Abstract
Mitotic cells experience double-strand breaks (DSBs) from both exogenous and endogenous sources. Since unrepaired DSBs can result in genome rearrangements or cell death, cells mobilize multiple pathways to repair the DNA damage. In the yeast Saccharomyces cerevisiae, mitotic cells preferentially use a homologous recombination repair pathway. However, when no significant homology to the DSB ends is available, cells utilize a repair process called non-homologous end joining (NHEJ), which can join ends with no homology through resection to uncover microhomologies of a few nucleotides. Although components of the homologous recombination repair system are also involved in NHEJ, the rejoining does not involve all of the homologous recombination repair genes. The SRS2 DNA helicase has been shown to be required for DSB repair when the homologous single-stranded regions are short. Here it is shown that SRS2 is also required for NHEJ, regardless of the cell mating type. Efficient NHEJ of sticky ends requires the Ku70 and Ku80 proteins and the silencing genes SIR2, SIR3 and SIR4. However, NHEJ of blunt ends, while very inefficient, is not further reduced by mutations in YKU70, SIR2, SIR3, SIR4 or SRS2, suggesting that this rejoining process occurs by a different mechanism.
INTRODUCTION
Mitotic double-strand breaks (DSBs) are repaired through homologous and non-homologous repair pathways. In the yeast Saccharomyces cerevisiae, DSBs are repaired through homologous recombination when there is sufficient homology between the broken ends and a target donor repair sequence. Homologous recombination involves the recombination repair genes RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, MRE11 and XRS2. Although not absolutely required for DSB repair, the SRS2 DNA helicase has been shown to be required for DSB repair when the homologous single-stranded segments that initiate homologous recombination are short (1). Since non-homologous end joining (NHEJ) often involves small homologies of a few nucleotides at the rejoined ends, it was of interest to determine whether the SRS2 DNA helicase had a role in NHEJ.
We were also interested in examining a possible interaction between SRS2 and the SIR genes in NHEJ. NHEJ requires the Ku70 and Ku80 proteins. Using a two-hybrid screen, Tsukamoto and co-workers (2) found an interaction between Ku70 and the silencing factor Sir4. Analysis of NHEJ efficiencies showed that the silencing genes SIR2, SIR3 and SIR4 were required for end joining (2,3). These experiments utilized sir null mutations in strains that carried silent mating type information at the HML and HMR loci. This combination renders the haploid cells non-mating, as loss of repression of the silent loci results in expression of MATa and MATα information in one cell, a situation similar to diploid cells. Since a/α diploids are more proficient in mitotic homologous recombination than either a/a or α/α diploids (4), we investigated whether NHEJ varied according to the mating potential of the sir mutant haploid strains. We found, as reported by other groups, that the SIR genes are not required for NHEJ in mating-competent sir mutant cells (5–7) when the DSB ends have small microhomologies. However, the SIR genes have no effect on NHEJ in mating-competent or non-mating sir mutant strains when the DSBs are blunt ends with no single-stranded microhomologies at the ends. Requirement for SRS2 in NHEJ in Sir– strains suggests that NHEJ in Sir– cells proceeds through a NHEJ pathway that differs from that used in Sir+ strains.
MATERIALS AND METHODS
Strains and media
Yeast strains used are listed in Table 1. Media were prepared as described (8).
Table 1. Efficiency of end joining.
