Abstract
Bempegaldesleukin (BEMPEG), a CD122-preferential IL2 pathway agonist, has been shown to induce proliferation and activation of NK cells. NK activation is dependent on the balance of inhibitory and excitatory signals transmitted by NK receptors, including Fc-gamma receptors (FCγRs) and killer immunoglobulin-like receptors (KIRs) along with their KIR-ligands. The repertoire of KIRs/KIR-ligands an individual inherits and the single-nucleotide polymorphisms (SNPs) of FCγRs can influence NK function and affect responses to immunotherapies. In this retrospective analysis of the single-arm PIVOT-02 trial, 200 patients with advanced solid tumors were genotyped for KIR/KIR-ligand gene status and FCγR SNP status and evaluated for associations with clinical outcome. Patients with inhibitory KIR2DL2 and its ligand (HLA-C1) observed significantly greater tumor shrinkage (TS, median change −13.0 vs. 0%) and increased PFS (5.5 vs. 3.3 months) and a trend toward improved OR (31.2 vs. 19.5%) compared to patients with the complementary genotype. Furthermore, patients with KIR2DL2 and its ligand together with inhibitory KIR3DL1 and its ligand (HLA-Bw4) had improved OR (36.5 vs. 19.6%), greater TS (median change −16.1 vs. 0%), and a trend toward prolonged PFS (8.4 vs. 3.6 months) as compared to patients with the complementary genotype. FCγR polymorphisms did not influence OR/PFS/TS.
These data show that clinical response to BEMPEG plus nivolumab treatment in the PIVOT-02 trial may be associated with the repertoire of KIR/KIR-ligands an individual inherits. Further investigation and validation of these results may enable KIR/KIR-ligand genotyping to be utilized prospectively for identifying patients likely to benefit from certain cancer immunotherapy regimens.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-023-03383-w.
Keywords: Nivolumab, Bempegaldesleukin, Killer immunoglobulin-like receptor (KIR), NK cells, Human leukocyte antigen (HLA)
Background
Within the past decade, significant advances in immunotherapy have led it to be an effective treatment option for a range of solid tumors [1–5]. Immunotherapeutic regimens comprised of immune checkpoint inhibitors (ICIs), such as anti-programmed death-1 (anti-PD-1) and high dose IL2 (HD-IL2) are among those that have shown clinical benefit, however, only in a select subset of patients [6, 7]. Thus, improved therapeutic agents and combination regimens are necessary to provide clinical benefit for a larger population of patients.
The PIVOT-02 Phase I/II clinical trial (NCT02983045) enrolled patients with advanced solid tumors to evaluate the efficacy of a combination therapy consisting of nivolumab (anti-PD-1) and a novel agent, bempegaldesleukin (BEMPEG). As a CD122-preferential IL2 pathway agonist, BEMPEG serves to address some of the limitations seen by ICIs and HD-IL2. Compared to HD-IL2, BEMPEG preferentially binds to the low-to-moderate affinity heterodimeric IL2βγ (CD122/132) receptors predominately expressed on NK and CD8 T cells, compared to the high-affinity trimeric IL2αR predominately expressed on immunosuppressive T regulatory cells (Tregs), and stimulates an anti-tumor immune response through the clonal expansion of NK and CD4 and CD8 T cells. A Phase I trial for BEMPEG monotherapy confirmed its ability to induce proliferation and activation of T cells and NK cells in the blood and tumor microenvironment (TME) [8]. Thus, in this retrospective analysis of data from the completed PIVOT-02 trial, considering BEMPEG can activate patients’ NK cells, we sought to investigate whether distinct immunogenotypes related to NK cell function were associated with patients’ clinical outcome from the combination therapy of BEMPEG plus nivolumab.
NK cell activation is dependent on the balance of inhibitory and excitatory signals transmitted by receptors expressed on NK cells, including killer immunoglobulin-like receptors (KIRs) and Fc-gamma receptors (FCγRs). Different KIRs can have an inhibitory or excitatory function as they interact with their corresponding HLA molecules (KIR-ligands) expressed on healthy or cancerous cells. FCγRs are expressed on NK cells, as well as other immune cells, and can influence NK cell function through binding to the fragment crystallizable (Fc) region of tumor-bound antibodies, subsequently leading to anti-tumor responses through triggering of antibody-dependent cellular cytotoxicity (ADCC). We, and others, have shown that the repertoire of KIRs and KIR-ligands an individual inherits and the single-nucleotide polymorphisms (SNPs) among FCγRs can influence NK cell function and affect responses to certain immunotherapies [9–14]. Here, we focus on the association between clinical response and the presence or absence of the four inhibitory KIRs and their ligands that we have previously found associated with clinical outcome [9–11]: KIR2DL1 with HLA-C2; KIR2DL2 and KIR2DL3 with HLA-C1; and KIR3DL1 with HLA-Bw4 epitopes. We acknowledge that the KIR2DL2 inhibitory receptor is in linkage disequilibrium with the KIR2DS2 activating receptor, and as such, some of our findings might reflect the influence of activating receptors, as we previously detailed [9]. We also report on our findings looking for potential associations of FCγRs, alone and in combinations, with clinical outcome. In this report, we found that there were no associations with FCγR combinations and clinical outcome, but similar to our prior observations with KIR/KIR-ligand genotypes, we found that certain combinations of KIR/KIR-ligand genotypes associate with clinical outcome.
Methods
Clinical trial and clinical samples
PIVOT-02, a phase I/II dose-escalation/expansion trial (NCT02983045), evaluated the safety and efficacy of BEMPEG in combination with nivolumab in selected advanced or metastatic solid tumors. All patients included in our analyses were treated with the recommended phase 2 dose (RP2D) of 0.006 mg/kg BEMPEG every 3 weeks plus 360 mg nivolumab every 3 weeks until disease progression, death, unacceptable toxicity, symptomatic deterioration, investigator decision to discontinue treatment, patient withdrawal of consent, loss to follow-up, or study termination by the sponsor. Responding patients were treated for a maximum of 2 years. For this retrospective analysis, we focused on a subset of patients from the PIVOT-02 trial who had not previously been treated with immunotherapy (IO-naïve). We excluded 13 patients previously treated with immunotherapy for whom we analyzed DNA, in order to have a more homogeneous population of 200 IO-naïve patients for our analysis. As these 13 patients previously treated with immunotherapy would be expected to have worse outcome than the IO-naïve patients, the random distribution of these few patients into the various genotyping groups evaluated here could influence the associations of clinical outcome with genotype. Supplementary Table S1 includes the patients who received prior immunotherapy (RP2D cohort, n = 213) and displays the results for the four associations reported on in this manuscript. A breakdown of the IO-naive patient subset according to tumor type is shown in Table 1. The clinical details of the PIVOT-02 trial and its phase I/II clinical conclusions have been reported by Diab et al. [15–18]. The study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. All patients provided written informed consent, and the protocol was approved by independent ethics committees and the institutional review board at each participating site.
