Abstract
Epidemics, such as COVID-19, have caused significant harm to human society worldwide. A better understanding of epidemic transmission dynamics can contribute to more efficient prevention and control measures. Compartmental models, which assume homogeneous mixing of the population, have been widely used in the study of epidemic transmission dynamics, while agent-based models rely on a network definition for individuals. In this study, we developed a real-scale contact-dependent dynamic (CDD) model and combined it with the traditional susceptible-exposed-infectious-recovered (SEIR) compartment model. By considering individual random movement and disease spread, our simulations using the CDD-SEIR model reveal that the distribution of agent types in the community exhibits spatial heterogeneity. The estimated basic reproduction number R0 depends on group mobility, increasing logarithmically in strongly heterogeneous cases and saturating in weakly heterogeneous conditions. Notably, R0 is approximately independent of virus virulence when group mobility is low. We also show that transmission through small amounts of long-term contact is possible due to short-term contact patterns. The dependence of R0 on environment and individual movement patterns implies that reduced contact time and vaccination policies can significantly reduce the virus transmission capacity in situations where the virus is highly transmissible (i.e., R0 is relatively large). This work provides new insights into how individual movement patterns affect virus spreading and how to protect people more efficiently.
Introduction
The outbreak of COVID-19 brought a profound disaster to human civilization worldwide, which has infected more than 600 million people and killed more than 6 million people by October 2022, and the virus is still in constant mutation [1, 2]. From the original strain to the Alpha, Delta, and Omicron variants, while the virulence of the virus decreased, the infectivity gradually increased. As an important indicator to characterize infectious diseases, the basic reproduction number (R0) has increased from 2.79 in the original strain to 9.5 in the Omicron variant [3–5], bringing new challenges to epidemic prevention and control.
In the theoretical models of studying the transmission of infectious disease, homogeneous mixing [6] is a classic assumption of the traditional compartmental model [7]. This approximation simplifies the model and thus can help to obtain epidemic transmission trends more efficiently. However, in real situations, heterogeneity, including differences between individuals [8–10] and differences in spatial distribution [11–14], is also an important factor in epidemic transmission that should be considered [8–14].
In recent years, agent-based models (ABMs) that focus on individual behavior (even considering the corresponding movement characteristics) have received increasing attention [15, 16]. This model examines the overall behavior and trends by modeling many individuals (called agents). In ABM, individuals are made autonomous and adaptive by defining them to choose behaviors based on their own decision rules (often derived from limited information in their immediate surroundings), and the decision rules could vary from agent to agent. Agents are social and interdependent. If an agent performs a certain action, other agents may be influenced and respond in some way. A central concept of ABM is that the overall characteristics of a group are not simply the sum of all individual characteristics in the group but are the result of interactions guided by individual characteristics. Interactions among individuals often result in a society with a different overall identity from that of the individuals. In the case of epidemic transmission, the spatial movement of individuals and the influence of spatial heterogeneity on the spread of an epidemic are important factors that cannot be ignored. Adopting the idea of ABM to study may be an appropriate idea [12, 14, 17–23].
In some ABMs, the location and interdependence between individuals are determined by a predefined social network (e.g., building a small-world network to simulate the social relationships of agents), and the spread of epidemics is demonstrated through the evolution of the network [24, 25]. Complex network models can better introduce heterogeneity among individuals but lack the spatial movement of individuals [19]. In addition, there are individual-based motion models that rely not on defined social networks but only on individual motion [26, 27]. These models suggest that characteristic quantities of motion, such as the velocity of individuals, can play an important role in the spread of the epidemic [19, 20]. However, these models often simplify factors related to epidemic transmission because of computational efficiency [12, 14, 17–22]. For example, the motion and the motion features of an individual at a real time-space scale are not fully considered.
In this work, we designed a contact-dependent dynamics (CDD) model based on the characteristics of epidemic transmission to mimic the motion of the population within a community. Our model consisted of alternating individual motions and contact between individuals (thus changing the direction of motion) in an ensemble of individuals. We combined this CDD model (to describe population movement) with the commonly used epidemic transmission model, the susceptible-exposed-infectious-recovered (SEIR) model (to describe infection between individuals), to study the characteristics of epidemic transmission by studying the movement of individuals at real time and space scales and epidemic transmission. We found that the CDD model has a larger proportion of short-term contacts as the velocity of agents decreases. To characterize the epidemic transmission capacity, we calculated the basic reproduction number R0 of the model. The calculated R0 with the CDD-SEIR model is nearly independent of the virulence of the virus and much less than that calculated by mean-field equations [19], which is attributed to the short exposure time ω of the CDD model when the velocity of all individuals in the population is less than a threshold value. This corroborates the effectiveness of prevention and control measures in the presence of highly infectious strains. Moreover, we found that R0 shows two stages of variation with the spatial heterogeneity of the system: that is, in the strong heterogeneity range, R0 is proportional to the logarithm of velocity, which is different from mean-field theory, and in the weak heterogeneity range, R0 reaches saturation. The above results provide us with a new understanding of the relationship between epidemic transmission and individual movement.
