Abstract
The Myc oncoprotein is implicated in transcriptional regulation of a variety of genes pertaining to cell cycle and neoplastic transformation. Examples of both positive and negative regulation have been reported that involve E-box and initiator (Inr) promoter elements, respectively. In both cases, Myc is thought to induce changes in transcription initiation. We have previously shown that overexpression of Myc causes down-regulation of the thrombospondin-1 (tsp-1) gene, an important negative modulator of tumor angiogenesis. In this study, we demonstrate that Myc in combination with Max can bind, albeit with low affinity, to an E-box-like element in the tsp-1 promoter. However, the 2.7 kb DNA segment containing both this non-canonical E-box and an Inr-like sequence does not constitute a Myc-responsive element in a transient expression system. Furthermore, Myc does not significantly affect the rate of initiation or elongation of the tsp-1 mRNA. Thus, in this instance Myc does not act as a canonical transcription factor. Instead, as demonstrated by blocking de novo RNA synthesis, down-regulation of the tsp-1 gene by Myc occurs through increased mRNA turnover. To our knowledge, this is the first example of gene regulation by Myc that involves mRNA destabilization.
INTRODUCTION
It is widely accepted that Myc contributes to neoplastic transformation by altering gene expression. Most Myc target genes pertain to cell growth, including telomerase (1–3) and the cdc25A phosphatase (4). However, there are a growing number of ‘orphan’ targets (5,6) whose contributions to neoplastic transformation remain to be determined. Moreover, despite extensive biochemical characterization, the molecular mechanisms of gene regulation by Myc often remain undeciphered. The likely reason for this complexity is that Myc does not bind to DNA on its own; instead it relies on interactions with a dazzling number of nuclear proteins (7) making the contribution of individual partners difficult to dissect.
We have previously reported that activation of the overexpressed Myc oncoprotein in avian and rodent fibroblasts results in down-regulation of thrombospondin-1 (tsp-1) mRNA (8). The tsp-1 gene is also down-regulated by other oncogenes such as jun (9,10) and src (11). Since tsp-1 is an important negative modulator of neovascularization (12,13), its down-regulation by Myc contributes to the angiogenic phenotype of Myc-overexpressing fibroblasts (14). Importantly, tsp-1 down-regulation could be observed within hours of addition of 4-hydroxytamoxifen (4-OHT), the estrogen analog capable of activating the Myc-estrogen receptor (MycER) chimera used in our study (8). Given the rapid response and since Myc is known to be a DNA binding protein, it seemed plausible that down-regulation of tsp-1 mRNA by Myc would involve direct interaction between Myc and the tsp-1 promoter. This scenario was supported by the fact that the tsp-1 promoter contains, at position –273 (15) the sequence 5′-CGCGTG-3′ that resembles binding sites for Myc, the canonical E-box (5′-CACGTG-3′) (16,17) and related elements (18,19). In the latter report, the authors observed binding of Myc/Max to the sequence 5′-CACGCG-3′ which is the inverse complement of the sequence present in the tsp-1 promoter. It was not clear whether Myc would bind to this inverted element or whether its binding would pertain to down-regulation of the tsp-1 gene, especially since this interaction is thought to result in gene activation, not repression (20,21).
Another promoter element implicated in regulation by Myc is the initiator (Inr) element present in many viral and cellular promoters (especially those lacking the TATA-box) at the site of transcription initiation. Inr is represented by a weak pyrimidine-rich consensus YYA+1NT/AYY (22) that is thought to position the RNA polymerase complex on DNA. Several Inr-containing promoters have been reported to be negatively influenced by Myc raising the possibility that the repressor activity of Myc is realized through Inr (23). In support of this notion, Myc was reported to interact with TFII-I, a component of the transcriptional machinery that binds to the initiator element (24,25). In addition, Myc forms complexes with and inhibits activity of YY1 (26), another Inr-binding transcription factor (27). The transcription start site in the human tsp-1 promoter includes the sequence CCA+1G_CC that lacks T or A at position +3 but otherwise resembles Inr. The presence of an E-box-like and an Inr-like element in the tsp-1 promoter supported the notion that Myc might down-modulate tsp-1 by interfering with transcription initiation.
To test this possibility, we studied molecular mechanisms of regulation of the tsp-1 gene by Myc. Although Myc appeared to bind, albeit with low affinity, to the E-box-like element in the tsp-1 promoter, it did not affect the activity of the 2.7 kb segment of the tsp-1 promoter in transient expression systems. These findings suggested that down-regulation of the tsp-1 gene by Myc might take place at a post-transcriptional level. Indeed, we have found that while Myc has minimal effect on transcription initiation or mRNA elongation, it profoundly increases the turnover of tsp-1 mRNA. In the presence of functional Myc, tsp-1 mRNA half-life is decreased to the extent that can account for its low steady-state levels in Myc-transformed cells.
MATERIALS AND METHODS
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using baculovirus-produced Max and the GST–Myc fusion protein, as described by Grandori et al. (19). Myc and Max were pre-mixed and allowed to form a complex for 10 min before adding DNA. Both sense and antisense oligonucleotides were labeled using T4 polynucleotide kinase (Boehringer Mannheim, Indianapolis, IN) and [γ-32P]ATP (NEN, Boston, MA) and purified via gel-filtration through Sephadex G-25 columns (Amersham Pharmacia, Piscataway, NJ). The complementary kinase reaction products were mixed, heated at 65°C and annealed to form double-stranded molecules by allowing the water bath to cool to room temperature. The compositions of the oligonucleotides were as follows (sense strand only): ACCCCCACCACGTGGTGCCTGA (CM1), ACCCCCACACCGGTGTGCCTGA (CM1*), CATGCCGCCACGCGGGCTGAAC (MM174), CCCGAGCCCGCGTGGCGCAAGA (TSP), CCCGAGCCCCGGGTGCGCAAGA (TSP*). Protein binding was performed by mixing 1 µl of 32P-labeled DNA (25 000 c.p.m. ∼250 pg), 2 µl of Myc/Max complex (25 and 15 ng, respectively) and 9 µl of the reaction mix. The final mix contained the following components: 50 mM KCl, 25 mM HEPES pH 7.5, 5 mM MgCl2, 0.5 mM EDTA, 0.1% NP-40, 1 mM DTT, 20 µg/ml single-stranded salmon sperm DNA, 0.5 mg/ml BSA, 5% glycerol. For competition experiments, unlabeled double-stranded oligonucleotides were included in the mix to attain 25 or 100 times molar excesses over 32P-labeled oligonucleotides. All binding reactions were incubated at room temperature for 30 min and analyzed by 4% PAGE containing 10 mM HEPES pH 7.5. Electrophoresis was performed at 4°C for 1 h at 200 V.
Cell culture
Rat-1A fibroblasts and their Myc-overexpessing derivatives were cultured in DMEM supplemented with 10% fetal calf serum. To activate the MycER fusion protein, cells were treated with the synthetic ER ligand 4-OHT (Sigma, St Louis, MO, catalog H7904 or H6278) at the final concentration of 250 nM. To block de novo RNA and protein syntheses, cells were treated with actinomycin D (ActD, 5 µg/ml; Sigma) and cycloheximide (CHX, 10 µM; Sigma), respectively.