Table 1.
PIVOT-02 tumor cohort enrollment. Patients with selected advanced solid tumors were enrolled in the PIVOT-02 Phase I/II trial. All patients analyzed in this retrospective study received the RP2D. All results reported here, unless otherwise specified, are of the IO-naïve cohort (n = 200). 1L = first-line therapy; 2L = second-line therapy
| Tumor Type | n |
|---|---|
| 1L melanoma (MEL) | 31 |
| Advanced melanoma progressing after adjuvant therapy | 7 |
| 1L renal cell carcinoma (RCC) | 37 |
| Renal cell carcinoma (other) | 3 |
| 1L Metastatic urothelial cancer (mUC) | 37 |
| 1-2L Triple-negative breast cancer (TNBC) | 33 |
| 1-2L non-small cell lung cancer (NSCLC) | 49 |
| Non-small cell lung cancer (other) | 3 |
| Immunotherapy (IO) Naïve | 200 |
The bold in Table 1 is highlighting the sum of all patients across all tumor types (n=200) being immunotherapy-naive
DNA isolation and whole-genome amplification
A total of 200 IO-naïve patients from the PIVOT-02 trial had DNA available for genotyping, along with clinical data for correlative analyses. Genomic DNA was isolated from peripheral blood mononuclear cells following the manufacturer’s protocol of the DNeasy Blood and Tissue Kit (Qiagen). If necessary, DNA was then whole-genome amplified using the REPLI-g Mini Kit as per the manufacturer’s protocol (Qiagen) and used for KIR/KIR-ligand and FCγR genotyping.
Genotyping
Frequencies for KIR/KIR-ligand and FCγR genotypes, shown in Supplementary Table S2, are similar to those reported by others for these genes [13, 19–26]. For simplicity, abbreviations for the specific genotype groups are used throughout the manuscript. The distinct genotypes comprising all the possible combinations assessed in these analyses are detailed in Supplementary Tables S3 and S4.
KIR/KIR-ligand
KIR gene status was assessed by real-time SYBR green PCR melt curve analyses as developed by Vilches et al. [27]. The KIR-ligand genotypes (HLA-C1, HLA-C2, HLA-Bw4) were determined by sequence specific primers-polymerase chain reaction (SSP-PCR) using the KIR HLA Ligand SSP Typing kit (Olerup) with GoTaq DNA Polymerase (Promega).
Fc-gamma receptor
FCγR SNP status for FCγR2A was determined using Taqman primers/probes. For FCγR3A and FCγR2C SNP status, RNaseH primers/probes were used with Taqman Fast Advanced master mix (ThermoFisher) [28]. For FCγR2C, a modified protocol was used to test for FCγR2C SNP rs759550223 as follows: A total reaction volume of 5uL included Taqman Fast Advanced master mix (ThermoFisher), 0.5uM each of the forward (5′-TCTCCCTCTCTCTTTATCCTTCTG-3′) and reverse (5′-TGTCAGAGTCACAGAGTCCTCrUTGGAC- C3spacer-3′) primers, both in TE buffer (pH 7.5); 0.5uM each of the FCγR2C-C (5′-ATTO532N CAC+T+G+GGG+CT-3′ _Iowa Black FQ Quencher) and FCγR2C-T (5′-FAM TCCAC+T+A+GGG+CT-3′ _Iowa Black FQ Quencher) probes, both in TE buffer (pH 8.0); 5 mU of RNase H2 Enzyme (IDT DNA); and 2.5 ng of DNA. The thermocycler conditions included a pre-read at 60C for 30 s, a 95C hold for 20 s, followed by 50 cycles of 95C for 1 s with 63C for 20 s, and a post-read at 60C for 30 s. FCγR2C SNP calls were made based on the amplification curves.
Data management
All KIR/KIR-ligand and FCγR genotyping was conducted in a blinded manner, whereby individuals who determined the genotype of the patients did not have access to the clinical outcome data. Genotyping results were collected and managed using REDCap electronic data capture tools hosted at the University of Wisconsin-Madison [29, 30]. The clinical outcome data from the PIVOT-02 study database were merged with the genotyping data in REDCap to create a SAS dataset for analysis.
Statistical analyses
Genotyping results were analyzed for association with the following clinical outcome parameters: (1) objective response (OR); (2) progression-free survival (PFS); and (3) maximum percentage tumor shrinkage (TS, defined as the best percentage change in tumor size from baseline). All 200 patients were included in the OR and PFS analysis. Three patients were excluded from TS analysis, as they did not have measurable disease per RECIST v1.1 at baseline and ≥ 1 post-baseline tumor response assessment. Tumor shrinkage as an additional parameter for clinical outcome has been utilized in our prior publications of associations between immunogenotypes and clinical outcome along with published clinical reports of this PIVOT-02 trial [10, 11, 17, 18]. The confirmed best overall response (CBOR) was evaluated by RECIST v1.1; PD, progressive disease; SD, stable disease; PR, partial response; CR, complete response. The OR rate (ORR) was defined as the percentage of the 200 patients with a CR or PR. The Kaplan–Meier method was used for estimation of the survival distribution for PFS. Comparative analyses were evaluated using a binomial test for OR, log-rank test for PFS, and Wilcoxon rank-sum test for tumor shrinkage. All tests conducted were two-sided. Analyses resulting in p values less than 0.05 were considered significant. Due to the relatively small patient population in this study, analyses resulting in p values less than 0.1 were considered trends. In the waterfall plots, dotted red boxes outline the data for the top third percent of patients with a positive clinical response (based on OR and tumor shrinkage parameters), to provide a visual “reference standard” to enable easier visual comparisons of the clinical benefit seen from each genotype at an individual patient level.
