ABSTRACT
Transgenesis is an essential technique for any genetic model. Tol2-based transgenesis paired with Gateway-compatible vector collections has transformed zebrafish transgenesis with an accessible modular system. Here, we establish several next-generation transgenesis tools for zebrafish and other species to expand and enhance transgenic applications. To facilitate gene regulatory element testing, we generated Gateway middle entry vectors harboring the small mouse beta-globin minimal promoter coupled to several fluorophores, CreERT2 and Gal4. To extend the color spectrum for transgenic applications, we established middle entry vectors encoding the bright, blue-fluorescent protein mCerulean and mApple as an alternative red fluorophore. We present a series of p2A peptide-based 3′ vectors with different fluorophores and subcellular localizations to co-label cells expressing proteins of interest. Finally, we established Tol2 destination vectors carrying the zebrafish exorh promoter driving different fluorophores as a pineal gland-specific transgenesis marker that is active before hatching and through adulthood. exorh-based reporters and transgenesis markers also drive specific pineal gland expression in the eye-less cavefish (Astyanax). Together, our vectors provide versatile reagents for transgenesis applications in zebrafish, cavefish and other models.
Keywords: Cavefish, Enhancer discovery, Flurophores, Trangenesis marker, Transgenesis, Zebrafish
Summary: The generation, validation, and application of several new plasmid vectors suitable for transgenesis in zebrafish and other species.
INTRODUCTION
Transgenesis is a central technique across model systems for investigating biological processes in vivo. Integration of a transgene into the genome of a model organism enables experiments including live imaging, lineage tracing, generating conditional mutants and disease modelling. Transgenic reporters permit labelling of cells, lineages and structures in vivo using fluorescent proteins, making transgenesis an imperative tool in the study of developmental processes. Transgenesis can be paired with spatio-temporally controlled or ectopic gene expression to investigate the role of a specific gene within a molecular pathway. The zebrafish is an ideal system for transgenic applications due to its rapid development, optically transparent embryos, fecundity and amenability to genetic manipulation. Numerous additions to the toolbox for creating transgenic lines in zebrafish and improving transferability of expression constructs between species have crucially enabled the study of evolutionarily conserved genes and their role in developmental processes and disease progression (Amsterdam et al., 1995; Carney and Mosimann, 2018; Fisher et al., 2006a; Kawakami, 2004, 2005; Kikuta and Kawakami, 2009; Long et al., 1997; Meng et al., 1997; White et al., 2013).
In particular, Tol2-based transgenesis and accessible vectors for its application have transformed the zebrafish community with a simple modular system for the efficient generation of transgene constructs (Kawakami, 2004, 2005). First isolated in medaka (Koga et al., 1996), Tol2-based transgenesis requires the delivery of transposase mRNA and a transgenesis vector containing a desired transgene cargo flanked by Tol2 transposon repeats, resulting in efficient random integration into the genome (Kawakami, 2005; Kawakami et al., 1998). This workflow is particularly suitable for use in zebrafish, which are injected with Tol2 transposase-encoding mRNA and a transgenesis vector at the one-cell stage (Kawakami, 2005, 2007; Kikuta and Kawakami, 2009; Urasaki et al., 2008). Tol2 transposon activity for transgenesis is not restricted to zebrafish, as injection or electroporation-based delivery methods have been successfully used in several fish species and in other vertebrate models, including Amphioxus, Xenopus and chicken embryos (Hamlet et al., 2006; Kozmikova and Kozmik, 2015; Sato et al., 2007).
Despite its versatility, taking full advantage of Tol2 transgenesis hinges upon the broad availability, applicability and functionality of its required components. The first iteration of the Tol2kit (Kwan et al., 2007) and other powerful plasmid collections (Don et al., 2017; Villefranc et al., 2007) provided several entry and destination vectors for Multisite Gateway cloning (Cheo et al., 2004; Hartley et al., 2000) or restriction enzyme-based assembly of [regulatory element(s)]-[effector coding sequence]-[3′ trailer] constructs into a Tol2 transposon-flanked backbone. However, as transgenesis applications have become more widely applied and complex, an expanded set of transgenesis components has become essential.
First, transgenes based on isolated regulatory elements need a basic complement of validated parts to constitute a gene expression unit. Most prominent is the need for a minimal promoter sequence that is inert to position effects upon random Tol2-based integration and that features reproducible, robust interaction with numerous enhancer elements (Carney and Mosimann, 2018; Felker and Mosimann, 2016). Second, the widespread use of EGFP, mCherry and other fluorophores that generate red and green fluorescent reporters has introduced a challenge in the generation of combinatorial transgenic lines (Don et al., 2017; Kwan et al., 2007; Villefranc et al., 2007). Third, marking the expression of a transgene carrying a gene of interest as a fusion protein is not always practical, and previous attempts to use internal ribosomal entry site (IRES) sequences to generate bi-cistronic reporters have been generally unsuccessful for practical use in zebrafish (Kwan et al., 2007). Finally, the limited collection of available destination backbones with validated transgenesis markers restricts combinatorial transgene use and at times limits observations in tissues dominated by common transgenesis markers, such as the heart (promoter of myl7, formerly cmlc2) and the eye lens (various crystallin gene promoters) (Hesselson et al., 2009; Huang et al., 2003; Kwan et al., 2007; Villefranc et al., 2007).
To expand on previously available transgenesis tool sets, including the Tol2kit (Kwan et al., 2007), we embarked on a collaborative effort to augment, diversify and increase the functionality of available plasmid sets for transgenesis. Building upon components and applications generated by many before us (including, but not restricted to, Amsterdam et al., 1995; Asaoka et al., 2002; Bessa et al., 2009; Distel et al., 2009; Fisher et al., 2006b; Hesselson et al., 2009; Huang et al., 2003; Kawakami et al., 2000; Koga et al., 1996; Mosimann et al., 2011; Tamplin et al., 2011; Woolfe et al., 2004), and with the goal of accessible documentation and distribution to the community, we present here several plasmid vectors compatible with MultiSite Gateway and traditional restriction enzyme cloning. These constructs are geared towards facilitating gene regulatory element discovery, transgene quality control, reporter analysis and an expanded fluorophore spectrum for imaging. For all germline-transmitted, stable transgenic lines, we selected multiple founders and quality controlled for expected Mendelian ratios in subsequent generations; we have been observing most transgenic lines for three or more generations if not noted otherwise. Although our new backbones are geared towards Tol2-based transgenesis in zebrafish, the individual modules are deployable for numerous applications in diverse species, such as cavefish. Together, our vectors (Table 1; all plasmid sequences and maps have been deposited in Addgene, accession link https://www.addgene.org/browse/article/28233546/) expand the available modules for reproducible, quality-controlled Tol2 transgenesis and gene-regulatory element testing in zebrafish and other model systems.
Table 1.
Plasmid list
RESULTS
The mouse beta-globin (Hbb-bt) minimal promoter for universal enhancer testing
The discovery and isolation of gene-regulatory elements, such as enhancer elements, has been instrumental in decoding gene-regulatory input and in the creation of new transgenic tools to label and manipulate cell types of choice. Enhancers require a minimal promoter region associated with the transgene reporter to initiate RNA Pol II-based transcription (Haberle and Stark, 2018). As reporter expression can be positively and negatively influenced by neighboring loci in the genome (Amsterdam et al., 1995; Jaenisch et al., 1981; Lalonde et al., 2022; Wilson et al., 1990), an expression construct designed to test an enhancer requires a minimal promoter that is not sensitive to other enhancers in the vicinity of the transgene insertion. Several minimal promoters have been applied in zebrafish transgenesis, including Ef1a, E1b, hsp70l, c-fos, krt4, TK and gata2a promoters that are several hundred to several kilobases in length (Amsterdam et al., 1995; Bessa et al., 2009; Johnson and Krieg, 1995; Kondrychyn et al., 2009; Li et al., 2010; Meng et al., 1997; Ogura et al., 2009; Scott et al., 2007; Shoji and Sato-Maeda, 2008; Weber and Köster, 2013).
The promoter activity of the mouse beta-globin minimal promoter (from the mouse Hbb-bt gene, 129 bp sequence including 54 bases of 5′ UTR) was initially characterized using HeLa cells (Myers et al., 1986). Coupling of the mouse beta-globin minimal promoter to a promoter-less enhancer enables the generation of stable Tol2-based zebrafish transgenics with tissue-specific reporter activity, including fluorescent reporters and CreERT2 and KalTA4 driver lines (D'Agati et al., 2019; Kaufman et al., 2016; Prummel et al., 2019; Tamplin et al., 2011; Woolfe et al., 2004). Reporters including the mouse beta-globin minimal promoter show minimal to no background in transient injections and drive stable transgene activity in zebrafish, including when paired with enhancers from other species (Prummel et al., 2019; Tamplin et al., 2011, 2015; Woolfe et al., 2004). To enable testing and application of candidate enhancers in 5′ entry vectors, we have generated middle entry vectors with mouse beta-globin minimal promoter-coupled mCerulean, EGFP, mCherry and mApple ORFs, as well as creERT2 and the codon-optimized Gal4-activator, KalTA4 (Distel et al., 2009) (Fig. 1A, Table 1 lists all generated plasmids used in this study).
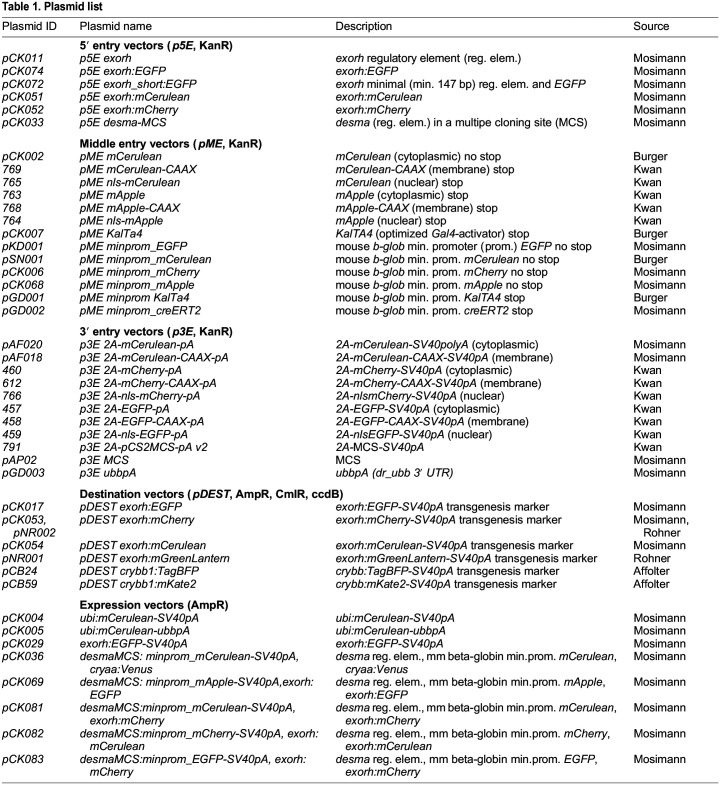
Fig. 1.
Incorporating the small mouse beta-globin minimal promoter into transgenic reporters. (A) Workflow of reporter generation or enhancer testing with Multisite Gateway recombination and Tol2 transgenesis. An enhancer fragment is cloned into a p5E vector and Gateway- or restriction enzyme-cloned into an expression construct composed of a 5′ fragment, a middle entry mouse beta-globin minimal promoter:fluorophore/KalTa4/creERT2, 3′ SV40 polyA and a transgenic marker (optional). The resulting Tol2 expression construct is injected along with Tol2 transposase-encoding mRNA into one-cell stage zebrafish, randomly integrated into the genome and screened to establish transgenic lines. Schematic created with BioRender.com (license through CU Anschutz). (B) Schematic of mBmp4ECR2_minprom:EGFP containing the distal mouse Bmp4ECR2 element, the mouse beta-globin (Hbb-bt) minimal promoter driving EGFP and the SV40 polyA. (C-G) mBmp4ECR2 reporter expression in a variety of zebrafish tissues linked to endogenous bmp4 expression. (C) Lateral view of a stable transgenic line of mBmp4ECR2_minprom:EGFP at the 20-somite stage; asterisk marks the developing heart; arrowhead indicates the olfactory bulb. (D-F) Lateral (D,E,G) and ventral (F) views of 3 dpf stable mBmp4ECR2_minprom:EGFP transgenic larva (D) show expression in the heart (asterisk) and pectoral fin (arrowhead), as well as in muscles and fins (D,E), parts of the head musculature (F), and in the mesothelium surrounding the yolk, the trunk musculature and fin fibroblasts (G). (H,I) The mHbb-bt minimal promoter has little to no background activity in transient injections. (H) Schematic of expression vector T7_minprom:mCerulean using the bacterial phage T7 promoter (inactive in zebrafish), followed by the mHbb-bt minimal promoter and mCerulean ORF as a reporter. An eye lens-specific cryaa:Venus cassette in cis was used as a transgenesis control. (I,J) Representative 2 dpf embryo of a control or reference injection repeat with T7_minprom:mCerulean, imaged for mCerulean (I) and with Venus as a reference for successful injection (J). There is no detectable mCerulean expression (representative among 136 individually injected embryos in this replicate). Scale bars: 500 μm (C,D,I,J); 200 μm (E-G).
To further illustrate the utility of the mouse beta-globin minimal promoter, we documented regulatory element testing of a mouse enhancer. The 668 bp-spanning downstream enhancer ECR2 in the mouse Bmp4 locus has previously been discovered to convey posterior lateral plate mesoderm activity when tested as a reporter in mouse embryos (Chandler et al., 2009). In zebrafish embryos, the ECR2 enhancer coupled with beta-globin-minprom:EGFP (mBmp4ECR2_minprom:EGFP) (Fig. 1B) as stable transgene resulted in EGFP reporter expression in a dynamic expression pattern in different cell types, including trunk and tail muscles, pectoral fins, the heart and the yolk-surrounding mesothelium (Fig. 1C). Notably, several of these reporter expression domains matched with the reported expression of zebrafish bmp4 (Fig. 1D-G) (Bisgrove et al., 2000; Lenhart et al., 2013; Nikaido et al., 1997; Stickney et al., 2007; Thisse et al., 2004). The mouse beta-globin minimal promoter appears inert to position effects or non-specific episomal expression upon injection: testing of a reference construct harboring the phage-derived T7 promoter that has no transcriptional activity in zebrafish (T7_minprom:mCerulean) reproducibly showed no discernible reporter expression at 2 dpf (n=136/136), while the transgenesis marker in cis revealed successful F0 injections (Fig. 1H-J). Together with previous work, these data illustrate the utility of the mouse beta-globin minimal promoter for zebrafish transgenes based on isolated regulatory elements.
