Abstract
Wildlife exhibits various sex-determination systems where sex chromosomes and environmental temperatures may both contribute to individual sexual development. The causes and consequences of this variability are important questions for evolutionary ecology, especially in light of ongoing environmental change. Amphibians and reptiles are emerging as a key group for studying these questions, with new data accumulating acceleratingly. We collected empirical data from earlier databases, reviews and primary literature to create the most up-to-date database on herpetological sex determination. We named our database HerpSexDet, which currently features data on genetic and temperature-dependent sex determination as well as reports on sex reversal for a total of 192 amphibian and 697 reptile species. This dataset, which we will regularly update in the future, facilitates interspecific comparative studies on the evolution of sex determination and its consequences for species-specific traits such as life history and conservation status, and may also help guiding future research by identifying species or higher taxa that are potentially most enlightening for the study of environmentally driven sex reversal.
Subject terms: Evolutionary developmental biology, Evolutionary ecology
Background & Summary
The diversity of sex-determination systems is stunning across eukaryotic organisms, and the causes and consequences of this diversity are important questions in evolutionary ecology1. This importance is heightened by contemporary human-induced environmental changes, because sex determination is often sensitive to environmental conditions, and thus the type of sex-determination system can influence how environmental change affects population genetics, demography, and extinction risk2–5. In gonochoristic taxa, two widespread systems are genetic and environmental sex determination, the latter often meaning temperature-dependent sex determination. Although these two types were conventionally considered mutually exclusive, they are now seen as two theoretical ends of a continuum, where high or low temperatures can override the sex otherwise defined by sex chromosomes6. As a result, sex reversal can occur, where genetically female individuals develop male gonads or the other way around. These topics have been attracting scientific attention at an accelerating rate in the past decade7–10, with ectothermic vertebrates emerging as a prominent “model taxon” due to their high variability in sex determination both across and within species11,12.
Nearly 10 years passed since the publication of the Tree of Sex database, the most comprehensive collection of sex-determination systems across vertebrates, invertebrates and plants13. Since then, Tree of Sex has been used for studying many research questions (e.g.8,10,14–16), and the “genomics era” has yielded exponential growth in novel data on sex-determination systems and revisions of molecular taxonomy; there is thus need for an up-to-date source of comparative data on sex-determination systems. Reflecting this need, two recent databases haven been published on amphibian karyotypes17 and sex-determination mechanisms in turtles18. Several authors also complemented the Tree of Sex database with additional sex-determination data for their own study objectives (e.g.8,10,14,16). Our goal was to gather the currently available data on the type of heterogamety (genetic sex determination, GSD) and temperature-dependent sex determination (TSD) across amphibians and reptiles, to provide a comprehensive database for future studies. Given the importance and recent growing interest in sex reversal8,19, we also collected all available data in these taxa on experimentally proven temperature-induced sex reversal and the occurrence and direction of sex reversal in wild populations. Because sex determination may be related to polyploidy20,21, we also collected data on the occurrence of polyploidy. We did not include fish, because the high diversity of sex determination in this class of ectothermic vertebrates is complicated by variation among artificially selected strains in economically relevant species, sequential hermaphroditism, and non-thermal triggers of environmental sex determination9, calling for a future database focusing on fish.
Altogether, HerpSexDet features sex-determination data on 192 amphibian and 697 reptile species, and two amphibian hybrids (891 rows and 32 columns; Tables 1, 2). Specifically, our dataset contains data on the type of heterogamety for 189 amphibian and 541 reptile species (vs. 99 and 351 species in Tree of Sex, respectively), and data on the effects of temperature on sex determination in 18 amphibian and 171 reptile species (vs. 0 and 134 in Tree of Sex, respectively). Besides the type of TSD, HerpSexDet includes detailed information on sex ratios at low, intermediate and high temperatures as well as temperature thresholds based on empirical data in 18 amphibian and 135 reptile species.
Table 1.
