Abstract
Background:
Emerging data suggest that metastasis is a spectrum of disease burden rather than a binary state, and local therapies, such as radiation, might improve outcomes in oligometastasis. However, current definitions of oligometastasis are solely numerical.
Objective:
To characterize the somatic mutational landscape across the disease spectrum of metastatic castration-sensitive prostate cancer (mCSPC) to elucidate a biological definition of oligometastatic CSPC.
Design, setting, and participants:
This was a retrospective study of men with mCSPC who underwent clinical-grade sequencing of their tumors (269 primary tumor, 25 metastatic sites). Patients were classified as having biochemically recurrent (ie, micrometastatic), metachronous oligometastatic (≤5 lesions), metachronous polymetastatic (>5 lesions), or de novo metastatic (metastasis at diagnosis) disease.
Outcome measurements and statistical analysis:
We measured the frequency of driver mutations across metastatic classifications and the genomic associations with radiographic progression-free survival (rPFS) and time to castrate-resistant prostate cancer (CRPC).
Results and limitations:
The frequency of driver mutations in TP53 (p = 0.01), WNT (p = 0.08), and cell cycle (p = 0.04) genes increased across the mCSPC spectrum. TP53 mutation was associated with shorter rPFS (26.7 vs 48.6 mo; p = 0.002), and time to CRPC (95.6 vs 155.8 mo; p = 0.02) in men with oligometastasis, and identified men with polymetastasis with better rPFS (TP53 wild-type, 42.7 mo; TP53 mutated, 18.5 mo; p = 0.01). Mutations in TP53 (incidence rate ratio [IRR] 1.45; p = 0.004) and DNA double-strand break repair (IRR 1.61; p < 0.001) were associated with a higher number of metastases. Mutations in TP53 were also independently associated with shorter rPFS (hazard ratio [HR] 1.59; p = 0.03) and the development of CRPC (HR 1.71; p = 0.01) on multivariable analysis. This study was limited by its retrospective nature, sample size, and the use of commercially available sequencing platforms, resulting in a limited predefined set of genes examined.
Conclusions:
Somatic mutational profiles reveal a spectrum of metastatic biology that helps in redefining oligometastasis beyond a simple binary state of lesion enumeration.
Patient summary:
Oligometastatic prostate cancer is typically defined as less than three to five metastatic lesions and evidence suggests that using radiation or surgery to treat these sites improves clinical outcomes. As of now, treatment decisions for oligometastasis are solely defined according to the number of lesions. However, this study suggests that tumor mutational profiles can provide a biological definition of oligometastasis and complement currently used numerical definitions.
Keywords: Oligometastatic prostate cancer, Next Generation Sequencing
1. Introduction
Metastasis has long been viewed as part of a binary state, not only for diagnosis but also for treatment decisions. However, emerging evidence suggests that metastatic disease is instead better described as a spectrum of metastatic proclivity [1] currently represented clinically by enumeration of lesions. From this spectrum hypothesis was born the idea of oligometastasis, a state whereby the biological behavior of the disease in patients with few lesions is more akin to locoregional disease rather than extensive systemic metastasis [1]. Seminal attempts to understand the biological underpinnings of the oligometastatic state have implicated basic biological programs of invasion and metastasis, such as epithelial-mesenchymal transition (EMT), innate and adaptive immunity, inactivation of tumor suppressor genes, and DNA repair pathways with metastatic potential [2–4].
One important implication of an attenuated metastatic phenotype postulated for an oligometastatic state is that local therapy to these macroscopic lesions might result in durable disease control and, in some cases, even cure [5]. A series of prospective randomized trials in lung cancer [6,7] and metachronous disease of varying histologies [8] has demonstrated that local therapy improves progression-free survival (PFS) and overall survival (OS) in oligometastatic disease. In metachronous oligometastatic castration-sensitive prostate cancer (oligomCSPC), metastasis-directed therapy (MDT) also prolongs androgen deprivation therapy–free survival and PFS compared to observation [9,10].
Several critical limitations remain in our understanding of the oligometastatic state. First, similar to the initial characterizations, metastasis is currently categorized according to a binary definition; and second, clinical characteristics alone are used to make decisions regarding treatment. Much of our understanding regarding the genomic landscape of metastatic PC comes from studies in the metastatic castration-resistant PC (mCRPC) state [11–13]. However, much less is known about the relative frequency and implications of mutations across the mCSPC spectrum, especially in oligomCSPC. Our hypothesis was that metastatic biology (oligometastases vs polymetastases) and metastatic onset (de novo vs metachronous) are driven by underlying genomic changes. The goal of this study was to better characterize the molecular landscape of oligomCSPC within the wider context of mCSPC to establish a more biological definition of the oligometastatic state and to understand the implications of this more refined description on prognosis and treatment outcomes.
