ABSTRACT
Our understanding of the molecular events driving cell specification in early mammalian development relies mainly on mouse studies, and it remains unclear whether these mechanisms are conserved across mammals, including humans. We have shown that the establishment of cell polarity via aPKC is a conserved event in the initiation of the trophectoderm (TE) placental programme in mouse, cow and human embryos. However, the mechanisms transducing cell polarity into cell fate in cow and human embryos are unknown. Here, we have examined the evolutionary conservation of Hippo signalling, which is thought to function downstream of aPKC activity, in four different mammalian species: mouse, rat, cow and human. In all four species, inhibition of the Hippo pathway by targeting LATS kinases is sufficient to drive ectopic TE initiation and downregulation of SOX2. However, the timing and localisation of molecular markers differ across species, with rat embryos more closely recapitulating human and cow developmental dynamics, compared with the mouse. Our comparative embryology approach uncovered intriguing differences as well as similarities in a fundamental developmental process among mammals, reinforcing the importance of cross-species investigations.
Keywords: Comparative embryology, Early development, Hippo signalling pathway, Human, Lineage specification, Placental programme
Highlighted Article: Analysis of the mechanisms involved in the first cell specification event in mouse, rat, cow and human embryos reveals that, despite some expected similarities, mice seem to diverge from other mammals at the molecular level.
INTRODUCTION
Our understanding of molecular mechanisms regulating cell specification during pre-implantation mammalian development relies mainly on mouse studies. Eight-cell mouse embryos undergo a morphological change in a process known as compaction (Ducibella and Anderson, 1975). After several rounds of cell division, two distinct cell populations are discernible at the morula stage: inner and outer cells. Upon reaching the blastocyst stage, inner cells give rise to the inner cell mass (ICM) and the outer cells become the trophectoderm (TE), a polarised epithelium that will form fetal components of the placenta. Subsequently, the ICM further segregates into the epiblast (EPI), which gives rise to the fetus, and the primitive endoderm, which primarily contributes to the yolk sac (Cockburn and Rossant, 2010).
Concomitant with compaction, cell polarity is established in the eight-cell mouse embryo. Inner and outer cells display different polarisation states, which influence their cell fate. Outer cells acquire an apical domain, enriched with atypical protein kinase C (aPKC), which, together with the proteins partitioning defective homolog 6B and 3 (PARD6B, PARD3), forms the PAR complex, whereas PAR1, E-cadherin (cadherin 1) and other cell adhesion molecules localise to the basolateral domain (Plusa et al., 2005; Vinot et al., 2005; Ohsugi et al., 1996; Alarcon, 2010). Angiomotin (AMOT), a modulator of the Hippo pathway, is sequestered to the apical domain of polar outer cells. This prevents activation of downstream Hippo pathway kinases, large tumor suppressor kinases 1/2 (LATS1/2) (Hirate et al., 2013). Consequently, yes-associated protein 1 (YAP1) and WW domain-containing transcription regulator protein 1 (WWTR1, also known as TAZ) accumulate in the nucleus where they bind TEA-domain family member 4 (TEAD4), to promote the expression of TE lineage-associated factors, such as caudal type homeobox 2 (Cdx2) and GATA binding protein 3 (Gata3) (Strumpf et al., 2005; Ralston et al., 2010; Nishioka et al., 2009). By contrast, AMOT is activated in apolar inner cells through phosphorylation by LATS1/2. Activation of the Hippo pathway results in YAP1 and WWTR1 phosphorylation and cytoplasmic retention, thus maintaining the inner cells in an unspecified state (Hirate et al., 2013; Cockburn et al., 2013; Frum et al., 2018; Leung and Zernicka-Goetz, 2013). SOX2 is a molecular marker of the EPI and is restricted to the inner cells at a comparatively early stage in the mouse morula (Wicklow et al., 2014). SOX2 expression is repressed by YAP1/WWTR1/TEAD4 in outer cells (Frum et al., 2018, 2019). These mechanistic insights derive from functional studies performed in the mouse embryo, and the extent to which these processes are conserved in other mammals is unclear.
In the mouse, the levels of CDX2 are higher in outer cells from the morula stage (Dietrich and Hiiragi, 2007), whereas in human and cow embryos, CDX2 is detectable only later, in TE cells of cavitating blastocysts (Niakan and Eggan, 2013; Goissis and Cibelli, 2014a; Berg et al., 2011). GATA3 has been shown to promote trophoblast development in parallel to CDX2 (Ralston et al., 2010). Like the mouse, GATA3 levels are higher in outer cells from morula stage in cow and human embryos (Ralston et al., 2010; Gerri et al., 2020). In our previous work, we showed that at the morula stage mouse, cow and human embryos acquire apical-basal polarity in outer cells. Thereby, aPKC expression becomes localised to the contact-free domain, and the Hippo pathway downstream effectors YAP1 and WWTR1 accumulate in the nucleus and restrict expression of TE-associated factors, including GATA3, which initiates a conserved TE programme across these species (Gerri et al., 2020). Furthermore, inhibition of aPKC activity impairs TE initiation in cow and human morula-stage embryos (Gerri et al., 2020), and it was subsequently shown that downregulation of the polarity regulators of phospholipase C signalling leads to decreased GATA3 levels in human embryos (Zhu et al., 2021).
Here, we expanded our morphokinetic and molecular analysis to another mammalian species, the rat. We found that dynamics of inner and outer cell molecular markers in the rat morula more closely resemble cow and human than its related rodent species, the mouse. We subsequently sought to understand the role and evolutionary conservation of the molecular cascade that drives TE fate downstream of aPKC, focusing on Hippo signalling pathway kinases. In all four species, inhibition of the LATS kinases promotes TE initiation in all cells, leading to ectopic nuclear enrichment of YAP1 and GATA3 in morula inner cells and in blastocyst ICM, accompanied by a reduction of SOX2 levels. Interestingly, LATS inhibition completely depleted SOX2 in mouse embryos; however, SOX2 levels were lower, but not completely abrogated in the other species. Altogether, these data indicate that although LATS kinases have a conserved role in preventing ectopic induction of TE in inner cells that go on to form the ICM, there are interesting species-specific differences in regulation of the pluripotency factor SOX2 that warrant further investigation.
RESULTS AND DISCUSSION
We initially performed morphokinetic analysis of rat embryos using similar criteria to our previous study (Gerri et al., 2020) (Movies 1-4). We calculated the duration of each morphological milestone as a percentage of time from the eight-cell stage to the end of cavitation. We observed an extended eight-cell-to-compaction transition with multiple cell divisions in rat, cow and human embryos, compared with the mouse (Fig. S1) (Gerri et al., 2020). We termed this stage ‘pre-compaction’. In addition, mouse embryos exhibited a comparatively longer morula stage, whereas cow, rat and human embryos showed a rapid transition from pre-compaction to cavitation (Fig. S1) (Gerri et al., 2020). These data provide a framework for comparative analysis of pre-implantation development in different species, as well as revealing important morphokinetic differences between the two rodent species considered in this study.
