Abstract
Diffuse midline gliomas (DMGs) are pediatric high-grade brain tumors in the thalamus, midbrain, or pons; the latter subgroup are termed diffuse intrinsic pontine gliomas (DIPG). The brain-stem location of these tumors limits the clinical management of DIPG, resulting in poor outcomes for patients. A heterozygous, somatic point mutation in one of two genes coding for the non-canonical histone H3.3 is present in most DIPG tumors. This dominant mutation in the H3-3A gene results in replacement of lysine 27 with methionine (K27M) and causes a global reduction of trimethylation on K27 of all wild-type histone H3 proteins, which is thought to be a driving event in gliomagenesis. In this study, we designed and systematically screened 2’-O-methoxyethyl phosphorothioate antisense oligonucleotides (ASOs) that direct RNase-H-mediated knockdown of H3-3A mRNA. We identified a lead ASO that effectively reduced H3-3A mRNA and H3.3K27M protein and restored global H3K27 trimethylation in patient-derived neurospheres. We then tested the lead ASO in two mouse models of DIPG: an immunocompetent mouse model employing transduced mutant human H3-3A cDNA, and an orthotopic xenograft with patient-derived cells. In both models, ASO treatment restored K27 trimethylation of histone H3 proteins and reduced tumor growth, promoted neural-stem-cell differentiation into astrocytes, neurons, and oligodendrocytes, and increased survival. These results demonstrate the involvement of the H3.3K27M oncohistone in tumor maintenance, confirm the reversibility of the aberrant epigenetic changes it promotes, and provide preclinical proof-of-concept for DMG antisense therapy.
One sentence summary
Antisense knockdown of H3.3K27M oncohistone restores H3K27 trimethylation, inhibits gliomagenesis, promotes differentiation and increases survival in mice.
Introduction
Pediatric high-grade gliomas (pHGGs) represent 10-15% of pediatric brain tumors, and have exceedingly poor outcomes (1, 2). About half of pHGGs, termed diffuse midline gliomas (DMGs), exhibit a diffuse pattern in the midline, including the thalamus, midbrain, and pons; the latter constitute an especially severe subgroup termed diffuse intrinsic pontine gliomas (DIPG). Most patients with DIPG die within two years of diagnosis, with a mean survival of nine months. The brainstem location limits the clinical management of DIPG, making complete surgical resection impossible. Moreover, localized chemotherapy is ineffective and causes severe side effects. Thus, new effective therapies are urgently needed.
A specific heterozygous point mutation that affects the non-canonical histone H3.3 is present in 70-80% of DIPG tumors. This dominant somatic mutation occurs in H3-3A—one of two genes encoding identical H3.3 proteins—replacing lysine 27 with methionine (K27M). H3.3K27M is an oncogenic gain-of-function mutation that inhibits the Enhancer of Zeste Homolog 2 (EZH2) methyltransferase subunit of the Polycomb Repressive Complex 2 (PRC2), leading to a global reduction in di- and tri-methylation of all histone H3 proteins (3, 4). This mutation thus activates many downstream genes, and appears to be a driving event in tumorigenesis (3, 4). Moreover, genetic knockout or knockdown studies suggested that the mutant histone could be a therapeutic target (5, 6). These observations led us to develop and test a pharmacological approach to directly target the mutant H3-3A mRNA. H3.3K27M gliomas are more aggressive than H3.1K27M (H3C2K27M) gliomas, which are less prevalent and drive distinct oncogenic programs (7). Importantly, these mutations correlate with poor patient outcomes, and can be diagnosed by MRI and stereotactic biopsy (8).
Targeted therapies for patients with the H3.3K27M mutation are currently in clinical trials, such as the HDAC inhibitor Panobinostat (LBH589) (NCT02717455) (9), but they target downstream genes that undergo epigenetic reprogramming. To achieve more direct gene-specific or allele-specific targeting for H3.3K27M gliomas, we sought to explore the potential of antisense oligonucleotides (ASOs) in H3.3K27M DIPG patient cells, using chemically modified “gapmer” ASOs that hybridize with complementary RNA sequences and mediate RNase-H cleavage of the target transcripts. The chemical composition of the gapmer wings is the same as that of nusinersen (Spinraza), a splice-modulating, uniformly modified ASO that promotes inclusion of exon 7 in Survival of Motor Neuron 2 (SMN2) mRNA (10, 11). Nusinersen was the first approved drug for spinal muscular atrophy, and the first disease-modifying therapy for neurodegeneration.
Here, we describe lead ASOs we identified in systematic screens, and show that they specifically delayed the growth of H3.3K27M patient-derived cells grown as neurospheres. Furthermore, intracerebroventricular (ICV) administration of a lead ASO in two different DIPG mouse models reduced tumor growth, promoted neural-stem-cell differentiation, and increased survival. These findings provide preclinical proof of principle for an antisense therapy for DMG.
Results
CRISPR-Cas9 depletion of H3.3K27M rescued H3K27 trimethylation and delayed the growth of patient-derived neurospheres and orthotopic xenografts
As a control for the biological consequence of complete genetic knockout of H3-3A in the models used in this study, we first used CRISPR-Cas9 with sgRNA targeting both mutant and wild-type alleles, but leaving H3-3B (which encodes the identical protein) unaffected in two DIPG-patient cell lines, SU-DIPG-XIII and XVII (9). The dominant-negative effect of the K27M mutation on the overall amounts of H3K27me3 is well documented (1-4). In both patient-cell lines harboring the H3-3A heterozygous mutation, H3.3K27M knockout restored the repressive H3K27me3 trimethylation mark and reduced the permissive H3K27ac acetylation mark, detected by immunoblotting (fig. S1A). Transduction of gRNA-resistant H3-3AK27M cDNA decreased the restored H3K27me3 mark, consistent with an on-target effect (fig. S1B). The knockout restored H2K27me3 trimethylation and reduced cell proliferation, detected by immunofluorescence, EdU staining (fig. S1 C, D and E), and soft-agar colony formation (fig. S1F and G). To determine whether H3.3K27M is required for tumor maintenance in vivo, we performed orthotopic transplantation of patient cells or H3-3AK27M knockout cells into immunocompromised Non-obese diabetic, Severe combined immunodeficiency disease, Gamma (NSG) mice at postnatal day 3 (P3) (9). We used the SU-DIPG-XIII line for this experiment, because its slower proliferation rate extends the therapeutic window for our treatment. The transplanted knockout cells showed delayed tumor latency, resulting in significantly (P = 0.039) increased survival (Fig. 1S H and I). The H3.3K27M xenograft tumors histologically resembled patient tumors, with a global reduction of the H3K27me3 mark, compared to normal adjacent tissue. This effect in H3.3K27M tumors was alleviated in the knockout tumors and correlated with elevated expression of the mature-neuron marker Neuronal Nuclei Antigen (NeuN) and the astrocyte marker Glial Fibrillary Acidic Protein (GFAP), detected by IF (fig. S1J), suggesting that the knockout cells are less proliferative and more committed to differentiated lineages in vivo. These results confirm and extend previous studies (5, 6) and underscore the potential of mutant H3.3K27M as a therapeutic target.
Gapmer ASO screen to reduce mutant H3-3A mRNA and H3.3K27M protein
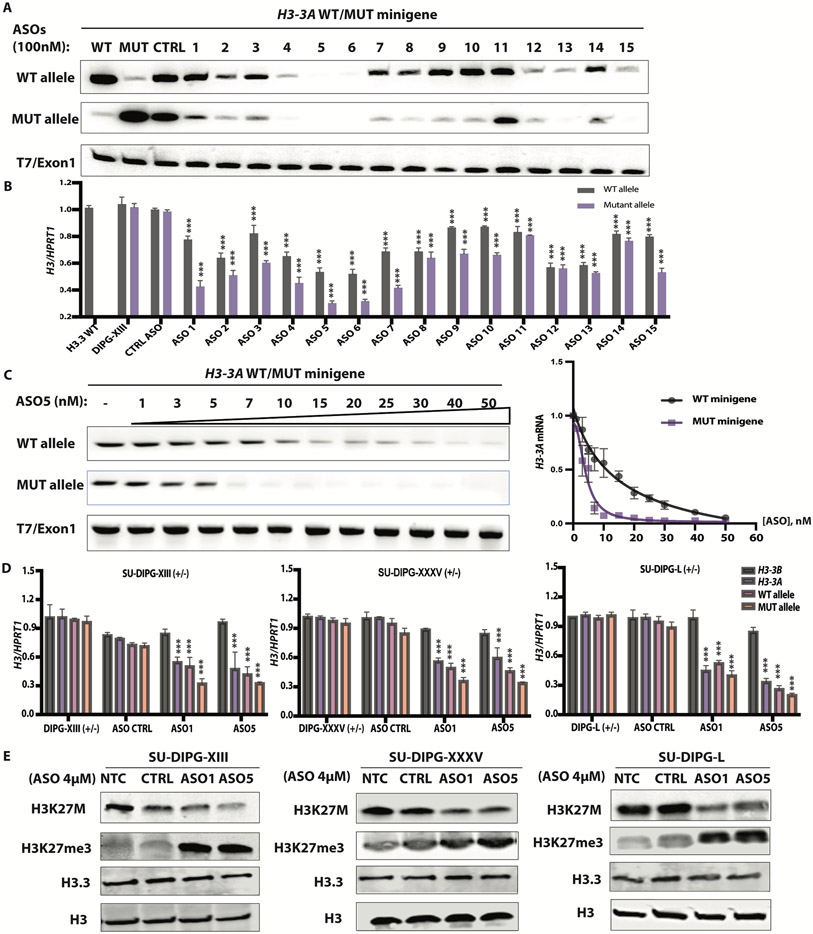
To pharmacologically target H3-3A mRNA, we explored the potential of ASOs in DIPG patient cells. We designed and tested gapmers with 2’-O-methoxyethyl (MOE) wings and phosphorothioate (PS) backbone (fig. S1A and B). These gapmer ASOs have a DNA-like central region that directs cleavage of the complementary mRNA or pre-mRNA target by endogenous RNase H, and chemically modified wings that promote tighter RNA binding, enhanced stability, and improved cellular uptake (fig. S1C) (12). First, to attempt to achieve targeted allele-specific knockdown, we designed 10 overlapping 20-mer PS-MOE-ASOs (#1-10) spanning the mutation site in H3-3A exon 2 and flanking nucleotides, with the mutation positioned across the ASO gap. These sequences were fully complementary to the H3-3AK27M allele, and had one mismatch to the H3-3AWT allele. Second, to achieve targeted gene-specific knockdown—considering that the wild-type H3.3 protein is still expressed from the H3-3B gene—we designed five additional overlapping 20-mer PS-MOE gapmer ASOs (#11-15) to target exonic regions upstream or downstream of the mutation site in exon 2 (fig. S1D). These ASOs were designed to target only H3-3A but not H3-3B transcripts, or other gene transcripts that encode H3.1 or H3.2 canonical histone proteins (fig. S1E). In addition, we generated a wild-type H3-3A minigene and a mutant H3-3A minigene with the A-to-T mutation by cloning genomic fragments comprising exons 1 to 3, with intact introns 1 and 2 (fig. S1F).
