Abstract
The outbreak of the corona virus disease (COVID-19) has changed the lives of most people on Earth. Given the high prevalence of this disease, its correct diagnosis in order to quarantine patients is of the utmost importance in the steps of fighting this pandemic. Among the various modalities used for diagnosis, medical imaging, especially computed tomography (CT) imaging, has been the focus of many previous studies due to its accuracy and availability. In addition, automation of diagnostic methods can be of great help to physicians. In this paper, a method based on pre-trained deep neural networks is presented, which, by taking advantage of a cyclic generative adversarial net (CycleGAN) model for data augmentation, has reached state-of-the-art performance for the task at hand, i.e., 99.60% accuracy. Also, in order to evaluate the method, a dataset containing 3163 images from 189 patients has been collected and labeled by physicians. Unlike prior datasets, normal data have been collected from people suspected of having COVID-19 disease and not from data from other diseases, and this database is made available publicly. Moreover, the method’s reliability is further evaluated by calibration metrics, and its decision is interpreted by Grad-CAM also to find suspicious regions as another output of the method and make its decisions trustworthy and explainable.
Keywords: COVID-19, CT scan, Deep learning, CycleGAN, Transfer learning
1. Introduction
The COVID-19 disease was initially spotted in December of 2019 in Wuhan, China, and was detected worldwide shortly after that [1]. In January 2020, the World Health Organization (WHO) stated its outbreak as a public health emergency and global concern, and later, a pandemic in March of 2020 [2]. SARS-CoV-2 is a novel strain of coronavirus that causes COVID-19; also, it has not been seen previously in humans [3]. Coronaviruses are common among animals, and some can infect humans [3], [4]. Bats are the natural hosts of these viruses, and several other species of animals have also been identified as sources [5], [6], [7]. For example, MERS-CoV-3 is transmitted from camels to humans, while SARS-CoV-14 is transmitted from intermediate hosts such as civet cats that were involved in the development of SARS-CoV-1 [8], [9]. The new coronavirus is genetically closely related to the SARS-CoV-1 virus [10].
The SARS-CoV-2 virus is transmitted mainly through respiratory droplets and aerosols from an infected person while sneezing, coughing, talking, or breathing in the presence of others [11], [12]. The virus can stay active at varying surfaces from a few hours to several days, and prior research has estimated the incubation period of this disease to be within 1 and 14 days [13]. However, the amount of live virus decreases over time and may not always be present in sufficient quantities to cause infection [14], [15].
The most frequent symptoms of COVID-19 include fever, dry cough, and fatigue. Pain, diarrhea, headache, sore throat, conjunctivitis, loss of taste or smell are other variable symptoms of the virus [16], [17]. Nevertheless, the most severe symptoms seen in COVID-19 patients include difficulty for breathing or shortness of breath, chest pain, and loss of movement or ability to speak [16], [17], [18].
Early diagnosis of this disease in the preliminary stages is vital. So far, various screening methods have been introduced for the diagnosis of COVID-19. At present, nucleic acid–based molecular diagnosis, by reverse transcriptase-polymerase chain reaction (RT-PCR) test, is considered the gold standard for early detection of COVID-19 [19], [20]. According to a WHO report, all diagnoses of COVID-19 must be verified by RT-PCR [21]. However, performing the RT-PCR test needs specialized equipment and equipped laboratories that are not available in most countries and takes at least 24 h to determine the test outcome. Also, the test result may not be accurate and may require to redo RT-PCR or other tests. Therefore, X-ray and CT-Scan imaging can be used as a primary diagnostic method for screening people suspected of having COVID-19 [22], [23].
X-ray imaging is one of the medical imaging techniques used to diagnose COVID-19 [24], [25]. X-ray imaging benefits include low cost and low risks of radiation that are dangerous to human health [26], [27]. In this imaging technique, the detection of COVID-19 is a relatively complicated task. An X-ray physician may also misdiagnose diseases such as pulmonary tuberculosis [28], [29].
CT-Scan imaging is used to reduce COVID-19 detection error. CT-scans have very high contrast and resolution, and are very successful in diagnosing lung diseases such as COVID-19 [30], [31], [32]. CT-Scan can also be used as a clinical feature of COVID-19 disease patients. CT scans of subjects with COVID-19 had shown marked destruction of the pulmonary parenchyma 6, such as interstitial inflammation and extensive consolidation [33]. During CT-Scan imaging of patients, multiple slices are recorded to diagnose COVID-19. This high number of CT-Scan images requires a high accuracy from specialists for accurate diagnosis of COVID-19. Factors such as eye exhaustion or a massive number of patients to interpret CT-Scan may lead to misdiagnosis of COVID-19 by specialists [34].
Due to the stated challenges, the use of artificial intelligence (AI) methods for accurate diagnosis of COVID-19 on CT-Scan or X-ray imaging modalities is of utmost importance. The design of computer aided diagnosis systems (CADS) based on AI using CT-Scan or X-ray images for precise diagnosis of COVID-19 has been highly regarded by researchers [35], [36], [37]. Deep learning (DL) is one of the fields of AI, and many research papers have been published on their application for diagnosing COVID-19 [38], [39].
In this paper, a new method of diagnosing COVID-19 from CT-Scan images using DL is presented. First, CT-Scan images of people with COVID-19 and normal people were recorded in Gonabad Hospital (Iran). Next, three expert radiologists have labeled the patients’ images. They have also selected informative slices from each scan. Then, after preprocessing data with a Gaussian filter, various deep learning networks were trained in order to separate COVID-19 from healthy patients. In this step, a CycleGAN [40], [41] architecture was first used for data augmentation of CT-Scan data; after that, a number of pre-trained deep networks [42] such as DenseNet [43], ResNet [44], ResNest [45], and ViT [46] have been used to classify CT-Scan images. Fig. 1 shows the block diagram of method. The results show that the proposed method of this study has promising and reliable results in detecting COVID-19 from CT-scan images of the lung.
Fig. 1.
Overall diagram of proposed method.
The rest of the paper is organized as follows. In the next section, we present a review of previous research on the diagnosis of COVID-19 from CT-Scan images using DL techniques. In Section 3, the proposed method of this research is presented. In Section 4, the evaluation process and the results of the proposed method are presented. Section 5 includes the discussion of paper and finally, the paper ends with the conclusion and future directions.
2. Related works
Prior research papers on the diagnosis of the COVID-19 disease using machine learning can be divided according to the algorithms used or the underlying modalities. Fig. 2 shows various types of methods that can be used for diagnosis of COVID-19. As can be seen in this figure, the methods based on medical imaging can be divided into two groups: CT scan and X-ray. The focus of this article is on CT scan modality. Also, machine learning algorithms can be divided into two categories: DL [47], [48], [49], [50] and conventional machine learning methods [51], [52], [53]. Due to the large number of machine learning papers for diagnosing COVID-19 disease from CT modality, we have only reviewed papers that have used deep learning methods for this imaging modality. Table 1 provides an overview of these papers, the datasets used by them, the components of methods, and finally, their performance. Moreover, some of those works are reviewed in the following:
Fig. 2.
Various criteria used for COVID-19 detection and their categories.
In [54], researchers introduced a method for detecting COVID-19 from CT images using pre-trained models. First, patches extraction was performed on CT images in the pre-processing step. Next, they used GoogleNet, VGG-16, and ResNet-50 models in the feature extraction step and then fused the feature together. Next, feature ranking and finally classification was done using SVM. In this work, they were able to achieve adequate results.
In another research, Yu et al. proposed a method for detecting COVID-19 from CT images using ResGNet-C architecture [55]. In this article, they used a clinical dataset to conduct experiments. The pre-processing of CT images for the diagnosis of COVID-19 is based on the visual inception method. Finally, the proposed ResGNet-C model has been selected for feature extraction and classification. In this work, in order to evaluate the results, K Fold with K=5 has also been used.
A new detection method for COVID-19 based on U-Net and dual-branch combination network architectures was introduced by Gao et al. [56]. In this work, all the experiments have been done on a clinical CT dataset containing the infected patients with COVID-19 and HC. They used a DA method to generate synthetic CT images in the pre-processing step. Next, U-Net and dual-branch combination network architectures were used to detect COVID-19. They also used attention maps as a post-processing step in their proposed method. The research results show the high efficiency of their proposed method in diagnosing COVID-19.
Next, Ouyang et al. presented a proposed 3D ResNet34 architecture for the detection of COVID-19 [57]. In this article, the tests are done on a clinical dataset of CT images. In the pre-processing step, segmentation and generating lung masks were performed on CT images. Next, the 3D architecture of ResNet34 along with attention layers was used to detect COVID-19. Also, the gradient-weighted class activation mapping (Grad-CAM) method was tested as a pre-processing step to detect suspected areas of COVID-19 in CT images accurately.