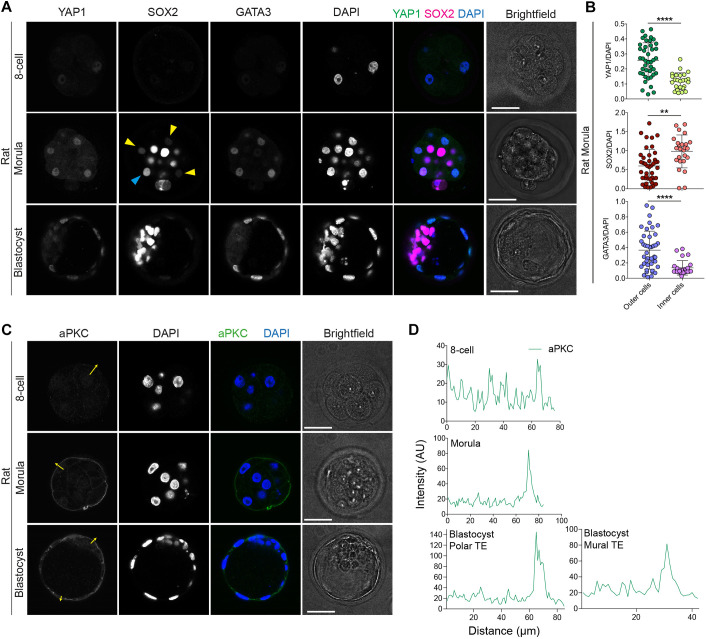
These intriguing results prompted us to analyse expression patterns of key differentiation and pluripotency factors in rat embryos. We observed weak nuclear localisation of YAP1 in all blastomeres in eight-cell-stage rat embryos (Fig. 1A,B, Fig. S2A), similar to mouse embryos (Nishioka et al., 2009). At the morula stage, YAP1 and GATA3 were present mainly in the nucleus of outer cells in rat embryos and were similarly nuclear localised in TE cells in rat blastocyst-stage embryos (Fig. 1A,B). Overall, the expression dynamics of YAP1 and GATA3 in rat embryos are similar to that reported in mouse, cow and human embryos (Gerri et al., 2020).
Fig. 1.
TE- and pluripotency-associated marker characterisation in rat embryos. (A) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in rat embryos at different developmental stages: eight-cell, morula and expanded blastocyst. Yellow arrowheads: cells with low SOX2; cyan arrowhead: outer cell with high SOX2. (B) Quantification of YAP1, GATA3 and SOX2 normalised fluorescence intensity in morula-stage rat embryos (n=67 cells from 5 embryos). Data are presented as mean±s.d. Two-tailed Mann–Whitney U-test: **P<0.01, ****P<0.0001. (C) Immunofluorescence analysis of aPKC and DAPI nuclear staining in rat embryos of different developmental stages: eight-cell, morula and expanded blastocyst. (D) Fluorescence intensity profiles of aPKC taken at the position of the yellow arrows in the rat embryos shown in C. AU, arbitrary units. Scale bars: 30 µm.
The transcription factor SOX2 is restricted to the nuclei of inner cells in mouse morula-stage embryos (Wicklow et al., 2014). By contrast, SOX2 is found more broadly in both inner and outer cells in cow and human morula-stage embryos and is restricted to ICM cells only at later blastocyst stages (Gerri et al., 2020; Cauffman et al., 2009; Goissis and Cibelli, 2014b). We therefore assessed SOX2 temporal and spatial dynamics in rat embryos, and observed nuclear localisation in both outer and inner cells at the morula stage and eventually nuclear enrichment in the ICM cells of expanded blastocysts (Fig. 1A,B, Fig. S2A). Moreover, we observed apical aPKC localisation in outer cells at the morula stage, and in both polar and mural TE cells in expanded-blastocyst rat embryos (Fig. 1C,D). Collectively, these data indicate conservation of aPKC protein localisation and expression dynamics of TE-associated markers at the morula stage in mouse, rat, cow and human embryos.
The downstream mechanisms linking cell polarity and TE initiation in mammals other than the mouse remain unclear. To investigate molecular mechanisms downstream of aPKC activity, we focused on the role of the Hippo pathway kinases LATS1 and LATS2. We used the small molecule inhibitor TRULI to test at a functional level the requirements for LATS1/2 kinases in regulation of early lineage initiation in rat, cow and human embryos (Kastan et al., 2021). We inhibited LATS1/2 in each species after embryonic genome activation, to avoid early developmental arrest. Embryonic genome activation is initiated at the two-cell stage in mouse and rat embryos (Aoki et al., 1997; Zernicka-Goetz, 1994), between the eight- and the 16-cell stages in cow embryos (Meirelles et al., 2004) and between the four- and eight-cell stages in human embryos (Braude et al., 1988; Tesarík et al., 1987). We first tested the inhibitor in mouse embryos by treating from pre-compaction at the four-cell stage to the morula stage. We performed a dose-response test and found the optimal concentration in mouse embryos, based on viability and phenotypic assessment (Fig. S3, Table S1). As expected (Cockburn et al., 2013; Nishioka et al., 2009), LATS inhibition led to nuclear enrichment of YAP1 and GATA3 in inner cells (Figs S4A-C, S2B). This contrasts with control embryos, in which YAP1 and GATA3 were nuclear localised in outer cells. Treated embryos also exhibited significant lower levels of SOX2 in the inner cells (Figs S4A-C, S2B), consistent with constitutively activated YAP1 (Frum et al., 2018). In addition, by treating embryos from the four-cell stage to expanded blastocysts we observed nuclear localisation of YAP1 and GATA3 in both TE and ICM cells, and a lack of detectable SOX2 in ICM cells (Figs S4A,D,E, S2B). These results demonstrated that LATS inhibition phenocopies results from genetic modification of the Hippo signalling pathway in the mouse and therefore represents a useful method to investigate the role of the Hippo pathway in early mammalian embryos (Nishioka et al., 2009). Interestingly, we observed a significant increase in nuclear YAP1 and GATA3 in outer cells (Fig. S4C), which may suggest some residual LATS activity in outer cells, which is blocked by LATS inhibitor treatment.