2. Patients and materials
After institutional review board approval, we retrospectively identified all men with mCSPC or biochemically recurrent (BCR, ie, micrometastatic) PC whose tumors had undergone standard-of-care somatic next-generation sequencing (NGS). Commercial NGS platforms with College of American Pathologists-Clinical Laboratory Improvement Amendments certification included the Foundation One CDx (324-gene panel) and Personal Genome Diagnostics CancerSELECT 125 (125-gene panel) assays. Both panels detect single-nucleotide variants (SNVs), small indels, amplifications, translocations, microsatellite instability, and tumor mutational burden (TMB). A total of 269 samples were from primary tumor and 25 samples were from metastatic sites. Putative driver mutations were alterations defined according to the commercial tests using the publicly available COSMIC tumor variant database. Variants of unknown significance (VUSs) or alterations not listed in COSMIC were not considered mutations. A complete list of genes profiled are listed at vendor websites, but specific genes and pathways of interest in our current manuscript were: TP53; DNA double-strand break (DDSB) repair genes: ATM, ATRX, CHEK2, MRE11, NBN, RAD51, FANCA, FANCD2, FANCG, and PALB2; WNT pathway genes: CTNNB1, APC, and RNF43; cell cycle genes: Rb1, CCND1–3, CDKN1B, and CDKN2A; and PIK3/AKT/mTOR pathway genes: PIK3R1, PIK3CA, PIK3CB, PTEN, mTOR, AKT1, TSC1, and TSC2.
Additional inclusion criteria were previous definitive treatment of the primary tumor for patients with BCR and metachronous disease. Available follow-up data from serial physical examination, imaging, and prostate-specific antigen (PSA) measurements were obtained; all follow-up procedures were generally, in accordance with National Comprehensive Cancer Network guidelines, conducted in a regimented fashion by a small group of oncologists at Johns Hopkins.
Patients were classified into four metastatic categories. The BCR (micrometastatic) category had rising PSA following definitive therapy without evidence of overt macrometastatic disease through current follow-up. The metachronous oligometastatic (met oligo) category had five or fewer sites of metastasis at first recurrence (pelvic node, extrapelvic nodes, bone, and visceral). The metachronous polymetastatic (met poly) category had more than five sites of metastatic recurrence. The de novo category had metastatic disease at first mCSPC diagnosis. Metastases were designated using conventional imaging (computed tomography, magnetic resonance imaging, and bone scintigraphy) for consistency.
Endpoints of interest were the distribution of genomic profiles across metastatic categories, radiographic PFS (rPFS; absence of new metastasis, growth of existing metastasis, or death), time to CRPC (according to Prostate Cancer Working Group 3 criteria [14]), and OS, all from the time of the original prostate cancer diagnosis. In addition, the pattern of failure at the time of metastatic disease was a primary endpoint. Patients without events were censored at the last patient contact for OS, the last imaging date for rPFS, and the last PSA measurement date for CRPC.
Differences in the frequency of putative driver mutations and patterns of failure within each metastatic category were compared using a Pearson’s χ2 test or Fisher’s exact test. Mutations in putative driver genes were predicted to be associated with higher number of metastatic lesions, which was assessed through negative binomial regression and incidence rate ratios (IRRs). In multivariable analysis (MVA) this model was adjusted for variables predicted to be associated with a higher number of lesions (Gleason grade group, PSA before identification of metastasis [pre-metastasis PSA], and metastatic status). High-risk mutations were predicted to be associated with poorer outcomes. This was assessed through Kaplan-Meier survival analysis reporting median time to rPFS, time to CRPC, and 8-yr OS, which were stratified by metastatic category and mutational profile and compared using log-rank and pairwise tests with the Bonferroni method. Cox proportional hazards regression was performed to calculate hazard ratios (HRs) and 95% confidence intervals (CIs), with adjustment for variables a priori felt to be associated with outcomes of interest (genomic mutations, pre-metastasis PSA, Gleason grade group, number of lesions, and metastatic category), all of which were included in an MVA. The proportionality of hazards was assessed for each variable, and Schoenfeld residuals were visually inspected for potential time-variant biases. A p value <0.05 was considered significant, except for MVA, which was adjusted to 0.05/3 to account for multiple comparisons. All analyses were conducted using R.
3. Results
3.1. Outcomes using clinical definitions of metastatic PC
A total of 294 patients underwent somatic sequencing: 45 with BCR at last follow-up, 102 in the met oligo category, 22 in the met poly category, and 125 in the de novo category of metastatic disease. Median follow-up from diagnosis for these categories was 77.7, 86.3, 82.2, and 27 mo, respectively. Baseline characteristics are reported in Supplementary Table 1. As expected, the de novo category tended to have differences in pathologic T and N stage and Gleason grade group. However, the distribution of variables was similar for the BCR, met oligo, and met poly categories.