In Loey et al.’s research [58], a method of detecting COVID-19 based on transfer learning was proposed along with the traditional DA technique and conditional GAN. In this work, pre-processing of CT images includes resizing and conventional DA steps. Next, the conditional GAN model was used along with ResNet50 to detect COVID-19. The results of the proposed method show that they have achieved outstanding results in diagnosing COVID-19 from CT images.
In another research, a method for detecting COVID-19 from CT images using GAN architecture was introduced by Goel et al. [59]. In this work, experiments have been performed on the SARS-CoV-2 CT-Scan dataset. In this dataset, all CT images are pre-processed. Therefore, the researchers applied the CT images of the dataset to the input of a proposed DL architecture. The proposed DL architecture of this paper includes a GAN model along with the Whale optimization method. In the classification section, researchers also used K Fold with K=10 to evaluate the results.
Next, In [60], researchers first used different datasets to demonstrate the effectiveness of the proposed method for detecting COVID-19. This paper’s feature extraction from CT images is based on ResNet-50 architecture. Next, a 3D CNN architecture was used to classify features. In addition, K Fold with K=10 has been used to evaluate the proposed model in this work. The research results show the proper performance of their proposed method in diagnosing COVID-19.
Moreover, In another study, Amyar et al. used a new multi-task learning architecture to detect COVID-19 [61]. In the first step, different datasets of CT images were selected for experiments. Next, resizing and intensity normalization steps were used to pre-process CT data. Finally, a new DL architecture based on multi-task learning was proposed for CT data segmentation and classification. The research results show that the method proposed in this article has reached a high accuracy in diagnosing COVID-19 from CT images.
Lastly, In [62], the authors presented a proposed DL architecture for the detection of COVID-19 called Covid CT-net. In this research, researchers used the Kaggle dataset to conduct experiments. The pre-processing step of this work includes resizing, normalization, and DA steps. Next, the proposed architecture of the Covid CT-net for accurate diagnosis of COVID-19 from CT images is presented. Also, they used a post-processing step in this article and displayed the suspected areas of COVID-19 in the CT images as a heat map. In this work, they were able to achieve promising results.
3. Materials and methods
This section of the paper is devoted to discussing the applied method and its components. In this paper, we have firstly collected a new CT scan dataset from COVID-19 patients; then, from each scan, the informative slices were selected by physicians. After that, several convolutional neural networks pre-trained on the ImageNet dataset [63] were fine-tuned to the task at hand. Here, we trained a Resnet-50 architecture [44], an EfficientNet B3 architecture [64], a Densenet-121 architecture [43], a ResNest-50 architecture [45], and a ViT architecture [46]. These networks were selected to include both attention-based models (ViT and ResNest) and conventional CNN networks (Resnet, Densenet, and EfficientNet). Also, in each category, we have selected networks that have reached more promising results than their predecessors. Several data augmentation techniques alongside a CycleGAN model were applied to improve the performance of each network further.
Here, the details of each step are presented; firstly, an explanation is given on the applied dataset. Specifications of each applied deep neural network (DNN) are discussed afterward. Finally, CycleGAN is explained in the last part alongside the overall proposed method.
Table 1.
Review of related works.
| Ref | Dataset | Modality | Number of cases | Pre-Processing | DNN | Post-Processing | Performance criteria |
|---|---|---|---|---|---|---|---|
| [54] | Clinical | CT | 3000 COVID-19 Images, 3000 Non-COVID-19 Images |
Patches Extraction |
VGG-16, GoogleNet, ResNet-50 |
Feature Fusion, Ranking Technique, SVM |
Acc 98.27 Sen 98.93 Spec 97.60 Prec 97.63 |
| [65] | Datasets from [66] & [67] |
CT | 460 COVID-19 Images, 397 Healthy Control (HC) Images |
Data Augmentation (DA) |
CNN Based on SqueezeNet |
Class Activation Mapping (CAM) |
Acc 85.03 Sen 87.55 Spec 81.95 Prec 85.01 |
| [68] | Various Datasets | CT | 2373 COVID-19 Images, 2890 Pneumonia Images, 3193 Tuberculosis Images, 3038 Healthy Images |
– | Ensemble DCCNs |
– | Acc 98.83 Sen 98.83 Spec 98.82 F1-Score 98.30 |
| [69] | Clinical | CT | 98 COVID-19 Patients, 103 Non-COVID-19 Patients |
Visual Inspection |
BigBiGAN | – | Sen 80 Spec 75 |
| [55] | Clinical | CT | 148 Images from 66 COVID-19 Patients, 148 Images from 66 HC Subjects |
Visual Inspection |
ResGNet-C | – | Acc 96.62 Sen 97.33 Spec 95.91 Prec 96.21 |
| [70] | COVID-CT Dataset |
CT | 349 COVID-19 Images, 397 Non-COVID-19 Images |
Scaling Process, DA |
Multiple Kernels-ELM -based DNN |
– | Acc 98.36 Sen 98.28 Spec 98.44 Prec 98.22 |
| [56] | Clinical | CT | 210,395 Images From 704 COVID-19 Patients and 498 Non-COVID-19 Subjects |
DA |
U-net Dual-Branch Combination Network |
Attention Maps | Acc 92.87 Sen 92.86 Spec 92.91 |
| [67] | Various Dataset | CT | 2933 COVID-19 Images | Deleting Outliers, Normalization, Resizing |
Ensemble DNN |
– | Acc 99.054 Sen 99.05 Spec 99.6 F1-Score 98.59 |
| [71] | Clinical | CT | 320 COVID-19 Images, 320 Healthy Control Images |
Histogram Stretching, Margin Crop, Resizing, Down Sampling |
FGCNet | Gradient- Weighted CAM (Grad-CAM) |
Acc 97.14 Sen 97.71 Spec 96.56 Prec 96.61 |
| [72] | Clinical | CT | 180 Viral Pneumonia, 94 COVID-19 Cases |
ROIs Extraction |
Modified Inception |
– | Acc 89.5 Sen 88 Spec 87 F1-Score 77 |
| [57] | Clinical | CT | 3389 COVID-19 Images, 1593 Non-COVID-19 Images |
Segmentation, Generating Lung Masks |
3D ResNet34 with Online Attention |
Grad-CAM | Acc 87.5 Sen 86.9 Spec 90.1 F1-Score 82.0 |
| [73] | COVIDx-CT Dataset |
CT | 104,009 Images From 1489 Patient Cases |
Automatic Cropping Algorithm, DA |
COVIDNet-CT | – | Acc 99.1 Sen 97.3 PPV 99.7 |
| [74] | Various Datasets | CT | 349 COVID-19 Images, 397 Non-COVID-19 Images |
Resizing, Normalization, Wavelet-Based DA |
ResNet18 | Localization of Abnormality |
Acc 99.4 Sen 100 Spec 98.6 |
| [58] | COVID-CT | CT | 345 COVID-19 Images, 397 Non-COVID-19 Images |
Resizing, DA | Conditional GAN ResNet50 |
– | Acc 82.91 Sen 77.66 Spec 87.62 |
| [75] | Clinical | CT | 151 COVID-19 Patient, 498 Non-COVID-19 Patient |
Resizing, Padding, DA |
3D-CNN | Interpretation by Two Radiologists |
AUC 70 |
| [59] | SARS-CoV-2 CT-Scan Dataset |
CT | 1252 CT COVID-19 Images, 1230 CT non-COVID-19 Images |
– | GAN with Whale Optimization Algorithm |
– | Acc 99.22 Sen 99.78 Spec 97.78 F1-score 98.79 |
| [66] | Various Datasets | CT | 1684 COVID-19 Patient, 1055 Pneumonia, 914 Normal Patients |
Resizing | Inception V1 | Interpretation by 6 Radiologists, t-SNE Method |
Acc 95.78 AUC 99.4 |
| [76] | Clinical | CT | 2267 COVID-19 CT Images, 1235 HC CT Images |
Compressing, Normalization, Cropping, Resizing |
ResNet50 | – | Acc 93 Sen 93 Spec 92 F1-Score 92 |
| [77] | Clinical | CT | 108 COVID-19 Patients, 86 Non-COVID-19 Patients |
Visual Inspection, Grey-Scaling, Resizing |
Various Networks |
– | Acc 99.51 Sen 100 Spec 99.02 |
| [60] | Various Datasets | CT | 413 COVID-19 Images, 439 Non-COVID-19 Images |
Feature Extraction with ResNet-50 |
3D-CNN | – | Acc 93.01 Sen 91.45 Spec 94.77 Prec 94.77 |
| [78] | Clinical | CT | 150 3D COVID-19 Chest CT, CAP and NP Patients (450 Patient Scans) |
Sliding Window, DA |
Multi-View U-Net 3D-CNN |
Weakly Supervised Lesions Localization, CAM |
Acc 90.6 Sen 83.3 Spec 95.6 Prec 74.1 |
| [61] | Various Datasets | CT | 449 COVID-19 Patients, 425 Normal, 98 Lung Cancer, 397 Different Kinds of Pathology |
Resizing, Intensity Normalization |
Autoencoder Based DNN |
– | Dice 88 Acc 94.67 Sen 96 Spec 92 |
| [79] | COVID-19 CT from [66] |
CT | 746 Images | – | GAN | – | Acc 84.9 Sen 85.33 Prec 85.33 |
| [80] | COVID-19 CT Datasets, Cohen |
CT | 345 COVID-19 CT Images, 375 Non-COVID-19 CT Image |
2D Redundant Discrete WT (RDWT) Method, Resizing |
ResNet50 | Grad-CAM, Occlusion Sensitivity Technique |
Acc 92.2 Sen 90.4 Spec 93.3 F1-Score 91.5 |
| [81] | SARS-CoV-2 CT Scan Dataset |
CT | 1262 COVID-19 Images, 1230 HC Images |
– | Convolutional Support Vector Machine (CSVM) |
– | Acc 94.03 Sen 96.09 Spec 92.01 Pre 92.19 |
| [82] | Chest CT and X-ray |
X-ray, CT |
5857 Chest X-rays, 767 Chest CTs |
– | Various Networks |
Heat Map | Acc 75 (CT) |
| [83] | medseg DlinRadiology |
CT | 10 Axial Volumetric CTS (Each Containing 100 Slices of COVID-19 Images) |