To test whether some of these ASOs mediate RNase-H1 cleavage of H3-3AK27M mRNA, or both mutant and wild-type H3-3A, we co-transfected individual ASOs (100 nM) with the H3-3AWT or H3-3AK27M minigene into HeLa cells. We also delivered these ASOs by free uptake (4 μM) into patient-derived neurosphere cultures. The latter method relies on receptor-mediated endocytosis and requires much higher ASO concentrations (12). Our initial minigene screen identified three consecutive ASOs (ASO4, 5, and 6) with allele-specific design, and two consecutive ASOs (ASO12 and 13) with gene-specific design, that achieved robust H3-3A knockdown (Fig. 1A). All the allele-specific ASOs achieved more robust knockdown of the mutant than the wild-type allele in patient-derived cells, whereas at this concentration, ASO4, 5, and 6 robustly knocked down both alleles in the minigene context. Similarly, ASOs delivered into neurosphere cultures by free uptake knocked down expression of the endogenous mutant allele by 50-70%, and expression of the WT allele by a lesser extent (30-40%) (Fig. 1B). Unexpectedly, one gene-specific ASO, ASO15, also promoted allele-specific knockdown, suggesting steric blocking of a putative regulatory RNA-binding protein that binds within its target region (Fig. 1B).
Fig. 1. ASO-mediated H3.3K27M depletion restored global H3K27me3.
(A) ASO screen using H3-3AWT and H3-3AK27M minigenes. HeLa cells were co-transfected with the minigenes, along with individual 20-mer PS-MOE ASOs, using Lipofectamine 2000; two days later, the extent of knockdown was quantified by radioactive RT-PCR with allele-specific primers; band intensities are shown below each band (N = 3); (B) Summary data for ASO screen by free uptake in patient-derived (SU-DIPG-XIII) neurosphere cultures, using RT-qPCR of total RNA extracted after 5 days (n = 3); (C) Dose-response experiment with co-transfected minigenes in HeLa cells, with representative RT-PCR gel on the left, and quantification on the right (N = 3); (D) Decrease of H3-3A mRNA measured by RT-qPCR (n = 3) in three patient-derived cell lines, detected with allele-specific primers to distinguish mutant and wild-type alleles (N = 3); (E) Immunoblot of acid-extracted histones from each patient cell line, with quantification of band intensities shown below each band. For RT-qPCR experiments, Welch’s two-sample t-test was used to compare two groups. For more than two groups, the measurements for each experimental group/treatment were analyzed by ANOVA, followed by pairwise comparisons using Welch’s two-sample t-tests. Family-wise error rate was adjusted using the Bonferroni-Holm method. Data are presented as means ± SEM. WT, wild type; mut, mutant. ***P < 0.0001
We then titrated the most potent ASO (ASO5) in HeLa cells and observed allele-specific knockdown between 3 to 40 nM in transfection experiments. We calculated an IC50 of 15 nM for the wild-type allele versus 4 nM for the mutant allele (Fig. 1C). We selected two allele-specific lead ASOs for testing in two additional patient-derived cell lines and developed primer pairs for H3-3B and for total H3-3A, amplifying a region downstream of the mutation. The two lead ASOs, delivered by free uptake at 4 μM, behaved similarly across the three different patient cell lines: they selectively knocked down H3-3A, but not H3-3B, and depleted the mRNA from the mutant allele to a greater extent than the wild-type allele (Fig. 1D).
Next, we measured the abundance of histone H3.3 protein by immunoblotting with an antibody specific for H3.3 with the K27M mutation and measured downstream epigenetic changes using antibodies to trimethylated H3K27, H3.3, and total H3 histone proteins as normalization controls. The two lead ASOs knocked down ~60%-70% of the mutant protein, resulting in 2-3-fold H3K27me3 elevation. We observed a slight decrease in H3.3 protein abundance across the three DIPG lines (Fig. 1E). In contrast, transfected ASO5 only slightly reduced H3-3A mRNA expression in the H3.3WT cell line, and had no effect on H3-3B mRNA expression. Consistently, H3.3 and total H3 protein abundance remained unchanged (fig. S3A and B).
ASO-mediated H3.3K27M depletion delayed neurosphere growth and changed cell morphology
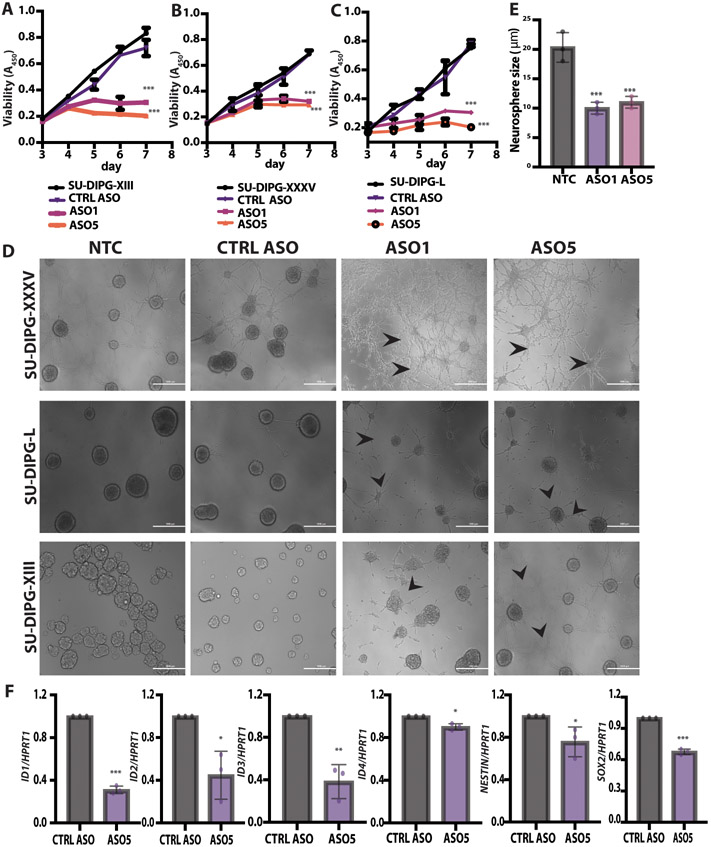
Using several H3.3K27M DIPG-patient-derived cell lines grown as neurospheres, we observed that the lead ASOs (ASO1 and 5) specifically delayed tumor-cell growth, compared to scrambled control ASO. We observed significantly (P < 0.001) slower proliferation of SU-DIPG-XIII, SU-DIPG-XXXV, and SU-DIPG-L neurospheres treated with 4 μM ASO1 or ASO5 (Fig. 2A, B and C). In contrast, neither the CTRL ASO nor ASO5 had measurable effects on the proliferation of the control H3.3WT glioma line (fig. S3C). At the 5-day time point, the cellular morphology of the three DIPG patient-derived lines changed, with neurite-like processes consistently observed (Fig. 2D). This morphological change is consistent with the differentiation phenotype we observed in the CRISPR-knockout orthotopic xenografts (fig. S1J). In addition, the SU-DIPG-XIII and SU-DIPG-L lines treated with the lead ASOs formed smaller neurospheres (Fig. 2E).
Fig. 2. ASO-mediated H3.3K27M depletion delayed neurosphere growth and changed cell morphology.
(A) Shown are cell-viability assays at each time point for the SU-DIPG-XIII H3.3K27M cell line (N = 3); (B) Shown are cell-viability assays at each time point for SU-DIPG-XXXV cell line; (C) Shown are cell-viability assays at each time point for SU-DIPG-L cell line; (D) Representative images of SU-DIPG-XIII, SU-DIPG-XXXV, and SU-DIPG-L patient cells treated with ASO1, ASO5, or control Scramble ASO by free uptake for 5 days. Arrowheads indicate neurite-like processes. Scale bars, 1000 μm; (E) Quantification of average neurosphere size (in μm) from the images in (D) (n = 3 random fields); (F) Real time RT-PCR (n = 3) of genes implicated in developmental processes in ASO-5 treated SU-DIPG-XIII cells, relative to expression of control HPRT1. For viability assays, P-values were adjusted for multiple comparisons by controlling family-wise error rate using the single-step method. For neurosphere size, the measurements for each experimental group/treatment were analyzed by ANOVA, followed by pairwise comparisons using two-sample t-tests. Data are presented as means ± SEM. ***P < 0.001, **P < 0.01, *P < 0.05.
To evaluate the state of cell differentiation, we assessed changes in expression of a panel of genes previously implicated in developmental processes. The Inhibitor of DNA Binding 1-4 (ID1-4) genes were significantly (P < 0.001 or P < 0.01) downregulated in ASO5-treated SU-DIPG-XIII cells (Fig. 2F). These ID genes normally inhibit differentiation and maintain self-renewal, as well as multipotency of stem cells (5). In addition, the neuroepithelial cell markers Nestin (NES) and Sex-Determining Region Y-Box Transcription Factor 2 (SOX2) were also significantly (P < 0.01) downregulated in response to ASO5 treatment (Fig. 2F). In summary, the ASO-driven reduction in H3.3K27M mutant protein had phenotypic consequences in patient cells, including slower proliferation and a differentiated morphology and gene-expression pattern.
