Abstract
Rationale
Idiopathic pulmonary fibrosis (IPF) is a devastating disease characterized by limited treatment options and high mortality. A better understanding of the molecular drivers of IPF progression is needed.
Objectives
To identify and validate molecular determinants of IPF survival.
Methods
A staged genome-wide association study was performed using paired genomic and survival data. Stage I cases were drawn from centers across the United States and Europe and stage II cases from Vanderbilt University. Cox proportional hazards regression was used to identify gene variants associated with differential transplantation-free survival (TFS). Stage I variants with nominal significance (P < 5 × 10−5) were advanced for stage II testing and meta-analyzed to identify those reaching genome-wide significance (P < 5 × 10−8). Downstream analyses were performed for genes and proteins associated with variants reaching genome-wide significance.
Measurements and Main Results
After quality controls, 1,481 stage I cases and 397 stage II cases were included in the analysis. After filtering, 9,075,629 variants were tested in stage I, with 158 meeting advancement criteria. Four variants associated with TFS with consistent effect direction were identified in stage II, including one in an intron of PCSK6 (proprotein convertase subtilisin/kexin type 6) reaching genome-wide significance (hazard ratio, 4.11 [95% confidence interval, 2.54–6.67]; P = 9.45 × 10−9). PCSK6 protein was highly expressed in IPF lung parenchyma. PCSK6 lung staining intensity, peripheral blood gene expression, and plasma concentration were associated with reduced TFS.
Conclusions
We identified four novel variants associated with IPF survival, including one in PCSK6 that reached genome-wide significance. Downstream analyses suggested that PCSK6 protein plays a potentially important role in IPF progression.
Keywords: IPF, genome-wide association study, genomics, survival, PCSK6 protein
At a Glance Commentary
Scientific Knowledge on the Subject
A host of gene variants that increase one’s risk of developing idiopathic pulmonary fibrosis (IPF) have been identified through genomic analyses. Few of these variants have been associated with differential outcomes, however, suggesting that genomic determinants of IPF susceptibility and progression may have limited overlap. To identify genomic determinants of IPF outcomes, a genome-wide association study of IPF survival was conducted.
What This Study Adds to the Field
We identified four novel gene variants associated with differential IPF survival, including one in PCSK6 (proprotein convertase subtilisin/kexin type 6) that reached genome-wide significance. Downstream analysis showed that PCSK6 was highly expressed in IPF lung parenchyma and that PCSK6 lung staining intensity, peripheral blood gene expression, and plasma concentration were associated with reduced transplantation-free survival. These findings suggest that PCSK6 may play a potentially important role in IPF progression.
Idiopathic pulmonary fibrosis (IPF) is a devastating disease characterized by progressive lung scarring and poor survival (1, 2). Two antifibrotic therapies were approved for the treatment of individuals with IPF after randomized controlled trials demonstrated efficacy in slowing lung function decline (3, 4). Despite this advance, outcomes remain poor, and antifibrotic therapy appears to provide only modest survival benefit (5). To improve IPF outcomes, novel therapeutic targets are needed.
We and others have identified molecular IPF risk factors through unbiased investigation of the genome, transcriptome, and proteome (6–15). Among the strongest molecular determinants of IPF is a common variant in the promoter region of MUC5B (mucin 5B, oligomeric mucus/gel-forming), which increases the odds of developing IPF by nearly fivefold per risk allele (6–9). Despite this strong association with IPF onset, the MUC5B promoter was paradoxically associated with improved survival (16), though this association was potentially confounded by index event bias (17). Few other susceptibility-associated gene variants have been shown to reliably predict differential IPF survival, suggesting that molecular determinants of IPF susceptibility and progression may have limited overlap.
To better understand molecular drivers of IPF progression and identify new therapeutic targets, we conducted a two-stage, multicenter, international genome-wide association study (GWAS) of IPF survival, followed by downstream analysis of genes and proteins associated with top survival-associated variants to determine whether these circulating proteins also predicted differential survival. Some results of this study have been previously reported in the form of abstracts at the American Thoracic Society (18) and British Thoracic Society (19) national meetings and in preprint form (https://www.medrxiv.org/content/10.1101/2022.05.06.22274705v1).
Methods
Cohorts and Case Selection
All patients provided informed consent for research blood draws in accordance with protocols approved by the institutional review board at each participating institution. GWAS stage I cases consisted of unrelated patients with IPF of European ancestry from three previously described case-control GWAS data sets from the United States (9), the United Kingdom (7), and a combined cohort from the United States, United Kingdom, and Spain (UUS) (6). Available outcome data were gathered for all cases meeting international consensus criteria for IPF (20), and survival was plotted for individual cohorts within each data set. Patients without available outcome data were excluded, as were clinical trial cohorts because of short follow-up (see the online supplement). Stage II cases consisted of previously described, unrelated patients with IPF of European ancestry from Vanderbilt University (21). Vital status was assessed in U.S.- and Spain-based cohorts by chart review and contact with family members and in United Kingdom–based cohorts by review of national vital status databases.
Genotyping and Quality Control
Genotypes were generated for stage I cases using SNP genotyping arrays according to previously described methods (6, 7, 9). A summary of arrays used for each cohort is provided in the online supplement. Imputation for stage I cases was performed using the Michigan Imputation Server using the Haplotype Reference Consortium panel (version 1.1 2016). Genotypes for the stage II cases were determined by whole-genome sequencing, as previously described (21). Stringent quality control measures were applied, with a two-tier variant-filtering scheme in which variants with minor allele frequencies (MAFs) of 0.5–1% were retained when imputation R2 was ⩾0.8 and those with MAFs ⩾1% were retained when imputation R2 was ⩾0.5. Variants deviating from Hardy-Weinberg equilibrium (P < 1.0 × 10−6) were removed.
Genome-wide Survival Analysis
The primary endpoint assessed was transplantation-free survival (TFS), defined as the time in months from the site-determined date of IPF diagnosis to event (death or lung transplantation) or censoring date. Variants associated with differential TFS were identified using a multivariable Cox proportional hazards regression model adjusted for age, sex, center, and first 10 genetic principal components, with principal components calculated separately for each cohort. Variant genotypes were treated as a continuous variable, with each patient having an imputed genotype dosage between zero and two risk alleles. To avoid considering results obtained for just one of the three individual studies (United States, United Kingdom, and UUS), only those variants with association results available for at least two data sets (United States, United Kingdom, and UUS) were meta-analyzed using a fixed-effect inverse variance–weighted meta-analysis (METAL [http://csg.sph.umich.edu/abecasis/metal/] version 2011-03-25) to generate stage I results.
