES 3.15.3. Genomic scaffolds were classified at the domain level using EukRep v 0.6.6 and eukaryotic genes predicted using AUGUSTUS 3.3.3 with the transcriptome assembly as training data.
Identification of TPS genes in E. caribaeorum
The E. caribaeorum transcriptome was analyzed using hidden Markov model (HMM) searches. An HMM profile was built by multiple aligning seven diterpene synthase protein sequences known to produce cembrane diterpenes using the clustal omega webserver (https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequences used for alignments represented full length proteins. These biochemically characterized proteins were selected on the basis that the proposed cyclization cascade involves a cembrane intermediate, thus similar enzyme function would confer sequence similarity13,28–31. The multiple sequence alignment was used to build an HMM profile with the hmmbuild tool implemented in the hmmer3 software v3.1b2. Additionally, publicly available HMM profiles were downloaded from the PFAM database (http://pfam.xfam.org/search#tabview=tab2). This included “Terpene synthase, N-terminal domain” (Accession # PF01397), “Terpene synthase family, metal binding domain” (Accession # PF03936) and “Terpene synthase family 2, C-terminal metal binding” (Accession # PF19086). These HMM profiles were used to query the proteins predicted from the transcriptome using the hmmsearch command in hmmer3. Searches using the profile for the N-terminal domain PF01397 provided seven hits with average E-value of 0.38 (zero hits above the 0.01 inclusion threshold) which did not resemble terpene synthases on the basis of PHMMER searches against the PFAM database or BLASTP (NCBI BLAST v2.11.0) searches against the nr database. The C-terminal metal binding profile PF19086 outperformed PF03936, providing thirteen hits with average E-values of 0.56, including five hits above the inclusion threshold ranging in E-value from 4.4 × 10−3 to 7.1 × 10−8 which resembled terpene cyclases on the basis of PHMMER and BLASTP searches. These hits included the functionally characterized EcTPS1 and EcTPS6 described herein. Searches using the profile built from cembrene synthase sequences provided 8 hits with an average E-value of 0.53, including six hits above the inclusion threshold. Analysis of these hits by BLASTP and PHMMER searches also indicated similarity to terpene cyclases, and the hits from this search included EcTPS1. The query HMM profiles based on cembrene synthase and terpene synthase metal binding C-terminal domain both gave clear alignment to the subject sequences at the DDXXE and NSE motifs, which represent detectable features of terpene cyclases from other taxa and extend this method toward animal terpene cyclases. All these initially identified TPSs were used as query to blastp search against the octocoral transcriptomes available in genbank SRA database. Hits meeting the criteria (perc_identity > 40%, query coverage >80% and subject coverage > 80%) were considered as TPSs derived from octocoral and selected to build a new HMM model. This new HMM model was then used to search against the E. caribaeorum transcriptome and genome sequences to find any missing TPSs. The partially assembled TPS hits from E. caribaeorum transcriptome were completed by aligning to the predicted genes from genome. Structural prediction and similarity searches were performed using the i-TASSER web server using the default parameters and only the protein sequence as input. The COACH and COFACTOR results from the i-TASSER pipeline were used to infer ligand binding and enzymatic function. For a detailed description of these computational methods, please refer to Zhang et. al32.
TPS genes in published octocoral transcriptomes
Sequence read archives (SRAs) were downloaded for eight other species of diterpene producing octocorals from the NCBI database and the transcriptomes assembled. These species included Renilla reniformis, Briareum asbestinum, Eunicia fusca, Dendronephthyea gigantea and Eleutherobia rubrum, all of which are known diterpene producers. These additional transcriptomes were further analyzed by BLAST, this time using sequences for octocoral putative TPSs from E. caribaeorum. This revealed eight additional TPS-like proteins, none of which were detected by BLAST search using plant, fungal, protist or bacterial TPS sequences.
Metagenome binning
The metagenome contigs were filtered by length (≥10K bp) and annotated by the module (make_taxonomy_table.py) in autometa (v2.0.0). The tetranucletotide composition of each contig was calculated using the perl script (tetramers.pl) described in YAMB package (v2.1.0.0). The coverage information in each contig (spades assembled) name was directly used for binning. t-SNE dimensionality reduction and sequential DBSCAN data clusterization were performed using the R script (tsne-clust.r) in YAMB with perplexities value=(42, 84 and 126). The clusters were examined by the taxonomy annotation result from autometa, only 90% pure clusters were accepted.
Phylogenetic analyses
Putative terpene synthases were extracted from the assembly of SRA data by both blastp searching using EcTPSs as query (qcov ≥80%, perc_identity > 30%) and hmmsearch (default threshold). A series of bacteria, fungi and plant TPS genes were also downloaded from GenBank database. TPS hits were aligned using t-Coffee v13.45.0.4846264 (-mode mcoffee -output = msf, fasta_aln). To remove poorly aligned regions, the resulting alignment of all octocoral TPSs was subsequently trimmed with trimAl v1.4 with a gap threshold of 0.6. For the alignment of TPSs including bacteria, fungi and plant the gap threshold was set to 0.4. The maximum likelihood tree was constructed using iqtree v1.6.12 (./iqtree -nt AUTO -st AA -alrt 1000 -bb 1000)33. Orthologous genes from 15 octocoral species were retrieved by OrthoFinder (v2.5.4) using standard parameters34. The topological congruence between the phylogenies of the OrthoFinder output and the TPS tree was evaluated by PACo parameters (nperm=100000, seed=12, method=‘r0’, symmetric = FALSE)18.
Putative cytochrome P450 genes were extracted from the E. caribaeorum transcriptome assembly by blastp searching using P450 sequences of enzymes related to terpenoid biosynthesis (Taxus cuspidata 5-alpha-taxadienol-10-beta-hydroxylase [AAK00946.1] and taxane 13-alpha-hydroxylase [AAL23619.1]) as query. Only sequences from this search with length between 350 and 600 amino acid residues were included for further analysis to eliminate gaps. Additional putative cytochrome P450 genes were extracted from the assembly of SRA data by blastp searching using all of the P450 genes identified from E. caribaeorum as query (qcov ≥80%, perc_identity > 20%). Hits selected from the E. caribaeorum transcriptome assembly and SRA data assemblies were filtered for duplicates using cd-hit v 4.8.1 (https://github.com/weizhongli/cdhit) to remove redundant, identical sequences ( -c 1.0 -t 1). Filtered sequences were aligned using the clustal omega webserver using default parameters. The alignment was trimmed with trimAl v1.4 with a gap threshold of 0.4 The maximum likelihood tree was constructed using iqtree v 1.6.12 (./iqtree -nt AUTO -st AA -alrt 1000 -bb 1000).
EcTPS gene cloning, expression, and purification
Total RNA was used to generate cDNA with a First Strand Synthesis Kit (Invitrogen) by reverse transcription using oligo(dT) primers. Five E. caribaeorum genes were amplified from the resulting cDNA pool using specific primers (Supplementary Table 7). PCR conditions were: initial denaturation at 98 °C, 30 sec; 3-step cycle: 98 °C, 10 sec; 56 °C, 30 sec; 72 °C,45 sec; repeated 35 times; final elongation at 72 °C, 10 min. The PCR product was checked by gel electrophoresis to verify amplicon size and purity. Restriction sites (NdeI/XhoI) were incorporated in a second round of PCR. The PCR product was digested with NdeI and XhoI (NEB) and purified on gel electrophoresis. Ligation was performed with linearized pET28-b(+) vector using T4 ligase (NEB), and the resulting plasmid was transformed into E. coli DH10-β on LB-kanamycin agar plates overnight at 37 °C. Single colonies were seeded into LB media with 50 mg/L and grown overnight at 30° C. Cells were harvested by centrifugation and plasmid DNA isolated by spin-column purification (Qiagen). Plasmids were screened by analytical restriction digest using NdeI and XhoI and the insert confirmed by Sanger sequencing (Genewiz).