| Strain | Genotype | Efficiency of end joining | Fidelity of repair | |||
|---|---|---|---|---|---|---|
| 5′-End repair (%) | 3′-End repair (%) | Blunt end repair (%) | 5′-End:blunt end | |||
| HKY579-10A | Wild-type | 100 ± 10 | 75 ± 10 | 1.7 ± 0.2 | 58.8 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HKY590-1D | srs2Δ | 30 ± 5 | 30 ± 5 | 0.8 ± 0.1 | 37.5 | Accurate |
| 30/30 | 30/30 | 26/30 | ||||
| HKY604-17A | rad50Δ | 7 ± 1 | 2 ± 1 | 0.2 ± 0.01 | 35.0 | Mostly accurate |
| 22/30 | 23/30 | 26/30 | ||||
| HKY580-5B | rad52Δ | 40 ± 5 | 20 ± 2 | 0.5 ± 0.01 | 80.0 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HFY2008-2D | yku70Δ | 2 ± 1 | 2 ± 1 | 1.8 ± 0.1 | 1.1 | Inaccurate |
| 21/30 | 24/30 | 15/30 | ||||
| VH1-4C | srs2Δ rad52Δ | 5 ± 1 | 3 ± 1 | 1.8 ± 0.1 | 2.8 | Mostly accurate |
| 29/30 | 30/30 | 27/30 | ||||
| VH2-2B | srs2Δ yku70Δ | 1 ± 0.1 | 1 ± 0.1 | 1.8 ± 0.1 | 0.6 | Mostly accurate |
| 29/30 | 30/30 | 24/30 | ||||
| HKY881-1C | sir2Δ non-mater | 5.9 ± 3.2 | 2.5 ± 0.7 | 1.4 ± 1.2 | 4.2 | Mostly accurate |
| 28/30 | 30/30 | 27/30 | ||||
| HKY867-5D | sir3Δ non-mater | 3.6 ± 2.6 | 4.9 ± 2.2 | 1.4 ± 0.4 | 2.6 | Mostly accurate |
| 30/30 | 30/30 | 26/30 | ||||
| HKY884-1D | sir4Δ non-mater | 2.8 | 4.0 | 1.5 | 1.9 | Mostly accurate |
| 26/30 | 27/30 | 25/30 | ||||
| HKY881-6D | sir2Δ MATα | 92.4 ± 9.5 | 52.5 ± 28.2 | 1.9 ± 0.7 | 48.6 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HKY867-12B | sir3Δ MATα | 72.3 ± 21.0 | 51.9 ± 23.6 | 2.1 ± 0.4 | 34.4 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HKY884-2B | sir4Δ MATα | 87.7 | 79.4 | 0.7 | 125.3 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HKY934-16B | srs2Δ sir3Δ non-mater | 1 ± 0.5 | 2 ± 0.5 | 0.8 ± 0.2 | 1.2 | Mostly accurate |
| 29/30 | 28/30 | 28/30 | ||||
| HKY934-18B | srs2Δ sir3Δ MATα | 77 ± 10 | 79 ± 5 | 2 ± 0.5 | 38.5 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HKY1000 | Wild-type MATα/MATa | 4.7 | 14.6 | 2.3 | 2.0 | Mostly accurate |
| 28/30 | 29/30 | 26/30 | ||||
| HKY1001 | Wild-type MATα/matΔ | 90.7 | 80.3 | 3.1 | 29.2 | Accurate |
| 30/30 | 30/30 | 30/30 | ||||
| HKY1002 | srs2Δ/srs2Δ MATα/MATa | 30 ± 10 | 25 ± 5 | 2 ± 0.5 | 15.0 | Accurate |
| 30/30 | 30/30 | 27/30 | ||||
All strains are in the RAD5 W303 background and are of the genotype leu2-3, 112 his3-11, 15 ade2-1 ura3-1 trp1-1 can1-100. All SIR strains are MATa. The srs2, rad50, rad52, yku70, sir2, sir3, sir4 and mat mutants are null alleles. The sir mating-competent strains are HMLα MATα HMR::TRP1 and the sir non-mater strains are HMLα MATa HMR::TRP1. Repair is shown as the ratio of number of transformants obtained using the indicated linear plasmid relative to the number obtained using circular plasmid. For each transformation, plasmid DNA was recovered from 30 transformants and transformants analyzed as described. The number of recovered plasmids that were accurately repaired and had regenerated the restriction enzyme site are shown beneath the percentage of repair data. When the restriction site was lost, ∼50% of those plasmids sustained deletions >100 bp in length.
Plasmid repair assay
Aliquots of 2–5 µg of the replicating yeast plasmid pRS316 (9), carrying the URA3 gene as a selectable marker, were digested to completion with restriction enzymes as indicated. After inactivation of the enzyme by incubation at 65°C for 20 min, the linearized DNA was used to transform yeast cells (10). Parallel transformations were performed using an equivalent amount of uncut plasmid DNA to provide a comparison for transformation efficiency. Cells were plated on synthetic complete medium lacking uracil and incubated at 30°C for 2 days. Colonies that grew on synthetic complete medium lacking uracil were verified to be true transformants carrying the pRS316 vector by streaking each transformant to be analyzed on medium containing 5-fluoroorotic acid (5-FOA). Ura+ colonies that could grow on 5-FOA medium contained a circular pRS316 plasmid while Ura+ colonies that could not grow on 5-FOA medium did not contain any plasmid and were not used in any analysis. Transformation numbers were corrected for the percentage of Ura+ colonies that contained plasmid pRS316. Plasmid DNA was isolated from individual transformants. A 1345 bp region flanking the polylinker region was amplified by PCR using the primers 5′-GTATCCGGTAAGCGGCAGGGT-3′ and 5′-AGGTGCCGTAAAGCACTAATC-3′. The amplified products were analyzed by restriction enzyme digestion.