Variants nominally associated with TFS in stage I were defined as those with Wald P values <0.05 in at least two data sets with the same direction of effect and P < 5.0 × 10−5 in stage I meta-analysis. Conditional analysis of these SNPs to deduce their independence was performed with GCTA-COJO version 1.26 (https://yanglab.westlake.edu.cn/software/gcta/#Overview). The proportional hazards assumption was then assessed for each independent variant meeting advancement criteria by testing whether Schoenfeld residual rank varied by genotype strata. Variants that satisfied the proportional hazards assumption were advanced for stage II testing. Stage I and II cases were then meta-analyzed using METAL with the genome-wide significance threshold set at P < 5.0 × 10−8. In silico assessments were used to infer the biological effect of variants associated with TFS after stage II testing.
PCSK6 Tissue Expression
Formalin-fixed paraffin-embedded human lung tissue sections obtained from patients with IPF undergoing surgical lung biopsy were compared with sections from control subjects undergoing lung resection for malignancy, with sections distal to areas of malignancy used. Immunohistochemistry was performed using standard methods (see the online supplement), and mean staining intensity of PCSK6 protein was compared between IPF cases and non-IPF control subjects using a Mann-Whitney U test. Single-cell RNA sequencing data from prior published data sets (11, 22, 23) were reanalyzed and jointly annotated using label transfer from an updated annotated version of GSE135893. Cell-type annotation was performed using the TransferAnchors function in Seurat version 4 (https://yanglab.westlake.edu.cn/software/gcta/#Overview) (24). Data visualization was performed using Scanpy version 1.7.2 (https://scanpy.readthedocs.io/en/stable/) (25). The code used for analysis and presentation is available at https://github.com/KropskiLab/ipf_survival_gwas.
PCSK6 Clinical Outcome Association
The association between circulating PCSK6 gene expression and TFS was assessed using three previously published microarray data sets from the COMET (Correlating Outcomes with Biochemical Markers to Estimate Time-Progression in IPF) trial, Imperial College, and the University of Chicago (UChicago) (see the online supplement) (26), which were analyzed separately, with results meta-analyzed and presented as a forest plot. Circulating plasma PCSK6 protein concentration was then determined in patients with IPF from the University of California, Davis (UC-Davis), and the University of Chicago (UChicago) (see the online supplement), log2 transformed, and tested for TFS association using Cox proportional hazards regression (27). The proportional hazards assumption was satisfied for all downstream survival analyses.
Results
Case Selection for Stage I
Patients constituting the U.S. cohort included those from UChicago (n = 118) and the University of Pittsburgh (n = 200) (see Figures E1 and E2). Those constituting the United Kingdom cohort included patients from the University of Edinburgh (n = 119), the Trent Lung Fibrosis Study (n = 210), a subset of those participating in the prospective, multicenter PROFILE (Prospective Study of Fibrosis In the Lung Endpoints) study (NCT 01134822) (n = 175), and aggregated patients from smaller United Kingdom centers (Hull and Papworth) (n = 61) (see Figures E1 and E3). Patients constituting the UUS cohort included those from UChicago (independent of those from the U.S. cohort; n = 187), the PROFILE study (independent of those in the United Kingdom data set; n = 299), the University of California (Davis and San Francisco) (n = 84), and aggregated patients from centers in Spain (n = 28) (see Figures E1 and E4).
Baseline characteristics and outcomes
After phenotypic exclusions, 1,481 patients constituting stage I were included in the analysis. These included 318 patients from the U.S. data set, 565 from the United Kingdom data set, and 598 from the UUS data set. Baseline characteristics for each data set are shown in Table 1. The mean age ranged from 67 to 72 years, and men constituted 71–75% of each data set. The mean percentage predicted FVC and diffusion capacity of the lung for carbon monoxide (DlCO) was lowest in patients constituting the U.S. data set and highest in the United Kingdom data set. A majority of patients in each data set were classified as gender, age, and physiology stage I or II (28). Median survival was highest in the United Kingdom cohort (53.2 mo), followed by the UUS cohort (40.6 mo) and the U.S. cohort (39.3 mo) (P = 0.001) (see Figure E2). Survival also varied substantially across the centers constituting each cohort (see Figures E3–E5). Median survival was 48 months in the Vanderbilt University validation cohort (see Figure E6). Survival was similar between stage I and II cases through 24 months of follow-up but could not be compared thereafter because of violation of the proportional hazards assumption (see Figure E6).
Table 1.
Baseline Characteristics and Outcomes of Stage I and II Data Sets
| Characteristic | Stage I (n = 1,481) |
Stage II (n = 397) |
||
|---|---|---|---|---|
| United States* (n = 318) |
United Kingdom† (n = 565) |
UUS‡ (n = 598) |
Vanderbilt§ | |
| Age, yr, mean (SD) | 67.3 (8.9) | 72.1 (8.4) | 69.8 (8.3) | 65.7 (9.0) |
| Male sex, n (%) | 234 (73.6) | 403 (71.3) | 449 (75.1) | 295 (74.3) |
| FVC, % predicted, mean (SD) | 62.7 (17.9) | 77.7 (18.5) | 70.2 (18.6) | 65.0 (16.1) |
| DlCO, % predicted, mean (SD) | 36.7 (14.6) | 43.3 (14.3) | 39.9 (14.6) | 39.1 (13.5) |
| GAP stage, n (%) | ||||
| I | 72 (23.1) | 106 (35.2) | 135 (24.2) | 115 (29.8) |
| II | 149 (47.8) | 144 (47.8) | 286 (51.4) | 201 (52.1) |
| III | 91 (29.2) | 51 (16.9) | 136 (24.4) | 70 (18.1) |
| Death, n (%) | 189 (59.4) | 366 (64.8) | 257 (43.0) | 202 (50.9) |
| Transplantation, n (%) | 52 (16.4) | 2 (0.4) | 26 (4.4) | 32 (8.1) |
| Death or transplantation, n (%) | 241 (75.8) | 366 (64.8) | 283 (47.3) | 234 (58.9) |
| Median survival, mo (IQR) | 39.3 (13.4–70.6) | 53.2 (24.8–92.5) | 40.6 (19.9–75.5) | 48 (15–105) |
Definition of abbreviations: GAP = gender, age, and physiology; IQR = interquartile range; SD = standard deviation; UUS = United States, United Kingdom, and Spain.
Missing data: FVC, n = 6; DlCO, n = 31: GAP stage, n = 6.
Missing data: FVC, n = 241; DlCO, n = 264: GAP stage, n = 241.