Components for fluorescent reporter generation
To expand available basic components for transgenic reporter generation, we revisited available fluorophores and 3′ UTR constructs. The prevalent use of green fluorescent avGFP derivatives and red-fluorescent proteins (Chalfie et al., 1994; Rodriguez et al., 2017; Tsien, 2003) has greatly expanded the number of available transgenic zebrafish reporters, yet has led to a bottleneck in combining reporters for multi-color imaging. To extend the color spectrum available for transgenic applications with accessible tools, we have generated middle entry clones with cytoplasmic, membrane and nuclear versions of mCerulean (Fig. 2D-F, Table 1). mCerulean is a monomeric, blue-fluorescent protein derived from avGFP and subsequent CFP with an excitation wavelength of 433 nm and an emission wavelength of 475 nm (Rizzo and Piston, 2005; Rizzo et al., 2004). Stable transgenes using mCerulean as reporter are well tolerated in zebrafish (Fig. 2A) (Hesselson et al., 2009; Trinh et al., 2011; Zuniga et al., 2011). Spectral separation of mCerulean from GFP is easily achievable on common confocal and light sheet setups, either by using a 445 nm or 458 nm laser or using features of typical confocal software platforms to minimize spectral overlap (e.g. ZEN software, Zeiss). We noted that, depending on genetic background, mCerulean imaging on a scanning confocal microscope resulted in auto-fluorescence of skin or pigment cells, particularly in the head of zebrafish embryos and larvae (48 hpf onwards) when visualized with an Argon laser (488 nm) (Fig. 2A-F). Use of the ideal excitation laser wavelengths greatly diminished this auto-fluorescence. Of note, mCerulean is easily visible on standard dissecting microscopes with epifluorescence equipment, ideally using blue fluorescence filters and excitation spectra. mCerulean is excited and detectable by standard setups used for GFP fluorescence, requiring care to avoid misinterpreting mCerulean versus GFP signal in double-transgenic embryos.
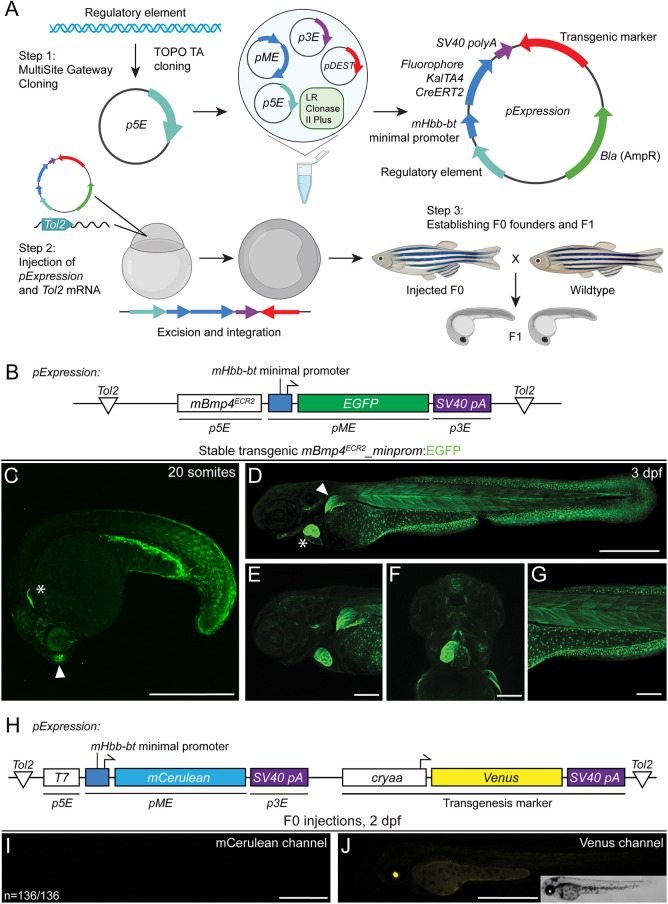
Fig. 2.
mCerulean as a versatile blue fluorophore for zebrafish transgenesis. (A) The stable transgenic line for HOPX4q12N3.1_minprom:mCerulean,crybb1:mKate2 shows mCerulean expression in both the vacuolar and sheath cells of the zebrafish notochord at 2 days post-fertilization and eye lens-specific mKate2 expression (asterisk) when crossed with the drl:mCherry transgenic, marking lateral plate mesoderm-derived lineages (cardiovascular system, blood at 2 dpf). (B,C) Transient injection of zebrafish ubiquitin (ubb) promoter-driven mCerulean with 3′ polyadenylation trailer (polyA) from SV40 (B) or ubb (C). (D-F) mCerulean constructs for different cellular localization. Transient expression of cytoplasmic mCerulean (D), nuclear nls-mCerulean (E) and membrane-bound mCerulean-CAAX (F). Scale bars: 500 μm (A-C); 50 μm (D-F).
To demonstrate the versatility of mCerulean, we imaged the transgenic reporter line Tg(Hs-HOPX4q12N3.1_minprom_mCerulean) (subsequently abbreviated as HOPX:mCerulean), which includes the upstream region of the human HOPX gene together with the mouse beta-globin minimal promoter and results in stable expression in the notochord at 48 hpf (Fig. 2A). This expression is consistent with mammalian expression of HOPX in the developing primitive streak, as well as in chordoma tumors (Loh et al., 2016; Nelson et al., 2012; Palpant et al., 2017). Notably, the human HOPX upstream region also drives expression in rhombomeres 3 and 5 in individual transgenic insertions (Chen et al., 2002). As a blue fluorescent reporter, HOPX:mCerulean combines well with mCherry-based reporters, such as drl:mCherry, which marks lateral plate mesoderm before restricting expression to cardiovascular lineages (Fig. 2A) (Mosimann et al., 2015; Sánchez-Iranzo et al., 2018). Together, mCerulean provides a practical alternative fluorophore to expand the reporter spectrum in transgenic experiments.
Transgene constructs require a 3′ UTR with polyadenylation signal (polyA) for productive mRNA transcription, stability and translation (Felker and Mosimann, 2016; Fink et al., 2006; Matoulkova et al., 2012; Okayama and Berg, 1992). The most common 3′ UTR with polyA sequences used for zebrafish transgenes are the bi-directional SV40 late polyA sequence (abbreviated as SV40 polyA) (Okayama and Berg, 1992) that was included in the original Tol2kit collection (vector 302) (Kwan et al., 2007) and the BGH polyA sequence (Higashijima et al., 1997). Several available Tol2 backbones for MultiSite Gateway assembly contain the SV40 polyA after the recombination cassette (Kwan et al., 2007); although practical, its presence can lead to assembly and sequencing issues when another SV40 polyA trailer is used in the 3′ position. To expand the available 3′ UTRs for transgene assembly, we isolated a 516 bp fragment of the 3′ UTR in the zebrafish ubiquitin B (ubb, ubi) gene. We designed pAP02 as p3E vector with versatile multiple cloning site to enable the generation of new 3′ vectors by restriction enzyme cloning; transferring the ubb 3′ region into pAP02 resulted in p3E_ubb-polyA. To test the p3E_ubb-polyA and compare it with the commonly used Tol2kit vector 302, we generated pCK005 ubi:mCerulean-ubb-polyA as well as pCK004 ubi:mCerulean-SV40-polyA, which provide convenient control vectors for co-injection along with newly generated reporter constructs of unknown activity (Fig. 2B,C; Table 1). In addition to this alternative SV40 polyA p3E vector, we also generated a p3E vector with the multiple cloning site from pBluescript KSII(+), providing single-cutter restriction sites for cloning inserts into the final plasmid (see Materials and Methods for details).
mCherry and multimeric dsRED versions are red fluorophores commonly used for zebrafish transgenesis (Davison et al., 2007; Kwan et al., 2007; Pisharath et al., 2007; Provost et al., 2007; Rodriguez et al., 2017; Traver et al., 2004). mApple (derived from Discosoma sp.) is a fast-maturing monomeric red fluorophore with an excitation maximum of 568 nm, which is therefore less red-shifted than mCherry and still excitable by standard lasers on confocal microscopes (543 or 561 nm) (Fig. 3A-G) (Shaner et al., 2008). mApple also has a better theoretical quantum yield and a fast-folding mechanism, rendering it a potentially suitable, additional red monomeric fluorophore for any transgenesis toolkit. mApple is well-tolerated in zebrafish and has been successfully pioneered for red-fluorescent transgenic applications (Heim et al., 2014; Lou et al., 2015), yet it remains less frequently used compared to mCherry or dsRED derivatives. Applied to label endoderm membranes, mApple expression in stable Tg(sox17:mApple-CAAX) embryos showed faithful and well-detectable expression (Fig. 3A,B) akin to Tg(sox17:EGFP-CAAX) (Fig. 3C,D), complementing the existing and widely used cytoplasmic sox17:EGFP to label endoderm progenitors (Chung and Stainier, 2008; Sakaguchi et al., 2006). To expand the application of red-fluorescent mApple in zebrafish, we further generated combinations of mApple with the mouse beta-globin minimal promoter as middle entry clones (see Materials and Methods for details) with nuclear as well as membrane-localized versions (Fig. 3E-G, Table 1). Together, our series of minimal promoter-coupled fluorophore vectors, multiple localization tags, plus p3E polyA with multiple cloning site vectors provide a basic set of reagents for versatile regulatory element discovery and simple fluorescent microscopy with standard equipment accessible in the field.
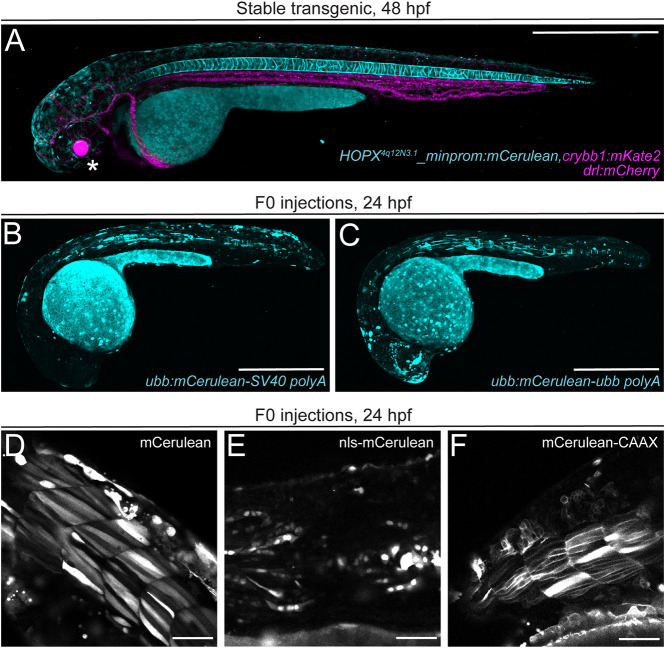
Fig. 3.
mApple as a bright red fluorophore for zebrafish transgenesis. (A-D) Comparison of membrane-localized mApple-CAAX with EGFP-CAAX in stable sox17:mAppleCAAX and sox17:EGFPCAAX lines show matching expression in the developing pharyngeal endoderm at 28 hpf. Pharyngeal pouches shown in greater detail with anterior towards the left in max projection (A,C) and single slice views (B,D). (E-G) mApple fusion proteins for different localizations. Transient expression of cytoplasmic mApple (E), nuclear nls-mApple (F) and membrane-bound mApple-CAAX (G). Scale bars: 50 µm.
2A peptide clones for fusion protein reporters
Transgenic expression of a protein of interest in a specific cell population greatly benefits from fluorescent labeling of transgene-expressing cells. One method to directly label the expressed protein of interest is by fusion with a fluorescent tag; yet the impact of adding a large fluorescent protein to the protein of interest can result in mis-localization, sequestration, degradation or inactivity of the fusion product (Giepmans et al., 2006). A widely deployed alternative is the translation of the protein of interest and a fluorescent reporter via a bi-cistronic message (Chan et al., 2011; Trichas et al., 2008). We have previously released a series of vectors, including IRES (internal ribosome entry site) sequences (Jang et al., 1988; Pelletier and Sonenberg, 1988), for such applications. However, IRES-containing constructs have been repeatedly reported to cause variable expression of the ORF (open reading frame) downstream of the IRES sequence (Ibrahimi et al., 2009; Wong et al., 2002). With IRES-containing constructs used in zebrafish, the ORF following the IRES (often a fluorescent reporter) is commonly expressed at a lower level than the first ORF. Individual users observed up to a 10-fold lower expression of the second ORF, while other data indicate that the presence of the IRES reporter can even dampen expression of the first ORF (Weber and Köster, 2013), severely limiting the practical utility of IRES constructs.
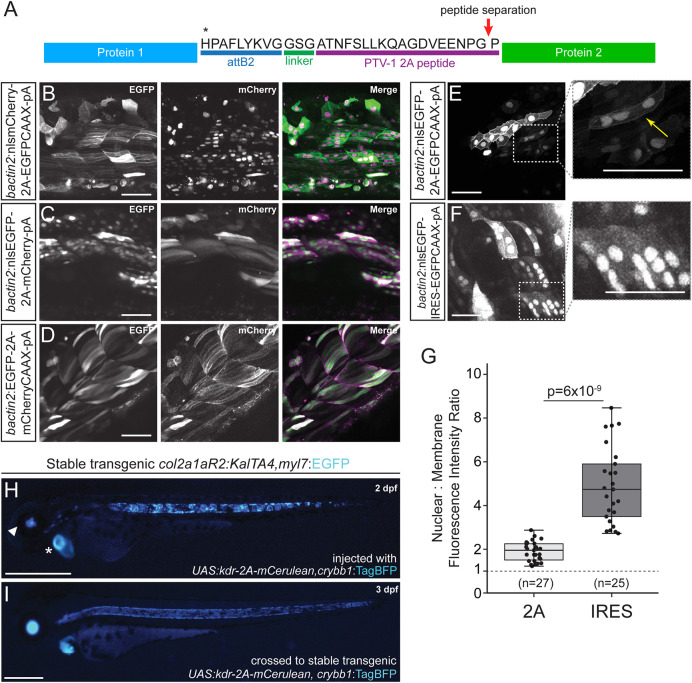
To provide an alternative strategy for fluorescent labeling, we have established 3′ entry (p3E) vectors that include the PTV-1 2A peptide sequence, a well-characterized viral peptide that results in production of two proteins from the same message across model systems (Kim et al., 2011; Provost et al., 2007; Ryan et al., 1991). 2A-mediated ‘self-cleavage’ takes advantage of the ribosome not forming a glycyl-prolyl peptide bond at the C-terminus of the 2A peptide, allowing the translational complex to release the first polypeptide and continue translation of the downstream product without independent ribosome recruitment or additional host factors (Donnelly et al., 2001a,b) (Fig. 4A, Table 1). The resulting equimolar protein amounts make the 2A peptide an ideal tool for the quantification of transgene expression or the controlled expression of multiple proteins from one transgene (Chi et al., 2008; Knopf et al., 2010, 2011). We generated 3′ entry constructs containing the 2A peptide-encoding sequence followed by different fluorophores (EGFP, mCherry and mCerulean) in cytoplasmic, membrane and nuclear forms (Fig. 4B-D, Table 1). These constructs are ready for use with middle entry clones that are in Gateway reading frame without a stop codon (see Materials and Methods, maps and Gateway manual for details on primer design).