This paper is organized as follows: we first propose a modified SEIR model to incorporate individual mobility. Then, we introduce the CDD model designed to characterize the stochastic motion of individuals within a community. Next, we describe the CDD-SEIR model, combining real-scale stochastic individual motion with epidemic transmission. To characterize the agent dynamics, the exposure time ω and mean free time τ will be discussed. Meanwhile, we show the characteristics of epidemic spreading described by this CDD-SEIR model, as well as the calculated basic reproduction number R0 and the spatial heterogeneity ζ, obtaining the effect of motion parameters (e.g., velocity) on these properties. We will then discuss the bipartite variation of R0 with the strength of spatial heterogeneity. Notably, R0 is nearly independent of the virulence of the virus when all individuals move at less than a threshold value and spatial heterogeneity is sufficiently strong. R0 is much less than that calculated by mean-field equations due to the short exposure time caused by the CDD model. Therefore, an in-depth understanding of heterogeneity is important for an accurate understanding of epidemic transmission, e.g., prevention and control measures are still effective when the virulent strain is highly infectious due to the presence of spatial heterogeneity. The short-term contact model can greatly suppress the epidemic when combined with control measures.
Methodology
The SEIR model
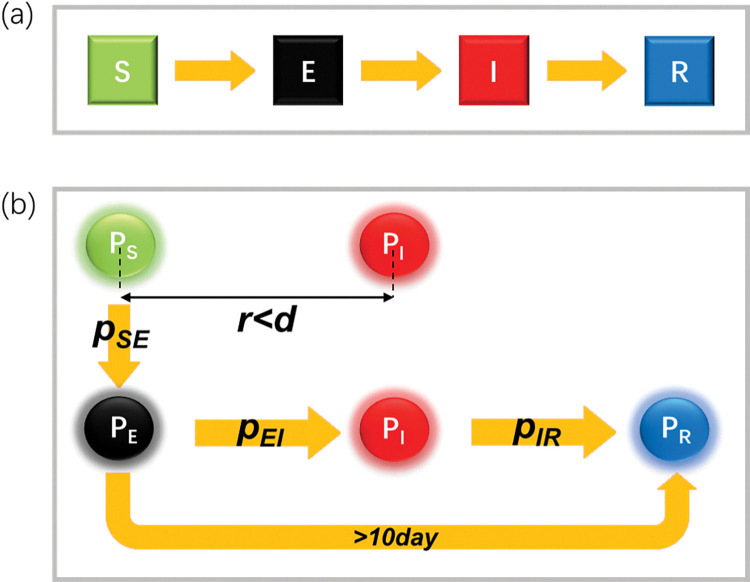
The SEIR model is one of the compartmental models that is commonly used to predict epidemic transmission [28]. Here, S, E, I and R stand for susceptible, exposed, infectious, and recovered, respectively, which are the four categories of people in epidemic transmission. The sum of these four is the population within the community. In the SEIR model, susceptible people who become infected come through exposed, infectious, and recovered compartments successively, as shown in Fig 1A. The latent period was considered when individuals had been infected but were not yet infectious themselves.
Fig 1. The classic SEIR model and the modified SEIR model.
(a) The classic SEIR model. Green, black, red, and blue squares represent susceptible, exposed, infectious, and recovered compartments, respectively. Susceptible individuals in the system successively go through these four stages. Transitions between adjacent stages occur according to certain probabilities. (b) The modified SEIR model. When the distance between the susceptible PS and PI is within the airborne transmission distance d, PS can be infected by PI and transformed into PE at the probability pSE. The latent period is within 4–10 days. During this period, PE converts with probability pEI to infectious PI that can transmit the virus. Any PE that remained unconverted to a PI after 10 days is classified as a recover PR. Meanwhile, PI could also be cured with probability pIR and transformed into PR.
COVID-19 has been reported to be transmitted mainly by proximity contact [29]. To introduce the mobility of individuals into the study of epidemic transmission, we proposed a modified SEIR model. According to the definition of the SEIR model, the agents here fall into four categories: susceptible, PS, healthy individuals who may be infected; exposed, PE, individuals who have been infected but do not cause infection after an incubation period; infectious, PI, individuals who have been infected and can transmit the virus to PS; and recovered, PR, individuals who have been cured or immunized and are thus removed from the transmission process.
In the modified SEIR model (Fig 1B), the PS has a probability pSE to convert to PE when the distance r between the susceptible PS and the infected PI is less than the airborne transmission distance d. The latent period for COVID-19 was estimated to be approximately 7 days in the literature [2, 30]. Accordingly, we set the latent period of PE between 4 and 10 days. Specifically, within 0–4 days, PE only acts as the virus carrier and does not have the ability to infect others; within 4–10 days, PE can transform with the probability pEI (Eq 1, where T is in the unit day) to PI, which can transmit the virus; otherwise, it remains as PE. After 10 days, PE that still does not convert to PI is classified as PR. Meanwhile, PI can be transformed into PR with the probability pIR.
| (1) |
The conversion probability from the exposed agent (PE) to the infectious agent (PI) pEI is reduced with the increase in Day T from latency, as shown in Eq 1. pEI,Day7 is the conversion probability of the 7th day of the latent period, and its value is shown in Table 1.
Table 1. The different mobility and propagation parameters tested in this model.