Resizing | VGG16, Resnet-50 U-net |
– | Acc 99.4 Spec 99.5 Sen 80.83 Dice 72.4 IOU 61.59 |
| [84] | BasrahDataset | CT | 50 Cases, 1425 Images | Gray-Scaling, Resizing |
VGG 16 | – | Acc 99 F1-Score 99 |
| [62] | Kaggle | CT | 1252 COVID-19 CT Images, 1240 non-COVID-19 CT Images |
Resizing, Normalization, DA |
Covid CT-net | heat map | Acc 95.78 Sen 96 Spec 95.56 |
| [85] | COVID-CT | CT | 708 CTs, 312 with COVID-19, 396 Non-COVID-19 |
Normalization | LeNet-5 | – | Acc 95.07 Sen 95.09 Prec 94.99 |
3.1. Dataset
In this paper, a new CT scans dataset of COVID-19 patients was collected from Gonabad Hospital in Iran; all data were recorded by radiologists between June 2020 and December 2020. The number of subjects with COVID-19 is 90, and 99 of the subjects are normal. In this study, three volumetric recordings were gathered from each patient in a 4-day duration (each recording two days apart from the last one); moreover, every recording contains at least 20 slices. It is noteworthy to mention that the normal subjects are patients with suspicious symptoms and not merely a control group; this makes this dataset unique compared to its prior ones, as they usually have used scans of other diseases for the control group. Patients with COVID-19 or normal subjects range in age from 2 to 88 years; 69 of which are female and 120 are male (both COVID-19 and normal classes). A total of 1766 slices of these scans were finally selected by specialist physicians from the abnormal class and 1397 from the normal class; the labeling of each CT image was performed by three experienced radiologists along with two infectious disease physicians. In addition, RT-PCR was taken from each subject to confirm labelings. All ethical approvals have been obtained from the hospital to use CT scans of COVID-19 patients and normal individuals for research purposes. Fig. 3 illustrates a few CT scans of healthy individuals and patients with COVID-19.
Fig. 3.
Examples of dataset, first two rows contain images from healthy subjects, whereas the last two rows contain images from COVID-19 patients.
3.2. Deep neural networks
3.2.1. Resnet
ResNet [44] architecture was introduced in 2015 with a depth of 152 layers; it is known as the deepest architecture up to that year and still is considered as one of the deepest. There are various versions of the architecture with different depths that are used depending on the need. This network’s main idea was to use a block called residual block, which tried to solve the problem of vanishing gradients, allowing the network to go deeper without reducing performance. Proving its capabilities by winning the Image Net Challenge in 2015; the ideas of this network have been applied in many others ever since. In this paper, a version of this network with a depth of 50 has been used, which is a wise choice given the considerably smaller amount of data compared to the ImageNet database.
3.2.2. EfficientNet
Three different criteria must be tested to design a convolutional neural network: the depth, width, and resolution of the input images. Choosing the proper values of the three criteria in such a way that they form a suitable network together is a challenging task. Increasing the depth can lead to finding complex patterns, but it can also cause problems such as vanishing gradients. More width can increase the quality of the features learned, but accuracy for such network tends to quickly saturate. Also, high image quality can have a detrimental effect on accuracy. The network was introduced in [64] with a study on how to scale the network in all three criteria properly. Using a step-by-step scheme, the network first finds the best structure for a small dataset and then scales that structure according to the activity. The network has been used for many tasks, including diagnosing autism [86] and schizophrenia [87].
3.2.3. Densenet
Introduced by Huang et al. [43], DenseNet, densely connected convolutional networks, has improved the baseline performance on benchmark computer vision task and shown its efficiency. Utilizing residuals in a better approach has allowed this network to exploit fewer parameters and go deeper. Also, by feature reuse, the number of parameters is reduced dramatically. Its building blocks are dense blocks and transition layers. Compared to ResNet, DenseNet uses concatenation in residuals rather than summing them up. To make this possible, each feature vector of each layer is chosen to have the same size for each dense block; also, training these networks has been shown to be easier than prior ones [43]. This is arguably due to the implicit deep supervision where the gradient is flowing back more quickly. The capability to have thin layers is another remarkable difference in DenseNet compared to other state-of-the-art techniques. The parameter K, the growth rate, determines the number of features for each layer’s dense block. These feature vectors are then concatenated with the preceding ones and given as input to the subsequent layer. Eliminating optimization difficulties for scaling up to hundreds of layers is another DenseNet superiority.
3.2.4. ViT
Arguably, the main problem with convolutional neural networks is their failure in encoding relative spatial information. In order to overcome this issue, researchers in [46] have adopted the self-attention mechanism from natural language processing (NLP) models. Basically, attention can be defined as trainable weights that model each part of an input sentence’s importance. Changing networks from NLP to computer vision, pixels are picked as parts of the image to train the attention model on them. Nevertheless, pixels are very small parts of an image; thus, one can pick a bigger segment of an image as one of its parts, i.e., a 16 by 16 block of images. ViT uses a similar idea; by dividing the image into smaller patches to train the attention model on them. Also, ViT-Large has 24 layers with a hidden size of 1,024 and 16 attention heads. Examination shows not only superior results but also significantly reduced training time and also less demand for hardware resources [46].
3.2.5. ResNest
Developed by researchers from Amazon and UC Davis, ResNest [45] is also another attention-based neural network that has also adopted the ideas behind ResNet structure. In its first appearance, this network has shown significant performance improvement without a large increase in the number of parameters, surpassing prior adaptations of ResNet such as ResNeXt and SEnet. In their paper, they have proposed a modular Split-Attention block that can distribute attention to several feature-map groups. The split-attention block is made of the feature-map group and split-attention operations; then, by stacking those split-attention blocks similar to ResNet, researchers were able to produce this new variant. The novelties of their paper are not merely introducing a new structure, but they also introduced a number of training strategies.
3.3. Data augmentation and training process
Generative adversarial networks were first introduced in 2014 [88] and found their way into various fields shortly after [47]. They have also been used as a method for data augmentation, and network pretraining [89] previously as well. A particular type of these networks is CycleGAN [40], a network created mainly for unpaired image-to-image translation. In this particular form of image-to-image translation, there is no need for a dataset containing paired images, which is itself a challenging task. The CycleGAN comprises of training of two generator discriminators simultaneously. One generator uses the first group of images as input and creates data for the second group, and the other generator does the opposite. Discriminator models are then utilized to distinguish the generated data from real ones and feed the gradients to generators subsequently.
The CycleGAN used in this paper has a similar structure to the one presented in the main paper [40]. Compared to other GAN paradigms, CycleGAN uses image-to-image translation, which simplifies the training process, especially where training data is limited, which also helps to create data of the desired class easily. However, using other GAN paradigms, such as conditional GAN [90], one can also create data of a specific class, yet training those methods is more complicated. A diagram of the CycleGAN is presented in Fig. 4, and also a few samples of generated data are illustrated in Fig. 5.
Fig. 4.
Overall diagram of applied CycleGAN.
Fig. 5.
Examples of generated data, the first column shows normal data from the main dataset. The second column shows the generated abnormal data from those images. The third column shows abnormal data from the main dataset. Lastly, the fourth column shows the generated normal data from those images.