Results
KIR-ligand present/missing status does not significantly influence clinical outcome in IO-naïve patients
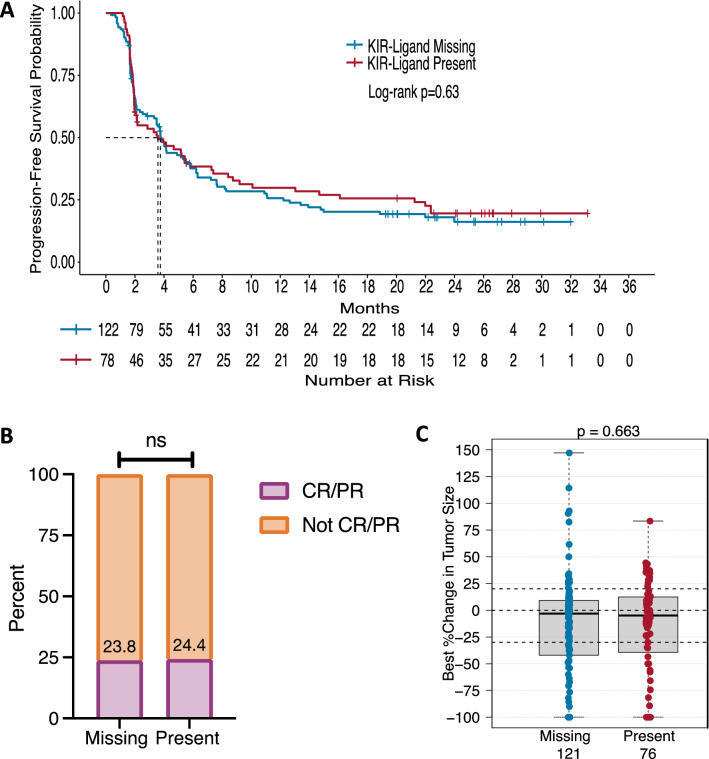
Mature NK cells that express inhibitory KIRs mediate reduced tumor-targeted direct killing or ADCC when the KIRs interact with their respective HLA molecules (KIR-ligands; refer to Supplementary Tables S3 and S4) present on the tumor [13, 31]. In previous reports [12, 14, 32, 33], NK-based immunotherapies resulted in improved clinical outcome for patients with a KIR-ligand missing genotype as compared to those with a KIR-ligand present genotype. “KIR-ligand present” is defined as all KIR-ligands present for each inhibitory KIR gene present, whereas “KIR-ligand missing” is defined as having at least one KIR-ligand absent for the inhibitory KIR genes present (Supplementary Table S3). We have previously presented data that did not show improved outcome for patients with KIR-ligand-missing in specific studies for follicular lymphoma patients treated with maintenance rituximab, neuroblastoma patients treated with anti-GD2 dinutuximab, and metastatic renal cell carcinoma patients treated with HD-IL2 [9, 11, 34]. Here, we found no associations for KIR-ligand present/missing status among the IO-naïve cohort. For patients with a KIR-ligand missing genotype (n = 122 or 121), there was no significant improvement in PFS, OR, or tumor shrinkage as compared to patients with KIR-ligand present (n = 78 or 76) (Fig. 1A–C).
Fig. 1.
KIR/KIR-ligand present/missing status does not influence clinical response to BEMPEG plus nivolumab therapy for IO-naïve patients. Kaplan–Meier curve for PFS a compares patients with a KIR-ligand missing genotype (blue line) to patients with a KIR-ligand present genotype (red line). OR is represented in (b) by the proportion of patients with a CR/PR (purple) compared to patients without a CR/PR (orange) based upon their KIR/KIR-ligand present/missing status. The box plots in (c) compare tumor shrinkage as indicated by the percent change in target lesion size from baseline for patients with a KIR-ligand missing genotype (blue) to patients with a KIR-ligand present genotype (red). Dotted horizontal lines indicate a +20% increase and −30% decrease in target lesion size from baseline
Triple-negative breast cancer patients may have greater clinical benefit when all KIR-ligands are present
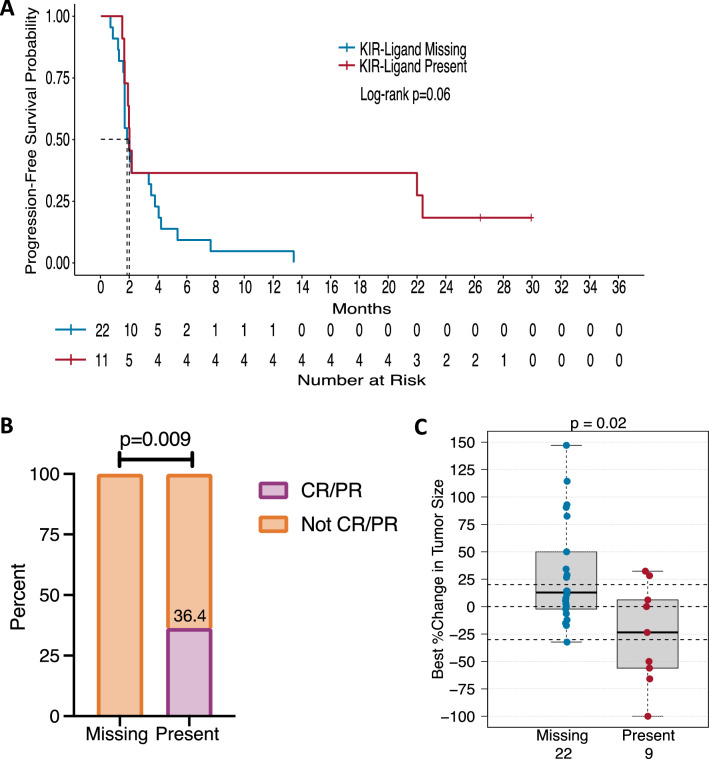
Although we observed no associations among the overall IO-naïve cohort for KIR-ligand present/missing status, we did observe that KIR-ligand present/missing status influenced clinical outcome in the triple-negative breast cancer (TNBC) patients (n = 33 or 31). Treatment with BEMPEG plus nivolumab showed a trend toward increased PFS for these KIR-ligand present TNBC patients (p = 0.06) (Fig. 2A) and significantly improved OR (p = 0.009; 36.4 vs. 0%) and tumor shrinkage (p = 0.02; median change −23.5 vs. +12.8%) as compared to their KIR-ligand missing counterparts (Fig. 2B, C).
Fig. 2.
TNBC patients with a KIR/KIR-ligand present genotype have improved clinical response to BEMPEG plus nivolumab therapy. Kaplan–Meier curve for PFS (a) compares patients with a KIR-ligand missing genotype (blue line) to patients with a KIR-ligand present genotype (red line). OR is represented in (b) by the proportion of patients with a CR/PR (purple) compared to patients without a CR/PR (orange) based upon their KIR/KIR-ligand present/missing status. The box plots in (c) compare tumor shrinkage as indicated by the percent change in target lesion size from baseline for patients with a KIR-ligand missing genotype (blue) to patients with a KIR-ligand present genotype (red). Dotted horizontal lines indicate a +20% increase and −30% decrease in target lesion size from baseline
KIR2DL2 in the presence of its HLA-C1 ligand is associated with clinical benefit
The KIR-ligand present/missing analysis takes all three KIR-ligands (HLA-C1, HLA-C2, and HLA-Bw4) into consideration as contributing equally to the inhibition or education of NK cells. However, with no associations found for the overall group of 200 IO-naïve patients when assessing all inhibitory KIR/KIR-ligands simultaneously, we sought to investigate whether specific inhibitory KIR/KIR-ligand pairs may have an influence on clinical outcome with BEMPEG plus nivolumab combination therapy. Thus, we individually assessed the inhibitory KIRs in the presence or absence of their ligands.
