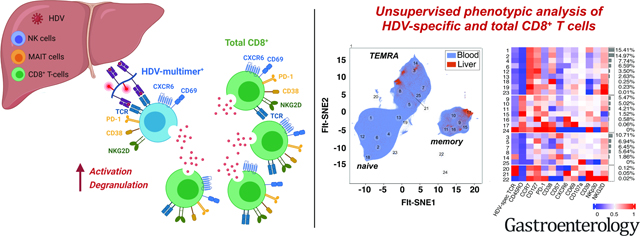
Abstract
Background & Aims:
The hepatitis D virus (HDV) causes the most severe form of chronic hepatitis, often progressing to cirrhosis within 5–10 years. There is no curative treatment and the mechanisms underlying the accelerated liver disease progression are unknown.
Methods:
Innate and adaptive immune responses were studied in blood and liver of 24 HDV-infected patients and 30 uninfected controls by multiparameter flow cytometry in correlation with disease severity and stage.
Results:
The two main intrahepatic innate immune-cell populations, MAIT cells and NK cells, were reduced in the livers of HDV-infected patients compared to those of uninfected controls but were more frequently activated in the liver compared to the blood. Most intrahepatic CD8+ T cells were memory cells or terminal effector memory cells, and the majority of activated and degranulating (CD107a+) HDV-specific and total CD8+ T cells were liver-resident (CD69+CXCR6+). Unsupervised analysis of flow cytometry data identified an activated, memory-like, tissue-resident HDV-specific CD8+ T-cell cluster with expression of innate-like NKp30 and NKG2D receptors. The size of this population correlated with liver enzyme activity (r=1.0). NKp30 and NKG2D expression extended beyond the HDV-specific to the total intrahepatic CD8+ T-cell population suggesting global bystander activation. This was supported by the correlations between NKG2D expression and degranulation of intrahepatic CD8+ T cells and between frequency of degranulating CD8+ T cells and liver enzyme activity, and APRI score and by in vitro demonstration of cytokine-induced NKDG2D-dependent cytotoxicity.
Conclusion:
Antigen-nonspecific activation of liver-resident CD8+ T cells may contribute to inflammation and disease stage in HDV infection.
Keywords: infection, cirrhosis
Lay summary
Activation and function of innate immune cells and, irrespective of their antigen-specificity, liver-resident CD8+ T cells contribute to inflammation and disease stage of chronic hepatitis delta.
Graphical Abstract
Introduction
Chronic hepatitis D virus (HDV) infection induces the most severe form of viral hepatitis,1 with accelerated progression to end-stage liver disease within 5–10 years of infection. Chronic HDV infection results from either simultaneous infection with the hepatitis B virus (HBV) and HDV or from HDV superinfection of HBsAg+ carriers, because HDV - a small, single-stranded RNA virus - requires the HBV surface antigen (HBsAg) to envelope its genome and infect cells.
The mechanisms that drive the rapid disease progression in chronically HBV/HDV-infected patients are not known. Histologic examination of liver biopsies of 863 patients with viral hepatitis who were referred to the NIH between 1981 and 1999 showed severe lobular inflammation in 88% of cases with chronic HDV infection as compared to 53% of cases with chronic HBV and 55% of cases with chronic HCV infection.2 Severe piecemeal necrosis was present in liver histology of 69% of patients with chronic HDV infection as compared to 34.2% with chronic HBV and 15.8% with chronic HCV infection. Likewise, 69% of patients with chronic HDV infection had bridging or multiacinar necrosis as compared to 38% with chronic HBV and 21% with chronic HCV infection.2 These findings point to a role of the intrahepatic immune response in accelerated disease pathogenesis.
Natural killer (NK) cells, mucosal-associated invariant T (MAIT) cells and CD8+ T cells are known as the major components of the intrahepatic immune cell infiltrate in the healthy liver.3 NK cells and MAIT cells constitutively express receptors for cytokines induced by HDV,4 in particular interferon-alpha/beta and the inflammatory cytokines IL-12 and IL-18. CD8+ T cells may additionally recognize HDV antigens presented by infected hepatocytes.
The activation, differentiation and function of these immune cells have mostly been studied in the blood and have revealed increased frequency and function of NK cells5 and activated but functionally impaired MAIT cells, with reduced frequency in more advanced hepatitis D.4 Likewise, HDV-specific CD8+ T cells are present at low frequency in the blood in HDV infection.6–8
To which extent results from the peripheral blood reflect those in the liver is not known. Recent studies in mice and in humans identified innate and adaptive immune cells that are tissue-resident and do not recirculate via the blood.9 Liver-resident NK cells and T cells are implicated in both immunosurveillance and front-line defense against pathogens that enter peripheral tissues.10, 11 Liver-resident memory CD8+ T cells for example, are preferentially expanded in patients with partial immune control of HBV12 and persist for an extended time after HBV infection is resolved.12 In contrast, liver-resident immune-cell populations are poorly described in chronic inflammatory conditions and the differential roles of antigen and cytokines in their activation and function are not known. To address their role in HDV infection, we characterized the innate and adaptive immune responses in paired blood and liver samples.
Materials and Methods
Patients
Innate and adaptive immune cells were studied ex vivo by multicolor flow cytometry in paired blood and liver samples of 24 patients with chronic HBV/HDV infection (Table 1). All patients were treated with nucleot(s)ide analogs and HBV viremia was effectively suppressed. Mononuclear cells from 23 research blood donors (NIH Department of Transfusion Medicine) and 7 livers that were not used for transplantation (Research Triangle Labs, Research Triangle Park, NC) were studied as non-viral controls. All were seronegative for HBV, hepatitis C virus (HCV), and human immunodeficiency virus. Liver biopsies were graded for inflammation using the histologic activity index (HAI)13 and for fibrosis using the Ishak scoring system.14 The aspartate aminotransferase-to-platelet-ratio index (APRI) was used as a noninvasive biomarker of liver fibrosis and cirrhosis. All patients gave written informed consent for research testing under protocol NCT03600714 (ClinicalTrials.gov) approved by the NIDDK/NIAMS institutional review board.
Table 1.