Missing data: FVC, n = 30; DlCO, n = 41; GAP stage, n = 30.
Missing data: FVC, n = 4; DlCO, n = 9; GAP stage, n = 4.
Genome-Wide Survival Analysis
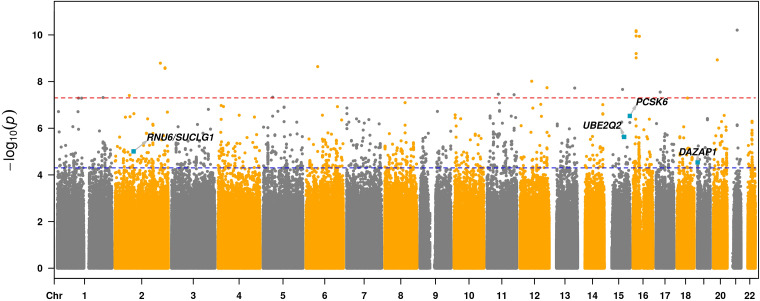
After filtering, 7,873,835 variants in the U.S. data set, 8,591,398 variants in the United Kingdom data set, and 8,620,496 variants in the UUS data set were tested for TFS association. Quantile-quantile plots for each stage I cohort suggested acceptable inflation (29) (see Figure E7). After stratifying stage I cohorts by MAF, inflation was higher for rare variants compared with low- and high-frequency variants, but within an acceptable range for each group (λ < 1.1) (see Figure E8). For meta-analysis, 9,075,629 variants were tested for TFS association in the aggregated stage I cohorts. One hundred sixty-one independent SNPs had Wald P values <0.05 in at least two data sets with the same direction of effect and P < 5.0 × 10−5 in stage I meta-analysis (Figure 1). Of those, 158 satisfied the proportional hazards assumption and advanced for stage II testing (see Table E1).
Figure 1.
Manhattan plot of stage I gene variants associated with idiopathic pulmonary fibrosis survival. Each dot represents a gene variant, arranged on the x-axis by chromosome. Variants falling above the blue line, which corresponds to P < 5 × 10−5, are considered to reach nominal significance, whereas those falling above the red line, which corresponds to P < 5 × 10−8, are considered to reach genome-wide significance. All variants crossing the nominal significance threshold were advanced for stage II testing. Chr = chromosome; DAZAP1 = deleted in azoospermia-associated protein 1; PCSK6 = proprotein convertase subtilisin/kexin type 6; RNU6 = U6 small nuclear RNA; SUCLG1 = succinate-CoA ligase GDP/ADP-forming subunit α; UBE2Q2 = ubiquitin-conjugating enzyme E2Q family member 2.
Genotype data were available in the Vanderbilt University cohort for 154 of the 158 variants advanced from stage I. Six variants were associated with TFS in the Vanderbilt University cohort at P < 0.05, including four with consistent effect direction that strengthened in TFS association after meta-analysis (Table 2; see Table E2). These four were rs184498750 near SUCLG1 (succinate-CoA ligase GDP/ADP-forming subunit α), rs60514164 near UBE2Q2 (ubiquitin-conjugating enzyme E2Q family member 2), rs35647788 in an intron of PCSK6 (proprotein convertase subtilisin/kexin type 6), and rs3893252 in an intron of DAZAP1 (deleted in azoospermia-associated protein 1) (Table 2). Of these, rs35647788 (PCSK6) showed the strongest TFS association across stage I (hazard ratio [HR], 4.76 [95% confidence interval (CI), 2.62–8.64]; P = 2.96 × 10−7) and stage II (HR, 3.12 [95% CI, 1.37–7.11]; P = 6.70 × 10−3) cohorts and crossed the genome-wide significance threshold in meta-analysis (HR, 4.11 [95% CI, 2.54–6.67]; P = 9.45 × 10−9) (Table 2). With the exception of rs60514164 (UBE2Q2) (MAF = 8%), these SNPs were low frequency, with MAFs of ∼1% in the study population. Regional association plots for each of the four variants are shown in Figure E9.
Table 2.
Results for the SNPs Nominally Associated across Stages and with Same Direction of Effects on Idiopathic Pulmonary Fibrosis Survival
| Variant Location (hg19) and Nearest Gene |
Stage I Results |
Stage II Results |
Meta-Analysis |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Position | SNP rsID | Gene | Ref | EA | R 2 | EAF (%) | HR (95% CI) | P Value | EAF (%) | HR (95% CI) | P Value | HR (95% CI) | P Value |
| 2 | 84291167 | rs184498750 | SUCLG1 | G | T | 0.86 | 1.1 | 3.11 (1.88–5.15) | 9.83 × 10−6 | 1.2 | 2.05 (1.79–4.18) | 0.049 | 2.71 (1.79–4.08) | 2.07 × 10−6 |
| 15 | 76081200 | rs60514164 | UBE2Q2 | C | T | 0.77 | 7.0 | 1.60 (1.32–1.95) | 2.35 × 10−6 | 7.7 | 1.42 (1.04–1.93) | 0.026 | 1.55 (1.31–1.83) | 2.23 × 10−7 |
| 15 | 101914234 | rs35647788 | PCSK6 | C | T | 0.93 | 0.8 | 4.76 (2.62–8.64) | 2.96 × 10−7 | 0.8 | 3.12 (1.37–7.11) | 6.72 × 10−3 | 4.11 (2.54–6.67) | 9.45 × 10−9 |
| 19 | 1412985 | rs3893252 | DAZAP1 | C | T | 0.82 | 0.6 | 3.57 (1.97–6.49) | 2.91 × 10−5 | 1.5 | 2.09 (1.05–4.15) | 0.036 | 2.84 (1.81–4.45) | 5.81 × 10−6 |
Definition of abbreviations: CI = confidence interval; DAZAP1 = deleted in azoospermia-associated protein 1; EA = effect allele; EAF = effect allele frequency; HR = hazard ratio; PCSK6 = proprotein convertase subtilisin/kexin type 6; R2 = lowest imputation quality value across studies; Ref = reference allele; SUCLG1 = succinate-CoA ligase GDP/ADP-forming subunit α; UBE2Q2 = ubiquitin-conjugating enzyme E2Q family member 2.