Transient transfection of mammalian cells and chloramphenicol acetyltransferase (CAT) and secreted alkaline phosphatase (SEAP) assays
Several methods were employed to introduce plasmid DNA into mammalian cells. To transfect Rat-1A cells already expressing Myc (or the parental cells) electroporation was used. Rat-1A, Rat-1A/LMycSN or Rat-1A/MycER cells (106) were mixed at room temperature in DMEM with 10 µg of the TSP1A-, 2A-, 12A- or MSV-CAT plasmids and 2 µg of the CMV-β-gal plasmid. The mixes were transferred into 0.4 cm cuvettes and electroporated in the Bio-Rad Gene Pulser apparatus (Hercules, CA). Electroporation conditions were as follows: 800 V/cm, 960 µF and 13 ms pulse time. In the case of Rat-1A/MycER cells, after electroporation cultures were split into two aliquots, one of which was maintained without and the other with 4-OHT.
For co-transfection experiments in 293 cells, standard calcium phosphate or lipofection techniques were employed. Precipitates were formed by adding to DNA 250 µl of 0.25 M CaCl2 and 250 µl of 2× BES-buffered saline (BBS) (pH 6.95). Lipofections were carried out using Lipofectamine Plus from Life Technologies (Rockville, MD). Typically, 5 × 105 293 cells were transfected with 1 µg of pTSP1A-CAT, pTSP-SEAP or pM4min-CAT, and increasing amounts of Myc-expressing vector pSPMyc (20). The total amount of DNA in each reaction was adjusted using irrelevant DNA (pGEM cloning vector from Promega, Madison, WI). All transfection mixes also contained 1 µg of pEQ176 (28) or pMAM-luc (Clontech, Palo Alto, CA). These two plasmids express β-gal and luciferase, respectively, using strong, constitutively active retroviral promoters. Prior to performing CAT/SEAP assays, all lysates were diluted to contain equal amounts of β-gal or luciferase (U/ml). Both CAT and β-gal assays have been described previously (8). CAT assays were quantitated using a phosphoimager by measuring the amount of chloramphenicol converted into monoacetylated forms. Luciferase and SEAP assays were performed using detection kits from Clontech and ICN Pharmaceuticals (Costa Mese, CA), respectively, and a luminometer from Promega.
Nuclear run-off transcription assay
For nuclei isolation, ∼108 cells were harvested, washed twice with PBS and resuspended in 2 ml of ice-cold Nuclei Isolation Buffer [10 mM Tris–Cl pH 7.5, 5 mM Mg(OAc)2, 1 mM DTT, 0.5% NP-40]. The cell suspension was incubated on ice for 5 min and transferred to a chilled 7 ml Dounce homogenizer. Cells were disrupted using 25–30 strokes with tight-fitting pestle A. Nuclei were spun for 5 min at 1000 r.p.m. at 4°C, and the pellets were resuspended in 1.1 ml of Nuclei Storage Buffer [50 mM Tris–Cl pH 7.5, 5 mM Mg(OAc)2, 25% glycerol]. Nuclei were frozen in liquid nitrogen in 210 µl aliquots and stored for up to several months.
To perform run-off assays, nuclei were thawed on ice and mixed with 60 µl of 5× High KCl buffer (24 mM Tris–HCl pH 8.0, 12 mM MgCl2, 720 mM KCl) and 27.5 µl of the nucleotide mixture {25 µl [32P]UTP (3000 Ci/mmol), 1 µl each of 100 mM ATP, CTP and GTP}. The samples were incubated at 29°C for 30 min and then treated with 50 µl RNase-free DNase I (Worthington, Lakewood, NJ) at 37°C for 10 min. Then 36 µl 10× SET (100 mM Tris–Cl pH 7.5, 50 mM EDTA, 10% SDS) and 10 µl Proteinase K (10 mg/ml) were added and the reactions were incubated for 1 h at 50°C. RNAs were extracted with phenol–chloroform, precipitated with ethanol and resuspended in 100 µl H2O. Unincorporated [32P]UTP and short oligonucleotides were removed by passing the RNAs through Sephadex G-50 columns (Boehringer Mannheim).
De novo synthesized [32P]UTP-labeled RNAs were hybridized to denatured DNA probes immobilized on nitrocellulose membranes at 65°C overnight in 1 ml Hybridization Buffer (10 mM TES pH 7.4, 10 mM EDTA, 300 mM NaCl, 0.25% dry milk, 1% SDS, 250 µg/ml Escherichia coli RNA, 1× Denhardt’s solution). After 12–18 h filters were rinsed briefly at room temperature in 2× SSC and then washed at 37°C for 30 min in 2× SSC containing 10 µg/ml RNase A, followed by two washes at 65°C for 15 min in 1× SSC/1% SDS. Washed filters were exposed to X-ray film or a phosphoimager screen.
RNase protection assay (RPA)
RPA itself and generation of riboprobes for the rat tsp-1 and gapdh genes were described in detail previously (8). The plasmid containing murine c-fos sequences from nucleotide –56 to +109 (p149; obtained from the laboratory of Dr Mark Groudine, Fred Hutchinson Cancer Research Center, Seattle, WA) was linearized with HindIII and transcribed using the T7 RNA polymerase. The resultant 225 nt probe protects a 110 nt fragment of the c-fos exon 1. Direct Protect Kit from Ambion Inc. (Austin, TX) was used in all experiments. In the experiment involving nuclear RNAs, nuclei were purified prior to RNA extraction as described in the previous section. Intensities of protected fragments were quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, and Bio-Rad).
Radioimmunoprecipitation
Detection of the Myc oncoprotein via radioimmunoprecipitation has been described earlier (29). For this work, we were using the antibody derived against human c-Myc protein kindly provided by Dr Robert Eisenman (Fred Hutchinson Cancer Research Center).
RESULTS
Myc binds with low affinity to the E-box-like element in the promoter region of the tsp-1 gene
To determine whether Myc is capable of binding to the tsp-1 gene promoter, we performed an EMSA on the oligonucleotide TSP containing the E-box-like sequence 5′-CGCGTG-3′ representing base pairs –273/–268 of the tsp-1 promoter (15). As positive and negative controls, respectively, we used the oligonucleotide CM1 containing the canonical Myc-binding site 5′-CACGTG-3′ (16) and its variant CM1* with the scrambled core sequence (5′-ACCGGT-3′). We have also used the oligonucleotide MM174 that contains a non-canonical Myc-binding sequence found in co-precipitates of Myc and genomic DNA (19). The protein components were represented by GST–Myc fusion protein and baculovirus-expressed Max, as described by Grandori et al. (19).
As expected, virtually no binding was detected to the scrambled CM1* variant or to any oligonucleotide by Myc alone (Fig. 1A). However, when combined with Max, Myc bound to CM1 avidly, as did Max alone. Importantly, the Myc/Max complex bound to both MM174 and TSP oligonucleotides, although the intensities of the shifted bands were diminished compared to that of CM1. Binding of Max alone was also greatly diminished. This suggested that the Myc/Max complex has lower affinity for non-canonical binding sites and would preferentially bind to the 5′-CACGTG-3′ hexamer. To confirm this, we performed competition experiments where both CM1 and TSP oligonucleotides were included in the reaction (Fig. 1B). When the CM1 oligonucleotide was 32P-labeled, ‘cold’ TSP, unlike ‘cold’ CM1, was not an effective competitor, even at a 100-fold molar excess (left, compare lanes 3 and 5). Similarly, when the TSP oligonucleotide was radioactively labeled, ‘cold’ CM1 competed more efficiently than ‘cold’ TSP (right, lanes 3 and 5). The fact that ‘cold’ TSP competed poorly even with the radiolabeled oligonucleotide of the same composition raised the concern that binding of Myc and Max to TSP was not specific. To rule out this possibility, we synthesized the TSP* oligonucleotide that differed from TSP only in that its E-box (but not the adjacent sequences) was scrambled (5′-CCGGGT-3′ instead of 5′-CGCGTG-3′). We found that binding of the Myc/Max protein complex to this oligonucleotide is greatly diminished, and the majority of residual DNA–protein complexes have lower electrophoretic mobilities (Fig. 1C) than TSP-, CM1- and MM174-specific complexes (Fig. 1A and B). Thus, binding of Myc/Max to TSP requires the presence of the intact E-box, and its weakness is likely to stem not from the random nature of DNA–protein interactions but rather from low affinity of binding to a non-canonical Myc-site. Therefore, in whole nuclei, where targets with canonical E-boxes are present, binding of Myc to the tsp-1 promoter might be too weak to affect its activity. To determine if this was the case, the transient expression experiment had to be performed.