Patient Characteristics
| Patient* | Age [years] | Sex | Race | ALT [U/L] | AST [U/L] | HAI total Inflammatory score (0–18) | Ishak fibrosis score (0–6) | HLA | HLA/HDV epitope multimer used to
stain HDV-speclflc CD8+ T cells |
HDV sequence present in
patient |
Determination of HLA/peptide
binding affinity |
Figures | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A-alleles | B-alleles | |||||||||||||||||
| Multimer | HLA | Epitope sequence | HLA binding [IC50, nmol/L]** | Peptide sequence | HLA binding [IC50, nmol/L] | Method | Reference | |||||||||||
|
| ||||||||||||||||||
| 1 | 37 | F | Asian | 33 | 40 | 10 | 1 | 2:01 | 13 | 1 | ||||||||
| 2 | 38 | M | White | 201 | 86 | 9 | 3 | 02:01, 03:01 | 27:02, 35:257/278 | Pentamer | B*27:05 | RRKALENKR | 109 | RRKALENKR | 109 | in vitro | 9 | 3A, 4, 5AB, 5CD, 6 |
| 3 | 66 | M | Multiple | 46 | 57 | 11 | 6 | 02:01 | 27:05, 40 | Pentamer | B*27:05 | RRKALENKR | 109 | RRKALENKS | 13584 | in vitro | 9 | 3A, 4, 5AB, 6 |
| 4 | 42 | M | Asian | 80 | 54 | 7 | 5 | 24, 31 | 15:18, 40:01 | Tetramer | B*15:01 | SMQGVPESPF | 22 | SMQGVPESPF | 22 | in silico | 17, 18 | 1, 2, 3A, 3BD, 4, 5, 6 |
| 5 | 42 | M | Asian | 107 | 98 | 11 | 6 | 02:03, 31 | 18:01, 46 | Tetramer | B*18:01 | DENPWLGNI | 149 | EDNPWLGNI | not binding | in vitro | 9 | 2, 3, 4, 5AB, 5CD, 6 |
| 6 | 37 | F | Asian | 62 | 43 | 6 | 2 | 68, 74 | 51, 55 | 1 | ||||||||
| 7 | 34 | F | Asian | 31 | 33 | 5 | 3 | 02:01, 03:01 | 51:01, 51:02 | 1, 4, 5AB, 5CD, 6CD | ||||||||
| 8 | 38 | M | Black | 57 | 56 | 9 | 5 | 23, 36 | 35, 51 | Dextramer | B*35:01 | FPWDILFPA | 5.7 | FPWDILFPS | 27 | in vitro | 9 | 1, 2, 3A, 3BD, 4, 5, 6 |
| 9 | 34 | M | White | 34 | 42 | 12 | 5 | 29:01, 32:01 | 07:05, 51:01 | Dextramer | B*07:05 | FPWDILFPA | 96 | FPWDILFPA | 96 | in vitro | 9 | 2, 3A, 3BD, 4, 5, 6 |
| 10 | 38 | F | Black | 155 | n.a. | 10 | 5 | 02:01, 02 | 35:48, 44 | Dextramer | B*35:01 | FPWDILFPA | 5.7 | FPWVSPGSPA*** | 722 | in silico | 17, 18 | 2, 3A, 3BD, 4, 5, 6, 7 |
| 11 | 35 | M | Black | 106 | 71 | 6 | 6 | 66:02, 68 | 35:01,42:01 | Dextramer | B*35:01 | FPWDILFPA | 5.7 | FPWDILFPS**** | 27 | in vitro | 9 | 2, 3A, 3BD, 4, 5 6, 7 |
| 12 | 36 | M | Asian | 166 | 66 | 10 | 4 | 24, 68 | 38, 77 | 1 | ||||||||
| 13 | 46 | M | White | 48 | 51 | 12 | 2 | 01, 26 | 38, 52 | 1 | ||||||||
| 14 | 64 | F | White | 34 | 32 | 8 | 1 | 11 | 14, 55 | 1 | ||||||||
| 15 | 39 | F | Asian | 37 | 27 | 10 | 3 | 01, 02:01 | 51:39, 54 | 1 | ||||||||
| 16 | 47 | M | Asian | 83 | 60 | 8 | 3 | 02:01, 23 | 13, 49 | 1 | ||||||||
| 17 | 29 | M | Asian | 37 | 45 | 10 | 5 | 1, 24:03 | 37:01, 50:55 | 1 | ||||||||
| 18 | 43 | F | Asian | 91 | 55 | 9 | 6 | 24:03, 24:03 | 46, 51 | 1 | ||||||||
| 19 | 23 | M | Asian | 26 | 20 | 9 | 0 | 01, 25 | 18:01, 52 | Tetramer | B*18:01 | DENPWLGNI | 149 | DDNPWLGNI | not binding | in vitro | 9 | 2, 3, 4, 5AB, 5CD, 6 |
| 20 | 62 | F | Asian | 32 | 32 | 5 | 2 | 02:01,11:40 | 13, 54 | 1 | ||||||||
| 21 | 39 | F | Asian | 237 | 118 | 10 | 3 | 33, 33 | 53, 78 | 1 | ||||||||
| 22 | 36 | M | Asian | 64 | 60 | 8 | 0 | 3, 31 | 35:01, 48:11 | Dextramer | B*35:01 | QGFPWDILF | 118 | QGFPWDILF | 118 | in vitro | 9 | 3A, 4, 5AB, 6 |
| 23 | 47 | M | Asian | 202 | 107 | 9 | 4 | 02:01, 24 | 35:01, 58 | Dextramer | B*35:01 | QGFPWDILF | 118 | QGFPWDLLF*** | 213 | in vitro | 9 | 3A, 4, 5AB, 6, 7 |
| 24 | 54 | M | Asian | 65 | 35 | 12 | 4 | 02:01, 30:02 | 18:01, 40:01 | Tetramer | B*18:01 | DENPWLGNI | 149 | EDNPWLGDI | not binding | in vitro | 9 | 3A, 4, 5AB, 6 |
| 25 | 34 | M | Asian | 126 | 51 | 8 | 4 | 02, 25 | 13, 48 | 1 | ||||||||
All patients were treated with nucleos(t)ide analogue at the time of the immunological analysis and were HBeAg negative.
IC50 represents the amount of peptide required for 50% inhibition of binding of a 125–1 radiolabeled reference peptide to each HLA molecule [8]. IC50 < 500 nmol/L: high binding affinity; IC50, 500–5000 nmol/L, intermediate binding affinity; IC50> 50,000 nmol/L: no binding.
not recognized in functional CD8+ T cell assay (Suppl. Fig. 7).
recognized in functional CD8+ T cell assay (Suppl. Fig. 7)). n.a., not available
Peptide/HLA binding
The NetMHC-4.0 program (http://www.cbs.dtu.dk/services/NetMHC-4.0) was used for in silico prediction of peptide-HLA binding affinity.15, 16
Peptide/HLA multimers
PE-conjugated HLA/peptide multimers were dextramers (Immudex, Copenhagen, Denmark), pentamers (Pro-immune, Oxford, UK) or tetramers (NIH tetramer facility, Atlanta, GA) and presented published HDV epitopes.6–8
Cytokine analysis
IL-12, IL-15 and IL-18 plasma concentrations were determined by mesoscale assay (Mesoscale, Rockville, MD).
HDV sequence analysis
RNA extraction from plasma or serum, reverse transcription, polymerase chain reaction and sequencing were performed as described.8
Isolation of mononuclear cells from liver biopsies and blood
Liver biopsies were placed in RPMI1640, mechanically homogenized in a pestle and washed with phosphate-buffered saline (PBS, Mediatech, Manassas, VA) with 2% fetal bovine serum (FBS, Serum Source International, Charlotte, NC). Peripheral blood mononuclear cells (PBMCs) were separated on Ficoll-Histopaque (Mediatech) density gradients and washed three times with PBS. Cells from paired blood and liver samples were used immediately for ex vivo analysis by flow cytometry. The remaining PBMCs were cryopreserved in 70% FBS (Serum Source International), 20% RPMI1640 (Mediatech) and 10% DMSO (Sigma-Aldrich, St. Louis, MO).
HLA-typing
Two million PBMCs were used for HLA-A/B typing using antibody-coated trays (One Lambda Inc., Canoga Park, CA). Two-digit resolution subtyping was performed as previously described.8
Ex vivo analyses of innate and adaptive immune cells by flow cytometry
Mononuclear cells from liver biopsies and blood were washed and stained with fixable viability dye V510 and antibody panel 1 (for innate immune cells) for 15 min at room temperature (Suppl. Table 1).
CD8+ T cells were enriched from PBMCs with magnetic beads (Miltenyi, San Diego, CA) or mononuclear cells isolated from liver biopsies were incubated with 50 nM dasatinib (Axon Medchem, Reston, VA) for 30 minutes at 37°C, stained with the respective HLA/epitope multimer for 10 min, and subsequently with fixable viability dye V510 and antibody panel 2 for 15 min at room temperature (Suppl. Table 1). An extended antibody panel 3 that also stained CD39, NKp30, NKG2D and CD107a was used on a subset of patients (Suppl. Table 1). Samples were acquired on LSR II or Symphony flow cytometers (BD Biosciences, San Diego, CA) and HDV-specific and total CD8+ T cells were analyzed using FlowJo version 10 (BD Biosciences). Manual gating was performed with fluorescent-minus-one controls. Naïve-like (CD45RO−CCR7+) T cells were included in the analysis.
Unsupervised analysis
For unsupervised analysis (antibody panel 3) FACS data were compensated using FlowJo version 10 (BD Biosciences), gated for HDV-specific and total CD8+ T cells and exported to fcs files, which were then read into R (version 4.0) using packages flowCore (version 2.0.0) and CATALYST (version 1.12.1), installed through Bioconductor (version 3.11).
Inverse hyperbolic sine transformation (mathematical function “arcsinh”) was performed in R with a factor of 150 as reported for flow cytometry data.17, 18 Data were not down-sampled at any point during downstream analysis. The median number of events included in the analysis was 1,090,121 per blood sample (IQR 620,393–1,619,494) and 4,680 per liver sample (IQR 1,114–11,134).
Using the reticulate library, data were exported to python (version 3.7). After selecting the appropriate markers for analysis (multimer, CD45RO, CCR7, CD127, PD-1, CD38, CD57, CXCR6, CD69, CD107a, CD39, NKp30, NKG2D) clustering was performed with the Phenograph library using the Leiden optimization (github.com/dpeerlab/phonograph).19, 20 Phenograph assigned each cell to a cluster based on the expression of the markers used for this analysis. In total, twenty-five clusters of cells were identified. Dimensionality reduction was performed by Fast Fourier transform-accelerated Interpolation-based t-Stochastic Neighbor Embedding (FIt-SNE, github.com/klugerlab/fit-sne) in python with late exaggeration.21 In FIt-SNE plots 4 main islands of CD8+ T cells (naïve, memory, TEMRA and outliers) were identified, to which the 25 Phenograph clusters were manually mapped according to the expression level of the selected markers.