We selected 9 different velocities v and 5 different values for the remaining parameters, including the initial proportion of PI nI,0, the airborne transmission distance d, the infection probability pSE, the conversion probability of Day 7 pEI,Day7 from PE to PI, and the recovery probability pIR. Those marked with * are the default values. In each change of one parameter, the default values were used for the other parameters. For each set of parameter combinations, we calculated 5 random motion trajectories of agents at 40 days length separately using the CDD model and postprocessed each trajectory using the SEIR model to obtain 5 independent modes of transmission. The final reported results were extracted from the average of 5*5 = 25 trajectories.
| Parameters | Values |
|---|---|
| v (10 -4 m/s) | 1, 2, 5, 10, 20, 50, 100, 200, 500 |
| n I,0 | 1%*, 3%, 5%, 7%, 9% |
| d (m) | 1, 2*, 3, 4, 5 |
| pSE (10−4) | 0.5, 1.0, 1.5, 2.0*, 2.5 |
| pEI,Day7 (10−6) | 1, 2*, 3, 4, 5 |
| pIR (10−6) | 1, 2*, 3, 4, 5 |
The CDD model
We introduce the CDD model to characterize the motion of agents. In CDD, the agents are described by points. Each agent i is assigned a velocity vi following a normal distribution with a mean of v, a standard deviation of , and a direction θi randomly generated from a uniform distribution. The motion of the agents consists of alternating individual motions and contact between individuals (thus changing the motion) in an ensemble of individuals.
When there are no other agents within the distance rc around agent i, the latter adopts uniform linear motion, and the evolution of its position is determined only by its velocity; otherwise, the direction of motion and the velocity of i will be regenerated, as shown in Fig 2A, when there are other agents within the distance rc around i (Eq 2). In this case, the velocity is randomly generated by the normal distribution, and the direction of motion is randomly reassigned by the normal distribution within the range of ±45 degrees from the original motion direction. To represent the real-scale motion, the units for length and time used in this work were in meters and seconds, respectively. We set rc to 2 m and used the time interval Δt of 1 s.
| (2) |
where vi(t) and θi(t) denote the velocity and direction of particle i at time t, respectively. vrnd is the reassigned velocity that satisfies a normal distribution with a mean of v and a standard deviation of . θrnd is the change in the reassigned direction (in the range of -45° to 45°), which satisfies a normal distribution with a mean of 0° and a standard deviation of 15°. The distributions of vrnd and θrnd are shown in Fig 2B.
Fig 2. Schematic diagram of the CDD model.
(a) Stochastic motion process characterized by the CDD model. The upper panel represents that the direction of motion of an agent varies within [-45°, 45°] of the original direction when there are other agents within rc. The lower panel gives an example of the change in motion state of two individuals before and after contact: i is the central agent, and the blue circle represents its contact range of radius rc. Individuals i and j fall in contact when their distance is less than rc, and the agents are in a uniform linear motion before contact. The red, green, and black solid vectors represent the velocities of i and j and the relative velocity j relative to i, respectively. The gray dashed line represents the trajectory of j relative to i. (b) The reassigned velocity vrnd that satisfies a normal distribution with a mean of v and a standard deviation of . θrnd is the change in the reassigned direction (in the range of -45° to 45°), which satisfies a normal distribution with a mean of 0° and a standard deviation of 15°.
Notably, this CDD model has some similarities to the stochastic rotation dynamics (SRD) model [31, 32], which has been widely used to characterize fluid flow. The main difference between the two models is that in the CDD model, the individuals change direction and velocity of motion when the distance between two individuals is less than a certain range, rather than waiting until they are in full contact and collide. Such a model setup provides the possibility to study the ability of the virus itself to spread in space for the spread of the epidemic and for social distancing interventions.
The CDD-SEIR model
We combined the CDD model (to describe population movement) with the modified SEIR model (to describe infection between individuals) to simulate epidemic transmission at real time and space scales.
As shown in Fig 3A, a community represented by a periodic square box with side length L has N individuals. The population is composed of susceptible PS, exposed PE, infectious PI and recovered PR. The number of each type is NS, NE, NI and NR, respectively, where N = NS + NE + NI + NR. In subsequent work, we take the early stage of the epidemic as the initial condition of the simulations, put only a small amount of PI in the system and set the rest of the particles as PS. It should be noted that other factors, such as repeated infections, vaccination, etc., have potential influence on the transmission mechanism, and can be easily added to the current model (by adding types of agents). Meanwhile, factors such as death can also be implemented in the current model by removing particles from the system or eliminating their movement. The motion of agents is driven by the CDD model, and the spread of the epidemic is determined by the modified SEIR model. Therefore, we obtained the temporal evolution of the agents’ populations (Fig 3B) and the dynamic progress of the infection spreading (Fig 3C). We can study the factors of epidemic spreading from the perspective of both spatial agent dynamics and epidemic dynamics.
Fig 3. System setup of the CDD-SEIR model.
(a) Schematic diagram of a representative system obtained from the simulations. The system contains 200 particles with a box length of m and density of (100 m2)-1. PS, PE, PI and PR are represented by green, black, red and blue particles, respectively. (b) The profiles of the temporal population of PS (in green), PE (in black), PI (in red) and PR (in blue) during epidemic transmission. (c) Representative snapshots of the infection within a community at days 0, 10, 20, 30, and 40 are shown.
Comparing the results obtained from the present CDD-SEIR model with the real epidemic data is a way to test the model’s reliability. However, limited by the current model setup, such a direct comparison is not easy: first, we used 1 s as the integration step of the equation of motion to not miss any of the exposure events, and thus currently, the model can only handle a few hundred agents in a few tens of days within an affordable computational cost; second, for a system containing hundreds of particles, a single initial infected agent already corresponds to a very high initial infection rate (e.g., 1 in 200 agents corresponds to an initial infection rate of 2.5%), while the ratio in the real situation is often less than 1 in 10,000. Therefore, the present work lies mainly in providing a simple model with clear physical meaning to study the dynamics of epidemic transmission.