Fig. 4.
2A peptide linkers for fluorescent protein tagging. (A) Schematic of protein product resulting from a Tol2 construct containing two protein sequences separated by a 2A peptide following the attB2 site of a Multisite Gateway expression construct. Translation of the first protein occurs, the first polypeptide is released at the site of peptide separation and translation of the second ORF proceeds with the same ribosome, resulting in equimolar protein amounts. (B-F) Transient injections of bactin2 promoter-driven fluorophore combinations separated by a 2A peptide tag. nlsmCherry-2A-EGFPCAAX (B), nlsEGFP-2A-mCherry (C) and EGFP-2A-mCherryCAAX (D). Transient injection of bactin2:nlsEGFP-2A-EGFPCAAX (E) demonstrates expression of EGFP in both the nucleus and membrane of cells, whereas injection of bactin2:nlsEGFP-IRES-EGFPCAAX (F) results in poor EGFP-CAAX detection in cells that are positive for nuclear EGFP. (G) Ratio of nuclear to membrane fluorescence intensity with n=total number of cells assayed (numbers at the base of the graph); for each condition, cells from five different embryos were analyzed. Welch's t-test (unequal variance); first and third quartiles are boxed, bars extend to the highest value within the 1.5× inter-quartile range. (H,I) Use of 2A fusions in Gal4/UAS transgene experiments. Injection of UAS:kdr-2A-mCerulean,crybb1:TagBFP into stable transgenic col2a1aR2:KalTA4,myl7:EGFP that expresses codon-optimized Gal4 in the developing notochord and subsequent cartilage lineages shows mosaic mCerulean expression in the developing notochord (H), with the secondary myl7:EGFP transgenic marker indicated with an asterisk and the crybb1:TagBFP marker indicated with an arrowhead. Homogenous notochord expression of mCerulean in stable F2 UAS:kdr-2A-mCerulean,crybb1:TagBFP crossed with col2a1aR2:KalTA4, myl7:EGFP (I). Scale bars: 50 μm (B-F); 500 μm (H,I).
To evaluate and compare stoichiometry of protein production using 2A and IRES, we generated transgene expression constructs in which the same fluorophore (EGFP) was targeted to two different subcellular locations in transient injections: nuclear-localized EGFP (nls-EGFP) was in the first position and membrane-targeted EGFP (EGFP-CAAX) was in the second (Fig. 4E, Table 1). When separated by the 2A peptide, EGFP was easily visualized in both nuclear and membrane locations, even in cells expressing low levels of the transgene (Fig. 4E). When separated by the IRES, in most cells, only nuclear-localized EGFP was detectable; EGFP-CAAX was detected only in the brightest cells, confirming that the 2A peptide yields effective production of two separate proteins targeted to distinct subcellular locations (Fig. 4F). We quantified this result by measuring the nuclear:membrane fluorescence intensity ratio in individual cells (see Materials and Methods for details). IRES constructs yielded a much higher ratio of nuclear:membrane fluorescence compared with the 2A (Fig. 4G), indicating a greater expression difference of the two EGFP fluorophores from the same messenger RNA.
The utility of 2A-based fusions becomes apparent when applied with the Gal4/UAS system. Gal4-dependent expression of genes of interest as UAS-controlled transgenes is a powerful approach to test individual gene function and for disease modeling (Akitake et al., 2011; Distel et al., 2009; Halpern et al., 2008; Köster and Fraser, 2001; Provost et al., 2007). The ability to fluorescently label the UAS transgene-expressing cells provides an immediate phenotype readout; as 2A-based fusions harbor the fluorescent reporter C-terminally of the ORF, the fluorescence signal additionally provides expression control of the entire coding sequence. Injection of UAS:kdr-2A-Cerulean into the cartilage-specific Gal4-driver transgenic line col2a1aR2:KalTA4 (D'Agati et al., 2019; Dale and Topczewski, 2011) resulted in mosaic Cerulean expression in the developing notochord (Fig. 4H). Stable UAS:kdr-2A-Cerulean transgenic insertions (selected for with the crybb1:TagBFP transgenesis marker in cis on the transgene backbone pCB24) crossed to the same driver resulted in homogenous notochord expression (Fig. 4I).
Of note, using the 2A peptide in the Gateway system results in the addition of a 28 aa ‘tag’ to the C-terminal end of the first protein (containing the translated att site and the 2A peptide) (Rothwell et al., 2010) (Fig. 4A). This residual amino acid sequence may disturb the protein of interest in the first position if it is sensitive to C-terminal tagging. To reverse the relative positions of the fluorophore and ORF of interest cloning, we also generated a 3′ 2A entry vector containing the MCS from pCS2 for cloning of genes of interest after the 2A peptide sequence that can be used with middle entry fluorescent reporters (see Materials and Methods for details). Together, our 2A-based vectors enable versatile tagging of proteins of interest in zebrafish transgenes and beyond.
A selective, inert transgenic marker labeling the pineal gland
Transgenesis requires rigorous quality control, which is facilitated by inclusion of an easy-to-screen, dominant reporter gene in cis with the intended transgene cargo. In practical terms, inclusion of an independent fluorescent transgene that is detectable for screening at a desired developmental stage enables identification of transgene-positive animals and tracking of the transgenic insertion across generations (Felker and Mosimann, 2016). The initial Tol2kit contained the destination vectors pDestTol2pA2 as an ‘empty’ destination vector and pDestTol2CG2 that included the myl7 (formerly cmlc2) promoter driving EGFP in the developing myocardium from late somitogenesis stages onwards, which is easily screenable by 24 hpf (Huang et al., 2003; Kwan et al., 2007). However, myl7-based transgenesis markers are not desirable for transgenes focused on heart development, which is of significant interest in the field. Furthermore, the heart becomes increasingly obscured by skin pigmentation in wild-type zebrafish strains, rendering transgene detection in adults challenging (White et al., 2008).
Eye lens-specific promoters of different crystallin genes have been used as powerful transgenesis markers (Davidson et al., 2003; Emelyanov and Parinov, 2008; Hesselson et al., 2009; Kurita et al., 2003; Marques et al., 2008; Waxman et al., 2008). Alpha- or beta-crystallin promoter-based transgenes (cryaa or crybb1) are visible throughout adulthood and are easily detected via UV flashlights and filter glasses (Felker and Mosimann, 2016). We have previously adopted cryaa:Venus (Hesselson et al., 2009) to continuously mark effector transgenics such as CreERT2 drivers with a yellow fluorescent transgenic marker (Mosimann et al., 2015). To complement existing reporter backbones, we generated pCB59 crybb1:mKate2 to serve as a red eye transgenesis marker (Fig. 2A) and pCB24 crybb1:TagBFP as a blue eye transgenesis marker (Fig. 4H,I). The far-red mKate2 (Lin et al., 2012; Shcherbo et al., 2009) is still detectable using standard red fluorescence filters on dissecting scopes with excitation at 588 nm and emission at 633 nm, and is complementary to blue, green or yellow fluorescent crystallin promoter-driven markers for combinatorial transgene experiments. TagBFP is a robust blue fluorophore with excitation at 402 nm and emission at 457 nm (Subach et al., 2008) that is also well tolerated in zebrafish (Singh et al., 2012). One drawback of crystallin-based reporters poses their expression initiation around 36-48 hpf (Emelyanov and Parinov, 2008; Hesselson et al., 2009; Kurita et al., 2003; Marques et al., 2008; Waxman et al., 2008), which is later than myl7-based reporters and close to the start of swimming that can render sorting of transgene-positive embryos challenging. Additionally, eye lens expression of fluorophores can interfere with imaging of head regions and is not desirable for experiments focused on development or regeneration of the visual system.
To establish a transgenesis marker with (1) early developmental expression to enable efficient screening, (2) persistent fluorescent reporter expression that is unobtrusive to tissues of interest, (3) focused expression in a small region in the zebrafish and (4) activity throughout the zebrafish life cycle, we sought to generate Tol2 backbones with pineal gland-specific reporter expression. The deeply conserved pineal gland, pineal organ or conarium is a small photosensitive endocrine gland that forms at the anterior dorsal midline on the head and is involved in circadian rhythm control (Erlich and Apuzzo, 1985; Gheban et al., 2019). Importantly, in zebrafish the pineal gland condenses during somitogenesis, remains in place for the lifespan of the animal, is small, and at a different anatomical location compared with commonly used transgenesis markers (Asaoka et al., 2002; Kazimi and Cahill, 1999; Ziv et al., 2005).
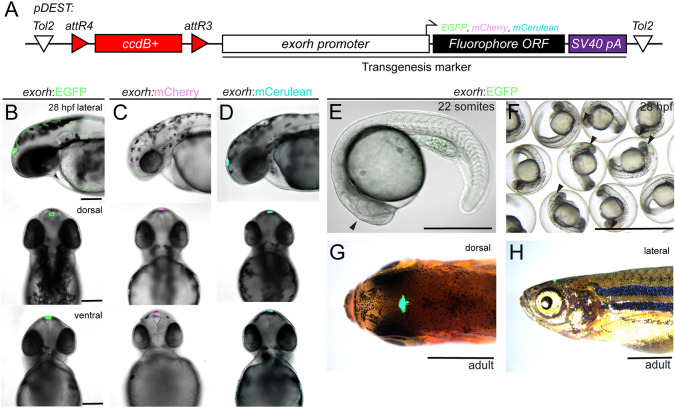
The 1055 bp upstream region of the exo-rhodopsin (exorh) gene harbors a pineal expression-promoting element (PIPE) that drives specific, persistent transgene expression in the pineal organ, as detected by stereo-microscopy from 22 somites onwards (Asaoka et al., 2002). We generated three destination vectors containing the exorh promoter driving EGFP, mCherry and mCerulean (pCK017 pDEST-exorh:EGFP, pCK053 pDEST-exorh:mCherry and pCK054 pDEST-exorh:mCerulean, respectively) and tested their performance as transgenesis backbones (Fig. 5A, Table 1).
Fig. 5.
Labeling the zebrafish pineal gland with the exorhodopsin (exorh) regulatory region as a transgenesis marker. (A) Schematic of the Tol2 secondary transgenic marker backbones pCK017 pDEST-exorh:EGFP, pCK053 pDEST-exorh:mCherry and pCK054 pDEST-exorh:mCerulean, including the attR4- and attR3-flanked ccdB cassette followed by the1055 bp exorh promoter driving EGFP, mCherry or mCerulean. (B-D) Stable transgenic lines for EGFP-, mCherry- and mCerulean-based exorh-driven fluorophore reporters; lateral, dorsal and ventral views at 2 dpf showing specific, easily detectable reporter activity in the pineal gland. (E-H) Stages and appearances of exorh-driven transgenesis reporters. Starting at the 22-somite stage (E), exorh:EGFP expression (black arrowhead) is easily detectable in individual embryos and in groups under a standard dissecting microscope (F), while embryos are still in their chorions. exorh-driven fluorophore expression persists into adulthood, as seen in dorsal (G) and lateral (H) views; dissecting microscope images. Scale bars: 200 μm (B-D); 500 μm (E); 0.25 cm (F); 0.5 cm (G,H).
Upon injection with Tol2 transposase mRNA, all three reporter backbones resulted in prominent, robust and reproducible pineal gland-restricted fluorophore expression, starting from around 22 hpf, in line with the initial reports characterizing exorh-based reporters (Fig. 5B-F) (Asaoka et al., 2002). As stable transgenic insertions over at least three generations, exorh:EGFP recapitulated the transient expression (Fig. 5B-D, Table 1) with reporter activity that is easy to screen under a standard fluorescent dissecting microscope from 24 hpf onwards, while embryos remained in their chorions (Fig. 5F). We observed stable integration and expression of exorh:EGFP throughout zebrafish development and adulthood, when the pineal gland-specific fluorescence can be detected by UV filter glasses for simple, non-intrusive screening of transgene-positive adult zebrafish (Fig. 5G,H, Movie 1). These data establish exorh-based transgenesis markers as a viable alternative to existing transgenesis markers in zebrafish.
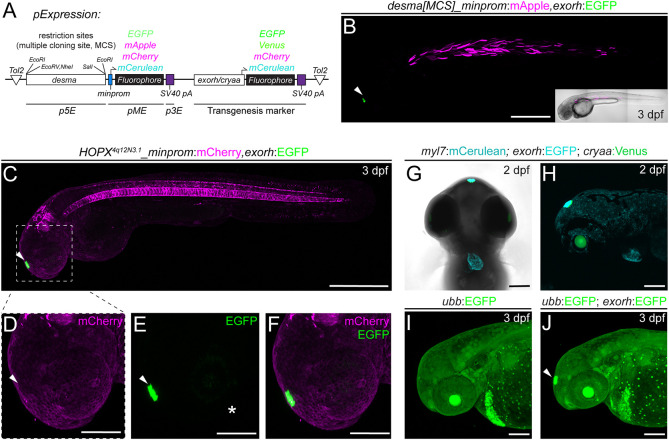
A key application benefiting from transgene markers in cis is testing of gene-regulatory elements by transient injections and subsequent generation of stable transgenic lines (Felker and Mosimann, 2016; Fisher et al., 2006a,b; Kague et al., 2010). To facilitate the use of exorh reporter-based backbones for regulatory element testing beyond Multisite Gateway, we introduced the 2.4 kb upstream regulatory region of zebrafish desmin a (desma) together with beta-globin-coupled EGFP, mApple, mCherry or mCerulean into Tol2 backbones carrying exorh:EGFP, mCherry or mCerulean (plus one version driving mCerulean in the cryaa:Venus-carrying Tol2 backbone) (Fig. 6A, Table 1). These vectors enable traditional cloning for replacement of the desma regulatory sequences (flanked 5′ by EcoRI, EcoRV and NheI, 3′ by SalI and EcoRI sites) with other regulatory elements of interest in vectors that include a minimal promoter and a transgenesis marker in cis. Upon transient injections into wild-type zebrafish embryos, we observed robust somatic muscle expression of pCK069 desmaMCS:minprom-mApple (Fig. 6B, Table 1). Taken together, these experiments establish exorh:fluorophore-carrying Tol2 backbones as a new tool for marking transgenes with different cargo.
Fig. 6.