In this paper, to train the networks properly, the following process is used. First, we preprocessed images by applying a Gaussian filter to remove any possible noise in images [91]. Then, we applied several data augmentation techniques [92], namely, by using random flips, rotations, zooms, warps, lighting transforms, and also presizing [93] to increase the size of training data and help the model to generalize better. Then, each network is trained on the augmented training dataset. After training, networks are evaluated on a test set. Also, to select hyperparameters of networks, the test dataset itself is split into test and validation sets, and the validation set is used to evaluate models with different hyperparameters and find the best setting for each network.
We also studied our models’ performance by training them using an augmented dataset generated by means of a CycleGAN model implemented using the UPIT library [94].
4. Results
4.1. Environment setup and hyper parameter selection
All models were trained using the FastAI library [93] and applying fine-tuning to the pre-trained models available at the timm repository [95] using a GPU Nvidia RTX 2080 Ti with 11 GB of RAM. Aside from CycleGAN data augmentation, other data augmentation methods were also implemented using FastAI, and their parameters are shown in Table 2. As for the CycleGAN implementation, the UPIT library [94] was used. To find the best hyperparameters, such as the learning rate for the task at hand, and to evaluate our models properly, we divided the data into three parts: the first one for training, the second one for validation, and the last one for testing. We used 70 percent of the data for training, 15 for validation, and 15 for testing. While this division is done on data points (images) and not subjects, we made sure that no two slices of any patient are presented in two different parts simultaneously to make the results trustworthy. In [96], it has been shown that a 70/30 split is the best train-to-test ratio. However, given that we needed two different sets for validation and test, we further split the data into 70/15/15. To set the learning rate for the architectures, we employed the two-stage procedure similar to the one presented in [64]; lastly, we applied early stopping in all the architectures to avoid overfitting. The final selected values for batch size and hyperparameters are all available in Table 3.
Table 2.
Data augmentation parameters.
| Technique parameter | Value |
|---|---|
| Resize | 640 |
| Size Crop | 512 |
| Minimum Scale Crop | 0.75 |
| Horizontal Flip | True |
| Maximum Degree of Rotation | 10 |
| Minimum Zoom | 1 |
| Maximum Zoom | 1.1 |
| Maximum Scale of Changing Brightness |
0.2 |
| Maximum Wrap | 0.2 |
| Probability of Affine Transform |
0.75 |
| Probability of Changing Brightness and Contrast |
0.75 |
Table 3.
Selected hyperparameters for each network.
| Network | Batch size | Learning rate |
|---|---|---|
| Densenet-121 | 16 | 1.00E−03 |
| EfficientNet-B3 | 16 | 1.00E−03 |
| Resnet-50 | 16 | 1.00E−03 |
| ResNeSt-50 | 16 | 1.00E−04 |
| ViT | 16 | 1.00E−05 |
4.2. Evaluation metrics
The evaluation of each network’s performance is measured by several different statistical metrics, considering that merely relying on one measure of accuracy, it is not possible to measure all the different aspects of the performance of a network. The metrics used in this article are accuracy, precision, recall, F1-score, and area under receiver operating characteristic (ROC) curve (AUC) [97]. How to calculate these metrics is also shown in Table 4. In this table, TP shows the number of positive cases that have been correctly classified, TN has shown the number of negative cases that have been correctly classified, and FP and FN are the numbers of positive and negative cases that have been misclassified, respectively. In addition, for each network, a learning curve is plotted that shows the speed of learning and how to converge.
Table 4.
Statistical metrics for performance evaluation.
| Performance evaluation parameter |
Mathematical equation |
|---|---|
| Accuracy | |
| Precision | |
| Recall | |
| F1-Score | |
| AUC | Area Under ROC Curve |
Nearly all the metrics mentioned before show discriminative performances, i.e., how each network has performed if it merely outputs a label for each input. This approach is quite common and considered de facto in evaluating results for ML methods; however, one can argue that by evaluating models this way, probabilities assigned to each class are totally ignored. Intuitively, probabilities given by models can be viewed as certainty models have assigned to each class, so it might be reasonable to work up a metric that considers these values as well. Nonetheless, these probabilities are considered to measure the reliability of the model.
Here, we have used Expected Calibration Error (ECE) [98] to evaluate the reliability of models; for calculation of this metric, firstly, confidence scores (probabilities assigned by models) are binned uniformly, the number of bins has been selected to be five here. So the first bin would be . is all the data points in Bin . For each bin, accuracy is calculated by this equation:
| (1) |
And confidence as:
| (2) |
Where is the true label of th sample, is the assigned label to th sample, and is the corresponding assigned probability.
Finally, the ECE is calculated by this equation:
| (3) |
To give an intuition over this metric, first, we must study what confidence scores show. A confidence of 0.9 indicates that we expect 90 percent of the outcome of the model to be correct if the model is calibrated. Now the distance of actual accuracy of the model with this confidence shows how much the model is calibrated. ECE is a weighted average of these calibration errors.
4.3. Performaces
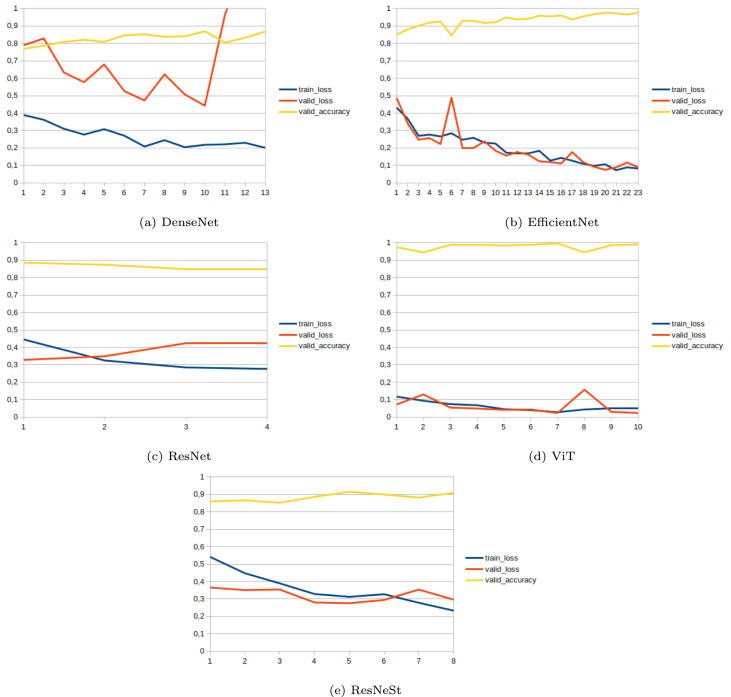
This part of the paper is dedicated to showing the results of networks. Each network is first trained without using the CycleGAN, and then the effect of adding CycleGAN is measured. Table 5, Table 6 demonstrate the network results without and with CycleGAN, and Fig. 6, Fig. 7 also show the networks’ learning curves. To make the results reliable, each network is evaluated ten times, and then the mean of performances, with confidence intervals, are presented. As observable in these tables, CycleGAN has improved the performance of EfficientNet, Resnet, and ResNeSt dramatically. Nevertheless, the ViT results show no sign of improvement in the presence of CycleGAN; this is arguably due to its robustness or indistinguishability of wrongly classified samples from the other class.
Table 5.
Results without CycleGAN.
| Network | Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) | AUC (%) | ECE |
|---|---|---|---|---|---|---|
| Densenet-121 | 88.05 ± 3.81 | 80.94 ± 5.55 | 93.18 ± 3.70 | 87.36 ± 3.63 | 96.71 ± 2.80 | 0.118 ± 0.130 |
| EfficientNet-B3 | 94.69 ± 2.04 | 92.55 ± 3.40 | 96.77 ± 1.57 | 94.09 ± 2.19 | 99.03 ± 0.66 | 0.067 ± 0.012 |
| Resnet-50 | 94.69 ± 1.15 | 90.31 ± 2.21 | 98.74 ± 0.48 | 94.25 ± 1.15 | 99.43 ± 0.20 | 0.060 ± 0.025 |
| ResNeSt-50 | 96.30 ± 2.31 | 93.96 ± 3.00 | 98.02 ± 2.43 | 95.97 ± 2.53 | 99.60 ± 1.18 | 0.020 ± 0.007 |
| ViT | 99.60 ± 0.79 | 99.46 ± 1.39 | 99.64 ± 0.38 | 99.55 ± 0.88 | 99.99 ± 0.10 | 0.014 ± 0.007 |
Table 6.