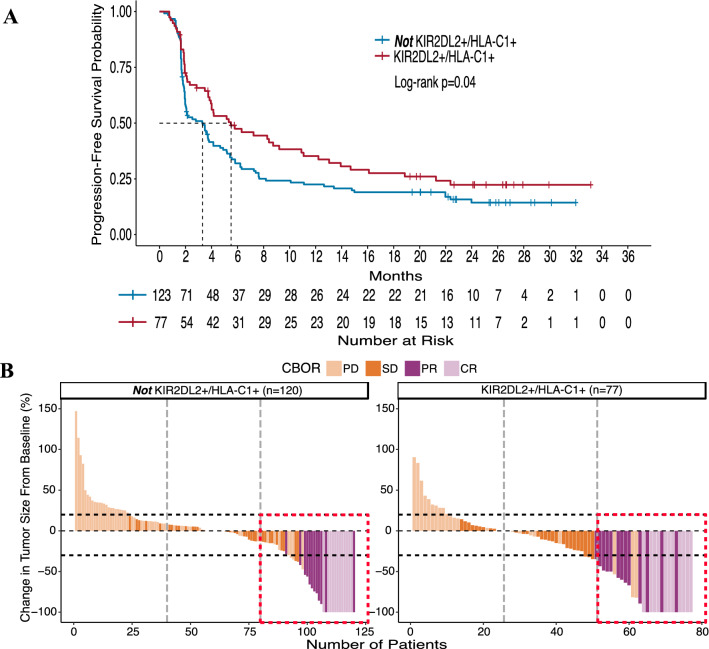
KIR2DL2 is an inhibitory KIR that recognizes the HLA-C1 ligand. Compared to KIR2DL3, which shares the same ligand, KIR2DL2 has a stronger affinity for the HLA-C1 ligand [35]. Therefore, we assessed the patients who have KIR2DL2 in the presence of its HLA-C1 ligand (KIR2DL2+/HLA-C1+; n = 77) compared to the patients who did not have this specific receptor-ligand pair (not KIR2DL2 + /HLA-C1+; n = 123 or 120) (Supplementary Table S4).
Patients who were KIR2DL2+/HLA-C1+ showed significantly prolonged PFS (median 5.5 months vs. 3.3 months; p = 0.04) as compared to patients who were not KIR2DL2+/HLA-C1+ (Fig. 3A). To visually represent the clinical benefit seen from this genotype at an individual patient level, the waterfall plots (Fig. 3B) display the percent change in target lesion size from baseline for each patient along with their CBOR. Patients represented in the right waterfall plot (KIR2DL2+/HLA-C1+) had significantly greater tumor shrinkage (median change −13.0 vs. 0%; p = 0.01) as compared to the patients represented in the left waterfall plot (not KIR2DL2+/HLA-C1+). Those same patients (KIR2DL2+/HLA-C1+) also showed a trend toward increased OR rate as indicated by the dotted red box representing a greater proportion of the top third percent of patients (31.2% with a CR/PR; right) having a CR/PR as compared to patients who were not KIR2DL2+/HLA-C1+ (19.5% with a CR/PR; left) (p = 0.07).
Fig. 3.
KIR2DL2+/HLA-C1+ patients have significantly greater clinical benefit compared to patients who are not KIR2DL2+/HLA-C1+. Kaplan–Meier curve for PFS (a) compares patients who are KIR2DL2+/HLA-C1+ (red line) to patients who are not KIR2DL2+/HLA-C1+ (blue line). Waterfall plots displaying OR and tumor shrinkage (b) compares patients who are KIR2DL2+/HLA-C1+ (right) with patients who are not KIR2DL2+/HLA-C1+ (left). CBOR is the confirmed best overall response by RECIST 1.1 criteria; PD, progressive disease (light orange); SD, stable disease (orange); PR, partial response (purple); CR, complete response (light purple). Vertical dotted lines divide the number of patients into thirds, and horizontal dotted lines indicate a +20% increase and −30% decrease in target lesion size from baseline. The dotted red box outlines the top third of patients, indicating a larger proportion of patients with a positive clinical response in the KIR2DL2+/HLA-C1+ group (right) than in the group who are not KIR2DL2+/HLA-C1+ (left)
KIR3DL1 and its HLA-Bw4 ligand do not influence clinical outcome
We also assessed the patients who have KIR3DL1 in the presence of its HLA-Bw4 ligand (KIR3DL1+/HLA-Bw4+; n = 59 or 58) compared to the patients who did not have this specific receptor-ligand pair (not KIR3DL1+/HLA-Bw4+; n = 141 or 139) (Supplementary Table S4). In contrast with some previous reports where the inhibitory KIR3DL1 gene in the presence of its HLA-Bw4 ligand was associated with improved clinical outcome from immunotherapy [9, 11], here, we found no association of KIR3DL1/HLA-Bw4 with clinical response in patients receiving BEMPEG plus nivolumab combination therapy (Supplementary Figure 1A, B).
Inhibitory KIR2DL2/HLA-C1+ interactions in combination with KIR3DL1/HLA-Bw4+ interactions improve outcome for patients receiving BEMPEG
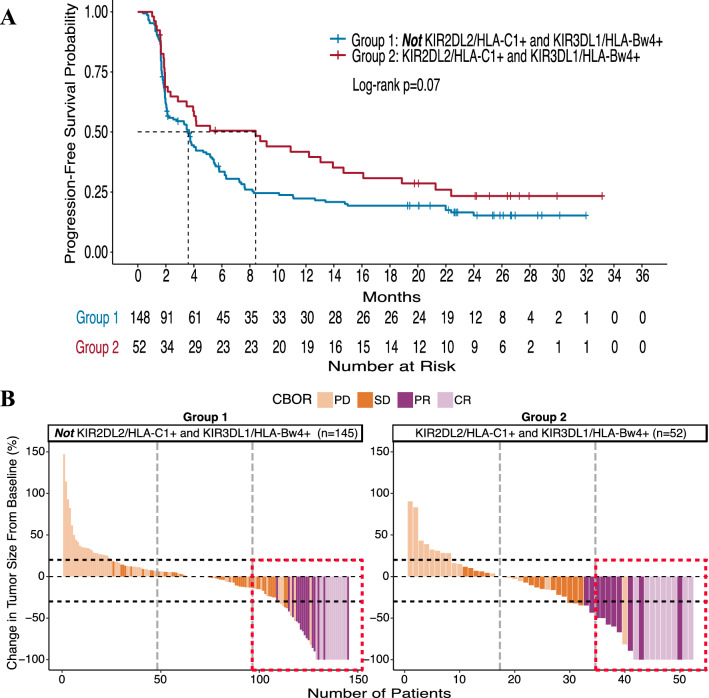
Although we did not observe any influence of KIR3DL1 and its HLA-Bw4 ligand on clinical outcome (Supplementary Figure 1A, B), we investigated whether KIR3DL1 in combination with KIR2DL2 and their ligands could further influence patient outcomes. We have previously shown that this interaction of inhibitory KIR2DL2 and KIR3DL1 with their ligands was associated with improved outcome in follicular lymphoma and neuroblastoma patients receiving rituximab maintenance therapy and dinutuximab monoclonal antibody (mAb) immunotherapy, respectively [9, 11]. Thus, we compared the group of patients who had KIR2DL2 with its HLA-C1 ligand as well as KIR3DL1 with its HLA-Bw4 ligand (Group 2: KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+; n = 52) to the remaining patients, i.e., those either lacking KIR2DL2, HLA-C1, KIR3DL1, and/or HLA-Bw4 (Group 1: not KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+; n = 148 or 145). The distinct genotypes comprising these two groups are detailed in Supplementary Table S4.