Short description of data collected for the HerpSexDet database.
| Category | Variable name | Short description |
|---|---|---|
| GSD | GSD_type | Type of heterogamety. Usually XX/XY, ZW/ZZ, complex XX/XY or complex ZW/ZZ. |
| Sex_chromosome_karyotype | Have sex chromosomes been identified by karyotyping? Values: homomorphic or identified. | |
| Size_difference | Size difference between the sex chromosomes is indicated by “yes”, no difference is indicated by “no”. Note that this information was collected non-systematically in this first version of HerpSexDet. | |
| Larger_chromosome | The relatively larger sex chromosome (X, Y, Z, or W). | |
| TSD | TSD_type | TSD reaction norm. Usually MF (type Ia), FM (type Ib), or FMF (type II). |
| TSD_type_detailed | More detailed categorization of the TSD reaction norm, including information on even sex ratios. | |
| Low_temperature_sex | The phenotypic sex in excess at the lower range of investigated temperatures: male, female, or even. | |
| Low_temperature_threshold | Temperature (°C) below which biased sex ratio is produced. | |
| Intermediate_temperature_sex | The phenotypic sex in excess at the intermediate range of investigated temperatures: male, female, or even. | |
| High_temperature_sex | The phenotypic sex in excess at the higher range of investigated temperatures: male, female, or even. | |
| High_temperature_threshold | Temperature (°C) above which biased sex ratio is produced. | |
| Sex reversal | Thermal_sex_reversal_proven | Is there experimental evidence for thermally induced sex reversal? Yes or no, if it was studied. |
| Sex_reversal_in_nature | Combinations of sexual genotypes and phenotypes found in wild populations. | |
| Other | Polyploid | Information on the occurrence of polyploidy. |
Detailed description including further column names for references, notes and curators is available at39.
Table 2.
Distribution of sex-determination and sex-reversal data across taxonomic orders in the HerpSexDet database (the number of species is shown).
| Class | Order | GSD | TSD1 | GSD and TSD1 | Thermal sex reversal proven | Sex reversal in nature |
|---|---|---|---|---|---|---|
| Amphibia | Anura | 135 | 112 | 7 | 2 | 6 |
| Caudata | 52 | 7 (1) | 6 (1) | 4 | — | |
| Gymnophiona | 2 | — | — | — | — | |
| Reptilia | Crocodylia | — | 14 | — | — | — |
| Rhynchocephalia | — | 1 | — | — | — | |
| Squamata | 509 | 58 (27) | 14 (6) | 3 | 2 | |
| Testudines | 32 | 98 (12) | 1 (1) | — | — | |
| Sum | 730 | 189 | 28 | 9 | 8 | |
1The number of species for which the existence of TSD is uncertain due to lack of reliable data (variable “TSD_type”, value “TSD uncertain”) is indicated in parentheses next to the total number of species with either certain or uncertain TSD.
29 species and two hybrids.
We hope that HerpSexDet and its future updates will significantly contribute to sex-related evolutionary ecology research worldwide. Sex-determination systems have been replacing each other frequently during ectotherm evolution12,14,22,23, and sex reversal has been suggested to play a role in this process2,24–26. However, the evolutionary forces and molecular mechanisms underlying these turnovers are poorly understood. Recent works suggested that aging rates may be higher in the heterogametic sex27,28 and species with different types of heterogamety might respond differently to anthropogenic environmental changes3,4,8,29. Therefore, our dataset may be of use for comparative studies on the evolution of sex determination, physiological traits and habitat adaptations. For example, HerpSexDet could be used together with other databases to assess questions on temperature-related developmental plasticity and heat tolerance30,31, or to evaluate if life-history traits32,33 differ by sex-determination systems. We expect that such studies will significantly improve our understanding of fundamental evolutionary processes as well as the potential consequences of the ongoing anthropogenic changes worldwide.