Results with CycleGAN.
| Network | Accuracy (%) | Precision (%) | Recall (%) | F1-score (%) | AUC (%) | ECE |
|---|---|---|---|---|---|---|
| Densenet-121 | 89.24 ± 3.78 | 81.67 ± 5.84 | 96.23 ± 2.45 | 88.64 ± 3.55 | 97.22 ± 1.89 | 0.060 ± 0.026 |
| EfficientNet-B3 | 98.25 ± 2.57 | 97.03 ± 3.51 | 99.28 ± 2.20 | 98.05 ± 2.80 | 99.79 ± 0.90 | 0.058 ± 0.185 |
| Resnet-50 | 96.20 ± 0.79 | 94.09 ± 1.33 | 97.49 ± 1.11 | 95.78 ± 0.87 | 99.43 ± 0.42 | 0.033 ± 0.028 |
| ResNeSt-50 | 98.89 ± 1.09 | 98.58 ± 1.34 | 99.10 ± 1.70 | 98.75 ± 1.24 | 99.95 ± 0.22 | 0.013 ± 0.006 |
| ViT | 99.20 ± 2.91 | 98.92 ± 3.97 | 98.92 ± 2.41 | 99.10 ± 3.19 | 99.95 ± 0.92 | 0.068 ± 0.037 |
Fig. 6.
Learning curve of networks without CycleGAN; axis shows the number of epoch and axis shows reached value for each metric (without any scaling).
Fig. 7.
Learning curve of networks with CycleGAN; axis shows the number of epoch and axis shows reached value for each metric (without any scaling).
As observable through results, ECEs are also consistent with other findings; they have improved by applying GAN, and they are mainly better for models with better performances. By this limited testing, applying CycleGAN can be viewed as a means to improve the calibration and reliability of models, yet this is arguably a loose deduction. A substantial literature has investigated methods for improvement of reliability, but as this is not the focus of our study, we have not investigated them here. An interested reader can study about different methods on [99], such as confidence penalty [100] and temperature scaling [101].
Lastly, ROC curve for one run of the networks is also plotted in Fig. 8.
Fig. 8.
ROC curve of networks without and with CycleGAN.
4.4. Qualitative analysis
In previous sections, each network was evaluated thoroughly by quantitative measures, both on performance and reliability. Nevertheless, aside from quantitative results, one expects any network not only to perform well but also to learn to extract meaningful information from data. These results are mainly considered qualitative outputs; one way to assess these performance types is to extract localized importance maps that show how much each part of the image has contributed more toward making the final decision [102].
Here, we have used Grad-CAM [103] to obtain them. This method uses the gradient of target outputs to visualize a heat map of activation from the last convolutional layer; in other words, it shows how much each part of the image is weighted for that specific class. Fig. 9 shows a sample of these types of results. Since showing results for each network would take immoderate space from the manuscript, we have visualized only for ViT and Resnet; this way, we have picked one network from attention-based types (ViT) and one from the conventional CNN types (Resnet). Notably, samples used for this figure are all from abnormal class.
Fig. 9.
Grad-CAM output for Resnet and ViT, with and without CycleGAN.
Fig. 9 demonstrates a few extraordinary points about how each network has learned. Firstly, we can observe that networks have put more weight on the more salient parts of images for the detection of abnormalities in CT images. Secondly, we can observe that attention-based methods have learned a better localization of important parts. It is worth mentioning that values closer to zero contribute the least, whilst the contribution is increased as we get further from zero. Lastly, the effect of applying GAN is also observable from this figure; after applying GAN, networks activations have concentrated more toward informative parts.
5. Discussion
In recent years, convolutional neural networks have revolutionized the field of image processing. Medical diagnoses are no exception, and today in numerous research papers in this field, the use of these networks to achieve the best accuracy is seen. Diagnosis of COVID-19 disease from CT images is also one of the applications of these networks. In this article, the performance of different networks in this task was examined, and also by applying a new method, an attempt was made to improve the performance of these networks. The networks used in this paper were Resnet, EfficientNet, Densenet, ViT, and ResNest, and the data augmentation method was based on CycleGAN. Table 7 summarizes the proposed method of previous papers. By comparing this table with our current work, the advantages of our work can be listed as using ViT, a transformer-based architecture that has achieved state-of-the-art performances; collecting a new dataset; and finally using CycleGAN for data augmentation.
Table 7.
Summary of related works.
| Ref | Dataset | Number of Cases (Images) | Pre-Processing | DNN | Performance (%) |
|---|---|---|---|---|---|
| [54] | SIRM | 3000 COVID-19, 3000 HC | Patches Extraction | PreTrain Networks | Acc 98.27 |
| [65] | Zhao et al | 460 COVID-19, 397 HC | DA | SqueezeNet | Acc 85.03 |
| [68] | Indian | 2373 COVID-19, 6321 HC | – | Ensemble DCCNs | Acc 98.83 |
| [69] | Different Datasets | – | Visual Inspection | BigBiGAN | – |
| [55] | Clinical | 148 COVID-19, 148 HC | Visual Inspection | ResGNet-C | Acc 96.62 |
| [70] | Clinical | 349 COVID-19, 397 HC | Scaling Process, DA | MKs-ELM-DNN | Acc 98.36 |
| [56] | COVID-CT | – | DA | U-net + DCN | Acc 92.87 |
| [67] | Public Dataset | 2933 COVID-19 | Normalization, Resizing | EDL_COVID | Acc 99.054 |
| [71] | Clinical | 320 COVID-19, 320 HC | HS, Margin Crop, Resizing | FGCNet | Acc 97.14 |
| [72] | Clinical | – | ROIs Extraction | Modified Inception | Acc 89.5 |
| [57] | Clinical | 3389 COVID-19, 1593 HC | Standard Preprocessing | 3D ResNet34 | Acc 87.5 |
| [73] | COVIDx-CT | 104,009 | DA | COVIDNet-CT | Acc 99.1 |
| [74] | Different Datasets | 349 COVID-19, 397 HC | Resizing, Normalization, DA | ResNet18 | Acc 99.4 |
| [58] | COVID-CT | 345 COVID-19, 397 HC | Resizing, DA | CGAN + ResNet50 | Acc 82.91 |
| [75] | Clinical | – | Resizing, Padding, DA | 3D-CNN | AUC 70 |
| [59] | SARS-CoV-2 | 1252 COVID-19, 1230 HC | – | GAN with WOA + InceptionV3 | Acc 99.22 |
| [66] | Different Datasets | – | Resizing | Inception V1 | Acc 95.78 |
| [76] | Clinical | 2267 COVID-19, 1235 HC | Normalization, Cropping, Resizing | ResNet50 | Acc 93 |
| [77] | Clinical | – | Visual Inspection, ROI, Cropping and Resizing |
ResNet 101 | Acc 99.51 |
| [60] | Different Datasets | 413 COVID-19, 439 HC | – | ResNet-50 + 3D-CNN | Acc 93.01 |
| [78] | Clinical | – | DA | Multi-View U-Net + 3D-CNN | Acc 90.6 |
| [61] | Different Datasets | – | Resized, Intensity Normalized | FCN | Acc 94.67 |
| [79] | Zhao et al | 746 | – | GAN + ShuffleNet | Acc 84.9 |
| [80] | COVID-CT | 345 COVID-19, 375 HC | 2D RDWT, Resizing | ResNet50 | Acc 92.2 |
| [81] | SARS-CoV-2 | 1262 COVID-19, 1230 HC | – | CSVM | Acc 94.03 |
| [82] | Different Datasets | 767 | – | Different PreTrain Methods | Acc 75 |
| [83] | MedSeg DII | – | Resizing | U-Net + VGG16 and Resnet-50 | Acc 99.4 |
| [84] | Basrah | 1425 | Resizing | VGG 16 | Acc 99 |
| [62] | Kaggle | 1252 COVID, 1240 HC | Resizing, Normalization, DA | COVID CT-net | Acc 95.78 |
| [85] | COVID-CT | 312 COVID-19, 396 HC | Normalization | LeNet-5 | Acc 95.07 |
| Ours | Clinical | 1766 COVID-19, 1397 HC | Filtering, DA using CycleGAN | Different PreTrain Methods | Acc 99.60 |
Eventually, the ViT network reached an accuracy of 99.60%, which shows its state-of-the-art performance and proves that it can be used as the heart of a CADS. By comparing the performances of our method compared to previous works in Table 1, our methods’ superiority is quite observable. The advantages of adding CycleGAN were also clearly displayed, and it was shown that this method could be used for this task by data augmentation to improve the performance of most deep neural networks. Nonetheless, the reliability of models was also asserted by calibration error, and the effect of applying CycleGAN on the improvement of reliability was demonstrated as well. Lastly, Grad-CAM was used to visualize the activation heatmap of a few samples to show how networks have learned to extract useful information.
Finally, this article’s achievements can be summarized in: first, introducing a new database and its public release; second, examining the performance and reliability of various neural networks on this database by standard evaluation metrics and calibration ones; thirdly, interpreting the decision of networks to find suspicious regions and finally, evaluating the use of CycleGAN for data augmentation and its impact on networks performances and reliability. Additionally, the performance of ViT was never previously studied for this task, which was investigated in this paper as well. To evaluate the method, a CT scan dataset was collected by physicians, which we also made available to researchers in public. Also, due to the fact that this dataset was collected from people suspected of having COVID-19, normal class data, unlike many previous datasets in this field, were collected from patients with suspicious symptoms and not from other diseases.