RESULTS
Requirement of SRS2 in NHEJ
NHEJ is strongly dependent on the RAD50 gene (5–7). The SRS2 gene encodes a DNA helicase that functions in a post-replication repair pathway (11,12). The double mutant srs2 rad50 exhibits extremely poor growth which is not seen in either single mutant. This could possibly reflect a synergistic effect on DSB repair. This fact, combined with the requirement for SRS2 in homologous recombination DSB repair when short tracts of homology are involved (1), led us to investigate whether a srs2 mutation had any effect on NHEJ. To examine NHEJ, we digested the replicating CEN plasmid pRS316 with EcoRI, KpnI or SmaI. These enzymes cut at one site each in the polylinker, which has no homology to the S.cerevisiae genome, yielding a 5′-overhang, a 3′-overhang or a blunt end, respectively. The efficiency of transformation with the linear plasmids was compared to the efficiency of transformation with an equal amount of circular plasmid. We observed a 3-fold reduction in end joining of sticky and blunt ends (Fig. 1 and Table 1) in the srs2 mutant strain. This was not as severe a reduction as that of a rad50 mutant strain, although both the srs2 mutant strain and the rad50 mutant strain reduce end joining of sticky and blunt ends without substrate preference. The ratio of the percent repair of a 5′-end to the percent repair of a blunt end was the same for the srs2 and rad50 strains and was close to the ratio observed in wild-type cells, given the low repair efficiency of the blunt end substrate. The poor growth of the srs2 rad50 strain prohibited examination of end joining in the double mutant. RAD50 is also required for telomeric length maintenance (3). The poor growth of the srs2 rad50 strain is not due to effects on telomeric maintenance as telomeres in a srs2 mutant are the same length as in a wild-type strain and the srs2 mutation does not increase the telomeric shortening of a rad50 mutant (data not shown).
Figure 1.
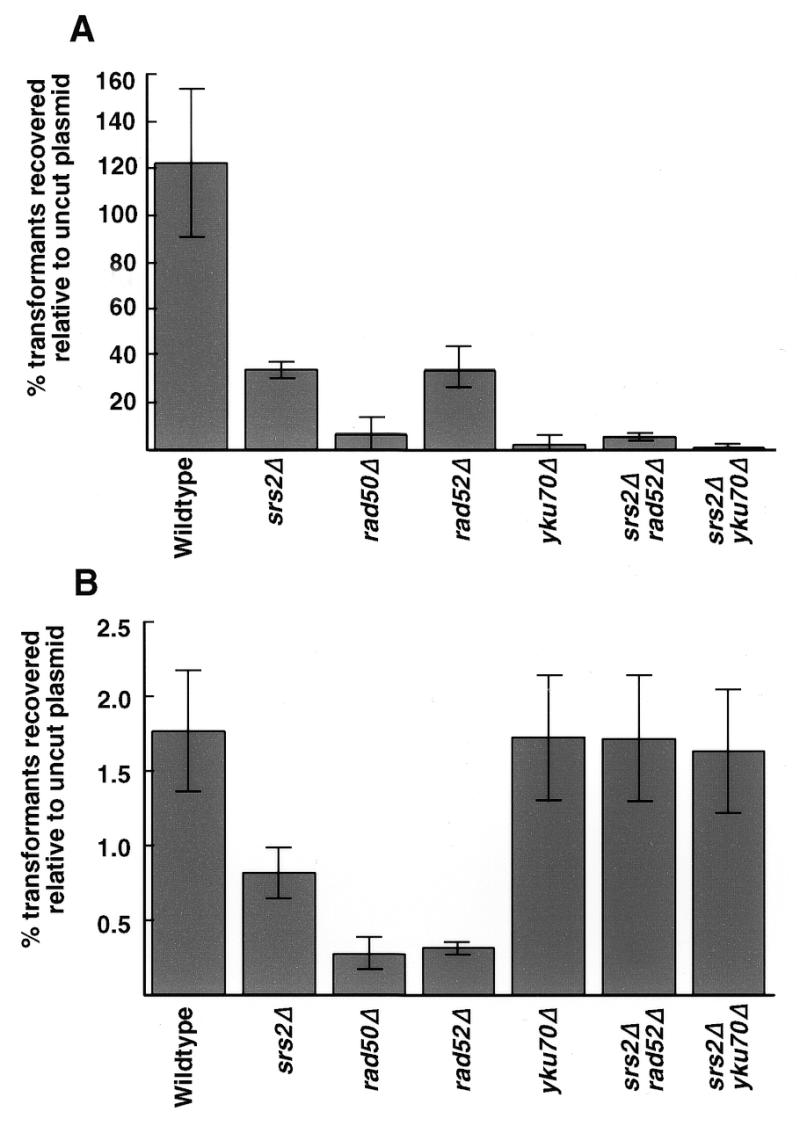
Comparison of NHEJ efficiencies in srs2 mutants to NHEJ efficiencies in rad50, rad52 and yku70 mutants. Cells of each indicated genotype were transformed with equal amounts of linear or supercoiled plasmid DNA. (A) Transformation efficiencies using plasmid pRS316 linearized with EcoRI. Similar results were obtained using KpnI-digested plasmid. (B) Transformation efficiencies using plasmid pRS316 linearized with SmaI. Each strain was transformed three or four times with the indicated plasmids.