Methods
Data collection
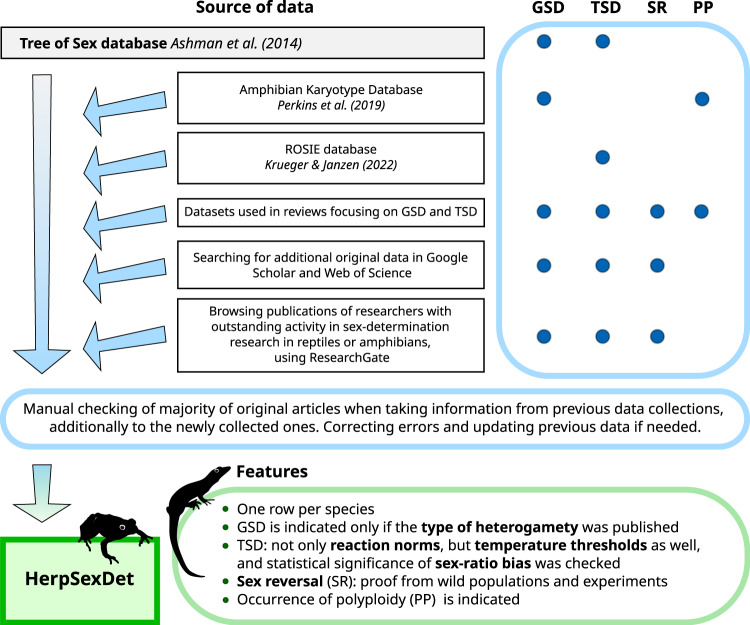
We collected data on sex determination in herpetofauna by relying on and revisiting previous data collections and adding more recent data (Fig. 1). As a starting point, we incorporated species-specific data on the type of heterogamety and the type of TSD reaction norm for amphibians and reptiles from the Tree of Sex database13. In case of those species for which data were shown in multiple rows in Tree of Sex (e.g. the same species occurring with different names, different subspecies, or different data sources), we merged the information into a single row, to represent the consensus data for each species. We preferred the one-row-per-species format because information on within-species variation in sex determination is very limited. However, in those few cases where we found such information, we noted it in the database (in variables “GSD_note”, “Size_note”, or “TSD_note”). Next, we collected sex-determination data for additional species, as well as more up-to-date data for the species already represented in the Tree of Sex database. For this purpose, we overviewed sex-determination data in two additional databases17,18 as well as the datasets used in review articles and book chapters focusing on sex determination or the temperature effects on sexual development in amphibians or reptiles8,14,34–38. We aimed to double-check the original sources cited in these collections and Tree of Sex, although this did not always happen. For example, in several cases, we could not identify the reference given in Tree of Sex, so we took these references unchanged, and noted this fact in the “Comment” column of the reference table39. Furthermore, we double-checked the cases where we found contradictions between the older and more recent data. During these double-checks, we revised the information that was given in the earlier databases whenever necessary. Finally, we searched for additional species-specific data in primary literature using Google Scholar (https://scholar.google.com) and Web of Science (https://www.webofscience.com). In the search phrases, we combined taxon names (class or order) with one of the following keywords: sex determination, heterogamety, sex chromosome, XY, ZW, TSD, or temperature-dependent sex determination. After reading the title and abstract of each article, we decided whether to read the article and extract data from it, based on whether the paper reported species-specific information either on the type of heterogamety or on the effect of temperature on sex determination or sex ratios. As we found that certain researchers were contributing especially actively to sex-determination research in reptiles or amphibians, we also browsed their articles on ResearchGate (https://www.researchgate.net). Furthermore, we collected data on the occurrence of sex reversal in wild populations as we encountered such reports (we did not do a targeted search for this because sex reversal in free-living herpetofauna is a young field of research which we have been continuously monitoring). After we finished collecting GSD data on the 21st of December 2022, we performed a final search in Google Scholar for TSD data in those families for which GSD data were present in our dataset, using search phrases combining temperature, sex ratio, and incubation (for reptiles) or larvae or tadpoles (for amphibians). We collected data on the occurrence of polyploidy from a database dedicated to amphibian karyotypes17 and a review on reptiles40.
Fig. 1.
Workflow of data collection and key features of HerpSexDet. The Tree of Sex database provided a baseline for species-specific data on genetic (GSD) and temperature-dependent sex determination (TSD), and was double-checked, complemented, and updated with new data. HerpSexDet also provides further details on TSD, including temperature thresholds, as well as sex-reversal (SR) data from wild populations and the occurrence of polyploidy (PP).