Figure 1.
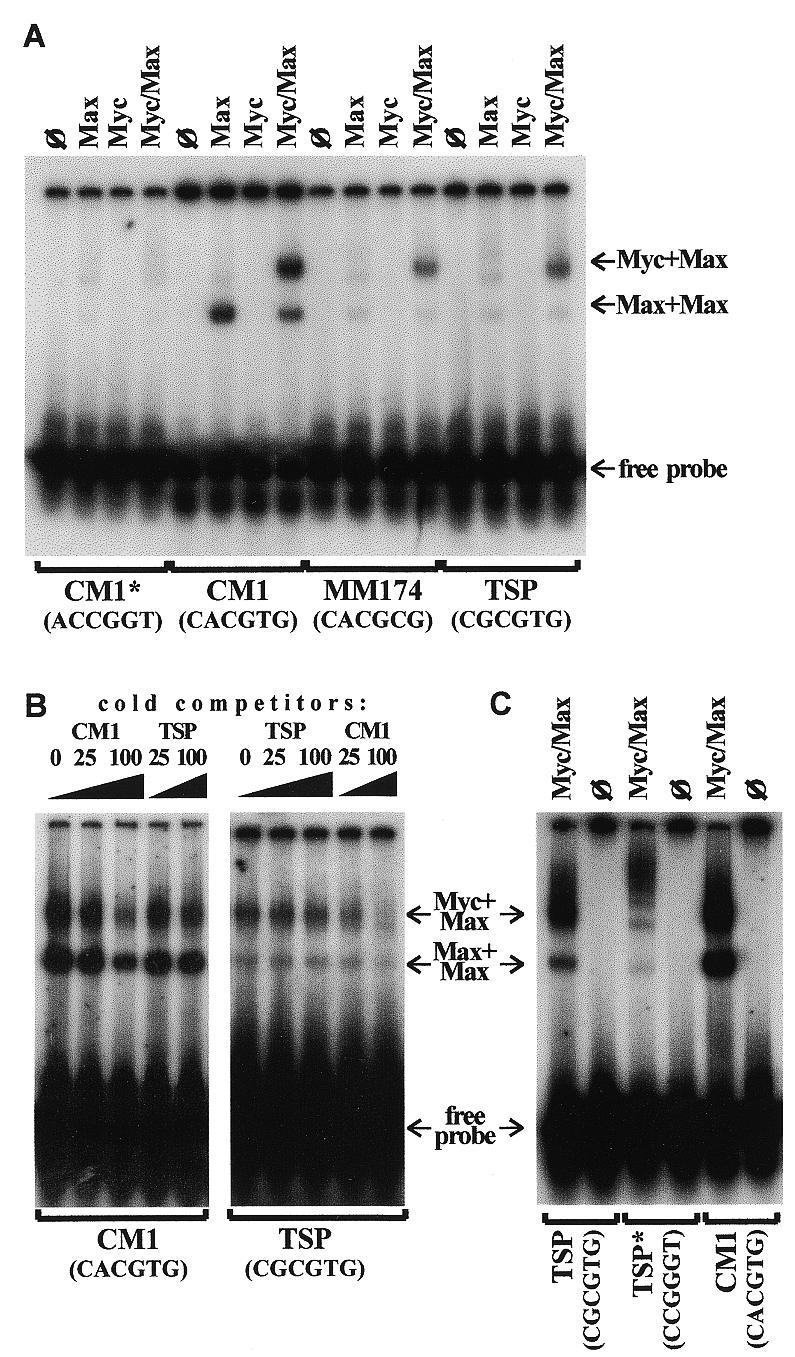
Physical interaction between Myc and the E-box-like element in the tsp-1 gene promoter. (A) EMSA performed on various oligonucleotides with Myc and Max proteins or Myc/Max protein complex. Proteins used in each reaction are denoted at the top; ø denotes reactions with no protein added. Oligonucleotides used in each reaction are denoted at the bottom, with core nucleotide sequences shown in parentheses. For complete sequences see Materials and Methods. Arrows, from top to bottom, show migrations of DNA-bound Myc/Max heterodimers, DNA-bound Max homodimers and free probes, respectively. (B) EMSA performed with the same reagents in the presence of competing unlabeled oligonucleotides. Labeled oligonucleotides used in each reaction are denoted at the bottom. The nature and the amount of ‘cold’ competitors are denoted at the top, with numbers referring to molar excesses over labeled oligonucleotides. (C) EMSA performed on the TSP derivative (TSP*) whose core E-box sequence is scrambled (see Materials and Methods). Oligonucleotides TSP and CM1 were used for comparison. ø denotes reactions with no protein added.
Myc overexpression has no direct effect on the activity of the tsp-1 promoter in a transient expression system
To determine whether overexpression of Myc influences activity of the tsp-1 promoter, we employed a series of plasmids where this promoter is driving expression of the CAT gene. The basic construct, TSP1A-CAT, included ~2 kb of upstream sequences as well as the untranslated exon 1 and intron 1; two shorter deletion variants (TSP2A-CAT and 12A-CAT, 30) were also used (Fig. 2A). These plasmids were transfected into either parental Rat-1A cells or their counterparts infected with the Myc retrovirus (LMycSN) (8,31). As a control construct, not expected to be affected by Myc, we used the MSV-CAT plasmid containing a strong, constitutively active promoter/enhancer of murine sarcoma retrovirus (32).
Figure 2.
