Abstract
In the present report we show that unknown DNA fragments are easily amplified in a single PCR reaction from an oligo-cassette library with a single genome-specific primer in combination with a cassette-specific primer. The novelty of the system, in comparison to the vectorette PCR method, lies in the use of unphosphorylated in contrast with phosphorylated oligo-cassettes in the ligation to the chromosomal DNA fragments. After denaturation of the DNA library, all chromosomal fragments carry a single-stranded linker attached to the 5′-end only. Therefore, the presence of the vectorette mismatched region is not required when unphosphorylated cassettes are used. As an example we report the amplification of the era gene from Lactococcus lactis.
INTRODUCTION
Among the available polymerase chain reaction (PCR) techniques for obtaining unknown DNA regions on either side of chromosomal regions of known nucleotide sequence (genomic walking), two methods are most widely used. Inverse PCR (1) uses two primers, located in each end of the known fragment, and pointing away from each other. The template for the amplification reaction is a library of circularised chromosomal DNA fragments. The other major genome walking method is called the vectorette PCR method (2). A vectorette is a partially double-stranded DNA cassette that is phosphorylated in its 5′-ends, and after ligation of the vectorette unit to a mixture of chromosomal DNA fragments, each strand has a vectorette unit attached to both ends. This library is used as a template for amplification with a genome-specific primer together with a vectorette-specific primer, which is identical but not complementary to a central mismatched region in the vectorette cassette. The central mismatched region is included to avoid first-strand synthesis by the vectorette primer. In the second round the vectorette primer can bind to the complementary region present on the product of the first synthesis round. Other methods circumvent the inherent background problem by using biotin-labelled chromosome-specific primers for the first synthesis round, followed by purification of the product using magnetic beads (3) or using nested primers (3,4). We show below that if an oligo-cassette without a vectorette-like mismatched region is left unphosphorylated, the desired DNA fragment can be amplified as the major product.
MATERIALS AND METHODS
Construction of oligo-cassette libraries
Annealing of the two primers MKP22 and MKP23 to form the double-stranded oligo-cassette MKD1 was performed by boiling a 100 µM solution of the primers, followed by slow cooling to room temperature. Primers were supplied by GATCopenhagen Aps (Symbion, Copenhagen, Denmark). For the construction of oligo-cassette library, EcoRI digested chromosomal DNA was ligated (5) to MKD1 oligo-cassette at a 10-fold molar excess of the cassette, followed by column purification of the ligation products, where unbound cassette DNA was removed (GFXTM PCR DNA and gel band purification kit, Amersham Pharmacia Biotech, Hørsholm, Denmark). In the same way the MKD3 oligo-cassette was ligated to HindIII digested chromosomal DNA.
PCR amplification
The amplification of the upstream and downstream parts of the era gene from Lactococcus lactis EcoRI and HindIII oligo-cassette libraries was performed with Taq DNA polymerase (Pharmacia Biotech) as recommended by the suppliers, in a RoboCycler Gradient 40 machine (Stratagene, GmbH, Heidelberg, Germany) using 30 cycles of the sequence 45 s denaturation at 94°C + 45 s annealing at 42–56°C + 2 min synthesis at 68°C. Amplification of the era gene from the intact Lactococcus chromosome was performed as above, but the proofreading Pfu polymerase (cloned, Stratagene) was used and with a longer synthesis time.
Nucleotide sequence determination
After purification of PCR products from agarose gels (GFXTM PCR DNA and gel band purification kit, Pharmacia Biotech), the nucleotide sequence was determined using the thermo-sequenase kit (Amersham Pharmacia Biotech). The nucleotide sequence could be unambiguously determined from the purified primary amplification products of the oligo-cassette libraries, and no background sequences were apparent.