Application examples of an exorh-based, pineal gland-specific transgenesis reporter. (A,B) Injection-based, transient testing and stable transgenesis of regulatory elements with Tol2 vectors that harbor a transgenesis marker in cis for quality control. The desmin a (desma) upstream region paired with the mouse beta-globin minimal promoter driving different fluorophores with exorh- or cryaa-based (eye lens) transgenesis markers in cis in Tol2 backbones (A, see also Table 1). In all vector versions, the desma regulatory region is flanked by restriction sites known as simple Multiple Cloning Sites (MCSs), enabling restriction enzyme-based replacement of desma with a regulatory element of interest (A). (B) Injection shows muscle-selective desma activity (mCherry) and pineal-specific transgenesis reporter expression (EGFP). (C-F) Regulatory element testing with exorh:EGFP as a transgenesis marker in cis. Stable transgene integration of HOPX4q12N3.1:minprom-mCherry,exorh:EGFP at 3 dpf shows mCherry expression in the notochord and rhombomeres, whereas EGFP expression is confined to the pineal gland (C, arrowhead). The transgenes do not cross-activate mCherry (D,F) or EGFP (E,F) (insets, white arrowheads), despite being encoded in cis and in close proximity, suggesting the exorh regulatory region is inert. The white asterisk (E) indicates faint patchy EGFP signal in the eye. (G,H) Combinatorial use of transgenesis markers. Ventral (G) and lateral (H) views of a stable myl7:mCerulean,exorh:EGFP transgenic crossed to cryaa:Venus and imaged at 2 dpf; there are distinct expression patterns of the secondary transgenic markers labeling the heart (myl7), pineal gland (exorh) and eye lens (cryaa), respectively. (I,J) exorh:EGFP fluorescence is discernible in ubiquitous green-fluorescent transgenics. Ubiquitous ubb:EGFP (ubi:Switch) expression (I) still allows detection of exorh:EGFP expression (white arrowhead) (J). Scale bars: 500 μm (B,C); 200 μm (D-J).
Enhancers can act at a distance on different promoters to activate transcription of target genes (Schaffner, 2015). This distant activation potential is detrimental when combining regulatory elements in cis on a confined transgenesis vector, such as a transgenesis reporter together with an enhancer-driven transgene. To assess the robustness and utility of the new exorh-based transgenesis backbones, we tested each exorh:fluorophore combination in assembled MultiSite Gateway constructs to determine whether the exorh promoter cross-reacts with any reporters cloned in cis, in particular minimal promoter-containing reporters. In stable transgene integrations of HOPX4q12N3.1_minprom-mCherry,exorh:EGFP, we observed faithful mCherry activity in the notochord and rhombomeres, and EGFP expression in the pineal gland, as anticipated (Fig. 6C,E,F); however, we did not observe any mCherry signal in the pineal (Fig. 6D) or notochord expression of EGFP (Fig. 6C). Although not comprehensive, this basic test indicates that both regulatory elements in our observed transgenic insertion were inert to the other's activities.
Crossing stable transgenics of myl7:mCerulean,exorh:EGFP with the hsp70l:Switch line that contains the cryaa:Venus lens marker in the backbone (Felker et al., 2018; Hesselson et al., 2009) further documented how the pineal gland marker can be used in combination with commonly applied transgene reporters (Fig. 6G,H). exorh-based fluorophore reporters, such as exorh:EGFP, are reproducibly strong and bright, enabling their use in ubiquitous fluorescent backgrounds, as documented with easily detectable exorh:EGFP expression and ubiquitous EGFP (Fig. 6I,J). Together, these observations establish exorh:fluorophore Tol2 backbones as new transgenesis vectors for a multitude of applications.
Zebrafish exorh:fluorophore reporters are functional in Astyanax
Establishing a new model organism greatly benefits from access to cross-compatible transgenesis reagents. The Mexican cavefish (Astyanax mexicanus) is an increasingly applied model system to study evolutionary selection and metabolism (McGaugh et al., 2020; Rohner, 2018). Although transgenesis has been successfully introduced in the system, including the use of originally zebrafish-derived regulatory sequences (Elipot et al., 2014; Lloyd et al., 2022; Stahl et al., 2019), a more widespread use has been hampered by the lack of functional transgenesis markers: myl7-based cardiac reporters seem not functional when tested in Astyanax (Elipot et al., 2014) and the diverse crystallin gene-based eye lens transgenesis markers cannot be used due to the eye degeneration phenotype in cavefish. We therefore sought to test the functionality of exorh-based reporters in the model.
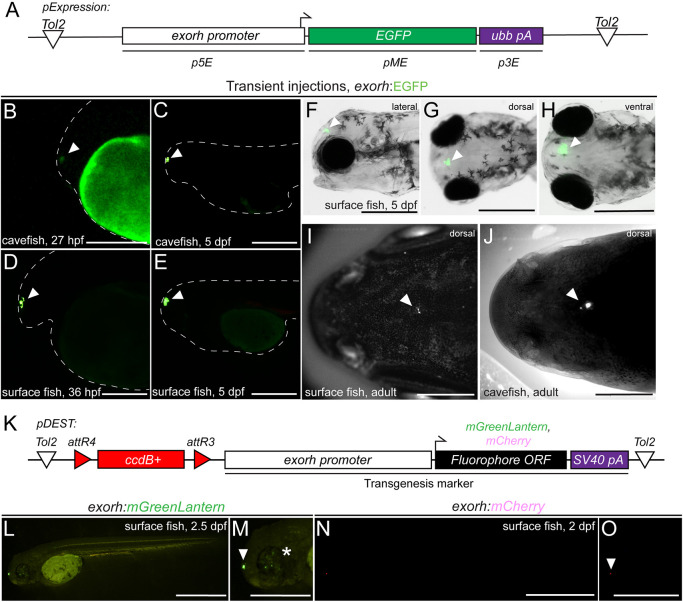
Injection of a Tol2 expression construct carrying exorh:EGFP (Fig. 7A) into one-cell stage embryos of different populations of Astyanax mexicanus (derived from surface and Tinaja cave populations) resulted in easily detectable pineal gland expression beginning around hatching stage (1 dpf) in both Tinaja cave and surface morphs (Fig. 7B-E). At 5 dpf, we continued to observe robust expression in both morphs (Fig. 7C,E), detectable from different imaging angles (lateral-dorsal-ventral) (Fig. 7F-H). Pineal expression of exorh-driven fluorophores continued throughout the larval stages of surface and cave morphs into adulthood (Fig. 7I,J). Tol2-based destination vectors engineered with the exorh promoter driving a fluorescent marker (tested here as mGreenLantern and mCherry) (Campbell et al., 2020) also recapitulated the pineal gland expression pattern in transient injections (Fig. 7K-O) and can be applied to generate transgenesis constructs using MultiSite Gateway. Of note, in our transient injections we observed weak mosaic expression in retinal cells of surface morph animals (Fig. 7M). Taken together, these observations document the activity of zebrafish exorh promoter-based reporters in Astyanax as a new transgenesis marker, as supported by the developmental retention of the pineal gland in cave morphs.
Fig. 7.
The exorh-based transgenesis marker functions in Mexican cavefish (Astyanax mexicanus). (A) Tol2 expression vector using the zebrafish exorh promoter driving the expression of EGFP; see also Fig. 5. (B-E) Transient injection of exorh:EGFP shows reporter activity in the pineal gland of both Tinaja cavefish at 27 hpf (B) and 5 dpf (C), as well as in surface fish at 36 hpf (D) and 5 dpf (E). Pineal expression is indicated with an arrowhead and embryo morphology traced with a dashed line. (F-H) Lateral, dorsal and ventral views at 5 dpf showing specific, easily detectable EGFP reporter activity in the pineal gland (arrowheads). (I,J) Grayscale images of live exorh:EGFP-injected animals, detection of EGFP (white dots) in surface and cavefish adults (arrowheads). (K) Schematic of Tol2-based transgenesis backbones pDEST-exorh:mGreenLantern and pDEST-exorh:mCherry, including the attR4 and attR3-flanked ccdB cassette, and the 1055 bp zebrafish exorh promoter driving mGreenLantern or mCherry for Multisite Gateway cloning. (L,M) Injection of the exorh:mGreenLantern-containing Tol2 Destination vector shows pineal gland expression (arrowhead) as well as patchy retinal expression in developing surface fish (asterisk). (N,O) Injection of the exorh:mCherry-expressing Tol2 destination vector, showing pineal gland expression in developing cavefish (arrowhead). Scale bars: 200 μm (B-E,M,O); 500 μm (F-H,L,N); 1 cm (I,J).
DISCUSSION
Tol2-based transgenesis and the wide accessibility of the required basic tools has led to a revolution of transgenic reporter and effector strains in the zebrafish field (Kawakami, 2005, 2007). The industriousness and collegiality of the zebrafish community has facilitated expansion beyond the basic tools as well as extensive sharing and distribution. Although by no means comprehensive, our data and reagents presented here augment, diversify and increase the functionality of the transgenesis toolkit for zebrafish and other systems. All plasmid sequences and maps have been deposited in Addgene, accession link https://www.addgene.org/browse/article/28233546/ (see Table 1).
The widespread use of green and red fluorophores, in particular EGFP, mCherry and dsRED2, increasingly hinders the combinatorial use of reporter transgenes. We provide a set of middle and 3′ entry vectors containing mCerulean, a blue-fluorescent avGFP derivative, which provides a complementary fluorophore to common green and red fluorophores. mCerulean is detectable with standard GFP filters on fluorescent dissecting microscopes as routinely used in the field, yet it is beneficial to use a dedicated blue filter set and matching excitation light source (Fig. 2). As an alternative to mCherry and dsRED2, mApple provides a complementary red fluorophore that is detectable with standard RFP filters (Fig. 3); we also found that antibodies against dsRED detect mApple to stain for its expression (data not shown). Combined with its better theoretical quantum yield and fast folding mechanism, mApple constructs provide a versatile red-fluorescent alternative to mCherry and dsRED derivatives.
Regulatory element testing is a powerful approach in zebrafish; the discovery and testing of regulatory elements from different species can reveal fundamental insights into upstream gene regulation as well as into the ontogeny of cell types (Fisher et al., 2006a,b). As enhancer sequences need to act on a promoter region to support assembly of productive RNA Pol II complexes, transgene constructs for enhancer testing require a sensitive, yet position effect-inert minimal promoter region that is also ideally short to keep assembled vector sizes smaller (Felker and Mosimann, 2016). Several minimal promoter regions have been successfully deployed in zebrafish, including sequences from other species. The Xenopus Ef1a minimal promoter has been repeatedly used for basic transgenes, yet remains sensitive to silencing (Amsterdam et al., 1995; Tamplin et al., 2011). The zebrafish gata2a promoter region has been successfully applied in a variety of settings, including enhancer trap vectors and reporters (Bessa et al., 2009; Meng et al., 1997); however, its comparatively large size (over 2 kb) can result in large transgene vectors. Previous work has provided solid evidence for the potency of the mouse beta-globin minimal promoter to test regulatory elements, including notochord and hematopoietic enhancers (Tamplin et al., 2011, 2015; Woolfe et al., 2004). We have previously incorporated this promoter for testing enhancers active in the lateral plate mesoderm and neural crest across species (Kaufman et al., 2016; Parker et al., 2019; Prummel et al., 2019). Here, we provide vectors pairing the mouse beta-globin minimal promoter with EGFP, mCherry, mApple and mCerulean for versatile gene-regulatory element testing (Fig. 1). Although required for enhancers, upstream regions that contain the transcription start of a gene of interest might not require an additional minimal promoter as the provided sequences might suffice to assemble a functional RNA Pol II complex. Incorporation of the mouse beta-globin minimal promoter is advisable when testing regulatory elements from divergent species or when establishing basic regulatory element activity (Bergman et al., 2022). Notably, we also present here the pAP02 p3E MCS 3′ entry vector that enables cloning of enhancer elements downstream of the reporter cassette, resulting in the enhancer being incorporated into the resulting mRNA before being terminated and polyadenylated. This vector design will allow the potential application of methods to discover active regulatory elements by sequencing, as successfully performed in cell culture systems such as STARR-seq (Arnold et al., 2013; Neumayr et al., 2019). The pAP02 vector can also be used to clone cassettes for 3′ barcoding (McKenna et al., 2016) or different polyadenylation sequences beyond the ones available to date.
Our collection of 2A peptide fusions with different fluorophores and localization tags is suitable for generating bi-protein constructs with minimal C-terminal tagging (Fig. 4). Previous attempts to broadly apply IRES sequences for bi-cistronic transgenes and fluorescent labeling have widely been unsuccessful for as of yet unclear reasons (Kwan et al., 2007). Viral p2A or similar peptides that break the emerging polypeptide chain during translation have been widely applied to express several genes of interests at equal protein levels in diverse cell culture systems, e.g. for cardiac reprogramming (Muraoka and Ieda, 2015; Song et al., 2012; Zhang et al., 2019). The direct fluorescent labeling of cells expressing a gene of interest for modifier studies, functional tests and disease modeling provides a potent phenotype readout and imaging tool (Fig. 4). The simple cloning requires omission of the stop codon in the N-terminal ORF of interest, cloned in-frame with the following attB2 Gateway repeat to connect with a 3′ p2A-fluorophore. However, the polypeptide breakage leaves a residual p2A sequence as well as the encoded Gateway repeat at the C-terminal end of the ORF of interest. Mechanistic tests are therefore warranted to ensure proper functionality of this tagging method in individual transgenes.
Versatile, easy-to-screen transgenesis markers greatly augment any model organism tool kit by enabling rigorous quality control and transgene identification. A frequently used transgenesis marker in zebrafish is based on the myl7 promoter that drives fluorescence in cardiomyocytes routinely from 20-24 hpf onward (Huang et al., 2003). However, the heart is obscured in the growing and adult zebrafish. crystallin gene-based reporters drive fluorescence in the eye lens, providing an elegant and simple reporter property for screening in adults using UV filter goggles and handheld flashlights. We here add two crybb1-based reporters driving mKate2 and TagBFP in Tol2-based Multisite backbones to the roster (Figs 2A and 4G,H). A drawback of crystallin-based reporters is their comparatively late onset of detectable fluorophore expression from around 36 hpf or later (Davidson et al., 2003; Kurita et al., 2003), which can render sorting of transgene-positive embryos challenging. Our exorh-based reporters documented here (Figs 5,6) circumvent this issue and provide reporters that are reproducibly expressed from 22 ss and stay active throughout adulthood (Asaoka et al., 2002); exorh-marked backbones are thereby screenable while embryos remain in their chorions as well as with UV filter goggles in adults (Movie 1). The confined small area of fluorescence dorsal and anterior in the head does not obscure major organs during early development and should still enable imaging of significant brain regions in fluorescent transgenic reporters or stained embryos. exorh:fluorophore cassettes are less than 2 kb in size, and we here document several successfully germline-transmitted transgenes using such Tol2 backbones (Figs 5 and 6). Notably, exorh-marked transgenes are easily combined with other transgenes with myl7, crystallin or even ubiquitously expressed reporters (Fig. 6E-H). Here, we have generated three distinct exorh:fluorophore combinations to facilitate planning and execution of such combinatorial experiments, and we provide versions that are compatible with restriction enzyme-based cloning and MultiSite Gateway (Fig. 5A). The provided backbones that harbor desma-driven fluorophores enable restriction site-based exchange of the desma regulatory region to test different regulatory elements (Fig. 6A). In addition, desma-driven reporters are well expressed in somatic muscle and are simple to detect (Fig. 6B), providing potent reagents to troubleshoot injections, to test Tol2 mRNA batches or to train new researchers in transgenesis basics.