Expression conditions were screened by transformation of plasmids (terpene cyclase ORF in pET28-b(+) backbone) into E. coli Rosetta™ (DE3) strain (Novagen) followed by growth under various induction conditions at the 50 mL scale and SDS-PAGE. Five EcTPS proteins were expressed in soluble protein form when grown in TB medium and induced at 16 °C with IPTG (0.5 mM) overnight. Thus, the five proteins were expressed in 1 L cultures for enzymatic screening using the following protocol: The EcTPS1 plasmid was transformed into chemically competent E. coli Rosetta™ strain (Novagen). Transformants were grown overnight on LB agar plates containing kanamycin and chloramphenicol. Single colonies were seeded into modified TB broth (10 mL) containing MgSO4 (1 g/L), Casamino acids (2 g/L), kanamycin (50 mg/L), and chloramphenicol (25 mg/L), then grown for approximately 6 hours at 37 °C and 220 rpm shaking. When the seed cultures appeared cloudy, they were transferred to the same medium (1 L) in 2.8 L baffled Fernbach flasks and grown to an OD600 ranging from 0.4 to 0.6 (approximately 3 hours) at 37 °C and 220 rpm shaking. Cultures were chilled on ice, induced with IPTG (0.5 mM), and incubated overnight at 16 °C with 220 rpm shaking. Cells were harvested by centrifugation then frozen at −80 °C until further use. Frozen cells were thawed, resuspended in lysis buffer (10 mL/g cell mass) containing 50 mM sodium phosphate (pH=8.0), 5 mM MgCl2, 150 mM NaCl, 10 mM imidazole, 10% v/v glycerol, 1 mg/mL lysozyme and 0.1 mg/mL DNase, and then the cell suspension was stirred for 1 hour at 4 °C. Cells were disrupted by sonication, and the resulting lysate was centrifuged at 50,000 × g, passed through a 0.45 micron filter, incubated over Ni-NTA resin (5 mL resin per 100 mL lysate) at 4 °C, and mixed with a rotator. The resin-lysate suspension was collected in a fritted glass column, washed with lysis buffer followed by wash buffer [50 mM sodium phosphate (pH=8.0), 5 mM MgCl2, 150 mM NaCl, 25 mM imidazole], and eluted using a stepwise gradient of imidazole in elution buffer [50 mM sodium phosphate (pH=8.0), 5 mM MgCl2, 150 mM NaCl, and 100, 200, 300 and 400 mM imidazole]. Initially, the purified protein product was desalted using 7,000 kDa MWCO dialysis cassettes by incubating overnight with stirring in 1 L of incubation buffer [20 mM sodium phosphate (pH=7.0), 5 mM MgCl2 and 10% v/v glycerol]. Protein concentrations were measured with the Bradford assay.
Enzyme reactions
For initial reaction screening, proteins in dialysis buffer (5 mL) were transferred to 20-mL scintillation vials without further dilution, and GGPP (200 μg in 500 μL of water) was added. Enzyme substrates were prepared using published methods (Supplementary Note 1). Reaction mixtures were shaken at 30 °C and 180 RPM overnight. The reaction mixture was extracted twice with hexane (5 mL), dried over anhydrous sodium sulfate, and evaporated under reduced pressure on a rotovap. The residue was dissolved in ethyl acetate (100 μL) and analyzed by gas chromatography-mass spectrometry (GCMS), performed on an Agilent 6890 equipped with a 5973 mass selective detector and an Agilent DB5-MS column (30 m x 0.25 mm x 0.25 μm) (Supplementary Fig. 2). All injections were performed in splitless mode with helium as the carrier gas (0.9 mL/min flow rate) using 1 μL of sample (inlet = 250 °C) with the following mass selective detector (MSD) parameters: TIC scan range = 40–500, quad = 150 °C, source = 250 °C, threshold = 150, and 3.15 scans/sec. The oven method was as follows: initial temperature 50 °C for 0.5 min followed by a 30 °C/min oven ramp to 280 °C then a 40 °C/min oven ramp to 330 °C with a 1 min hold (S.A. Bell and J.M. Winter, personal communication). All data were analyzed using the HP (Agilent) ChemStation Data Analysis software v A.05.01 or Agilent Mass Hunter v B.07.00. A negative control consisting of GGPP incubated overnight in dialysis buffer without protein was analyzed in an identical manner for comparison. Only EcTPS1 and EcTPS6 provided GCMS traces containing additional peaks, indicating enzymatic products. The electron impact mass spectra of these peaks were consistent with the presence of diterpenoids (Supplementary Fig. 3).
Optimization of the enzymatic reaction was done using EcTPS1 dialyzed against a buffer containing 10% glycerol, 5 mM MgCl, 5 mM DTT and either 20 mM sodium phosphate, 50 mM Tris or 50 mM HEPES. Solutions of EcTPS1 in each buffer were adjusted to a pH ranging from 6.5 to 8.0 and 50 mM GGPP was added. After 16 h of incubation, reaction products were extracted once with hexane (500 μL) spiked with caryophyllene. The organic layer was analyzed directly by GCMS as described above, and the amount of product measured as the ratio of product peak area to standard peak area. It was determined that the best condition used sodium phosphate buffer at pH 6.5–7.0, so this condition was selected for further enzymatic reactions.
Kinetic analysis of EcTPS1
Kinetic measurements were performed with three biological replicates. The enzymatic reaction was done using enzyme reaction mixtures (400 μL) consisting of 9.8 μM EcTPS1, 10% glycerol and 5 mM MgCl buffered with 20 mM sodium phosphate (pH 6.8). The reaction was initiated by addition of GGPP (100 μL) at a final concentration ranging from 20 to 60 μM with thorough mixing. After 10 min of incubation at 28 °C, the reaction was quenched with EDTA (100 μL, 0.5 M, pH 8.0) products were extracted once with hexane (200 μL) spiked with caryophyllene. The organic layer was analyzed directly by GCMS as described above, and the peak areas, normalized with respect to internal standard peak area, were used to calculate the concentration of product by comparison to a standard curve generated using purified klysimplexin R (Extended Data Fig. 5). The increase in concentration of klysimplexin R over 10 min (in nM/s) with respect to concentration of GGPP added (in μM) was plotted and analyzed by non-linear curve fitting using the enzyme kinetics Michaelis-Menten model in GraphPad Prism v9. Calibration curves were generated by plotting integration versus concentration in Excel v2201.
Extended Data
Extended Data Fig. 1. Predicted structure of EcTPS1.
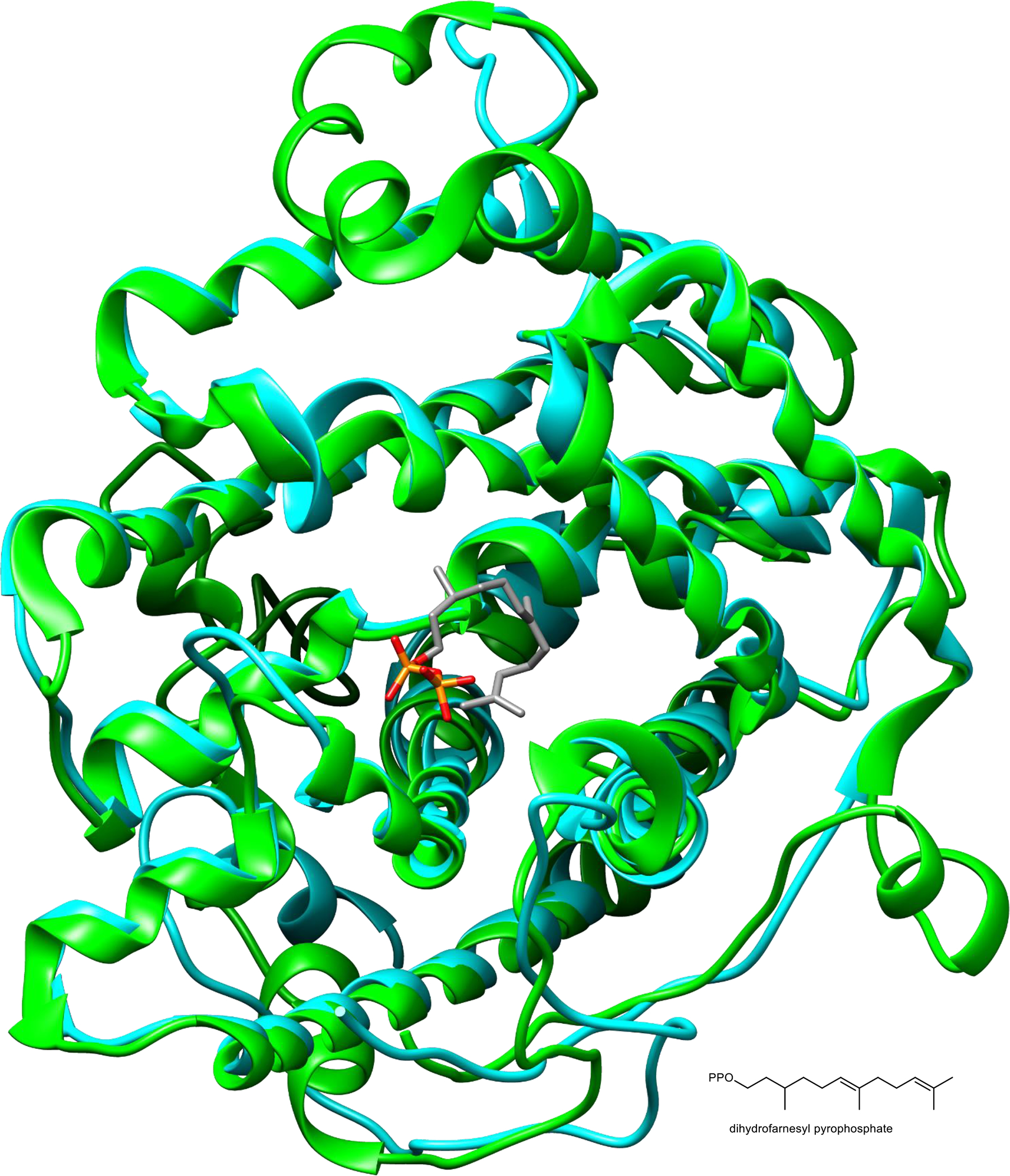
3D model of EcTPS1 protein structure (blue ribbon) superimposed on selinadiene synthase (green ribbon, PDB# 4OKZ) in complex with dihydrofarnesyl pyrophosphate (carbon = grey, phosphorus=orange, oxygen=red). The structural comparison and alignment were performed in i-TASSER. This figure reveals the predicted close structural similarity between EcTPS1 and other type I TPS proteins despite a lack of sequence similarity.