RESULTS
Construction of a universal oligo-cassette and oligo-cassette libraries of chromosomal DNA from L.lactis
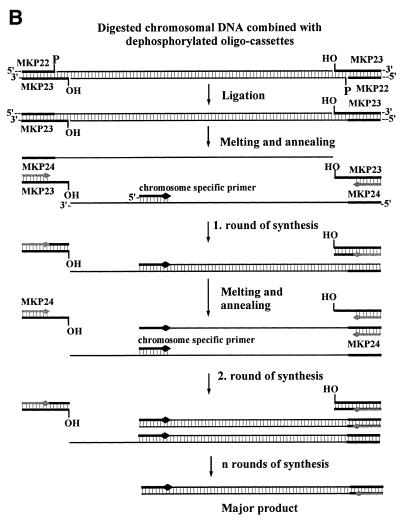
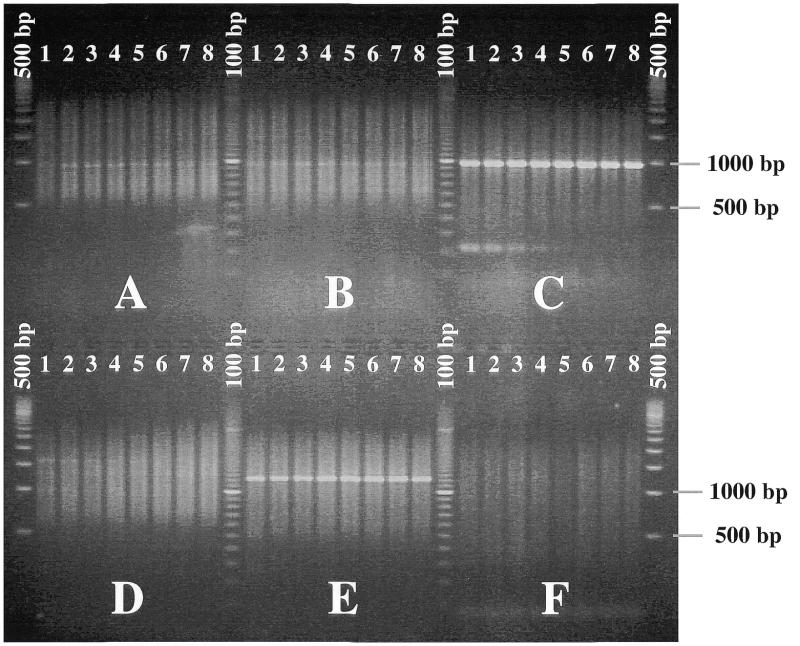
To facilitate the ligation of the oligo-cassette to different types of DNA fragments, three restriction sites, EcoRI, Sau3AI and HindIII, were incorporated in one end of the cassette (Fig. 1A). The EcoRI site was only added as a TTAA overhang which can combine with EcoRI digested ends to form a full EcoRI site. Three additional restriction sites, SacI, SphI and PstI were incorporated in the other end of the oligo-cassette for the subsequent digestion of the amplified fragment with different restriction enzymes. The resulting double-stranded oligo-cassette, MKD1, which contains six restriction sites, was constructed by annealing of the two oligonucleotides MKP22 and MKP23 (Fig. 1A). An oligo-cassette-specific primer MKP24, which spans the SacI–PstI region on MKD1 was synthesised for amplification of DNA ligated to the oligo-cassette. Upon digestion of MKD1 with HindIII and de-phosphorylation using calf intestinal phosphatase (5), the HindIII-specific oligo-cassette MKD3 (Fig. 1A) was constructed. A HindIII-specific oligo-cassette could preferentially be constructed directly from unphosphorylated primers, but the present approach was included to document that dephosphorylation of an existing cassette is a possible alternative. For use in the present study, we constructed two libraries using either EcoRI or HindIII digested chromosomal DNA from L.lactis MG1363. Amplification of DNA from the two libraries gave rise to a smear when the cassette-specific primer MKP24 was used alone, at annealing temperatures from 42 to 56°C (Fig. 2A and D). Faint bands could be detected above the background at low temperatures, with decreasing intensities at annealing temperatures from 48 to 52°C (Fig. 2A and D). As the melting point of MKP24 (78.5°C) is significantly higher, the products are probably due to partial annealing of MKP24 to chromosomal DNA.
Figure 1.
(A) Primers and oligo-cassettes used in the creation of oligo-cassette libraries. MKD1 was constructed by annealing of MKP22 and MKP23. MKD3 was constructed by HindIII digestion of MKD1 followed by dephosphorylation. (B) Amplification of oligo-cassette libraries with the cassette-specific primer MKP24 and a chromosome-specific primer.
Figure 2.
Amplification products from MG1363/EcoRI-MKD1 (A, B and C) and MG1363/HindIII-MKD3 (D, E and F) oligo-cassette libraries. The amplification was performed with the cassette-specific primer MKP24 alone (A and D) or in combination with the era-specific primers MKP198 (B and E) or MKP199 (C and F). The annealing temperatures were 42, 44, 46, 48, 50, 52, 54 and 56°C for lanes 1–8 in all panels, respectively. The DNA size markers were 100 bp extended-range ladder (marked 100 bp) and 500 bp ladder (marked 500 bp) from FMC BioProducts Europe (Denmark).