Use of our vector tools is not confined to zebrafish, as many can be applied in other models and Gateway-compatible cloning reactions. We have previously used mouse beta-globin-containing enhancer constructs used for zebrafish transgenes without modifications for electroporation in chick (Prummel et al., 2019), underscoring the utility of the mouse beta-globin minimal promoter for enhancer testing. Relevant to other aquatic models, exorh-based reporter utility expands to Astyanax mexicanus, an emerging model for the study of evolutionary adaptation (Fig. 7) (Elipot et al., 2014; McGaugh et al., 2020; Rohner, 2018). Although the cave forms of Astyanax have lost their eyes, they do form functional pinealocytes and a pineal gland, although their impact on behavior and circadian rhythms remains to be fully elucidated (Lloyd et al., 2022; Mack et al., 2021; Omura, 1975; Yoshizawa and Jeffery, 2008). exorh-based reporter transgenes have the potential to facilitate transgene quality control, visual genotyping and combinatorial transgene experiments, such as Cre/lox approaches in the model. As the pineal gland remains visible on the skull of fishes and amphibians, exorh-based reporters have the potential to support transgenesis experiments in a wide variety of aquatic models.
MATERIALS AND METHODS
Zebrafish husbandry and procedures
Animal care and procedures were carried out in accordance with the veterinary office of the University of Zürich, Switzerland; the IACUC of the University of Colorado School of Medicine (protocol 00979), Aurora, Colorado, USA; and the IACUC of the University of Utah (protocol 21-01007), Salt Lake City, Utah, USA. All zebrafish embryos were incubated at 28.5°C in E3 medium unless indicated otherwise. Staging was performed as described by Kimmel et al. (1995).
Cavefish husbandry and procedures
Astyanax husbandry and care was conducted as described previously (Xiong et al., 2022) and as approved by the Institutional Animal Care and Use Committee (IACUC) of the Stowers Institute for Medical Research on protocol 2021-122. All methods described here are approved on protocol 2021-129. Housing conditions meet federal regulations and are accredited by AAALAC International.
Plasmid construction
First-time users of Tol2 technology are asked to connect with Dr Koichi Kawakami (Division of Molecular and Developmental Biology, National Institute of Genetics, Shizuoka, Japan) to set up a material transfer agreement (MTA). Plasmid sequences and maps have been deposited in Addgene (https://www.addgene.org/browse/article/28233546/).
Entry clones
All entry clones were verified by colony PCR and sequencing with M13 fw 5′-GTAAAACGACGGCCAG-3′ and M13 rv 5′-CAGGAAACAGCTATGAC-3′.
p5E
For p5E entry clones, all PCR products were amplified using Roche Expand High Fidelity PCR kit (Sigma, 11732641001), which contains a 3′-adenine overhang-depositing Taq. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, 28104) or the QIAquick Gel Extraction Kit (Qiagen, 28706) using deionised H2O elution as the last step. PCR products were then TOPO-cloned into MultiSite Gateway entry vectors with the pENTR5′-TOPO TA Cloning Kit (Invitrogen, K59120): 4 μl of purified PCR product was immediately combined with 1 μl pENTR5′-TOPO TA and 1 μl of salt solution, and incubated at room temperature for a minimum of 2 h to overnight to ligate. 3 µl of the TOPO reactions were then transformed with One Shot TOP10 Chemically Competent E. coli (Invitrogen, C404010) and plated on Kanamycin-infused agar plates. Half reactions can also be used. Primer sequences for pE5′ enhancer/promoter fragments are as follows: for pCK079 p5E Mm BMP4ECR2, oBuS014 Mm BMP4 ECR2 fw (5′-GGGGATGAAAGTAGCATCCTG-3′) and oBuS015 Mm BMP4 ECR2 rv (5′-TTCCACTTTGCTTCCCAAACTGG-3′); for pAB019 pE5 Hs HOPX_4q12N3.1, oAB019 Hs HOPX fw (5′-GTGTTGGGTTAGTTTGAGC-3′) and oAB019 Hs HOPX rv (5′-GTTCTGTTGGGGATATGTCC-3′); for pCK033 p5E desma MCS, oCK024 Dr desma MCS fw (5′-gatatcACTGATgctagcTCCTTGAGGCACTTTCGG-3′) and oCK025 Dr desma MCS rv (5′-gtcgacTACGCTGTGTGAATGCTGG-3′).
pME, pENTR/D, mouse beta-globin minimal promoter and fluorophore ORFs
For middle entry vectors, blunt-end PCR products were generated using Phusion High Fidelity DNA Polymerase (NEB M0530S) and forward primers were designed, including 5′-CACC-3′ on the 5′ end for directional cloning and then ligated into MultiSite Gateway-compatible pME entry vectors using pENTR/D-TOPO (Invitrogen, K240020). 4 μl of purified PCR product was combined with 1 μl pENTR/D-TOPO and 1 μl of salt solution and incubated at room temperature for a minimum of 2 h to overnight to ligate. 3 µl of the TOPO reactions were then transformed with One Shot TOP10 Chemically Competent E. coli and plated Kanamycin-containing agarose plates. Half reactions can also be used.
The mouse beta-globin (Hbb-bt) minimal promoter sequence spans 129 base pairs: 5′-CCAATCTGCTCAGAGAGGACAGAGTGGGCAGGAGCCAGCATTGGGTATATAAAGCTGAGCAGGGTCAGTTGCTTCTTACGTTTGCTTCTGAGTCTGTTGTGTTGACTTGCAACCTCAGAAACAGACATC-3′, as annotated in the reference genome NC_000073.7:103463077-103463205 Mus musculus strain C57BL/6J chromosome 7, GRCm39; the underlined sequence stretch represents the 5′ UTR.
The mCerulean ORF sequence can be amplified using mCerulean fw: 5′-ATGGTGAGCAAGGGCGAGGAGC-3′ and mCerulean rv: 5′-CAGCTCGTCCATGCCGAGAGTG-3′. The mApple ORF sequence can be amplified using mApple fw: 5′-ATGGTGAGCAAGGGCGAGG-3′ and mApple rv: 5′-CTTGTACAGCTCGTCCATGC-3′.
p3E
pAP02_p3E-MCS was generated by amplifying the multiple cloning site from pBluescript KSII(+) with the primers AP13fw: 5′-GGGGACAGCTTTCTTGTACAAAGTGGGCGGCCGCTCTAGAA-3′ and AP14rv: 5′-GGGGACAACTTTGTATAATAAAGTTGGGTACCGGGCCCCCC-3′, and inserted into pDONRP2R-P3 via BP reaction.
p3E_ubb-polyA was cloned by amplifying a 516 bp fragment of the zebrafish ubiquitinB (ubb, ubi) locus using the primers ubb polyA fw: 5′-TAGAACCGACAGTCTTAGGGATGG-3′ and ubb polyA rv: 5′-GAATTCATTGCCATCAAGTGTTAGC-3′ with Phusion High Fidelity DNA Polymerase (NEB M0530S), subcloned into the Zero Blunt TOPO PCR Cloning vector (Invitrogen, K283020), digested out with BglII and ligated into the single cutter BamHI site in pAP02_p3E-MCS.
Of note, frequent users of both the #302 p3E-polyA from the original Tol2kit (Kwan et al., 2007) and p3E_ubb-polyA introduced here have experienced decreased Multisite Gateway reaction efficiencies after multiple freeze-thaw cycles of these p3E vectors. For optimal colony numbers and reaction efficiency, we recommend freshly preparing minipreps, facilitated by frequent retransformations of the vector and storage of the plasmid in small aliquots. The reason for this reduction in cloning efficiency with prolonged storage of these p3E vectors is currently unknown, but could be due to structural factors, such as secondary structure formation, that are detrimental to Multisite Gateway assembly or bacteria-based propagation.
2A constructs were built via a two-step process. First, the 2A peptide-encoding sequence was added to the N-terminus of fluorescence protein ORFs using PCR and inclusion of the 2A peptide sequence in the 5′ upstream primer. The fusion product was conventionally cloned into pCS2FA using the FseI and AscI sites (enzymes from NEB). This placed the fusion product upstream of the SV40 polyA signal sequence. Ligation reactions were transformed using Subcloning Efficiency DH5α chemically competent E. coli (Invitrogen/ThermoFisher) and plated onto ampicillin- or carbenicillin-containing agar plates. Cloning was confirmed by restriction digest and subsequent sequencing.
This 2A-FP-polyA cassette was then cloned into the 3′ entry vector using common att PCR primers (2A-attB2F and CS2pA-attB3R). For p3E-2A-CS2MCS-pA, pCS2P+ was used as a template for a PCR reaction in which the 2A peptide-encoding sequence was included in the att2F forward primer. Care was taken to ensure that all primers used ‘Gateway reading frame’, in which bases listed in the manual as ‘N’ in the primer were included as ‘G’. This ensures maintenance of the reading frame between the middle entry clone (which must not include a stop codon) and the 3′ clone containing the 2A peptide sequence and reporter. PCR products were gel-purified (Qiagen Gel Extraction Kit) and quantified. For the recombination, equimolar amounts (usually 50 fmoles each) of purified PCR product were combined with pDONR-P2R-P3 donor vector and Gateway BP Clonase II Enzyme Mix (Invitrogen/ThermoFisher). Reactions were incubated at room temperature for several hours and transformed using Subcloning Efficiency DH5α chemically competent E. coli (Invitrogen/ThermoFisher) and plated onto Kanamycin-containing agar plates. Cloning was confirmed by restriction digest and subsequent sequencing.
For 2A-FP cloning into pCS2FA (restriction sites in bold, sequence complementary to template underlined): 2AEGFP5Fse, 5′-gaacGGCCGGCCGGATCCGGAGCCACGAACTTCTCTCTGTTAAAGCAAGCAGGAGACGTGGAAGAAAACCCCGGTCCTATGGTGAGCAAGGGCGAGGAGCTG-3′; 2AmCherry5Fse, 5′-gaacGGCCGGCCGATCCGGAGCCACGAACTTCTCTCTGTTAAAGCAAGCAGGAGACGTGGAAGAAAACCCCGGTCCTATGGTGAGCAAGGGCGAGGAGGAC-3′; 2Anls5Fse, 5′-gaacGGCCGGCCGGATCCGGAGCCACGAACTTCTCTCTGTTAAAGCAAGCAGGAGACGTGGAAGAAAACCCCGGTCCTATGGCTCCAAAGAAGAAGCGTAAGG-3′; EGFP3Asc (rev), 5′-gaacGGCGCGCCTCACTATAGGGCTGCAGAATCTAGAGG-3′; mCherry3Asc (rev), 5′-gaacGGCGCGCCTTACTTGTACAGCTCGTCCATGCCGC-3′; and CAAX3Asc (rev), 5′-gaacGGCGCGCCTCAGGAGAGCACACACTTGCAGCTC-3′. For p3E cloning from pCS2FA: 2A-attB2F, 5′-ggggACAGCTTTCTTGTACAAAGTGGGGGGATCCGGAGCCACGAACTTCTCTC-3′; 2A-CS2PMCS-attB2F, 5′-ggggACAGCTTTCTTGTACAAAGTGGGGGGATCCGGAGCCACGAACTTCTCTCTGTTAAAGCAAGCAGGAGACGTGGAAGAAAACCCCGGTCCTAGGGATCCCATCGATTCGAATTCAAGGCCTC-3′; and CS2pA-attB3R (rev), 5′-ggggACAACTTTGTATAATAAAGTTGGAAAAAACCTCCCACACCTCCCCCTG-3′.
Expression constructs
Multisite Gateway recombination reactions were principally performed as described in the Invitrogen Multisite Gateway Manual, with minor modifications. 10 fmol of pE5′, pME and pE3′ vectors were combined with 20 fmol of destination vector, 1 μl of LR Clonase II Plus Enzyme Mix (Invitrogen, 12538120; vortexed twice for 2 s prior to use) and deionised H2O for a final reaction volume of 5 μl. Vector calculations for molarity were performed using the Multisite Gateway Excel spreadsheet (Mosimann, 2022). Reactions were incubated at 25°C overnight and treated the following day with 1 μl 2 μg/μl Proteinase K for 15 min at 37°C and 3 μl of the reaction was transformed with One Shot TOP10 Chemically Competent E. coli (Invitrogen C404010) and plated on Ampicillin selection plates. T4 ligations into multiple cloning site plasmids were performed by amplifying fragments via PCR with Roche Expand High Fidelity Polymerase (Sigma 11732641001) with primers containing 5′ restriction enzyme sequences for digestion after PCR purification. Digested backbones were treated with shrimp alkaline phosphatase (NEB M0371S) for 2 h at 37°C before gel extraction. T4 ligations were all performed with a 1:3 pmol ratio of backbone to insert at 16°C overnight. 7.5 μl of T4 reactions were then transformed with One Shot TOP10 Chemically Competent E. coli (Invitrogen, C404010) and ampicillin selection. All assembled gateway reactions were tested by diagnostic digest and confirmed by sequencing using the following primers (Mosimann, 2022): attB1 fw, 5′-CAAGTTTGTACAAAAAAGCAGGCT-3′; attB1 rv, 5′-AGCCTGCTTTTTTGTACAAACTTG-3′; attB2 fw, 5′-ACCCAGCTTTCTTGTACAAAGTGG-3′; and attB4 fw, 5′-CAACTTTGTATAGAAAAGTTG-3′.