Extended Data Fig. 2. Metagenomic analysis of E. caribaeorum.
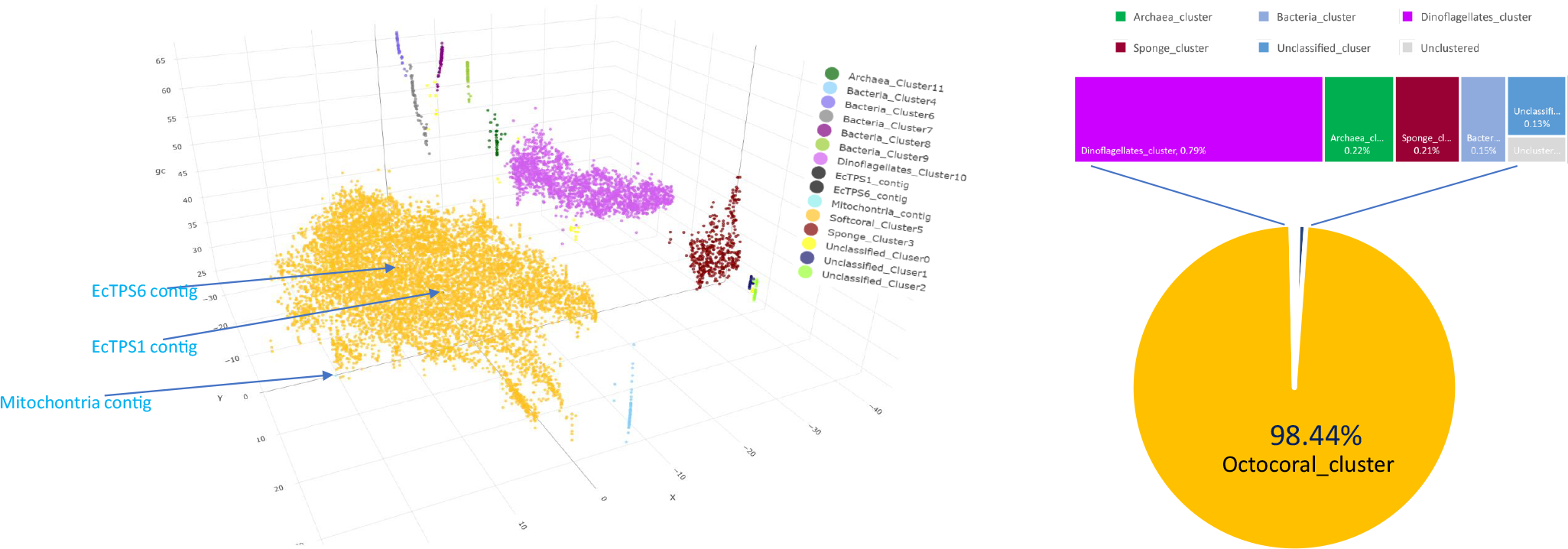
Binning plot of E. caribaeorum metagenome. Each points represent a unique assembled contig from the metagenome. They are plotted on the two dimensions that result from dimension-reduction by BH-tSNE and GC content of the contig. The points were grouped by DBSCAN and supervised by taxonomic identification of each contig. The elu BGC containing contig is part of the E. caribaeorum genome. Only a small percentage of the metagenome originates in bacteria and dinoflagellates. A small amount of sponge DNA is present because the coral forms a tight association with a sponge.
Extended Data Fig. 3. Phylogenetic analysis of octocoral terpene cyclases.
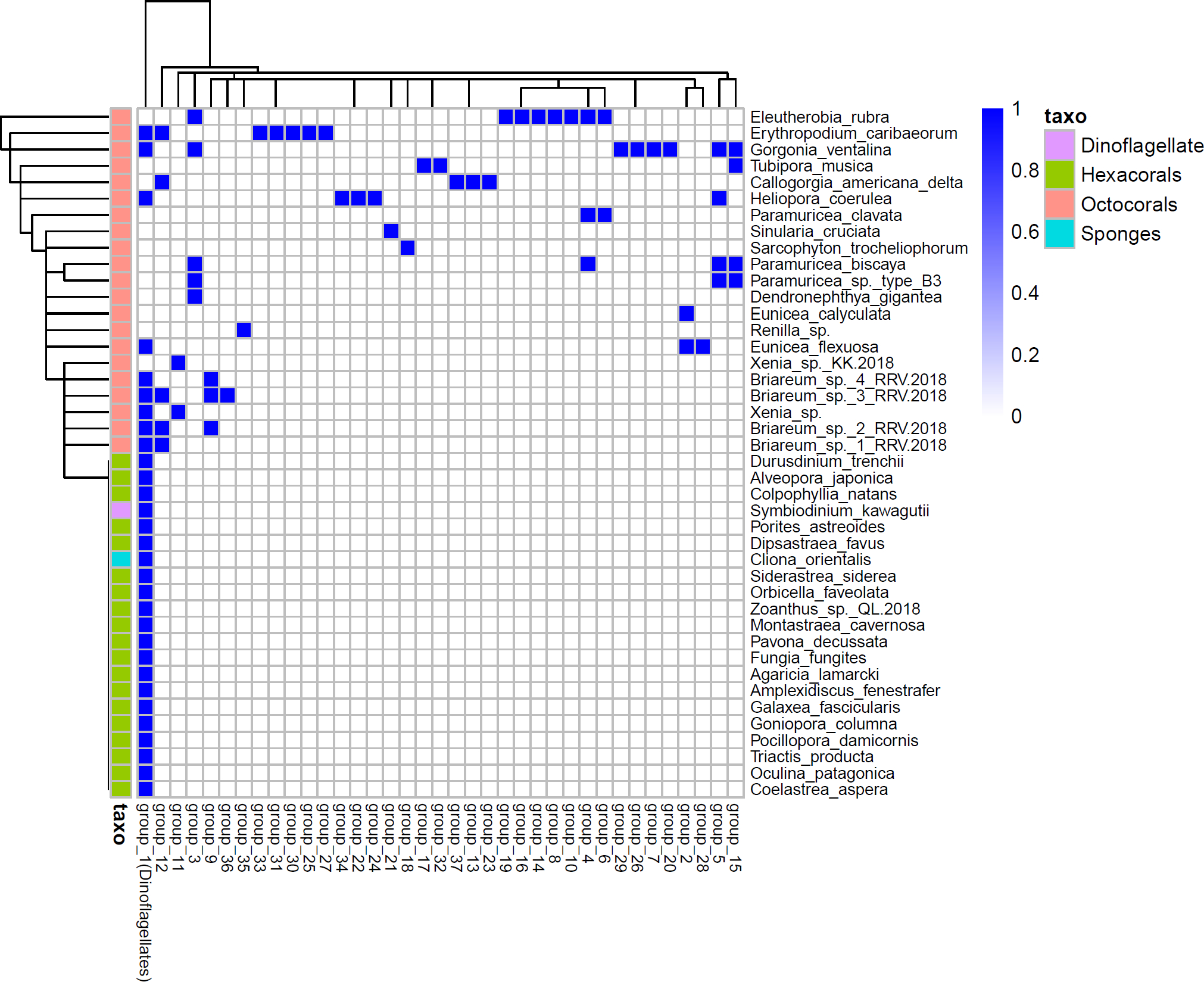
TPS gene distributions in marine invertebrate transcriptomes. The TPS genes were grouped by a threshold of 65% protein sequence identity. A single TPS group (group 1) is found in soft corals, hard corals, sponges, and dinoflagellates, and represents a dinoflagellate-encoded TPS group. The remaining groups (TPS groups 2–36) are only detected in octocorals genomes and transcriptomes and thus represent octocoral terpene cyclases such as EcTPS1 and EcTPS6.