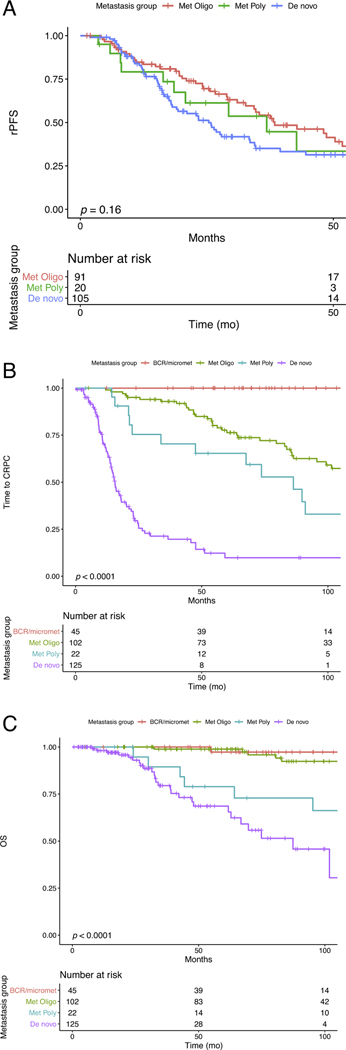
Median rPFS was 38.4 mo (n = 47 events) for the met oligo category, 36.8 mo (n = 11 events) for the met poly category, and 25.4 mo (n = 65 events) for the de novo category (log-rank p = 0.16; Fig. 1A). Median follow-up for patients without an rPFS event was 48.7 mo. There were significant differences in median time to CRPC metastatic category: BCR, not reached (n = 0 events); met oligo, 127.6 mo (n = 41 events); met poly, 86.3 mo (n = 15 events); and de novo, 15.6 mo (n = 81 events); log-rank p < 0.0001 (Fig. 1B). There were also significant differences in median OS: BCR, not reached (n = 1 event); met oligo, not reached (n = 10 events); met poly, 141.4 mo (n = 8 events); and de novo, 87.4 mo (n = 26 events); log rank p < 0.0001 (Fig. 1C). The median follow-up for patients not experiencing a CRPC or OS event was 62.8 and 61.2 mo, respectively. Finally, the number of lesions was also associated with significant differences in rPFS (log-rank p = 0.01), time to CRPC (log-rank p < 0.0001), and OS (log-rank p < 0.0001; Supplementary Fig. 1A–C). Pairwise comparisons between groups are presented in Supplementary Table 2.
Fig. 1.
– Metastatic categories are associated with time to castrate-resistance and overall survival. Metastatic categories of biochemically recurrent (BCR, i.e., micrometastatic), metachronous oligometastatic (≤ 5 lesions), metachronous polymetastatic (> 5 lesions), and de novo (metastasis at diagnosis). (A) Radiographic progression-free survival (rPFS) stratified by metastatic category (p = 0.16). (B) Time to CRPC (per Prostate Cancer Working Group 3) by metastatic category (p < 0.0001). (C) Overall survival (OS) from original diagnosis by metastatic category (p < 0.0001).
3.2. Genomic profiles of metastatic categories
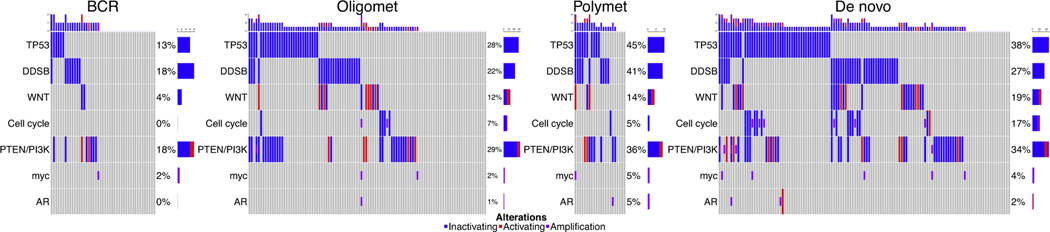
Driver mutations in genes of interest were significantly different across metastatic categories and represented a spectrum of frequency for several genes and pathways (Fig. 2, Supplementary Fig. 2), including TP53 (p = 0.01) and the WNT (p = 0.08) and cell cycle (p = 0.004) pathways (Supplementary Table 3). TMB was similar between all groups (p = 0.3). The met oligo category had a frequency of driver mutations between those of the BCR and met poly categories. The frequency of mutations in the met poly category tended to be similar to the de novo metastatic category (all p > 0.05).
Fig. 2.
– Cancer-relevant gene mutations correlate directly across the spectrum of metastatic castration-sensitive prostate cancer. An oncoprint of metastatic categories shows that the mutation frequency increases with increasing metastatic tendency and burden, including TP53 (p = 0.01), WNT (p = 0.04), and cell cycle (p = 0.004) genes. The metachronous oligometastatic (Oligomet) category had a frequency of driver mutations intermediate between those of BCR (micrometastatic) and metachronous polymetastatic (Polymet). The frequency of mutations in the Polymet category tended to mirror or were higher than those of the de novo metastatic category (no significant differences according to p values).