For heatmaps, data in the outer quantiles (<1% and >99%) were trimmed and rescaled to a [0, 1] range for each marker in R using the CATALYST library. Data for each marker were then aggregated by cluster and the median for each marker and cluster was calculated. The function Heatmap from the library complexHeatmap (version 2.4.2) was used to generate heatmaps.
Analysis of CD8+ T-cell effector function after TCR-dependent and -independent activation
For assessment of HDV-specific effector functions, PBMCs were stimulated in 96-well plates (4 × 105 cells/well) in complete AIM-V (containing 5% FBS, 50 μg/mL streptomycin sulfate, 10 μg/mL gentamicin sulfate, 1% L-glutamine, 1% HEPES [all from Mediatech]) with 10 μg/mL prototype HDV peptide, 10 ng/mL IL-7 (PeproTech, Rocky Hill, NJ), and 300 pg/mL IL-12 (R&D Systems, Minneapolis, MN). On days 3 and 7, 100 μL complete AIM-V with 20 IU/mL IL-2 (Prometheus, San Diego, CA) was added. On day 10, cells were restimulated with or without the respective prototype and variant peptide for 5 hours, washed, and stained with live/dead fixable aqua dye (Invitrogen, Waltham, MA), anti-CD3-A700, anti-CD8-PerCP (Biolegend, San Diego, CA) and anti-CD4-eF450 (Invitrogen) for 15 minutes at room temperature. Cells were fixed, permeabilized, and incubated with anti-IFN-γ phycoerythrin (BD Biosciences) for 30 minutes at 4°C. Data were acquired on a Symphony flow cytometer (BD Biosciences) and analyzed using FlowJo version 10 (BD Biosciences).
For further expansion, CD8+ T cells were negatively selected from the day-10 cultures with magnetic beads (Miltenyi), stained with HLA/epitope multimer (dextramer) and subsequently with live/dead fixable aqua dye (Invitrogen) and anti-CD8-PerCP (Biolegend). Dextramer+ cells were sorted on a Fusion (BD Biosciences) and restimulated in complete AIM-V with 10 ng/mL IL-7, 300 pg/mL IL-12, 3000 IU/mL IL-2, 50 ng/mL anti-CD3 (clone X35, Beckman-Coulter, Atlanta, GA) and a 100-fold excess of irradiated (9000 rad) PBMCs. Complete AIM-V with 3000 IU/mL IL-2 was added biweekly until sufficient cells were grown for cytotoxicity assays.
Cytotoxicity was assessed in a 51Cr release assays as described22 using as target cells C1R-B*35:01 or the HLA-B*3501-homozygous EBV-B cell line BVR (both provided by Dr. A. Sette, La Jolla Institute for Immunology) that had been pulsed with or without the indicated HLA-B*3501-restricted HDV peptides overnight.
For assessment of bystander CD8+ T-cell function, cryopreserved PBMCs from HDV-uninfected blood donors that did not share HLA class I allele with HuH7 hepatoma cells were thawed, depleted of CD56+ cells using MicroBeads (MIltenyi) and either assessed directly for cytotoxicity as described below or stimulated in 96-well flat-bottom plates (3 × 105 cells/well) in complete RPMI1640 (containing 5% human AB-serum, 1% penicillin/streptomycin, 2 mM L-glutamine (Mediatech) with 20 ng/mL IL-15 (R&D Systems). On day 3 and 7, 100 μL complete medium with 20 ng/mL IL-15 were added. On day 7 or 9, the IL-15-stimulated cells were washed and either stained with viability dye, anti-CD3, CD4, CD8, CD56, NKG2D and NKp30 as described in panel 3 (Suppl. Table 1) for flow cytometry or assessed for cytotoxicity against 51Cr-labeled HuH7 hepatoma cells in a 4h-coculture at effector:target ratio of 50:1 with or without anti-NKG2D (clone 1D11) or mouse IgG1 isotype control antibody (both from BD Biosciences).
Analysis of NKG2D-ligands on HDV-expressing hepatoma cells
The HDV-replicating and -secreting HuH7-END cell line was cultured as previously described23, stained with live/dead cell dye, fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer set (eBioscience) and stained with an HDV-specific monoclonal mouse antibody (provided by Dr. Stephan Urban). After washing with permeabilization buffer, cells were stained with an Alexa 647-conjugated goat anti-mouse secondary antibody, followed by staining with PE-conjugated antibodies against either MICA/B (Biolegend), ULBP-1 or ULBP-2/5/6 (both R&D Systems). Samples were acquired on a Symphony flow cytometer (BD Biosciences) and MICA/B and ULBP expression was compared on HDV-positive and HDV-negative cells.
Statistical analyses
Wilcoxon matched-pairs signed rank test, Mann-Whitney U test, Spearman correlation analyses, paired t-tests and nonparametric Friedman and Kruskal-Wallis test with Dunn’s multiple comparisons test were performed with GraphPad Prism version 8.0 (GraphPad Software Inc., San Diego, CA). P < .05 was considered significant.
Results
Innate immune cells are activated in chronic HDV infection
We started the immunological analysis by characterizing the two major innate immune cell populations, MAIT cells and NK cells by flow cytometry (Suppl. Fig. 1). The frequency of MAIT cells was significantly lower in blood and liver of HDV-infected patients than in blood and liver of uninfected controls (P = .0011 and P = .0002, respectively, Fig. 1A, left panel). However, in both HDV-infected patients and uninfected controls MAIT cells were enriched in the immune-cell compartment of the liver compared to that of the blood and more frequently HLA-DR+ (activated) and CD107a+ (degranulating) (Fig. 1A, middle and right panel). Because degranulation is associated with release of intracellular perforin and cytotoxic effector functions,24 cell surface CD107a expression is considered a biomarker of cytotoxic effector functions. MAIT-cell degranulation correlated negatively with HDV viremia (r = −0.5653, P = .0303) and with Ishak fibrosis score (r = −0.5715, P = .0283) (Fig. 1B).
Fig. 1: Activation and degranulation of innate immune cells in blood and liver of HDV-infected patients and uninfected controls.
(A) Frequency of total (left panel), activated (HLA-DR+, middle panel) and degranulating (CD107a+, right panel) MAIT cells in blood and liver of HDV-infected patients and uninfected controls.
(B) Spearman correlation between the frequency of degranulating CD107a+ intrahepatic MAIT cells and HDV viremia (left panel) and Ishak fibrosis score (right panel) of HDV-infected patients.
(C) Frequency of total (left panel), activated (HLA-DR+, middle panel) and degranulating (CD107a+, right panel) NK cells in blood and liver of HDV-infected patients and uninfected controls.
(D) Spearman correlation between the frequency of activated (HLA-DR+) liver NK cells and HDV viremia (left panel), and the frequency of degranulating (CD107a+) blood NK cells and Ishak fibrosis score (right panel). Median and interquartile range are shown in A and C. Wilcoxon matched pairs signed rank test for comparing paired blood and liver samples of HDV-infected patients, Mann Whitney test for all other comparisons.
Like MAIT cells, NK cells were less frequent in the livers of HDV-infected patients than in those of uninfected controls (Fig. 1C, left panel) but the relative proportions of activated blood and liver NK cells were increased (Fig. 1C, middle panel). As for MAIT cells, NK-cell activation and degranulation were significantly more frequent in the liver than in the blood (Fig. 1C, middle and right panel). The frequency of activated NK cells in the liver correlated negatively with HDV viremia (r = −0.5452, P = .0111, Fig. 1D, left panel) and the frequency of degranulating NK cells in the blood correlated negatively with Ishak fibrosis score (r = − 0.5223, P = .0482, Fig. 1D, right panel).
These results demonstrate that the size and activation status of two major innate immune-cell populations are altered in chronic HDV infection, and their activation and degranulation status correlates inversely with viremia and fibrosis.
Plasma IL-12 concentration is increased in chronic HDV infection
To evaluate cytokines in MAIT-cell and NK-cell activation, we assessed the plasma concentration of IL-12, IL-15 and IL-18, whose receptors are constitutively expressed on innate immune cells. Indeed, the concentration of IL-12p40 and IL-12p70 was increased in HDV-infected patients as compared to uninfected controls. IL-15 and IL-18 concentrations did not differ in the blood of patients and controls (Suppl. Fig. 2).