ICV injection of lead ASO promoted H3.3K27M depletion, lower tumor grade, and differentiation in an RCAS-Tva mouse model
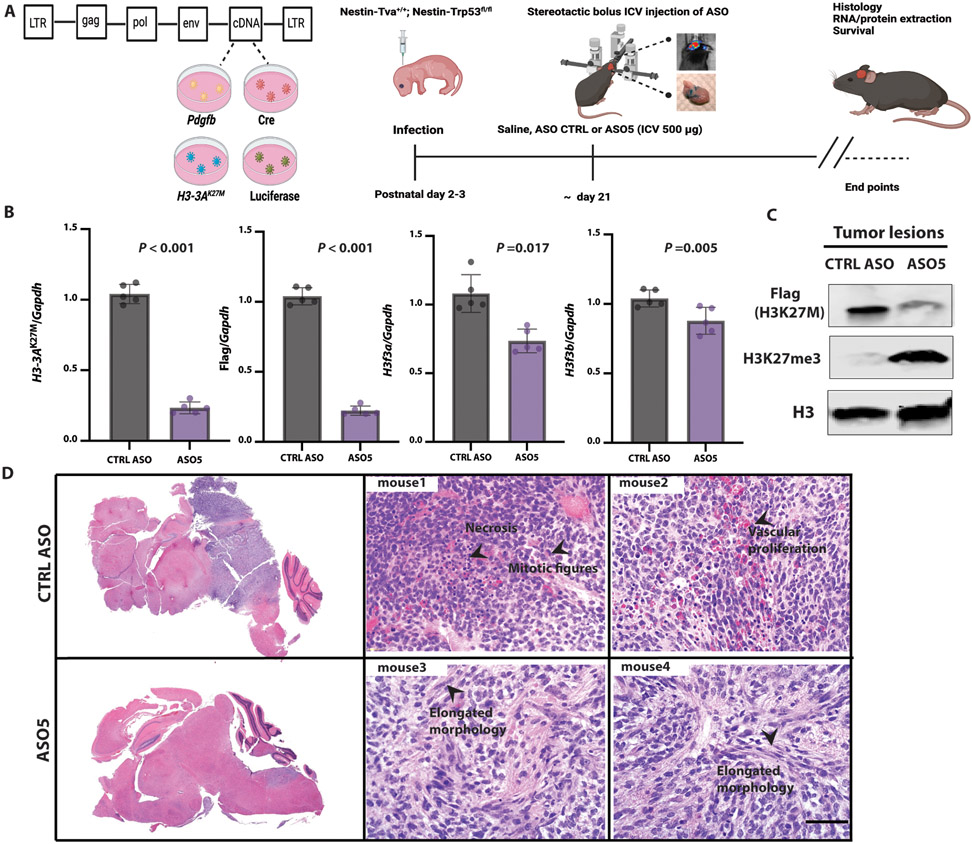
To test our lead ASOs in vivo, we first employed the RCAS-Tva system to establish a mouse model of DIPG. RCAS, or Replication-Competent Avian sarcoma-leukosis virus long terminal repeat (LTR) with Splice acceptor, is a viral vector that only infects cells expressing the avian Tva retrovirus receptor (13). This system was previously used to show that murine histone H3.3K27M or H3.1K27M accelerates gliomagenesis, consistent with results in another genetic mouse model (14-16). We adapted the system by introducing instead a human H3-3AK27M cDNA, whose transcripts can be targeted by our human-specific ASOs. We delivered chicken DF1 cells producing four viruses encoding P1 bacteriophage Cre recombinase, human H3.3K27M, murine Platelet-Derived Growth Factor B (PDGFB), and firefly luciferase, into the brainstem of neonate mice with Tva driven by the murine Nestin promoter, and a Transformation-related protein 53 (Trp53)-floxed allele (Fig. 3A). Mice developed high-grade gliomas within 3-6 weeks, localized in the midline region. FLAG-tagged H3.3K27M was present in the glioma lesions, as seen by histological analysis (fig. S4A). Approximately 90% of H3.3K27M tumors showed global H3K27me3 reduction, compared to the normal adjacent tissue, a pattern that resembled patient-tumor histology (fig. S4B). Tumor cells also stained positive for expression of the oligodendroglial lineage marker Oligodendrocyte transcription factor 2 (OLIG2) (15) (fig. S4C).
Fig. 3. ICV administration of ASO at the time of tumor onset in the RCAS-Tva mouse model.
(A) Diagram of RCAS plasmids; 105 RCAS-Pdgfb, RCAS-Cre, and RCAS-H3-3AK27M cDNA-expressing producer chicken cells (DF1) were injected into the brainstem of Nestin-Tva; P53fl/fl mice at postnatal day 3 (P3); a single dose (500 μg) of lead ASO or CTRL ASO in saline was stereotactically injected ICV at P21; RNA, protein, and histology samples were collected at the end points when the mice were symptomatic, including an enlarged head, ataxia, or >25% weight loss; (B) Summary data of mRNA expression of H3-3AK27M allele, flag tag, endogenous murine H3f3a and H3f3b as compared to Gapdh (N = 5); (C) Immunoblot of acid-extracted histones from two representative mice; quantification of average band intensities is shown below each band; (D) Representative H&E-stained tumors confirming their location in the midline region of the brain (top left, CTRL ASO-treated; bottom left, ASO5-treated); representative H&E-stained high-grade tumors from control-ASO-treated mice (top, middle and right) and lower-grade tumors with elongated morphology from ASO5-treated mice (bottom, middle and right); scale bar, 500 μm (N = 5). For RT-qPCR experiments, the measurements for each experimental group/treatment were analyzed by Welch’s two-sample t-test to compare the H3-3AK27M expression normalized to the Gapdh loading control between CTRL ASO and ASO5 treatments. Data are presented as means ± SEM.
To test the effectiveness of the lead ASO in vivo, we performed stereotactic ICV injections to deliver it directly into the cerebrospinal fluid (CSF) (17, 18). We injected a single dose (500 μg) of lead ASO5 or CTRL ASO in saline into a lateral ventricle, at the time of tumor onset, as detected by bioluminescence imaging (Fig. 3A). We extracted RNA and protein from the tumors or normal adjacent tissue at preset time points. ASO5 significantly (P < 0.001) knocked down the human H3-3AK27M mRNA and FLAG-tag, and to a lesser extent the endogenous murine wild-type H3f3a, but not H3f3b (Fig. 3B). At the protein level, ASO5 significantly (P < 0.001) knocked down human FLAG-tagged H3.3K27M, and increased H3K27me3 abundance (Fig. 3C). We further observed the disappearance of FLAG-tag IF staining, such that in ~80% of treated cells, the FLAG signal was below detection (fig. S4D).
Mice treated with the control ASO developed highly proliferative and aggressive gliomas, with numerous mitotic figures, extensive vascular proliferation, occasional necrosis, and pseudo-palisades around necrotic areas. These tumors were non-encapsulated and poorly demarcated, with some tumor invasion at the tumor-brain interface. In contrast, mice treated with ASO5 showed an extended latency of tumor growth, and the tumors exhibited elongated morphology (Fig. 3D). In these tumor lesions, mitotic figures were rare, and necrosis was not prominent. Moreover, the cells in ASO5-treated tumor lesions morphologically resembled glia and mature neurons. We conclude that the knockdown of mutant H3-3A elicited by the lead ASO in vivo resulted in lower-grade tumor formation and a more differentiated appearance.
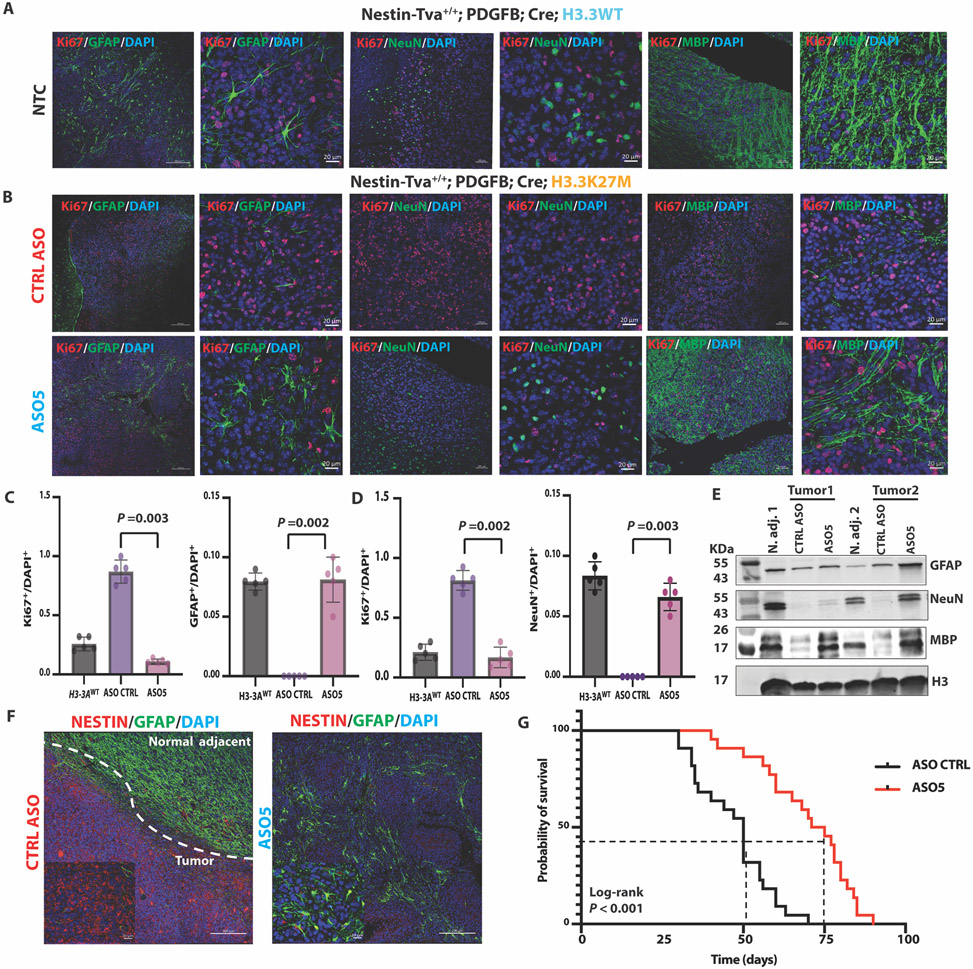
To further characterize the phenotypes observed by histology, we performed IF staining for known differentiation markers. In mice of the same genetic background, but bearing H3.3WT tumors, we detected markers of mature astrocytes (GFAP) (19), neurons (NeuN) (20), and oligodendrocytes (myelin basic protein (MBP) (21)), suggesting the occurrence of neurogenesis and gliogenesis in H3.3WT gliomas (Fig. 4A). In contrast, there were few detectable GFAP+, NeuN+, and MBP+ cells within tumor lesions in the presence of H3.3K27M (Fig. 4B, top panels). After a single ICV dose of ASO5, we detected numerous GFAP+, NeuN+, and MBP+ cells, correlating with a reduction in proliferating cells marked by Ki67, a nuclear cell-proliferation-associated antigen expressed in all active stages of the cell cycle (Fig. 4B, bottom panels). The reduction in Ki67 and increase in GFAP and NeuN markers were statistically significant (P = 0.002 or 0.003) (Fig. 4C and D). We confirmed these results by western blotting of total protein extracted from normal adjacent tissue and tumor lesions (Fig. 4E). We conclude that H3.3K27M blocked astrocyte and neuron differentiation, and ASO-mediated H3.3K27M depletion restored the differentiation programs.