Standards for data inclusion
We extracted data from each source manually. We extracted data with caution and evaluated the text of the article overall before including the data in our database, keeping only those records that provided reliable information on the sex-determination system in a species (see the paragraphs below for more specific criteria). When we found contradiction between reports in well-established journals and grey zone publications or mentions of non-accessible resources, we disregarded the latter.
We included GSD data only for those species in which the type of heterogamety (“GSD_type”, see Table 1) was identified: mainly by karyotypes or sex-linked molecular markers, but in some studies in the previous century, heterogamety was assessed based on breeding experiments following artificial sex reversal. Species for which GSD was assumed only based on an observed lack of temperature effects on sex ratios or anecdotical information were excluded from our database. Based on our knowledge on related works, we judged that information on sex-linked DNA loci mapped to a reference species´ genome was available only for a fraction of species with identified sex chromosomes. So, because we assumed that chromosome identity for most species would not be comparable with that of others, we did not indicate which chromosome pair is named as the sex chromosome in each species. Instead, we indicated if the sex chromosomes were identified (variable: “Sex_chromosome_karyotype”, as described in Table 1). The published chromosome numbers can be found in the references for GSD.
If sex-ratio data were available for a species from thermal manipulation experiments, we evaluated TSD by testing whether the ratio of male and female offspring deviated significantly from 1:1 at each applied temperature within each study by performing binomial tests. When significantly biased sex ratio was found at some temperature(s) but not at other(s), we categorized the species into the type of TSD reaction norm that best fit the data (variable “TSD_type”) and we recorded the temperature thresholds below and/or above which sex-ratio bias was significant. Because statistical tests of sex ratios are not reliable at small sample sizes, we added a warning on small sample sizes whenever appropriate (variable “TSD_note”) and marked uncertain data by question marks (see metadata at39). For a few TSD species where many studies were done at many different temperatures, we used the estimated boundaries of the transitional range of temperatures that produce both sexes41 (see also metadata at39). When there was not enough information for formally assessing TSD, e.g. because the original data were not accessible, we categorized the species as “TSD uncertain”.
We indicated the presence of sex-reversed individuals in nature only if it was proven by mismatching sexual genotypes and phenotypes in free-living animals (variable: “Sex_reversal_in_nature”). Potential occurrence of sex reversal in further species was also suggested earlier, based on findings that both GSD and TSD was detected in the same species, although not in the same individuals19. We did not include such circumstantial data as evidence for sex reversal in our database (Table 2). However, by combining information present in our database (i.e. if species in the taxon feature both GSD and TSD), researchers may get information on the suggestive occurrence of temperature-induced sex reversal, even if it has not yet been reported in their species of interest (see Table 2); this may help identifying taxa worthwhile for future studies.
Taxonomy
We extracted species lists manually from two taxon-specific databases42,43. Using these lists, we searched for species names present in our dataset and added taxonomic classification (family, order, class) to them using a custom code (see Supplementary File 1). If a species could not be identified automatically, we corrected the entry manually after searching for relevant synonyms on NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy). To enable the identification of species names that had been used in our data sources, we listed synonyms as well as subspecies names in the “Synonym” variable. Our dataset contains two rows with TSD data on amphibian hybrids, because TSD data were available in the literature only for these two hybrids within their respective taxonomic families.
Data Records
The database consists of three files. Data and references are included in two separate text files, while detailed metadata for the database is provided as a pdf file in the Figshare repository39. The most important variables are listed and briefly explained in Table 1, but for detailed information see the metadata file.
The database is provided as tabulator-separated plain text (character encoding: UTF-8) named HerpSexDet_v1-1.tsv. Since all data (except for the detailed references) are incorporated in a single file, usage of the database is straightforward. For example, users can simply import the whole dataset to R44, and based on the metadata file, they can easily decide which variables are relevant for their study. In the main database file, references are indicated as identifiers (numbers in square brackets), and the full citation for each identifier is available in the References_for_HerpSexDet_v1_1.tsv file. To help users reading our database, we also provided a readme file and an example R code39.