Blood and liver CD8+ T cells cluster into naïve, memory and TEMRA cells
As IL-12 is known to drive the differentiation and function of T cells and to act as a third signal for TCR-mediated CD8+ T-cell activation,25 we compared the phenotype of HDV-specific and total CD8+ T cells in chronic HDV infection. Based on the HLA-haplotype of each patient, we stained lymphocytes from paired blood samples and liver biopsies ex vivo with fluorescently labeled HLA multimers that present dominant HDV epitopes6–8 and with antibodies against cell surface molecules. Flow cytometry data from blood and liver samples were combined and analyzed using Fast Fourier Transform-accelerated Interpolation-based t-Stochastic Neighbor Embedding (Flt-SNE) and Phenograph clustering. This approach did not require down-sampling of the analyzed events and therefore allowed the comparison of low-frequency HDV-specific CD8+ T cells and abundant total CD8+ T cells in the same biosamples.
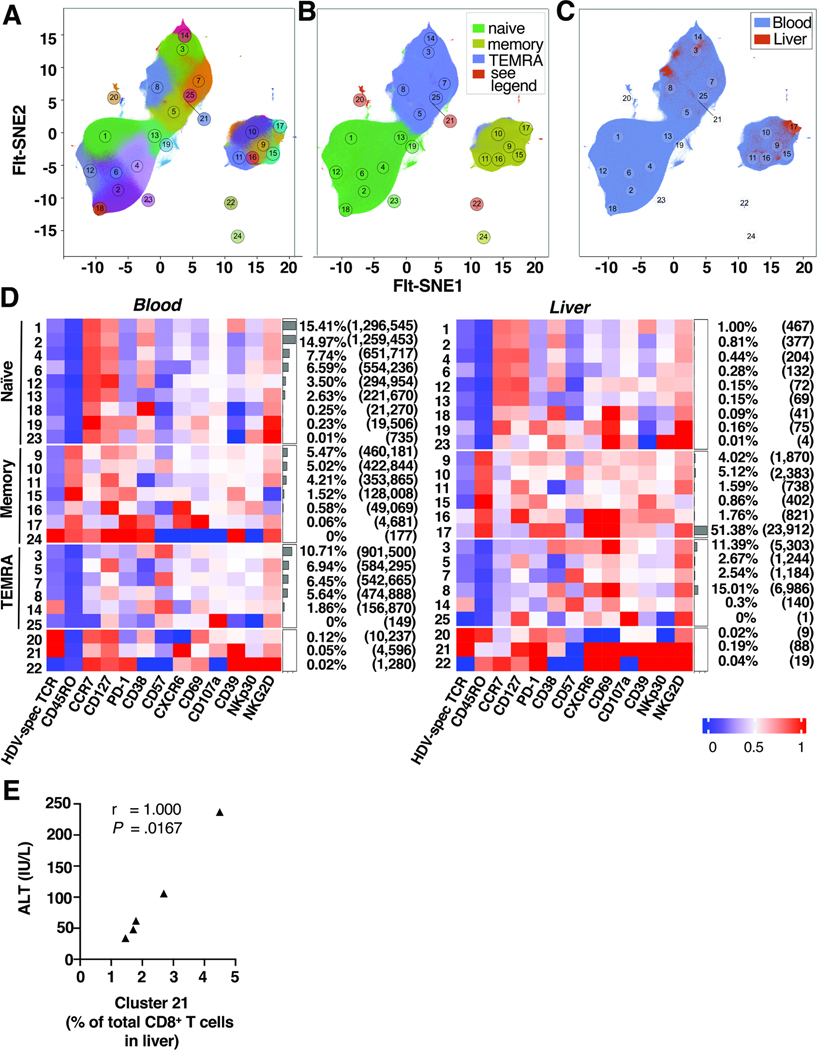
CD8+ T cells from blood and liver samples grouped into 25 phenotypic clusters based on cell surface marker expression (Fig. 2A). These clusters formed the three major islands of naïve, memory and terminal effector memory re-expressing CD45RA (TEMRA) cells (Fig. 2B). Three additional clusters were categorized separately because they differed for blood CD8+ T cells (clusters 20, 21 and 22 represented a naïve phenotype) and for liver CD8+ T cells (clusters 20 and 22 represented a memory phenotype and cluster 21 a TEMRA phenotype). Liver CD8+ T cells were exclusively found in the memory and TEMRA islands (Fig. 2C) and were almost never in the naïve island, as also confirmed by manual gating (Suppl. Fig. 3A-C).
Fig. 2: HDV-specific and total CD8+ T cells form distinct phenotypic clusters in blood and liver of HDV-infected patients.
FIt-SNE embedding of the full data set of 8,461,832 CD8+ T cells from blood and liver of HDV-infected patients.
(A) CD8+ T cells form 25 distinct clusters based on the expression of cell surface markers shown in panel F. Cluster 21 is located next to cluster 25 (as indicated by the line).
(B) CD8+ T cells from blood and liver form islands of naïve-like, memory and TEMRA cells. Clusters 20, 21 and 22 have a naïve-like phenotype in the blood, whereas clusters 20 and 22 have a memory phenotype and cluster 21 a TEMRA phenotype in the liver.
(C) Cells colored according to the biosample source: liver CD8+ T cells (red) and blood CD8+ T cells (blue).
(D) Heatmap displaying median expression values of cell surface markers in the 25 clusters of CD8+ T cells in blood (left panel) and liver (right panel).
(E) Spearman correlation between the frequency of cells present in cluster 21 and ALT activity. Two patients without cells in cluster 21 were excluded. FIt-SNE: Fast Fourier Transform-accelerated Interpolation-based t-Stochastic Neighbor Embedding; TEMRA: terminal effector memory re-expressing CD45RA.
Differential phenotypes of HDV-specific and total CD8+ T cells in blood and liver of HDV-infected patients
Analysis based on the source of the biosamples showed that most blood CD8+ T cells were naïve (51.33%), about a third were TEMRA (31.6%) and the minority were memory cells (16.86%, Fig. 2D, left panel). Blood CD8+ T cells grouped mainly with clusters 1 (15.41%), 2 (14.97%) and 3 (10.71%). Clusters 1 and 2 represented naïve CD38+ cells with high expression of the IL-7 receptor CD127, the innate-like activation receptor NKG2D and (for cluster 1 only) the activation/exhaustion marker CD39. Cluster 3 contained activated, terminally differentiated (CD57+) TEMRA cells.
In contrast, most liver CD8+ T cells exhibited a memory or TEMRA phenotype (Fig. 2D, right panel). Clusters 17 (51.38%), 8 (15.01%) and 3 (11.39%), combined accounting for more than 75% of liver CD8+ T cells. Cluster 17 was an effector memory subset that expressed markers of tissue residency (CD69+CXCR6+), activation (CD38+) and degranulation (CD107a+) but was found in an exhausted state (PD-1+CD39+). Cluster 8 was a tissue-resident TEMRA population with increased degranulation (CD107a+) and expression of the innate-like receptor NKG2D which is associated with TCR-independent bystander activation. Cluster 3 from the liver was similar to cluster 3 from the blood, but expressed the tissue-residency markers CD69 and CXCR6 and high levels of CD107a and NKG2D. Most blood CD8+ T cells were naïve, whereas most intrahepatic CD8+ T cells were activated and/or terminally differentiated liver-resident cells.
HLA/epitope multimer staining for HDV-specific CD8+ T cells in the blood gave the greatest signal in clusters 24 (memory cells), 14 (TEMRA), 20 and 21 (naïve-like cells Fig. 2D, left panel). Cluster 24 contained memory-like CD127+PD-1+CD38+CD39+NKG2D+ cells that were unique to the blood and not represented in the liver. Cluster 14 contained TEMRA cells with an activated and terminally differentiated CD38+CD57+CD69+CD39+NKp30+NKG2D+ phenotype. Clusters 20 and 21 contained cells that were activated (CD38+) and degranulated (CD107a+) cells with high CD127 expression. Notably, these cells also expressed the innate-like receptors NKG2D and NKp30.
HLA/epitope multimer staining of liver samples was greatest in clusters 14, 20 and 21 (Fig. 2D, right panel). The phenotype of cells in cluster 14 in the liver resembled that of cluster 14 in the blood, but the expression level of CD39 and NKG2D was higher in the liver. Cells in clusters 20 and 21 had an activated memory-like phenotype (CD127+PD1+CD38+CD107a+CD39+NKG2D+) with higher levels of PD-1 expression than in the corresponding cluster in the blood. Of note, the size of cluster 21 within the intrahepatic CD8+ T-cell population correlated perfectly with alanine aminotransferase activity (r = 1.00, P = .0157, Fig. 2E).
In conclusion, the intrahepatic CD8+ T-cell population was less diverse than in the blood and highly enriched for HDV-specific and total CD8+ T cells with an effector memory or TEMRA phenotype. These cells displayed increased expression of markers of tissue-residency and innate-like activation, degranulation and exhaustion.