Fig. 4. ASO-mediated H3.3K27M depletion induced astrocyte, neuron, and oligodendrocyte differentiation, decreased tumor cell proliferation and the NESTIN+ cell population, and extended the latency of tumor growth in the Nestin-Tva mouse model.
(A) Representative IF images of no-treatment control (NTC) tumors stained for markers of differentiation (GFAP, NeuN, MBP; green) or proliferation (Ki67; red), and for nuclei with DAPI (blue) in Nestin-Tva mice transduced with H3-3AWT cDNA; (low-magnification images: scale bar, 200 μm; high-magnification images: scale bar, 20 μm); (B) Representative IF images of tumor sections from control (CTRL) ASO-treated (top panels) and ASO5-treated (bottom panels) mice; IF and DAPI staining, and magnification bars are as in (A); (C) Summary data from (A) and (B) of Ki67+ and GFAP+ cell numbers normalized to DAPI+ nuclei for each treatment group (n = 5 random fields); (D) Same as (C) but for Ki67+ and NeuN+ cells; (E) Immunoblot of differentiation markers (GFAP, NeuN, and MBP) in samples prepared from normal adjacent tissue and tumor lesions for each treatment group of mice; (F) Representative IF images showing NESTIN+ cells (red) and GFAP+ cells (green) in CTRL ASO-treated tumor and normal-adjacent tissue (left) and ASO5-treated tumor lesion (right); DAPI staining shows nuclei (blue); magnification-scale bars are as in (A); (G) Kaplan-Meier survival analysis of mouse cohorts following CTRL ASO treatment (N = 22) or ASO5 treatment (N = 21). Cell counts were analyzed by ANOVA, followed by t-tests for the pairwise comparisons, and data are presented as means ± SEM. Probability of survival was compared using log-rank survival estimate.
In addition, we observed significantly (P <0.001) elevated expression of the microglia-activation marker Aif1 induced by ASO5 compared to the control ASO in mRNA from tumor tissue, but not in mRNA from normal adjacent tissue (fig. S5A). We further observed a significant (P < 0.001) elevation of several murine genes related to cytokines and A2-specific reactive astrocytes (19) (fig. S5B), suggesting that the ASO5 treatment somehow triggers a tumor-intrinsic immune response. To investigate the functional role of the activated microglia, we measured the expression of mRNAs encoding a panel of pro-inflammatory cytokines that inhibit tumor growth, including IL-1β, TNF-α, INF-γ, IL-12α, CXCL10, and CXCL11 (22-25). We observed significant (P < 0.0121 to < 0.0001) upregulation of these markers, consistent with anti-tumor immune activity of the activated microglia (fig. S5C).
ICV injection of ASO5 decreased the NESTIN+ cell population and extended the latency of tumor growth
DMGs arise within defined spatial-temporal contexts and occur during middle childhood, with an incidence peak at six to nine years of age. The cellular origin and the microenvironment are essential for tumor growth (26, 27). Retrospective clonal analysis previously revealed that NESTIN+ cells are enriched in the human midbrain, pons, and medulla throughout childhood, with peak density in the ventral pons; thus, the NESTIN+ cell population corresponds precisely with the spatial and temporal incidence of DIPG (27). In our mouse model, the NESTIN+ cells were enriched at the location of tumor lesions, relative to the normal adjacent brain tissue. After ASO5 treatment, the NESTIN+ cell population was visibly reduced, inversely correlating with the high number of GFAP+ cells (Fig. 4F). Moreover, ASO5-treated mice had significantly (P < 0.0001) longer survival than control-ASO-treated mice (Fig. 4G). These results suggest that H3.3K27M tumors originate from neural stem cells (NESTIN+) in the context of TRP53 loss and PDGF signaling.
The differentiated cells resulting from ASO5 treatment in the RCAS-Tva model are of tumor origin.
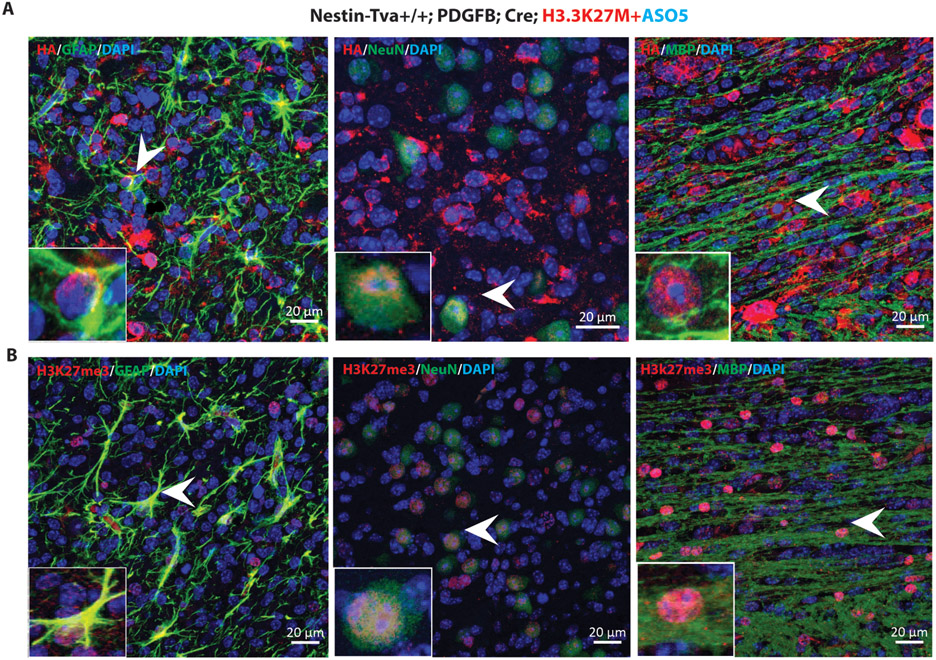
To address whether the differentiated cells seen after ASO5 treatment were of tumor origin, as opposed to murine cells recruited to the lesions, we took advantage of the hemagglutinin (HA)-tagged Pdgfb and Cre cDNAs expressed in the RCAS-Tva model. We performed IF staining with HA antibody, and observed that a large fraction of the tumor cells were HA+. When we double-labeled the cells with HA and individual differentiation markers, we observed numerous HA+ astrocytes, neurons, and oligodendrocytes (Fig. 5A; individual channels are shown in fig. S6A).
Fig. 5. ASO5 treatment promoted differentiation of tumor cells in the RCAS-Tva model.
(A) Representative tumor sections were co-stained with HA-tagged antibody (red for PDGFB and Cre), either GFAP, NeuN, or MBP antibodies (green), and DAPI for nuclei (blue). Arrowheads point to representative cells with co-localization of the green and red signals; (B) Representative tumor sections co-stained with H3K27me3 (red) and either GFAP, NeuN or MBP (green), plus DAPI (blue). Scale bars, 20 μm.
Next, to investigate whether H3K27me3 is restored in the differentiated cells, we performed double IF labeling of H3K27me3 with each of the above differentiation makers. We observed H3K27me3+ signal in GFAP+, NeuN+, and MBP+ cells (Fig. 5B). We conclude that ASO5 treatment inhibited proliferation of the tumor cells and promoted their differentiation.