Given the similarities in mutation frequency between the met poly and de novo categories, we tested whether met poly was a lag-time bias manifestation of de novo disease. When measured from first metastasis, the time to CRPC was similar between the met poly and de novo category (16.4 mo [n = 15 events] vs 15.6 mo [n = 81 events]; pairwise p = 0.8), while time to CRPC in the met oligo category was longer (43 mo [n = 43 events]; p = 0.002 for met oligo vs met poly; p < 0.001 for met oligo vs de novo).
Univariable negative binomial regression identified covariates associated with increasing number of metastatic lesions in men with metachronous disease (Supplementary Table 4). On MVA, TP53 mutation (IRR 1.45; p = 0.004), DDSB pathway mutations (IRR 1.61; p < 0.001), pre-metastasis PSA (IRR 1.001; p < 0.001), and the de novo metastatic category (IRR 2.2; p < 0.001) were associated with a greater number of lesions (Table 1). These findings remained after sensitivity analysis including only patients with sequencing of the primary tumor (Supplementary Table 5). In addition, when performing the analysis using multivariable logistic regression with the same variables, DDSB mutations remained associated with the development of met poly versus met oligo disease (odds ratio 3.49; p = 0.03).
Table 1.
– Multivariable negative binomial for factors associated with increasing number of lesions in men with metachronous metastatic castration-sensitive prostate cancer
| IRR (95% CI) | p value | |
|---|---|---|
|
| ||
| TP53 | 1.45 (1.13–1.86) | 0.004 |
| DNA double-strand break | 1.61 (1.23–2.12) | <0.001 |
| WNT | 1.31 (0.94–1.84) | 0.12 |
| Cell cycle | 1.18 (0.82–1.74) | 0.40 |
| PTEN/PI3K | 1.09 (0.84–1.43) | 0.50 |
| Gleason grade | 1.01 (0.91–1.13) | 0.80 |
| Pre-metastasis prostate-specific antigen | 1.001 (1.0003–1.002) | <0.001 |
| De novo vs metachronous | 2.20 (1.69–2.88) | <0.001 |
IRR = incidence rate ratio; CI = confidence interval.
3.3. Genomic profiles associated with clinical outcomes across metastatic categories
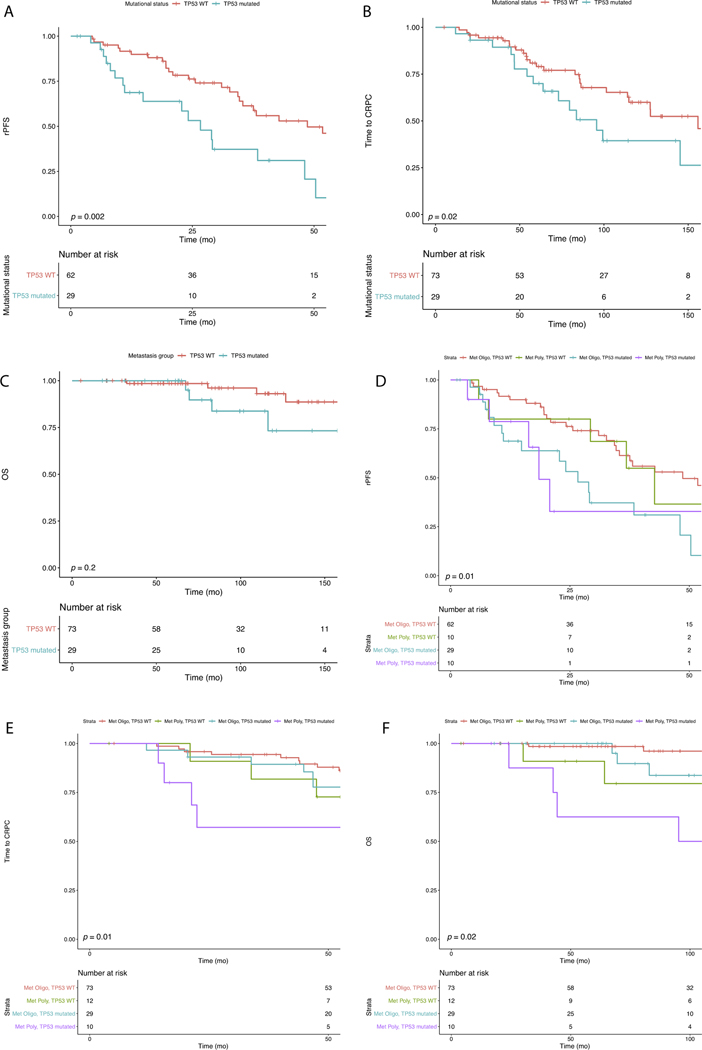
Mutations in TP53 were associated with shorter median rPFS in the whole population (23.4 mo [n = 52 events] vs 36.8 mo [n = 71 events]; p = 0.02). For the met oligo category, TP53 mutations remained associated with shorter median rPFS (26.7 mo [n = 18 events] vs 48.6 mo [n = 28 events]; log-rank p = 0.002; Fig. 3A] and median time to CRPC (95.6 mo [n = 17 events] vs 155.8 mo [n = 24 events]; log-rank p = 0.02; Fig. 3B). In addition, the 8-yr OS rate was lower for men with TP53 mutation, but the difference was not significant (82.8% [n = 3 events] vs 96.1% [n = 2 events]; log-rank p = 0.2; Fig. 3C). Mutations in DDSB, WNT, cell cycle, and PTEN/PI3K pathways did not impact rPFS, time to CRPC, or OS, but the power was limited because of low mutation frequencies (Supplementary Fig. 3A–L).