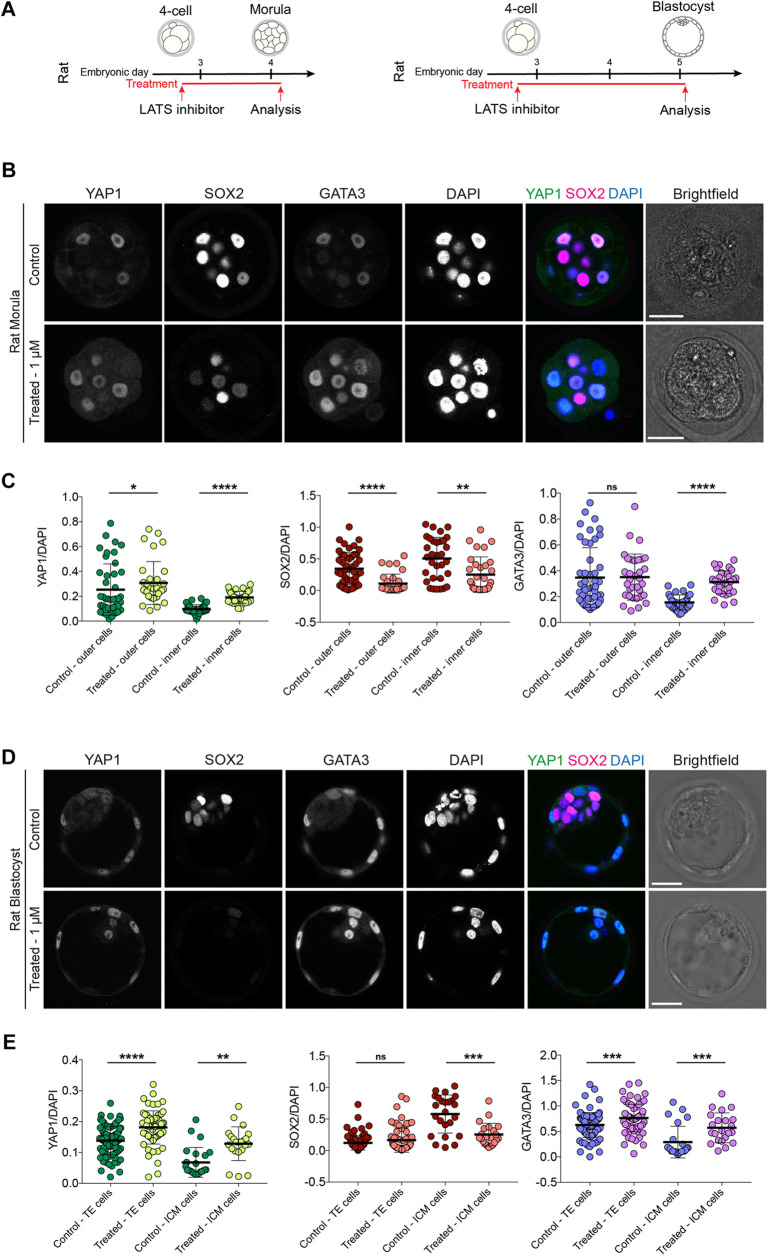
We extended our functional analysis to investigate the role of LATS kinases in rat, cow and human embryos by initially performing dose-response experiments in rat embryos (Fig. S5, Table S2). Similar to the mouse, treatment of rat embryos with the LATS inhibitor led to nuclear localisation of YAP1 and GATA3 in inner cells at the morula stage (Fig. 2A-C, Fig. S2C). SOX2 expression was significantly reduced in both outer and inner cells (Fig. 2A-C, Fig. S2C); however, it was not entirely abolished, in contrast to mouse (Fig. S4). SOX2 expression was undetectable in rat embryos treated with LATS inhibitor in expanded blastocysts, whereas YAP1 and GATA3 were nuclear localised in both TE and presumptive ICM cells (Fig. 2A,D,E, Fig. S2C). Moreover, although we observed a significant increase in nuclear YAP1 levels in outer cells, GATA3 was unaffected in rat morula-stage embryos (Fig. 2C), indicating that mouse embryos exhibit more rapid changes in the localisation of these factors following inhibition of Hippo signalling pathway components.
Fig. 2.
LATS inhibitor treatment in rat embryos. (A) Schematics of LATS inhibitor treatments in rat embryos. (B) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in control and LATS inhibitor-treated, morula-stage rat embryos. (C) Quantification of YAP1, SOX2, GATA3 normalised fluorescence intensity in outer and inner cells in control and LATS inhibitor-treated, morula-stage rat embryos (n=80 cells from 8 control embryos and 64 cells from 6 treated embryos). (D) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in control and LATS inhibitor-treated, expanded blastocyst-stage rat embryos. (E) Quantification of YAP1, SOX2, GATA3 normalised fluorescence intensity in TE and ICM cells in control and LATS inhibitor-treated, expanded blastocyst-stage rat embryos (n=117 cells from 6 control embryos and 90 cells from 5 treated embryos). Data are presented as mean±s.d. Two-tailed Mann–Whitney U-test: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. ns, not significant. Scale bars: 30 µm.
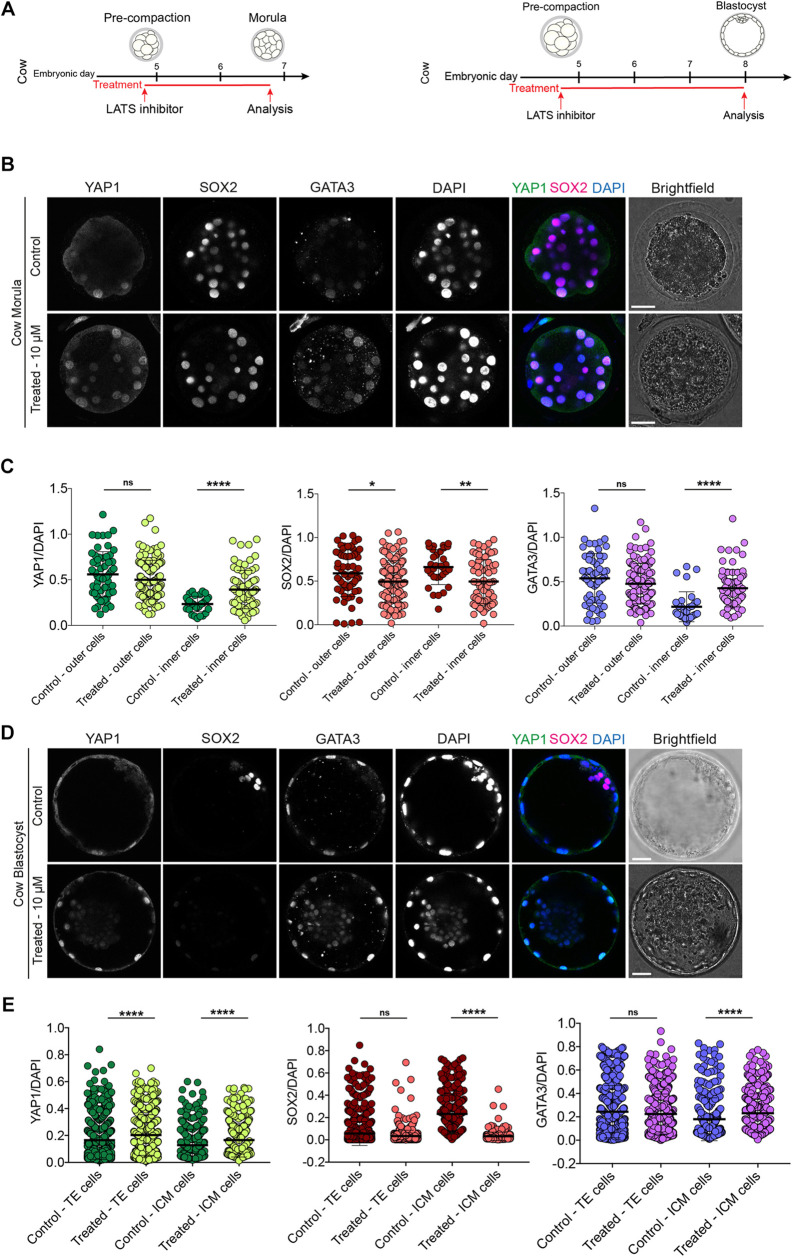
Cow embryos treated from pre- to post-compaction with LATS inhibitor showed significant nuclear localisation of YAP1 and GATA3 in inner cells (Fig. 3A-C, Figs S2D, S6, Table S3). Upon LATS inhibition, SOX2 expression decreased, but was not completely abolished, in both outer and inner cells at the morula stage (Fig. 3A-C, Fig. S2D), an expression pattern similar to that observed in rats (Fig. 2). Longer treatment with the LATS inhibitor until the blastocyst stage resulted in sustained nuclear enrichment of YAP1 and GATA3 in both the ICM and TE, with significantly lower levels of SOX2 (Fig. 3A,D,E, Fig. S2D).