Most activated and degranulating CD8+ T cells in the liver are tissue-resident
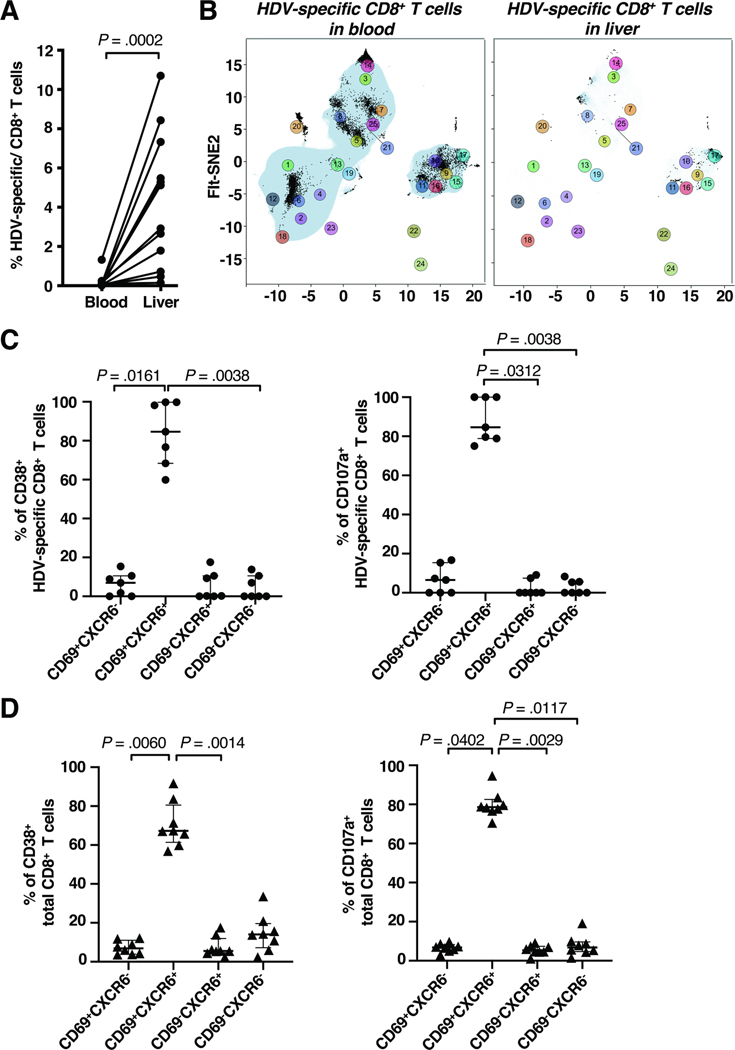
Next, we analyzed the phenotype of HDV-specific CD8+ T cells in liver and blood compared to that of total CD8+ T cells (for manual gating strategy see Suppl. Fig. 3, 4). HDV-specific CD8+ T cells were significantly more frequent in the liver than in the blood (median 80-fold enrichment, P = .0002), with up to 11% of the intrahepatic CD8+ T-cell population targeting individual HDV epitopes (Fig. 3A).
Fig. 3: Most activated and degranulating CD8+ T cells in the liver are tissue-resident.
(A) Frequency of HDV-specific CD8+ T cells in liver and blood.
(B) Flt-SNE visualization of the distribution of HDV-specific CD8+ T cells (black dots) relative to total blood (left panel) and liver CD8+ T cells (right panel).
(C, D) Activated (left) and degranulating (right) HDV-specific CD8+ T cells (C) and total CD8+ T cells (D) were assessed for markers of liver residency (CD69 and CXCR6).
Statistics: Wilcoxon matched-pairs signed-rank test, Friedman test with Dunn’s multiple comparisons test. FIt-SNE: Fast Fourier Transform-accelerated Interpolation-based t-Stochastic Neighbor Embedding.
In the blood, 42% of HDV-specific CD8+ T cells were located in cluster 20 (naïve-like cells), 17.5% in cluster 14 (TEMRA cells), 13.7% in cluster 21 (naïve-like cells), 6.3% in cluster 9 (memory cells) and 4.3% in cluster 6 (naïve-like cells, Fig. 3B left panel). In the liver, 75.9% of HDV-specific CD8+ T cells present were in cluster 17 (memory cells), 7.6% in cluster 8 (TEMRA cells), 3.9% in cluster 21 (TEMRA cells), 3.4% in cluster 9 (memory cells) and 3% in cluster 3 (TEMRA cells, Fig. 3B right panel). This confirms the results of the unsupervised analysis that HDV-specific CD8+ T cells in the blood were mostly naïve-like, whereas HDV-specific CD8+ T cells in the liver were mostly effector memory cells.
More than 80% of the activated or degranulating HDV-specific CD8+ T cells in the liver expressed the tissue residency markers CD69 and CXCR6 (Fig. 3C). This liver--residency phenotype extended to the activated and degranulating total CD8+ T-cell population (Fig. 3D), thus appeared to be independent from T cell receptor specificity.
The liver microenvironment affects the phenotype of HDV-specific and total CD8+ T cells
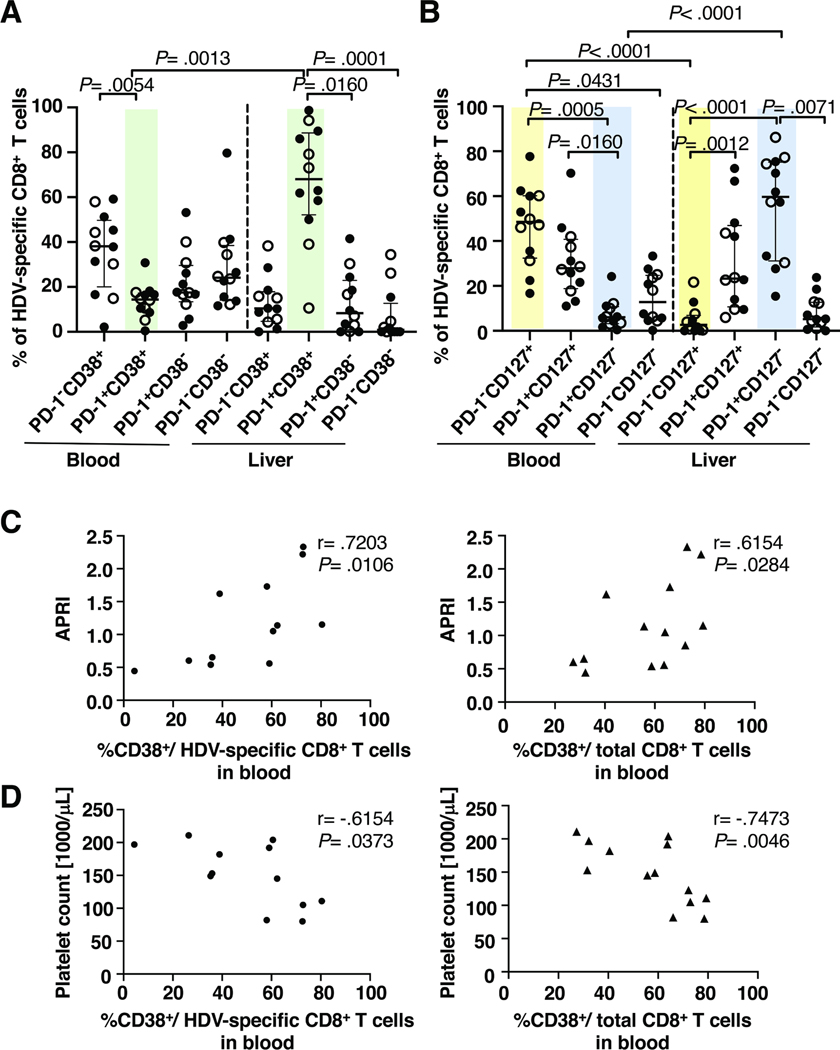
To further assess the effect of the liver environment, we quantitated via manual gating the frequency of HDV-specific and total CD8+ T cells that express specific markers of interest. The median percentage of cells expressing individual markers or combinations thereof is presented in Fig. 4 for both HDV-specific and total CD8+ T cells in blood and liver. Bar graphs with individual data points are shown in Suppl. Fig. 5 and 6.