ICV injection of ASO5 promoted astrocyte, neuron, and oligodendrocyte differentiation, and decreased tumor proliferation in a DIPG patient-derived xenograft model
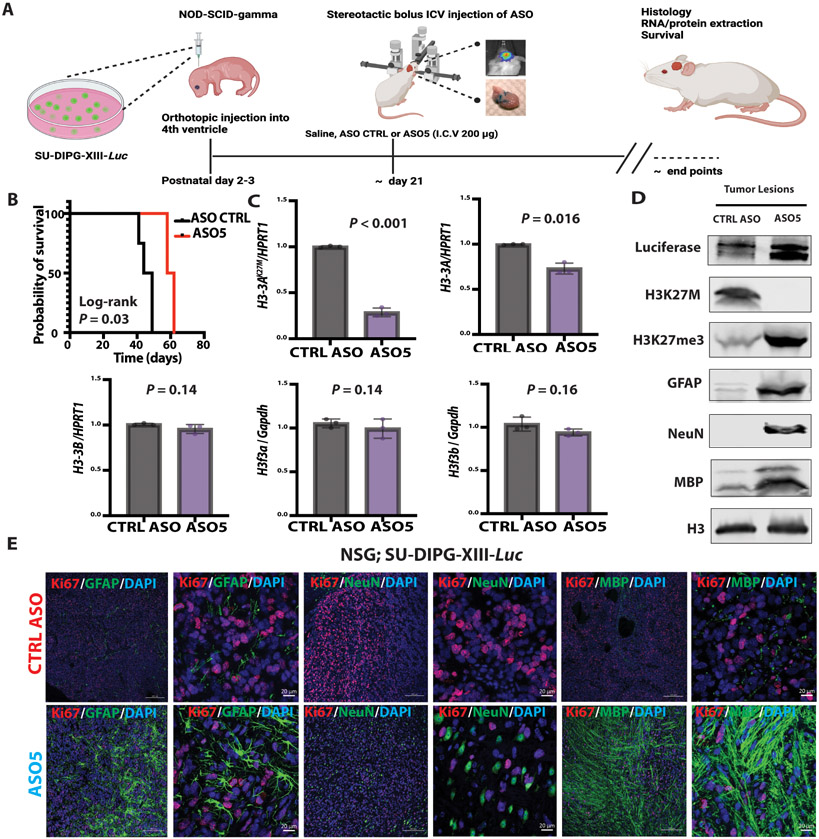
To confirm and extend the above observations, we also tested ASO5 in an orthotopic xenograft mouse model, as described (9), using one of the SU-DIPG patient lines shown in Fig. 3. We injected 105 luciferase-expressing SU-DIPG-XIII cells into the 4th ventricle of P3 immunocompromised NSG mice and used bioluminescence imaging to follow tumor onset. Because these immunocompromised mice did not tolerate the high ASO concentration used in the RCAS-Tva model, we administered a single ICV injection of 200 μg of control ASO or ASO5 (Fig. 6A). The mice treated with ASO5 survived significantly (P = 0.03) longer than the control-ASO-treated cohort (Fig. 6B). Similar to the RCAS-Tva mouse model, ASO5 significantly (P < 0.001; P = 0.016) knocked down the human H3-3AK27M mRNA and total human H3-3A mRNA, but did not affect endogenous murine wild-type H3f3a or H3f3b (Fig. 6C). Moreover, ASO5 knocked down human H3.3K27M protein and increased the extent of H3K27me3 modification. ASO5 treatment also resulted in elevated GFAP, NeuN, and MBP protein abundance in the Luciferase-labeled SU-DIPG-XIII line (Fig. 6D). The ASO5-treated mice exhibited a differentiated phenotype in the tumor lesions (GFAP+, NeuN+, and MBP+), with fewer proliferating cells (Ki67+), whereas neurogenesis and gliogenesis were compromised in the control-ASO-treated cohort, and most of the tumor cells remained in a proliferative state (Fig. 6E). Similar to the RCAS-Tva model, the cells expressing one of the various differentiation markers were also H3K27me3+ (fig. S6B).
Fig. 6. ICV administration of ASO at the time of tumor onset induced human-specific astrocyte, neuron, and oligodendrocyte differentiation, decreased tumor cell proliferation, and extended the latency of tumor growth in a patient-derived xenograft mouse model.
(A) A single dose (200 μg) of lead ASO or CTRL ASO in saline was stereotactically injected ICV at the time of tumor onset (~day 21) in SU-DIPG-XIII-Luc xenografted mice; (B) Kaplan-Meier survival analysis after CTRL ASO (N = 5) or ASO5 (N = 5) treatment; (C) Summary of data of mRNA expression detected by RT-qPCR (n = 3) of H3-3AK27M allele, total H3-3A, and H3-3B, normalized to HPRT1, and of endogenous murine H3f3a and H3f3b normalized to Gapdh expression (N = 3 mice per group); (D) Immunoblot for histones, transduced luciferase, and differentiation markers (GFAP, NeuN, and MBP) in tissue samples prepared from CTRL ASO-treated or ASO5-treated tumor lesions; (E) Representative IF images showing GFAP+, NeuN+ and MBP+ cells (green) and proliferation by Ki67 staining (red) after CTRL ASO (top row) or ASO5 treatment (bottom row); DAPI staining shows nuclei (blue) (low-magnification images: scale bar, 200 μm; high-magnification images: scale bar, 20 μm). For RT-qPCR experiments, the measurements for each experimental group/treatment were analyzed by Welch’s two-sample t-test, and data are presented as means ± SEM. Probability of survival was compared using log-rank survival estimate.
We conclude that treatment with ASO5 resulted in effective depletion of H3.3K27M protein, leading to restored neurogenesis and gliogenesis, longer latency of tumor growth, and increased survival in these mouse models. (fig. S7).
Discussion
The recognition that H3.3K27M-mediated aberrant gene activation or de-repression is an oncogenic driver in DMG motivated us to develop a direct strategy to deplete the mutant histone H3.3. We confirmed that H3.3K27M is required by tumors, using genetic knockout with CRISPR-Cas9 and sgRNA targeting both H3-3A alleles. In agreement with previous work (5, 6), we showed that H3-3A-knockout DIPG cells remain viable, but become less proliferative and more differentiated, thus extending survival of an orthotopic-xenograft mouse model. The H3-3AK27M mutation is dominant-negative, whereas the H3-3AWT allele might be redundant in the tumors and normal cells. This is because H3-3A and its paralog, H3-3B, encode identical H3.3 histone proteins, and are ubiquitously and similarly expressed across different cell lineages, including in the central nervous system (CNS). Moreover, single knockout H3f3a or H3f3b male and female mice are normal and fertile; only double-knockout mice show developmental retardation and embryonic lethality (28). Thus, targeting H3-3A would still allow H3-3B to express normal H3.3 protein to carry out its functions in various tissues. This assumption led us to explore both allele-specific and gene-specific targeting of H3-3A, using chemically modified gapmer ASOs. Our systematic ASO screen with allele-specific design showed preferential targeting of the mutant allele. In contrast, the gene-specific design had similar effects on both alleles, except for one ASO that somehow preferentially knocked down the mutant allele.
Depleting mutant H3.3K27M restored the balance of post-translational modifications (PTMs) at K27 on all histone H3 proteins. Brown et al. demonstrated the specificity of the K27M mutation, as different amino-acid substitutions of, or surrounding, K27M failed to inhibit EZH2 in vitro—the proposed mechanism for K27M’s dominant-negative toxicity (29). Furthermore, Grasso et al. reported that the HDAC inhibitor Panobinostat restores K27 trimethylation, inhibits proliferation in vitro, and increases survival in a DIPG orthotopic xenograft model (9). Our work confirms and extends these results, demonstrating that direct targeting of H3-3AK27M mRNA also restores K27 trimethylation, inhibits tumorigenesis, and promotes differentiation. The differentiated phenotype we observed in H3.3K27M-depleted patient cells and mouse models treated with gapmer ASO suggests that some downstream genes are associated with neurogenesis and/or gliogenesis.
A previous study employed genetically engineered mice to show that H3.3K27M enhances neural-stem-cell self-renewal and accelerates spontaneous brain-stem gliomas (16). The same group also reported a paired isogenic comparison using shRNA knockdown of H3.3K27M to address the underlying mechanism of the mutation-specific effect on the transcriptome and epigenome (6). Silveira et al. used ChIP-seq and RNA-seq analyses to identify highly enriched genes associated with neurogenesis and nervous-system development upon H3.3K27M shRNA knockdown. Likewise, another study reported that H3K27M gliomas are derived from oligodendrocyte precursor cells (OPC) (26). Single-cell RNA-seq of primary patient cells showed that large undifferentiated OPC-like cells are over-represented in H3K27M gliomas and exhibit more proliferation and oncogenic properties than their more differentiated counterparts, in the presence of PDGFRA signaling. Our ASO-mediated H3.3K27M depletion rescued the impaired differentiation programs, resulting in slower glioma proliferation. Known H3K27me3-marked genes that inhibit differentiation and maintain self-renewal, such as those encoding ID proteins (5), were downregulated in ASO-treated DIPG-patient cells. Our study, together with the previous work, suggests that the H3.3K27M oncohistone drives tumorigenesis by enhancing the self-renewal capacity of neural stem cells and blocking neural/glial differentiation.
Administration of the lead ASO to an immunocompetent DIPG mouse model generated by viral transduction reduced tumor growth, promoted neural-stem-cell differentiation, and increased survival. The RCAS-Nestin Tva mouse model develops midline high-grade gliomas by viral infection of endogenous NESTIN+ cells in a relevant brain-development window and environment (14, 15). Here we adopted this model by incorporating human H3-3AK27M cDNA—whose transcripts can be targeted by our human-specific ASOs—in addition to Pdgfb cDNA and TRP53 depletion. NESTIN marks a neuroepithelial stem-cell population with self-renewal capacity and the potential to generate differentiated cells. In the mouse model, we observed that NESTIN+ cells are abundant in the tumor lesions, reflecting the self-renewal capacity of neural stem cells, but are absent in the normal adjacent tissues. Thus, NESTIN+ cells are overrepresented in tumor lesions that maintain a highly proliferative state in the presence of the H3.3K27M mutation. When we treated mice with ASO5 to deplete H3.3K27M, there was a decrease in NESTIN+ cells and a corresponding increase in GFAP+ cells.
Several neural stem-cell markers, including NESTIN, are expressed in tumor-forming cells in patients (27). Moreover, DIPG-like gliomas can develop from induced pluripotent stem cell (iPSC)-derived induced neural stem cells (NSC), when overexpressing H3.3K27M with TP53 depletion (30). These studies showed that H3.3K27M can block neural stem-cell differentiation and keep the NESTIN+ tumors in the self-renewal state. Consistent with this notion, ASO5 treatment of the RCAS-Tva mice extended survival by converting highly proliferative neural stem cells into more differentiated cells.
We confirmed this differentiation process using a different mouse model generated by orthotopic transplantation of DIPG patient cells. The xenografts were generated with neurospheres passaged in culture and derived from tumors that were lethal to the patients, compared to acute viral transduction with two driver oncogenes plus knockout of a tumor suppressor in the immunocompetent RCAS-Tva mice. The immunocompromised mice used for the xenografts did not tolerate the maximum dose (500 μg) we used in the RCAS-Tva mice. Therefore, we used a lower dose (200 μg) for a single ICV bolus injection. These procedural differences probably account for the stronger effect of ASO5 on survival in the RCAS-TVA
We demonstrated that the differentiated cells are of tumor origin in the RCAS-Tva mouse model, by double labeling the tumor cells with HA antibody against the viral-vector tag for the Pdgfb and Cre cDNAs, plus antibodies to various differentiation markers, which labeled the same cells. In addition, the differentiated cells were also co-labeled with H3K27me3 and the three lineage differentiation markers in both mouse models, indicating that the restored H3K27me3 mark correlated with tumor-cell differentiation. Given that the impairment of normal cellular differentiation may contribute to the development of cancer, therapeutic agents that facilitate or restore normal differentiation represent a promising approach, which to date has been primarily explored for hematological malignancies (31).