Parameter selection of the CDD-SEIR model
Considering the computational efficiency, in the simulations, we used the following parameters for the CDD model: number of particles N = 200, density of particles ρ = (100 m2)-1, and contact distance rc = 2 m. These parameters remained intact in the simulations. Forty-day trajectories were accumulated separately under the different combinations of parameters.
Based on the case of COVID-19 propagation [2], we set the default values of some propagation parameters in the SEIR model to be close to the real situation. For example, we assumed the airborne transmission distance d = 2 m. This is considered a safe social distance to prevent the spread of the virus [33]. In addition, we set pEI,Day7 = pIR = 2*10−6, which determines the duration of the incubation and disease periods, and assumed that pSE = 2*10−4, which determines R0 to be close to the real situation based on the reported data [30, 34, 35].
Meanwhile, we carefully examined the effects of the rest of the propagation parameters in the combined CDD-SEIR model, including the mobility v, the initial proportion of PI nI,0, the airborne transmission distance d, the infection probability pSE, the conversion probability of Day 7 from the exposed agent (PE) to the infectious agent (PI) pEI,Day7, and the recovery probability pIR. By changing the value of the parameters at a time (keeping the other parameters at their default values), we systematically studied the effect of that parameter and the combination of different parameters on the spread of the epidemic. The values of the parameters are shown in Table 1.
Results and discussion
Agent dynamics and contact pattern
To investigate the effect of contact-dependent dynamics, we estimate the exposure time ω (the time that two particles stay in contact) and the mean free time τ (the time interval between two consecutive contacts) in the CDD model.
By counting the time that particles i and j remain in contact in trajectories, we can estimate the exposure time ω of different velocities in the CDD model. As shown in Fig 4A, the average of ω is inversely proportional to v, while the median decreases slightly as v increases. This means that the agents do not intend to stay together for a long time as v decreases. We classify contacts into short-term contact (ω < 0.5*rc/v), medium-term contact (0.5*rc /v ≤ ω < 4*rc /v) and long-term contact (ω ≥ 4*rc/v) by the value of ω (Fig 4B). The results show that the CDD model has a larger proportion of short-term contacts as the velocity of agents decreases.
Fig 4. Contact pattern of the CDD model.
(a) The mean (blue) and median (red) of the contact time ω influenced by the velocity v of agents in the CDD model. (b) The percentages of ω in different time ranges for the velocity v of agents in the CDD model. The blue line represents ω < 0.5*rc /v, the red line represents 0.5*rc /v ≤ ω < 4*rc /v, and the green line represents ω ≥ 4*rc/v. (c) The mean free time τ in the CDD model (solid line) and the CDD-NC model (dashed line) for different velocities.
In addition, by dividing the total trajectory time by the average number of particle contacts obtained from the statistics of the CDD model, we can estimate the mean free time τ. As a control, we also set up a contact-dependent dynamics with no contact (CDD-NC) model (S1 and S2 Figs in S1 File, please refer to the S1 File for more details), in which the velocities and directions of the individuals follow the same distribution in CDD and the motion does not change when two particles are in contact, as in the no contact case of Eq 2. We further estimated the mean free time τ (Eq 3) of the CDD-NC model.
| (3) |
where rc is the contact distance, as defined before, is the average relative velocity and was set as , and ρ is the density of the system. With respect to τ, we found that there was no difference between the two models (Fig 4C).
Therefore, the CDD model does not change the frequency of contact but only makes the contact pattern of individuals more inclined to short-term contact as the velocity of agents decreases. In the following, we found that this contact pattern caused strong spatial heterogeneity and determined the transmission pattern at low velocity.
Epidemic dynamics and the basic reproduction number R0
We examined the effect of five propagation parameters, including nI,0, d, pSE, pEI,Day7 and pIR on epidemic evolution. For example, different pSE agents show different patterns of epidemic spread as shown in Fig 5A and 5B.
Fig 5. Infection spreading in different pSE and the basic reproduction number R0 influenced by pSE and the velocity v of agents in the system.
The dynamic progress of the infection spreading when pSE = 1.0*10−4 (a) and pSE = 2.0*10−4 (b). Snapshots of Days 0, 20, and 40 are shown in order. (c) The influence of pSE and v of agents on R0 in the system. The points represent the calculated data, and the solid line represents the fitting curve to eliminate noise.
To reasonably characterize the epidemic transmission capacity obtained from the current CDD-SEIR model simulations, we defined the basic reproduction number R0. Traditional models commonly use the basic reproduction number R0 to indicate how contagious an infectious disease is. R0 is the number of secondary infections an infectious PI can cause in a fully susceptible environment [36]. We calculated the basic reproduction number R0 by the definition of R0 in ordinary differential equations. Similar to SEIR model,
| (4) |
| (5) |
where NS, NI, NR and N represent the susceptible, infectious, recovered and total populations of agents, and β and γ represent the transmission rate and recovery rate (which equals to pIR in our model), respectively. Then R0 can be calculated by
| (6) |
We calculated the basic reproduction number R0 in the CDD-SEIR model by
| (7) |
where ΔNS represents the variation of NS in the integration time step Δt (please refer to the S1 File for more details).
Fig 5C shows how R0 is influenced by pSE and the velocity v of agents in the system. In addition to R0, we also counted the peak proportion of infections in the system, nI,max, and the infection rate p of the system when the epidemic had developed for 40 days to characterize spreading (S4 Fig in S1 File).
