Abstract
Primary biliary cholangitis (PBC) is a rare autoimmune disease with a clear predisposition for human leukocyte antigen (HLA)-DR/DQ-associated loss of immune tolerance for the E2 component of the pyruvate dehydrogenase complex. Three-field-resolution HLA imputation of 1,670 Japanese PBC patients and 2,328 healthy controls was conducted using Japanese population-specific HLA reference panels. Eighteen previously reported Japanese PBC-associated HLA alleles were confirmed and extended to 3-field-resolution, including HLA-DRB1*08:03 to HLA-DRB1*08:03:02, HLA-DQB1*03:01 to HLA-DQB1*03:01:01, HLA-DQB1*04:01 to HLA-DQB1*04:01:01 and HLA-DQB1*06:04 to HLA-DQB1*06:04:01. In addition, additional significant novel HLA alleles were identified, including 3 novel susceptible HLA-DQA1 alleles: HLA-DQA1*03:03:01, HLA-DQA1*04:01:01, HLA-DQA1*01:04:01 and 1 novel protective HLA-DQA1 allele, HLA-DQA1*05:05:01. In addition, PBC patients carrying HLA-DRB1*15:01:01 and HLA-DQA1*03:03:01 would have a higher predisposition toward developing concomitant autoimmune hepatitis (AIH). Further, late-stage and symptomatic PBC shared the same susceptible HLA alleles of HLA-A*26:01:01, HLA-DRB1*09:01:02 and HLA-DQB1*03:03:02. Lastly, HLA-DPB1*05:01:01 was identified as a potential risk HLA allele for development of hepatocellular carcinoma (HCC) in PBC patients. In conclusion, we have extended the current knowledge of HLA allele associations to 3-field resolution and identified novel HLA allele associations with predisposition risk, staging, symptomatic state, and AIH and HCC events for Japanese PBC patients.
Keywords: primary biliary cholangitis, HLA, autoimmune hepatitis, hepatocellular carcinoma, Scheuer staging system
Introduction
Primary biliary cholangitis (PBC), previously known as primary biliary cirrhosis, is a complex autoimmune disease that predominantly affects middle-aged women, with a sex ratio of 9 females to 1 male (1). Clinical manifestations of PBC include chronic and progressive destruction of the small bile duct, granulomatous lymphocytic cholangitis with seroreactivity for antimitochondrial antibodies (AMA) against the E2 component of the pyruvate dehydrogenase complex (2). Common symptoms of PBC include, but are not limited to, fatigue (80% of cases) (3), pruritus (20–70% of cases) (4) and jaundice (25% of end-stage cases)(5). Other complications of PBC (6) include portal hypertension (ascites, varices, hepatic encephalopathy), fat malabsorption, fat deposits and osteoporosis/osteomalacia.
The etiopathogenesis of PBC indicates a genetically predisposed autoimmune disease with the possibility of involving still-unknown xenobiotics or infections (7). An international genome-wide meta-analysis (8, 9) of 5 European and 2 East-Asian cohorts (PBC Consortia, Canadian PBC Consortium, Chinese PBC Consortium, Italian PBC Study Group, Japan PBC-GWAS Consortium, US PBC Consortium, and UK-PBC Consortium) reported human leukocyte antigen (HLA) genes as the strongest associated genes. HLA-targeted gene approach studies have reported the strongest susceptible HLA allele associations for HLA class II genes, with HLA-DRB1*08:03 and HLA-DQB1*06:01 in Japanese PBC (10–12) and HLA-DRB1*08:01 in Italian PBC (13), US PBC (14) and UK PBC (15).
Autoimmune hepatitis (AIH) is a disease of the hepatic parenchyma (16) that sometimes arises in patients with PBC. According to the Japan AIH National Survey(17), the prevalence of AIH is 23.9 per 100,000 individuals. HLA-DRB1*04:01 has been reported as the HLA allele most closely associated with both European (17) and Japanese (18) AIH, while HLA-DRB1*04:05 associations have been reported for Japanese (19), Korean (20) and Latin-American (21) AIH.