Fig. 3.
LATS inhibitor treatment in cow embryos. (A) Schematics of LATS inhibitor treatments in cow embryos. (B) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in control and LATS inhibitor-treated, morula-stage cow embryos. (C) Quantification of YAP1, SOX2, GATA3 normalised fluorescence intensity in outer and inner cells in control and LATS inhibitor-treated, morula-stage cow embryos (n=93 cells from 4 control embryos and 201 cells from 7 treated embryos). (D) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in control and LATS inhibitor-treated, expanded blastocyst-stage cow embryos. (E) Quantification of YAP1, SOX2, GATA3 normalised fluorescence intensity in TE and ICM cells in control and LATS inhibitor-treated, expanded blastocyst-stage cow embryos (n=1347 cells from 10 control embryos and 1223 cells from 9 treated embryos). Data are presented as mean±s.d. Two-tailed Mann–Whitney U-test: *P<0.05, **P<0.01, ****P<0.0001. ns, not significant. Scale bars: 30 µm.
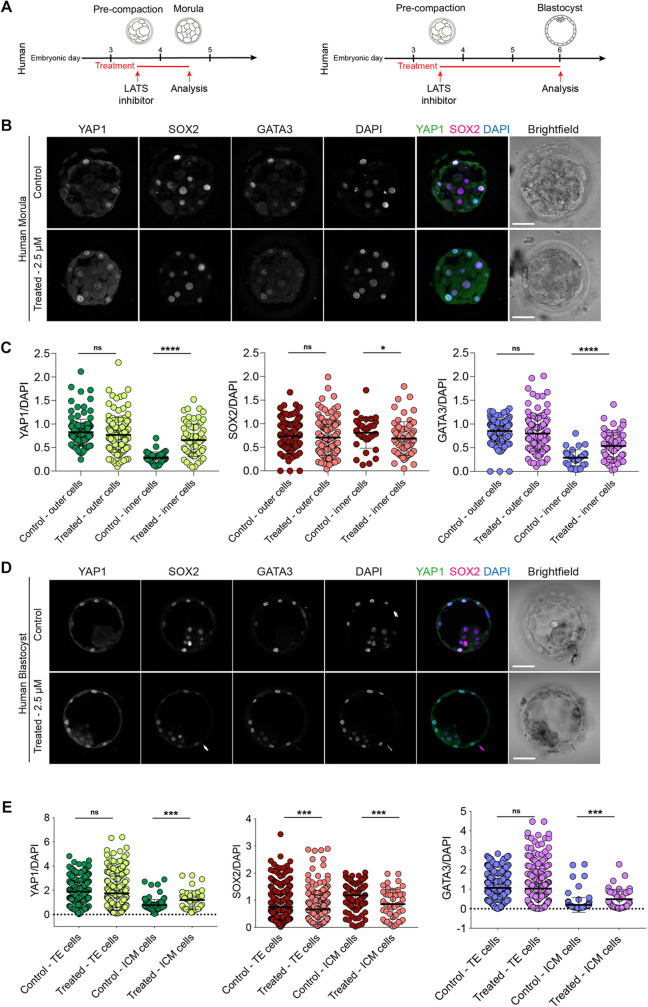
Next, we treated human embryos with the LATS inhibitor from the pre-compaction until the post-compaction morula stage (Fig. 4A, Fig. S7, Table S4). Upon LATS inhibition, the inner cells of human embryos exhibited nuclear enrichment of YAP1 and GATA3 (Fig. 4B,C, Fig. S2E). Similar to cow and rat embryos, LATS inhibition resulted in reduced, but not abolished, SOX2 expression in inner cells of morula-stage embryos, compared with controls (Fig. 4B,C, Fig. S2E). Moreover, in both cow and human morula embryos, nuclear expression of YAP1 and GATA3 in the outer cells at the morula stage was unaffected (Figs 3B,C, 4B,C), unlike in mouse (Fig. S4). Interestingly, we observed low levels of cytoplasmic YAP1 in rat, cow and human morula inner cells (Figs 2B, 3B, 4B), whereas YAP1 was clearly retained in the cytoplasm of treated mouse embryos (Fig. S4B). LATS treatment up to the blastocyst stage led to nuclear enrichment of YAP1 and GATA3, and reduction of SOX2, in human embryo ICM cells (Fig. 4A,D,E).
Fig. 4.
LATS inhibitor treatment in human embryos. (A) Schematic of LATS inhibitor treatment in human embryos. (B) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in control and LATS inhibitor-treated, morula-stage human embryos. (C) Quantification of YAP1, SOX2, GATA3 normalised fluorescence intensity in outer and inner cells in control and LATS inhibitor-treated, morula-stage human embryos (n=173 cells from 5 control embryos and 195 cells from 8 treated embryos). (D) Immunofluorescence analysis of YAP1, SOX2, GATA3 and DAPI nuclear staining in control and LATS inhibitor-treated, expanded blastocyst-stage human embryos. (E) Quantification of YAP1, SOX2, GATA3 normalised fluorescence intensity in TE and ICM cells in control and LATS inhibitor-treated, expanded blastocyst-stage human embryos (n=664 cells from 9 control embryos and 311 cells from 5 treated embryos). Data are presented as mean±s.d. Two-tailed Mann–Whitney U-test: *P<0.05, ***P<0.001, ****P<0.0001. ns, not significant. Scale bars: 30 µm.
Our findings suggest that mouse, rat, cow and human embryos show an evolutionarily conserved requirement for the Hippo signalling pathway mediated through LATS kinases. This regulates protein levels and localisation differences between inner and outer cells, leading to TE and ICM divergence (Fig. S8). Interestingly, the TE lineage-associated factor CDX2 is present in the mouse morula, and detectable in human and cow embryos only at later stages of development (Goissis and Cibelli, 2014a; Niakan and Eggan, 2013). This suggests that, although we report a conserved role for aPKC-mediated cell polarity and Hippo signalling, not all the downstream factors involved in TE specification are conserved in their expression dynamics across species. This might reflect differences in the timing or mechanism of lineage specification that necessitates further investigation. Consistent with this, blastocyst chimera, re-aggregation experiments and live-imaging analysis suggest that the timing of irreversible commitment of the TE occurs earlier in the mouse compared with human embryos (De Paepe et al., 2013; Posfai et al., 2017; Strnad et al., 2016). Further comparative analysis of putative developmental regulators across more species and functional studies of upstream and downstream regulators of early lineage specification would be informative.
The more restricted localisation of SOX2 in morula-stage mouse embryos compared with the broader expression throughout the morula in rat, cow and human embryos warrants further investigation. Sox2-null mouse embryos arrest soon after implantation and exhibit abnormal ICM development (Avilion et al., 2003). However, SOX2 is dispensable for initiation of OCT4 (POU5F1) and NANOG expression in mouse blastocysts (Wicklow et al., 2014). Interestingly, SOX2 is required for OCT4 and NANOG expression in cow embryos (Luo et al., 2022). Moreover, human SOX2 knockdown studies suggest a requirement for embryo genome activation (Gao et al., 2018). The YAP1/TEAD axis has been shown to function directly upstream of SOX2 in the mouse, but whether YAP1 and TEAD4 function similarly in the other species is unclear (Frum et al., 2018, 2019). We observed differences in the detection of cytoplasmic YAP1 at the morula stage across these mammals. We speculate that this intriguing discrepancy between the mouse and the other species may reflect differences in the regulation of YAP1 between species; however, future detailed studies are required to quantify the levels of YAP1 in the different cellular compartments. Together with the comparatively more rapid changes in SOX2 and GATA3 levels following LATS kinase inhibition in the mouse, this suggests molecular differences in regulation of these factors and/or temporal differences in protein turnover across mammals, as suggested for other biological processes (Rayon et al., 2020; Matsuda et al., 2020). In the future, it will be interesting to investigate whether the prolonged presence of SOX2 in cow and human blastocyst TE cells is mechanistically linked to the late appearance of CDX2, as these two transcription factors, considered to be master regulators of early lineage specification, might possess species-specific functions in a fundamental developmental process.