To determine whether SRS2 functions in a homologous recombination or non-homologous end joining pathway for DSB repair, accuracy of repair in srs2 rad52 and srs2 yku70 mutants was examined. While the rad52 and srs2 single mutants reduced rejoining of 5′-ends to 30–40% of the wild-type level, the double mutant showed a synergistic reduction in end joining to the level of the yku70 mutant. A similar reduction was seen in rejoining of 3′-ends. Further evidence of the srs2 rad52 double mutant behaving similarly to the yku70 single mutant can be seen in the ratio of repair of the 5′-end to the blunt end substrate. However, the srs2 rad52 double mutant differs from the yku70 mutant in that the fidelity of repair in the double mutant is very high. This suggests that SRS2 and RAD52 act synergistically in the main haploid end joining pathway.
For rejoining of 5′ or 3′ overlapping ends, the Ku70 mutation yku70 was epistatic to the srs2 mutation. However, the repair was far more accurate than repair in the single mutant yku70 (Table 1; χ2 = 12.2, P < 0.001), suggesting that the srs2 mutation channels the DSB repair into an alternative pathway that is different from the Ku-dependent pathway used in wild-type strains.
Relationship of SRS2 to the SIR-dependent end joining process
To examine a possible role for SRS2 in NHEJ events influenced by the SIR genes, we constructed isogenic strains that carried a deletion of the HMR locus, α information at the HML locus and either a or α information at the MAT locus. The MATa strains were non-mating in sir null mutant backgrounds while the MATα strains were mating competent in the sir mutant backgrounds. sir2, sir3 and sir4 null mutants in non-mating strains reduce NHEJ 25- to 35-fold (2,3,13–15; Fig. 2A and Table 1). The ends are rejoined accurately for the most part. However, when the DSB ends do not contain short sticky ends, the reduction in rejoining is comparable in wild-type and sir mutant strains (Fig. 2B and Table 1). This result suggests that blunt ends are rejoined through a process that is different from the one that rejoins ends with small single-stranded homologous tails. The rejoining of blunt ends is an inefficient process that is immune to the mating status of the cell and the presence or absence of the Sir proteins. This effect is most apparent in the ratio of end joining efficiency of 5′-ends to blunt ends. If rejoining of both substrates were equally affected by the mating status of the cell, then the ratio should remain unchanged when comparing mating-competent and non-mating strains. This is not the case (Table 1).
Figure 2.