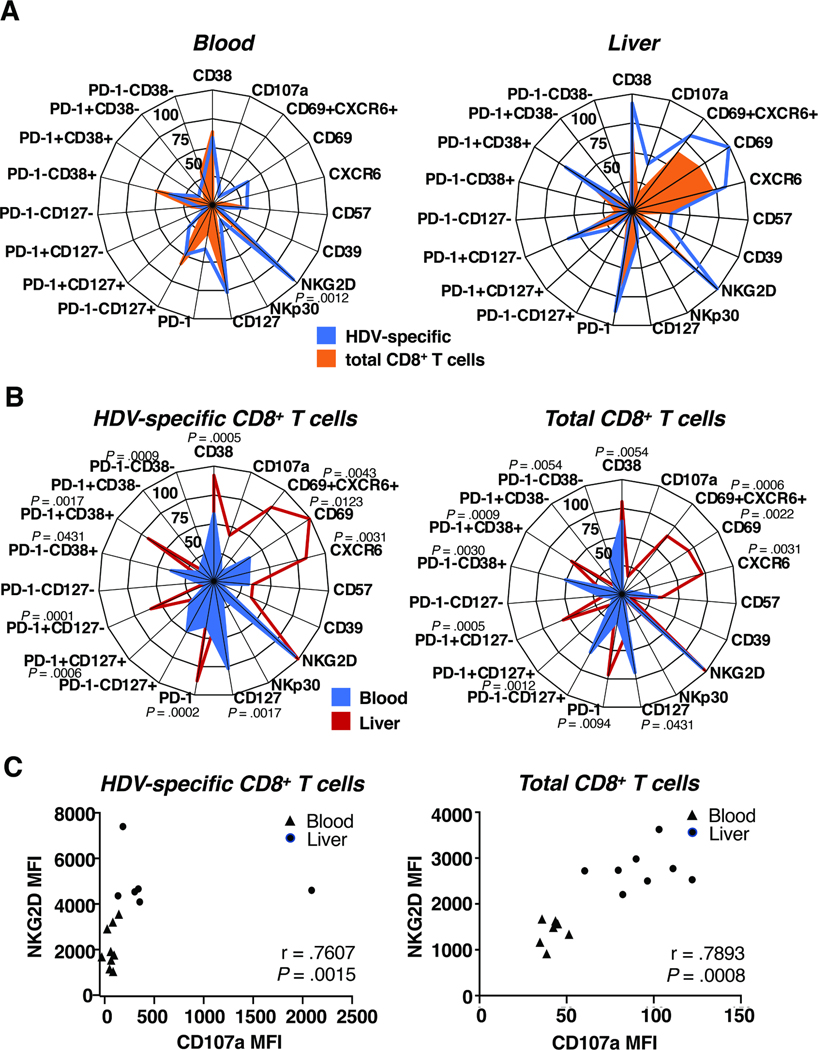
Fig. 4: The liver microenvironment affects the phenotype of HDV-specific and total CD8+ T cells.
(A, B) Radar plots depicting the median frequency of subsets of HDV-specific and total CD8+ T cells in paired blood samples and liver biopsies of HDV-infected patients based on their localization (A) and antigen specificity (B) (raw data shown in Suppl. Fig. 5, 6). Statistics: Friedman test with Dunn’s multiple comparisons test.
(C) Spearman correlation between the NKG2D and CD107a median fluorescence intensity (MFI) on HDV-specific (left) and total CD8+ T cells (right).
The phenotype of HDV-specific CD8+ T cells did not differ significantly from that of the total CD8+ T cell population (Fig. 4A, Suppl. Fig. 5 and 6). Activated/exhausted cells were equally frequent among HDV-specific compared to total CD8+ T cells (Fig. 4A). About half of the HDV-specific and total CD8+ T cells were PD-1-CD127+ (48.5% and 57.8% respectively, Fig. 4A), confirming the results of the unsupervised Flt-SNE analysis. Solely the frequency of NKp30+ cells was significantly higher among HDV-specific than among total CD8+ T-cells in the blood (P = .0012, Fig. 4A).
When comparing HDV-specific or total CD8+ T cells between the blood and the liver compartment, there were many more phenotypic differences detected (Fig. 4B). HDV-specific and total CD8+ T cells were much more activated and exhausted in the liver than in the blood (P = .0005 and P = .0054 for CD38+, P =.0002 and P = .0094 for PD-1+, P = .0431 and P = .0030 for PD-1−CD38+, P = .0017 and P = .0009 for PD-1+CD38+, P = .0009 and P = .0054 for PD-1−CD38− cells respectively, Fig. 4B), with around 80% of intrahepatic HDV-specific CD8+ T cells being activated (CD38+). In the liver, 56.5% of HDV-specific cells and 52.6% of total CD8+ T cells were PD-1+CD38+, compared to only 14.4% and 6.6% in the blood. In the liver, there was a significant enrichment of cells with the CD69+CXCR6+ tissue residency phenotype, with 81.5% of HDV-specific and 64.2% of total CD8+ T cells falling in this category compared to 9.44% of the HDV-specific CD8+ T cells in the blood. (P = .0123 and P = .0022 for the CD69+, P = .0031 for the CXCR6+ and P = .0043 and P = .0006 for the CD69+CXCR6+ population respectively, Fig. 4B). More HDV-specific and total CD8+ T cells in the liver than in the blood were PD-1+CD127− (57.4% and 56.1% compared to 6% and 7.2%, P = .0001 and P = 0.0005 respectively, Fig. 4B). There was a trend for HDV-specific and total CD8+ T cells degranulating more in the liver than in the blood (Fig. 4B, C). The extent of degranulation as determined by median fluorescence intensity of cell surface CD107a expression, correlated with expression of the innate-like receptor NKG2D on HDV-specific and total CD8+ T cells (r = .7607 and r=.7803, respectively, Fig. 4C). These data suggest that innate-like, bystander mechanisms contribute to CD8+ T-cell activation.
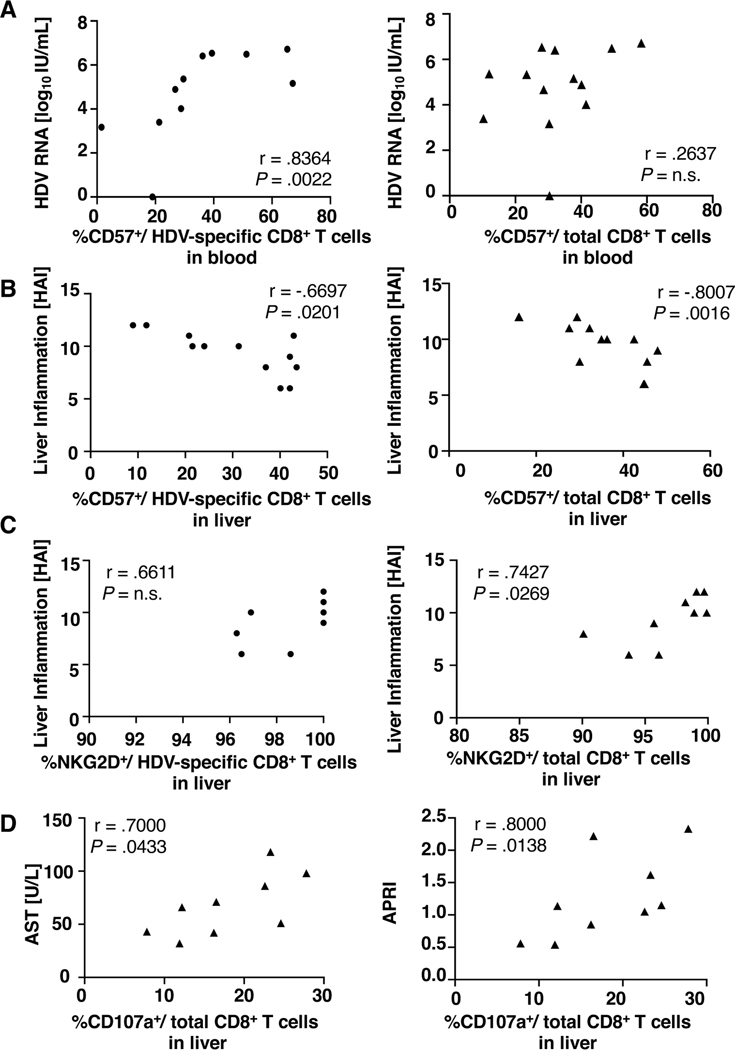
The phenotype of HDV-specific and total CD8+ T cells correlates with disease parameters
As shown in figure 5A, the frequency of terminally differentiated (CD57+) HDV-specific CD8+ T cells correlated with HDV viremia (r = .8364, P = .0022). This was not observed for total CD8+ T cells, consistent with the notion that HDV antigen recognition drives T-cell differentiation. However, when correlating the phenotype of CD57+ CD8+ T cells in the liver with liver inflammation (HAI inflammatory score), a significant correlation was found not only for the HDV-specific but also for the total CD8+ T-cell population (Fig. 5B). The HAI inflammatory score also correlated with the percentage of total CD8+ T cells expressing the innate-like receptor NKG2D (Fig. 5C, left panel). Likewise, the percentage of degranulating CD107a+ cells in the total CD8+ T-cell population correlated with serum AST activity (Fig. 5D, left panel), a biomarker of hepatocyte death, and with AST-to-platelet-ratio index (APRI, Fig. 5D, right panel), a biomarker of liver fibrosis and cirrhosis.