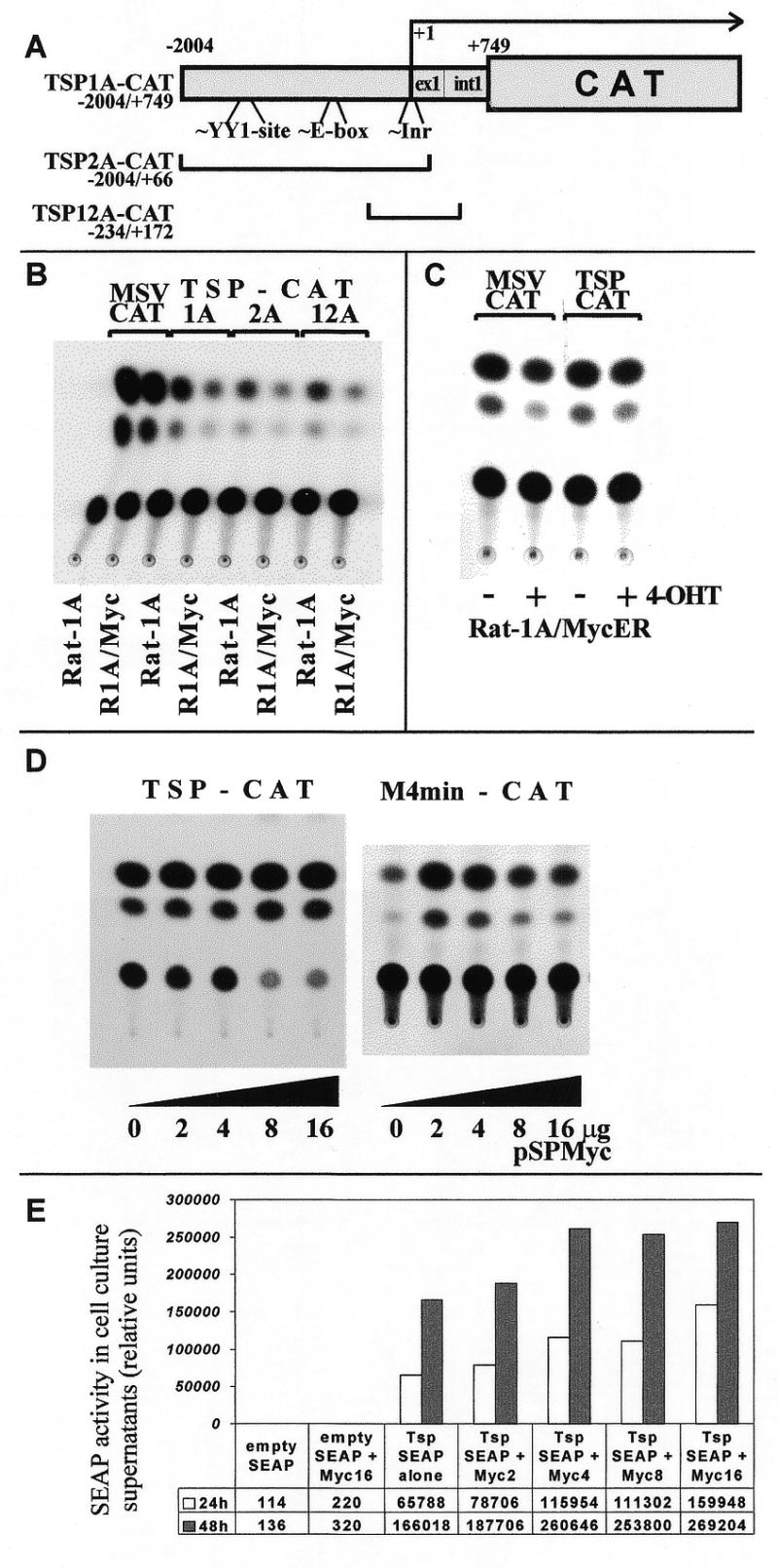
Transient expression of reporter constructs driven by the tsp-1 promoter in Myc-overexpressing cells. (A) Reporter constructs utilizing various segments of the tsp-1 promoter. Locations of the YY-1 binding site, the non-canonical E-box and the putative initiator element are shown. All numbers refer to the distance from the transcription start site (+1). (B) TLC detection of CAT in lysates of Rat-1A or Rat-1A/LMycSN (R1A/Myc) cells transiently transfected with TSP1A-, 2A- and 12A-CAT, or MSV-CAT (positive control). (C) TLC assay performed on lysates of Rat-1A/MycER cells transfected with TSP1A- or MSV-CAT and cultured in the presence (+) or in the absence (–) of 4-OHT to maintain Myc in the active or the inactive state, respectively. (D) TLC assay performed on lysates of 293 kidney epithelial cells co-transfected with 1 µg of either pTSP1A-CAT (left) or pM4min-CAT (right) and increasing (from 0 to 16 µg) amounts of the Myc-expressing construct, pSPMyc. (E) SEAP assay performed on supernatants of 293 cells co-transfected with 1 µg of either the ‘empty’ SEAP vector or the TSP-SEAP plasmid and increasing (from 0 to 16 µg) amounts of the Myc-expressing construct, pSVMyc.
We have previously reported that expression levels of TSP1A-CAT (but not MSV-CAT) are on average 3/5-fold lower in Rat-1A/LMycSN cells than in parental Rat-1A cells, as measured by the amount of radiolabeled chloramphenicol converted into monoacetylated forms (8). In the experiment depicted in Figure 2B, we observed a 3.8-fold reduction in the enzymatic activity with TSP1A-CAT and very similar levels of down-regulation with TSP2A-CAT and TSP12A-CAT (4.0- and 3.9-fold, respectively). Since these two constructs retain nucleotides –2004/+66 and –234/+172 of the tsp-1 promoter, respectively, the element responsible for diminished expression in Myc-overexpressing cells most likely resides between nucleotides –234 and +66. This segment lacks the E-box but does contain Inr, a potential Myc-responsive element. However, low levels of TSP-CAT expression could be due to secondary changes associated with the neoplastic phenotype, not the activity of Myc per se. To determine whether overexpression of Myc is sufficient to down-regulate the tsp-1 promoter in the absence of any changes in cell physiology, the following two experiments were performed.
In the first experiment (Fig. 2C) we employed Rat-1A cells infected by a retrovirus encoding the Myc protein fused to the ligand binding domain of the mutated form of the ER (ER‘; 33). Consequently, the MycER chimera is always produced but only when a synthetic ligand for ER‘, 4-OHT, is present in the medium is Myc capable of nuclear translocation and gene regulation. To determine whether the tsp-1 promoter is negatively influenced by Myc, Rat1A–MycER cells were transfected with pTSP1A-CAT and maintained either in the presence or in the absence of 4-OHT for 48 h. No reproducible differences in CAT levels between untreated and 4-OHT-treated cultures were apparent (Fig. 2C), suggesting that Myc per se is not capable of down-regulating the tsp-1 promoter in a transient expression system. At the same time, Myc was fully competent to down-regulate the endogenous tsp-1 mRNA (8, and data not shown), proving the functionality of this system.
To corroborate this notion, we used 293 kidney epithelial cells (34) where high frequency of co-expression could be achieved following calcium phosphate transfection or lipofection. 293 cells were transfected with pTSP1A-CAT and increasing amounts of Myc-expression vector, pSPMyc. Even when the pSPMyc:TSP1A-CAT ratio reached 16:1 (Fig. 2D, left) no diminished expression of pTSP1A-CAT was apparent. Since in this experiment Myc is not overexpressed in every cell, it was not possible to confirm its functionality by analyzing the endogenous tsp-1 mRNA. Thus we have transfected 293 cells with pM4min-CAT, the plasmid containing the reporter gene under the control of the minimal TK gene promoter and four copies of the Myc-response element. On its own, this construct expresses very small amounts of CAT; however, inclusion of moderate amounts of pSPMyc DNA (3-fold excess over the reporter plasmid) resulted in sharply increased CAT activity in 3T3 fibroblasts (20). We observed that this was also the case in 293 cells (Fig. 2D, right). Thus, transiently overexpressed Myc is functional when co-transfected with a reporter construct in 293 cells. When the pSPMyc:pM4min-CAT ratio was further increased, slight but reproducible down-regulation of the promoter became apparent. Such a bi-phasic effect (activation followed by repression) can be explained by ‘squelching’ of putative co-activators via protein–protein interactions. Interestingly, similar effects have also been reported in the study on the adenoviral major late (AdML) promoter. The AdML promoter lacks the TATA-box but contains a Myc-responsive Inr element (23) which is thought to mediate bi-phasic response. This parallelism suggests that apparently different mechanisms of gene regulation by Myc might, under certain circumstances, result in similar effects on gene expression.