Extended Data Fig. 4. GCMS analysis of enzymatic terpene cyclase reactions.
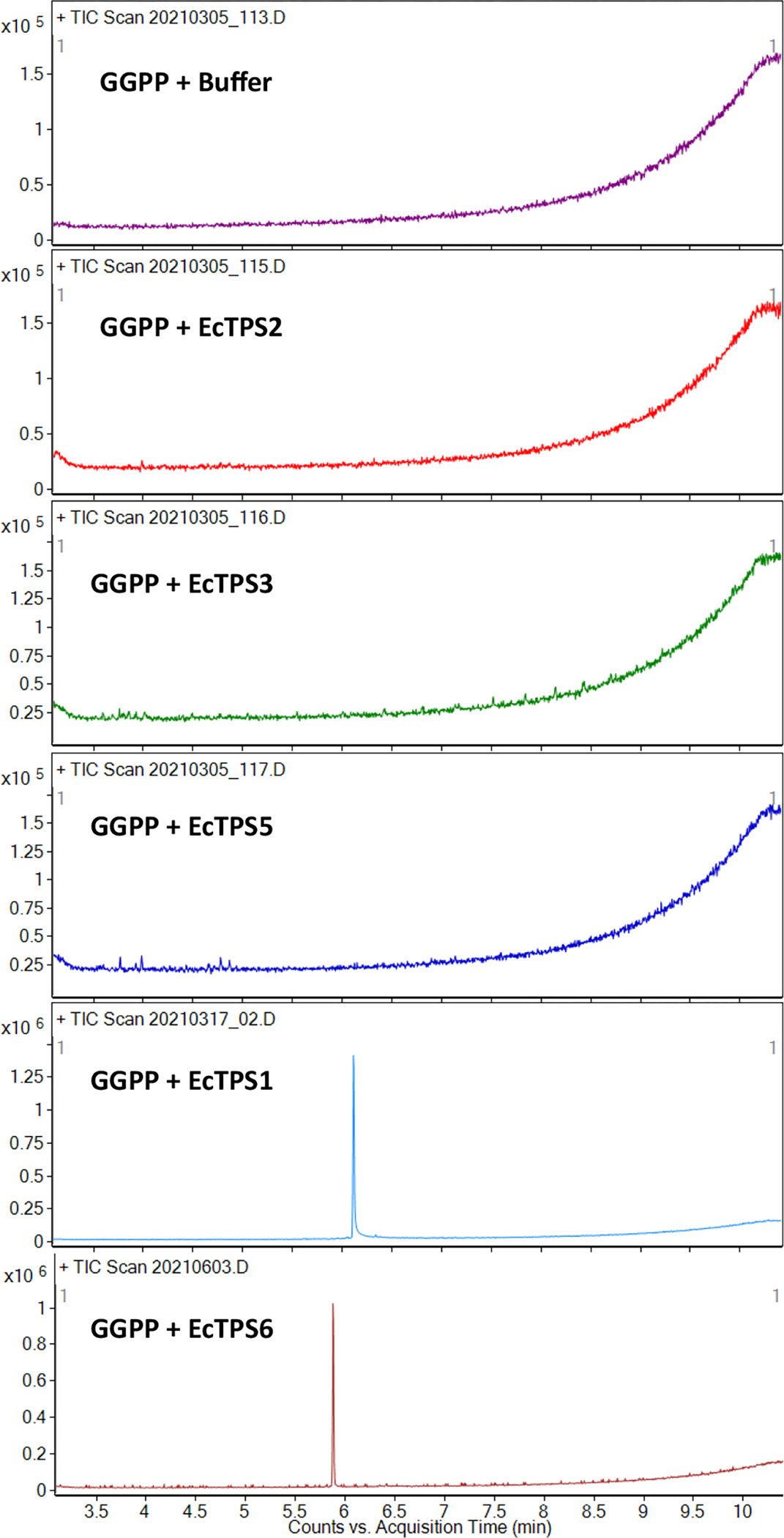
Raw data GCMS total ion current chromatograms of control and sample enzyme assays. The x-axis is time, while the y-axis is the ion counts. Note that because of the robust synthesis of 4and 5 by EcTPS1 and EcTPS6, respectively, the scale is zoomed in by 10-fold in the control experiments.
Extended Data Fig. 5. Characterization and kinetic analysis of terpene cyclases.
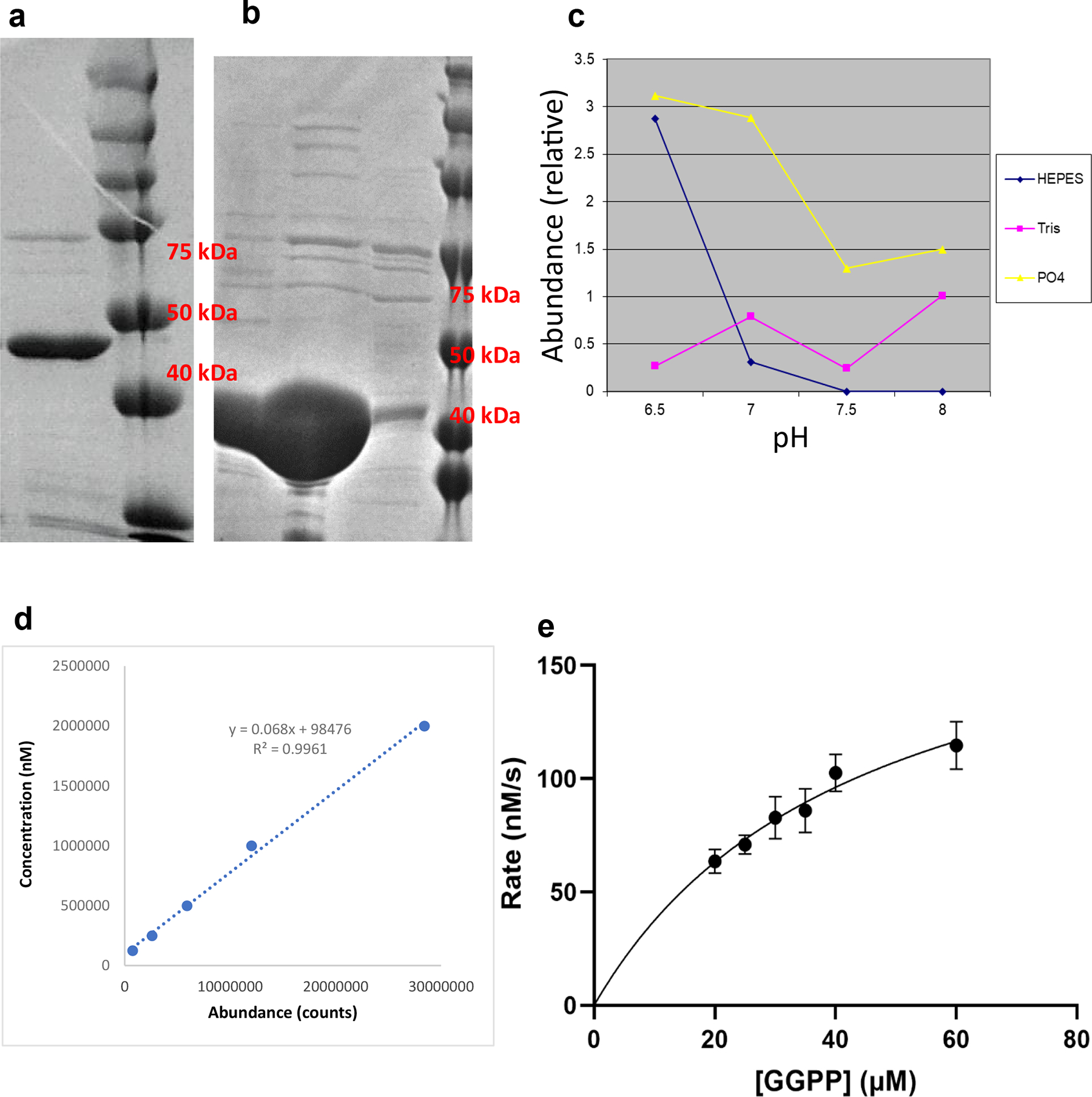
SDS-PAGE analysis of EcTPS1 elution fraction. EcTPS1 was expressed and purified over five times, providing a similar result. B) SDS-PAGE analysis of EcTPS6 elution fraction. EcTPS6 was expressed and purified three times, providing a similar result. C) Relationship between production of 4 and buffer pH D) GCMS calibration curve for quantification of 4. E) Michaelis-Menten analysis of EcTPS1 upon incubation with GGPP. Independent reactions were run in triplicate (n = 3 biological replicates) and analyzed once by GCMS (n = 1 technical replicate) and normalized to an internal standard. Data are presented as mean values +/− SD.