6. Conclusion, and future works
The new virus COVID-19 or nCOV-2019 is a hazardous infection that enters the human body through the respiratory system. This disease is contagious and is spreading among people all over the world. In recent years, several methods have been tested for rapid diagnosis of COVID-19, including medical tests and medical imaging methods. The most important medical imaging methods in diagnosing COVID-19 include CT, X-ray, and ultrasound. According to the WHO report, people are diagnosed with COVID-19 who have a positive PCR test and a medical imaging method at the same time. Diagnosing COVID-19 from medical images poses many challenges for specialist physicians. To resolve these challenges, in recent years, extensive research has been done in the field of diagnosing COVID-19 from medical images using DL techniques. The main goal of all these research papers is to achieve a DL-based method to help quickly diagnose COVID-19 from medical images.
In this paper, a DL-based CADS is proposed for the diagnosis of COVID-19 from CT images. The proposed CADS consists of dataset creation, pre-processing, feature extraction, and post-processing stages. In this work, the COVID-19 dataset has been gathered clinically by some authors of the paper and made available publicly, and this is the first outcome of the paper. As stated in Section 3 of the manuscript, this dataset contains 1397 images of HC subjects and 1766 images of subjects with COVID-19. CT data pre-processing, including image denoising and data augmentation, is performed in the second step of the proposed CADS. At this stage, a new GAN architecture called CycleGAN was used for data augmentation. In [104], researchers used the CycleGAN method to detect COVID-19. In their research, the CycleGAN model is used in the pre-processing step only for X-ray images [104]. Until now, CycleGAN has not been used to generate artificial CT images in the diagnosis of COVID-19. In our research, for the first time, the CycleGAN model has been utilized to generate artificial CT images in the diagnosis of COVID-19, and therefore this is the second novelty part of this paper. In the third step, the latest pre-trained and transformer models were tested to detect COVID-19 from CT images. In this section, the ViT architecture (vision transformer architecture) is used for the first time for the diagnosis of COVID-19 along with the CycleGAN model, and therefore it is the third and final novelty of the manuscript. The results show that the combination of the CycleGAN can help deep learning models in improving their performances by data augmentation and also that the ViT architecture is well suited for the task at hand. In order to show the performance of the proposed CADS, various statistical parameters, including accuracy, precision, recall, F1-score, and AUC, have been calculated for various DL models. In addition, for the first time, the ECE parameter, which is the most important calibration criterion, has also been calculated for each of the DL models. Finally, a post-processing step based on explainable AI has been done to show the suspected areas of COVID-19 in CT images. At this stage, the researchers used the Grad-CAM method as a post-processing step in the proposed CADS. This step will help the specialist physicians to see the suspected areas of COVID-19 in the CT images for the diagnosis of COVID-19.
For future work, several different paths can be considered; first, more complicated methods in deep neural networks can be used, such as deep metric, few-shot learning, or feature fusion solutions. Also, the combination of different datasets to improve the accuracy and evaluate its impact on the training of different networks can be examined. Finally, combining different modalities to increase accuracy can also be a direction for future research.
CRediT authorship contribution statement
Navid Ghassemi: Conceptualization, Methodology, Software, Validation, Formal analysis, Resources, Writing – original draft, Writing – review & editing, Visualization. Afshin Shoeibi: Conceptualization, Methodology, Software, Validation, Formal analysis, Resources, Writing – original draft, Writing – review & editing, Visualization. Marjane Khodatars: Validation, Formal analysis, Resources, Writing – original draft, Writing – review & editing. Jonathan Heras: Software, Visualization. Alireza Rahimi: Software, Formal analysis. Assef Zare: Writing – review & editing. Yu-Dong Zhang: Methodology, Visualization. Ram Bilas Pachori: Methodology, Visualization. J. Manuel Gorriz: Conceptualization, Methodology, Software, Validation, Formal analysis, Resources, Writing – original draft, Writing – review & editing, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
All authors have read and agreed to the published version of the manuscript.
Footnotes
All data and codes for this work have been made available publicly on the following link: github.com/afshin0919/Shoeibi-COVID19-Dataset.
References
- 1.Jamshidi M., Lalbakhsh A., Talla J., Peroutka Z., Hadjilooei F., Lalbakhsh P., Jamshidi M., La Spada L., Mirmozafari M., Dehghani M., et al. Artificial intelligence and COVID-19: Deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaid S., Kalantar R., Bhandari M. Deep learning COVID-19 detection bias: Accuracy through artificial intelligence. Int. Orthopaed. 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perchetti G.A., Nalla A.K., Huang M.-L., Zhu H., Wei Y., Stensland L., Loprieno M.A., Jerome K.R., Greninger A.L. Validation of SARS-CoV-2 detection across multiple specimen types. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopez-Rincon A., Tonda A., Mendoza-Maldonado L., Mulders D.G., Molenkamp R., Perez-Romero C.A., Claassen E., Garssen J., Kraneveld A.D. Classification and specific primer design for accurate detection of SARS-CoV-2 using deep learning. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-020-80363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shoeibi A., Khodatars M., Alizadehsani R., Ghassemi N., Jafari M., Moridian P., Khadem A., Sadeghi D., Hussain S., Zare A., et al. 2020. Automated detection and forecasting of Covid-19 using deep learning techniques: A review. arXiv preprint arXiv:2007.10785. [Google Scholar]
- 6.Kumar A., Gupta P.K., Srivastava A. A review of modern technologies for tackling COVID-19 pandemic. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14(4):569–573. doi: 10.1016/j.dsx.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammadpoor M., Shoeibi A., Shojaee H., et al. A hierarchical classification method for breast tumor detection. Iran. J. Med. Phys. 2016;13(4):261–268. doi: 10.22038/IJMP.2016.8453. [DOI] [Google Scholar]
- 8.Albahri O., Zaidan A., Albahri A., Zaidan B., Abdulkareem K.H., Al-Qaysi Z., Alamoodi A., Aleesa A., Chyad M., Alesa R., et al. Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: Taxonomy analysis, challenges, future solutions and methodological aspects. J. Infect. Public Health. 2020 doi: 10.1016/j.jiph.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., et al. Hospital outbreak of middle east respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan S., Shaker B., Ahmad S., Abbasi S.W., Arshad M., Haleem A., Ismail S., Zaib A., Sajjad W. Towards a novel peptide vaccine for middle east respiratory syndrome coronavirus and its possible use against pandemic COVID-19. J. Mol. Liq. 2021;324 doi: 10.1016/j.molliq.2020.114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals. 2020 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alouani D.J., Rajapaksha R.R., Jani M., Rhoads D.D., Sadri N. Deep learning analysis improves specificity of SARS-CoV-2 real time PCR. J. Clin. Microbiol. 2021 doi: 10.1128/JCM.02959-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benameur N., Mahmoudi R., Zaid S., Arous Y., Hmida B., Bedoui M.H. SARS-CoV-2 diagnosis using medical imaging techniques and artificial intelligence: A review. Clin. Imaging. 2021 doi: 10.1016/j.clinimag.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghoshal B., Tucker A. 2020. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769. [Google Scholar]