Amplification of an internal fragment of the L.lactis era gene by PCR using degenerate primers
To amplify the era gene [era = Escherichia coli RAS-like GTP binding protein (6)] from the L.lactis wild-type strain MG1363 (7) the sequences of the known era homologues from Streptococcus pneumoniae (GenBank accession no. AF07281) and Streptococcus pyrogenes (GenBank accession no. SPU31915) were first used to identify the homologous regions from Streptococcus mutans and Enterococcus faecalis using computer aided searches in the available partial genome sequences (Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org ). From the alignment of the era genes present in these four close relatives of L.lactis, it appeared that the N-terminal regions were very similar, but the C-terminal regions showed a high degree of variation (data not shown). When all the DNA sequences were compared, it was possible to find two regions, ∼550 bp apart, which showed particularly high sequence similarity. From these regions two degenerate primers were designed. The upstream primer mixture MKP196 (TAATCWGGHTTYGTAGC) contained 12 different sequences and the downstream primer mixture MKP197 (TCYGGATGATCMGTRATYTG) was 18-fold degenerated. A dominant product of 550 bp was produced by amplification of MG1363 chromosomal DNA using MKP196 and MKP197 (data not shown). Sequence determination from both ends showed that the amplified DNA was indeed an era homologue (data not shown). Using this sequence information two primers were synthesised. Primer MKP198 (TTTGACCCATCACGTGATTC) reads from the upstream end of the fragment towards the promoter, and MKP199 (CAAGGAAATAACACCGAAAG) reads from the downstream end of the fragment towards the end of the gene.
Amplification of the entire era gene by genome walking using amplification of the L.lactis oligo-cassette libraries
To obtain the rest of the era gene from the MG1363 chromosome, DNA fragments from the MKD1 (EcoRI) and MKD3 (HindIII) libraries were amplified using the cassette-specific primer MK24 together with either MK198 or MK199. The upstream region was amplified as a 1.2-kb fragment using the MKD3 library and era-specific primer MKP198 (Fig. 2E), and the downstream region as a 1-kb fragment using the MKD1 library and primer MKP199 (Fig. 2C). The two fragments were produced at all annealing temperatures from 42 to 56°C, while the few non-specific background products were eliminated at annealing temperatures >50°C (Fig. 2C and E). No distinct products were produced with other library/primer combinations (Fig. 2B and F), possibly because the restriction sites were located too far away from the era gene to be amplified with the present PCR protocol. Using the nucleotide sequence of the ends of era structural gene, determined from the amplified products with primers MK198 and MK199, the entire era gene was amplified on a 1-kb fragment from the MG1363 chromosome with the specific upstream primer MK213 (AATCGAAACCTTGTTTCGTC) and downstream primer MK214 (CAAAAAATAGCGATAATTCG). The nucleotide sequence, which was determined from within this amplified fragment is submitted under the GenBank accession number AF233268.
CONCLUSION
In the present report we have shown that unknown DNA fragments are easily amplified in a single PCR reaction from an oligo-cassette library with a single genome-specific primer in combination with a cassette-specific primer. As an example, the amplification of the era gene of L.lactis was reported. The applicability of the technique to the study of more complicated genomes and on larger DNA distances has recently been tested by Dr Zoran Gojkovic, who amplified a 20-kb fragment containing the TRP1 upstream region from an MKD1 oligo-cassette library of partially EcoRI digested Saccharomyces kluyveri chromosomal DNA (personal communication).
Acknowledgments
ACKNOWLEDGEMENTS
We want to thank Dr Zoran Gojkovic for comments on the manuscript. This project was funded by a grant from the Danish FØTEK program.
REFERENCES
- 1.Triglia T., Peterson,M.G. and Kemp,D.J. (1988) Nucleic Acids Res., 16, 8186. [DOI] [PMC free article] [PubMed]
- 2.Riley J., Butler,R., Ogilvie,D., Finniear,R., Jenner,D., Powell,S., Anand,R., Smith,J.C. and Markham,A.F. (1990) Nucleic Acids Res., 18, 2887–2890. [DOI] [PMC free article] [PubMed]
- 3.Rosenthal A. and Jones,D.S.C. (1990) Nucleic Acids Res., 18, 3095–3096. [DOI] [PMC free article] [PubMed]
- 4.Harrison R.W., Miller,J.C., D’Souza,M.J. and Kampo,G. (1997) Biotechniques, 22, 650–653. [DOI] [PubMed]
- 5.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Sping Harbor, New York, NY.
- 6.Ahnn J., March,P.E., Takiff,H.E. and Inouye,M. (1986) Proc. Natl Acad. Sci. USA, 83, 8849–8853. [DOI] [PMC free article] [PubMed]
- 7.Gasson M.J. (1983) J. Bacteriol., 154, 1–9. [DOI] [PMC free article] [PubMed]