Technical Validation
Data were extracted manually from the referred sources. Subsets of both the GSD and TSD data records were evaluated by both authors, and in most cases the authors agreed in data entries and categorizations. In those few cases when inconsistency occurred, decision was made based on double-checking the original sources and mutual agreement of the two authors.
We encourage researchers to let us know if they find any error in our dataset or if they publish new sex-determination data that should be included in future versions of HerpSexDet.
Usage Notes
References for GSD, TSD, sex-reversal data from nature, occurrence of polyploidy and taxonomy are listed under five respective variables: “GSD_reference”, “TSD_reference”, “Sex_reversal_reference”, “Polyploid_reference” and “Taxonomy_reference”. Each variable may contain the identifiers of multiple references, separated by commas. Full citations for the reference identifiers are listed in the References_for_HerpSexDet_v1_1.tsv file. To help the users find and check our data sources upon interest, we indicated the DOI identifier of each source if available. Otherwise, we aimed to provide the most unequivocal reference that was possible.
We recommend users to check for updates on https://hsd-project.eu before starting a new analysis based on HerpSexDet, because we are planning to release updates under new version numbers on a regular basis.
Supplementary information
Acknowledgements
E.N. was supported by the Austrian Science Fund (FWF; grant ESP 239-B). V.B. was supported by the National Research, Development and Innovation Office of Hungary (NKFIH K-135016).
Author contributions
E.N. formulated the main concept of the database and wrote the first draft of the manuscript. V.B. contributed with new ideas to the concept, performed statistical analyses of sex-ratio bias data and revised the manuscript. The authors contributed equally to data curation.
Funding
Open access funding provided by ELKH Centre for Agricultural Research.
Code availability
The custom code that we used for taxonomic identification is available as Supplementary File 1. Additionally, an example code for reading our database in R is provided at39.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Edina Nemesházi, Email: nemeshazi.edina@protonmail.com.
Veronika Bókony, Email: bokony.veronika@atk.hu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-023-02268-y.
References
- 1.Bachtrog D, et al. Sex determination: Why so many ways of doing it? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossen C, Neuenschwander S, Perrin N. Temperature-dependent turnovers in sex-determination mechanisms: a quantitative model. Evolution (N. Y). 2011;65:64–78. doi: 10.1111/j.1558-5646.2010.01098.x. [DOI] [PubMed] [Google Scholar]
- 3.Bókony V, Kövér S, Nemesházi E, Liker A, Székely T. Climate-driven shifts in adult sex ratios via sex reversals: the type of sex determination matters. Philos. Trans. R. Soc. B Biol. Sci. 2017;372:20160325. doi: 10.1098/rstb.2016.0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemesházi E, Kövér S, Bókony V. Evolutionary and demographic consequences of temperature-induced masculinization under climate warming: the effects of mate choice. BMC Ecol. Evol. 2021;21:16. doi: 10.1186/s12862-021-01747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwanz LE, Ezaz T, Gruber B, Georges A. Novel evolutionary pathways of sex-determining mechanisms. J. Evol. Biol. 2013;26:2544–2557. doi: 10.1111/jeb.12258. [DOI] [PubMed] [Google Scholar]
- 6.Sarre SD, Georges A, Quinn A. The ends of a continuum: genetic and temperature-dependent sex determination in reptiles. BioEssays. 2004;26:639–645. doi: 10.1002/bies.20050. [DOI] [PubMed] [Google Scholar]