Extended Data Fig. 6. Proposed cyclization cascade for EcTPS1.
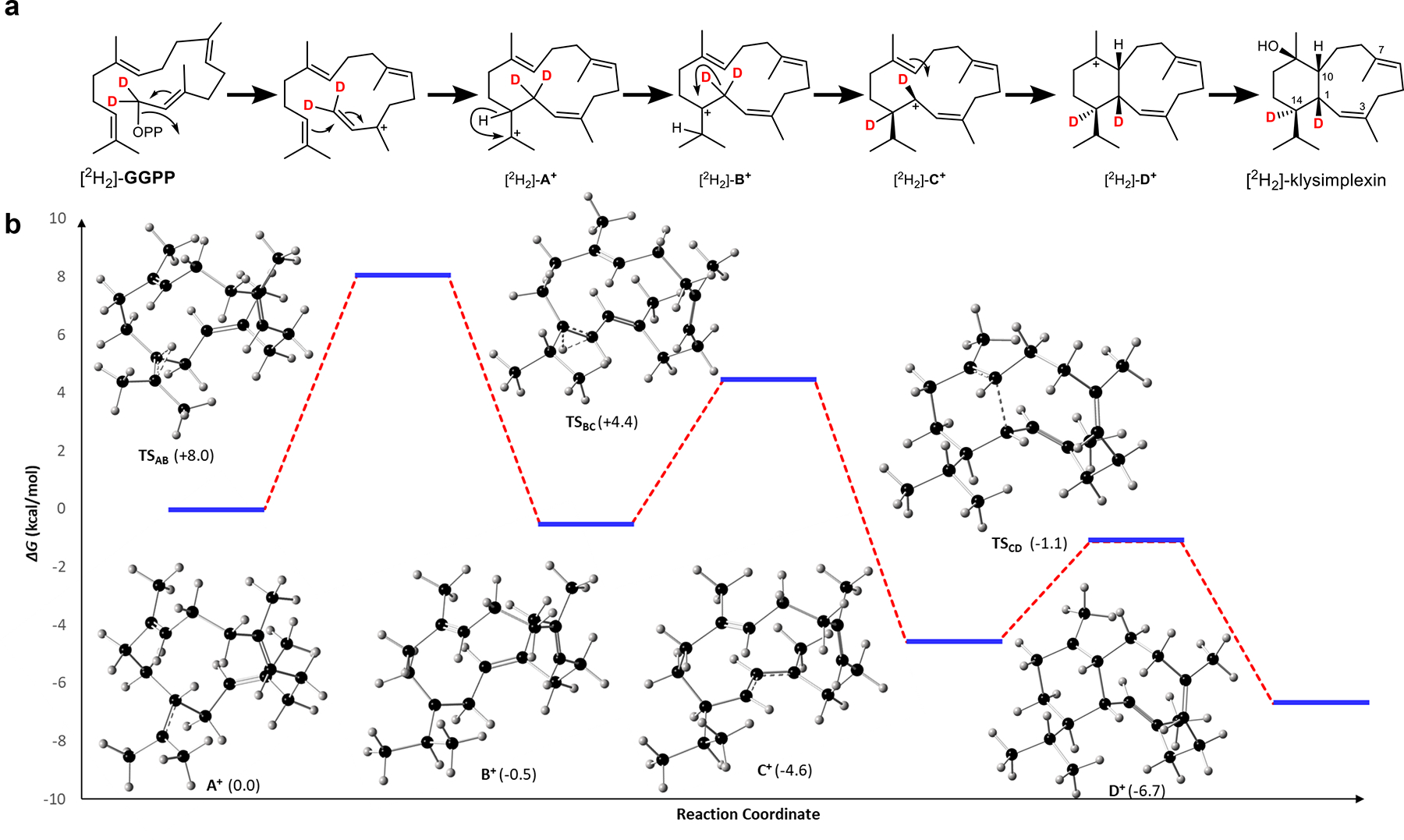
A) Proposed cyclization cascade starting from GGPP to form [2H2]-klysimplexin R (4). B) Relative free energies of intermediates and transition state structures in kcal mol−1, calculated with mPW1PW91/6–311 + G(d,p) in a water CPCM model.
Extended Data Fig. 7. Phylogenetic analysis of marine terpene cyclases.
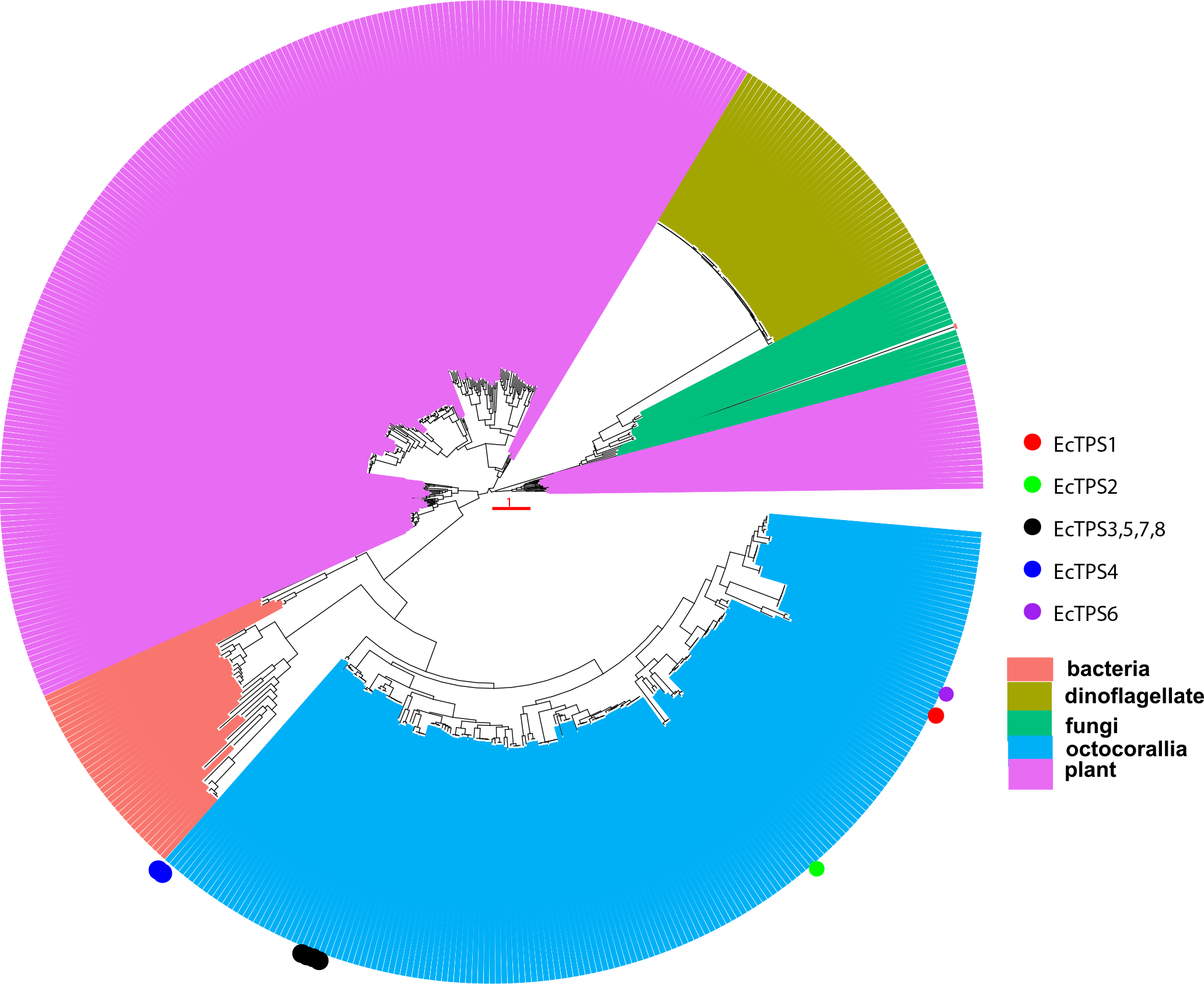
Maximum likelihood phylogenetic tree of TPS protein sequences. The dinoflagellate series includes transcripts from dinoflagellates, as well as from corals and sponges but attributed to dinoflagellates. The tree shows that octocoral TPS sequences form a clade that is distinct from terpene cyclases of other origin. All orders of octocorals are included, and thus the results reveal that these genes do not result from horizontal transfer after the split of the octocoral orders.