Hepatocellular carcinoma (HCC) is the most common form of liver cancer, with hepatitis B virus (HBV) and hepatitis C virus (HCV) infections as two of the most prominent risk factors for HCC development (22). Studies have suggested that cirrhotic PBC represents a rare precursor for HCC development (23–25). Among cases of late-stage PBC, HCC shows an incidence of around 5.9% and occurs predominantly in males (26). A meta-analysis of 8 different studies reported HLA-DRB1*07 and HLA-DRB1*12 associations with the risk of HCC, while HLA-DRB1*07, HLA-DRB1*12 and HLA-DRB1*15 alleles were associated with significantly increased risks of HCC in Asians (27). HLA-DQB1*03:01 and HLA-DQB1*06:02 are reportedly associated with Taiwanese HCV-HCC (28), while HLA-A*33:03 is reportedly associated with Japanese HBV-HCC (29).
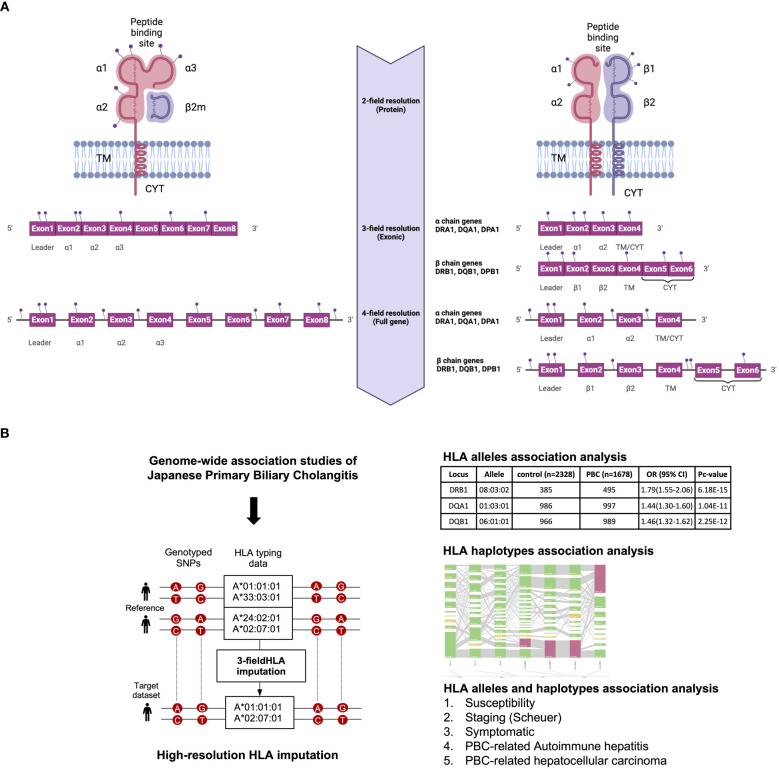
All the above-mentioned HLA studies have reported a maximum of 2-field-resolution HLA association studies that mainly focused on variations in the core exons of HLA genes, namely exons 2 and 3 for HLA class I genes and exon 2 for HLA class II genes (Figure 1). The present study investigated 3-field-resolution HLA allele associations (taking into consideration all variants in the exonic regions of HLA class I and class II genes) with susceptibility for PBC, staging of PBC, symptomatic state of PBC, PBC-associated AIH (PBC-AIH) and PBC-associated HCC (PBC with HCC) (Figure 1).
Figure 1.
(A) Schematic representation of HLA allele resolutions in relation to the mutation sites on the HLA class I and II genes. The 2-field resolution covers the changes in amino acid sequences of the encoded protein for HLA alleles while the 3-field resolution of HLA alleles takes into account changes in all exons that differ by synonymous nucleotide substitutions. Finally, the 4-field resolution encompasses differences in sequence polymorphism in the 5’ or 3’ untranslated regions and introns. α1/2/3, alpha chain; β1, beta chain; β2m, beta2 microglobulin; TM, transmembrane region; CYT, cytoplasmic tail. (B) Schematic representation of the study design. 3-field HLA imputation were performed using the SNP genotyping data from GWAS of Japanese PBC. HLA alleles association test were carried out by stratifying based on susceptibility, staging (Scheuer), symptomatic, PBC with AIH and PBC with HCC.