- 15.Sharifrazi D., Alizadehsani R., Roshanzamir M., Joloudari J.H., Shoeibi A., Jafari M., Hussain S., Sani Z.A., Hasanzadeh F., Khozeimeh F., et al. Fusion of convolution neural network, support vector machine and sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomed. Signal Process. Control. 2021 doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Syeda H.B., Syed M., Sexton K.W., Syed S., Begum S., Syed F., Prior F., Yu F., Jr. Role of machine learning techniques to tackle the COVID-19 crisis: Systematic review. JMIR Med. Inform. 2021;9(1) doi: 10.2196/23811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong D., Tang Z., Wang S., Hui H., Gong L., Lu Y., Xue Z., Liao H., Chen F., Yang F., et al. The role of imaging in the detection and management of COVID-19: A review. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 18.Albahri A., Hamid R.A., Alwan J.K., Al-Qays Z., Zaidan A., Zaidan B., Albahri A., AlAmoodi A., Khlaf J.M., Almahdi E., et al. Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (COVID-19): A systematic review. J. Med. Syst. 2020;44:1–11. doi: 10.1007/s10916-020-01582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah V., Keniya R., Shridharani A., Punjabi M., Shah J., Mehendale N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021:1–9. doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharyya A., Bhaik D., Kumar S., Thakur P., Sharma R., Pachori R.B. A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images. Biomed. Signal Process. Control. 2022;71 doi: 10.1016/j.bspc.2021.103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaudhary P.K., Pachori R.B. FBSED based automatic diagnosis of COVID-19 using X-ray and CT images. Comput. Biol. Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: A comprehensive study. Biomed. Signal Process. Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karakanis S., Leontidis G. Lightweight deep learning models for detecting COVID-19 from chest X-ray images. Comput. Biol. Med. 2021;130 doi: 10.1016/j.compbiomed.2020.104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blain M., Kassin M.T., Varble N., Wang X., Xu Z., Xu D., Carrafiello G., Vespro V., Stellato E., Ierardi A.M., et al. Determination of disease severity in COVID-19 patients using deep learning in chest X-ray images. Diagn. Interv. Radiol. 2021;27(1):20. doi: 10.5152/dir.2020.20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lassau N., Ammari S., Chouzenoux E., Gortais H., Herent P., Devilder M., Soliman S., Meyrignac O., Talabard M.-P., Lamarque J.-P., et al. Integrating deep learning CT-scan model, biological and clinical variables to predict severity of COVID-19 patients. Nature Commun. 2021;12(1):1–11. doi: 10.1038/s41467-020-20657-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alizadehsani R., Sharifrazi D., Izadi N.H., Joloudari J.H., Shoeibi A., Gorriz J.M., Hussain S., Arco J.E., Sani Z.A., Khozeimeh F., et al. 2021. Uncertainty-aware semi-supervised method using large unlabelled and limited labeled COVID-19 data. arXiv preprint arXiv:2102.06388. [Google Scholar]
- 32.Gaur P., Malaviya V., Gupta A., Bhatia G., Pachori R.B., Sharma D. COVID-19 disease identification from chest CT images using empirical wavelet transformation and transfer learning. Biomed. Signal Process. Control. 2022;71 doi: 10.1016/j.bspc.2021.103076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19) Eur. Radiol. 2021:1–9. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J., et al. Weakly supervised deep learning for Covid-19 infection detection and classification from ct images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 35.Islam M.M., Karray F., Alhajj R., Zeng J. A review on deep learning techniques for the diagnosis of novel Coronavirus (Covid-19) IEEE Access. 2021;9:30551–30572. doi: 10.1109/ACCESS.2021.3058537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salehi A.W., Baglat P., Gupta G. Review on machine and deep learning models for the detection and prediction of Coronavirus. Mater. Today: Proc. 2020;33:3896–3901. doi: 10.1016/j.matpr.2020.06.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhary P.K., Pachori R.B. Automatic diagnosis of COVID-19 and pneumonia using FBD method. 2020 IEEE International Conference on Bioinformatics and Biomedicine; BIBM; IEEE; 2020. pp. 2257–2263. [DOI] [Google Scholar]
- 38.Swapnarekha H., Behera H.S., Nayak J., Naik B. Role of intelligent computing in COVID-19 prognosis: A state-of-the-art review. Chaos Solitons Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for Covid-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 40.J.-Y. Zhu, T. Park, P. Isola, A.A. Efros, Unpaired image-to-image translation using cycle-consistent adversarial networks, in: Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 2223–2232.
- 41.Bar-El A., Cohen D., Cahan N., Greenspan H. Medical Imaging 2021: Image Processing, Vol. 11596. International Society for Optics and Photonics; 2021. Improved CycleGAN with application to COVID-19 classification; p. 1159614. [DOI] [Google Scholar]
- 42.Ghassemi N., Mahami H., Darbandi M.T., Shoeibi A., Hussain S., Nasirzadeh F., Alizadehsani R., Nahavandi D., Khosravi A., Nahavandi S. 2020. Material recognition for automated progress monitoring using deep learning methods. arXiv preprint arXiv:2006.16344. [Google Scholar]
- 43.G. Huang, Z. Liu, L. Van Der Maaten, K.Q. Weinberger, Densely connected convolutional networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 4700–4708.
- 44.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 45.Zhang H., Wu C., Zhang Z., Zhu Y., Zhang Z., Lin H., Sun Y., He T., Mueller J., Manmatha R., et al. 2020. Resnest: Split-attention networks. arXiv preprint arXiv:2004.08955. [Google Scholar]
- 46.Dosovitskiy A., Beyer L., Kolesnikov A., Weissenborn D., Zhai X., Unterthiner T., Dehghani M., Minderer M., Heigold G., Gelly S., et al. 2020. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929. [Google Scholar]
- 47.Goodfellow I., Bengio Y., Courville A., Bengio Y. MIT press Cambridge; 2016. Deep Learning, Vol. 1, no. 2. [Google Scholar]
- 48.Górriz J.M., Ramírez J., Ortíz A., Martínez-Murcia F.J., Segovia F., Suckling J., Leming M., Zhang Y.-D., Álvarez-Sánchez J.R., Bologna G., et al. Artificial intelligence within the interplay between natural and artificial computation: Advances in data science, trends and applications. Neurocomputing. 2020;410:237–270. doi: 10.1016/j.neucom.2020.05.078. [DOI] [Google Scholar]
- 49.Shoeibi A., Ghassemi N., Khodatars M., Jafari M., Moridian P., Alizadehsani R., Khadem A., Kong Y., Zare A., Gorriz J.M., et al. 2021. Applications of epileptic seizures detection in neuroimaging modalities using deep learning techniques: Methods, challenges, and future works. arXiv preprint arXiv:2105.14278. [Google Scholar]
- 50.Shoeibi A., Khodatars M., Jafari M., Moridian P., Rezaei M., Alizadehsani R., Khozeimeh F., Gorriz J.M., Heras J., Panahiazar M., Nahavandi S., Acharya U.R. Applications of deep learning techniques for automated multiple sclerosis detection using magnetic resonance imaging: A review. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104697. [DOI] [PubMed] [Google Scholar]
- 51.Bishop C.M. springer; 2006. Pattern Recognition and Machine Learning. [Google Scholar]
- 52.Shoeibi A., Ghassemi N., Khodatars M., Moridian P., Alizadehsani R., Zare A., Khosravi A., Subasi A., Acharya U.R., Gorriz J.M. Detection of epileptic seizures on EEG signals using ANFIS classifier, autoencoders and fuzzy entropies. Biomed. Signal Process. Control. 2022;73 doi: 10.1016/j.bspc.2021.103417. [DOI] [Google Scholar]
- 53.Jiménez-Mesa C., Ramírez J., Suckling J., Vöglein J., Levin J., Górriz J.M., ADNI A.D.N.I., DIAN D.I.A.N. 2021. Deep learning in current neuroimaging: A multivariate approach with power and type I error control but arguable generalization ability. arXiv preprint arXiv:2103.16685. [Google Scholar]
- 54.Özkaya U., Öztürk Ş., Barstugan M. Big Data Analytics and Artificial Intelligence Against COVID-19: Innovation Vision and Approach. Springer; 2020. Coronavirus (Covid-19) classification using deep features fusion and ranking technique; pp. 281–295. [DOI] [Google Scholar]
- 55.Yu X., Lu S., Guo L., Wang S.-H., Zhang Y.-D. ResGNet-C: A graph convolutional neural network for detection of COVID-19. Neurocomputing. 2020 doi: 10.1016/j.neucom.2020.07.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao K., Su J., Jiang Z., Zeng L.-L., Feng Z., Shen H., Rong P., Xu X., Qin J., Yang Y., et al. Dual-branch combination network (DCN): Towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B., et al. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39(8):2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 58.Loey M., Manogaran G., Khalifa N.E.M. A deep transfer learning model with classical data augmentation and Cgan to detect Covid-19 from chest Ct radiography digital images. Neural Comput. Appl. 2020:1–13. doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goel T., Murugan R., Mirjalili S., Chakrabartty D.K. Automatic screening of COVID-19 using an optimized generative adversarial network. Cogn. Comput. 2021:1–16. doi: 10.1007/s12559-020-09785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S. Deep transfer learning based classification model for COVID-19 disease. Irbm. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amyar A., Modzelewski R., Li H., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation. Comput. Biol. Med. 2020;126 doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swapnarekha H., Behera H.S., Nayak J., Naik B. Covid CT-net: A deep learning framework for COVID-19 prognosis using CT images. J. Interdisc. Math. 2021:1–26. doi: 10.1080/09720502.2020.1857905. [DOI] [Google Scholar]
- 63.A. Sharif Razavian, H. Azizpour, J. Sullivan, S. Carlsson, CNN features off-the-shelf: An astounding baseline for recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, 2014, pp. 806–813.