Several molecular differences between mouse and other mammalian species have been reported over recent years, yet whether these were rodent- or mouse-specific characteristics remained unclear. Our data show that some mouse-specific phenotypes are indeed not conserved with the rat, which conforms more to cow and human embryos. However, our study has some limitations. Based on our dose-response analysis, the optimal concentration of the LATS inhibitor was different in the species considered (5 μM mouse, 1 μM rat, 10 μM cow, 2.5 μM human). This may reflect differences in drug efficiency across species, leading to incomplete LATS inhibition. Diverse culture conditions and possible different antibody affinities between species could also have influenced our results. In addition, we focused on only three markers: YAP1, SOX2 and GATA3. In the future, we will employ alternative methods, such as CRISPR/Cas9-mediated genome editing or Trim-Away knockdown, to alter Hippo signalling activity. A more extensive analysis of lineage markers could provide further insights into the regulation and conservation of the first lineage specification event across species. Finally, we cannot exclude the possibility that additional kinases might function in parallel. In conclusion, our results highlight the importance of cross-species comparisons to elucidate conserved and divergent developmental mechanisms and highlight the importance of the mouse in uncovering fundamental principles that can be investigated in other species.
MATERIALS AND METHODS
Ethics statement
Experiments performed in the UK
This study was approved by the UK Human Fertilisation and Embryology Authority (HFEA; research licence number 0162) and the Health Research Authority's Research Ethics Committee (Cambridge Central reference number 19/EE/0297).
The process of licence approval entailed independent peer review along with consideration by the HFEA Licence and Executive Committees and the Cambridge Central Research Ethics Committee. Our research is compliant with the HFEA Code of Practice and has undergone inspections by the HFEA since the licence was granted. Research donors were recruited from patients at Bourn Hall clinic, Create Fertility, Hewitt Fertility Centre and Homerton University Hospital.
Informed consent was obtained from all couples that donated spare embryos following IVF treatment. Before giving consent, people donating embryos were provided with all the necessary information about the research project, an opportunity to receive counselling and the conditions that apply within the licence and the HFEA Code of Practice. Donors were informed that embryos used in the experiments would be stopped before 14 days post-fertilisation and that subsequent biochemical and genetic studies would be performed. Informed consent was also obtained from donors for all the results of these studies to be published in scientific journals. No financial inducements were offered for donation. Consent was not obtained to perform genetic tests on patients and no such tests were performed. The patient information sheets and consent document provided to patients are publicly available (https://www.crick.ac.uk/research/a-z-researchers/researchers-k-o/kathy-niakan/hfea-licence/). Embryos surplus to the patient's IVF treatment were donated cryopreserved and were transferred to the Francis Crick Institute where they were thawed and used in the research project.
Experiments performed in Belgium
The use of human embryos donated to research was approved by the Local Ethical Committee of UZ Brussel (BUN 1432021000526) and the Belgian Federal Committee for research on human embryos (AdV087). The embryos were surplus after IVF treatment in Brussels IVF at UZ Brussel. The embryos were cryopreserved and donated to research following informed consent and after the legally determined period of 5 years of cryopreservation.
Human embryo thawing
For embryos thawed in the UK, slow frozen human cleavage-stage embryos (between the two- and eight-cell stages) were thawed using Quinn's Advantage thaw kit (Origio, ART-8016). Briefly, with Quinn's Advantage thaw kit, after thawing the embryos were transferred to 0.5% sucrose thawing medium and incubated for 5 min at 37°C, followed by 0.2% sucrose thawing medium for 10 min at 37°C. The embryos were then washed through seven drops of diluent solution before culture. For vitrification, cleavage-stage embryos were warmed using a vitrification thaw kit (Irvine Scientific, 90137-SO) following the manufacturer's recommendations. In brief, embryos were warmed in TS thawing solution for 1 min and transferred to DS thawing solution for 4 min at room temperature (RT). Then, embryos were washed twice in washing solution for 4 min at RT before culture. For embryos donated in Belgium, the embryos were warmed as previously described (De Paepe et al., 2019).
Mouse zygote collection
All animal research was performed in accordance with UK Home Office regulations under project licence PP8826065, which passed ethical review by the Francis Crick Institute Animal Welfare Review Board in 2019. Within the Biological Research Facility animal units of the Francis Crick institute, mice had ad libitum access to feed and water, and were housed in individually ventilated cages maintained at 22°C with 60% humidity on a 12 h light-dark cycles. Four to eight-week-old (C57BL6×CBA) F1 female mice were super-ovulated using injection of 5 IU of pregnant mare serum gonadotrophin (PMSG; Sigma-Aldrich). Forty-eight hours after PMSG injection, 5 IU of human chorionic gonadotrophin (Sigma-Aldrich) was administered. Super-ovulated females were set up for mating with eight-week-old or older (C57BL6×CBA) F1 males. Mouse zygotes were isolated in flushing and holding medium (FHM) under mineral oil (Origio, ART-4008-5P) and cumulus cells were removed with hyaluronidase (Sigma-Aldrich, H4272).
Human and mouse embryo culture
Mouse embryos or human embryos were cultured in drops of pre-equilibrated Global medium (LifeGlobal, LGGG-20) supplemented with 10% human serum albumin (LifeGlobal, GHSA-125) and overlaid with mineral oil (Origio, ART-4008-5P). Pre-implantation embryos were incubated at 37°C and 5% CO2 in an EmbryoScope+ time-lapse incubator (Vitrolife) and cultured until the day of analysis. For embryos donated in Belgium, the embryos were cultured as previously described (De Paepe et al., 2019).
Rat embryo collection and culture
Super-ovulated and mated 7- to 9-week-old Sprague Dawley females were purchased by Charles River UK Limited. Rat two-cell embryos were isolated in FHM and then cultured in drops of pre-equilibrated rat embryo culture medium (mR1ECM, Cosmo Bio Co. Ltd., CSR-R-N174) and overlaid with mineral oil (Origio, ART-4008-5P). Embryos were incubated at 37°C and 5% CO2 and cultured until the day of analysis.