Methods
Population and SNP genotyping
A total of 1,670 Japanese PBC patients and 2,328 Japanese healthy controls were recruited through the Japan PBC-GWAS Consortium, which comprises 31 hospitals participating in the Japanese National Hospital Organization Study Group for Liver Disease in Japan (NHOSLJ) and 24 university hospitals participating in the gp210 Working Group in Intractable Liver Disease Research Project Team of the Ministry of Health and Welfare in Japan, as described previously (30, 31). The extensive collection of clinical data for Japanese PBC patients and SNP genotyping of Japanese PBC patients and healthy controls are carried out through research undertaken as Japan PBC-GWAS Consortium activities.
Single Nucleotide Polymorphism (SNP) genotyping was performed using the Affymetrix Axiom Japonica v1 (Toshiba, Tokyo, Japan) (32) and Affymetrix Axiom Genome-Wide ASI 1 (Affymetrix, Santa Clara, California, USA) genotyping arrays, as described in previous publications (31, 33). SNP-level and individual-level quality controls were provided using PLINK v1.90 (www.cog-genomics.org/plink/1.9/) (34) with individual call rate ≥98%, SNP call rate ≥99%, minor allele frequency ≥5%, and consistency with Hardy–Weinberg equilibrium (P ≥1.0E-6). Duplicate or closely related samples were removed when identity by descent for each sample pair (pi-hat > 0.1).
Scoring of chronic hepatitis
Of the total 1,670 Japanese PBC patients, liver biopsies were performed in 1,115 patients and histologically evaluated based on the Scheuer’s classification (35): stage 1, florid duct lesion; stage 2, destruction of limiting plate with piecemeal necrosis or ductular proliferation; stage 3, presence of fibrous septa which is called scarring; stage 4, cirrhosis (Table 1).
Table 1.
Clinical characteristics of Japanese PBC cases.
| PBC (n=1670) | |
|---|---|
| Age (years), median(range) | 57 (22-89) |
| Female (%) | 85.9 |
| AST (IU/L), (mean ± SD) | 55 ± 88 |
| ALP (IU/L), (mean ± SD) | 580 ± 411 |
| IgM (mg/dL), (mean ± SD) | 353 ± 257 |
| IgG (mg/dL), (mean ± SD) | 1640 ± 519 |
| Anti-gp210 antibodies positive (%) | 25.9 |
| Anti-mitochondrial antibodies positive (%) | 88.3 |
| Anti-nuclear antibodies positive (%) | 72.5 |
| Scheuer stage 1-2/3-4 (%) | 81.8/18.2 |
| Severity | |
| aPBC/s0,1, 2 (%) | 66.4/33.6 |
| Concomitant disease | |
| autoimmune hepatitis (%) | 7.2 |
| other autoimmune diseases (%) | 20.9 |
| Clinical outcome | |
| hepatocellular carcinoma (%) | 3.1 |
| liver transplantation and/or death by hepatic failure (%) | 8.4 |
Symptom classification of PBC
Of the total of 1,670 Japanese PBC patients, 1,337 were further classified into the different stages of severity based on clinical symptoms (Table 1): asymptomatic PBC (aPBC) = PBC without pruritus, without varices, without ascites, without encephalopathy, and without jaundice; s0 = positive symptoms of portal hypertension, such as esophageal varices; s1 = positive symptoms of pruritus; and s2 = positive symptoms of jaundice (T.bil. > 2.5 mg/dl).
Clinical diagnosis of PBC-AIH and PBC with HCC
The diagnosis of PBC-AIH is based on the criteria defined by Poupon et al., (36, 37): i) ALT >5 times the upper limit of normal (ULN); ii) IgG >1.5 times ULN; iii) The positive anti-smooth muscle antibody (ASMA), iv) liver biopsy specimen showing moderate to severe periportal or periseptal lymphocytic piecemeal necrosis The presence of HCC was evaluated by ultrasound sonography (UST), computed tomography (CT) or magnetic Resonance Imaging (MRI).