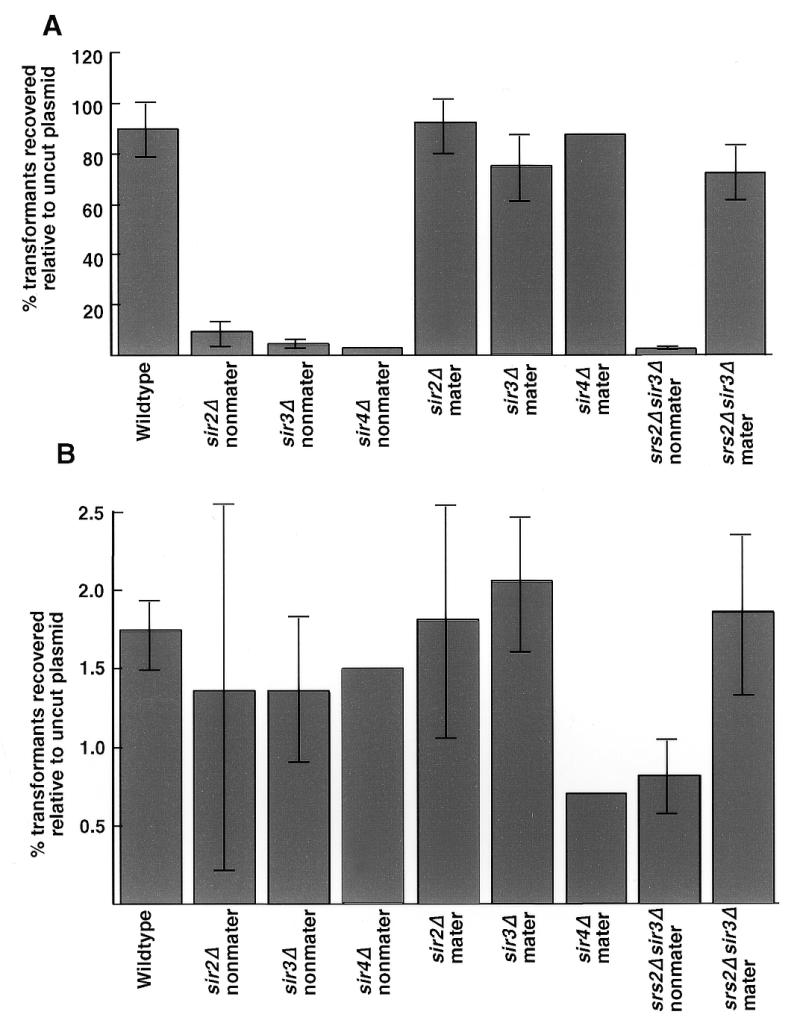
NHEJ of linear plasmid DNA is impaired in non-mating sir mutant strains, but is normal in mating sir mutant strains. Cells of each indicated genotype were transformed with equal amounts of linear or supercoiled plasmid DNA. (A) Transformation efficiencies using plasmid pRS316 linearized with EcoRI. Similar results were obtained using KpnI-digested plasmid. (B) Transformation efficiencies using plasmid pRS316 linearized with SmaI. Each strain was transformed three or four times with the indicated plasmids, with the exception of the sir4Δ and the wild-type diploid strains, which were transformed twice with the indicated plasmids.
The SIR dependence of end joining does not necessarily reflect an active role of the Sir proteins at the DSBs. For example, we find that NHEJ is not reduced in mating sir mutant strains (Fig. 2A and Table 1). This suggests that the role of the Sir proteins in NHEJ in yeast may be indirect and may act through regulation of the homologous recombination potential of the haploid sir mutant strains. Similar conclusions have been reached in other studies (13–15).
To determine whether the effect of the SIR genes was at the level of mating type heterozygosity or the ploidy of the strain, we examined NHEJ in diploids expressing both mating types (MATα/MATa). NHEJ of 5′- and 3′-ends was reduced to the same extent as that seen in the sir non-mater haploid strains (Table 1; 13,15). We also examined NHEJ efficiency in a MATα mating diploid strain (MATα/mat::hisG). In this case NHEJ of 5′- and 3′-ends remained as efficient as in the mating wild-type haploid strain (Table 1). Neither strain showed any reduction in joining of blunt ends compared to the wild-type haploid strain (Table 1).