Fig. 3.
– TP53 mutational status is prognostic for metachronous castration-sensitive prostate cancer. TP53 mutations are associated with outcomes for the metachronous oligometastatic (Met Oligo) category for: (A) rPFS (26.7 vs 48.6 mo; p = 0.002); (B) time to CRPC (95.6 vs 155.8 mo; p = 0.02); and (C) 8-yr OS (82.8% vs 96.1%; p = 0.2). TP53 mutation also stratifies the metachronous polymetastatic (Met Poly) category. (D) rPFS for men in the Met Poly category with TP53 WT (42.7 mo) or TP53 mutated tumors (18.5 mo; p = 0.01) (E) Time to CRPC for the Met Poly category with TP53 WT (86.3 mo) or TP53 mutated tumors (73.7 mo; p = 0.01); (F) 8-yr OS for the Met Poly category with TP53 WT (83.7%) or TP53 mutated tumors (50.0%; p = 0.02). rPFS = radiographic progression-free survival; CRPC = castrate-resistant prostate cancer; OS = overall survival; WT = wild type.
TP53 mutation also stratified the met poly category and identified patients with outcomes that mirrored those for patients in the met oligo category. Median rPFS was similar for men with met poly/TP53 wild-type (WT) tumors (42.7 mo; n = 5 events) and men with met oligo/TP53 WT tumors (48.6 mo; n = 29 events), while median rPFS for men with met poly/TP53 mutated tumors (18.5 mo; n = 6 events) was more similar to that for men with met oligo/TP53 mutated tumors (26.7 mo; n = 18 events; log-rank p = 0.01; Fig. 3D). In addition, men with met poly/TP53 WT tumors had a median time to CRPC (86.3 mo; n = 7 events) that was closer to that for met oligo/TP53 mutated tumors (95.6 mo; n = 17 events) than for met poly/TP53 mutated tumors (73.7 mo; n = 8 events; log-rank p = 0.01; Fig. 3E). Finally, the 8-yr OS rate for men with met poly/TP53 WT tumors (79.5%; n = 2 events) was more similar to that for men with met oligo/TP53 mutated tumors (96.1%; n = 2 events) compared to men with met poly/TP53 mutated tumors (50.0%; n = 4 events; log-rank p = 0.02; Fig. 3G).
Univariable Cox regression analysis for rPFS and CRPC is presented in Supplementary Table 6. On MVA (Table 2) TP53 mutation was associated with rPFS (HR 1.59; p = 0.03) and CRPC (HR 1.71; p = 0.01). These findings remained after performing a sensitivity analysis including only patients with sequencing of the primary tumor (Supplementary Table 7).
Table 2.
– Multivariable Cox regression for factors associated with rPFS and time to CRPC
| rPFS |
CRPC |
|||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
|
| ||||
| TP53 | 1.59 (1.04–2.41) | 0.03 | 1.71 (1.16–2.52) | 0.01 |
| DNA double-strand break | 1.07 (0.68–1.73) | 0.7 | 1.22 (0.80–1.86) | 0.4 |
| WNT | 1.24 (0.65–2.36) | 0.5 | 0.94 (0.53–1.67) | 0.8 |
| Cell cycle | 1.08 (0.59–1.98) | 0.8 | 0.95 (0.50–1.81) | 0.9 |
| PTEN/PI3K | 0.92 (0.59–1.44) | 0.7 | 0.95 (0.63–1.43) | 0.8 |
| Pre-metastasis prostate-specific antigen | 1.0006 (1–1.001) | 0.2 | 1.01 (1.001–1.003) | <0.001 |
| Number of lesions | 1.05 (1.01–1.10) | 0.03 | 1.08 (1.04–1.12) | <0.001 |
| Gleason grade group | 1.08 (0.90–1.29) | 0.4 | 1.52 (1.24–1.86) | <0.001 |
| Location (vs abdominal node) | 0.6 | 0.9 | ||
| Pelvic node | 1.004 (0.47–2.14) | 1.28 (0.57–2.87) | ||
| Bone | 0.74 (0.40–1.38) | 1.002 (0.53–1.90) | ||
| Visceral | 0.64 (0.30–1.39) | 1.12 (0.51–2.45) | ||
| De novo vs metachronous | 1.05 (0.63–1.72) | 0.9 | 5.39 (3.18–8.88) | <0.001 |
rPFS = radiographic progression-free survival; CRPC = castrate-resistant prostate cancer; HR = hazard ratio; CI = confidence interval.