ASOs with appropriate chemical modifications have a long duration of action in the CNS. For example, nusinersen (Spinraza), which targets SMN2 pre-mRNA, maintains its effect for >6 months after ICV infusion or injection in adult SMN2-transgenic mice (17, 18). Similarly, our lead gapmer ASO maintained H3-3A knockdown for at least 90 days after a single 500-μg dose in the RCAS-Tva mouse model. We performed ICV injection to directly deliver the ASO into CSF, bypassing the blood-brain barrier (BBB). This route allows ASO penetration into the brain and other CNS tissues, with less exposure in peripheral tissues (which express only wild-type H3.3 histone). Subsequently, ASO is cleared through the CSF flow tracts to the venous blood (32). Some ASO in the blood circulation may then return to the tumor through its vasculature and compromised BBB, potentially contributing to the overall knockdown effects in the tumors. Indeed, we observed that robust vascular proliferation dissects the tumors into pseudo-lobules in our RCAS-Tva mouse model, suggesting angiogenesis in the tumor lesions.
Microglia, the most abundant innate immune cell in the CNS, can influence BBB function; when microglia are activated, they cause either BBB repair or disruption during inflammation, the latter increasing BBB permeability (33, 34). However, DMGs are “cold” tumors, characterized by immunosuppression (35). We observed that ASO5, but not control ASO, treatment triggered neuroinflammation in the RCAS-Tva mouse model, as seen by Aif1 elevation. However, ASO5 did not cause microglia activation in normal adjacent tissue, so the response in the tumor tissue is not directed to the ASO. Upregulation of genes related to cytokines and A2-specific reactive astrocytes further suggested that the ASO treatment triggers a tumor-intrinsic immune response.
Activated microglia can have dual roles in high-grade gliomas: they can exhibit anti-tumor activity by phagocytosis and release of pro-inflammatory cytotoxic factors; or they can be polarized into tumor-supportive cells by releasing immunosuppressive cytokines (22-25). In our study, microglia activation was associated with anti-tumor activity involving the release of the pro-inflammatory cytokines IL-1β, TNF-α, INF-γ, and IL-12α.
Our study had some limitations. First, we employed two acute DIPG mouse models that developed brain tumors in approximately three weeks. Although this rapid rate of tumor growth accelerated the study, it may have limited the overall efficacy of the experimental therapy. Second, we used only one patient-derived line to establish the orthotopic xenograft model. It will be of interest to test additional lines, so as to establish the general effectiveness of the antisense therapy, and also to compare the results in H3-3AK27M tumors with different additional oncogenic drivers and growth rates. Third, we used single doses of ASO close to the maximum tolerability for these mouse strains. Although the long CNS half-life of this type of ASO exceeds the observed survival, detailed dose-response and multiple-dose studies should be conducted in the future. Finally, the differentiation of ASO-treated neurospheres could only be followed over a short time course, because neurospheres grown in culture for more than seven days fuse together and form necrotic cores, whereas passaging selects for proliferative cells. In addition, neurosphere cultures and mouse brain tissues were collected in bulk for the RNA and protein analyses. Single-cell RNA-seq may provide the resolution for detailed characterization of treatment-induced tumor-cell differentiation along neuronal and glial lineage pathways in vitro and in vivo.
In summary, we developed gapmer ASOs to directly target an oncohistone mRNA in vitro and in vivo. The lead ASO efficiently degraded H3-3AK27M mRNA, reducing H3.3K27M protein in patient-derived cells and mouse models. The decrease in H3.3K27M protein abundance resulted in restored neurogenesis and gliogenesis, a longer latency of tumor growth, and increased survival in mouse models. The pharmacological intervention was less effective than complete genetic knockout, which was expected, because ASO treatment reduces but does not eliminate expression of the mutant protein. Furthermore, unlike the pre-implantation genetic knockout, therapeutic ASO is administered after tumor onset, which represents a more realistic clinical scenario. We anticipate that maximal clinical efficacy will likely require combination therapy involving ASO administration with, for example, radiotherapy (36) or CAR-T cell immunotherapy (37, 38).
Materials and Methods
Study Design
The aim of this preclinical translational project was to develop lead ASOs as a targeted therapy for H3.3K27M pediatric brain cancers. For in vitro experiments, RT-qPCR, radioactive RT-PCR, viability, EdU staining, and other experiments were performed in biological triplicates. For in vivo experiments, tumor growth was monitored via bioluminescence imaging. Mice were monitored daily and euthanized with CO2 when they became symptomatic (including an enlarged head, ataxia, or weight loss up to 25%) or at 6 months post-injection if they remained asymptomatic. Mice were excluded when hydrocephalus was observed. For both models, mice of both sexes were randomized to each group (CTRL ASO or ASO5) before treatment (N = 5 for IHC/IF, RNA, and protein analysis; CTRL ASO (N = 22) or ASO5 (N = 21) for the survival study in the RCAS-TVA model; and CTRL ASO (N = 5) or ASO5 (N = 5) for the survival study in the DIPG orthotopic xenograft mouse model). IF/IHC analyses were performed blinded. All animal procedures were performed with approval from Cold Spring Harbor Laboratory’s Institutional Animal Care and Use Committee (IACUC).
RCAS-Tva mouse model.
DF1 cells (ATCC Catalog #CRL-12203) were cultured in DMEM supplemented with 10% FBS, 2 mM L-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin, and incubated at 39 °C and 5% CO2. 5 μg of each RCAS plasmid was transfected into DF1 cells using X-TremeGENE 9 (Roche) following the manufacturer’s instructions. The Nestin-Tva; p53fl/fl mouse strain was a generous gift from Dr. Oren Becher at Northwestern University. For the generation of midline gliomas, transfected cells were passaged at least three times prior to injection; 105 virus-producing DF1 cells were injected intracranially into a depth of 1-3 mm below the lambda suture of neonatal N-tva; p53fl/fl pups (P3–5) in 1 μL, using a Hamilton syringe (7659-01) and a 30-gauge needle. Four virus-producing cell lines expressing RCAS-Pdgfb, RCAS-Cre, RCAS-H3-3AK27M, and Rcas-Luciferase, were injected together in equal amounts.
DIPG orthotopic xenograft mouse model.
This procedure was carried out as described (9). Briefly, we prepared a single-cell suspension of luciferase-transduced SU-DIPG-XIII-luc neurospheres (pLenti PGK V5-LUC Neo (w623-2); Addgene, plasmid #21471), and injected 105 cells (50,000 cells/μL) into the fourth ventricle/pons of immunocompromised NOD-SCID-gamma (NSG) (strain 005557; The Jackson Laboratory), cold-anesthetized, P3 mouse pups through a 30-gauge burr hole (stereotactic coordinates: 3 mm posterior to the lambda suture and 3 mm deep).
Antisense oligonucleotides.
PS-MOE ASOs were purchased from IDT. ASOs synthesized in large scale for animal work were purified by HPLC. We dissolved the ASOs in water and diluted them in saline before use. A list of oligonucleotide sequences is provided in Supplementary Table S1. The ASOs tested in mice were: ASO5 (MOE/PS-DNA/PS-MOE/PS: 5-10-5; GGCGCACTCATGCGAGCGGC) and Control ASO (MOE/PS-DNA/PS-MOE/PS: 5-10-5; CCTTCCCTGAAGGTTCCTCC).
Primary pediatric human glioma cell lines.
SU-DIPG-XIII, SU-DIPG-XXXV, and SU-DIPG-L patient cells heterozygous for the H3-3A mutation (A>T) and derived from autopsy tissue, were a generous gift from Dr. M. Monje at Stanford University, in accordance with informed-consent protocols and in compliance with Stanford University and Cold Spring Harbor Laboratory Institutional Review Board human-subject protocols. The cells were grown as neurospheres in tumor stem media (TSM) consisting of DMEM/F12 (Invitrogen), Neurobasal (-A) (Invitrogen), B27 (-A) (Invitrogen), human-bFGF (20 ng/mL; Protech), human- EGF (20 ng/mL; Peprotech), human PDGF-AB (20 ng/mL; Peprotech), and heparin (10 ng/mL; Stemcell). The point mutation in H3-3AK27M was confirmed by Sanger sequencing using primers listed in Supplementary Table S2.
Lentivirus preparation and Infection.
Two 20-nucleotide gRNA pairs against human H3-3A were annealed and cloned in the pSpCas9 (BB) vector (pX459; Addgene plasmid #62988) expressing Cas9 lentiviral constructs; Cas9-resistance gRNA pairs were annealed and cloned in the lentiV-neo vector (Addgene plasmid #108101). Lentiviral particles were generated by co-transfection of lentiviral-expressing constructs with packaging plasmids (pspAX2, VSV-G) into HEK-293T cells, and then concentrated by polyethylene glycol (PEG-it) precipitation (SBI). For lentiviral infection, dissociated DIPG cells were seeded on a 1% Matrigel-coated plate (Corning), and incubated with gRNA-expressing lentivirus for 12 h before replacing with fresh medium. Puromycin (0.5 μg/ml) was added at 48 h post-infection to select infected cells. After 7 days, puromycin was removed and the cells were allowed to recover in regular growth medium. Bulk cells were used by immunostaining, western blotting, and functional assays.
Transfection and free uptake of ASOs.
HeLa cells were grown to 70-80% confluence in 12-well plates, and transfected for 3 days with 2 μL of Lipofectamine 2000 transfection reagent (Invitrogen) and different amounts of ASOs, ranging from 30 nM to 150 nM, following the manufacturer’s recommendations. For free uptake of ASOs, patient cells were dissociated into single cells using TrypLE Express (Invitrogen), and 15,000/well cells were seeded in a 96-well plate and incubated at 37 °C for 1 h; 4-10 μM ASO was then added for 3 to 5 days, cell medium was replaced with fresh medium, and a second ASO dose was added on day 3.