We found that patients who were in Group 2 had significantly improved OR and tumor shrinkage, p = 0.02 and p = 0.04, respectively. Although not significant, Group 2 patients also showed a trend toward prolonged PFS, p = 0.07 (Fig. 4A). Visualized at the individual patient level, the waterfall plots of the percent change in target lesion size from baseline (Fig. 4B) demonstrate that within the Group 2 genotype (right), a greater proportion of the top third percent of patients, outlined in the dotted red box, had a CR or PR (19/52 patients with CR/PR vs. 29/148) along with greater tumor shrinkage (median change −16.1% vs. 0%) compared to Group 1 (left).
Fig. 4.
IO-naïve patients observe greater clinical benefit with a KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+ genotype. Kaplan–Meier curve for PFS (a) compares patients who are KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+ (Group 2, red line) to patients who are not KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+ (Group 1, blue line). Waterfall plots displaying OR and tumor shrinkage (b) compares patients who are KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+ (Group 2, right) with patients who are not KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+ (Group 1, left). CBOR is the confirmed best overall response by RECIST 1.1 criteria; PD, progressive disease (light orange); SD, stable disease (orange); PR, partial response (purple); CR, complete response (light purple). Vertical dotted lines divide the number of patients into thirds, and horizontal dotted lines indicate a + 20% increase and −30% decrease in target lesion size from baseline. The dotted red box outlines the top third of patients, indicating a larger proportion of patients with a positive clinical response in Group 2 (right) than in Group 1 (left)
Overall associations of KIR/KIR-ligand genotypes with outcome for the specific groups of cancer patients tested
The specific genotype groupings shown for the overall 200 IO-naïve patients shown in Figs. 1, 3, 4 and S1 are shown for each of the separate disease groups tested, in Supplementary Table S5A-D. In addition to the association of KIR-ligands present with improved outcome for TNBC patients, shown above in Fig. 2, we note that the KIR2DL2/HLA-C1 associations (Supplementary Table S5B) are conserved in some of the clinical parameters for metastatic urothelial cancer (mUC; OR p = 0.05, tumor shrinkage p = 0.003) and non-small cell lung cancer (NSCLC; tumor shrinkage p = 0.05). Similar significance is observed in the KIR2DL2/HLA-C1 and KIR3DL1/HLA-Bw4 associations (Supplementary Table S5D) for mUC (OR p = 0.02, tumor shrinkage p = 0.005) as well. Although the primary results for all cohorts have been presented in various forms, not all cohorts have had the primary results published. Additionally, it is worth noting the small patient populations that these results are comprised of when analyzed as distinct cohorts. Thus, these results for the separate disease groups should be interpreted with caution due to multiplicity of testing without adjustment until primary results have been published for all cohorts.
SNPs among FCγR genes (2A, 2C, 3A) were not associated with clinical outcome
In our prior report for metastatic renal cell carcinoma patients treated with HD-IL2, we observed that improved clinical response may be correlated with specific “high-affinity” SNPs within the three FCγR genes assessed here, including FCγR2A, 2C, and 3A [10]. However, in this study, we did not find any significant associations between FCγR and clinical outcome (Supplementary Tables S6 and S7).
Discussion
In this retrospective analysis of patients with advanced solid tumors who received BEMPEG plus nivolumab combination therapy as part of the PIVOT-02 trial, we assessed the potential associations of KIR/KIR-ligand genotypes with clinical outcome. Here, we show that the repertoire of KIR/KIR-ligands that an individual inherits is associated with their clinical response to BEMPEG plus nivolumab treatment; consistent with a role for NK cells in the anti-tumor efficacy of this combination therapy.
Unlike some prior reports [12, 14, 32, 33], we found no evidence of improved clinical response for IO-naïve patients with the KIR-ligands missing genotype compared to the KIR-ligands present genotype (Fig. 1). Interestingly, however, we did find that for TNBC patients, response to BEMPEG plus nivolumab combination therapy was influenced by their KIR-ligand present/missing status. Namely, those with KIR-ligands present had greater clinical benefit in both OR and tumor shrinkage and a trend toward prolonged PFS as compared to those with KIR-ligands missing (Fig. 2). TNBC has been shown to be susceptible to NK cell-induced lysis supporting a role for NK cells in the anti-tumor response toward TNBC [36]. Yet, these prior reports [12, 14, 32, 33] mentioned above have shown the KIR-ligand missing genotype as beneficial for patients’ responses, in part due to NK-mediated killing of tumor cells under conditions in which the inhibitory KIRs on the NK cells are not interacting with their corresponding inhibitory ligands on the tumor cells, thus being less inhibited [13]. In the case of tumors that have no or low expression of KIR-ligands (namely HLA Class I), KIR-mediated inhibition would not be relevant, as the KIRs would not be seeing their corresponding ligand. This may be the case for TNBC, which has recently been shown to have prominent HLA-I loss, with more than half of patients having subclonal or diffuse HLA-I loss [26]. In this setting, the impact of NK cell licensing should also be taken into consideration.
The interactions of the KIRs on the NK cells with their corresponding HLA ligands on healthy cells during NK development have been shown to be associated with greater downstream NK cell function as a result of licensing [37–40]. If these properly licensed NK cells become activated through immune stimulatory signaling (i.e., through BEMPEG plus nivolumab), they can readily kill non-HLA-expressing tumor cells. In Wang et al., we previously reported that KIR/KIR-ligand genotypes known to directly drive licensing of NK cells can influence the functionality of NK cells following ex vivo immune stimulatory signaling when expanding NK cells [41]. Here, the combination treatment of BEMPEG and nivolumab might provide similar immune stimulatory signaling on endogenously licensed NK cells in the TME. Therefore, we hypothesize that patients with the KIR-ligands present genotype may have more ‘licensed’ (namely more potent) NK cells that, in this setting, would not be inhibited due to the low level of HLA on their tumors and thus be associated with a greater anti-tumor effect. In addition to TNBC, HLA-I downregulation or loss has been observed in 40–90% of human tumors, including those tumor types enrolled in this trial (melanoma, urothelial, NSCLC, and RCC) [42–48]. Thus, when combined with an immunotherapy regimen that activates NK cell activity and function (BEMPEG) together with an agent that represses checkpoint inhibition (nivolumab), in patients with cancers that have low or no HLA-I expression, these licensed NK cells may elicit enhanced tumor killing. In this scenario for patients with KIR-ligands present, this effective immunotherapy combination of BEMPEG plus nivolumab may take advantage of these licensed NK cells to overcome the inhibitory signaling from KIR/KIR-ligand interactions that may be present within the TME, such that NK cells can have persistent responses to reduce the tumor burden. Thus, our findings in the TNBC cohort may indicate a role for licensed NK cells in the anti-tumor response against TNBC following BEMPEG plus nivolumab therapy.