3.4. Patterns of failure in the metachronous metastatic categories
Patterns of first metastasis for patients in the met oligo and met poly categories were associated with the genomic profile (Table 3). Tumors recurring in abdominal nodes, bone, or viscera had a higher incidence of driver mutations in DDSB (p = 0.04) and WNT pathway genes (p = 0.004). Among patients with pelvic-only nodal recurrence, one patient (3%) had a mutation in the WNT pathway, compared to 13% who had bone and 33% who had visceral recurrence. At 5 yr, the cumulative incidence of visceral metastasis was 36% among men with WNT pathway mutations versus 7% among individuals without a mutation (p < 0.001, Supplementary Fig. 4).
Table 3.
– Patterns of failure at the time of first recurrence
| Pathogenic alteration | Patients, n (%) |
p value | |||
|---|---|---|---|---|---|
| Pelvic n = 30 | Abdominal node n = 18 | Bone n = 55 | Visceral n = 21 | ||
|
| |||||
| TP53 | 7 (23) | 9 (50) | 19 (35) | 4 (19) | 0.14 |
| DNA double-strand break | 4 (13) | 5 (28) | 12 (22) | 10 (48) | 0.04 |
| WNT | 1 (3) | 0 (0) | 7 (13) | 7 (33) | 0.004 |
| PTEN/PI3K | 8 (27) | 6 (33) | 17 (31) | 7 (33) | 0.9 |
4. Discussion
Here we report the mutational landscape across the spectrum of mCSPC and demonstrate that the frequency of specific gene mutations correlates directly with increasing metastatic proclivity, consistent with the biologic basis implied by the spectrum theory of metastasis. We found that mutations were lowest among men in the BCR category and highest among those in the met poly and de novo metastatic categories. Importantly, we found that mutations in genes such as TP53 and WNT pathway genes can risk-stratify outcomes for men with metachronous recurrence of their CSPC and predict patterns of metastatic dissemination.
Most studies investigating the genomic landscape of PC have focused on localized CSPC or mCRPC [15–19]. Only a limited number of studies have investigated the mCSPC state [12,20,21]. Hamid et al [12] included 43 men with mCSPC and documented the important observation that an increasing number of mutated tumor suppressor genes were associated with higher risk of progression. Stopsack et al [21] recently demonstrated that individuals with mCSPC and high-volume disease experience shorter times to CRPC and OS, and tend to have a higher frequency of mutations within NOTCH and cell-cycle pathway genes. In addition, mutations in the androgen receptor (AR) gene, cell cycle pathways, and TP53 appeared to be associated with the development of CRPC.
Our report adds significantly to the current body of knowledge: First, we add a large number of sequenced cases for patients who ultimately developed recurrence spanning the mCSPC spectrum. Second, our study had much longer follow-up of 6–7 yr for patients with metachronous metastasis, which is necessary to follow the natural history of the disease. Third, we profiled the whole mCSPC spectrum by grouping men into four disease categories according to the number and timing of metastases, which probably led to the finding that the frequency of TP53, DDSB, WNT, and cell-cycle pathway mutations varies with the metastatic disease category. Fourth, we note that gene mutations in TP53 and the DDSB pathway are strongly associated with the number of metastatic lesions, and mutations within the WNT developmental pathway are associated with patterns of failure at first metastatic recurrence. We and others have shown that EMT developmental plasticity programs are sufficient and appear to be important for prostate cancer metastasis [22,23]. Fifth, we found a similar mutational profile between men in the met poly and de novo mCSPC categories, and the two groups had a comparable time to CPRC when defined from the time of first metastasis. This suggests that these two disease entities share similar disease biology and are only separated by a leadtime bias from earlier manifestation of macrometastatic disease in the de novo category. Lastly, we confirmed the importance of TP53 mutation in the prognosis of mCSPC in general, and show for the first time that TP53 mutational status can stratify outcomes for those with metachronous metastatic recurrence.
These findings have several important research and clinical implications; primary among them is that the data may help in redefining our current definition of oligometastasis, which relies simply on clinical features, namely, the number of lesions detected on imaging [24]. There is no consensus on the definition of oligomCSPC, but three to five lesions is a common criterion [9,10,25,26]. A numerical definition of oligomCSPC has several drawbacks, one of which is the dependence on the sensitivity of the imaging used [27]. Thus, a better understanding of the molecular heterogeneity of oligometastatic disease will be important for future trial design and ultimately patient treatment [28].