Cell viability and proliferation assay.
Primary patient cells were starved in TSM base with B27 for 3 days. Then, 3,000 cells/well were plated in a 96-well plate in TSM base with normal growth medium with EGF, FGF, and PDGF-AB, as described (39). Cell viability and growth were measured using a cell-counting kit (CCK-8; Sigma #96992) at set time points.
Bioluminescence imaging.
D-Luciferin was reconstituted as per the manufacturer’s protocol (Goldbio, LUCK-100) and administered intraperitoneally (10 μg/g body weight) into isoflurane-anesthetized animals, 12 min prior to imaging. Animals were excluded if no tumors were present.
Intracerebroventricular injection of ASOs.
After confirming the presence of tumors, tumor-bearing mice were randomized and treated with a single ICV injection of CTRL ASO or ASO5 (500 μg for the RCAS-TVA mouse model; 200 μg for the orthotopic xenograft model) using a Hamilton syringe with a 28-gauge burr hole needle in isoflurane-anesthetized animals (stereotactic coordinates: 1.0 mm posterior to the bregma, 0.2 mm lateral, and 3 mm in depth).
Immunofluorescence and immunohistochemistry.
Tumor tissue was fixed in 4% paraformaldehyde, cut into 5-μm sections, and embedded in paraffin. IHC was performed using heat-induced antigen retrieval with sodium citrate buffer, followed by primary antibodies to GFAP (1:1000; Millipore Sigma, rabbit polyclonal, AB5804), NeuN (1:100; Sigma, rabbit, monoclonal, 13E6), MBP (1:5000; Abcam, rabbit monoclonal, EPR21188), Ki67 (1:50; BD biosciences, mouse monoclonal, B56), NESTIN (1:100; R&D, mouse monoclonal MAB2736), HA tag (1:300;Thermo Fisher, mouse monoclonal, 2-2.2.14), OLIG2 (1:500; Millipore, rabbit polyclonal), or H3K27me3 (1:1000; CST rabbit polyclonal, C36B11; or 1:500; Abcam, mouse monoclonal, ab6002 for co-staining experiments). For IHC, the signal was visualized with HRP-labeled anti-rabbit polyclonal (1:200; Agilent, P0448) and DAB (Agilent, K346711). Slides were counterstained with hematoxylin (Sigma) and captured on a Zeiss Observer microscope. For IF, the signal was visualized with a fluoro-conjugated secondary antibodies (Thermo Fisher, Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed, Alexa Fluor Plus 555, A32727 or Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed, Alexa Fluor Plus 488, A32731) and captured on a Zeiss LSM780 confocal laser-scanning microscope.
EdU-staining assays.
Primary human glioma cells (SU-DIPG-XIII, 5×103 cells/well) were seeded onto 1% matrigel-coated 8-well chamber slides (Falcon) and treated with 4 μM ASO by free uptake for five days. On day 5, 10 μM EdU was added to the cells, and incubated at 37 °C for 2 h. EdU incorporation was measured using a Click-it Plus EdU Alexa Fluor 594 Imaging Kit (Invitrogen) in accordance with the manufacturer’s instructions. Images were captured on a Zeiss Observer microscope. All images within the same figure panel were taken with the same exposure setting, and identically processed using Image J software.
Soft-agar assay.
Primary human glioma cells (SU-DIPG-XIII, 103 cells/well) were incubated in an upper layer of 0.3% agar (ThermoFisher Scientific) in TSM. The bottom layer consists of the same medium with supplements, but 0.6% solidified basal agar, in a 12-well plate. Plates were incubated at 37 °C/5% CO2 for at least 3 weeks, before staining with crystal violet. Visible colonies were then counted.
RNA and protein extraction.
Cells or tissues were harvested at the end points and snap-frozen in liquid nitrogen. For RNA extraction, 1 mL of Trizol (Invitrogen, 15596-018) was added to homogenized brain tissue or cells, following the standard Trizol protocol with chloroform extraction, isopropanol precipitation, and 70% EtOH RNA-pellet wash. RNA was resuspended in 20-40 μL of nuclease-free water. For protein extraction, cells or tissues were harvested and lysed on ice using Triton Extraction Buffer (TEB: PBS containing 0.5% Triton X 100 (v/v), protease inhibitor cocktail (Roche)), followed by centrifugation at 6,500 x g for 10 min at 4 °C to spin down the nuclei; the supernatant was removed and discarded; the pellet was resuspended in 0.2 N HCl to perform acid extraction overnight; the supernatant was collected after centrifugation at 6,500 x g for 10 min at 4 °C; and the protein concentration was measured by Bradford assay (Bio-Rad).
Radioactive RT-PCR and RT-qPCR.
Total RNA was extracted from cells or tissues as described above, and reverse-transcribed with ImProm-II reverse transcriptase (Promega) using oligo-dT primers. Total H3-3A cDNA was amplified with AmpliTaq DNA polymerase (Thermo Fisher) using Fwd 5’- GGACTTTAAAACAGATCTGCGCTT and Rev 5’- GTCTTTTGGCATAATTGTTACACGT primers that sit on exon 3 and exon 4 (downstream of the H3-3A mutation site in exon 2), respectively. The H3-3AWT allele was amplified using Fwd 5’-GCTACAAAAGCCGCTCTCAA; the H3-3AK27M mutant allele was amplified using Fwd 5’- GCTACAAAAGCCGCTCGAAT; and the same Rev 5’- CCAGACGCTGGAAGGGAAGT primer was used for both. cDNA from minigenes was amplified using vector-specific (pcDNA3.1) primers, listed in Supplementary Table 2. For radioactive PCR, 0.16 μL of 250uCi [α-32P]-dCTP (PerkinElmer, NEG-013H) was added to a 20-μL PCR reaction. Amplicons were separated by 5% native PAGE (Bio-Rad), followed by phosphorimage analysis on a Typhoon 9410 phosphorimager (GE Healthcare). Band intensities were quantified using Image J, and the values normalized for the G+C content according to the DNA sequence. For RT-qPCR, 2x SYBR green master mix (Applied Biosystems) was used, and the cDNA was analyzed on a QuantStudio 6 Flex Real-Time PCR system (ThermoFisher Scientific). Fold changes were calculated using the ΔΔCq method.
Western Blotting.
One microgram of acid-extracted protein (6) was run on a 8-20% precast protein gel (Bio-Rad), transferred onto a nitrocellulose membrane, and probed with rabbit polyclonal anti-H3K27M (1:1000; ABE419, Millipore), rabbit monoclonal anti–H3K27me3 (1:1000; C36B11, CST) or rabbit polyclonal H3 (1:1000, ab1791, Abcam) antibodies. The membranes were incubated with infrared-dye-conjugated secondary antibody (1:10000; LI-COR Biosciences, 926-32211), and protein bands were visualized by quantitative fluorescence using Odyssey software (LI-COR Biosciences). Molecular weight markers confirmed the sizes of the bands. Band intensities were quantified using Image J and normalized to total H3 protein.
Statistical analyses
were performed with advice from a biostatistician. Raw data for experiments in which n < 20 are presented in Data File S1. For IHC/IF experiments, cells were counted in 5 randomly picked fields at 40x magnification. The measurements for each experimental group/treatment were analyzed by ANOVA, followed by pairwise comparisons using two-sample t-tests. For viability assays, P-values were adjusted for multiple comparisons by controlling for family-wise error rate using the single-step method. For RT-qPCR experiments, Welch’s two-sample t-test was used to compare two groups. For more than two groups, the measurements for each experimental group/treatment were analyzed by ANOVA, followed by pairwise comparisons using Welch’s two-sample t-tests. Family-wise error rate was adjusted using the Bonferroni-Holm method. For IHC, IF, viability and RT-qPCR, the results are displayed as means ± SEM. For in vivo experiments, we used the Kaplan-Meier estimator to calculate survival differences between cohorts, using a log-rank test and median-survival rate. P values are reported in the figure legends.
Supplementary Material
Acknowledgments
We thank M. Monje for generously sharing DIPG patient cell lines, and O. Becher for sharing the RCAS-Tva mouse strain and protocol. We thank T. Ha for help with statistical analyses. We are grateful to K. O’Connell and J. Lombard for bringing DIPG to our attention, and for encouragement during the course of this research.
Funding
We gratefully acknowledge support to A.R.K. from the Cure Starts Now Foundation, the Simons Foundation (552716), the V Foundation (T2021-001), and the St. Giles Foundation. This work was performed with assistance from the CSHL Animal, Histology, and Microscopy Shared Resources, funded in part by an NCI Cancer Center Support Grant (5P30CA045508).
Footnotes
Publisher's Disclaimer: * This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior written permission of AAAS.
Competing interests
Related to this work: Q.Z. and A.R.K. are inventors in a patent application (Antisense oligonucleotide therapy for H3.3 K27M diffuse midline gliomas; PCT/US2022/041273). Unrelated to this work: A.R.K. is a co-founder, Director, Chair of the SAB, and equity holder of Stoke Therapeutics; a member of the SABs of Skyhawk Therapeutics, Envisagenics, Autoimmunity Biologic Solutions, and Assembl.cns; and a consultant for Biogen.
Data and materials availability
All data associated with this study are present in the paper or the Supplementary Materials. DIPG patient-derived lines were provided by Dr. M. Monje under an MTA with Stanford University. The Nestin-Tva; p53fl/fl transgenic mouse strain was provided by Dr. O. Becher under an MTA with Northwestern University.
References
- 1.Phillips RE, Soshnev AA, Allis CD, Epigenomic reprogramming as a driver of malignant glioma. Cancer Cell 38, 647–660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. http://www.danafarberbostonchildrens.org/conditions/brain-tumor/diffuse-pontine-glioma.aspx .