We examined the effect of five propagation parameters, including nI,0, d, pSE, pEI,Day7, and pIR, on R0 when the agents in the system had a specific mobility. As shown in Fig 6A, except for the parameters nI,0 and pEI,Day7, R0 shows a significant dependence on the other three parameters. Specifically, the ability of the virus to spread/infect is positively correlated with d, and pSE and negatively correlated with pIR. This observation is consistent with our common perception: the larger the infection range (d), the more virulent the virus; the lower the recovery rate, the higher the severity of the outbreak. Meanwhile, it should be noted that these three parameters also showed different effects: in the speed range studied in the current work, R0 shows a clear dependence on the infection range d, i.e., a larger d led to a larger R0; in contrast, R0 is not significantly affected by different pSE and pIR when the system is under low mobility and only reflects notable differences when the agents are in high mobility.
Fig 6.
The basic reproduction number R0 (a), the heterogeneity variable ζ (b) and the critical velocity vc of R0 and ζ (c) influenced by five parameters (nI,0, d, pSE, pEI,Day7, and pIR) and the velocity v of agents in the system. In each plot, the results corresponding to the five values used for the parameter that changed are represented by five different colored curves. The points represent the calculated data, and the solid line represents the fitting curve to eliminate noise. The horizontal coordinates in (a) and (b) are on a logarithmic scale.
We fit the curves for each of the two phases of R0 linearly and define the critical velocity vc of R0 at which R0 reaches 90% of the saturated reproduction number (please refer to the S1 File for more details). The calculations show that when the mobility of the system is small (i.e., the individual’s v is less than vc), R0 is influenced almost exclusively by d, independent of other propagation parameters. When v > vc, the system is close to the scenario obtained from the classical epidemic model assuming uniform mixing [6]. For example, in classical models, an infectious agent can come into contact with susceptible individuals at a probability proportional to NS. Thus, the mobility of the infectious is not directly reflected in the process of virus transmission. However, when v < vc, this is not the case. Epidemics transmitted by contact are closely related to the type of individuals who can be reached locally, the so-called spatial heterogeneity [12, 14].
Heterogeneity variable ζ and spatial heterogeneity
Fig 6A clearly shows that the epidemic varies linearly with v when v < vc, indicating that heterogeneity significantly influences the spread of the epidemic; in contrast, the spread of the epidemic is no longer dependent on the variation of v when v > vc [20]. Therefore, it is necessary to explore the factors influencing vc. As shown in Fig 6C, we found that vc of R0 is tightly associated with certain propagation parameters. Specifically, vc is positively correlated with pSE, and nearly independent of d, initial nI,0, pEI,Day7, and pIR. It should be noted that the independence between vc and the initial nI,0 indicates that the epidemic is determined by its inherent properties. On the other hand, pEI, Day7 and pIR do not affect the distribution of PS around PI, which is associated with heterogeneity and thus vc.
Apparently, vc indicates the boundary between heterogeneity and homogeneity. To further elucidate the above trend, we calculated the relative neighbor concentration RNCS of the system for characterizing spatial heterogeneity: RNCS was defined as the average of the ratio of the local concentration of PS within the infection range d of a single PI in the system to the global concentration of PS in the whole system. The heterogeneity variable ζ was defined as 1- RNCS (Eq 8).
| (8) |
Here, N´S represents the total number of PS within radius d of a PI, N´ represents the total number of all individuals within radius d of this PI, and the ratio of these two corresponds to the local concentration of PS; NS/N represents the global concentration of PS (please refer to the S1 File for more details).
As shown in Fig 6B, ζ gradually converges to 0 with increasing v, indicating that the system gradually transitions from a strongly heterogeneous system to a weakly heterogeneous system (therefore satisfying the assumption of homogeneous mixing). We can also define the critical velocity vc of ζ at which ζ reaches 0.1(RNCS reaches 0.9). As shown in Fig 6C, the saturation velocity of ζ is comparable to that of R0, and both are positively correlated with pSE, and less correlated with d, initial nI,0, pEI,Day7, and pIR. This is easy to understand. When pSE increases, the number of PS decreases while PI hardly reaches new susceptible particles, so the spatial heterogeneity is further enhanced. The spatial heterogeneity of agents other than PS is not strongly associated with PS. When d increases, the distribution of agents within d (local distribution) is closer to the distribution of agents within the whole system (global distribution), so the spatial heterogeneity decreases. However, this effect seems not significant to vc. Therefore, the trend of the two phases of R0 under different velocity conditions is caused by spatial heterogeneity.
Fig 6 shows that the variation in R0 with velocity shows two different phases, with the boundary of vc. We further investigated the relationship between the maximum slope k obtained by fitting the data before vc and the saturated reproduction number Rs after vc and other propagation parameters (Fig 7). The two indexes, k and RS, have consistent variability, i.e., they are both positively correlated with d and pSE, negatively correlated with pIR, and independent of the initial nI,0 and pEI,Day7. For RS, we further find that the data can be approximately proportional to d2pSE/pIR. In this case, the result obtained from the CDD model is close to that of the mean field method.
Fig 7.
The slope k (a) and the saturated reproduction number RS (b) influenced by five parameters (nI,0, d, pSE, pEI,Day7, and pIR) in the system.
Notably, the results obtained from the CDD-SEIR model show that R0 is linearly related to log(v). This is not the same trend as that obtained by mean-field theory [17, 19]. One possible reason for the discrepancy obtained by the two models is that the treatment of the mean field method does not consider spatial heterogeneity, but other factors may also play a role.