We also considered the possibility that, during transient expression, silencing of the tsp-1 promoter could occur hours and even days after activation of Myc, and since the CAT protein is extremely stable, a decrease in its levels might be difficult to detect. We thus used another construct where a similar fragment of the murine tsp-1 promoter was controlling expression of SEAP (9). SEAP is a highly labile protein and its concentration in the conditioned medium reflects current expression levels, not the amount of protein accumulated during the course of the experiment. We have co-transfected 293 cells with TSP-SEAP and pSPMyc and measured the SEAP levels at 24 and 48 h time points. As with TSP1A-CAT construct, no negative effect of Myc on the tsp-1 promoter was apparent; in fact a slight increase in expression levels was observed (Fig. 2E). This finding suggested that the tsp-1 promoter might not be responsive to Myc, prompting us to check whether Myc affects transcription initiation of tsp-1 mRNA.
Myc has a minimal effect on initiation or processivity of the tsp-1 gene transcription
To determine if down-regulation of tsp-1 mRNA occurs at the level of transcription, nuclear run-off experiments have been performed. In the first experiment, we isolated nuclei from either parental or Myc-transformed cells, performed run-off reactions and hybridized the labeled products to either complete tsp-1 cDNA (to determine the overall rate of transcription) or to the promoter-proximal exon 1 (to determine the rate of transcription initiation). The rat gapdh and vector (pSL1180) DNAs were used as internal and negative controls, respectively (Fig. 3). Intensities of tsp-1 signals were normalized to that produced by the gapdh probe. Using the full-length probe, the rate of transcription in Myc-transformed cells was found to be 66% of that observed in parental cells (left). Using the exon 1 probe, it was found to vary from 106% (experiment 1, left) to 59% (experiment 2, right). Despite these variations in transcription levels, we have never observed, with either probe, >40% decrease in transcription rates. This difference, even if meaningful, is unlikely to account for the 7–10-fold decrease in tsp-1 mRNA levels in Myc-transformed cells (8) suggesting that other levels of regulation ought to exist. Interestingly, when the probe representing the 3′-terminal exon 22 was used, we actually observed markedly higher (3.9-fold) transcription rates in Rat-1A/LMycSN cells (right) although interpretation of results obtained with promoter-distal probes is difficult, due to low processivity of transcription in vitro and consequently weak signals.
Figure 3.
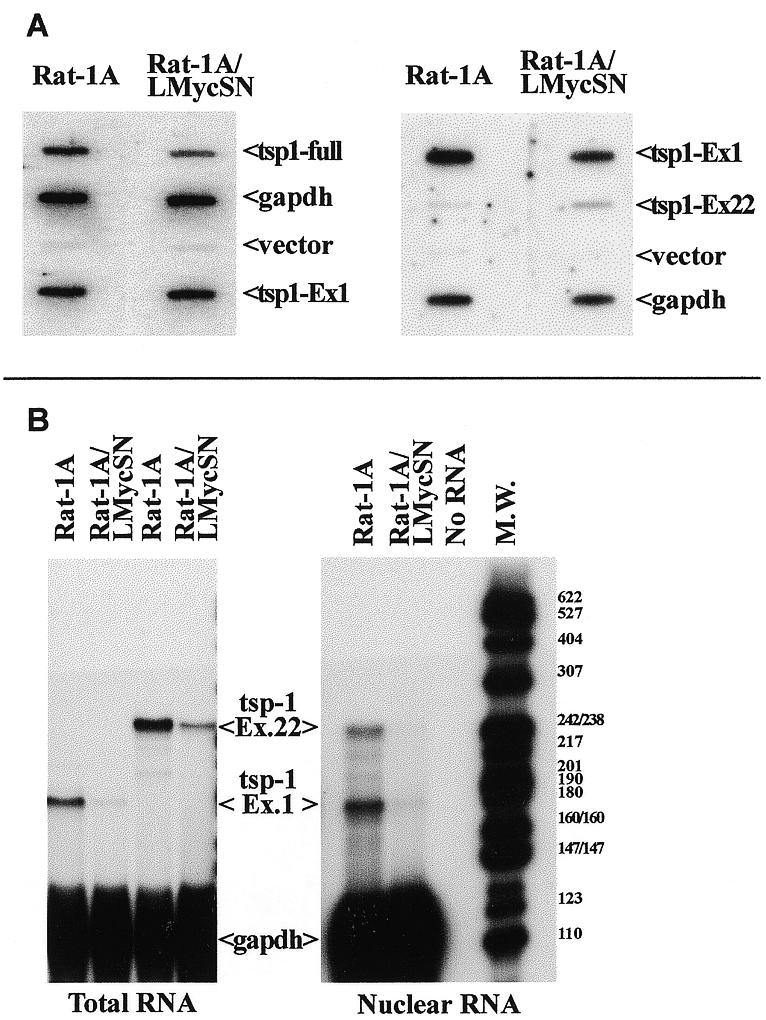
Myc-independent initiation and elongation of tsp-1 mRNA. (A) Nuclear run-off assay performed with Rat-1A (left columns) and Rat-1A/LMycSN (right columns) cells. Probes used were the full-length human tsp-1 cDNA (‘tsp1-full’), exons 1 (‘tsp1-Ex1’) and 22 (‘tsp1-Ex22’) of the rat tsp-1 gene, rat gapdh cDNA (‘gapdh’) and the empty cloning vector pGEM (‘vector’). (B) RNase protection analyses performed on total (left) and nuclear (right) RNAs from Rat-1A and Rat-1A/LMycSN cells. The probes represented exons 1 and 22 of the rat tsp-1 gene and the rat gapdh gene (loading control). Electrophoretic mobility of the molecular mass markers (MspI fragments of the plasmid pBR322) is shown on the right.
To conclusively rule out regulation at the level of elongation, we performed RNase protection analyses to measure the steady-state levels of tsp-1 mRNA using probes representing 5′- and 3′-termini (exons 1 and 22, respectively). If elongation of tsp-1 mRNA is blocked in Myc-transformed cells, we would expect to detect a higher signal with the 5′ probe than with the 3′ probe. This, however, was not the case: with both 5′ and 3′ probes, similar levels of down-regulation were observed. Using total RNA (Fig. 3B, left), we detected 7.1- and 6.6-fold reduction, respectively, and using nuclear RNA (right) 7.7- and 6.1-fold reduction, respectively. Hence neither elongation nor import from the nucleus of tsp-1 mRNA were grossly affected by Myc overexpression. This finding implicated mRNA stability as the only remaining mechanism that could account for decreased tsp-1 mRNA levels in Myc-transformed cells.
Activation of Myc results in decreased tsp-1 RNA stability
To determine whether tsp-1 mRNA is indeed destabilized upon activation of Myc, we performed a series of experiments with ActD that inhibits de novo transcription and thus allows one to follow the fate of pre-existing mRNA molecules. Rat-1A/MycER cells were treated with ActD and subjected to RNase protection and radioimmunoprecipitation analyses as described in Materials and Methods.