Cow embryo generation and culture
Cow ovaries were obtained from the abattoir Dunbia Cardington, Bedford, UK. Frozen bull sperm was obtained from UK Sire Services. Oocyte isolation, culture and fertilisation were performed using the IVF Bioscience media suite following the manufacturer's protocol, with small changes. In brief, cumulus–oocyte complexes (COCs) were aspirated from antral follicles using an 18-gauge needle mounted on a 5 ml disposable syringe. The aspirated follicular fluid was transferred to a 50 ml Falcon tube kept at 38.5°C. Only fully grown oocytes with a homogeneous cytoplasm and at least three to five complete layers of compact cumulus cells were selected for the experiments. COCs were washed and transferred to pre-warmed and equilibrated BO-IVM media (IVF Biosciences) and incubated at 38.8°C with 5% CO2. After 20 h, frozen semen was thawed and transferred in 4 ml of pre-warmed BO-Semen media (IVF Bioscience) and centrifuged for 5 min at 328 g. The pellet was resuspended in 4 ml of pre-warmed BO-Semen media and centrifuged for 5 min at 328 g. The supernatant was discarded, and sperm cells were gently resuspended in 1 ml of BO-IVF (IVF Bioscience) and counted using a Bürker chamber. The semen was diluted with BO-IVF (IVF Bioscience) to a concentration of 1×106 spermatozoa and 500 μl wells of diluted semen were prepared. COCs were partially denuded using a P200 pipette and washed in BO-WASH media (IVF Biosciences). COCs were washed into pre-warmed and equilibrated BO-IVF media and then placed in the wells with the diluted semen. Insemination was performed at 38.5°C, 5% CO2 for 20 h. Zygotes were retrieved after insemination and gently denuded of cumulus cells in pre-warmed BO-WASH medium. Zygotes were washed into pre-warmed and equilibrated BO-IVC media (IVF Biosciences) and incubated at 38.5°C, 5% CO2 and 5% O2.
Morphokinetic analysis and embryo staging
Time-lapse imaging was performed using an EmbryoScope+. Stages were defined following our morphological embryo benchmarking reported in extended figure 1 in our previous work (Gerri et al., 2020). Briefly, the eight-cell stage was considered to begin when the embryos displayed eight obvious blastomeres. The beginning of compaction was defined as when blastomeres started to flatten and spread onto their neighbours. The end of compaction was defined as when no visible cell boundaries were observed. The morula stage starts when the embryos appear as a compacted group of cells until the formation of small microlumens (small cavities appearing between blastomeres). We considered the blastocyst stage to start when embryos showed a single dominant blastocoel cavity. In our analysis, cavitation was considered from the end of the morula stage until the formation of an expanded blastocyst, when TE cells touch and start to stretch the zona pellucida, causing zona pellucida thinning. In addition, from time-lapse imaging we manually counted the number of cell divisions occurring between the eight-cell stage and the beginning of compaction. Data on mouse, cow and human embryos have already been reported in our previous publication (Gerri et al., 2020). We compared these data with our new analysis on rat embryos.
Mouse embryos, once they reach the eight-cell stage, undergo compaction without any additional cell divisions. Whereas in rat, cow and human embryos, we observed an extended period of time between the eight-cell stage and compaction. In this transition period, which we named ‘pre-compaction’, the embryos undergo multiple cell divisions before compacting (see Fig. S1C). The pre-compaction stage differs from cleavage stage, which is the period from the two- to the eight-cell stage.
Immunofluorescence
Embryos were fixed with freshly prepared 4% paraformaldehyde in PBS that was pre-chilled at 4°C. Embryo fixation was performed for 20 min at RT and then the embryos were transferred through three washes of 1× PBS with 0.1% Tween-20 to remove residual paraformaldehyde. Embryos were permeabilised with 1× PBS with 0.5% Triton X-100 and then blocked in blocking solution (3% bovine serum albumin in 1× PBS with 0.2% Triton X-100) for 2 h at RT on a rotating shaker. Then, embryos were incubated with primary antibodies diluted in blocking solution overnight at 4°C on rotating shaker. The antibodies and the concentrations used are reported in Table S5. The following day, embryos were washed in 1× PBS with 0.2% Triton X-100 for 20 min at RT on a rotating shaker and then incubated with secondary antibodies diluted in blocking solution for 1 h at RT on a rotating shaker in the dark. Next, embryos were washed in 1× PBS with 0.2% Triton X-100 for 20 min at RT on rotating shaker. Finally, embryos were placed in 1× PBS with 0.1% Tween-20 with Vectashield and DAPI mounting medium (Vector Laboratories, H-1200; 1:30). Embryos were placed on µ-Slide 8-well dishes (ibidi, 80826) for confocal imaging.
Quantification of YAP1, GATA3 and SOX2 expression
For morula-stage embryos, measurement of YAP1, SOX2 and GATA3 expression was performed using the 3D Fiji/ImageJ Suite. Briefly, the nuclei of immunofluorescently stained embryos were segmented automatically based on DAPI signal using Fiji, and YAP1, GATA3 and SOX2 fluorescence intensities were measured within the nuclei. We manually verified that the 3D segmentation did not count the same nucleus twice or fused together two adjacent nuclei, and in that case the measurement was excluded from the quantification.
For blastocyst-stage embryos, the quantification required a more sophisticated approach because of the high number of small cells very close to each other. Briefly, first the StarDist Fiji plugin was used to segment each cell and create a mask for each image. Then, CellProfiler used the mask files to measure the intensity of each cell, in each channel and in respect to the DAPI channel. These objects where then filtered to remove any objects that were touching the edge of the image. The size and shape of the objects were then measured. Objects were then filtered again, this time to exclude any objects that were less than 300 pixels, or over 6000 pixels (pre-determined parameters after some trial and error). This filtering therefore excluded any potential errors in the segmentation by StarDist, such as any cells that had fused together, dividing cells (nuclear DNA fragments), or other artefacts.
Individual objects were then tracked through the z-stack, so all z-stack images were confirmed to correspond with an individual blastomere. The final object intensity for each channel was then measured. In addition, we manually verified that the 3D segmentation did not count the same nucleus twice or fused together two adjacent nuclei, and in that case the measurement was excluded from the quantification.
Quantification of aPKC expression at the apical membrane
The fluorescence intensity profile of aPKC at the apical domain of outer blastomeres was determined using the plot profile tool in Fiji. We drew a ∼1-μm-thick line on the apical domain and measured the mean fluorescence intensity of aPKC. We selected confocal slices that transected the cell in the middle, in order to cover the whole apical domain, as previously performed (Gerri et al., 2020; Zhu et al., 2017; Rodriguez et al., 2017; Maître et al., 2015).
Confocal imaging
Confocal immunofluorescence images were taken with a Leica SP8 confocal microscopes with a Leica HC PL APO 63×/1.3 NA glycerol objective and 1-μm-thick optical sections were collected. For embryos imaged in Belgium, images were taken with a LSM800 (Zeiss) confocal microscope with an LD C-Apochromat 40×/1.1 NA water immersion objective and 1-μm-thick optical sections were collected.
Inhibitor treatment
The LATS inhibitor (Z730688380, Enamine) was dissolved in DMSO to 100 mM stock concentration and diluted at the required concentrations in pre-equilibrated embryo culture media. A dose-response experiment with analysis at the morula stage was performed in all the species. The following concentrations were used for the characterisation experiments: 5 μM for mouse embryos, 1 μM for rat embryos, 10 μM for cow embryos and 2.5 μM for human embryos. The optimal concentration was selected from the dose-response experiment by assessing the effects of the inhibitor on embryo viability (see Tables S1-S4) and phenotypic results based on YAP1, GATA3 and SOX2 expression at the morula stage (shown in Figs S3, S5-S7). The optimal concentration was then used to perform the experiments shown in Figs 2-4 and Fig. S4. Control embryos were developed in pre-equilibrated media to which the same volume of DMSO was added. It is important to note that for the treatment dishes, we diluted our inhibitor stock in order to add a maximum of 1 μl of inhibitor, and consequently 1 μl of DMSO for the control dishes, diluted in 500 μl of culture medium each.