Effect of the CDD model on R0
As shown in Fig 6, when v < vc, the basic reproduction number R0 is nearly independent of the virulence pSE. In the work of the mean field models [19], , where ω is the exposure time, τT is the characteristic time of infection, represents the probability of infection in each contact, σ0 is the scattering cross section of the agents, ρ is the density, and τI is the transition time from infectious to recovered.
When the mobility of the agents is low, ω is much larger than τT, and therefore, infection must occur in each contact, resulting in R0 that is independent of viral virulence. From this perspective, the results obtained by the CDD-SEIR model are consistent with those obtained by the mean-field model. However, there are naturally obvious differences between the two. Spatial heterogeneity caused by the contact model of the CDD model plays an important role in the infection.
As shown in Fig 4, the CDD model prefers short-term contact at low velocity, while the proportion of a long contact time is low, indicating that even though the CDD-SEIR model and the mean-field model obtain qualitatively consistent trends, they are fundamentally different. The reason for this lies in the fact that differences in individual motion are considered in the CDD model so that spatial heterogeneity can be described, while the mean field treatment ignores such differences. To further illustrate the difference between these two models, we selected the cases of a relatively low velocity (v = 0.001 m/s, where v < vc) and a medium velocity case (v = 0.02 m/s, where v > vc) and calculated the total time tSE required for the PS agents to contact PI agents to become infected. The number of contacts between PS and PI before infection TSI and the mean and median of contact time per contact between PS and PI before infection ωSI (please refer to the S1 File for more details) varied with the probability of infection pSE (Fig 8).
Fig 8. Contact time required for infection tSE at low velocity (a, v = 0.001 m/s) and medium velocity (b, v = 0.02 m/s), number of contacts between PS and PI agents before infection TSI, the mean (blue) and median (orange) of the average time per contact between PS and PI agents before infection ωSI with the probability of infection pSE compared with the contact time ω calculated by the CDD model.
At a low velocity (v = 0.001 m/s), infection tends to occur through a small number of long-term contacts.
At low velocities (v = 0.001 m/s), the PS agent only has 2–3 contacts with PI agents before becoming infected, and the duration of each contact ωSI tends to be relatively long. This observation indicates that at low velocities, long-term contact is essential for the infection, and short-term contact is usually not infectious. This can be understood by the longer mean free time τ at low velocities (Fig 4C). From Eq 3, we can obtain τ ≈ 0.22 days when v = 0.001 m/s. In this case, even if all the surroundings are PI agents, it would take several days for PS agents to accumulate enough short exposures to become infected. However, in the real situation, the disease state of the infected person cannot be maintained indefinitely, and the whole system cannot maintain the state of many infected persons for a long time, so the contact time tSE obtained is often less than the ideal value of 1/pSE (Fig 8A), and there may even be a simultaneous decrease in pSE and tSE. Therefore, infections caused by multiple short-term contacts that take a long time to occur at low velocity are in fact very difficult to occur. This means that only a small number of prolonged exposures under the CDD model can achieve the conditions for infection occurrence in the mean field model. The long-term contact for transmission may account for strong spatial heterogeneity at low velocity.
When the velocity increases (v = 0.02 m/s), it is known by Eq 3 that τ will decrease significantly. In this case, the particles do not have to sift through longer collisions used for infection to occur, and infection can occur through more contacts. Meanwhile, ωSI is also closer to an ordinary contact (Fig 8B), which is close to the results obtained by the mean-field model.
Presumably, allowing short contact exposure with good protective measures can significantly reduce R0. Especially when control measures are strong, it is difficult for infections to reach new susceptible particles, and the effective ability of the virus to spread will be limited.
Comparison with other models
Various epidemiological models that consider multiple types of compartments beyond the basic SIR framework have been proposed. In this study, we compare our CDD-SEIR method to two recently reported methods. One is a random walker’s approach that incorporates the memory effect into a compartment model [37], where recovered agents are assumed to stay immune for a random duration. In contrast, our work considers the scenario where the susceptible agents become carriers after being infected and combines a modified SEIR model with contact-dependent dynamics. Both methods are based on the traditional SIR model and explore the influence of other factors on space-time dynamics of epidemics. The random walker’s method is efficient for characterizing epidemics in the long term, while our CDD-SEIR model introduces more dynamical details by considering patterns of contacts. As the two methods consider different impacts of other factors on disease transmission, direct comparison of results is challenging. Future studies can further incorporate an immunity impact on transmission in our model.
The SEIRS model considers the effects of birth, death, and incubation period on infectious disease dynamics [38]. Both this model and our work take into account the latency effect, although in different ways. However, the SEIRS model integrates multiple influences, such as the length of the latent period, the various routes of transmission, and age-specific transmission. Additionally, the introduction of repeated infection in the SEIRS and random walker’s models enables the epidemic equilibrium, allowing for longer timescale characterization of the disease spreading. While our CDD-SEIR model can incorporate these factors by adding agent types to simulate more realistic scenarios, we primarily focused on the impact of contact due to motion on epidemic transmission. Although some details of the epidemic transmission process are excluded, our CDD-SEIR model captures the fundamental information and physical laws.
Conclusion
In this work, our proposed CDD-SEIR model, which combines the real-scale random motion of individuals with the traditional compartment model, discloses that epidemic transmission depends on spatial heterogeneity and contact patterns. The CDD model has a larger proportion of short-term contacts as the velocity of agents decreases. However, we demonstrate that the contact pattern of the CDD model with short contact times at low velocity makes the virus more likely to spread through a small number of long contacts, which causes strong spatial heterogeneity and low R0. Due to the presence of spatial heterogeneity, R0 will be proportional to the logarithm of the group mobility in the strongly heterogeneous range and saturated in the weakly heterogeneous range, and R0 is approximately independent of the virulence of the virus when the mobility of the group is low. This demonstrates that in the case of a high reproduction number, the vaccination policy and reduced contact time can still significantly inhibit the virus transmission capacity.