In multivariable analysis, each variant except rs3893252 (DAZAP1) maintained survival association after adjustment for relevant confounders of IPF survival (see Table E3). Among patients with the rs35647788 (PCSK6) variant, all were heterozygotes (see Table E4) and were evenly distributed across centers constituting the United Kingdom and UUS cohorts. No rs35647788 (PCSK6) variants were observed in the U.S. cohort despite good imputation quality (r2 = 0.74). In sensitivity analysis of the PCSK6 variant, results were consistent when censoring transplantations (see Table E5). In silico testing revealed functional effects for each of the four variants associated with TFS after stage II meta-analysis (see Table E6), none of which had a known association with fibrotic lung disease. Using Genotype-Tissue Expression, we found multiple common sentinel PCSK6 expression quantitative trait loci in high linkage disequilibrium (D′ = 1) with rs35647788 (see Table E7).
PCSK6 Tissue Expression
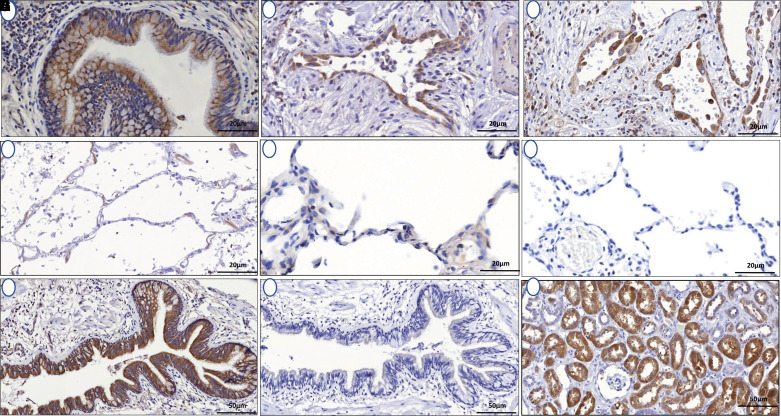
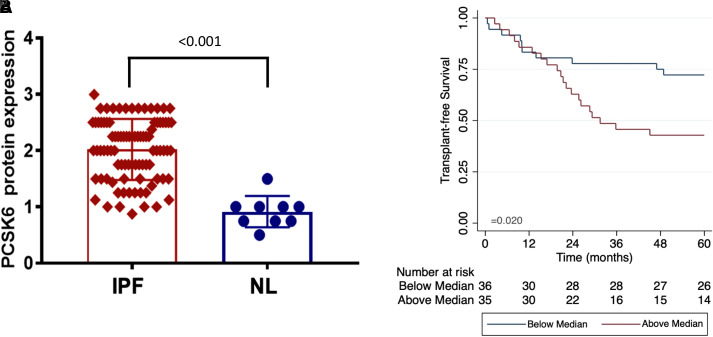
Morphologic assessment of histological sections from lung tissue of patients with IPF and control subjects without fibrotic lung disease was performed. In IPF lung, cytoplasmic PCSK6 expression localized to ciliated epithelial cells and alveolar epithelial cells and was markedly higher than PCSK6 expression in non-IPF control sections (Figure 2). Western blot confirmed the presence of only a single PCSK6 band (see Figure E10). Relative staining intensity was twofold higher in IPF lung samples (n = 86) compared with non-IPF control samples (n = 9) (P < 0.001) (Figure 3A). Increased PCSK6 protein staining score was associated with reduced TFS in those with available survival data (n = 71), with staining scores above the median associated with greater than twofold increased risk of death or lung transplantation (HR, 2.41 [95% CI, 1.12–5.16]; P = 0.024) (Figure 3B). Interrogating previously published single-cell RNA sequencing data from IPF and control lungs, PCSK6 expression was highest in lymphatic endothelial cells and adventitial fibroblasts, while broad expression in the airway epithelium was observed (see Figure E11).
Figure 2.
(A–F) PCSK6 immunohistochemistry showed increased cytoplasmic PCSK6 (proprotein convertase subtilisin/kexin type 6) expression in ciliated epithelial cells (A) and alveolar epithelial cells (B and C) compared with normal lung control sections (D–F). (G and H) Parallel idiopathic pulmonary fibrosis sections confirm increased PCSK6 staining (G) compared with control section (H). (I) Human kidney positive control is provided for reference. Scale bars, 20 μm (A–F) and 50 μm (G–I).
Figure 3.
(A) Comparison of relative PCSK6 (proprotein convertase subtilisin/kexin type 6) staining intensity between idiopathic pulmonary fibrosis (IPF) cases and non-IPF control subjects demonstrated significantly higher median intensity in IPF lungs (P < 0.001). (B) Survival was lower among IPF cases with PCSK6 staining intensity above the median. NL = non-IPF control subjects.
PCSK6 Clinical Outcome Association
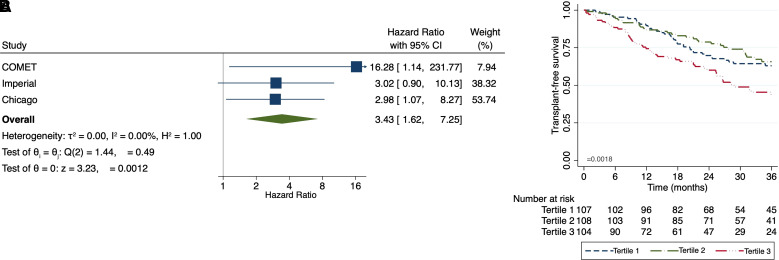
When assessing PCSK6 gene expression in the COMET (n = 75), Imperial College (n = 55), and UChicago (n = 45) cohorts, increasing PCSK6 expression was associated with increased mortality risk in each cohort, with each one-unit increased associated with greater than threefold increased risk of death or lung transplantation in meta-analysis (HR, 3.43 [95% CI, 1.62–7.25]; P = 0.0012) (Figure 4A). When assessing PCSK6 plasma concentration in patients with IPF from UC-Davis (n = 138) and UChicago (n = 181), increasing plasma PCSK6 concentration was associated with reduced TFS, with each one-unit change in log-transformed plasma concentration associated with a nearly 50% increase in outcome risk (HR, 1.47 [95% CI, 1.14–1.89]; P = 0.0031). These results were consistent across UC-Davis (HR, 1.47 [95% CI, 1.14–1.89]; P = 0.08) and UChicago (HR, 1.34 [95% CI, 0.92–1.94]; P = 0.12) cohorts. After stratification of the combined cohort by tertiles, those with PCSK6 concentration in the highest tertile displayed significantly worse survival than those in the second and third tertiles (P = 0.0018) (Figure 4B).
Figure 4.
Relationship between proprotein convertase subtilisin/kexin type 6 and clinically relevant idiopathic pulmonary fibrosis endpoints. (A and B) Higher peripheral blood gene expression (A) and circulating plasma protein concentration (B) are associated with reduced transplantation-free survival. CI = confidence interval; COMET = Correlating Outcomes with Biochemical Markers to Estimate Time-Progression in IPF.