Fig. 5: The phenotype of HDV-specific and total CD8+ T cells correlates with inflammation.
Spearman correlation between (A) the frequency of terminally differentiated (CD57+) HDV-specific (left panel) and total CD8+ T cells (right panel) in the blood and HDV viremia, (B) the frequency of terminally differentiated (CD57+) HDV-specific (left panel) and total CD8+ T cells (right panel) in the liver and HAI score, (C) the frequency of NKG2D+ HDV-specific CD8+ T cells (left panel) and total CD8+ T cells (right panel) in the liver and HAI score, (D) the frequency of degranulating (CD107a+) CD8+ T cells in the liver and AST activity (left panel) and APRI score (right panel). AST: Aspartate aminotransferase; APRI: AST to platelet ratio index; HAI: histologic activity index; N.s: not significant.
Overall, differences in immune cell phenotypes between liver and blood compartments by far exceeded the difference between HDV-specific and total CD8+ T cells in each compartment. Correlations with disease activity were not only observed for the HDV-specific CD8+ T-cell population but also for the total CD8+ T-cell population in the liver.
Antigen-nonspecific bystander activation induces cytotoxicity
Previous studies reported that HDV induces antigen-processing and presentation26 but escapes from CD8+ T-cell recognition via mutation.6–8 Consistent with this notion, 6 of the 12 patients that were studied for HDV-specific CD8+ T-cell responses, harbored HDV escape mutants in the current study. In 5 of these 6 cases, viral escape was due to >100-fold reduction or complete abrogation of binding of the respective T cell epitopes to the patients’ HLA. In the 6th case, HLA-binding was preserved but T-cell receptor stimulation was abrogated, as confirmed with functional assays for T-cell IFN-γ production and cytotoxicity (Suppl. Fig. 7A, B). Collectively, these results suggest that TCR-mediated activation of HDV-specific CD8 T cells is reduced in chronic HDV infection.
To evaluate whether CD8+ T cell activation and function is induced in a TCR- and HLA-independent bystander manner, we cocultured IL-15-activated CD8+ T cells from HDV-uninfected blood donors with Huh7 hepatoma cells. As shown in Suppl. Fig. 7, the IL-15-activated CD8+ T cells lysed the hepatoma cells even though the T cells were not HDV-specific and did not share any HLA with the hepatoma cells. Cytotoxicity was reduced in the presence of anti-NKG2D (Suppl. Fig. 7C), suggesting innate-like effector function. Indeed, IL-15 increased NKG2D expression on CD8+ T cells (Suppl. Fig. 7D) and HDV induced the expression of NKG2D ligands on hepatoma cells (Suppl. Fig. 7E). Thus, cytokine-mediated activation of bystander CD8+ T cells may contribute to liver injury.
Consistent with bystander activation, the PD-1/CD38 or PD-1/CD127 phenotype of CD8 T cells in blood and liver was not affected by viral escape (Figure 6A, B, comparing filled and open circles respectively). The frequency of activated CD38+ cells correlated with APRI score (Fig. 6C) and inversely with platelet count (Fig. 6D) and this correlation held true for both HDV-specific and for total CD8 T cells. Thus, HDV-specific CD8+ T cells and nonspecific CD8+ T cells in the liver share many phenotypic features. Cytokine-mediated bystander activation may contributes to their effector function.
Fig. 6: The liver environment has a greater effect on the phenotype of HDV-specific CD8+ T cells than viral sequence variation.
(A, B) Phenotype of HDV-specific CD8+ T cells that do (filled circle) or do not (open circle) recognize the sequence of the respective patients’ HDV strain. Shaded backgrounds mark cell populations with significant differences in frequency between liver and blood. Statistics: Friedman test with Dunn’s multiple comparisons test.
(C, D) Spearman correlation between the frequency of activated (CD38+) HDV-specific (left panel) or total blood CD8+ T cells (right panel) and APRI score (C) and between the frequency of activated (CD38+) HDV-specific (left panel) or total CD8+ T cells (right panel) in the blood and platelet count (D).
Discussion
This is the first report to describe the immune-cell landscape in the liver during chronic HDV infection. We found that the two major innate immune-cell populations in the liver, MAIT cells and NK cells were similarly affected by HDV infection. Compared to healthy livers from uninfected controls, their frequency was decreased in the livers of HDV-infected patients. Both MAIT cells and NK cells were more frequently activated and degranulated in the liver than in the blood. This recapitulates the cytokine-driven phenotype previously described in HCV infection.27, 28 Because HDV induces the same inflammatory cytokines as HCV4, 29 and because HBV replication is suppressed in the patients studied (Table 1), this phenotype is likely attributed to HDV.
The characterization of intrahepatic CD8+ T cells was the main focus of this study. Using HLA/epitope multimers for ex vivo staining of lymphocytes, we found HDV-specific CD8+ T cells significantly enriched in the liver with up to 11% of the intrahepatic CD8+ T-cell population specific for a single HDV epitope. This is comparable to the frequency of intrahepatic HBV-specific CD8+ T cells in chronic hepatitis B30, and more than 10-fold higher than reported for HCV-specific CD8+ T cells in chronic hepatitis C.31–33 The majority of HDV-specific CD8+ T cells were liver-resident (CD69+CXCR6+). This is again consistent with CXCR6 expression in chronic hepatitis B,12 where HBV-specific CD8+ T cells have been shown to persist in transplanted livers for more than a decade.10 As CD69 expression can be driven by TCR- or cytokine-mediated stimulation34, HDV-specific CD8+ T cells may have either seeded in the liver during acute inflammation or been primed in the liver.
As in HBV30 and HCV infection33, almost all intrahepatic HDV-specific CD8+ T cells displayed increased PD-1 and decreased CD127 expression, indicating impaired ability for homeostatic proliferation in response to IL-7. As shown by both unsupervised Flt-SNE analysis and manual gating of flow cytometry data, this phenotype extended to the total intrahepatic CD8+ T-cell population. HDV-specific and total CD8+ T cell populations did not differ in CD38 expression and HDV-specific CD8+ T-cells had the same PD-1/CD38 or PD-1/CD127 phenotype irrespective of viral escape from T-cell recognition.
CD8+ T cells expressed the innate-like activation receptor NKG2D, and NKG2D expression correlated with degranulation (CD107a expression), a biomarker for cytotoxicity. NKG2D expression on CD8+ T cells was increased in the liver as compared to the blood, and can be induced by IL-15 as shown in this study and in a recent report on HAV infection.35 Moreover, increased NKG2D expression on intrahepatic CD8+ T cells has also been observed in chronic HBV and HCV infection.36 Because IL-15 is locally trans-presented by the IL-15-producing cell to the IL-15-responding cell, systemic IL-15 levels may not always be increased as shown here and in HDV-superinfected chimpanzees.37 This upregulation of NKG2D on intrahepatic CD8 T cells can result in innate-like cytotoxicity of CD8+ T cells, irrespective of the TCR-specificity. Accordingly, intrahepatic NKG2D mRNA levels are highest in patients with severe hepatitis38. This process may be particularly relevant in HDV infection, because HDV upregulates NKG2D ligands on hepatoma cells (Suppl. Fig. 7E).
The proposed concept of a role of bystander activation in chronic liver inflammation and disease progression is relevant for new therapies that aim to induce immune-mediated viral control because pre-existing bystander activation impairs memory T-cell development.39 As recently proposed,40 it may be feasible to develop blocking reagents that reduce immune-mediated pathology by interfering with the recognition of innate-like receptor ligands by CD8+ T cells. For example, antibody-mediated blocking of NKG2D engagement was shown to reduce virus-induced bystander memory-cell activation, effector function and immunopathology in a mouse model of leishmaniosis.41 It would therefore be of interest to explore this approach in chronic inflammatory liver disease.
Supplementary Material
What you need to know.
BACKGROUND AND CONTEXT
Superinfection of chronic hepatitis B with the hepatitis D virus accelerates progression to liver cirrhosis. The role of the innate and adaptive immune response in liver disease pathogenesis are unknown.