In the selection of parameters, we tried to make the individual parameters more adapted to the real scale, which allowed us to obtain a more accurate description of the distribution of exposure times, thus helping to explain how the movement model and spatial heterogeneity can influence the spread of epidemics from a microscopic perspective. This is something that has been relatively lacking in previous work. Such data also fully illustrate the existence of heterogeneity in the CDD-SEIR model and the difference with the mean-field model.
Overall, we used a real-scale contact-dependent motion model to reveal the effects of spatial heterogeneity and infection patterns on disease transmission from a microscopic perspective. The insight obtained from this work provides us with a new understanding of how movement patterns affect the ability of viruses to spread and how to prevent them more effectively. We hope to improve this CDD-SEIR model in the future to make the movement of individuals more realistic and thus be able to simulate some of the transmission that is difficult to characterize by numerical or network models.
Supporting information
(DOCX)
Data Availability
All relevant data are accessible through this DOI:(10.5061/dryad.qrfj6q5mp).
Funding Statement
This work was supported by the National Natural Science Foundation of China (Grant Nos. 12090052, 21833002), the National Key R&D Program of China (2021ZD201302), and the “Fundamental Research Funds for the Central Universities” (021514380018). Parts of the calculations were performed using computational resources on an IBM Blade cluster system from the High-Performance Computing Center (HPCC) of Nanjing University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382(13):1199–207. Epub 2020/01/30. doi: 10.1056/NEJMoa2001316 ; PubMed Central PMCID: PMC7121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2):taaa021. Epub 2020/02/14. doi: 10.1093/jtm/taaa021 ; PubMed Central PMCID: PMC7074654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Rocklöv J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J Travel Med. 2021;28(7):taab124. Epub 2021/08/10. doi: 10.1093/jtm/taab124 ; PubMed Central PMCID: PMC8436367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Rocklöv J. The effective reproductive number of the Omicron variant of SARS-CoV-2 is several times relative to Delta. J Travel Med. 2022;29(3):taac037. Epub 2022/03/10. doi: 10.1093/jtm/taac037 ; PubMed Central PMCID: PMC8992231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson RM, May RM. Infectious diseases of humans: dynamics and control. Oxford: Oxford university press; 1992. [Google Scholar]
- 7.Kermack WO, McKendrick AG, Walker GT. A contribution to the mathematical theory of epidemics. Proc R Soc Lond A Math Phys Sci. 1927;115(772):700–21. doi: 10.1098/rspa.1927.0118 [DOI] [Google Scholar]
- 8.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438(7066):355–9. doi: 10.1038/nature04153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apolloni A, Poletto C, Colizza V. Age-specific contacts and travel patterns in the spatial spread of 2009 H1N1 influenza pandemic. BMC Infect Dis. 2013;13:176. Epub 2013/04/17. doi: 10.1186/1471-2334-13-176 ; PubMed Central PMCID: PMC3644502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Britton T, Ball F, Trapman P. A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science. 2020;369(6505):846–9. Epub 2020/06/25. doi: 10.1126/science.abc6810 ; PubMed Central PMCID: PMC7331793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley S. Large-scale spatial-transmission models of infectious disease. Science. 2007;316(5829):1298–301. Epub 2007/06/02. doi: 10.1126/science.1134695 . [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Wang X, Wang M. Inhomogeneity of epidemic spreading. Chaos. 2010;20(2):023128. Epub 2010/07/02. doi: 10.1063/1.3445630 ; PubMed Central PMCID: PMC7117601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poletto C, Tizzoni M, Colizza V. Heterogeneous length of stay of hosts’ movements and spatial epidemic spread. Sci Rep. 2012;2:476. Epub 2012/06/29. doi: 10.1038/srep00476 ; PubMed Central PMCID: PMC3384080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen-Jie Z, Xing-Yuan W. Inhomogeneity of epidemic spreading with entropy-based infected clusters. Chaos. 2013;23(4):043105. Epub 2014/01/07. doi: 10.1063/1.4824316 ; PubMed Central PMCID: PMC7112474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macal CM, North MJ, editors. Tutorial on agent-based modeling and simulation. Proceedings of the Winter Simulation Conference, 2005; 2005 4–4 Dec; Orlando, FL, USA: IEEE. [Google Scholar]
- 16.Silva PCL, Batista PVC, Lima HS, Alves MA, Guimarães FG, Silva RCP. COVID-ABS: An agent-based model of COVID-19 epidemic to simulate health and economic effects of social distancing interventions. Chaos Solitons Fractals. 2020;139:110088. Epub 2020/08/25. doi: 10.1016/j.chaos.2020.110088 ; PubMed Central PMCID: PMC7340090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González MC, Herrmann HJ. Scaling of the propagation of epidemics in a system of mobile agents. Physica A. 2004;340(4):741–8. [Google Scholar]