Extended Data Fig. 8. TPS BGCs in octocorals.
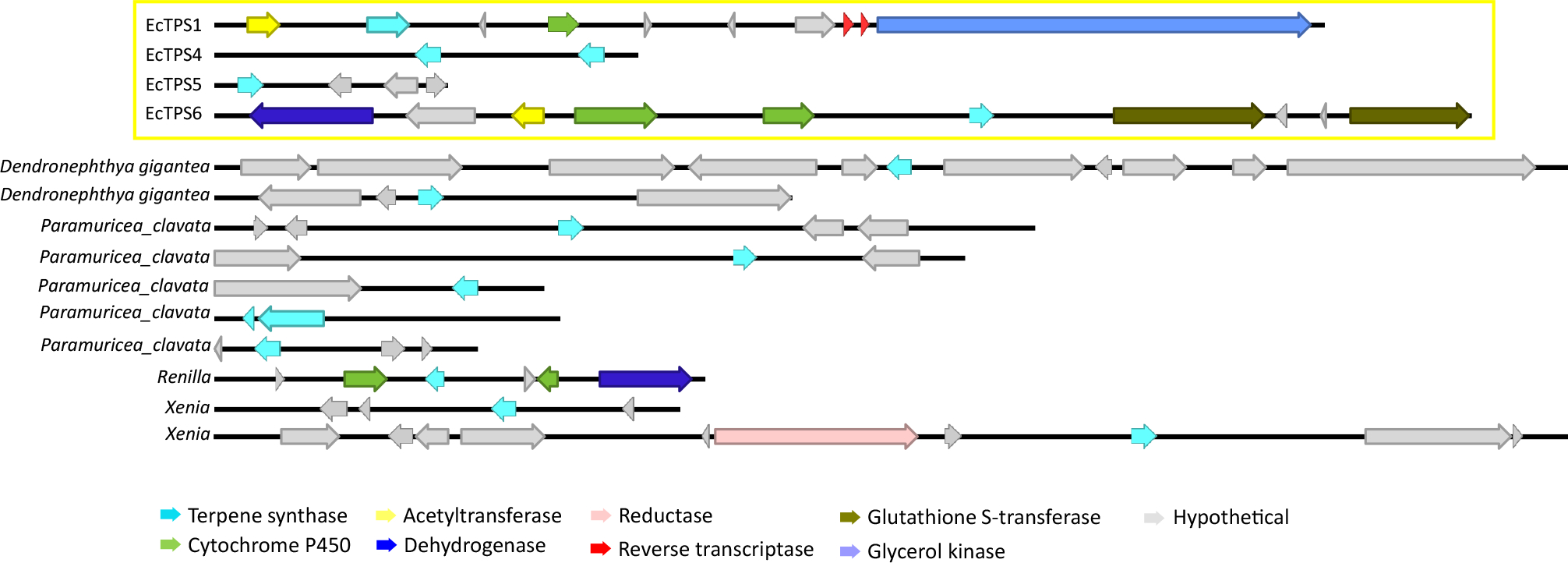
TPS-containing contigs, presumably harboring biosynthetic gene clusters (BGCs), identified in previously published octocoral genomes.
Extended Data Fig. 9. Phylogenetic analysis of octocoral cytochrome P450 genes.
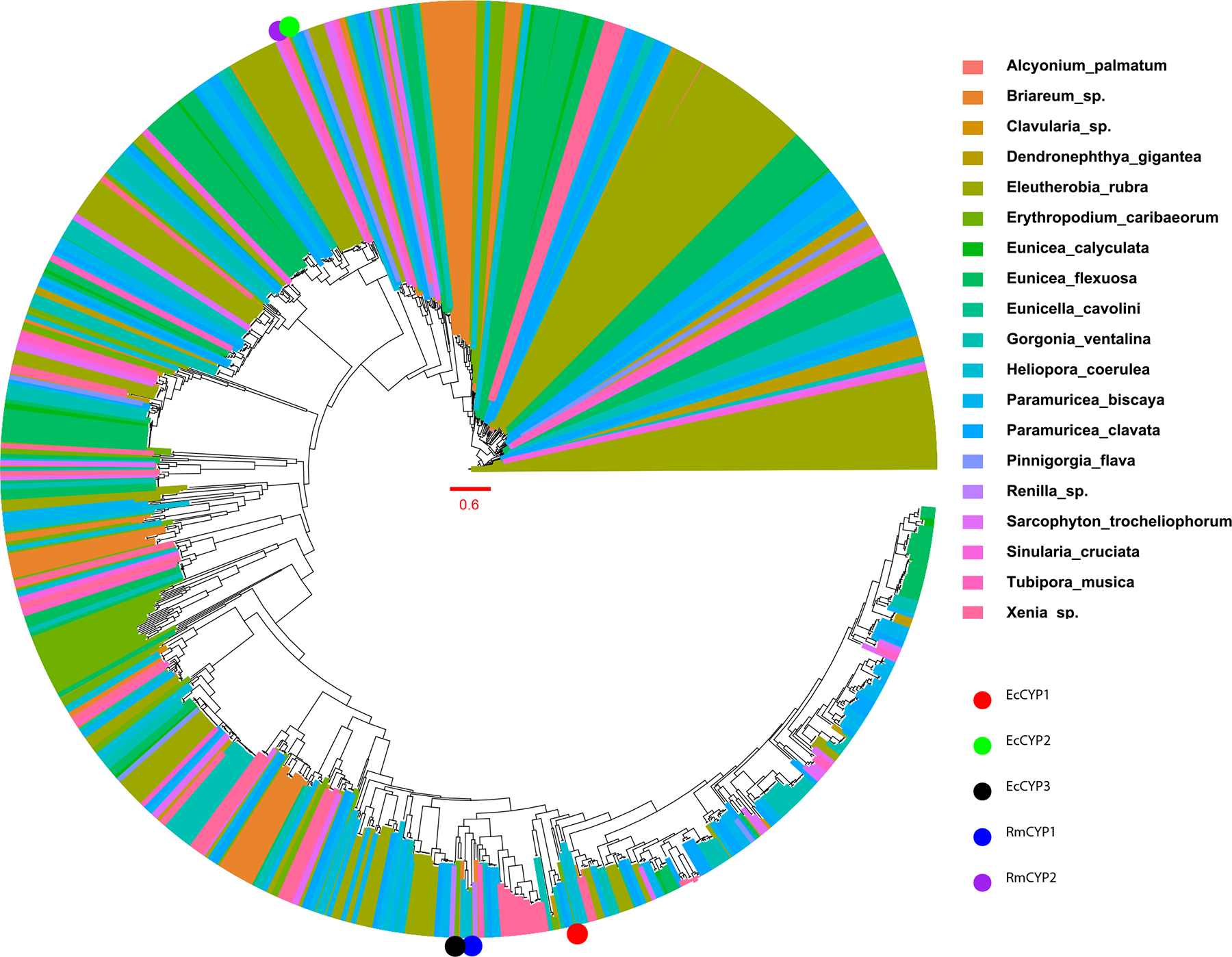
Maximum likelihood phylogenetic analysis of octocoral cytochrome P450 genes. Many TPS-linked P450s cluster together, indicating a potential common origin in octocoral specialized metabolism.
Extended Data Fig. 10. Predicted protein structure of EcAT1.