For rat embryos, it is important to highlight that SOX2 levels were strongly abrogated at 2.5 and 5 μM of LATS inhibitor (Fig. S5). However, this phenotype was also accompanied by a strong decrease of viability, with, respectively, 44% and 64% of the rat embryos arresting their development at the morula stage (Table S2). For this reason, we believe that 2.5 and 5 μM were toxic to rat development, and we chose 1 μM of LATS inhibitor as our working concentration.
Statistical analysis
No statistical methods were used to predetermine sample size. The experiments were not randomised. The investigators were aware of group allocation during experiments and outcome assessment. All statistical analyses in this study were performed using GraphPad Prism 6.0. The number of cells or embryos analysed (n), statistical tests, and P-values are all stated in each figure or figure legend. Data are represented as mean±s.d. Unless otherwise noted, each experiment was performed at least three times.
Supplementary Material
Acknowledgements
We thank the individuals and couples whose donations have supported this research; M. Wilding, H. Premannandan and A. Srikantharajah for the coordination and donation of embryos to our research project; N. Tapon Lab for suggesting the LATS inhibitor used in this work; members of the laboratories of K.K.N., J. M. A. Turner, R. Lovell-Badge, J. Briscoe, as well as M. Marass, N. Tapon and A.M. Romao for advice and feedback on the manuscript; the Advanced Light Microscopy and Biological Research Facilities for their technical support (Francis Crick Institute).
Footnotes
Author contributions
Conceptualization: C.G., K.K.N.; Methodology: C.G., A.M., G.M.S., M.R., S.B., C.S.S., K.K.N.; Validation: C.G., G.M.S., M.R., P. Stamatiadis; Investigation: C.G., G.M.S., M.R., P. Stamatiadis; Resources: J.L., C.M., C.H., S.H., D.H., K.E., P. Snell, L.C., A.A.F.-N., H.V.d.V., K.K.N.; Data curation: C.G., K.K.N.; Writing - original draft: C.G.; Writing - review & editing: C.G., K.K.N.; Supervision: C.G., K.K.N.; Funding acquisition: K.K.N.
Funding
Work in the laboratory of H.V.d.V. was funded by the Fonds Wetenschappelijk Onderzoek Flanders (FWO G075222) and the Wetenschappelijk Fonds Willy Gepts (WFWG, UZ Brussel, G142). Work in the laboratory of A.A.F.-N. was supported by Comparative Biomedical Sciences Departmental fund from the Royal Veterinary College. Work in the laboratory of K.K.N. was supported by the Wellcome Trust (221856/Z/20/Z). Work in the laboratory of K.K.N. was also supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (CC2074), the UK Medical Research Council (CC2074) and the Wellcome Trust (CC2074). Open access funding provided by the Francis Crick Institute. Deposited in PMC for immediate release.
Data availability
All relevant data can be found within the article and its supplementary information.
Contributor Information
Claudia Gerri, Email: gerri@mpi-cbg.de.
Kathy K. Niakan, Email: kkn21@cam.ac.uk.
References
- Alarcon, V. B. (2010). Cell polarity regulator PARD6B is essential for trophectoderm formation in the preimplantation mouse embryo. Biol. Reprod. 83, 347-358. 10.1095/biolreprod.110.084400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, F., Worrad, D. M. and Schultz, R. M. (1997). Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181, 296-307. 10.1006/dbio.1996.8466 [DOI] [PubMed] [Google Scholar]
- Avilion, A. A., Nicolis, S. K., Pevny, L. H., Perez, L., Vivian, N. and Lovell-Badge, R. (2003). Multipotent cell lineages in early mouse development depend on Sox2 function. Genes Dev. 17, 126-140. 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, D. K., Smith, C. S., Pearton, D. J., Wells, D. N., Broadhurst, R., Donnison, M. and Pfeffer, P. L. (2011). Trophectoderm lineage determination in cattle. Dev. Cell 20, 244-255. 10.1016/j.devcel.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Braude, P., Bolton, V. and Moore, S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459-461. 10.1038/332459a0 [DOI] [PubMed] [Google Scholar]
- Cauffman, G., De Rycke, M., Sermon, K., Liebaers, I. and Van De Velde, H. (2009). Markers that define stemness in esc are unable to identify the totipotent cells in human preimplantation embryos. Hum. Reprod. 24, 63-70. 10.1093/humrep/den351 [DOI] [PubMed] [Google Scholar]
- Cockburn, K., Biechele, S., Garner, J. and Rossant, J. (2013). The hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195-1201. 10.1016/j.cub.2013.05.044 [DOI] [PubMed] [Google Scholar]
- Cockburn, K. and Rossant, J. (2010). Making the blastocyst: lessons from the mouse. J. Clin. Invest. 120, 995-1003. 10.1172/JCI41229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe, C., Cauffman, G., Verloes, A., Sterckx, J., Devroey, P., Tournaye, H., Liebaers, I. and Van De Velde, H. (2013). Human trophectoderm cells are not yet committed. Hum. Reprod. 28, 740-749. 10.1093/humrep/des432 [DOI] [PubMed] [Google Scholar]
- De Paepe, C., Aberkane, A., Dewandre, D., Essahib, W., Sermon, K., Geens, M., Verheyen, G., Tournaye, H. and Van De Velde, H. (2019). Bmp4 plays a role in apoptosis during human preimplantation development. Mol. Reprod. Dev. 86, 53-62. 10.1002/mrd.23081 [DOI] [PubMed] [Google Scholar]
- Dietrich, J. E. and Hiiragi, T. (2007). Stochastic patterning in the mouse pre-implantation embryo. Development 134, 4219-4231. 10.1242/dev.003798 [DOI] [PubMed] [Google Scholar]
- Ducibella, T. and Anderson, E. (1975). Cell shape and membrane changes in the eight-cell mouse embryo: prerequisites for morphogenesis of the blastocyst. Dev. Biol. 47, 45-58. 10.1016/0012-1606(75)90262-6 [DOI] [PubMed] [Google Scholar]
- Frum, T., Murphy, T. M. and Ralston, A. (2018). Hippo signaling resolves embryonic cell fate conflicts during establishment of pluripotency in Vivo. eLife 7, e42298. 10.7554/eLife.42298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum, T., Watts, J. L. and Ralston, A. (2019). Tead4, Yap1 and Wwtr1 prevent the premature onset of pluripotency prior to the 16-cell stage. Development 146, dev179861. 10.1242/dev.179861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L., Wu, K., Liu, Z., Yao, X., Yuan, S., Tao, W., Yi, L., Yu, G., Hou, Z., Fan, D.et al. (2018). Chromatin accessibility landscape in human early embryos and its association with evolution. Cell 173, 248-259.e15. 10.1016/j.cell.2018.02.028 [DOI] [PubMed] [Google Scholar]
- Gerri, C., Mccarthy, A., Alanis-Lobato, G., Demtschenko, A., Bruneau, A., Loubersac, S., Fogarty, N. M. E., Hampshire, D., Elder, K., Snell, P.et al. (2020). Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature 587, 443-447. 10.1038/s41586-020-2759-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goissis, M. D. and Cibelli, J. B. (2014a). Functional characterization of Cdx2 during bovine preimplantation development in Vitro. Mol. Reprod. Dev. 81, 962-970. 10.1002/mrd.22415 [DOI] [PubMed] [Google Scholar]