- 64.Tan M., Le Q. International Conference on Machine Learning. PMLR; 2019. Efficientnet: Rethinking model scaling for convolutional neural networks; pp. 6105–6114. [Google Scholar]
- 65.Polsinelli M., Cinque L., Placidi G. A light cnn for detecting Covid-19 from ct scans of the chest. Pattern Recognit. Lett. 2020;140:95–100. doi: 10.1016/j.patrec.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang K., Liu X., Yang Y., Liao X., Wang R., Zeng X., Wang Y., Zhang M., Zhang T. 2020. End-to-end COVID-19 screening with 3D deep learning on chest computed tomography. [Google Scholar]
- 67.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh D., Kumar V., Kaur M. Densely connected convolutional networks-based COVID-19 screening model. Appl. Intell. 2021:1–8. doi: 10.1007/s10489-020-02149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song J., Wang H., Liu Y., Wu W., Dai G., Wu Z., Zhu P., Zhang W., Yeom K.W., Deng K. End-to-end automatic differentiation of the Coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur. J. Nucl. Med. Mol. Imaging. 2020;47(11):2516–2524. doi: 10.1007/s00259-020-04929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turkoglu M. COVID-19 detection system using chest CT images and multiple kernels-extreme learning machine based on deep neural network. IRBM. 2021 doi: 10.1016/j.irbm.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S.-H., Govindaraj V.V., Górriz J.M., Zhang X., Zhang Y.-D. Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf. Fusion. 2021;67:208–229. doi: 10.1016/j.inffus.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., et al. A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19) Eur. Radiol. 2021:1–9. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gunraj H., Wang L., Wong A. Covidnet-ct: A tailored deep convolutional neural network design for detection of Covid-19 cases from chest ct images. Front. Med. 2020;7 doi: 10.3389/fmed.2020.608525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahuja S., Panigrahi B.K., Dey N., Rajinikanth V., Gandhi T.K. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl. Intell. 2021;51(1):571–585. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pu J., Leader J., Bandos A., Shi J., Du P., Yu J., Yang B., Ke S., Guo Y., Field J.B., et al. Any unique image biomarkers associated with COVID-19? Eur. Radiol. 2020;30:6221–6227. doi: 10.1007/s00330-020-06956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Z., Xingming Z., Tao G., Dan T., Li J., Chen X., Li Y., Zhou Z., Zhang X., Zhou J., et al. Classification of COVID-19 by compressed chest CT image through deep learning on a large patients cohort. Interdisc. Sci. Comput. Life Sci. 2021;13(1):73–82. doi: 10.1007/s12539-020-00408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J., et al. Weakly supervised deep learning for Covid-19 infection detection and classification from ct images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 79.Khalifa N.E.M., Taha M.H.N., Hassanien A.E., Taha S.H.N. Big Data Analytics and Artificial Intelligence Against COVID-19: Innovation Vision and Approach. Springer; 2020. The detection of COVID-19 in CT medical images: A deep learning approach; pp. 73–90. [DOI] [Google Scholar]
- 80.Matsuyama E., et al. A deep learning interpretable model for novel Coronavirus disease (COVID-19) screening with chest CT images. J. Biomed. Sci. Eng. 2020;13(07):140. doi: 10.4236/jbise.2020.137014. [DOI] [Google Scholar]
- 81.Özkaya U., Öztürk Ş., Budak S., Melgani F., Polat K. 2020. Classification of COVID-19 in chest CT images using convolutional support vector machines. arXiv preprint arXiv:2011.05746. [Google Scholar]
- 82.Deng X., Shao H., Shi L., Wang X., Xie T. A classification–detection approach of COVID-19 based on chest X-ray and CT by using keras pre-trained deep learning models. CMES Comput. Model. Eng. Sci. 2020;125(2):579–596. doi: 10.32604/cmes.2020.011920. [DOI] [Google Scholar]
- 83.Bhargavi V., Rubi R.D., Subramanian R., Yadav S. Automatic identification of Covid-19 regions on CT-images using deep learning. Eur. J. Mol. Clin. Med. 2021;7(3):668–676. [Google Scholar]
- 84.Khalaf Z.A., Hammadi S.S., Mousa A.K., Ali H.M., Alnajar H.R., Mohsin R.H. 2020. Coronavirus disease (COVID-19) detection using deep features learning. [Google Scholar]
- 85.Carvalho E.D., Carvalho E.D., de Carvalho Filho A.O., de Araújo F.H.D., Rabêlo R.d.A.L. Diagnosis of COVID-19 in CT image using CNN and XGBoost. 2020 IEEE Symposium on Computers and Communications; ISCC; IEEE; 2020. pp. 1–6. [DOI] [Google Scholar]
- 86.Khodatars M., Shoeibi A., Ghassemi N., Jafari M., Khadem A., Sadeghi D., Moridian P., Hussain S., Alizadehsani R., Zare A., et al. 2020. Deep learning for neuroimaging-based diagnosis and rehabilitation of Autism spectrum disorder: A review. arXiv preprint arXiv:2007.01285. [DOI] [PubMed] [Google Scholar]
- 87.Sadeghi D., Shoeibi A., Ghassemi N., Moridian P., Khadem A., Alizadehsani R., Teshnehlab M., Gorriz J.M., Nahavandi S. 2021. An overview on artificial intelligence techniques for diagnosis of schizophrenia based on magnetic resonance imaging modalities: Methods, challenges, and future works. arXiv preprint arXiv:2103.03081. [DOI] [PubMed] [Google Scholar]
- 88.Goodfellow I.J., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., Bengio Y. 2014. Generative adversarial networks. arXiv preprint arXiv:1406.2661. [Google Scholar]
- 89.Ghassemi N., Shoeibi A., Rouhani M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control. 2020;57 doi: 10.1016/j.bspc.2019.101678. [DOI] [Google Scholar]
- 90.Mirza M., Osindero S. 2014. Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784. [Google Scholar]
- 91.Sallay H., Bourouis S., Bouguila N. Online learning of finite and infinite gamma mixture models for COVID-19 detection in medical images. Computers. 2020;10(1):6. doi: 10.3390/computers10010006. [DOI] [Google Scholar]
- 92.Simard P.Y., Steinkraus D., Platt J.C., et al. Icdar, Vol. 3, no. 2003. Citeseer; 2003. Best practices for convolutional neural networks applied to visual document analysis. [Google Scholar]
- 93.Howard J., Gugger S. O’Reilly Media; 2020. Deep Learning for Coders with Fastai and PyTorch. [Google Scholar]
- 94.Abraham T., et al. 2021. Unpaired image-to-image translation. https://github.com/tmabraham/UPIT. [Google Scholar]
- 95.Wightmann R. 2021. PyTorch image models. https://github.com/rwightman/pytorch-image-models/ [Google Scholar]
- 96.Nguyen Q.H., Ly H.-B., Ho L.S., Al-Ansari N., Le H.V., Tran V.Q., Prakash I., Pham B.T. Influence of data splitting on performance of machine learning models in prediction of shear strength of soil. Math. Probl. Eng. 2021;2021 doi: 10.1155/2021/4832864. [DOI] [Google Scholar]
- 97.Shoeibi A., Ghassemi N., Alizadehsani R., Rouhani M., Hosseini-Nejad H., Khosravi A., Panahiazar M., Nahavandi S. A comprehensive comparison of handcrafted features and convolutional autoencoders for epileptic seizures detection in EEG signals. Expert Syst. Appl. 2021;163 doi: 10.1016/j.eswa.2020.113788. [DOI] [Google Scholar]
- 98.Küppers F., Kronenberger J., Schneider J., Haselhoff A. Bayesian confidence calibration for epistemic uncertainty modelling. 2021 IEEE Intelligent Vehicles Symposium; IV; IEEE; 2021. pp. 466–472. [DOI] [Google Scholar]
- 99.Guo C., Pleiss G., Sun Y., Weinberger K.Q. International Conference on Machine Learning. PMLR; 2017. On calibration of modern neural networks; pp. 1321–1330. [Google Scholar]
- 100.Pereyra G., Tucker G., Chorowski J., Kaiser Ł., Hinton G. 2017. Regularizing neural networks by penalizing confident output distributions. arXiv preprint arXiv:1701.06548. [Google Scholar]
- 101.Z. Ding, X. Han, P. Liu, M. Niethammer, Local temperature scaling for probability calibration, in: Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021, pp. 6889–6899.
- 102.Alqaraawi A., Schuessler M., Weiß P., Costanza E., Berthouze N. Proceedings of the 25th International Conference on Intelligent User Interfaces. 2020. Evaluating saliency map explanations for convolutional neural networks: A user study; pp. 275–285. [DOI] [Google Scholar]
- 103.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE International Conference on Computer Vision. 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [DOI] [Google Scholar]
- 104.Bargshady G., Zhou X., Barua P.D., Gururajan R., Li Y., Acharya U.R. Application of cyclegan and transfer learning techniques for automated detection of COVID-19 using X-ray images. Pattern Recognit. Lett. 2022;153:67–74. doi: 10.1016/j.patrec.2021.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]