- 3.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD, Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A, Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–1070 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harutyunyan AS, Krug B, Chen H, Papillon-Cavanagh S, Zeinieh M, De Jay N, Deshmukh S, Chen CCL, Belle J, Mikael LG, Marchione DM, Li R, Nikbakht H, Hu B, Cagnone G, Cheung WA, Mohammadnia A, Bechet D, Faury D, McConechy MK, Pathania M, Jain SU, Ellezam B, Weil AG, Montpetit A, Salomoni P, Pastinen T, Lu C, Lewis PW, Garcia BA, Kleinman CL, Jabado N, Majewski J, H3K27M induces defective chromatin spread of PRC2-mediated repressive H3K27me2/me3 and is essential for glioma tumorigenesis. Nat. Commun 10, 1262 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silveira AB, Kasper LH, Fan Y, Jin H, Wu G, Shaw TI, Zhu X, Larson JD, Easton J, Shao Y, Yergeau DA, Rosencrance C, Boggs K, Rusch MC, Ding L, Zhang J, Finkelstein D, Noyes RM, Russell BL, Xu B, Broniscer A, Wetmore C, Pounds SB, Ellison DW, Zhang J, Baker SJ, H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 137, 637–655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castel D, Philippe C, Calmon R, Dret LL, Truffaux N, Boddaert N, Pagès M, Taylor KR, Saulnier P, Lacroix L, Mackay A, Jones C, Sainte-Rose C, Blauwblomme T, Andreiuolo F, Puget S, Grill J, Varlet P, Debily MA, Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 130, 815–827 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, Corcoran A, Jones DT, Sturm D, Johann P, Tomita T, Goldman S, Nagib M, Bendel A, Goumnerova L, Bowers DC, Leonard JR, Rubin JB, Alden T, Browd S, Geyer JR, Leary S, Jallo G, Cohen K, Gupta N, Prados MD, Carret AS, Ellezam B, Crevier L, Klekner A, Bognar L, Hauser P, Garami M, Myseros J, Dong Z, Siegel PM, Malkin H, Ligon AH, Albrecht S, Pfister SM, Ligon KL, Majewski J, Jabado N, Kieran MW, Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat. Genet 46, 462–466 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, Quist MJ, Davis LE, Huang EC, Woo PJ, Ponnuswami A, Chen S, Johung TB, Sun W, Kogiso M, Du Y, Qi L, Huang Y, Hütt-Cabezas M, Warren KE, Dret LL, Meltzer PS, Mao H, Quezado M, van Vuurden DG, Abraham J, Fouladi M, Svalina MN, Wang N, Hawkins C, Nazarian J, Alonso MM, Raabe EH, Hulleman E, Spellman PT, Li XN, Keller C, Pal R, Grill J, Monje M, Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat. Med 21, 555–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR, Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet 82, 834–848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett CF, Krainer AR, Cleveland DW, Antisense oligonucleotide therapies for neurodegenerative diseases. Annu. Rev. Neurosci 42, 385–406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts TC, Langer R, Wood MJA, Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov 19, 673–694 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahronian LG, Lewis BC, In vivo delivery of RCAS virus to mice. Cold Spring Harb. Protoc 2014, 1167–1169 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Hoeman CM, Cordero FJ, Hu G, Misuraca K, Romero MM, Cardona HJ, Nazarian J, Hashizume R, McLendon R, Yu P, Procissi D, Gadd S, Becher OJ, ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat. Commun 10, 1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordero FJ, Huang Z, Grenier C, He X, Hu G, McLendon RE, Murphy SK, Hashizume R, Becher OJ Histone H3.3K27M represses p16 to accelerate gliomagenesis in a murine model of DIPG. Mol. Cancer Res 15, 1243–1254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larso JD, Kasper LH, Paugh BS, Jin HJ, Wu G, Kwon CH, Fan YP, Shaw TI, Silveira A, Qu CX, Xu R, Zhu XY, Zhang JY, Russel HR, Peters J, Finkelstein D, Xu BS, Lin T, Tinkle CL, Patay Z, Onar-Thomas A, Pounds SB, McKinnon PJ, Ellison DW, Zhang JH, Baker SJ, Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell 35, 140–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR, Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR, Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478, 123–126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, nch AE, Chung WS, Peterson TC, Wilton DK, Frouin A, Napier BA, Panicker N, Kumar M, Buckwalter MS, Rowitch DH, Dawson VL, Dawson TM, Stevens B, Barres BA, Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, Zhang X, Xue Y, Maimon R, Dowdy SF, Devaraj NK, Zhou Z, Mobley WC, Cleveland DW, Fu XD, Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature 582, 550–556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarava N, Chen-Yu C, Molesworth K, Baskakov IV, Posttranslational modifications define course of prion strain adaptation and disease phenotype. J. Clin. Invest 130, 4382–4395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.France R, Fernández-Suárez D, Alternatively activated microglia and macrophages in the central nervous system. Prog. Neurobiol 131, 65–86 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Yu H, Pardoll D, Jove R, STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei J, Gabrusiewicz K, Heimberger A, The controversial role of microglia in malignant gliomas. Clin. Dev. Immunol 2013, 285246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowling EA, Wang JH, Gong F, Wu W, Neill NJ, Kim IK, Tyagi S, Orellana M, Kueley SJ, Dominguez-Vidan R, Chung HC. Hsu TYT, Dubrulle J, Saltzman AB, Li HY, Meena JK, Canlas GM, Chamakuri S, Singh S, Simon LM, Olson CM, Dobrolecki LE, Lewis MT, Zhang B, Golding I, Rosen JM, Young DW, Malovannaya A, Stossi F, Miles G, Ellis MJ, Yu LH, Buonamici S, Lin CY, Karlin KL, Zhang XH-F, Westbrook TF, Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell 184, 384–403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, Haberler C, Yizhak K, Gojo J, Egervari K, Mount C, Galen PV, Bonal DM, Nguyen QD, Beck A, Sinai C, Czech T, Dorfer C, Goumnerova L, Lavarino C, Carcaboso AM, Mora J, Mylvaganam R, Luo CC, Peyrl A, Popović M, Azizi A, Batchelor TT, Frosch MP, Martinez-Lage M, Kieran MW, Bandopadhayay P, Beroukhim R, Fritsch G, Getz G, Rozenblatt-Rosen O, Wucherpfennig KW, Louis DN, Monje M, Slavc I, Ligon KL, Golub TR, Regev A, Bernstein BE, Suvà ML, Development and oncogenic programs in H3K27M gliomas dissected by single-cell-RNA-seq. Science 360, 331–335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monje M, Mitra S, Freret ME, Raveh TB, Kim J, Masek M, Attema JL, Li G, Haddix T, Edwards MSB, Fisher PG, Weissman IL, Rowitch DH, Vogel H, Wong AJ, Beachy PA, Hedgehog-responsive candidate cell of origin for diffuse intrinsic pontine glioma. Proc. Natl. Acad. Sci. USA 108, 4453–4458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang CW, Shibata Y, Starmer J, Yee D, Magnuson T, Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29, 1377–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown ZZ, Müller MM, Jain SU, Allis CD, Lewis PW, Muir TW, Strategy for “detoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J. Am. Chem. Soc 136, 13498–13501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haag D, Mack N, Silva PBGD, Statz B, Clark J, Tanabe K, Sharma T, Jäger N, Jones DTW, Kawauchi D, Wernig M, Pfister SM, H3.3-K27M drives neural stem cell-specific gliomagenesis in a human iPSC-derived model. Cancer Cell 39, 407–422 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Eytan S, Yen K, Targeted differentiation therapy with mutant IDH inhibitors: Early experiences and parallels with other differentiation agents. Annu. Rev. Cancer Biol 1, 379–401 (2017). [Google Scholar]
- 32.Laterra J, Keep R, Betz LA, Goldstein GW, Blood—Cerebrospinal Fluid Barrier. In Basic Neurochemistry: Molecular, Cellular and Medical Aspects, 6th edition. Siegel GJ, Agranoff BW, Albers RW et al. , eds. (Philadelphia: Lippincott-Raven; 1999). [Google Scholar]
- 33.Iadecola C, The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron 96, 17–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haruwaka K, Ikegami A, Tachibana Y, Ohno N, Konishi H, Hashimoto A, Matsumoto M, Kato D, Ono R, Kiyama H, Moorhouse AJ, Nabekura J, Wake H, Dual microglia effects on blood brain barrier permeability induced by systemic inflammation. Nat. Commun 10, 5816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang SS, Bandopadhayay P, Jenkins MR, Towards immunotherapy for pediatric brain tumors. Trends Immunol. 40, 748–761 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Deland K, Starr BF, Mercer JS, Byemerwa J, Crabtree DM, Williams NT, Luo L, Ma Y, Chen M, Becher OJ, Kirsch DG, Tumor genotype dictates radiosensitization after Atm deletion in primary brainstem glioma models. J. Clin. Invest 131, e142158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mount CW, Majzner RG, Sundaresh S, Arnold EP, Kadapakkam M, Haile S, Labanieh L, Hulleman E, Woo PJ, Rietberg SP, Vogel H, Monje M, Mackall CL, Potent antitumor efficacy of anti-GD2 carT cells in H3-K27M+diffuse midline gliomas. Nat. Med 24, 572–579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Majzner RG, Ramakrishna S, Yeom KW, Patel S, Chinnasamy H, Schultz LM, Richards RM, Jiang L, Barsan V, Mancusi R, Geraghty AC, Good Z, Mochizuki AY, Gillespie SM, Toland AMS, Mahdi J, Reschke A, Nie EH, Chau IJ, Rotiroti MC, Mount CW, Baggott C, Mavroukakis S, Egeler E, Moon J, Erickson C, Green S, Kunicki M, Fujimoto M, Ehlinger Z, Reynolds W, Sreevidya Kurra S, Katherine E, Warren KE, Prabhu S, Vogel H, Rasmussen L, Cornell TT, Partap S, Fisher PG, Campen CJ, Filbin M, Grant G, Sahaf B, Davis KL, Feldman SA, Crystal L, Mackall CL, Monje M, GD2-CAR T cell therapy for H3K27M-mutated diffuse midline gliomas. Nature 603, 934–941 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagaraja S, Quezada MA, Gillespie SM, Arzt M, Lennon JJ, Woo PJ, Hovestadt V, Kambhampati M, Filbin MG, Suva ML, Nazarian J, Monje M, Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol. Cell 76, 965–980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.