Zebrafish exorh promoter element
The exorh promoter:EGFP was amplified from the pCR2.1 TOPO vector containing a 1055 bp upstream region and short coding sequence of the zebrafish exorh gene driving EGFP (a kind gift from Dr Yoshitaka Fukuda and Dr Daisuke Kojima, University of Tokyo, Japan) (Asaoka et al., 2002) with a 5′ Asp718l (Acc651) restriction site containing primers. Of note, exorh lies on the edge of a genomic contig in the sequenced zebrafish genome, rendering the promoter isolation challenging without the pioneering work by Asaoka et al.. The Asp718l-flanked exorh:EGFP fragment was subcloned into pENTR5′-TOPO as stated in the Entry Clone section above to generate pCK074 pE5′ exorh:EGFP. The shorter 147 bp exorh promoter fragment driving EGFP was amplified from the same vector (Asaoka et al., 2002) and subcloned in the same way to generate pCK072 pE5′ 147 bp exorh:EGFP. The exorh promoter alone was also subcloned into pENTR5′-TOPO to generate pCK011 pE5′ exorh. The vectors containing the exorh promoter driving mCerulean and mCherry ORFs were cloned using PCR fusion of Asp718l- flanked fragments and subcloned into pENTR5′-TOPO to generate pCK051 pE5′ exorh:mCerulean and pCK052 pE5′ exorh:mCherry, respectively. Of note, the exorh promoter sequence contains a GT-rich repeat region, resulting in challenging PCR amplification. Primer sequences are as follows: pCK011 p5E exorh, oCK001 exorh fw 5′- GCTCAGCTGGCAGTACTACC-3′ and oCK009 exorh rv 5′-GATGGAGAAGTGGACGATCG-3′; pCK072 p5E 147bp exorh:EGFP, oCK018 ASP7181-147bpexorh:EGFP fw 5′-agatctggtaccGCGAGCGCTGCTGTGTCTCC-3′ and oCK017 ASP7181-exorh:EGFP Rv 5′-agatctggtaccTTACTTGTACAGCTCGTCC-3′; pCK074 p5E Exorh:EGFP, oCK016 ASP7181-exorh:EGFP fw 5′-agatctggtaccGCTCAGCTGGCAGTACTACC-3′ and oCK017 ASP7181-exorh:EGFP rv 5′-agatctggtaccTTACTTGTACAGCTCGTCC-3′; pCK051 p5E Exorh:mCerulean, oCK016 ASP7181-exorh:EGFP fw 5′-agatctggtaccGCTCAGCTGGCAGTACTACC-3′ and oCK054 mcerulean-STOP-Asp7181 rv 5′-GGTACCtaCAGCTCGTCCATGCCGAGAG-3′; and pCK052 p5E Exorh:mCherry, oCK016 ASP7181-exorh:EGFP fw 5′-agatctggtaccGCTCAGCTGGCAGTACTACC-3′ and oCK056 mcherry-STOP-Asp7181 rv 5′-GGTACCtaCTTGTACAGCTCGTCCATGC-3′ (lowercase letters indicate introduced spacers and bases for restriction sites).
Destination vectors
To generate destination vectors containing the exorh promoter driving various fluorophores as secondary transgenic markers, digested Asp718l-exorh:fluorophore-Asp718l fragments were T4-ligated as described above into the Asp718l-digested and dephosphorylated pCM326 pDEST_cryaa:Venus (Mosimann et al., 2015) after gel extraction of the digested backbone minus the cryaa:Venus cassette to generate pCK017 pDEST exorh:EGFP, pCK053 pDEST exorh:mCherry and pCK054 pDEST exorh:mCerulean. Clones were verified by test digest and full plasmid sequencing. All destination vectors were transformed with One Shot ccdB Survival 2 T1R Chemically Competent Cells (Invitrogen, A10460) with both ampicillin and chloramphenicol selection.
pCB59_crybb1:mKate2 was amplified from pDEST-Tol2_Fli1ep-mCherry-GM130 (a kind gift from Dr Holger Gerhard, MDC Berlin, Germany) (Franco et al., 2015) including the zebrafish crybb1 promoter, parts of the minimal CMV promoter and the mKate2 ORF. The PCR product was cloned into the Tol2-based Multisite Gateway backbone pDestTol2CG2 (Tol2kit 395) (Kwan et al., 2007) via the BglII restriction site. Of note, the reporter uses the terminal Tol2 repeat downstream of the insert as possible polyadenylation signal.
pCB54_crybb1:TagBFP, containing the zebrafish crybb1 promoter and parts of the minimal CMV promoter expressing a zebrafish codon-optimized TagBFP ORF flanked by BglII restriction sites, was synthesized (GENEWIZ). After sequence verification, the fragment was cloned into the Tol2-based Multisite Gateway backbone pDestTol2CG2 (Tol2kit 395) (Kwan et al., 2007) via the BglII restriction site.
The construction of cavefish-tested pDEST_exorh:mGreenLantern and pDEST_exorh:mCherry was performed using the NEBuilder HiFi DNA Assembly Cloning Kit (NEB E5520S). The original vector pDestTol2A2 (Tol2kit 394) (Kwan et al., 2007) was digested with BglII (NEB R0144S) and HpaI (NEB R0105S) restriction enzymes. The digested plasmid was purified using the Wizard SV Gel and PCR Clean-Up System (Promega A9281). The zebrafish exorh promoter region was amplified from pCK029 (this manuscript) using the following primers: HpaIBgl2_NEB_exorhprom, o fwd 5′-GCCACAGGATCAAGAGCACCCGTGGCCGTATCTTCGCAGCTCAGCTGGCAGTACTACCGC-3′ and o rev 5′-cccttgctcaccatggtggcGATGGAGAAGTGGACGATCGG-3′ (lowercase letters indicate added bases for spacers and restriction sites).
The mGreenLantern and mCherry fluorescent protein-coding sequences were amplified from pTol2_tg:mGreen Lantern/mCherry (a kind gift from Dr Sumeet Pal Singh, IRIBHM, Anderlecht, Belgium) using the following primers: HpaIBgl2_NEB_mGL_, o fwd 5′- CGATCGTCCACTTCTCCATCgccaccatggtgagc-3′ and o rev 5′- TATTTGTAACCATTATAAGCTGCAATAAACAAGTTttacttgtacagctcgtccatgtca-3′; HpaIBgl2_NEB_mCherry, o fwd 5′-CGATCGTCCACTTCTCCATCgccaccatggtgagc-3′ and o rev 5′-TGCTTTATTTGTAACCATTATAAGCTGCAATAAACAAGTTttacttgtacagctcgtccatgcc-3′ (lowercase letters indicate added bases for spacers and restriction sites). Purified amplicons were ligated to the digested pDestTol2A2 according to the NEB HiFi DNA assembly protocol.
RNA synthesis
Tol2 transposase-encoding capped mRNA was created by in vitro transcription using the SP6 mMessage mMachine Kit (Ambion, AM1340) from pCS2+Tol2 plasmid (Kwan et al., 2007) linearized with NotI restriction digest. RNA was purified with lithium chloride precipitation followed by the Megaclear Kit (Ambion AM1908).
Zebrafish injections and transgenesis
All plasmids were miniprepped with the ZymoPURE plasmid miniprep kit (ZYMO D4212). Embryos were injected into the early cell of one-cell-stage wild-type embryos (AB or TU) with 1 nl injection mix containing plasmid DNA concentrations of 25 ng/µl and Tol2 mRNA at 25 ng/µl. Adult F0 animals mosaic for the transgene were outcrossed to wild type and their progeny analyzed for stable transgene expression. Tol2-based zebrafish transgenics were generated using standard experimental protocols as specified in Felker and Mosimann (2016) to achieve single-insertion transgenics and reproducible quality control. Our transgenesis rate is routinely between 15 and 50% for each injected Tol2 transgenesis vector. We investigate several clutches from at least three independent F1 founders. All presented stable transgenic lines are at least F3 generation or beyond and were selected to transmit the transgenes at Mendelian ratios as outlined in Felker and Mosimann (2016).
Imaging
Embryos or larvae were anesthetized with 0.016% Tricaine-S (MS-222, Pentair Aquatic Ecosystems, Apopka, Florida, NC0342409) in E3 embryo medium. Dissecting microscope fluorescence imaging was performed on a Leica M205FA with a DFC450 C camera and 1.0× PlanApo M-Series objective, illuminated with a TL5000 light base and CoolLED pE-300white Illumination System. We used the following Leica filter sets for fluorescence: ET GFP 10447408, ET CFP 10447409 and ET DSR 10447412 for imaging and sorting; and TXR LP 10450590 for routine sorting of red fluorescence. Laser scanning confocal microscopy was performed on a Zeiss LSM880 following embedding in E3 with 1% low-melting-point agarose (Sigma Aldrich, A9045) on glass bottom culture dishes (Greiner Bio-One, Kremsmunster, Austria, 627861). Images were collected with a 10/0.8 air-objective lens swith all channels captured sequentially with maximum speed in bidirectional mode, with the range of detection adjusted to avoid overlap between channels. Maximum projections of acquired z-stacks were made using ImageJ/Fiji (Schindelin et al., 2012) and cropped and rotated using Adobe Photoshop 2022.
Nulear:membrane fluorescence intensity ratio quantification
Fluorescence intensity of the membrane and nuclear EGFP, comparing the 2A and IRES was assessed by drawing a line across the cell in ImageJ/Fiji and measuring the fluorescence intensity profile along the line. Signal maxima in the nucleus and at the membrane were recorded; measurements were not used if signal was saturated. The fluorescence intensity ratio (nucleus:membrane) was calculated for each individual cell. Welch's t-test was used to calculate significance. Box and whisker plots were generated using the ggplot2 package in RStudio. The upper and lower hinges correspond to the first and third quartiles. The upper whisker extends from the upper hinge to the highest value within (1.5×IQR), where IQR is the inter-quartile range. The lower whisker extends from the lower hinge to the lowest value within (1.5×IQR). Data points outside of the ends of the whiskers are outliers.
Supplementary Material
Acknowledgements
We thank Christine Archer, Molly Waters, Nikki Tsuji, Ainsley Gilbert and Olivia Gomez for zebrafish husbandry support, and all members of the Mosimann, Burger, Kwan and Lovely labs for critical input on tool design, cloning and the manuscript. We thank Dr Stephanie Woo, Dr Sumeet Pal Singh and Dr Holger Gerhard for sharing plasmids. We are especially grateful to Dr Yoshitaka Fukuda and Dr Daisuke Kojima for sharing the isolated zebrafish exorh promoter sequence with us.
Footnotes
Author contributions
Conceptualization: K.M.K., A.B., C.M.; Methodology: M.A., N.R., C.B.L.; Validation: C.L.K., H.R.M., B.F.M., A.S., J.R.K., R.L.E., E.L., S. Bertho, S.N., S. Burger, G.D., C.B., A.-C.P., A.F., K.D., S. Bötschi; Formal analysis: C.L.K., H.R.M., B.F.M., A.S., J.R.K., S. Bertho, S.N., S. Burger, G.D., C.B., A.-C.P., A.F., K.D., S. Bötschi, C.B.L., K.M.K., A.B., C.M.; Investigation: C.L.K., H.R.M., B.F.M., A.S., J.R.K., R.L.E., E.L., S. Bertho, S.N., S. Burger, G.D., C.B., A.-C.P., A.F., K.D., S. Bötschi; Resources: M.A., N.R., C.B.L., K.M.K., A.B., C.M.; Data curation: C.L.K., H.R.M., B.F.M., A.S., J.R.K., R.L.E., E.L., S. Bertho, S.N., S. Burger, G.D., C.B., A.-C.P., A.F., K.D., S. Bötschi, N.R., C.B.L., K.M.K., A.B., C.M.; Writing - original draft: C.L.K., H.R.M., C.M.; Writing - review & editing: C.L.K., H.R.M., S. Bertho, G.D., C.B., M.A., N.R., C.B.L., K.M.K., A.B., C.M.; Visualization: C.L.K., H.R.M., S. Bertho, G.D., C.B.L., K.M.K., A.B., C.M.; Supervision: M.A., N.R., C.B.L., K.M.K., A.B., C.M.; Project administration: K.M.K., A.B., C.M.; Funding acquisition: K.M.K., A.B., C.M.
Funding
This work was supported by the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney (NIH/NIDDK) (1R01DK129350-01A1 to A.B.); the Swiss Bridge Foundation and the University of Colorado School of Medicine, Anschutz Medical Campus (A.B. and C.M.); the Children's Hospital Colorado Foundation, the National Science Foundation (2203311) and the Swiss National Science Foundation (Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung) (Sinergia grant CRSII5_180345) (to C.M.); the National Institutes of Health/National Eye Institute (NIH/NEI) (R01EY025378 and R01EY025780 to K.M.K.); the National Institutes of Health/National Institute on Alcohol Abuse and Alcoholism (NIH/NIAAA) (R00AA023560 to C.B.L.); the Stowers Institute for Medical Research (to N.R.); and the National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) 1T32GM141742-01 and 3T32GM121742-02S1 (to H.R.M.). Open access funding provided by the National Institutes of Health. Deposited in PMC for immediate release.
Contributor Information
Kristen M. Kwan, Email: kmkwan@genetics.utah.edu.
Alexa Burger, Email: alexa.burger@cuanschutz.edu.