- 7.Schwanz LE, Georges A. Sexual development and the environment: Conclusions from 40 years of theory. Sex. Dev. 2021;15:7–22. doi: 10.1159/000515221. [DOI] [PubMed] [Google Scholar]
- 8.Nemesházi E, Bókony V. Asymmetrical sex reversal: Does the type of heterogamety predict propensity for sex reversal? BioEssays. 2022;47:2200039. doi: 10.1002/bies.202200039. [DOI] [PubMed] [Google Scholar]
- 9.Baroiller JF, D’Cotta H. The reversible sex of gonochoristic fish: Insights and consequences. Sex. Dev. 2016;10:242–266. doi: 10.1159/000452362. [DOI] [PubMed] [Google Scholar]
- 10.Edmands S. Sex ratios in a warming world: thermal effects on sex-biased survival, sex determination, and sex reversal. J. Hered. 2021;112:155–164. doi: 10.1093/jhered/esab006. [DOI] [PubMed] [Google Scholar]
- 11.Ospina-Álvarez N, Piferrer F. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS One. 2008;3:2–4. doi: 10.1371/journal.pone.0002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarre SD, Ezaz T, Georges A. Transitions between sex-determining systems in reptiles and amphibians. Annu. Rev. Genomics Hum. Genet. 2011;12:391–406. doi: 10.1146/annurev-genom-082410-101518. [DOI] [PubMed] [Google Scholar]
- 13.Ashman TL, et al. Tree of Sex: A database of sexual systems. Sci. Data. 2014;1:140015. doi: 10.1038/sdata.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma WJ, Veltsos P. The diversity and evolution of sex chromosomes in frogs. Genes (Basel). 2021;12:1–17. doi: 10.3390/genes12040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornejo-Páramo P, et al. Sex determination systems in reptiles are related to ambient temperature but not to the level of climatic fluctuation. BMC Evol. Biol. 2020;20:1–14. doi: 10.1186/s12862-020-01671-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sultanova Z, Downing PA, Carazo P. Genetic sex determination, sex chromosome size and sex‐specific lifespans across tetrapods. J. Evol. Biol. 2022;36:480–494. doi: 10.1111/jeb.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perkins RD, et al. A database of amphibian karyotypes. Chromosom. Res. 2019;27:313–319. doi: 10.1007/s10577-019-09613-1. [DOI] [PubMed] [Google Scholar]
- 18.Krueger CJ, Janzen FJ. ROSIE, a database of reptilian offspring sex ratios and sex-determining mechanisms, beginning with Testudines. Sci. Data. 2022;9:1–6. doi: 10.1038/s41597-021-01108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holleley CE, Sarre SD, O’Meally D, Georges A. Sex reversal in reptiles: reproductive oddity or powerful driver of evolutionary change? Sex. Dev. 2016;10:279–287. doi: 10.1159/000450972. [DOI] [PubMed] [Google Scholar]
- 20.Kobel HR, Pasquier D. L. Genetics of polyploid Xenopus. Trends Genet. 1986;2:310–315. doi: 10.1016/0168-9525(86)90286-6. [DOI] [Google Scholar]
- 21.Stöck M, et al. Sex chromosomes in meiotic, hemiclonal, clonal and polyploid hybrid vertebrates: Along the ‘extended speciation continuum’. Philos. Trans. R. Soc. B Biol. Sci. 2021;376:20200103. doi: 10.1098/rstb.2020.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Augstenová B, et al. ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution. 2018;72:1701–1707. doi: 10.1111/evo.13543. [DOI] [PubMed] [Google Scholar]
- 23.Pla S, Benvenuto C, Capellini I, Piferrer F. Switches, stability and reversals in the evolutionary history of sexual systems in fish. Nat. Commun. 2022;13:1–13. doi: 10.1038/s41467-022-30419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castelli MA, et al. Evolving thermal thresholds explain the distribution of temperature sex reversal in an Australian dragon lizard. Divers. Distrib. 2021;27:427–438. doi: 10.1111/ddi.13203. [DOI] [Google Scholar]
- 25.Schwanz LE, Georges A, Holleley CE, Sarre SD. Climate change, sex reversal and lability of sex-determining systems. J. Evol. Biol. 2020;33:270–281. doi: 10.1111/jeb.13587. [DOI] [PubMed] [Google Scholar]