Measurement of autoantibody testing
Anti-mitochondrial and anti-gp210 antibodies were measured by ELISA, while anti-nuclear antibodies were measured by immunofluorescence test (IF), as described elsewhere (38). 1:100 diluted sera were used for ELISA and 1/40 to 5120 diluted sera were used for IF.
HLA genotyping and HLA imputation
The 3-field-resolution HLA imputation reference consisting of 418 Japanese healthy individuals was generated using the HIBAG R package (v3.16; Xiuwen Zheng) (39, 40) using the SNP genotyping data Affymetrix Axiom Japonica v1 (Toshiba) (32) and Affymetrix Axiom Genome-Wide ASI 1 (Affymetrix) genotyping arrays and high-resolution HLA genotyping data by AllType assay (One Lambda, West Hills, CA, USA). The AllType assay (One Lambda) was designed to cover the full length of five HLA genes (HLA-A, -C, -B, -DPA1, -DQA1). High-resolution HLA genotyping was carried out following vendor instructions for HLA gene-specific amplification, HLA library preparation, HLA template preparation, HLA template purification and loading of the HLA template onto an ion 530v1 chip (Thermo Fisher Scientific, Waltham, MA, USA) in the ion chef (Thermo Fisher Scientific) followed by final sequencing on the ion S5 machine (Thermo Fisher Scientific). HLA genotype assignments were carried out using HLATypeStream Visual (TSV v2.0; One Lambda) and NGSengine® (v2.18.0.17625; GenDX, Utrecht, the Netherlands). Demultiplexing of barcodes and base-calling was carried out in Torrent Suite version 5.8.0 (Thermo Fisher Scientific). Raw fastq reads were extracted using the FileExporter function in Torrent Suite version 5.8.0. HLA genotype assignments were carried out using two different types of software, namely HLATypeStream Visual (TSV v2.0; One Lambda) as the default software for the Alltype™ NGS Assay and NGSengine® (v2.18.0.17625; GenDX). The default analysis parameters and healthy metrics threshold was applied for TSV2.0 while we applied the “ignore regions” function in NGSengine to eliminate known sequencing error sites such as homopolymer sites in the ion S5 system. Three- and 4-field HLA allele genotyping was determined by comparing reads with IPD-IMGT/HLA database release 3.40.0 (https://www.ebi.ac.uk/ipd/imgt/hla/).
SNP data from 3,998 samples were extracted from an extended MHC (xMHC) region ranging from 25,759,242 to 33,534,827 bp based on the hg19 position. Three-field HLA imputation was carried out using the HIBAG R package (39) and our in-house 3-field Japanese HLA imputation reference (40). Post-imputation quality control of call threshold (call threshold > 0.5) was applied to remove low-quality samples. In total, we successfully imputed the number of unique HLA alleles as follows: HLA-A, n=14; HLA-C, n=19; HLA-B, n=33; HLA-DRB1, n=29; HLA-DQB1, n=14; HLA-DQA1, n=18; HLA-DPA1, n=7; and HLA-DPB1, n=14.
Statistical analysis
Case-control and clinical phenotype HLA allele association tests, HLA haplotype estimations, case-control HLA haplotype association tests and Hardy–Weinberg equilibrium tests were prepared and analyzed using the Bridging ImmunoGenomics Data Analysis Workflow Gaps (BIGDAWG) R package (v3.0.3; Derek Pappas, Steve Mack, Jill Hollenbach)(41). The default parameters of BIGDAWG were used except for the manual specification of HLA haplotypes for testing. Rare HLA alleles with an expected count < 5 were combined into a common class.
Results
Clinical characteristics of the 1,670 Japanese PBC cases
A total of 1,670 Japanese PBC patients with extensive clinical information and 2,328 healthy controls were recruited through Japan PBC-GWAS Consortium (Table 1). To identify HLA alleles potentially associated with the severity of PBC (please refer to the Methods for classification definitions), patients showing Scheuer 0, 1 and 2 were grouped as the early-stage PBC group, while patients showing Scheuer 3 and 4 were grouped as the late-stage PBC group, resulting in 808 of 982 patients (82.3%) showing early-stage PBC and 174 of 982 patients (17.5%) showing late-stage PBC. Similarly, HLA profiles were compared between 892 of 1,337 (66.7%) asymptomatic PBC patients and 445 of 1,337 (33.3%) symptomatic PBC patients (please refer to the Methods for classification definitions). In addition, 116 of 1,670 (7.0%) PBC-AIH cases and 49 of 1,670 (2.9%) PBC with HCC cases were compared with their respective negative group.