Overlap between IPF Risk and TFS
Variants previously associated with IPF risk (6–10) were investigated for outcome association (see the online supplement). None of the 15 genetic variants previously associated with IPF risk (6–10) was associated with TFS after Bonferroni correction (P = 0.0033) (see Table E8). As previously reported (16, 17), individuals with the MUC5B promoter polymorphism (rs3570590) displayed better overall survival, though this did not reach significance after adjustment for multiple testing. None of the four validated survival variants was associated with differential IPF risk (see Table E9). When combing the effect of thousands of IPF risk variants in a polygenic risk score, this risk score was not significantly associated with TFS for any significance threshold used (see Figure E12), again suggesting that variants that affect disease risk may have little impact on survival times after diagnosis.
Discussion
In this investigation, we conducted the first GWAS of IPF survival, identifying a variant intronic to PCSK6 that associated with differential TFS at genome-wide significance in two independent IPF cohorts totaling nearly 2,000 patients. We subsequently found that PCSK6 protein was highly expressed in IPF lung tissue, localizing to the airway epithelium, which plays a key role in IPF onset and progression (30). Finally, we found that PCSK6 lung staining, peripheral blood gene expression, and circulating plasma concentration were negatively correlated with TFS across independent IPF cohorts. To our knowledge, this study is the first to systematically identify gene variants associated with IPF survival and the first to identify PCSK6 as a potentially relevant mediator of IPF progression.
PCSK6, also called PACE4, encodes a widely expressed calcium-dependent serine endoprotease, which we demonstrate is expressed most highly in the airway epithelium, adventitial fibroblasts, and lymphatic endothelial cells in the lung. PCSK6 is a critical mediator of TGF-β (transforming growth factor-β) processing and is crucial for reproduction, embryological development, and blood pressure regulation (31–35). A PCSK6 gene variant has been implicated in the development of hypertension (35), and dysregulated PCSK6 gene expression has been linked to vascular disease (36–38), and cardiac remodeling after myocardial ischemia (39). These cardiovascular remodeling effects make PCSK6 of particular relevance to IPF, as PCSK6 overexpression can lead to increased collagen I and III deposition, TGF-β activation, and extracellular matrix formation (39, 40), which are cardinal features of IPF pathogenesis (2). In addition, PCSK6 may bind tissue inhibitors of metalloproteinases (41), potentially counteracting the antimetalloproteinase activity of tissue inhibitors of metalloproteinases (42).
PCSK6 dysregulation has also been implicated in the development of cancer of the lung (43), breast (44), ovary (45), thyroid (46), and prostate (47). PCSK6 has been shown to regulate apoptosis in prostate cancer (48) and pancreatic cancer (49) and is also linked to increased cancer cell invasiveness by enhancing bioactivity of matrix metalloproteinases and cytokines (50). Accordingly, PCSK6 has been proposed as an antitumor therapeutic target (51, 52), and a bioavailable formulation of an anti-PCSK6 molecule is currently under investigation (53). In vitro PCSK6 inhibition has already been shown to reduce fibroblast proliferation, migration, and invasion in rheumatoid arthritis–associated synovitis (54). Given its relatively high expression in adventitial (including airway-associated) fibroblasts and the airway epithelium, PCSK6 could serve as a potential therapeutic in patients with pulmonary fibrosis. However, additional research is needed to better characterize the role PCSK6 may play in IPF progression before therapeutic blockade is considered.
Despite being the largest genomic analysis of IPF survival performed to date, the modest size of this cohort limited our ability to identify higher frequency SNPs with modest effect sizes. In addition, the rare nature of the PCSK6 variant identified in this study suggests that it is unlikely to singularly explain subsequent gene expression and protein findings. In silico analyses identified several nearby regulatory elements that may include common functional variants, with smaller effects, in linkage disequilibrium with this PCSK6 variant. Further research is needed to determine whether rs35647788 and other rare variants in PCSK6 have additive effects on gene expression, together with investigation of regulatory elements, expression quantitative trait loci, and more complex structural variants that may be contributing to our findings.
These results, and others recently published by our group (55), demonstrate the value of case-only GWAS to identify genes associated with a relevant trait or outcome within a disease state. This approach is subject to temporal selection bias, however, as the timing of blood draw is likely associated with outcome when using the date of blood draw as the starting point for determining survival time (56). This approach was necessary given difficulties defining the date of diagnosis, together with variable periods of preclinical disease in patients with IPF, but variability in survival across cohorts suggests that temporal selection bias was likely present in our study. This study also suggests potentially important differences between genomic determinants of IPF susceptibility and survival, whereby those involved in disease onset may be independent of those driving disease progression. No survival-associated variant identified in this analysis was associated with IPF risk. Although we replicate prior IPF risk association for the MUC5B promoter polymorphism, we found only a weak association with favorable survival, an observation that may be influenced by index event bias (17). As none of the survival-associated variants showed an association with disease risk, it is unlikely that these survival results are affected by index event bias. These findings have potential implications for drug development, as genes associated with IPF survival may represent more effective therapeutic targets than those associated with IPF onset.
This study has several limitations. Given sample size constraints for this rare disease, we pursued a two-stage approach with meta-analysis of candidate variants rather than a discovery and replication approach, which would have required substantially higher sample sizes in each cohort. Furthermore, there was heterogeneity in the distribution of the PCSK6 variant, as this variant was not detected in any individuals from the U.S. cohort. This was likely driven by the rare nature of this variant, but consistent effect association in the stage II cohort and genome-wide significance for the PCSK6 variant after meta-analysis increase confidence that this represents a true association, as does the downstream clinical outcome analysis showing PCSK6 gene expression and protein concentration to be associated with differential TFS. Next, we relied on all-cause mortality when modeling these data, leaving it unclear what proportion of subjects died of IPF. This could have biased results to some extent if variants were associated with death events unrelated to IPF. There were also substantial differences in transplantation across cohorts, which could have biased results when modeling TFS. This did not appear to be an issue for the PCSK6 variant, which maintained strong association with death when censoring transplantation events (see Table E5). In addition, a large proportion of patients constituting the UUS data set were recruited after the U.S. approval of pirfenidone and nintedanib, which may affect survival (57), and patients recruited before 2012 may have been exposed to potentially harmful immunosuppression (58). Finally, to facilitate accurate imputation, we focused on individuals of European ancestry, leaving it unclear whether these findings would extend to patients of non-European ancestry. The PCSK6 variant was not present in any of the 123 patients excluded because of non-European ancestry, but dedicated analysis in this population is warranted. Additional variants of interest that failed to reach genome-wide significance were also identified, including one in DAZAP1, which lies close to PCSK4 on chromosome 19. Additional research is needed to ascertain the role these genes nearby these variants may play in IPF progression.