NEW FINDINGS
We observed global activation and degranulation of all liver-resident CD8+ T cells irrespective of their antigen-specificity and presence of HDV escape mutations. Expression of the innate-like receptor NKG2D on CD8+ T cells correlated with the degree of degranulation, suggesting bystander activation.
LIMITATIONS
The small number of cells isolated from liver biopsies limited the functional analysis of intrahepatic immune cells to the ex vivo assessment of degranulation, a biomarker of cytotoxicity.
IMPACT
Activation and function of all, rather than just HDV-specific, CD8+ T cells contribute to liver disease pathogenesis. It may be feasible to develop blocking reagents that reduce immune-mediated pathology by interfering with bystander T-cell activation.
Acknowledgement
We thank Dr. Alessandro Sette, La Jolla Institute for Allergy and Immunology, for HLA-B*35:01-positive target cell lines for 51Cr release assays, Dr. Stephan Urban, University of Heidelberg Germany for HuH7-END cells and an HDV-specific antibody, the NIH Tetramer Core Facility at Emory University, Atlanta, GA for tetramer synthesis and the NIDDK/NHLBI Flow Cytometry Facility for sorting HDV-specific CD8+ T cells. Part of the graphical abstract was created with BioRender.com (Toronto, ON, Canada).
Grant support:
This work was supported by the intramural research program of NIDDK, NIH.
Abbreviations:
- AST
aspartate aminotransferase
- APRI
aspartate aminotransferase to platelet ratio index
- CD
cluster of differentiation
- CXCR6
C-X-C motif chemokine receptor 6
- DNA
deoxyribonucleic acid
- FACS
fluorescent activated cell sorting
- FBS
fetal bovine serum
- Flt-SNE
fast Fourier transform-accelerated interpolation-based t-stochastic neighbor embedding
- HAI
histologic activity index
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HDV
hepatitis D virus
- HLA
human leucocyte antigen
- IL
interleukin
- MAIT cell
mucosal-associated invariant T-cell
- MHC
major histocompatibility complex
- NK cell
natural killer cell
- NKG2D
natural killer group 2D
- NKp30
natural killer protein 30
- PBMCs
peripheral blood mononuclear cells
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PD-1
programmed cell-death 1
- PE
Phycoerythrin
- RNA
ribonucleic acid
- TCR
T-cell receptor
- TEMRA
terminal effector memory re-expressing CD45RA
Footnotes
Conflict of interest statement: The authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lempp FA, Ni Y, Urban S. Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options. Nat Rev Gastroenterol Hepatol 2016;13:580–9. [DOI] [PubMed] [Google Scholar]
- 2.Kleiner DE. Pathology of hepatitis C. In: Liang TJ, Hoofnagle JH, eds. Hepatitis C: Academic Press, 2000:107–124. [Google Scholar]
- 3.Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology 2006;43:S54–62. [DOI] [PubMed] [Google Scholar]
- 4.Dias J, Hengst J, Parrot T, et al. Chronic hepatitis delta virus infection leads to functional impairment and severe loss of MAIT cells. J Hepatol 2019;71:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lunemann S, Malone DF, Grabowski J, et al. Effects of HDV infection and pegylated interferon alpha treatment on the natural killer cell compartment in chronically infected individuals. Gut 2015;64:469–82. [DOI] [PubMed] [Google Scholar]
- 6.Karimzadeh, Kiraithe MM, Kosinska AD, et al. Amino acid substitutions within HLAB*27-Rrestricted T Cell epitopes prevent recognition by hepatitis Delta virus-specific CD8(+) T Cells. J Virol 2018;92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karimzadeh H, Kiraithe MM, Oberhardt V, et al. Mutations in hepatitis D virus allow It to escape detection by CD8(+) T cells and evolve at the population level. Gastroenterology 2019;156:1820–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kefalakes H, Koh C, Sidney J, et al. Hepatitis D virus-specific CD8(+) T cells have a memory-like phenotype associated with viral immune escape in patients with chronic hepatitis D virus infection. Gastroenterology 2019;156:1805–1819 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity 2014;41:886–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pallett LJ, Burton AR, Amin OE, et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J Exp Med 2020;217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinbach K, Vincenti I, Merkler D. Resident-memory T cells in tissue-restricted immune responses: for better or worse? Front Immunol 2018;9:2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pallett LJ, Davies J, Colbeck EJ, et al. IL-2(high) tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. J Exp Med 2017;214:1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knodell RG, Ishak KG, Black WC, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1981;1:431–5. [DOI] [PubMed] [Google Scholar]
- 14.Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol 1995;22:696–9. [DOI] [PubMed] [Google Scholar]
- 15.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 2003;12:1007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber LM, Robinson MD. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytometry A 2016;89:1084–1096. [DOI] [PubMed] [Google Scholar]
- 19.Levine JH, Simonds EF, Bendall SC, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 2015;162:184–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 2019;9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun 2019;10:5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehermann B, Fowler P, Sidney J, et al. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J Exp Med 1995;181:1047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni Y, Zhang Z, Engelskircher L, et al. Generation and characterization of a stable cell line persistently replicating and secreting the human hepatitis delta virus. Sci Rep 2019;9:10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betts MR, Brenchley JM, Price DA, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods 2003;281:65–78. [DOI] [PubMed] [Google Scholar]
- 25.Valenzuela J, Schmidt C, Mescher M. The roles of IL-12 in providing a third signal for clonal expansion of naive CD8 T cells. J Immunol 2002;169:6842–9. [DOI] [PubMed] [Google Scholar]
- 26.Tham CYL, Kah J, Tan AT, et al. Hepatitis Delta Virus Acts as an Immunogenic Adjuvant in Hepatitis B Virus-Infected Hepatocytes. Cell Rep Med 2020;1:100060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolte FJ, O’Keefe AC, Webb LM, et al. Intra-hepatic depletion of mucosal-associated invariant T cells in hepatitis C virus-induced liver inflammation. Gastroenterology 2017;153:1392–1403 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serti E, Chepa-Lotrea X, Kim YJ, et al. Successful interferon-free therapy of chronic hepatitis C virus infection normalizes natural killer cell function. Gastroenterology 2015;149:190–200 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Filzmayer C, Ni Y, et al. Hepatitis D virus replication is sensed by MDA5 and induces IFN-beta/lambda responses in hepatocytes. J Hepatol 2018;69:25–35. [DOI] [PubMed] [Google Scholar]
- 30.Fisicaro P, Valdatta C, Massari M, et al. Antiviral intrahepatic T-cell responses can be restored by blocking programmed death-1 pathway in chronic hepatitis B. Gastroenterology 2010;138:682–93, 693 e1–4. [DOI] [PubMed] [Google Scholar]
- 31.He XS, Rehermann B, Lopez-Labrador FX, et al. Quantitative analysis of hepatitis C virus-specific CD8(+) T cells in peripheral blood and liver using peptide-MHC tetramers. Proc Natl Acad Sci U S A 1999;96:5692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroy DC, Ciuffreda D, Cooperrider JH, et al. Liver environment and HCV replication affect human T-cell phenotype and expression of inhibitory receptors. Gastroenterology 2014;146:550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radziewicz H, Ibegbu CC, Fernandez ML, et al. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J Virol 2007;81:2545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiow LR, Rosen DB, Brdickova N, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 2006;440:540–4. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Chang DY, Lee HW, et al. Innate-like cytotoxic function of bystander-activated CD8(+) T cells is associated with liver injury in acute hepatitis A. Immunity 2018;48:161–173 e5. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy PT, Gehring AJ, Nowbath A, et al. The expression and function of NKG2D molecule on intrahepatic CD8+ T cells in chronic viral hepatitis. J Viral Hepat 2008;15:901–9. [DOI] [PubMed] [Google Scholar]
- 37.Engle RE, De Battista D, Danoff EJ, et al. Distinct cytokine profiles correlate with disease severity and outcome in longitudinal studies of acute hepatitis B virus and hepatitis D virus infection in chimpanzees. mBio 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Wang W, Shen C, et al. NKG2D modulates aggravation of liver inflammation by activating NK cells in HBV infection. Sci Rep 2017;7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stelekati E, Shin H, Doering TA, et al. Bystander chronic infection negatively impacts development of CD8(+) T cell memory. Immunity 2014;40:801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prajapati K, Perez C, Rojas LBP, et al. Functions of NKG2D in CD8(+) T cells: an opportunity for immunotherapy. Cell Mol Immunol 2018;15:470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crosby EJ, Goldschmidt MH, Wherry EJ, et al. Engagement of NKG2D on bystander memory CD8 T cells promotes increased immunopathology following Leishmania major infection. PLoS Pathog 2014;10:e1003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.