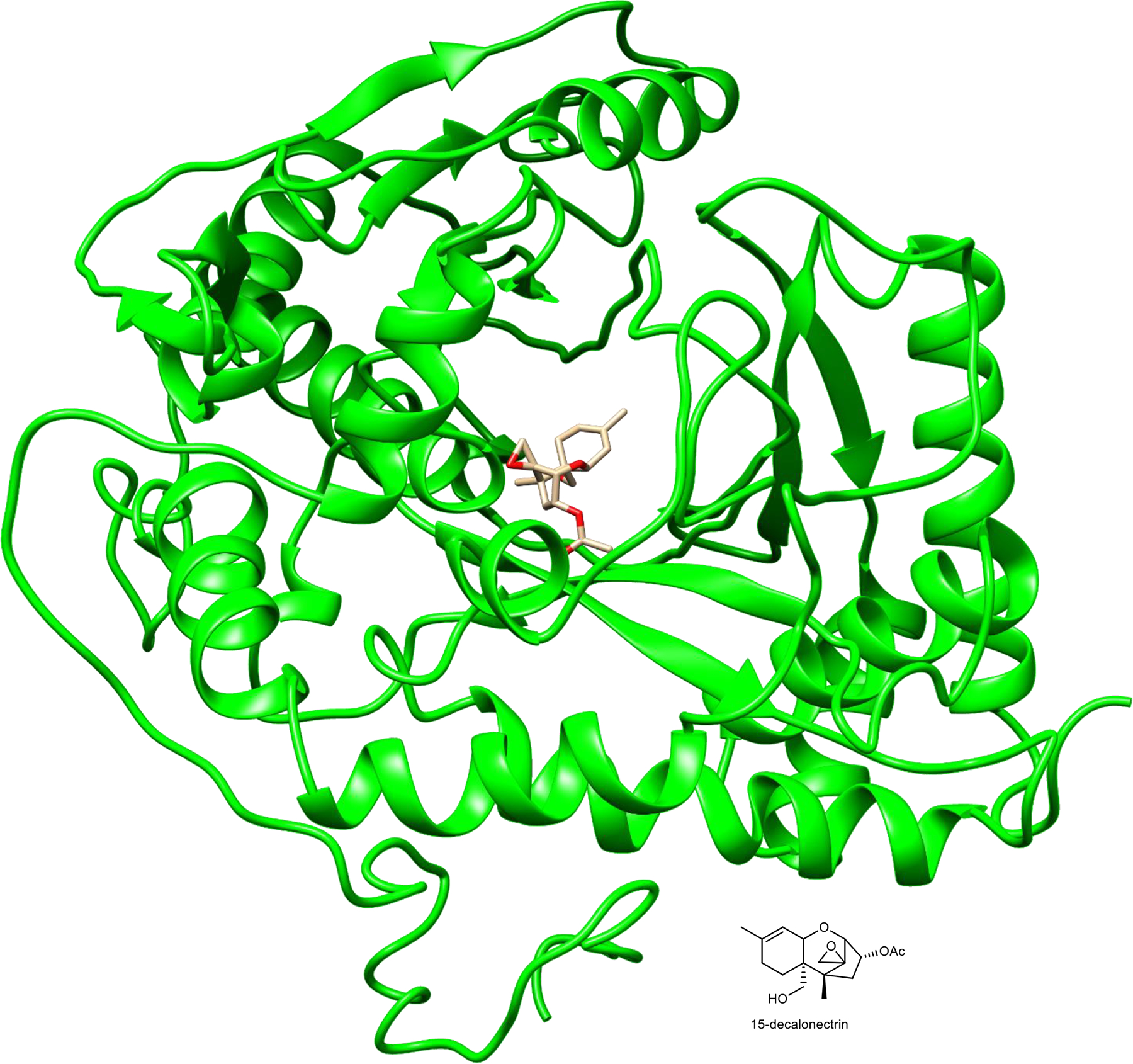
3D model of EcAT1 protein structure (green ribbon) showing sesquiterpene alcohol substrate, 15-decalonectrin, bound to the active site. This structure and ligand binding model were obtained using the i-TASSER web server on the basis of similarity to trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides (TRI3, PDB# 3fp0).
Supplementary Material
Acknowledgments
The coral photos in the graphical abstract and Fig. 1 were provided by Bailey Miller and Jonathan Simpson, respectively, and are used with permission. We thank J.M. Winter for helpful discussions, J. Skalicky for assistance with NMR data acquisition, and J.A. Maschek for assistance with GCMS data acquisition. NMR, MS, and sequencing data were acquired at University of Utah core facilities. This work was funded by National Institutes of Health grant GM122521 and the ALSAM Foundation.
Footnotes
Competing interests Authors declare that they have no competing interests.
Data availability:
All supporting data are described in the publication and associated raw data files are available upon request. All sequencing data (raw reads and assembled TPS genes and contigs) were deposited in GenBank (accession numbers, Supplementary Information Table 3). Sequencing data reported in this study: SRR15783032: SRR15817518, SRR15817517, OK081311, OK081312, OK081313, OK081314, OK081315, OK081316, OK081317, OK081318, OK081319, OK081320; Octocoral SRA data obtained from NCBI: DRR253190, SRR8506632, SRR12021959, SRR6782832, SRR6820379, SRR5123105, ERR2192493, SRR14295591, ERR2190350, ERR2190370, ERR2191368, SRR4449115, SRR14295593, SRR9330360, SRR14295588, SRR10873896, SRR12904788, SRR14295605, SRR14295600, SRR8297742, SRR7585363, SRR7174588, SRR7174589, SRR7174590, SRR7174591, SRR13925246, SRR8293935, SRR8486075, SRR8486076, SRR8486077, SRR8486078, SRR8486079, SRR8486080, SRR8486081, SRR8486082, SRR8486083, SRR6039601, SRR6039602, SRR6039603, SRR6039604, SRR6039605, SRR6039606, SRR12876609, SRR12876610, SRR12876613, SRR12876614, SRR12876620, SRR12876621, SRR12876622, SRR12876624, SRR12876625, SRR12876628, SRR12876629, SRR12876631, SRR12876632, SRR12876633, SRR12876635, SRR12876636, SRR12876637, SRR12876638, SRR12876639, SRR12876640, SRR12876644, SRR12876647, SRR12876648, SRR12876649, SRR12876650, SRR12876653, SRR12876654, SRR12876657, SRR12876658, SRR12876659, SRR12876660, SRR12876661, SRR12876663, SRR12876664, SRR935078, SRR935079, SRR935080, SRR935081, SRR935082, SRR935083, SRR935084, SRR935085, SRR935086, SRR935087, SRR935088, SRR935089, ERR3040053, ERR3040054, SRR12587798, SRR12587799, SRR12587800, SRR12587801, SRR12587803, SRR12587805, SRR12587806, SRR12587807, SRR12587808, SRR5949848, SRR7521178, SRR7521179, SRR7521180, SRR7521181, ERR3664727, ERR3664728, ERR3664729, ERR3664730, SRR13925244, SRR12573942, SRR12573944, SRR12573945, SRR12573946, ERR3026434, ERR3026435, SRR9278440, SRR9278441, SRR9278446, SRR9278447, SRR8113906, SRR8113907, SRR8113908, SRR8113909; Proteins used for HMM: BAM78698.1, BAM78697.1, WP_030430753.1, ADI87447.1, ADI87448.1, AXN72980.1; PDB accessions for EcTPS1 structural homologues: 4okmA, 6tbdA, 3kb9A, 3v1vA, 6vkzA, 5a0iA, 4zq8A, 1hm7B, 6q4sA, 5uv0A; PDB accessions for EcAT1 structural homologues: 7kvw, 3fp0, 6n8e, 6ad3, 5u89, 6mfz, 4zxh, 5t3e, 5isw, 4jn3.