Conclusions
Here we present results from the first GWAS of IPF survival conducted to date. This study sheds important light on the genetics of IPF progression and identified novel variants that may contribute to this process, including rs35647788 in an intron of PCSK6. Downstream analysis demonstrated PCSK6 protein lung staining, peripheral blood gene expression, and circulating plasma concentration to be associated with reduced IPF survival, suggesting that PCSK6 may serve as a potential therapeutic target in patients with IPF.
Acknowledgments
Acknowledgment
The authors thank all the patients who contributed biologic samples and clinical data for this research.
Footnotes
Supported by NHLBI grants K23HL138190 (J.M.O.), R56HL158935 (J.M.O.), K23HL150301 (J.S.K.), K23HL146942 (A.A.), T32HL007605 and Pulmonary Fibrosis Foundation (C.T.L.), R01HL145372 (J.A.K. and N.E.B.), RO1HL130796 (I.N.), and UG3HL145266 (I.N.); Ministerio de Ciencia e Innovación grants RTC-2017-6471-1 and AEI/FEDER UE (C.F.); Instituto de Salud Carlos III cofinanced by the European Regional Development Fund “A Way of Making Europe” from the European Union (grant PI20/00876 to C.F.); Instituto Tecnológico y de Energías Renovables grant OA17/008 (C.F.); Consejería de Educación-Gobierno de Canarias and Cabildo Insular de Tenerife grant BOC n.° 173, 24/08/2017 (J.M.L.-S.); National Institute for Health Research (NIHR) Research Professorship RP-2017-08-ST2-014 (R.G.J.); Medical Research Council grants G0901226 and MR/V00235X/1 (R.G.J.); NIHR Clinician Scientist Fellowship CS-2013-13-017 (T.M.M.); NIHR Senior Investigator Award NIHR201371 (M.D.T.); the British Lung Foundation Chair in Respiratory Research (C17-3 to T.M.M.); the GlaxoSmithKline/British Lung Foundation Chair in Respiratory Research (C17-1 to L.W.W.); Wellcome Trust grants WT202849/Z/16/Z (M.D.T.) and 221680/Z/20/Z (B.G.-G.); BREATHE — The Health Data Research Hub for Respiratory Health grant MC_PC_19004 (L.W.W.); and Action for Pulmonary Fibrosis Research Fellows (R.J.A. and P.L.M.). This research used the ALICE and SPECTRE High Performance Computing Facilities at the University of Leicester and Teide-HPC (https://teidehpc.iter.es/en/home) at Instituto Tecnológico y de Energías Renovables. The views expressed are our own and not necessarily those of the National Health Service, the NIHR, or the UK Department of Health. This study was funded by the agencies listed above, which had no role in the design or conduct of the study. All authors had full access to all data, and the corresponding author had final responsibility for the decision to submit for publication.
Author Contributions: J.M.O., R.J.A., J.M.L.-S., S.-F.M., R.G.J., C.F., L.V.W., and I.N. designed and supervised the study. J.M.O., P.L.M., J.A.K., A.A., J.V.P., M.E.S., R.H., N.H., M.K.B.W., S.H., H.P., N.K., P.J.W., M.M.-M., F.J.M., T.M.M., T.S.B., R.G.J., and I.N. recruited patients for the study. J.M.O., P.L.M., S.-F.M., J.A.K., A.L.L., V.V., J.J., Y.Z., A.S., N.K., P.J.W., I.P.H., M.D.T., W.A.F., T.M.M., T.S.B., B.L.Y., R.G.J., C.F., and I.N. contributed to DNA extraction and genotyping. C.J., A.G.N., D.R., W.W., and R.G.J. secured lung tissue and performed the immunohistochemistry analysis. N.E.B., J.A.K., and T.S.B. contributed single-cell sequencing data. J.M.O., R.J.A., J.M.L.-S., C.J., J.S.K., B.G.-G., T.H.-B., J.A.K., Y.H., A.M.-B., L.A.R.-R., J.M., A.S., N.E.B., T.S.B., B.L.Y., R.G.J., C.F., and L.V.W. performed the analyses. J.M.O., P.L.M., J.A.K., C.T.L., A.A., J.V.P., M.E.S., L.L., E.V., Y.Z., N.K., P.J.W., M.M.-M., F.J.M., T.M.M., R.G.J., and I.N. collected paired clinical data. J.M.O., J.M.L.-S., R.J.A., R.G.J., C.F., L.W., and I.N. wrote the manuscript with the input of all authors. All authors approved the final version of this manuscript.