The impact of NK cell licensing may further explain our findings when we individually assessed the inhibitory KIRs in the presence of their ligands. For the inhibitory KIR2DL2 and its HLA-C1 ligand, patients who inherited both (KIR2DL2+/HLA-C1+) observed greater clinical responses (significant for PFS and tumor shrinkage and a trend for OR) as compared to the responses seen in the complementary group of patients (not KIR2DL2+/HLA-C1+) (Fig. 3). Conversely, we did not see similar associations with the inhibitory KIR3DL1 and its HLA-Bw4 ligand when assessed alone (Supplementary Figure S1). When we further assessed the potential interactions between these two inhibitory KIRs simultaneously, we found that patients who inherited both KIRs in the presence of their ligands (Group 2: KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+) had greater clinical benefit (significant for OR and tumor shrinkage and a trend for PFS) as compared to the complementary group of patients (Group 1: not KIR2DL2+/HLA-C1+ and KIR3DL1+/HLA-Bw4+) (Fig. 4). In separate randomized studies of anti-GD2 mAb and rituximab maintenance immunotherapy for neuroblastoma and follicular lymphoma patients, respectively, we found similar results. Namely, those patients with a Group 2 genotype had improved outcome from the anti-GD2 or rituximab maintenance immunotherapy (vs. no immunotherapy for neuroblastoma or non-maintenance for follicular lymphoma) [9, 11]. Altogether, these similar findings, which include different patient populations and disease types, provide some degree of validation for the influence of KIR/KIR-ligand genotypes on clinical outcome with immunotherapy regimens. Yet, the associations found in the IO-naïve patients in the PIVOT-02 trial need further evaluation prior to any clinical application, especially considering the limitation of the single-arm nature of this trial.
The associations of KIR/KIR-ligand genotypes with clinical outcome in the subset of IO-naïve patients in this trial suggest that either the immunologic effect of BEMPEG, or of nivolumab, or of their combined effect, may involve NK function in a way that influences outcome in this group of patients with solid tumors receiving this combination regimen. A prior study has reported that there was no association of KIR genotype, nor of KIR/KIR-ligand genotype with outcome in a group of 112 patients with advanced melanoma treated with single agent nivolumab [49]. This suggests, but does not prove, that the associations of KIR/KIR-ligand genotype with outcome observed here may be due to the effect of BEMPEG or due to an interaction of BEMPEG and nivolumab, rather than due to the action of nivolumab alone. Nivolumab and BEMPEG should synergize to drive a persistent anti-tumor response, via activation of immune cells (BEMPEG) while overcoming inhibitory signaling (nivolumab). NK cells primarily express CD122/CD132 (IL2βγ receptor). Since BEMPEG was designed to preferentially bind to this IL2βγ receptor (as compared to the IL2αβγ receptor found on Tregs), it should lead to NK cell activation and enhanced expansion of NK cells and CD8 T cells compared to Tregs. Yet, recent findings by Hashimoto and colleagues suggest that the ability of IL-2 to interact with CD25 (IL2α receptor) may influence the efficacy of IL-2 therapies given in combination with anti-PD-1. Data showed that with a mutated version of IL-2 that does not bind to CD25 (like BEMPEG), the synergy between IL-2 cytokine and anti-PD-1 (like nivolumab) treatment was abrogated [50]. Furthermore, upon activation, NK cells can also express CD25 [51]. This can increase NK cells’ affinity for IL-2, thereby enhancing their cytotoxic capabilities but also allowing NK cells to compete with Tregs for available IL-2 [52, 53]. Thus, the potential role that NK cells and KIR/KIR-ligand genotypes have in influencing response to IL-2 therapeutic combinations, including anti-PD-1 nivolumab, may be dependent on how that IL-2 therapy can bind to the IL2αβγ versus the IL2βγ receptors.
In the aforementioned studies for neuroblastoma and follicular lymphoma patients, both were two-arm, randomized clinical trials which allowed us to further suggest, based upon the results, a potential to discriminate between patients who are likely to benefit from anti-GD2 mAb or rituximab maintenance immunotherapy and those who are not. Such findings, once validated in the respective disease/treatment settings, may enable these genotypes to be used as predictive biomarkers to select which patients are most likely to benefit from immunotherapy [54]. This type of genotype-based patient selection for therapy may have greater clinical application, as many of the standard immunotherapy regimens are accompanied by toxic side effects. Thus, the ability to discern which patients may benefit from immunotherapy will in turn limit the number of patients we subject to these toxic regimens. In contrast, all patients evaluated in this current non-randomized PIVOT-02 trial received the combination therapy of BEMPEG and nivolumab. Therefore, the genotypes we identified as associated with improved outcome, if validated, would potentially be prognostic of improved outcome among patients receiving this combination therapy.
In summary, our evaluation of these immunogenotypes suggests that the repertoire of KIR/KIR-ligands that an individual inherits is associated with their clinical response to BEMPEG plus nivolumab therapy among those who are IO-naïve in the PIVOT-02 trial, which further supports a role for NK cells in the anti-tumor efficacy of this combination therapy. Further clinical validation in other studies is needed to determine whether these immunogenotypes may provide prospective data that could be clinically actionable.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank all patients and their families who participated in the PIVOT-02 clinical trial and Dr. Matthew Hellmann for his role as site PI for this trial while he was at Memorial Sloan Kettering Cancer Center. The PIVOT-02 trial was sponsored by Nektar Therapeutics and Bristol Myers Squibb.
Abbreviations
- ADCC
Antibody-dependent cellular cytotoxicity
- BEMPEG
Bempegaldesleukin
- CBOR
Confirmed best overall response
- CR
Complete response
- FCγRs
Fc-gamma receptors
- Fc
Fragment crystallizable
- HD-IL2
High dose IL2
- ICIs
Immune checkpoint inhibitors
- IO-naïve
Immunotherapy naïve
- KIRs
Killer immunoglobulin-like receptors
- mAb
Monoclonal antibody
- mUC
Metastatic urothelial cancer
- NSCLC
Non-small cell lung cancer
- OR
Objective response
- ORR
Objective response rate
- PD
Progressive disease
- PD-1
Anti-programmed death-1
- PFS
Progression-free survival
- PR
Partial response
- RP2D
Recommended phase 2 dose
- SD
Stable disease
- SNPs
Single-nucleotide polymorphisms
- SSP-PCR
Sequence specific primers-polymerase chain reaction
- TME
Tumor microenvironment
- TNBC
Triple-negative breast cancer
- Tregs
T regulatory cells
- TS
Tumor shrinkage
Author contributions
ASF and AKE genotyped the patients, interpreted the patient data regarding the clinical associations with outcome, and were major contributors to the writing of the manuscript. JB, KMK, and DY performed statistical analysis and inference of clinical outcome in association with genotype status and were major contributors to the writing of the manuscript. UH, SLC, TN, AOS-R, NMT, SMT and AD led the clinical study and provided the clinical samples and data. PMS led the genotyping data collection portion of this data set, interpreted the patient data regarding the clinical associations with outcome, and was a major contributor to the writing of the manuscript. All authors participated in the writing and editing of this manuscript.