References
- 1.McFadden CS, Sánchez JA & France SC Molecular Phylogenetic Insights into the Evolution of Octocorallia: A Review. Integrative and Comparative Biology 50, 389–410 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Hoang B, Sawall Y, Al-Sofyani A & Wahl M Chemical versus structural defense against fish predation in two dominant soft coral species (Xeniidae) in the Red Sea. Aquat. Biol. 23, 129–137 (2015). [Google Scholar]
- 3.Department of Chemistry, University of Canterbury. MarinLit database. MarinLit http://www.chem.canterbury.ac.nz/marinlit/marinlit.shtml. [Google Scholar]
- 4.Rocha J, Peixe L, Gomes NCM & Calado R Cnidarians as a Source of New Marine Bioactive Compounds—An Overview of the Last Decade and Future Steps for Bioprospecting. Marine Drugs 9, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X-T et al. The Total Synthesis of Eleutherobin. J. Am. Chem. Soc. 121, 6563–6579 (1999). [Google Scholar]
- 6.Mydlarz LD, Jacobs RS, Boehnlein J & Kerr RG Pseudopterosin Biosynthesis in Symbiodinium sp., the Dinoflagellate Symbiont of Pseudopterogorgia elisabethae. Chemistry & Biology 10, 1051–1056 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Barsby T & Kubanek J Isolation and structure elucidation of feeding deterrent diterpenoids from the sea pansy, Renilla reniformis. J. Nat. Prod. 68, 511–516 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Stierle DB, Carte B, Faulkner DJ, Tagle B & Clardy J The asbestinins, a novel class of diterpenes from the gorgonian Briareum asbestinum. J. Am. Chem. Soc. 102, 5088–5092 (1980). [Google Scholar]
- 9.Yamada Y et al. Terpene synthases are widely distributed in bacteria. Proc Natl Acad Sci USA 112, 857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field B & Osbourn AE Metabolic Diversification—Independent Assembly of Operon-Like Gene Clusters in Different Plants. Science 320, 543 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Morita M & Schmidt EW Parallel lives of symbionts and hosts: chemical mutualism in marine animals. Nat. Prod. Rep. 35, 357–378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen B-W et al. Klysimplexins I–T, eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Org. Biomol. Chem. 9, 834–844 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Meguro A, Tomita T, Nishiyama M & Kuzuyama T Identification and Characterization of Bacterial Diterpene Cyclases that Synthesize the Cembrane Skeleton. ChemBioChem 14, 316–321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderah DJ, Rutledge N, Schmitz FJ & Ciereszko LS Marine natural products: cembrene-A and cembrene-C from a soft coral, Nephthea species. J. Org. Chem. 43, 1614–1616 (1978). [Google Scholar]
- 15.Roethle PA & Trauner D The chemistry of marine furanocembranoids, pseudopteranes, gersolanes, and related natural products. Nat. Prod. Rep. 25, (2008). [DOI] [PubMed] [Google Scholar]
- 16.Rinkel J et al. Mechanisms of the Diterpene Cyclases β-Pinacene Synthase from Dictyostelium discoideum and Hydropyrene Synthase from Streptomyces clavuligerus. Chem. Eur. J. 23, 10501–10505 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Conci N, Vargas S & Wörheide G The Biology and Evolution of Calcite and Aragonite Mineralization in Octocorallia. Frontiers in Ecology and Evolution 9, 81 (2021). [Google Scholar]
- 18.Hutchinson MC, Cagua EF, Balbuena JA, Stouffer DB & Poisot T paco: implementing Procrustean Approach to Cophylogeny in R. Methods Ecol Evol 8, 932–940 (2017). [Google Scholar]
- 19.Li G, Dickschat JS & Guo Y-W Diving into the world of marine 2,11-cyclized cembranoids: a summary of new compounds and their biological activities. Nat. Prod. Rep. 10.1039.D0NP00016G (2020) doi: 10.1039/D0NP00016G. [DOI] [PubMed] [Google Scholar]
- 20.Frenz JL Terpene Biosynthesis in the Octocorals Erythropodium caribaeorum and Plexaurella spp. (Florida Atlantic University, 2006). [Google Scholar]
- 21.Al-Footy KO, Alarif WM, Zubair MS, Ghandourah MA & Aly MM Antibacterial and cytotoxic properties of isoprenoids from the red sea soft coral, Lobophytum sp. Trop. J. Pharm Res 15, 1431 (2016). [Google Scholar]
- 22.Garvey GS, McCormick SP, Alexander NJ & Rayment I Structural and functional characterization of TRI3 trichothecene 15-O-acetyltransferase from Fusarium sporotrichioides. Protein Science 18, 747–761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britton R, Roberge M, Berisch H & Andersen RJ Antimitotic diterpenoids from Erythropodium caribaeorum: isolation artifacts and putative biosynthesis intermediates. Tetrahedron Lett. 42, 2953–2956 (2001). [Google Scholar]
- 24.Faddeeva-Vakhrusheva A et al. Coping with living in the soil: the genome of the parthenogenetic springtail Folsomia candida. BMC Genomics 18, 493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkpatrick M How and Why Chromosome Inversions Evolve. PLoS Biol 8, e1000501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino T & McLysaght A Interacting Gene Clusters and the Evolution of the Vertebrate Immune System. Molecular Biology and Evolution 25, 1855–1862 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Takos AM & Rook F Why biosynthetic genes for chemical defense compounds cluster. Trends in Plant Science 17, 383–388 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Rinkel J, Lauterbach L, Rabe P & Dickschat JS Two Diterpene Synthases for Spiroalbatene and Cembrene A from Allokutzneria albata. Angewandte Chemie International Edition 57, 3238–3241 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Ennajdaoui H et al. Trichome specific expression of the tobacco (Nicotiana sylvestris) cembratrien-ol synthase genes is controlled by both activating and repressing cis-regions. Plant Molecular Biology 73, 673–685 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Li X-L et al. Rapid discovery and functional characterization of diterpene synthases from basidiomycete fungi by genome mining. Fungal Genetics and Biology 128, 36–42 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Rinkel J, Köllner TG, Chen F & Dickschat JS Characterisation of three terpene synthases for β-barbatene, β-araneosene and nephthenol from social amoebae. Chem. Commun. 55, 13255–13258 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Roy A, Kucukural A & Zhang Y I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trifinopoulos J, Nguyen L-T, von Haeseler A & Minh BQ W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emms DM & Kelly S OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supporting data are described in the publication and associated raw data files are available upon request. All sequencing data (raw reads and assembled TPS genes and contigs) were deposited in GenBank (accession numbers, Supplementary Information Table 3). Sequencing data reported in this study: SRR15783032: SRR15817518, SRR15817517, OK081311, OK081312, OK081313, OK081314, OK081315, OK081316, OK081317, OK081318, OK081319, OK081320; Octocoral SRA data obtained from NCBI: DRR253190, SRR8506632, SRR12021959, SRR6782832, SRR6820379, SRR5123105, ERR2192493, SRR14295591, ERR2190350, ERR2190370, ERR2191368, SRR4449115, SRR14295593, SRR9330360, SRR14295588, SRR10873896, SRR12904788, SRR14295605, SRR14295600, SRR8297742, SRR7585363, SRR7174588, SRR7174589, SRR7174590, SRR7174591, SRR13925246, SRR8293935, SRR8486075, SRR8486076, SRR8486077, SRR8486078, SRR8486079, SRR8486080, SRR8486081, SRR8486082, SRR8486083, SRR6039601, SRR6039602, SRR6039603, SRR6039604, SRR6039605, SRR6039606, SRR12876609, SRR12876610, SRR12876613, SRR12876614, SRR12876620, SRR12876621, SRR12876622, SRR12876624, SRR12876625, SRR12876628, SRR12876629, SRR12876631, SRR12876632, SRR12876633, SRR12876635, SRR12876636, SRR12876637, SRR12876638, SRR12876639, SRR12876640, SRR12876644, SRR12876647, SRR12876648, SRR12876649, SRR12876650, SRR12876653, SRR12876654, SRR12876657, SRR12876658, SRR12876659, SRR12876660, SRR12876661, SRR12876663, SRR12876664, SRR935078, SRR935079, SRR935080, SRR935081, SRR935082, SRR935083, SRR935084, SRR935085, SRR935086, SRR935087, SRR935088, SRR935089, ERR3040053, ERR3040054, SRR12587798, SRR12587799, SRR12587800, SRR12587801, SRR12587803, SRR12587805, SRR12587806, SRR12587807, SRR12587808, SRR5949848, SRR7521178, SRR7521179, SRR7521180, SRR7521181, ERR3664727, ERR3664728, ERR3664729, ERR3664730, SRR13925244, SRR12573942, SRR12573944, SRR12573945, SRR12573946, ERR3026434, ERR3026435, SRR9278440, SRR9278441, SRR9278446, SRR9278447, SRR8113906, SRR8113907, SRR8113908, SRR8113909; Proteins used for HMM: BAM78698.1, BAM78697.1, WP_030430753.1, ADI87447.1, ADI87448.1, AXN72980.1; PDB accessions for EcTPS1 structural homologues: 4okmA, 6tbdA, 3kb9A, 3v1vA, 6vkzA, 5a0iA, 4zq8A, 1hm7B, 6q4sA, 5uv0A; PDB accessions for EcAT1 structural homologues: 7kvw, 3fp0, 6n8e, 6ad3, 5u89, 6mfz, 4zxh, 5t3e, 5isw, 4jn3.