We then examined srs2 sir3 double mutants for NHEJ efficiencies. If SRS2-independent DSB repair events were proceeding through some type of recombination repair process, we would expect repair to be inhibited in non-mating cells that can only use a recombinational repair process, while in mating-competent cells, repair would be higher. This is what is observed (Fig. 2A). However, repair of the 5′-ends is reduced in mating srs2 SIR strains, while it is highly efficient in the mating srs2 sir3 strains (Figs 1A and 2A). Similar results were obtained with 3′-ends. This suggests that the repair of sticky ends in sir mating mutants strains is not identical to repair in SIR mating strains. The SIR genes could be acting indirectly to regulate the type of repair process used, instead of acting directly in the repair process, and the Srs2 DNA helicase would be needed more in the Sir+ pathway than the Sir– pathway.
Role of RAD5 in NHEJ
Some NHEJ assays have been done in the W303 strain (3,13), which has a mutation in the RAD5 gene (16). rad5 mutants have been reported to have an inhibitory effect on the repair of DSBs through NHEJ (17). We therefore examined end joining efficiencies in RAD5 and rad5 null derivatives of all strains reported in Table 1 and found no difference between the RAD5 and rad5 strains, in terms of both the efficiency and accuracy of repair.
DISCUSSION
The Srs2 DNA helicase has been proposed to function as an anti-recombinase, channeling spontaneous damage away from a recombination repair pathway into the RAD6 post-replication repair pathway (11,12). Srs2 may act to channel repair of DSBs into non-recombinational repair pathways, regardless of any available homologies to the broken ends. For NHEJ, we suggest that Ku70 may function together with Srs2 and Rad52 in a repair pathway of DSBs with some end homology (the 5′ and 3′ sticky end substrates), perhaps by protecting the ends from degradation. In the srs2 yku70 double mutant, end joining occurs through an alternative repair process that is less efficient, perhaps due to less protection of the DSB ends or inefficiency in forming open regions at the DSB ends. Those ends that are not degraded are rejoined accurately. This explanation predicts that SRS2-independent DSB repair proceeds through a repair pathway that requires the recombination repair gene RAD52 (Figs 1 and 3 and Table 1).
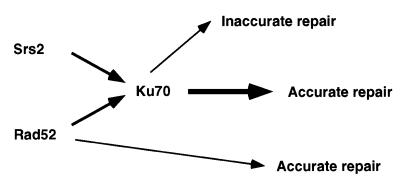
Figure 3.
Proposed end joining pathways for sticky ends in Sir+ haploid cells. The DNA helicase Srs2 and the Rad52 protein act first to process the DSB end to be rejoined via the action of the Ku70 and Ku80 proteins. This results in accurate repair. When either Srs2 or Rad52 is missing, repair still proceeds through the Ku pathway, but with reduced efficiency. Absence of both Srs2 and Rad52 reduces the efficiency of repair, but repair still proceeds through the Ku pathway and is accurate. In the absence of the Ku proteins, repair is channeled into an inaccurate repair pathway that is not efficient or has a reduced amount of substrate available for end joining. When both Srs2 and Ku70 are absent, repair is diverted into a minor pathway that is accurate. The reduced efficiency could result from a reduced amount of substrate available for end joining or inefficient use of this pathway.
Curiously, end joining of DSBs with no overlapping homology appears to occur through a different pathway. Rejoining of blunt ends is inefficient even in wild-type strains (Figs 1B and 2B) and is independent of the SIR2, SIR3 and SIR4 genes, regardless of the cell mating capability, suggesting that there is a separate non-recombinational pathway used in diploid cells to repair these types of broken ends. Rejoining of blunt ends has a slight dependence on SRS2 and a more significant dependence on RAD50 and RAD52. However, blunt end rejoining does not require the YKU70 gene and the YKU70-independent pathway is highly inaccurate (Fig. 1B and Table 1). This could be the result of lack of protection of the ends in the yku70 mutant. Blunt end joining in the srs2 rad52 double mutant is as efficient as the wild-type strain, suggesting that RAD52-independent blunt end repair is different in a SRS2 strain from the srs2 mutant strain. However, repair of blunt ends in the srs2 rad52 double mutant differs from repair of blunt ends in the Ku70 mutant in that the end joining is mostly accurate (Table 1; χ2 = 9.6, P < 0.01).