Christian Mosimann, Email: christian.mosimann@cuanschutz.edu.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.201531.reviewer-comments.pdf
References
- Akitake, C. M., Macurak, M., Halpern, M. E. and Goll, M. G. (2011). Transgenerational analysis of transcriptional silencing in zebrafish. Dev. Biol. 352, 191-201. 10.1016/j.ydbio.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam, A., Lin, S. and Hopkins, N. (1995). The aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev. Biol. 171, 123-129. 10.1006/dbio.1995.1265 [DOI] [PubMed] [Google Scholar]
- Arnold, C. D., Gerlach, D., Stelzer, C., Boryń, Ł. M., Rath, M. and Stark, A. (2013). Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science (80-.) 339, 1074-1077. 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Asaoka, Y., Mano, H., Kojima, D. and Fukada, Y. (2002). Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 99, 15456-15461. 10.1073/pnas.232444199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, D. T., Jones, T. R., Liu, V., Ray, J., Jagoda, E., Siraj, L., Kang, H. Y., Nasser, J., Kane, M., Rios, A.et al. (2022). Compatibility rules of human enhancer and promoter sequences. Nature 607, 176-184. 10.1038/s41586-022-04877-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa, J., Tena, J. J., de la Calle-Mustienes, E., Fernández-Miñán, A., Naranjo, S., Fernández, A., Montoliu, L., Akalin, A., Lenhard, B., Casares, F.et al. (2009). Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 238, 2409-2417. 10.1002/dvdy.22051 [DOI] [PubMed] [Google Scholar]
- Bisgrove, B. W., Essner, J. J., Yost, H. J., Bisgrove, B. W. and Yost, H. J. (2000). Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development 127, 3567-3579. 10.1242/dev.127.16.3567 [DOI] [PubMed] [Google Scholar]
- Campbell, B. C., Nabel, E. M., Murdock, M. H., Lao-Peregrin, C., Tsoulfas, P., Blackmore, M. G., Lee, F. S., Liston, C., Morishita, H. and Petsko, G. A. (2020). mGreenLantern: a bright monomeric fluorescent protein with rapid expression and cell filling properties for neuronal imaging. Proc. Natl. Acad. Sci. USA 117, 30710-30721. 10.1073/pnas.2000942117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney, T. J. and Mosimann, C. (2018). Switch and trace: recombinase genetics in zebrafish. Trends Genet. 34, 362-378. 10.1016/j.tig.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W. W. and Prasher, D. C. (1994). Green fluorescent protein as a marker for gene expression. Science (80-.). 263, 802-805. 10.1126/science.8303295 [DOI] [PubMed] [Google Scholar]
- Chan, H. Y., Sivakamasundari, V., Xing, X., Kraus, P., Yap, S. P., Ng, P., Lim, S. L. and Lufkin, T. (2011). Comparison of IRES and F2A-based locus-specific multicistronic expression in stable mouse lines. PLoS ONE 6, e28885. 10.1371/journal.pone.0028885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, K. J., Chandler, R. L. and Mortlock, D. P. (2009). Identification of an ancient Bmp4 mesoderm enhancer located 46 kb from the promoter. Dev. Biol. 327, 590-602. 10.1016/j.ydbio.2008.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Kook, H., Milewski, R., Gitler, A. D., Lu, M. M., Li, J., Nazarian, R., Schnepp, R., Jen, K., Biben, C.et al. (2002). Hop is an unusual homeobox gene that modulates cardiac development. Cell 110, 713-723. 10.1016/S0092-8674(02)00932-7 [DOI] [PubMed] [Google Scholar]
- Cheo, D. L., Titus, S. A., Byrd, D. R. N., Hartley, J. L., Temple, G. F. and Brasch, M. A. (2004). Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 14, 2111-2120. 10.1101/gr.2512204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, N. C., Shaw, R. M., De Val, S., Kang, G., Jan, L. Y., Black, B. L. and Stainier, D. Y. R. R. (2008). Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734-739. 10.1101/gad.1629408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, W.-S. and Stainier, D. Y. R. (2008). Intra-endodermal interactions are required for pancreatic β cell induction. Dev. Cell 14, 582-593. 10.1016/j.devcel.2008.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agati, G., Cabello, E. M., Frontzek, K., Rushing, E. J., Klemm, R., Robinson, M. D., White, R. M., Mosimann, C. and Burger, A. (2019). Active receptor tyrosine kinases, but not Brachyury, are sufficient to trigger chordoma in zebrafish. Dis. Model. Mech. 12, dmm039545. 10.1242/dmm.039545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, R. M. and Topczewski, J. (2011). Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev. Biol. 357, 518-531. 10.1016/j.ydbio.2011.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, A. E., Balciunas, D., Mohn, D., Shaffer, J., Hermanson, S., Sivasubbu, S., Cliff, M. P., Hackett, P. B. and Ekker, S. C. (2003). Efficient gene delivery and gene expression in zebrafish using the Sleeping Beauty transposon. Dev. Biol. 263, 191-202. 10.1016/j.ydbio.2003.07.013 [DOI] [PubMed] [Google Scholar]
- Davison, J. M., Akitake, C. M., Goll, M. G., Rhee, J. M., Gosse, N., Baier, H., Halpern, M. E., Leach, S. D. and Parsons, M. J. (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811-824. 10.1016/j.ydbio.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel, M., Wullimann, M. F. and Köster, R. W. (2009). Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. USA 106, 13365-13370. 10.1073/pnas.0903060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don, E. K., Formella, I., Badrock, A. P., Hall, T. E., Morsch, M., Hortle, E., Hogan, A., Chow, S., Gwee, S. S. L., Stoddart, J. J.et al. (2017). A Tol2 gateway-compatible toolbox for the study of the nervous system and neurodegenerative disease. Zebrafish 14, 69-72. 10.1089/zeb.2016.1321 [DOI] [PubMed] [Google Scholar]
- Donnelly, M. L. L., Hughes, L. E., Luke, G., Mendoza, H., Ten Dam, E., Gani, D. and Ryan, M. D. (2001a). The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J. Gen. Virol. 82, 1027-1041. 10.1099/0022-1317-82-5-1027 [DOI] [PubMed] [Google Scholar]
- Donnelly, M. L. L., Luke, G., Mehrotra, A., Li, X., Hughes, L. E., Gani, D. and Ryan, M. D. (2001b). Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol. 82, 1013-1025. 10.1099/0022-1317-82-5-1013 [DOI] [PubMed] [Google Scholar]
- Elipot, Y., Legendre, L., Père, S., Sohm, F. and Rétaux, S. (2014). Astyanax transgenesis and husbandry: how cavefish enters the laboratory. Zebrafish 11, 291-299. 10.1089/zeb.2014.1005 [DOI] [PubMed] [Google Scholar]
- Emelyanov, A. and Parinov, S. (2008). Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 320, 113-121. 10.1016/j.ydbio.2008.04.042 [DOI] [PubMed] [Google Scholar]
- Erlich, S. S. and Apuzzo, M. L. J. (1985). The pineal gland: anatomy, physiology, and clinical significance. J. Neurosurg. 63, 321-341. 10.3171/jns.1985.63.3.0321 [DOI] [PubMed] [Google Scholar]
- Felker, A. and Mosimann, C. (2016). Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 135, 219-244. 10.1016/bs.mcb.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Felker, A., Prummel, K. D., Merks, A. M., Mickoleit, M., Brombacher, E. C., Huisken, J., Panáková, D. and Mosimann, C. (2018). Continuous addition of progenitors forms the cardiac ventricle in zebrafish. Nat. Commun. 9, 2001. 10.1038/s41467-018-04402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, M., Flekna, G., Ludwig, A., Heimbucher, T. and Czerny, T. (2006). Improved translation efficiency of injected mRNA during early embryonic development. Dev. Dyn. 235, 3370-3378. 10.1002/dvdy.20995 [DOI] [PubMed] [Google Scholar]
- Fisher, S., Grice, E. A., Vinton, R. M., Bessling, S. L., Urasaki, A., Kawakami, K. and McCallion, A. S. (2006a). Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 1, 1297-1305. 10.1038/nprot.2006.230 [DOI] [PubMed] [Google Scholar]
- Fisher, S., Grice, E. A., Vinton, R. M., Bessling, S. L. and McCallion, A. S. (2006b). Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312, 276-279. 10.1126/science.1124070 [DOI] [PubMed] [Google Scholar]
- Franco, C. A., Jones, M. L., Bernabeu, M. O., Geudens, I., Mathivet, T., Rosa, A., Lopes, F. M., Lima, A. P., Ragab, A., Collins, R. T.et al. (2015). Dynamic endothelial cell rearrangements drive developmental vessel regression. PLoS Biol. 13, e1002125. 10.1371/journal.pbio.1002125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheban, B. A., Rosca, I. A. and Crisan, M. (2019). The morphological and functional characteristics of the pineal gland. Med. Pharm. Rep. 92, 226. 10.15386/mpr-1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giepmans, B. N. G., Adams, S. R., Ellisman, M. H. and Tsien, R. Y. (2006). The fluorescent toolbox for assessing protein location and function. Science (80-.) 312, 217-224. 10.1126/science.1124618 [DOI] [PubMed] [Google Scholar]
- Haberle, V. and Stark, A. (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621-637. 10.1038/s41580-018-0028-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern, M. E., Rhee, J., Goll, M. G., Akitake, C. M., Parsons, M. and Leach, S. D. (2008). Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97-110. 10.1089/zeb.2008.0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlet, M. R. J., Yergeau, D. A., Kuliyev, E., Takeda, M., Taira, M., Kawakami, K. and Mead, P. E. (2006). Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis 44, 438-445. 10.1002/dvg.20234 [DOI] [PubMed] [Google Scholar]
- Hartley, J. L., Temple, G. F. and Brasch, M. A. (2000). DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788-1795. 10.1101/gr.143000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, A. E., Hartung, O., Rothhämel, S., Ferreira, E., Jenny, A. and Marlow, F. L. (2014). Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development 141, 842-854. 10.1242/dev.090449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselson, D., Anderson, R. M., Beinat, M. and Stainier, D. Y. R. (2009). Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc. Natl. Acad. Sci. USA 106, 14896-14901. 10.1073/pnas.0906348106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima, S.-I., Okamoto, H., Ueno, N., Hotta, Y. and Eguchi, G. (1997). High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 192, 289-299. 10.1006/dbio.1997.8779 [DOI] [PubMed] [Google Scholar]
- Huang, C.-J., Tu, C.-T., Hsiao, C.-D., Hsieh, F.-J. and Tsai, H.-J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30-40. 10.1002/dvdy.10356 [DOI] [PubMed] [Google Scholar]
- Ibrahimi, A., Velde, G. V., Reumers, V., Toelen, J., Thiry, I., Vandeputte, C., Vets, S., Deroose, C., Bormans, G., Baekelandt, V.et al. (2009). Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum. Gene Ther. 20, 845-860. 10.1089/hum.2008.188 [DOI] [PubMed] [Google Scholar]
- Jaenisch, R., Jähner, D., Nobis, P., Simon, I., Löhler, J., Harbers, K. and Grotkopp, D. (1981). Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell 24, 519-529. 10.1016/0092-8674(81)90343-3 [DOI] [PubMed] [Google Scholar]
- Jang, S. K., Kräusslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C. and Wimmer, E. (1988). A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636-2643. 10.1128/jvi.62.8.2636-2643.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. D. and Krieg, P. A. (1995). A Xenopus laevis gene encoding EF-1αS, the somatic form of elongation factor 1α: Sequence, structure, and identification of regulatory elements required for embryonic transcription. Dev. Genet. 17, 280-290. 10.1002/dvg.1020170313 [DOI] [PubMed] [Google Scholar]
- Kague, E., Weber, C. and Fisher, S. (2010). Mosaic zebrafish transgenesis for evaluating enhancer sequences. J. Vis. Exp. 41, 10.3791/1722-v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, C. K., Mosimann, C., Fan, Z. P., Yang, S., Thomas, A. J., Ablain, J., Tan, J. L., Fogley, R. D., van Rooijen, E., Hagedorn, E. J.et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197. 10.1126/science.aad2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K. (2004). Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 77, 201-222. 10.1016/S0091-679X(04)77011-9 [DOI] [PubMed] [Google Scholar]
- Kawakami, K. (2005). Transposon tools and methods in zebrafish. Dev. Dyn. 234, 244-254. 10.1002/dvdy.20516 [DOI] [PubMed] [Google Scholar]
- Kawakami, K. (2007). Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8 Suppl 1, S7. 10.1186/gb-2007-8-s1-s7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., Koga, A., Hori, H. and Shima, A. (1998). Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene 225, 17-22. 10.1016/S0378-1119(98)00537-X [DOI] [PubMed] [Google Scholar]
- Kawakami, K., Shima, A. and Kawakami, N. (2000). Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97, 11403-11408. 10.1073/pnas.97.21.11403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazimi, N. and Cahill, G. M. (1999). Development of a circadian melatonin rhythm in embryonic zebrafish. Dev. Brain Res. 117, 47-52. 10.1016/S0165-3806(99)00096-6 [DOI] [PubMed] [Google Scholar]
- Kikuta, H. and Kawakami, K. (2009). Transient and stable transgenesis using tol2 transposon vectors. Methods Mol. Biol. 546, 69-84. 10.1007/978-1-60327-977-2_5 [DOI] [PubMed] [Google Scholar]
- Kim, J. H., Lee, S.-R., Li, L.-H., Park, H.-J., Park, J.-H., Lee, K. Y., Kim, M.-K., Shin, B. A. and Choi, S.-Y. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6, e18556. 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Knopf, F., Schnabel, K., Haase, C., Pfeifer, K., Anastassiadis, K. and Weidinger, G. (2010). Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 107, 19933-19938. 10.1073/pnas.1007799107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf, F., Hammond, C., Chekuru, A., Kurth, T., Hans, S., Weber, C. W., Mahatma, G., Fisher, S., Brand, M., Schulte-Merker, S.et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713-724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Koga, A., Suzuki, M., Inagaki, H., Bessho, Y. and Hori, H. (1996). Transposable element in fish. Nature 383, 30. 10.1038/383030a0 [DOI] [PubMed] [Google Scholar]
- Kondrychyn, I., Garcia-Lecea, M., Emelyanov, A., Parinov, S. and Korzh, V. (2009). Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics 10, 418. 10.1186/1471-2164-10-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster, R. W. and Fraser, S. E. (2001). Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233, 329-346. 10.1006/dbio.2001.0242 [DOI] [PubMed] [Google Scholar]
- Kozmikova, I. and Kozmik, Z. (2015). Gene regulation in amphioxus: an insight from transgenic studies in amphioxus and vertebrates. Mar. Genomics 24, 159-166. 10.1016/j.margen.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Kurita, R., Sagara, H., Aoki, Y., Link, B. A., Arai, K.-I. and Watanabe, S. (2003). Suppression of lens growth by αA-crystallin promoter-driven expression of diphtheria toxin results in disruption of retinal cell organization in zebrafish. Dev. Biol. 255, 113-127. 10.1016/S0012-1606(02)00079-9 [DOI] [PubMed] [Google Scholar]
- Kwan, K. M., Fujimoto, E., Grabher, C., Mangum, B. D., Hardy, M. E., Campbell, D. S., Parant, J. M., Yost, H. J., Kanki, J. P. and Chien, C.-B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088-3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Lalonde, R. L., Kemmler, C. L., Riemslagh, F. W., Aman, A. J., Kresoja-Rakic, J., Moran, H. R., Nieuwenhuize, S., Parichy, D. M., Burger, A. and Mosimann, C. (2022). Heterogeneity and genomic loci of ubiquitous transgenic Cre reporter lines in zebrafish. Dev. Dyn. 251, 1754-1773. 10.1002/dvdy.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart, K. F., Holtzman, N. G., Williams, J. R. and Burdine, R. D. (2013). Integration of nodal and BMP signals in the heart requires FoxH1 to create left-right differences in cell migration rates that direct cardiac asymmetry. PLoS Genet.. 9, e1003109. 10.1371/journal.pgen.1003109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Ritter, D., Yang, N., Dong, Z., Li, H., Chuang, J. H. and Guo, S. (2010). A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev. Biol. 337, 484-495. 10.1016/j.ydbio.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.-F., Swinburne, I. and Yelon, D. (2012). Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev. Biol. 362, 242-253. 10.1016/j.ydbio.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, E., McDole, B., Privat, M., Jaggard, J. B., Duboué, E. R., Sumbre, G. and Keene, A. C. (2022). Blind cavefish retain functional connectivity in the tectum despite loss of retinal input. Curr. Biol. 32, 3720-3730.e3. 10.1016/j.cub.2022.07.015 [DOI] [PubMed] [Google Scholar]