Data sharing statement: Genome-wide association study summary statistics for this study are available at https://github.com/genomicsITER/PFgenetics.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202205-0845OC on February 13, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Raghu G, Remy-Jardin M, Myers JL, Richeldi L, Ryerson CJ, Lederer DJ, et al. American Thoracic Society, European Respiratory Society, Japanese Respiratory Society, and Latin American Thoracic Society Diagnosis of idiopathic pulmonary fibrosis: an official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care Med . 2018;198:e44–e68. doi: 10.1164/rccm.201807-1255ST. [DOI] [PubMed] [Google Scholar]
- 2. Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med . 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 3. Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, et al. INPULSIS Trial Investigators Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med . 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 4. King TE, Jr, Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, et al. ASCEND Study Group A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med . 2014;370:2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 5. Dempsey TM, Sangaralingham LR, Yao X, Sanghavi D, Shah ND, Limper AH. Clinical effectiveness of antifibrotic medications for idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2019;200:168–174. doi: 10.1164/rccm.201902-0456OC. [DOI] [PubMed] [Google Scholar]
- 6. Allen RJ, Guillen-Guio B, Oldham JM, Ma SF, Dressen A, Paynton ML, et al. Genome-wide association study of susceptibility to idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2020;201:564–574. doi: 10.1164/rccm.201905-1017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Allen RJ, Porte J, Braybrooke R, Flores C, Fingerlin TE, Oldham JM, et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med . 2017;5:869–880. doi: 10.1016/S2213-2600(17)30387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet . 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med . 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dhindsa RS, Mattsson J, Nag A, Wang Q, Wain LV, Allen R, et al. FinnGen Consortium Identification of a missense variant in SPDL1 associated with idiopathic pulmonary fibrosis. Commun Biol . 2021;4:392. doi: 10.1038/s42003-021-01910-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv . 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Todd JL, Neely ML, Overton R, Durham K, Gulati M, Huang H, et al. IPF-PRO Registry investigators Peripheral blood proteomic profiling of idiopathic pulmonary fibrosis biomarkers in the multicentre IPF-PRO Registry. Respir Res . 2019;20:227. doi: 10.1186/s12931-019-1190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DePianto DJ, Chandriani S, Abbas AR, Jia G, N’Diaye EN, Caplazi P, et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax . 2015;70:48–56. doi: 10.1136/thoraxjnl-2013-204596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sivakumar P, Ammar R, Thompson JR, Luo Y, Streltsov D, Porteous M, et al. Integrated plasma proteomics and lung transcriptomics reveal novel biomarkers in idiopathic pulmonary fibrosis. Respir Res . 2021;22:273. doi: 10.1186/s12931-021-01860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O’Dwyer DN, Norman KC, Xia M, Huang Y, Gurczynski SJ, Ashley SL, et al. The peripheral blood proteome signature of idiopathic pulmonary fibrosis is distinct from normal and is associated with novel immunological processes. Sci Rep . 2017;7:46560. doi: 10.1038/srep46560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peljto AL, Zhang Y, Fingerlin TE, Ma SF, Garcia JG, Richards TJ, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA . 2013;309:2232–2239. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dudbridge F, Allen RJ, Sheehan NA, Schmidt AF, Lee JC, Jenkins RG, et al. Adjustment for index event bias in genome-wide association studies of subsequent events. Nat Commun . 2019;10:1561. doi: 10.1038/s41467-019-09381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allen R, Lorenzo-Salazar JM, Ma SF, Huang Y, Zhang Y, Kaminski N, et al. Genome-wide association study of survival in patients with idiopathic pulmonary fibrosis [abstract] Am J Respir Crit Care Med . 2017;195:A7649. [Google Scholar]
- 19. Allen R, Oldham J, Lorenzo-Salazar J, Molyneaux P, Ma S, Stockwell A, et al. S65 Genome-wide association study of survival times after diagnosis of idiopathic pulmonary fibrosis [abstract] Thorax . 2021;76:A43. [Google Scholar]
- 20. Raghu G, Remy-Jardin M, Myers J, Richeldi L, Wilson KC. The 2018 diagnosis of idiopathic pulmonary fibrosis guidelines: surgical lung biopsy for radiological pattern of probable usual interstitial pneumonia is not mandatory. Am J Respir Crit Care Med . 2019;200:1089–1092. doi: 10.1164/rccm.201907-1324ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dressen A, Abbas AR, Cabanski C, Reeder J, Ramalingam TR, Neighbors M, et al. Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study. Lancet Respir Med . 2018;6:603–614. doi: 10.1016/S2213-2600(18)30135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bui LT, Winters NI, Chung MI, Joseph C, Gutierrez AJ, Habermann AC, et al. Human Cell Atlas Lung Biological Network Chronic lung diseases are associated with gene expression programs favoring SARS-CoV-2 entry and severity. Nat Commun . 2021;12:4314. doi: 10.1038/s41467-021-24467-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv . 2020;6:eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, III, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell . 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol . 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang Y, Oldham JM, Ma SF, Unterman A, Liao SY, Barros AJ, et al. Blood transcriptomic predicts progression of pulmonary fibrosis and associates natural killer cells. Am J Respir Crit Care Med . 2021;204:197–208. doi: 10.1164/rccm.202008-3093OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang Y, Ma SF, Vij R, Oldham JM, Herazo-Maya J, Broderick SM, et al. A functional genomic model for predicting prognosis in idiopathic pulmonary fibrosis. BMC Pulm Med . 2015;15:147. doi: 10.1186/s12890-015-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ley B, Ryerson CJ, Vittinghoff E, Ryu JH, Tomassetti S, Lee JS, et al. A multidimensional index and staging system for idiopathic pulmonary fibrosis. Ann Intern Med . 2012;156:684–691. doi: 10.7326/0003-4819-156-10-201205150-00004. [DOI] [PubMed] [Google Scholar]
- 29.Rivadeneira F, Uitterlinden AG. In: Marcus and Feldman’s osteoporosis. 5th ed. Dempster DW, Cauley JA, Bouxsein ML, Cosman F, editors. San Diego, CA: Academic Press; 2021. Chapter 18: genetics of osteoporosis; pp. 405–451. [Google Scholar]
- 30. Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med . 2018;379:797–798. doi: 10.1056/NEJMc1807508. [DOI] [PubMed] [Google Scholar]
- 31. Diaz FJ, Sugiura K, Eppig JJ. Regulation of Pcsk6 expression during the preantral to antral follicle transition in mice: opposing roles of FSH and oocytes. Biol Reprod . 2008;78:176–183. doi: 10.1095/biolreprod.107.063537. [DOI] [PubMed] [Google Scholar]