Funding
The genotyping research was supported by the Midwest Athletes Against Childhood Cancer; Stand Up To Cancer; St. Baldrick’s Foundation; American Association for Cancer Research; University of Wisconsin Carbone Cancer Center; Nektar Therapeutics; and public health service grants CA014520, CA166105, and CA197078 from the National Cancer Institute, the National Institutes of Health, and the Department of Health and Human Services. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute.
Declarations
Conflict of interest
UH, DY, SLC, and TN are, or were, employees of Nektar Therapeutics, the sponsor of the clinical trial.
Ethics approval
The study was conducted in accordance with Good Clinical Practice guidelines and the Declaration of Helsinki. The protocol was approved by independent ethics committees and the institutional review board at each participating site.
Consent to participate
Written informed consent was obtained from all patients who participated in this study.
Footnotes
S. L. Currie: Formerly Nektar Therapeutics, San Francisco, CA, USA.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. New England J Med. 2015;373:1803. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cortes J, Cescon DW, Rugo HS, Nowecki Z, Im S-A, Yusof MM, et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. The Lancet. 2020;396:1817. doi: 10.1016/S0140-6736(20)32531-9. [DOI] [PubMed] [Google Scholar]
- 3.Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18:312. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 4.Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim S-W, Carcereny Costa E et al. (2019) Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. New England J Med, 381 [DOI] [PubMed]
- 5.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD et al. (2019) Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. New England J Med, 381 [DOI] [PubMed]
- 6.Haslam A, Prasad V. Estimation of the percentage of us patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open. 2019;2:1–9. doi: 10.1001/jamanetworkopen.2019.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bright R, Coventry BJ, Eardley-Harris N, Briggs N. Clinical response rates from interleukin-2 therapy for metastatic melanoma over 30 years’ experience: a meta-analysis of 3312 patients. J Immunother. 2017;40:21–30. doi: 10.1097/CJI.0000000000000149. [DOI] [PubMed] [Google Scholar]
- 8.Bentebibel SE, Hurwitz ME, Bernatchez C, Haymaker C, Hudgens CW, Kluger HM, et al. A first-in-human study and biomarker analysis of NKTR-214, a novel IL2Rβγ-biased cytokine, in patients with advanced or metastatic solid tumors. Cancer Discov. 2019;9:711–721. doi: 10.1158/2159-8290.CD-18-1495. [DOI] [PubMed] [Google Scholar]
- 9.Erbe AK, Wang W, Carmichael L, Kim KM, Mendoņca EA, Song Y, et al. Neuroblastoma patients’ KIR and KIR-ligand genotypes influence clinical outcome for dinutuximab-based immunotherapy: A report from the children’s oncology group. Clin Cancer Res. 2018;24:189–196. doi: 10.1158/1078-0432.CCR-17-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erbe AK, Wang W, Goldberg J, Gallenberger M, Kim KM, Carmichael L, et al. FCGR polymorphisms influence response to IL2 in metastatic renal cell carcinoma. Clin Cancer Res. 2017;23:2159–2168. doi: 10.1158/1078-0432.CCR-16-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erbe AK, Wang W, Carmichael L, Hoefges A, Grzywacz B, Reville PK, et al. Follicular lymphoma patients with KIR2DL2 and KIR3DL1 and their ligands (HLA-C1 and HLA-Bw4) show improved outcome when receiving rituximab. J Immunother Cancer. 2019;7:1–12. doi: 10.1186/s40425-019-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado DC, Hank JA, Kolesar J, Lorentzen D, Gan J, Seo S, et al. Genotypes of NK cell KIR receptors, their ligands, and Fcγ receptors in the response of neuroblastoma patients to Hu14.18-IL2 immunotherapy. Cancer Res. 2010;70:9554–9561. doi: 10.1158/0008-5472.CAN-10-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarek N, le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Investig. 2012;122:3260–3270. doi: 10.1172/JCI62749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung NKV, Cheung IY, Kushner BH, Ostrovnaya I, Chamberlain E, Kramer K, et al. Murine anti-GD2 monoclonal antibody 3F8 combined with granulocyte- macrophage colony-stimulating factor and 13-cis-retinoic acid in high-risk patients with stage 4 neuroblastoma in first remission. J Clin Oncol. 2012;30:3264–3270. doi: 10.1200/JCO.2011.41.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolany S, Baldini C, Spira A, Cho D, Grignani G, Racca F et al. (2019) Clinical activity of BEMPEG plus NIVO observed in metastatic TNBC: preliminary results from the TNBC cohort of the Ph1/2 PIVOT-02 study [abstract]. 5th CRI-CIMT-EATI-AACR international cancer immunotherapy conference. p. A001.