- Loh, K. M., Chen, A., Koh, P. W., Deng, T. Z., Sinha, R., Tsai, J. M., Barkal, A. A., Shen, K. Y., Jain, R., Morganti, R. M.et al. (2016). Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell 166, 451-467. 10.1016/j.cell.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, Q., Meng, A., Wang, H., Jessen, J. R., Farrell, M. J. and Lin, S. (1997). GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development 124, 4105-4111. 10.1242/dev.124.20.4105 [DOI] [PubMed] [Google Scholar]
- Lou, S. S., Diz-Muñoz, A., Weiner, O. D., Fletcher, D. A. and Theriot, J. A. (2015). Myosin light chain kinase regulates cell polarization independently of membrane tension or Rho kinase. J. Cell Biol. 209, 275-288. 10.1083/jcb.201409001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, K. L., Jaggard, J. B., Persons, J. L., Roback, E. Y., Passow, C. N., Stanhope, B. A., Ferrufino, E., Tsuchiya, D., Smith, S. E., Slaughter, B. D.et al. (2021). Repeated evolution of circadian clock dysregulation in cavefish populations. PLoS Genet. 17, e1009642. 10.1371/journal.pgen.1009642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, S. R., Lee, Y., Poss, K. D. and Yelon, D. (2008). Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev. Biol. 321, 397-406. 10.1016/j.ydbio.2008.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoulkova, E., Michalova, E., Vojtesek, B. and Hrstka, R. (2012). The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 9, 563-576. 10.4161/rna.20231 [DOI] [PubMed] [Google Scholar]
- McGaugh, S. E., Kowalko, J. E., Duboué, E., Lewis, P., Franz-Odendaal, T. A., Rohner, N., Gross, J. B. and Keene, A. C. (2020). Dark world rises: the emergence of cavefish as a model for the study of evolution, development, behavior, and disease. J. Exp. Zool. B. Mol. Dev. Evol. 334, 397-404. 10.1002/jez.b.22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A., Findlay, G. M., Gagnon, J. A., Horwitz, M. S., Schier, A. F. and Shendure, J. (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. 10.1126/science.aaf7907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, A., Tang, H., Ong, B. A., Farrell, M. J. and Lin, S. (1997). Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc. Natl. Acad. Sci. USA 94, 6267-6272. 10.1073/pnas.94.12.6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann, C. (2022). Multisite Gateway Calculations: Excel spreadsheet. protocols.io. 10.17504/protocols.io.b4xdqxi6 [DOI]
- Mosimann, C., Kaufman, C. K., Li, P., Pugach, E. K., Tamplin, O. J. and Zon, L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169-177. 10.1242/dev.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann, C., Panáková, D., Werdich, A. A., Musso, G., Burger, A., Lawson, K. L., Carr, L. A., Nevis, K. R., Sabeh, M. K., Zhou, Y.et al. (2015). Chamber identity programs drive early functional partitioning of the heart. Nat. Commun. 6, 8146. 10.1038/ncomms9146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka, N. and Ieda, M. (2015). Stoichiometry of transcription factors is critical for cardiac reprogramming. Circ. Res. 116, 216-218. 10.1161/CIRCRESAHA.114.305696 [DOI] [PubMed] [Google Scholar]
- Myers, R. M., Tilly, K. and Maniatis, T. (1986). Fine structure genetic analysis of a β-globin promoter. Science (80-.). 232, 613-618. 10.1126/science.3457470 [DOI] [PubMed] [Google Scholar]
- Nelson, A. C., Pillay, N., Henderson, S., Presneau, N., Tirabosco, R., Halai, D., Berisha, F., Flicek, P., Stemple, D. L., Stern, C. D.et al. (2012). An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J. Pathol. 228, 274-285. 10.1002/path.4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayr, C., Pagani, M., Stark, A. and Arnold, C. D. (2019). STARR-seq and UMI-STARR-seq: assessing enhancer activities for genome-wide-, high-, and low-complexity candidate libraries. Curr. Protoc. Mol. Biol. 128, e105. 10.1002/cpmb.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido, M., Tada, M., Saji, T. and Ueno, N. (1997). Conservation of BMP signaling in zebrafish mesoderm patterning. Mech. Dev. 61, 75-88. 10.1016/S0925-4773(96)00625-9 [DOI] [PubMed] [Google Scholar]
- Ogura, E., Okuda, Y., Kondoh, H. and Kamachi, Y. (2009). Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev. Dyn. 238, 641-655. 10.1002/dvdy.21863 [DOI] [PubMed] [Google Scholar]
- Okayama, H. and Berg, P. (1992). A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. 1983. Biotechnology 24, 270-279. [PubMed] [Google Scholar]
- Omura, Y. (1975). Influence of light and darkness on the ultrastructure of the pineal organ in the blind cave fish, Astyanax mexicanus. Cell Tissue Res. 160, 99-112. 10.1007/BF00219844 [DOI] [PubMed] [Google Scholar]
- Palpant, N. J., Wang, Y., Hadland, B., Zaunbrecher, R. J., Redd, M., Jones, D., Pabon, L., Jain, R., Epstein, J., Ruzzo, W. L.et al. (2017). Chromatin and transcriptional analysis of mesoderm progenitor cells identifies HOPX as a regulator of primitive hematopoiesis. Cell Rep. 20, 1597-1608. 10.1016/j.celrep.2017.07.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, H. J., De Kumar, B., Green, S. A., Prummel, K. D., Hess, C., Kaufman, C. K., Mosimann, C., Wiedemann, L. M., Bronner, M. E. and Krumlauf, R. (2019). A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nat. Commun. 10, 1189. 10.1038/s41467-019-09197-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J. and Sonenberg, N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320-325. 10.1038/334320a0 [DOI] [PubMed] [Google Scholar]
- Pisharath, H., Rhee, J. M., Swanson, M. A., Leach, S. D. and Parsons, M. J. (2007). Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech. Dev. 124, 218-229. 10.1016/j.mod.2006.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost, E., Rhee, J. and Leach, S. D. (2007). Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis 45, 625-629. 10.1002/dvg.20338 [DOI] [PubMed] [Google Scholar]
- Prummel, K. D., Hess, C., Nieuwenhuize, S., Parker, H. J., Rogers, K. W., Kozmikova, I., Racioppi, C., Brombacher, E. C., Czarkwiani, A., Knapp, D.et al. (2019). A conserved regulatory program initiates lateral plate mesoderm emergence across chordates. Nat. Commun. 10, 3857. 10.1038/s41467-019-11561-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, M. A. and Piston, D. W. (2005). High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys. J. 88, L14-L16. 10.1529/biophysj.104.055442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, M. A., Springer, G. H., Granada, B. and Piston, D. W. (2004). An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22, 445-449. 10.1038/nbt945 [DOI] [PubMed] [Google Scholar]
- Rodriguez, E. A., Campbell, R. E., Lin, J. Y., Lin, M. Z., Miyawaki, A., Palmer, A. E., Shu, X., Zhang, J. and Tsien, R. Y. (2017). The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem. Sci. 42, 111. 10.1016/j.tibs.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner, N. (2018). Cavefish as an evolutionary mutant model system for human disease. Dev. Biol. 441, 355-357. 10.1016/j.ydbio.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Rothwell, D. G., Crossley, R., Bridgeman, J. S., Sheard, V., Zhang, Y., Sharp, T. V., Hawkins, R. E., Gilham, D. E. and McKay, T. R. (2010). Functional expression of secreted proteins from a bicistronic retroviral cassette based on foot-and-mouth disease virus 2A can be position dependent. Hum. Gene Ther. 21, 1631-1637. 10.1089/hum.2009.197 [DOI] [PubMed] [Google Scholar]
- Ryan, M. D., King, A. M. Q. and Thomas, G. P. (1991). Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72, 2727-2732. 10.1099/0022-1317-72-11-2727 [DOI] [PubMed] [Google Scholar]
- Sakaguchi, T., Kikuchi, Y., Kuroiwa, A., Takeda, H. and Stainier, D. Y. R. (2006). The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development 133, 4063-4072. 10.1242/dev.02581 [DOI] [PubMed] [Google Scholar]
- Sánchez-Iranzo, H., Galardi-Castilla, M., Minguillón, C., Sanz-Morejón, A., González-Rosa, J. M., Felker, A., Ernst, A., Guzmán-Martínez, G., Mosimann, C. and Mercader, N. (2018). Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat. Commun. 9, 428. 10.1038/s41467-017-02650-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, Y., Kasai, T., Nakagawa, S., Tanabe, K., Watanabe, T., Kawakami, K. and Takahashi, Y. (2007). Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev. Biol. 305, 616-624. 10.1016/j.ydbio.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Schaffner, W. (2015). Enhancers, enhancers - from their discovery to today's universe of transcription enhancers. Biol. Chem. 396, 311-327. 10.1515/hsz-2014-0303 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., et al. , (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E. K., Mason, L., Arrenberg, A. B., Ziv, L., Gosse, N. J., Xiao, T., Chi, N. C., Asakawa, K., Kawakami, K. and Baier, H. (2007). Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods 4, 323-326. 10.1038/nmeth1033 [DOI] [PubMed] [Google Scholar]
- Shaner, N. C., Lin, M. Z., McKeown, M. R., Steinbach, P. A., Hazelwood, K. L., Davidson, M. W. and Tsien, R. Y. (2008). Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545-551. 10.1038/nmeth.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo, D., Murphy, C. S., Ermakova, G. V., Solovieva, E. A., Chepurnykh, T. V., Shcheglov, A. S., Verkhusha, V. V., Pletnev, V. Z., Hazelwood, K. L., Roche, P. M.et al. (2009). Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 418, 567-574. 10.1042/BJ20081949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji, W. and Sato-Maeda, M. (2008). Application of heat shock promoter in transgenic zebrafish. Dev. Growth Differ. 50, 401-406. 10.1111/j.1440-169X.2008.01038.x [DOI] [PubMed] [Google Scholar]
- Singh, S. P., Holdway, J. E. and Poss, K. D. (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879-886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K., Nam, Y.-J., Luo, X., Qi, X., Tan, W., Huang, G. N., Acharya, A., Smith, C. L., Tallquist, M. D., Neilson, E. G.et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599-604. 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, B. A., Peuß, R., McDole, B., Kenzior, A., Jaggard, J. B., Gaudenz, K., Krishnan, J., McGaugh, S. E., Duboue, E. R., Keene, A. C.et al. (2019). Stable transgenesis in Astyanax mexicanus using the Tol2 transposase system. Dev. Dyn. 248, 679-687. 10.1002/dvdy.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickney, H. L., Imai, Y., Draper, B., Moens, C. and Talbot, W. S. (2007). Zebrafish bmp4 functions during late gastrulation to specify ventroposterior cell fates. Dev. Biol. 310, 71-84. 10.1016/j.ydbio.2007.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach, O. M., Gundorov, I. S., Yoshimura, M., Subach, F. V., Zhang, J., Grüenwald, D., Souslova, E. A., Chudakov, D. M. and Verkhusha, V. V. (2008). Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15, 1116-1124. 10.1016/j.chembiol.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin, O. J., Cox, B. J. and Rossant, J. (2011). Integrated microarray and ChIP analysis identifies multiple Foxa2 dependent target genes in the notochord. Dev. Biol. 360, 415-425. 10.1016/j.ydbio.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Tamplin, O. J., Durand, E. M., Carr, L. A., Childs, S. J., Hagedorn, E. J., Li, P., Yzaguirre, A. D., Speck, N. A. and Zon, L. I. (2015). Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell 160, 241-252. 10.1016/j.cell.2014.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse, B., Heyer, V., Lux, A., Alunni, V., Degrave, A., Seiliez, I., Kirchner, J., Parkhill J.-P. and Thisse C. (2004). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505-519. 10.1016/s0091-679x(04)77027-2 [DOI] [PubMed] [Google Scholar]
- Traver, D., Winzeler, A., Stern, H. M., Mayhall, E. A., Langenau, D. M., Kutok, J. L., Look, A. T. and Zon, L. I. (2004). Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood 104, 1298-1305. 10.1182/blood-2004-01-0100 [DOI] [PubMed] [Google Scholar]
- Trichas, G., Begbie, J. and Srinivas, S. (2008). Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 6, 40. 10.1186/1741-7007-6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh, L. A., Hochgreb, T., Graham, M., Wu, D., Ruf-Zamojski, F., Jayasena, C. S., Saxena, A., Hawk, R., Gonzalez-Serricchio, A., Dixson, A.et al. (2011). A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 25, 2306-2320. 10.1101/gad.174037.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien, R. Y. (2003). The green fluorescent protein. Annu. Rev. Biochem. 67, 509-544. 10.1146/annurev.biochem.67.1.509 [DOI] [PubMed] [Google Scholar]
- Urasaki, A., Asakawa, K. and Kawakami, K. (2008). Efficient transposition of the Tol2 transposable element from a single-copy donor in zebrafish. Proc. Natl. Acad. Sci. USA 105, 19827-19832. 10.1073/pnas.0810380105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc, J. A., Amigo, J. and Lawson, N. D. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077-3087. 10.1002/dvdy.21354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman, J. S., Keegan, B. R., Roberts, R. W., Poss, K. D. and Yelon, D. (2008). Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev. Cell 15, 923-934. 10.1016/j.devcel.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, T. and Köster, R. (2013). Genetic tools for multicolor imaging in zebrafish larvae. Methods 62, 279-291. 10.1016/j.ymeth.2013.07.028 [DOI] [PubMed] [Google Scholar]
- White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., Bourque, C., Dovey, M., Goessling, W., Burns, C. E.et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183-189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R., Rose, K. and Zon, L. (2013). Zebrafish cancer: the state of the art and the path forward. Nat. Rev. Cancer 13, 624-636. 10.1038/nrc3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C., Bellen, H. J. and Gehring, W. J. (1990). Position effects on eukaryotic gene expression. Annu. Rev. Cell Biol. 6, 679-714. 10.1146/annurev.cb.06.110190.003335 [DOI] [PubMed] [Google Scholar]
- Wong, E.-T., Ngoi, S.-M. and Lee, C. G. L. (2002). Improved co-expression of multiple genes in vectors containing internal ribosome entry sites (IRESes) from human genes. Gene Ther. 9, 337-344. 10.1038/sj.gt.3301667 [DOI] [PubMed] [Google Scholar]
- Woolfe, A., Goodson, M., Goode, D. K., Snell, P., McEwen, G. K., Vavouri, T., Smith, S. F., North, P., Callaway, H., Kelly, K.et al. (2004). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7. 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, S., Wang, W., Kenzior, A., Olsen, L., Krishnan, J., Persons, J., Medley, K., Peuß, R., Wang, Y., Chen, S.et al. (2022). Enhanced lipogenesis through Pparγ helps cavefish adapt to food scarcity. Curr. Biol. 32, 2272-2280.e6. 10.1016/j.cub.2022.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa, M. and Jeffery, W. R. (2008). Shadow response in the blind cavefish Astyanax reveals conservation of a functional pineal eye. J. Exp. Biol. 211, 292. 10.1242/jeb.012864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Zhang, A. D., Kim, L. J. and Nam, Y.-J. (2019). Ensuring expression of four core cardiogenic transcription factors enhances cardiac reprogramming. Sci. Rep. 9, 6362. 10.1038/s41598-019-42945-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv, L., Levkovitz, S., Toyama, R., Falcon, J. and Gothilf, Y. (2005). Functional development of the zebrafish pineal gland: light-induced expression of period2 is required for onset of the circadian clock. J. Neuroendocrinol. 17, 314-320. 10.1111/j.1365-2826.2005.01315.x [DOI] [PubMed] [Google Scholar]
- Zuniga, E., Rippen, M., Alexander, C., Schilling, T. F. and Crump, J. G. (2011). Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development 138, 5147-5156. 10.1242/dev.067785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.