Associations of HLA alleles and HLA haplotypes with Japanese PBC cases
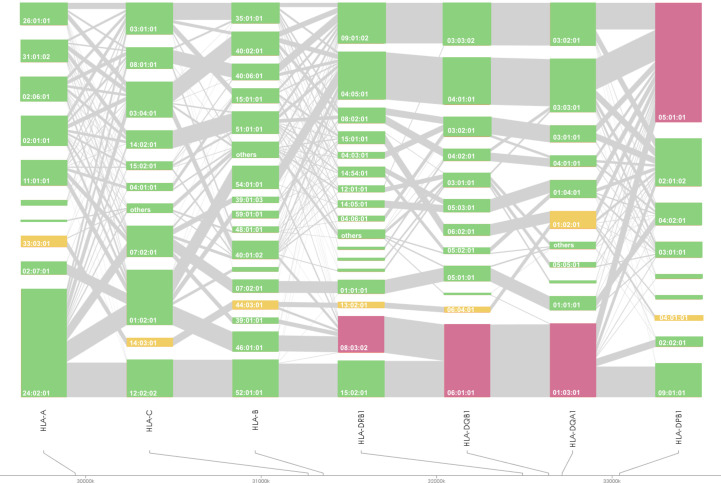
HLA association studies for the 8 HLA genes were carried out comparing HLA allele frequencies in the 1,670 Japanese PBC patients and 2,328 healthy controls (Table 2). A total of 1 HLA-A, 2 HLA-C, 5 HLA-B, 10 HLA-DRB1, 8 HLA-DQA1, 7 HLA-DQB1, 1 HLA-DPA1 and 3 HLA-DPB1 alleles were significantly associated with Japanese PBC after correction for multiple testing. The most significant susceptible HLA alleles were HLA-DRB1*08:03:02, HLA-DQB1*06:01:01, HLA-DQA1*01:03:01 and HLA-DPB1*05:01:01 (Table 2). HLA haplotype analysis confirmed strong linkage among the above-mentioned HLA alleles, with the strongest HLA haplotype associations (Table 3; Figure 2) for HLA-DRB1*08:03:02-HLA-DQA1*01:03:01-HLA-DQB1*06:01:01 but weaker HLA haplotype association (Table 3; Figure 2) after adding HLA-DPB1 allele with HLA-DRB1*08:03:02-HLA-DQA1*01:03:01-HLA-DQB1*06:01:01-HLA-DPB1*05:01:01. In contrast, the most significant protective HLA alleles (Table 2; Figure 2) were HLA-DRB1*13:02:01, HLA-DQB1*06:04:01, HLA-DQA1*01:02:01 and HLA-DPB1*04:01:01. All these protective HLA alleles formed strong HLA haplotypes (Table 3; Figure 2) with HLA-DRB1-13:02:01-HLA-DQA1*01:02:01-HLA-DQB1*06:04:01, showing a weaker association after including HLA-DPB1*04:01:01 with HLA-DRB1*13:02:01-HLA-DQA1*01:02:01-HLA-DQB1*06:04:01-HLA-DPB1*04:01:01 (Table 3).
Table 2.
Association analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients and Japanese healthy controls.