- 18.González MC, Lind PG, Herrmann HJ. System of mobile agents to model social networks. Phys Rev Lett. 2006;96(8):088702. Epub 2006/04/12. doi: 10.1103/PhysRevLett.96.088702 . [DOI] [PubMed] [Google Scholar]
- 19.Peruani F, Sibona GJ. Dynamics and steady states in excitable mobile agent systems. Phys Rev Lett. 2008;100(16):168103. Epub 2008/06/04. doi: 10.1103/PhysRevLett.100.168103 . [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez JP, Ghanbarnejad F, Eguíluz VM. Particle velocity controls phase transitions in contagion dynamics. Sci Rep. 2019;9(1):6463. Epub 2019/04/25. doi: 10.1038/s41598-019-42871-x ; PubMed Central PMCID: PMC6478726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levis D, Diaz-Guilera A, Pagonabarraga I, Starnini M. Flocking-enhanced social contagion. Phys Rev Res. 2020;2(3):032056. doi: 10.1103/PhysRevResearch.2.032056 [DOI] [Google Scholar]
- 22.Norambuena A, Valencia FJ, Guzmán-Lastra F. Understanding contagion dynamics through microscopic processes in active Brownian particles. Sci Rep. 2020;10(1):20845. Epub 2020/12/02. doi: 10.1038/s41598-020-77860-y ; PubMed Central PMCID: PMC7705763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sajjadi S, Hashemi A, Ghanbarnejad F. Social distancing in pedestrian dynamics and its effect on disease spreading. Phys Rev E. 2021;104(1–1):014313. Epub 2021/08/21. doi: 10.1103/PhysRevE.104.014313 . [DOI] [PubMed] [Google Scholar]
- 24.Du M. Mitigating COVID-19 on a small-world network. Sci Rep. 2021;11(1):20386. Epub 2021/10/16. doi: 10.1038/s41598-021-99607-z ; PubMed Central PMCID: PMC8516975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen BF, Simonsen L, Sneppen K. COVID-19 Superspreading Suggests Mitigation by Social Network Modulation. Phys Rev Lett. 2021;126(11):118301. Epub 2021/04/03. doi: 10.1103/PhysRevLett.126.118301 . [DOI] [PubMed] [Google Scholar]
- 26.Merler S, Ajelli M, Fumanelli L, Gomes MF, Piontti AP, Rossi L, et al. Spatiotemporal spread of the 2014 outbreak of Ebola virus disease in Liberia and the effectiveness of non-pharmaceutical interventions: a computational modelling analysis. Lancet Infect Dis. 2015;15(2):204–11. Epub 2015/01/13. doi: 10.1016/S1473-3099(14)71074-6 ; PubMed Central PMCID: PMC4409131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeAngelis DL, Grimm V. Individual-based models in ecology after four decades. F1000Prime Rep. 2014;6:39. Epub 2014/07/06. doi: 10.12703/P6-39 ; PubMed Central PMCID: PMC4047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hethcote HW. The Mathematics of Infectious Diseases. SIAM Review. 2000;42(4):599–653. doi: 10.1137/s0036144500371907 [DOI] [Google Scholar]
- 29.Howard J, Huang A, Li Z, Tufekci Z, Zdimal V, van der Westhuizen HM, et al. An evidence review of face masks against COVID-19. Proc Natl Acad Sci U S A. 2021;118(4). Epub 2021/01/13. doi: 10.1073/pnas.2014564118 ; PubMed Central PMCID: PMC7848583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Zeng Z, Wang K, Wong SS, Liang W, Zanin M, et al. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J Thorac Dis. 2020;12(3):165–74. Epub 2020/04/11. doi: 10.21037/jtd.2020.02.64 ; PubMed Central PMCID: PMC7139011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malevanets A, Kapral R. Mesoscopic model for solvent dynamics. J Chem Phys. 1999;110(17):8605–13. doi: 10.1063/1.478857 [DOI] [Google Scholar]
- 32.Kapral R. Multiparticle Collision Dynamics: Simulation of Complex Systems on Mesoscales. In: Rice SA, editor. Advances in Chemical Physics. 140. Hoboken: John Wiley & Sons; 2008. p. 89–146. [Google Scholar]
- 33.Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395(10242):1973–87. Epub 2020/06/05. doi: 10.1016/S0140-6736(20)31142-9 ; PubMed Central PMCID: PMC7263814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689–97. Epub 2020/02/06. doi: 10.1016/S0140-6736(20)30260-9 ; PubMed Central PMCID: PMC7159271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian L, Li X, Qi F, Tang QY, Tang V, Liu J, et al. Harnessing peak transmission around symptom onset for non-pharmaceutical intervention and containment of the COVID-19 pandemic. Nat Commun. 2021;12(1):1147. Epub 2021/02/21. doi: 10.1038/s41467-021-21385-z ; PubMed Central PMCID: PMC7895830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallinga J, Lipsitch M. How generation intervals shape the relationship between growth rates and reproductive numbers. Proc Biol Sci. 2007;274(1609):599–604. Epub 2007/05/04. doi: 10.1098/rspb.2006.3754 ; PubMed Central PMCID: PMC1766383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bestehorn M, Michelitsch TM, Collet BA, Riascos AP, Nowakowski AF. Simple model of epidemic dynamics with memory effects. Phys Rev E. 2022;105(2–1):024205. Epub 2022/03/17. doi: 10.1103/PhysRevE.105.024205 . [DOI] [PubMed] [Google Scholar]
- 38.Bjørnstad ON, Shea K, Krzywinski M, Altman N. The SEIRS model for infectious disease dynamics. Nat Methods. 2020;17(6):557–8. Epub 2020/06/06. doi: 10.1038/s41592-020-0856-2 . [DOI] [PubMed] [Google Scholar]