In the first pilot experiment we confirmed that ActD is functional in Rat-1A/MycER cells. To this end, we detected c-fos mRNA which is known to be extremely short-lived (35) and should rapidly disappear from ActD-treated cells. Indeed, c-fos mRNA was readily detectable in untreated cells but not in cells treated with the drug for only 2 h (Fig. 4A). tsp-1 mRNA, on the other hand, was quite stable and no decrease in its steady-state levels was apparent even after 6 h. Moreover, the retroviral genomic RNA, off which MycER is translated, is also quite stable (data not shown) allowing the rather labile MycER protein to be continuously synthesized. To confirm that a reduction in MycER protein levels does not occur even after prolonged (>6 h) exposure to ActD, we performed radioimmunoprecipitation with an anti-human c-Myc antibody. As predicted, only after 8 h of treatment with ActD were MycER protein levels detectably reduced, and the reduction did not exceed 50% (Fig. 4B). It is worth noting that, since regulation of ER fusion proteins by estrogens is post-translational, there was no difference in MycER levels between untreated and 4-OHT-treated cells. Therefore this system is suitable for the studies of the effect of MycER on tsp-1 mRNA stability.
Figure 4.
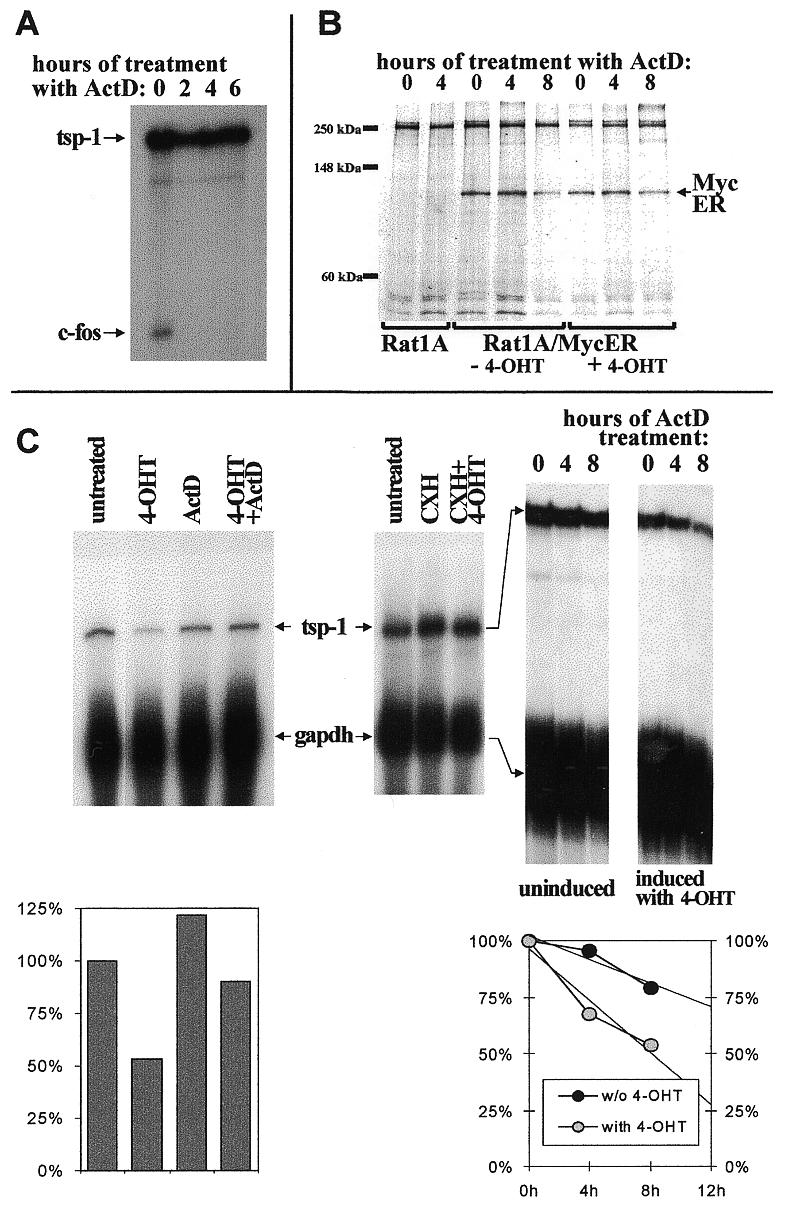
Increased turnover of tsp-1 mRNA in Myc overexpressing cells. (A) RNase protection analysis detecting tsp-1 and c-fos mRNAs in Rat1A/MycER cells treated with ActD for the indicated number of hours. (B) Radioimmunoprecipitation of the MycER fusion protein from lysates of parental Rat-1A and Rat1A/MycER cells, untreated or treated with 4-OHT. Cells were also treated with ActD for the indicated number of hours. MycER protein has the apparent molecular mass of ~95 kDa. Electrophoretic mobility of the molecular mass markers is shown on the left. (C) RNase protection analyses detecting tsp-1 and gapdh (loading control) mRNAs in Rat1A/MycER cells treated with ActD, CHX, 4-OHT or combinations of these drugs. Left, Rat1A/MycER cells were treated with the indicated drug for 12 h. Middle, cells were treated with the indicated drug for 6 h. Right, cells were left untreated (first three lanes) or pre-treated with 4-OHT for 12 h (last three lanes) and then treated with ActD for the indicated number of hours. In the two graphs, tsp:gapdh ratios corresponding to different treatments are plotted as percentages of those observed in untreated cells. In the right graph, the left and right y-axes correspond to untreated and 4-OHT-treated cells, respectively.
We compared the levels of tsp-1 mRNA in untreated, 4-OHT-, ActD- and doubly treated Rat-1A/MycER cells, as detailed in Materials and Methods. As an internal standard, the stable transcript encoding the housekeeping protein GAPDH was used. After 12 h of treatment with ActD, no decrease in tsp-1 mRNA levels was apparent, reflecting its relative stability. In contrast, treatment with 4-OHT for the same period resulted in a 2-fold down-regulation of tsp-1 mRNA (Fig. 4C, left and graph below) but only in cells expressing MycER (8, and data not shown). This indicates that tsp-1 mRNA is actively destabilized upon activation of Myc and does not merely cease being synthesized. When both drugs were added simultaneously, Myc was not effective indicating that other proteins might be involved in tsp-1 mRNA regulation. To verify this notion, it would have been useful to determine whether continuous protein synthesis is required. However, treatment with the protein synthesis inhibitor CHX resulted, regardless of the presence or absence of Myc, in 4-fold up-regulation of tsp-1 mRNA (Fig. 4C, middle). Such a positive effect of CHX on mRNA expression, although well-documented for many genes including c-myc itself (36) and some of its putative targets (19), makes it difficult to assess whether tsp-1 mRNA destabilization is a direct function of Myc.
To further quantify this effect, we pre-treated Rat-1A/MycER cells with 4-OHT or solvent alone (ethanol) and then added ActD for 4 or 8 h. Pre-treatment was necessary because the tsp-1 gene is not down-regulated by Myc in the absence of de novo RNA synthesis (see above). In mock-treated cells down-regulation of tsp-1 mRNA was minimal: 5% after 4 h and 21% after 8 h. In contrast, in 4-OHT-treated cells down-regulation was apparent as early as 4 h after ActD-treatment (33%) and reached 47% after 8 h. (Fig. 4C, right and graph below). By 12 h, cultures treated with both 4-OHT and ActD are not viable; by extrapolating existing data to the 12 h time point (trend lines), we concluded that after 12 h of exposure to 4-OHT, reduction in steady-state tsp-1 mRNA levels would be >3-fold. Thus in the presence of active, overexpressed Myc tsp-1 mRNA half-life is decreased to the extent that can account for its low steady-state levels in Myc-transformed cells.