| Locus | Allele | control (2n=4656) | % | PBC (2n=3306) | % | OR (95%CI) | p.value | Pc-value |
|---|---|---|---|---|---|---|---|---|
| A | 33:03:01 | 427 | 9.2 | 139 | 4.2 | 0.43 (0.35–0.53) | 2.22E-16 | 2.66E-15 |
| C | 01:02:01 | 849 | 18.2 | 695 | 21.0 | 1.19 (1.07–1.34) | 1.93E-03 | 3.29E-02 |
| C | 14:03:01 | 419 | 9.0 | 106 | 3.2 | 0.33 (0.27–0.42) | 2.22E-16 | 3.77E-15 |
| B | 07:02:01 | 324 | 7.0 | 165 | 5.0 | 0.70 (0.58–0.86) | 3.30E-04 | 9.24E-03 |
| B | 15:01:01 | 412 | 8.8 | 185 | 5.6 | 0.61 (0.51–0.73) | 6.08E-08 | 1.70E-06 |
| B | 40:02:01 | 308 | 6.6 | 301 | 9.1 | 1.42 (1.20–1.68) | 3.56E-05 | 9.96E-04 |
| B | 44:03:01 | 418 | 9.0 | 106 | 3.2 | 0.34 (0.27–0.42) | 2.22E-16 | 6.22E-15 |
| B | 46:01:01 | 258 | 5.5 | 246 | 7.5 | 1.37 (1.14–1.65) | 5.72E-04 | 1.60E-02 |
| DRB1 | 01:01:01 | 336 | 7.2 | 175 | 5.3 | 0.72 (0.59–0.87) | 6.51E-04 | 1.56E-02 |
| DRB1 | 04:05:01 | 619 | 13.3 | 596 | 18.1 | 1.44 (1.27–1.63) | 4.69E-09 | 1.13E-07 |
| DRB1 | 04:06:01 | 170 | 3.7 | 68 | 2.1 | 0.56 (0.41–0.74) | 4.32E-05 | 1.04E-03 |
| DRB1 | 08:02:01 | 190 | 4.1 | 192 | 5.8 | 1.45 (1.18–1.80) | 3.31E-04 | 7.94E-03 |
| DRB1 | 08:03:02 | 385 | 8.3 | 456 | 13.8 | 1.78 (1.54–2.06) | 1.71E-15 | 4.11E-14 |
| DRB1 | 11:01:01 | 110 | 2.4 | 36 | 1.1 | 0.46 (0.30–0.67) | 3.28E-05 | 7.86E-04 |
| DRB1 | 12:01:01 | 189 | 4.1 | 88 | 2.7 | 0.65 (0.50–0.84) | 8.88E-04 | 2.13E-02 |
| DRB1 | 13:02:01 | 395 | 8.5 | 80 | 2.4 | 0.27 (0.21–0.34) | 2.22E-16 | 5.33E-15 |
| DRB1 | 14:03:01 | 68 | 1.5 | 14 | 0.4 | 0.29 (0.15–0.52) | 6.75E-06 | 1.62E-04 |
| DRB1 | 15:01:01 | 352 | 7.6 | 152 | 4.6 | 0.59 (0.48–0.72) | 1.10E-07 | 2.64E-06 |
| DQA1 | 01:01:01 | 335 | 7.2 | 176 | 5.3 | 0.73 (0.60–0.88) | 8.11E-04 | 1.30E-02 |
| DQA1 | 01:02:01 | 736 | 15.9 | 221 | 6.7 | 0.38 (0.32–0.45) | 2.22E-16 | 3.55E-15 |
| DQA1 | 01:03:01 | 986 | 21.3 | 924 | 28.1 | 1.45 (1.30–1.61) | 2.60E-12 | 4.16E-11 |
| DQA1 | 01:04:01 | 233 | 5.0 | 224 | 6.8 | 1.38 (1.14–1.68) | 7.86E-04 | 1.26E-02 |
| DQA1 | 03:03:01 | 714 | 15.4 | 673 | 20.4 | 1.41 (1.26–1.59) | 5.20E-09 | 8.32E-08 |
| DQA1 | 04:01:01 | 114 | 2.5 | 139 | 4.2 | 1.75 (1.35–2.27) | 1.04E-05 | 1.66E-04 |
| DQA1 | 05:05:01 | 172 | 3.7 | 62 | 1.9 | 0.50 (0.37–0.67) | 2.25E-06 | 3.60E-05 |
| DQB1 | 03:01:01 | 450 | 9.7 | 182 | 5.5 | 0.54 (0.45–0.65) | 1.32E-11 | 1.72E-10 |