We suggest that usage of alternative DSB repair pathways is dependent on both the presence of the Sir proteins and the Rad52 protein and the diploid MAT information of a1/α2. The diploid MAT information activates diploid-specific functions, including a homolog-mediated recombination repair process, and suppresses haploid-specific functions such as the NHEJ pathway. In the haploid non-mating sir mutants, the recombination repair pathway would be preferentially used, but since the site of the plasmid DSB in the polylinker has no homology to the yeast genome, recombination repair fails and the plasmid is lost due to its linear nature. In the haploid mating-competent sir mutants, the recombination repair pathway is not activated and the NHEJ pathway is functional, allowing efficient and accurate repair of the plasmid DSB. However, the results with the srs2 mutant suggest that this process is not identical to end joining in wild-type mating cells and that some homology-related process is affected. The Srs2 helicase could also have an active role in NHEJ. If its role were only to regulate usage of a recombination repair pathway, we would not expect to find a reduction in end joining efficiencies in srs2 haploids since these strains are mating-competent haploids and as such should effectively use a NHEJ pathway. The fact that a reduction in end joining is seen in srs2 mutants suggests that the DNA helicase functions in processing the DSB ends for rejoining, whether this is through a recombination repair process or the NHEJ repair process.
Lastly, we note that either end joining is different in true haploid and diploid cells or that the Srs2 helicase has an important role in end joining, regardless of the mating status or ploidy of the cell. NHEJ of a 5′-end is reduced to 30% in a srs2 mating haploid and a srs2 non-mating diploid (Table 1). Which repair pathway is used is less clear. We would expect the wild-type diploid and the srs2 diploid to show similar reductions in end joining since the cell would attempt to use a recombinational repair process. However, 5′-end joining is reduced to 5% in the wild-type diploid and to 30% in the srs2 diploid. This suggests that in diploids the Srs2 helicase has an additional function in channeling repair into a recombination pathway. In its absence, repair proceeds through an alternative pathway that does not greatly depend on the Srs2 helicase to act at the ends. Perhaps another helicase takes over in the diploid or perhaps there may be a greater reliance on the Rad52 protein in diploids. The repair efficiency of the srs2 diploid is much higher than repair in the srs2 sir3 non-mating haploid. This could mean that, as stated above, repair of a broken end is different in true diploids from non-mating haploids.
Acknowledgments
ACKNOWLEDGEMENTS
We thank members of the laboratory and Tom Petes for discussions and comments on the manuscript. We thank Jim Haber and Frederic Paques for the mat null plasmid. This work was supported by PHS grant GM53738.
REFERENCES
- 1.Paques F. and Haber,J.E. (1997) Mol. Cell. Biol., 17, 6765–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsukamoto Y., Kato,J. and Ikeda,H. (1997) Nature, 388, 900–903. [DOI] [PubMed] [Google Scholar]
- 3.Boulton S.J. and Jackson,S.P. (1998) EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heude M. and Fabre,F. (1993) Genetics, 133, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schiestl R.H., Zhu,J. and Petes,T.D. (1994) Mol. Cell. Biol., 14, 4493–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore J.K. and Haber,J.E. (1996) Mol. Cell. Biol., 16, 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukamoto Y., Kato,J. and Ikeda,H. (1997) Mol. Gen. Genet., 255, 543–547. [DOI] [PubMed] [Google Scholar]
- 8.Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Sikorski R.S. and Hieter,P. (1989) Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill J., Ian,K.A., Donald,G. and Griffiths,D.E. (1992) Nucleic Acids Res., 19, 5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aboussekhra,A., Chanet,R., Zgaga.Z., Cassier-Chauvat,C., Heude,M. and Fabre,F. (1989) Nucleic Acids Res., 17, 7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong L., Palladino,F., Aguilera,A. and Klein,H.L. (1991) Genetics, 127, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Astrom S.U., Okamura,S.M. and Rine,J. (1999) Nature, 397, 310. [DOI] [PubMed] [Google Scholar]
- 14.Mills K.D., Sinclair,D.A. and Guarente,L. (1999) Cell, 97, 609–620. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.E., Paques,F., Sylvan,J. and Haber,J.E. (1999) Curr. Biol., 9, 767–770. [DOI] [PubMed] [Google Scholar]
- 16.Fan H.Y., Cheng,K.K. and Klein,H.L. (1996) Genetics, 142, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahne F., Jha,B. and Eckardt-Schupp,F. (1997) Nucleic Acids Res., 25, 743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]