DISCUSSION
Here we demonstrate that activation of the Myc oncoprotein leads to down-modulation of the tsp-1 gene via a mechanism involving increased mRNA turnover. The exact nature of this mechanism remains to be elucidated. It is possible that Myc exerts its effect through direct interactions, at either DNA or mRNA levels, with specific sequences of the tsp-1 gene. Alternatively, Myc might regulate expression of an intermediate gene whose product, perhaps an RNA-binding protein, affects tsp-1 mRNA turnover.
The standard approach to choose between these two scenarios is to determine whether new protein synthesis is required for the regulation to take place. Apparent requirement for protein synthesis is usually interpreted to mean that intermediate gene products are involved. Alternatively, regulation might still be direct but involve short-lived co-factors that are eliminated during treatment with CHX. Down-regulation of the tsp-1 gene by Myc might be the case in point. Admittedly, no tsp-1 down-regulation takes place when 4-OHT and CHX are added simultaneously. However, interpretation of this result is complicated by the fact that inhibition of protein synthesis on its own increases tsp-1 mRNA steady-state levels. This increase invokes the involvement of labile co-factors. It would also mask any direct, protein synthesis-independent effect that Myc might have on the stability of tsp-1 mRNA.
The result of the ActD experiment also allows two interpretations. Apparently, continuous mRNA synthesis is required for Myc to exert its effect on tsp-1 mRNA stability, consistent with an indirect mode of regulation. Still, if mRNAs encoding putative Myc co-factors are short-lived, treatment with ActD might make tsp-1 mRNA less prone to destabilization and preclude down-regulation by Myc. Thus the data presented in this study do not allow us to distinguish unequivocally between direct and indirect regulation.
Our previous study (8) has demonstrated that a single amino acid substitution in the DNA-binding domain of Myc abolishes regulation of the tsp-1 gene. It is conceivable that binding of Myc to DNA (inside or outside the promoter region) is required to somehow alter the nascent tsp-1 message and target it for rapid, perhaps co-transcriptional, turnover. Co-transcriptional modification of messenger RNA is a well-established mechanism of gene regulation, best exemplified by the tat protein of HIV which recruits to the transcription machinery a kinase that phosphorylates RNA polymerase II and increases its processivity (37). However, the apparent involvement of the DNA-binding domain is more consistent with the idea that in order to down-regulate tsp-1 mRNA, Myc has to induce/repress some co-factors, and sequence-specific DNA-binding is required for this auxiliary event.
Regardless of the exact molecular mechanism, the association between Myc activation and increased tsp-1 mRNA turnover is unexpected since Myc was not known to influence expression of its downstream genes at a post-transcriptional level. One of the more recent addition to the growing list of c-Myc-repressed genes (6,38), gadd45, has been shown to be repressed at the level of transcription and this regulation could be readily recapitulated in a transient expression setting (39). However, the role of Myc in gene regulation might be more complex as evidenced by negative auto-regulation of the c-myc gene itself. It has been well established that the c-Myc protein down-modulates expression of its own gene (40); this regulation, at least in part, stems from events occurring at the P2 promoter (41). At the same time, c-myc mRNA is also regulated at the level of elongation (36,42), and its instability is known to contribute to down-regulation of Myc during terminal differentiation as well (43,44). Our finding that Myc is capable of influencing mRNA turnover suggests that an additional, post-transcriptional mechanism for c-myc autoregulation might exist and that turnovers of c-myc and tsp-1 mRNAs might be regulated in a similar fashion.
tsp-1 and c-myc mRNAs indeed resemble each other in that both possess 3′-terminal, AU-rich untranslated regions (UTRs) (45) that are known to contribute to rapid mRNA turnover (46). The tsp-1 UTR, extending through the entire exon 22, is particularly long (>2 kb) and AU-rich (47). The long, non-coding 3′-exon is also present in the closely related thrombospondin-2 mRNA (48) that has recently been shown to be destabilized in the presence of Myb, another nuclear oncoprotein (49). These 3′-terminal elements could conceivably mediate Myc-dependent tsp-1 mRNA turnover. However, precise identification of such destabilization signals in tsp-1 mRNA would be a difficult endeavor, given the size of the UTR, the lack of well-defined RNA degradation motifs and the unavailability of a transient expression system. In addition, coding sequences are also known to contribute to mRNA degradation (50). Therefore, further insights into the mechanisms of tsp-1 down-regulation by Myc are likely to emerge from biochemical characterization of this oncoprotein, its target genes and in particular its dimerization partners. Continuing emergence of new Myc partners (7) might yet yield a candidate implicated in RNA turnover and, by inference, in the tsp-1 gene regulation.
Acknowledgments
ACKNOWLEDGEMENTS
We are pleased to acknowledge our former colleagues Drs Carol Laherty and Carla Grandori (Fred Hutchinson Cancer Research Center) who provided us with TSP-CAT expression constructs and purified Myc and Max proteins, respectively, and also with the expert advice on the use of these reagents. The TSP-SEAP expression construct was a kind gift of Drs Florence Cabon and Bernard Binetruy (Institut de Recherche sur le Cancer, Villejuif, France). We are indebted, for many insightful comments, to Drs Maxine Linial, Anton Krumm and Mark Groudine (Fred Hutchinson Cancer Research Center) and Dr Narayan Avadhani (University of Pennsylvania). This work was supported by the National Cancer Institute grant R29 CA 71881 and a Special Fellowship of the Leukemia Society of America to A.T.-T.