- 26.Quinn AE, Sarre SD, Ezaz T, Marshall Graves JA, Georges A. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol. Lett. 2011;7:443–448. doi: 10.1098/rsbl.2010.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cayuela H, et al. Sex‐related differences in aging rate are associated with sex chromosome system in amphibians. Evolution (N. Y). 2021;76:346–356. doi: 10.1111/evo.14410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xirocostas, Z. A., Everingham, S. E. & Moles, A. T. The sex with the reduced sex chromosome dies earlier: a comparison across the tree of life. Biol. Lett. 16, (2020). [DOI] [PMC free article] [PubMed]
- 29.Whiteley S L, Castelli M A, Dissanayake D S B. Temperature-induced sex reversal in reptiles: prevalence, discovery, and evolutionary implications. Sex. Dev. 2021;15:148–156. doi: 10.1159/000515687. [DOI] [PubMed] [Google Scholar]
- 30.While GM, et al. Patterns of developmental plasticity in response to incubation temperature in reptiles. J. Exp. Zool. Part A Ecol. Integr. Physiol. 2018;329:162–176. doi: 10.1002/jez.2181. [DOI] [PubMed] [Google Scholar]
- 31.Pottier P, et al. A comprehensive database of amphibian heat tolerance. Sci. Data. 2022;9:1–15. doi: 10.1038/s41597-022-01704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira BF, São-Pedro VA, Santos-Barrera G, Penone C, Costa GC. AmphiBIO, a global database for amphibian ecological traits. Sci. Data. 2017;4:1–7. doi: 10.1038/sdata.2017.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimm A, Ramírez AMP, Moulherat S, Reynaud J, Henle K. Life-history trait database of European reptile species. Nat. Conserv. 2014;9:45–67. doi: 10.3897/natureconservation.9.8908. [DOI] [Google Scholar]
- 34.Chardard, D., Penrad-Mobayed, M., Chesnel, A., Pieau, C. & Dournon, C. in Temperature-dependent sex determination in vertebrates (eds. Valenzuela, N. & Lance, V.) Ch. 7 (Smithsonian Books, 2004).
- 35.Schmid M, Steinlein C. Chromosome banding in amphibia. XXXVII. Y-autosome translocations in Anura. Cytogenet. Genome Res. 2018;154:153–180. doi: 10.1159/000487907. [DOI] [PubMed] [Google Scholar]
- 36.Viets BE, Ewert MA, Talent LG, Nelson CE. Sex‐determining mechanisms in squamate reptiles. J. Exp. Zool. 1994;270:45–56. doi: 10.1002/jez.1402700106. [DOI] [Google Scholar]
- 37.Harlow, P. S. in Temperature Dependent Sex Determination in Vertebrates (eds. Valenzuela, N. & Lance, V. A.) Ch. 5 (Smithsonian Institution, 2004).
- 38.Green, D. M. & Sessions, S. K. Amphibian cytogenetics and evolution. (Academic Press, Inc., 1991).
- 39.Nemesházi E, Bókony V. 2023. HerpSexDet: the herpetological database of sex determination and sex reversal. Figshare. [DOI] [PMC free article] [PubMed]
- 40.Kearney, M., Fujita, M. K. & Ridenour, J. in Lost Sex - The Evolutionary Biology of Parthenogenesis (eds. Schön, I., Martens, K. & van Dijk, P.) Ch. 21 (Springer Netherlands, 2009).
- 41.Abreu-Grobois FA, et al. Recent advances on the estimation of the thermal reaction norm for sex ratios. PeerJ. 2020;8:e8451. doi: 10.7717/peerj.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.AmphibiaWeb. University of California, Berkeley, CA, USA Accessed 14 April 2022. https://amphibiaweb.org (2022).
- 43.Uetz, P., Freed, P., Aguila, R. & Hošek, J. The Reptile Database. Accessed 06 June 2022. http://www.reptile-database.org (2022).
- 44.R Core Team. R: A language and environment for statistical computing. R ver. 4.2.1. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org (2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Nemesházi E, Bókony V. 2023. HerpSexDet: the herpetological database of sex determination and sex reversal. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The custom code that we used for taxonomic identification is available as Supplementary File 1. Additionally, an example code for reading our database in R is provided at39.