| DQB1 | 04:01:01 | 618 | 13.3 | 597 | 18.1 | 1.44 (1.27–1.63) | 4.90E-09 | 6.37E-08 |
| DQB1 | 04:02:01 | 141 | 3.0 | 154 | 4.7 | 1.56 (1.23–1.99) | 1.48E-04 | 1.93E-03 |
| DQB1 | 05:01:01 | 358 | 7.7 | 196 | 5.9 | 0.76 (0.63–0.91) | 2.35E-03 | 3.05E-02 |
| DQB1 | 06:01:01 | 966 | 20.7 | 916 | 27.7 | 1.46 (1.32–1.63) | 5.90E-13 | 7.67E-12 |
| DQB1 | 06:02:01 | 342 | 7.3 | 142 | 4.3 | 0.57 (0.46–0.69) | 1.99E-08 | 2.59E-07 |
| DQB1 | 06:04:01 | 388 | 8.3 | 75 | 2.3 | 0.26 (0.20–0.33) | 2.22E-16 | 2.89E-15 |
| DPA1 | 02:02:02 | 1260 | 37.7 | 644 | 42.1 | 1.20 (1.06–1.36) | 3.23E-03 | 1.61E-02 |
| DPB1 | 02:01:02 | 1143 | 24.5 | 616 | 18.6 | 0.70 (0.63–0.79) | 3.94E-10 | 5.12E-09 |
| DPB1 | 04:01:01 | 308 | 6.6 | 60 | 1.8 | 0.26 (0.19–0.35) | 2.22E-16 | 2.89E-15 |
| DPB1 | 05:01:01 | 1697 | 36.4 | 1476 | 44.7 | 1.41 (1.28–1.54) | 1.52E-13 | 1.97E-12 |
OR, odds ratio; 95%CI, 95% confidence interval; Pc-value, multiple testing-corrected p-value; binned, rare HLA alleles with expected count < 5, combined into a common class.
Table 3.
HLA haplotype analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients and Japanese healthy controls.
| A~B | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
|---|---|---|---|---|---|---|---|
| 33:03:01~44:03:01 | 375 | 8.1 | 93 | 2.8 | 0.33 (0.26–0.42) | 2.22E-16 | 4.88E-15 |
| 24:02:01~07:02:01 | 270 | 5.8 | 134 | 4.1 | 0.69 (0.55–0.85) | 4.70E-04 | 1.03E-02 |
| 11:01:01~15:01:01 | 130 | 2.8 | 55 | 1.7 | 0.59 (0.42–0.82) | 9.89E-04 | 2.18E-02 |
| 24:02:01~40:02:01 | 82 | 1.8 | 93 | 2.8 | 1.61 (1.18–2.21) | 1.61E-03 | 3.54E-02 |
| C~B | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
| 14:03:01~44:03:01 | 418 | 9.0 | 106 | 3.2 | 0.34 (0.27–0.42) | 2.22E-16 | 3.77E-15 |
| 04:01:01~15:01:01 | 158 | 3.4 | 62 | 1.9 | 0.54 (0.40–0.74) | 4.66E-05 | 7.92E-04 |
| 07:02:01~07:02:01 | 324 | 7.0 | 165 | 5.0 | 0.70 (0.58–0.85) | 3.14E-04 | 5.33E-03 |
| 01:02:01~46:01:01 | 254 | 5.5 | 246 | 7.4 | 1.39 (1.16–1.68) | 3.20E-04 | 5.43E-03 |
| 03:04:01~40:02:01 | 260 | 5.6 | 248 | 7.5 | 1.37 (1.14–1.65) | 5.62E-04 | 9.55E-03 |
| 08:01:01~40:06:01 | 171 | 3.7 | 171 | 5.2 | 1.43 (1.14–1.79) | 1.14E-03 | 1.95E-02 |
| A~C~B | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
| 33:03:01~14:03:01~44:03:01 | 375 | 9.3 | 93 | 3.2 | 0.33 (0.26–0.42) | 2.22E-16 | 4.44E-15 |
| 11:01:01~04:01:01~15:01:01 | 120 | 3.0 | 37 | 1.3 | 0.43 (0.29–0.63) | 4.00E-06 | 8.00E-05 |