- Goissis, M. D. and Cibelli, J. B. (2014b). Functional characterization of Sox2 in bovine preimplantation embryos. Biol. Reprod. 90, 30. 10.1095/biolreprod.113.111526 [DOI] [PubMed] [Google Scholar]
- Hirate, Y., Hirahara, S., Inoue, K., Suzuki, A., Alarcon, V. B., Akimoto, K., Hirai, T., Hara, T., Adachi, M., Chida, K.et al. (2013). Polarity-dependent distribution of angiomotin localizes hippo signaling in preimplantation embryos. Curr. Biol. 23, 1181-1194. 10.1016/j.cub.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan, N., Gnedeva, K., Alisch, T., Petelski, A. A., Huggins, D. J., Chiaravalli, J., Aharanov, A., Shakked, A., Tzahor, E., Nagiel, A.et al. (2021). Small-molecule inhibition of lats kinases may promote yap-dependent proliferation in postmitotic mammalian tissues. Nat. Commun. 12, 3100. 10.1038/s41467-021-23395-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, C. Y. and Zernicka-Goetz, M. (2013). Angiomotin prevents pluripotent lineage differentiation in mouse embryos via hippo pathway-dependent and -independent mechanisms. Nat. Commun. 4, 2251. 10.1038/ncomms3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L., Shi, Y., Wang, H., Wang, Z., Dang, Y., Li, S., Wang, S. and Zhang, K. (2022). Base editing in bovine embryos reveals a species-specific role of Sox2 in regulation of pluripotency. PLoS Genet. 18, e1010307. 10.1371/journal.pgen.1010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maître, J. L., Niwayama, R., Turlier, H., Nédélec, F. and Hiiragi, T. (2015). Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol. 17, 849-855. 10.1038/ncb3185 [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Hayashi, H., Garcia-Ojalvo, J., Yoshioka-Kobayashi, K., Kageyama, R., Yamanaka, Y., Ikeya, M., Toguchida, J., Alev, C. and Ebisuya, M. (2020). Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 369, 1450-1455. 10.1126/science.aba7668 [DOI] [PubMed] [Google Scholar]
- Meirelles, F. V., Caetano, A. R., Watanabe, Y. F., Ripamonte, P., Carambula, S. F., Merighe, G. K. and Garcia, S. M. (2004). Genome activation and developmental block in bovine embryos. Anim. Reprod. Sci. 82-83, 13-20. 10.1016/j.anireprosci.2004.05.012 [DOI] [PubMed] [Google Scholar]
- Niakan, K. K. and Eggan, K. (2013). Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev. Biol. 375, 54-64. 10.1016/j.ydbio.2012.12.008 [DOI] [PubMed] [Google Scholar]
- Nishioka, N., Inoue, K.-i., Adachi, K., Kiyonari, H., Ota, M., Ralston, A., Yabuta, N., Hirahara, S., Stephenson, R. O., Ogonuki, N.et al. (2009). The hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398-410. 10.1016/j.devcel.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Ohsugi, M., Hwang, S. Y., Butz, S., Knowles, B. B., Solter, D. and Kemler, R. (1996). Expression and cell membrane localization of catenins during mouse preimplantation development. Dev. Dyn. 206, 391-402. [DOI] [PubMed] [Google Scholar]
- Plusa, B., Frankenberg, S., Chalmers, A., Hadjantonakis, A.-K., Moore, C. A., Papalopulu, N., Papaioannou, V. E., Glover, D. M. and Zernicka-Goetz, M. (2005). Downregulation of Par3 and Apkc function directs cells towards the icm in the preimplantation mouse embryo. J. Cell Sci. 118, 505-515. 10.1242/jcs.01666 [DOI] [PubMed] [Google Scholar]
- Posfai, E., Petropoulos, S., De Barros, F. R. O., Schell, J. P., Jurisica, I., Sandberg, R., Lanner, F. and Rossant, J. (2017). Position- and hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. eLife 6, e22906. 10.7554/eLife.22906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, A., Cox, B. J., Nishioka, N., Sasaki, H., Chea, E., Rugg-Gunn, P., Guo, G., Robson, P., Draper, J. S. and Rossant, J. (2010). Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 137, 395-403. 10.1242/dev.038828 [DOI] [PubMed] [Google Scholar]
- Rayon, T., Stamataki, D., Perez-Carrasco, R., Garcia-Perez, L., Barrington, C., Melchionda, M., Exelby, K., Lazaro, J., Tybulewicz, V. L. J., Fisher, E. M. C.et al. (2020). Species-specific pace of development is associated with differences in protein stability. Science 369, eaba7667. 10.1126/science.aba7667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J., Peglion, F., Martin, J., Hubatsch, L., Reich, J., Hirani, N., Gubieda, A. G., Roffey, J., Fernandes, A. R., St Johnston, D.et al. (2017). Apkc cycles between functionally distinct par protein assemblies to drive cell polarity. Dev. Cell 42, 400-415.e9. 10.1016/j.devcel.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad, P., Gunther, S., Reichmann, J., Krzic, U., Balazs, B., De Medeiros, G., Norlin, N., Hiiragi, T., Hufnagel, L. and Ellenberg, J. (2016). Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139-142. 10.1038/nmeth.3690 [DOI] [PubMed] [Google Scholar]
- Strumpf, D., Mao, C. A., Yamanaka, Y., Ralston, A., Chawengsaksophak, K., Beck, F. and Rossant, J. (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093-2102. 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Tesarík, J., Kopecný, V., Plachot, M. and Mandelbaum, J. (1987). High-resolution autoradiographic localization of DNA-containing sites and rna synthesis in developing nucleoli of human preimplantation embryos: a new concept of embryonic nucleologenesis. Development 101, 777-791. 10.1242/dev.101.4.777 [DOI] [PubMed] [Google Scholar]
- Vinot, S., Le, T., Ohno, S., Pawson, T., Maro, B. and Louvet-Vallée, S. (2005). Asymmetric distribution of par proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev. Biol. 282, 307-319. 10.1016/j.ydbio.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Wicklow, E., Blij, S., Frum, T., Hirate, Y., Lang, R. A., Sasaki, H. and Ralston, A. (2014). Hippo pathway members restrict Sox2 to the inner cell mass where it promotes Icm fates in the mouse blastocyst. PLoS Genet. 10, e1004618. 10.1371/journal.pgen.1004618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernicka-Goetz, M. (1994). Activation of embryonic genes during preimplantation rat development. Mol. Reprod. Dev. 38, 30-35. 10.1002/mrd.1080380106 [DOI] [PubMed] [Google Scholar]
- Zhu, M., Leung, C. Y., Shahbazi, M. N. and Zernicka-Goetz, M. (2017). Actomyosin polarisation through Plc-Pkc triggers symmetry breaking of the mouse embryo. Nat. Commun. 8, 921. 10.1038/s41467-017-00977-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, M., Shahbazi, M., Martin, A., Zhang, C., Sozen, B., Borsos, M., Mandelbaum, R. S., Paulson, R. J., Mole, M. A., Esbert, M.et al. (2021). Human embryo polarization requires Plc signaling to mediate trophectoderm specification. eLife 10, e65068. 10.7554/eLife.65068 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.