- 32. Mujoomdar ML, Hogan LM, Parlow AF, Nachtigal MW. Pcsk6 mutant mice exhibit progressive loss of ovarian function, altered gene expression, and formation of ovarian pathology. Reproduction . 2011;141:343–355. doi: 10.1530/REP-10-0451. [DOI] [PubMed] [Google Scholar]
- 33. Constam DB, Robertson EJ. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev . 2000;14:1146–1155. [PMC free article] [PubMed] [Google Scholar]
- 34. Bessonnard S, Mesnard D, Constam DB. PC7 and the related proteases Furin and Pace4 regulate E-cadherin function during blastocyst formation. J Cell Biol . 2015;210:1185–1197. doi: 10.1083/jcb.201503042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen S, Cao P, Dong N, Peng J, Zhang C, Wang H, et al. PCSK6-mediated corin activation is essential for normal blood pressure. Nat Med . 2015;21:1048–1053. doi: 10.1038/nm.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perisic L, Hedin E, Razuvaev A, Lengquist M, Osterholm C, Folkersen L, et al. Profiling of atherosclerotic lesions by gene and tissue microarrays reveals PCSK6 as a novel protease in unstable carotid atherosclerosis. Arterioscler Thromb Vasc Biol . 2013;33:2432–2443. doi: 10.1161/ATVBAHA.113.301743. [DOI] [PubMed] [Google Scholar]
- 37. Röhl S, Suur BE, Lengquist M, Seime T, Caidahl K, Hedin U, et al. Lack of PCSK6 increases flow-mediated outward arterial remodeling in mice. Cells . 2020;9:1009. doi: 10.3390/cells9041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rykaczewska U, Suur BE, Röhl S, Razuvaev A, Lengquist M, Sabater-Lleal M, et al. IMPROVE Study Group PCSK6 is a key protease in the control of smooth muscle cell function in vascular remodeling. Circ Res . 2020;126:571–585. doi: 10.1161/CIRCRESAHA.119.316063. [DOI] [PubMed] [Google Scholar]
- 39. Kuhn TC, Knobel J, Burkert-Rettenmaier S, Li X, Meyer IS, Jungmann A, et al. Secretome analysis of cardiomyocytes identifies PCSK6 (proprotein convertase subtilisin/kexin type 6) as a novel player in cardiac remodeling after myocardial infarction. Circulation . 2020;141:1628–1644. doi: 10.1161/CIRCULATIONAHA.119.044914. [DOI] [PubMed] [Google Scholar]
- 40. Tsuji A, Sakurai K, Kiyokage E, Yamazaki T, Koide S, Toida K, et al. Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix: potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim Biophys Acta . 2003;1645:95–104. doi: 10.1016/s1570-9639(02)00532-0. [DOI] [PubMed] [Google Scholar]
- 41. Nour N, Mayer G, Mort JS, Salvas A, Mbikay M, Morrison CJ, et al. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol Biol Cell . 2005;16:5215–5226. doi: 10.1091/mbc.E05-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Craig VJ, Zhang L, Hagood JS, Owen CA. Matrix metalloproteinases as therapeutic targets for idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol . 2015;53:585–600. doi: 10.1165/rcmb.2015-0020TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin YE, Wu QN, Lin XD, Li GQ, Zhang YJ. Expression of paired basic amino acid-cleaving enzyme 4 (PACE4) correlated with prognosis in non-small cell lung cancer (NSCLC) patients. J Thorac Dis . 2015;7:850–860. doi: 10.3978/j.issn.2072-1439.2015.05.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lapierre M, Siegfried G, Scamuffa N, Bontemps Y, Calvo F, Seidah NG, et al. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells’ malignant phenotypes: role of tissue inhibitors of metalloproteinase-1. Cancer Res . 2007;67:9030–9034. doi: 10.1158/0008-5472.CAN-07-0807. [DOI] [PubMed] [Google Scholar]
- 45. Longuespée R, Couture F, Levesque C, Kwiatkowska A, Desjardins R, Gagnon S, et al. Implications of proprotein convertases in ovarian cancer cell proliferation and tumor progression: insights for PACE4 as a therapeutic target. Transl Oncol . 2014;7:410–419. doi: 10.1016/j.tranon.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fradet L, Temmar R, Couture F, Belzile M, Fortier PH, Day R. Evaluation of PACE4 isoforms as biomarkers in thyroid cancer. J Otolaryngol Head Neck Surg . 2018;47:63. doi: 10.1186/s40463-018-0311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Delic S, Lottmann N, Jetschke K, Reifenberger G, Riemenschneider MJ. Identification and functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma invasion-associated candidate genes. Neuropathol Appl Neurobiol . 2012;38:201–212. doi: 10.1111/j.1365-2990.2011.01207.x. [DOI] [PubMed] [Google Scholar]
- 48. Yao Z, Sun B, Hong Q, Yan J, Mu D, Li J, et al. PACE4 regulates apoptosis in human prostate cancer cells via endoplasmic reticulum stress and mitochondrial signaling pathways. Drug Des Devel Ther . 2015;9:5911–5923. doi: 10.2147/DDDT.S86881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tian XF, Huang GM, Zang HL, Cao H. PACE4 regulates apoptosis in human pancreatic cancer Panc-1 cells via the mitochondrial signaling pathway. Mol Med Rep . 2016;14:5205–5210. doi: 10.3892/mmr.2016.5885. [DOI] [PubMed] [Google Scholar]
- 50. Bassi DE, Fu J, Lopez de Cicco R, Klein-Szanto AJ. Proprotein convertases: “master switches” in the regulation of tumor growth and progression. Mol Carcinog . 2005;44:151–161. doi: 10.1002/mc.20134. [DOI] [PubMed] [Google Scholar]
- 51. Levesque C, Fugère M, Kwiatkowska A, Couture F, Desjardins R, Routhier S, et al. The Multi-Leu peptide inhibitor discriminates between PACE4 and furin and exhibits antiproliferative effects on prostate cancer cells. J Med Chem . 2012;55:10501–10511. doi: 10.1021/jm3011178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levesque C, Couture F, Kwiatkowska A, Desjardins R, Guérin B, Neugebauer WA, et al. PACE4 inhibitors and their peptidomimetic analogs block prostate cancer tumor progression through quiescence induction, increased apoptosis and impaired neovascularisation. Oncotarget . 2015;6:3680–3693. doi: 10.18632/oncotarget.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kwiatkowska A, Couture F, Ait-Mohand S, Desjardins R, Dory YL, Guérin B, et al. Enhanced anti-tumor activity of the Multi-Leu peptide PACE4 inhibitor transformed into an albumin-bound tumor-targeting prodrug. Sci Rep . 2019;9:2118. doi: 10.1038/s41598-018-37568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang F, Wang L, Jiang H, Chang X, Pan J. Inhibition of PCSK6 may play a protective role in the development of rheumatoid arthritis. J Rheumatol . 2015;42:161–169. doi: 10.3899/jrheum.140435. [DOI] [PubMed] [Google Scholar]
- 55. Allen RJ, Oldham JM, Jenkins DA, Leavy OC, Guillen-Guio B, Melbourne CA, et al. Longitudinal lung function and gas transfer in individuals with idiopathic pulmonary fibrosis: a genome-wide association study. Lancet Respir Med . 2022;11:65–73. doi: 10.1016/S2213-2600(22)00251-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kehl KL, Schrag D, Hassett MJ, Uno H. Assessment of temporal selection bias in genomic testing in a cohort of patients with cancer. JAMA Netw Open . 2020;3:e206976. doi: 10.1001/jamanetworkopen.2020.6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dempsey TM, Payne S, Sangaralingham L, Yao X, Shah ND, Limper AH. Adoption of the antifibrotic medications pirfenidone and nintedanib for patients with idiopathic pulmonary fibrosis. Ann Am Thorac Soc . 2021;18:1121–1128. doi: 10.1513/AnnalsATS.202007-901OC. [DOI] [PubMed] [Google Scholar]
- 58. Idiopathic Pulmonary Fibrosis Clinical Research Network. Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med . 2012;366:1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]