Increasing evidence suggests a benefit from consolidation of oligometastatic disease with local therapies [8–10] and additional evidence suggests that men with low-volume metastatic disease might also benefit from local consolidation of the primary tumor [29]. An understanding of the genomic determinants affecting prognosis might aid in deciding how to most effectively incorporate consolidative MDT for individuals with metastatic disease. For example, we noted a trend whereby patients in the met poly category with TP53 WT tumors had rPFS, CRPC, and OS intervals similar to those in the met oligo category, and thus these individuals might also benefit from MDT. These genomic findings, in conjunction with other readily available prognostic indicators such as pre-metastasis PSA, should be studied further to risk-stratify patients and assist in determining the optimal therapy for these individuals such as the use of local consolidative therapy.
The use of primary tumor samples in our study allows for easy clinical translation given the general accessibility of this tissue. However, as noted by Mateo et al [30], increases in the frequency of mutations in AR, TP53, RB1, and the PI3K/AKT pathway are seen in matched mCRPC and primary tumor specimens. This lends credence to our study findings that TP53 is associated with metastasis category and time to CRPC, and highlights the importance of continued investigation of the genomic characteristics of metastatic castration-sensitive lesions.
At present, little therapeutic personalization is carried out for mCSPC in routine practice, with almost all patients receiving luteinizing hormone–releasing hormone agonists/antagonists. There is a trend to also incorporate additional agents targeting the AR axis (abiraterone, enzalutamide, and apalutamide) or chemotherapy, with clinical rather than molecular factors used to aid in this decision. However, given the more aggressive phenotypes and worse prognosis for men with metachronous mCSPC and TP53 mutations, treatment intensification may be preferentially used for these individuals, sparing toxicity for those who are less likely to derive benefit. In addition, while mutations in other pathways did not appear to be prognostic, alterations were found in upwards of 20–30% of men and these might represent actionable targets for therapy [31,32].
This study has several limitations. The cohort was retrospectively identified and thus imaging, treatment, endpoints, and sequencing of tissue were not prospectively defined, collected, or performed. Thus, the cohort is susceptible to all the caveats of such a retrospective study, but patients were managed in a regimented fashion by a small group of oncologists at a single institution. In addition, while 300 patients were included, the sample size for the different metastatic categories was in some cases small and thus the study could be underpowered for detection of some genomic differences. We made additional attempts to account for potential bias via MVA, but all the confounders cannot be completely accounted for and prospective validation is necessary. The multiple comparisons made can also confound MVA, but we adjusted for this in our analysis. Similarly, there are several limitations that arise from the use of these NGS panels. Genes were not predefined at study entry and were limited to those on two commercial NGS panels, and certain deletions or other complicated copy number alterations are not detected, which limits the interrogation and discovery of other important genomic alterations, particularly those with canonical tumor suppressor genes. These clinical-grade NGS panels do not always necessitate comparison with normal tissue, which likely resulted in higher frequency of mutation calls. Furthermore, most samples were from the primary tumor rather than metastatic castration-sensitive sites which may miss mutations acquired during disease progression. Finally, issues related to primary prostate cancer multifocality and sampling of tissue via biopsies may have led to missing of relevant clonal alterations.
5. Conclusions
In conclusion, we found that the frequency of cancer-relevant gene mutations correlates directly across the spectrum of mCSPC and can stratify outcomes for men with mCSPC. These findings, if validated prospectively, may help in redefining oligometastatic disease beyond a simple binary state of lesion enumeration.
Supplementary Material
Funding/Support and role of the sponsor:
PT Tran recieved support from Ronald Rose and Joan Lazar, Nesbitt-McMaster Foundation, Barbara’s Fund, National Capital Cancer Research Fund, Prostate Cancer Foundation, Movember Foundation and the NIH/NCI (U01CA212007, U01CA231776 and R21CA223403).
Footnotes
Financial disclosures: Phuoc T. Tran certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.eururo.2020.12.040.