REFERENCES
- 1.Wang J., Xie,L.Y., Allan,S., Beach,D. and Hannon,G.J. (1998) Genes Dev., 12, 1769–1774. [DOI] [PMC free article] [PubMed]
- 2.Falchetti M.L., Falcone,G., D’Ambrosio,E., Verna,R., Alema,S. and Levi,A. (1999) Oncogene, 18, 1515–1519. [DOI] [PubMed]
- 3.Greenberg R.A., O’Hagan,R.C., Deng,H., Xiao,Q., Hann,S.R., Adams,R.R., Lichtsteiner,S., Chin,L., Morin,G.B. and DePinho,R.A. (1999) Oncogene, 18, 1219–1226. [DOI] [PubMed]
- 4.Galaktionov K., Chen,X. and Beach,D. (1996) Nature, 382, 511–517. [DOI] [PubMed]
- 5.Dang C.V. (1999) Mol. Cell. Biol., 19, 1–11. [DOI] [PMC free article] [PubMed]
- 6.Facchini L.M. and Penn,L.Z. (1998) FASEB J., 12, 633–651. [PubMed]
- 7.Sakamuro D. and Prendergast,G.C. (1999) Oncogene, 18, 2942–2954. [DOI] [PubMed]
- 8.Tikhonenko A.T., Black,D.J. and Linial,M.L. (1996) J. Biol. Chem., 271, 30741–30747. [DOI] [PubMed]
- 9.Mettouchi A., Cabon,F., Montreau,N., Vernier,P., Mercier,G., Blangy,D., Tricoire,H., Vigier,P. and Binetruy,B. (1994) EMBO J., 13, 5668–5678. [DOI] [PMC free article] [PubMed]
- 10.Dejong V., Degeorges,A., Filleur,S., Ait,S.A., Mettouchi,A., Bornstein,P., Binetruy,B. and Cabon,F. (1999) Oncogene, 18, 3143–3151. [DOI] [PubMed]
- 11.Slack J.L. and Bornstein,P. (1994) Cell Growth Differ., 5, 1373–1380. [PubMed]
- 12.Good D.J., Polverini,P.J., Rastinejad,F., Le Beau,M.M., Lemons,R.S., Frazier,W.A. and Bouck,N.P. (1990) Proc. Natl Acad. Sci. USA, 87, 6624–6628. [DOI] [PMC free article] [PubMed]
- 13.Iruela-Arispe M.L., Bornstein,P. and Sage,H. (1991) Proc. Natl Acad. Sci. USA, 88, 5026–5030. [DOI] [PMC free article] [PubMed]
- 14.Ngo C., Gee,M.S., Akhtar,N., Yu,D., Volpert,O.V., Auerbach,R. and Thomas-Tikhonenko,A.T. (2000) Cell Growth Differ., 11, 201–210 . [PMC free article] [PubMed]
- 15.Donoviel D.B., Framson,P., Eldridge,C.F., Cooke,M., Kobayashi,S. and Bornstein,P. (1988) J. Biol. Chem., 263, 18590–18593. [PubMed]
- 16.Blackwell T.K., Kretzner,L., Blackwood,E.M., Eisenman,R.N. and Weintraub,H. (1990) Science, 250, 1149–1151. [DOI] [PubMed]
- 17.Prendergast G.C. and Ziff,E.B. (1991) Science, 251, 186–190. [DOI] [PubMed]
- 18.Blackwell T.K., Huang,J., Ma,A., Kretzner,L., Alt,F.W., Eisenman,R.N. and Weintraub,H. (1993) Mol. Cell. Biol., 13, 5216–5224. [DOI] [PMC free article] [PubMed]
- 19.Grandori C., Mac,J., Siebelt,F., Ayer,D.E. and Eisenman,R.N. (1996) EMBO J., 15, 4344–4357. [PMC free article] [PubMed]
- 20.Kretzner L., Blackwood,E.M. and Eisenman,R.N. (1992) Nature, 359, 426–429. [DOI] [PubMed]
- 21.Amati B., Dalton,S., Brooks,M.W., Littlewood,T.D., Evan,G.I. and Land,H. (1992) Nature, 359, 423–426. [DOI] [PubMed]
- 22.Roy A.L., Malik,S., Meisterernst,M. and Roeder,R.G. (1993) Nature, 365, 355–359. [DOI] [PubMed]
- 23.Li L., Nerlov,C., Prendergast,G.C. and Ziff,E.B. (1994) EMBO J., 13, 4070–4079. [DOI] [PMC free article] [PubMed]
- 24.Roy A.L., Carruthers,C., Gutjahr,T. and Roeder,R.G. (1993) Nature, 365, 359–361. [DOI] [PubMed]
- 25.Roy A.L., Du,H., Gregor,P.D., Novina,C.D., Martinez,E. and Roeder,R.G. (1997) EMBO J., 16, 7091–7104. [DOI] [PMC free article] [PubMed]
- 26.Shrivastava A., Saleque,S., Kalpana,G.V., Artandi,S., Goff,S.P. and Calame,K. (1993) Science, 262, 1889–1892. [DOI] [PubMed]
- 27.Austen M., Cerni,C., Henriksson,M., Hilfenhaus,S., Luscher-Firzlaff,J.M., Menkel,A., Seelos,C., Sommer,A. and Luscher,B. (1997) Curr. Top. Microbiol. Immunol., 224, 123–130. [DOI] [PubMed]
- 28.Schleiss M.R., Degnin,C.R. and Geballe,A.P. (1991) J. Virol., 65, 6782–6789. [DOI] [PMC free article] [PubMed]
- 29.Tikhonenko A.T., Hartman,A.-R. and Linial,M. (1993) Mol. Cell. Biol., 13, 3623–3631. [DOI] [PMC free article] [PubMed]
- 30.Laherty C.D., Gierman,T.M. and Dixit,V.M. (1989) J. Biol. Chem., 264, 11222–11227. [PubMed]
- 31.Blackwood E.M., Lugo,T.G., Kretzner,L., King,M.W., Street,A.J., Witte,O.N. and Eisenman,R.N. (1994) Mol. Biol. Cell, 5, 597–609. [DOI] [PMC free article] [PubMed]
- 32.Laimins L.A., Gruss,P., Pozzatti,R. and Khoury,G. (1984) J. Virol., 49, 183–189. [DOI] [PMC free article] [PubMed]
- 33.Littlewood T.D., Hancock,D.C., Danielian,P.S., Parker,M.G. and Evan,G.I. (1995) Nucleic Acids Res., 23, 1686–1690. [DOI] [PMC free article] [PubMed]
- 34.Graham F.L., Smiley,J., Russell,W.C. and Nairn,R. (1977) J. Gen. Virol., 36, 59–74. [DOI] [PubMed]
- 35.Chen C.Y. and Shyu,A.B. (1995) Trends Biochem. Sci., 20, 465–470. [DOI] [PubMed]
- 36.Linial M., Gunderson,N. and Groudine,M. (1985) Science, 203, 1126–1132. [DOI] [PubMed]
- 37.Jones K.A. (1997) Genes Dev., 11, 2593–2599. [DOI] [PubMed]
- 38.Claassen G.F. and Hann,S.R. (1999) Oncogene, 18, 2925–2933. [DOI] [PubMed]
- 39.Marhin W.W., Chen,S., Facchini,L.M., Fornace,A.J. and Penn,L.Z. (1997) Oncogene, 14, 2825–2834. [DOI] [PubMed]
- 40.Penn L.J.Z., Brooks,M.W., Laufer,E.M. and Land,H. (1990) EMBO J., 9, 1113–1121. [DOI] [PMC free article] [PubMed]
- 41.Facchini L.M., Chen,S., Marhin,W.W., Lear,J.N. and Penn,L.Z. (1997) Mol. Cell. Biol., 17, 100–114. [DOI] [PMC free article] [PubMed]
- 42.Krumm A., Hickey,L.B. and Groudine,M. (1995) Genes Dev., 9, 559–572. [DOI] [PubMed]
- 43.Yeilding N.M., Procopio,W.N., Rehman,M.T. and Lee,W.M. (1998) J. Biol. Chem., 273, 15749–15757. [DOI] [PubMed]
- 44.Yeilding N.M. and Lee,W.M. (1997) Mol. Cell. Biol., 17, 2698–2707. [DOI] [PMC free article] [PubMed]
- 45.Brewer G. (1991) Mol. Cell. Biol., 11, 2460–2466. [DOI] [PMC free article] [PubMed]
- 46.Ross J. (1996) Trends Genet., 12, 171–175. [DOI] [PubMed]
- 47.Hennessy S.W., Frazier,B.A., Kim,D.D., Deckwerth,T.L., Baumgartel,D.M., Rotwein,P. and Frazier,W.A. (1989) J. Biol. Chem., 108, 729–736. [DOI] [PMC free article] [PubMed]
- 48.Shingu T. and Bornstein,P. (1993) Genomics, 16, 78–84. [DOI] [PubMed]
- 49.Bein K., Ware,J.A. and Simons,M. (1998) J. Biol. Chem., 273, 21423–21429. [DOI] [PubMed]
- 50.Yeilding N.M., Rehman,M.T. and Lee,W.M. (1996) Mol. Cell. Biol., 16, 3511–3522. [DOI] [PMC free article] [PubMed]