- 16.Tannir NM, Cho DC, Diab A, Sznol M, Bilen MA, Balar A V, et al (2022) Bempegaldesleukin plus nivolumab in first-line renal cell carcinoma: results from the PIVOT-02 study. J Immunother Cancer. 10 [DOI] [PMC free article] [PubMed]
- 17.Diab A, Tannir NM, Bentebibel SE, Hwu P, Papadimitrakopoulou V, Haymaker C, et al. Bempegaldesleukin (NKTR-214) plus nivolumab in patients with advanced solid tumors: Phase I dose-escalation study of safety, effi cacy, and immune activation (PIVOT-02) Cancer Discov. 2020;10:1158–1173. doi: 10.1158/2159-8290.CD-19-1510. [DOI] [PubMed] [Google Scholar]
- 18.Diab A, Tykodi SS, Daniels GA, Maio M, Curti BD, Lewis KD et al. (2021) Bempegaldesleukin plus nivolumab in first-line metastatic melanoma. J Clin Oncol. JCO.21.00675. [DOI] [PMC free article] [PubMed]
- 19.van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, Kuijpers TW. Phenotypic variation in IgG receptors by nonclassical FCGR2C alleles. J Immunol. 2012;188:1318. doi: 10.4049/jimmunol.1003945. [DOI] [PubMed] [Google Scholar]
- 20.Lejeune J, Piègu B, Gouilleux-Gruart V, Ohresser M, Watier H, Thibault G (2012) FCGR2C genotyping by pyrosequencing reveals linkage disequilibrium with FCGR3A V158F and FCGR2A H131R polymorphisms in a Caucasian population. MAbs, 4 [DOI] [PMC free article] [PubMed]
- 21.Breunis WB, van Mirre E, Geissler J, Laddach N, Wolbink G, van der Schoot E, et al. (2009) Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat, 30 [DOI] [PubMed]
- 22.Ernst LK, Metes D, Herberman RB, Morel PA (2002) Allelic polymorphisms in the FcγRIIC gene can influence its function on normal human natural killer cells. J Mol Med, 80. [DOI] [PubMed]
- 23.Weng W-K, Levy R (2003) Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol, 21. [DOI] [PubMed]
- 24.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G et al. (2008) Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of Trastuzumab-based therapy in patients with HER-2/neu–positive metastatic breast cancer. J Clin Oncol, 26. [DOI] [PubMed]
- 25.Keating SE, Ní Chorcora C, Dring MM, Stallings RL, O’Meara A, Gardiner CM (2015) Increased frequencies of the killer immunoglobulin-like receptor genes KIR2DL2 and KIR2DS2 are associated with neuroblastoma. Tissue Antigens, 86. [DOI] [PubMed]
- 26.Dusenbery AC, Maniaci JL, Hillerson ND, Dill EA, Bullock TN, Mills AM. MHC Class i loss in triple-negative breast cancer: a potential barrier to PD-1/PD-L1 checkpoint inhibitors. Am J Surg Pathol. 2021;45:701–707. doi: 10.1097/PAS.0000000000001653. [DOI] [PubMed] [Google Scholar]
- 27.Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 28.Erbe AK, Wang W, Gallenberger M, Hank JA, Sondel PM. Genotyping single nucleotide polymorphisms and copy number variability of the FCGRs expressed on NK cells. Methods Mol Biol. 2016;1441:43–56. doi: 10.1007/978-1-4939-3684-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform. 2019;95:103208. doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM (2015) NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol. [DOI] [PMC free article] [PubMed]
- 32.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SGE, et al. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol. 2014;192:4592–4600. doi: 10.4049/jimmunol.1302517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, et al. HLA-C–dependent prevention of leukemia relapse by donor activating KIR2DS1. New England J Med. 2012;367:805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Erbe AK, Gallenberger M, Kim KM, Carmichael L, Hess D, et al. Killer immunoglobulin-like receptor (KIR) and KIR–ligand genotype do not correlate with clinical outcome of renal cell carcinoma patients receiving high-dose IL2. Cancer Immunol, Immunother. 2016;5:1523–1532. doi: 10.1007/s00262-016-1904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 36.Engel JB, Honig A, Kapp M, Hahne JC, Meyer SR, Dietl J, et al. Mechanisms of tumor immune escape in triple-negative breast cancers (TNBC) with and without mutated BRCA 1. Arch Gynecol Obstet. 2014;289:141. doi: 10.1007/s00404-013-2922-9. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 38.Jonsson AH, Yokoyama WM (2009) Chapter 2 natural killer cell tolerance. Licensing and other mechanisms. Adv Immunol. p. 27–79. [DOI] [PubMed]
- 39.Yokoyama WM, Kim S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev. 2006;214:143. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim S, Sunwoo JB, Yang L, Choi T, Song Y-J, French AR, et al. (2008) HLA alleles determine differences in human natural killer cell responsiveness and potency [Internet]. Available from: www.pnas.org/cgi/content/full/ [DOI] [PMC free article] [PubMed]
- 41.Wang W, Erbe AK, Alderson KA, Phillips E, Gallenberger M, Gan J, et al. Human NK cells maintain licensing status and are subject to killer immunoglobulin-like receptor (KIR) and KIR-ligand inhibition following ex vivo expansion. Cancer Immunol Immunother. 2016;65:1047–1059. doi: 10.1007/s00262-016-1864-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornel AM, Mimpen IL, Nierkens S. MHC class I downregulation in cancer: underlying mechanisms and potential targets for cancer immunotherapy. Cancers (Basel) 2020;12:1760. doi: 10.3390/cancers12071760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Méndez R, Rodríguez T, del Campo A, Monge E, Maleno I, Aptsiauri N, et al. Characterization of HLA class I altered phenotypes in a panel of human melanoma cell lines. Cancer Immunol Immunother. 2008;57:719–729. doi: 10.1007/s00262-007-0411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slotta-Huspenina J, Schwamborn K, Steiger K, Simon R, Kirchhoff FP, Büchler JW, et al. MHC I expression predicts response to checkpoint inhibitors in metastatic urothelial carcinoma but lacks prognostic value in localized disease. Bladder Cancer. 2022;8:269–276. doi: 10.3233/BLC-211604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koukourakis IM, Giatromanolaki A, Mitrakas A, Koukourakis MI. Loss of HLA-class-I expression in non-small-cell lung cancer: association with prognosis and anaerobic metabolism. Cell Immunol. 2022;373:104495. doi: 10.1016/j.cellimm.2022.104495. [DOI] [PubMed] [Google Scholar]
- 47.Atkins D, Ferrone S, Schmahl GE, Störkel S, Seliger B. Down-regulation of HLA class I antigen processing molecules: an immune escape mechanism of renal cell carcinoma? J Urol. 2004;171:885–889. doi: 10.1097/01.ju.0000094807.95420.fe. [DOI] [PubMed] [Google Scholar]
- 48.Yuan J, Liu S, Yu Q, Lin Y, Bi Y, Wang Y, et al. Down-regulation of human leukocyte antigen class I (HLA-I) is associated with poor prognosis in patients with clear cell renal cell carcinoma. Acta Histochem. 2013;115:470–474. doi: 10.1016/j.acthis.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 49.Ishida Y, Nakashima C, Kojima H, Tanaka H, Fujimura T, Matsushita S, et al. Killer immunoglobulin-like receptor genotype did not correlate with response to anti-PD-1 antibody treatment in a Japanese cohort. Sci Rep. 2018;8:1. doi: 10.1038/s41598-018-34044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hashimoto M, Araki K, Cardenas MA, Li P, Jadhav RR, Kissick HT, et al. PD-1 combination therapy with IL-2 modifies CD8+ T cell exhaustion program. Nature. 2022;610:173–181. doi: 10.1038/s41586-022-05257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S-H, Fragoso MF, Biron CA. Cutting edge: a novel mechanism bridging innate and adaptive immunity: il-12 induction of CD25 To form high-affinity IL-2 receptors on NK cells. J Immunol. 2012;189:2712–2716. doi: 10.4049/jimmunol.1201528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasteiger G, Hemmers S, Firth MA, le Floc’h A, Huse M, Sun JC, et al. IL-2–dependent tuning of NK cell sensitivity for target cells is controlled by regulatory T cells. J Exp Med. 2013;210:1167–1178. doi: 10.1084/jem.20122462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. doi: 10.3389/fimmu.2018.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sechidis K, Papangelou K, Metcalfe PD, Svensson D, Weatherall J, Brown G. Distinguishing prognostic and predictive biomarkers: an information theoretic approach. Bioinformatics. 2018;34:3365–3376. doi: 10.1093/bioinformatics/bty357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.