References
- [1].Hellman S, Weichselbaum RR. Oligometastases. J Clin Oncol 1995;13:8–10. [DOI] [PubMed] [Google Scholar]
- [2].Pitroda SP, Khodarev NN, Huang L, et al. Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat Commun 2018;9:1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Khodarev NN, Pitroda SP, Weichselbaum RR. microRNAs and oligometastasis. Aging 2015;7:146–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Uppal A, Wightman SC, Mallon S, et al. 14q32-encoded microRNAs mediate an oligometastatic phenotype. Oncotarget 2015;6:3540–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Adson MA, van Heerden JA, Adson MH, Wagner JS, Ilstrup DM. Resection of hepatic metastases from colorectal cancer. Arch Surg 1984;119:647–51. [DOI] [PubMed] [Google Scholar]
- [6].Gomez DR, Tang C, Zhang J, et al. Local consolidative therapy vs. maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer: long-term results of a multi-institutional, phase II, randomized study. J Clin Oncol 2019;37:1558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Iyengar P, Wardak Z, Gerber DE, et al. Consolidative radiotherapy for limited metastatic non-small-cell lung cancer: a phase 2 randomized clinical trial. JAMA Oncol 2018;4:e173501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Palma DA, Olson R, Harrow S, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): a randomised, phase 2, open-label trial. Lancet 2019;393:2051–8. [DOI] [PubMed] [Google Scholar]
- [9].Ost P, Reynders D, Decaestecker K, et al. Surveillance or metastasis-directed therapy for oligometastatic prostate cancer recurrence: a prospective, randomized, multicenter phase II trial. J Clin Oncol 2018;36:446–53. [DOI] [PubMed] [Google Scholar]
- [10].Phillips R, Shi WY, Deek M, et al. Outcomes of observation vs stereotactic ablative radiation for oligometastatic prostate cancer: the ORIOLE phase 2 randomized clinical trial. JAMA Oncol 2020;6:650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Abida W, Armenia J, Gopalan A, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017;1, PO.17.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hamid AA, Gray KP, Shaw G, et al. Compound genomic alterations of TP53, PTEN, and RB1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol 2019;76:89–97. [DOI] [PubMed] [Google Scholar]
- [13].Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Scher HI, Morris MJ, Stadler WM, et al. Trial design and objectives for castration-resistant prostate cancer: updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol 2016;34:1402–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Grasso CS, Wu YM, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Espiritu SMG, Liu LY, Rubanova Y, et al. The evolutionary landscape of localized prostate cancers drives clinical aggression. Cell 2018;173, 1003–13.e15. [DOI] [PubMed] [Google Scholar]
- [18].Fraser M, Sabelnykova VY, Yamaguchi TN, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017;541:359–64. [DOI] [PubMed] [Google Scholar]
- [19].Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vandekerkhove G, Struss WJ, Annala M, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol 2019;75:667–75. [DOI] [PubMed] [Google Scholar]
- [21].Stopsack KH, Nandakumar S, Wibmer AG, et al. Oncogenic genomic alterations, clinical phenotypes, and outcomes in metastatic castration-sensitive prostate cancer. Clin Cancer Res 2020;26:3230–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lo UG, Lee CF, Lee MS, Hsieh JT. The role and mechanism of epithelial-to-mesenchymal transition in prostate cancer progression. Int J Mol Sci 2017;18:2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Malek R, Gajula RP, Williams RD, et al. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis.Cancer Res 2017;77:3181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Singh D, Yi WS, Brasacchio RA, et al. Is there a favorable subset of patients with prostate cancer who develop oligometastases? Int J Radiat Oncol Biol Phys 2004;58:3–10. [DOI] [PubMed] [Google Scholar]
- [25].Kneebone A, Hruby G, Ainsworth H, et al. Stereotactic body radiotherapy for oligometastatic prostate cancer detected via prostate-specific membrane antigen positron emission tomography. Eur Urol Oncol 2018;1:531–7. [DOI] [PubMed] [Google Scholar]
- [26].Siva S, Bressel M, Murphy DG, et al. Stereotactic abative body radiotherapy (SABR) for oligometastatic prostate cancer: a prospective clinical trial. Eur Urol 2018;74:455–62. [DOI] [PubMed] [Google Scholar]
- [27].Perera M, Papa N, Christidis D, et al. Sensitivity, specificity, and predictors of positive 68Ga-prostate-specific membrane antigen positron emission tomography in advanced prostate cancer: a systematic review and meta-analysis. Eur Urol 2016;70:926–37. [DOI] [PubMed] [Google Scholar]
- [28].Chang DT, Pollom EL, Keane FK, Wo JY. Treating oligometastatic disease with SABR: more than just a numbers game? Int J Radiat Oncol Biol Phys 2020;107:257–60. [DOI] [PubMed] [Google Scholar]
- [29].Parker CC, James ND, Brawley CD, et al. Radiotherapy to the primary tumour for newly diagnosed, metastatic prostate cancer (STAM-PEDE): a randomised controlled phase 3 trial. Lancet 2018;392:2353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mateo J, Seed G, Bertan C, et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest 2020;130:1743–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Isaacsson Velho P, Qazi F, Hassan S, et al. Efficacy of radium-223 in bone-metastatic castration-resistant prostate cancer with and without homologous repair gene defects. Eur Urol 2019;76:170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nizialek E, Antonarakis ES. PARP inhibitors in metastatic prostate cancer: evidence to date. Cancer Manag Res 2020;12:8105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.