| 24:02:01~07:02:01~07:02:01 | 270 | 6.7 | 135 | 4.7 | 0.69 (0.56–0.86) | 5.97E-04 | 1.19E-02 |
| DRB1~DQB1 | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
| 13:02:01~06:04:01 | 388 | 8.3 | 75 | 2.3 | 0.26 (0.20–0.33) | 2.22E-16 | 3.55E-15 |
| 08:03:02~06:01:01 | 385 | 8.3 | 456 | 13.8 | 1.77 (1.53–2.05) | 2.73E-15 | 4.37E-14 |
| 04:05:01~04:01:01 | 617 | 13.3 | 596 | 18.0 | 1.44 (1.27–1.63) | 5.10E-09 | 8.17E-08 |
| 15:01:01~06:02:01 | 342 | 7.3 | 142 | 4.3 | 0.57 (0.46–0.69) | 1.99E-08 | 3.19E-07 |
| 08:02:01~04:02:01 | 85 | 1.8 | 108 | 3.3 | 1.82 (1.35–2.45) | 3.78E-05 | 6.05E-04 |
| 04:06:01~03:02:01 | 170 | 3.7 | 68 | 2.1 | 0.55 (0.41–0.74) | 3.84E-05 | 6.15E-04 |
| 12:01:01~03:01:01 | 151 | 3.2 | 62 | 1.9 | 0.57 (0.42–0.77) | 1.94E-04 | 3.10E-03 |
| 01:01:01~05:01:01 | 336 | 7.2 | 176 | 5.3 | 0.72 (0.60–0.88) | 6.92E-04 | 1.11E-02 |
| DRB1~DQB1~DPB1 | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
| 13:02:01~06:04:01~04:01:01 | 268 | 6.1 | 40 | 1.3 | 0.20 (0.14–0.28) | 2.22E-16 | 4.44E-15 |
| 04:05:01~04:01:01~05:01:01 | 345 | 7.9 | 389 | 12.4 | 1.67 (1.43–1.95) | 3.55E-11 | 7.10E-10 |
| 08:03:02~06:01:01~05:01:01 | 197 | 4.5 | 254 | 8.1 | 1.88 (1.55–2.29) | 5.17E-11 | 1.03E-09 |
| 15:01:01~06:02:01~02:01:02 | 144 | 3.3 | 40 | 1.3 | 0.38 (0.26–0.55) | 3.59E-08 | 7.18E-07 |
| 04:06:01~03:02:01~02:01:02 | 121 | 2.8 | 44 | 1.4 | 0.51 (0.35–0.72) | 9.10E-05 | 1.82E-03 |
| 01:01:01~05:01:01~04:02:01 | 242 | 5.5 | 115 | 3.7 | 0.66 (0.52–0.83) | 2.60E-04 | 5.20E-03 |
| 08:03:02~06:01:01~02:02:01 | 79 | 1.8 | 95 | 3.0 | 1.71 (1.25–2.35) | 4.01E-04 | 8.03E-03 |
| DRB1~DQA1~DQB1 | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
| 13:02:01~01:02:01~06:04:01 | 388 | 8.3 | 75 | 2.3 | 0.26 (0.20–0.33) | 2.22E-16 | 3.33E-15 |
| 08:03:02~01:03:01~06:01:01 | 385 | 8.3 | 456 | 13.8 | 1.77 (1.53–2.05) | 2.73E-15 | 4.10E-14 |
| 04:05:01~03:03:01~04:01:01 | 617 | 13.3 | 595 | 18.0 | 1.44 (1.27–1.63) | 6.30E-09 | 9.44E-08 |
| 15:01:01~01:02:01~06:02:01 | 342 | 7.3 | 142 | 4.3 | 0.57 (0.46–0.69) | 1.99E-08 | 2.99E-07 |
| 04:06:01~03:01:01~03:02:01 | 170 | 3.7 | 68 | 2.1 | 0.55 (0.41–0.74) | 3.84E-05 | 5.77E-04 |
| 08:02:01~04:01:01~04:02:01 | 85 | 1.8 | 107 | 3.2 | 1.80 (1.34–2.43) | 5.25E-05 | 7.88E-04 |
| 01:01:01~01:01:01~05:01:01 | 336 | 7.2 | 176 | 5.3 | 0.72 (0.60–0.88) | 6.92E-04 | 1.04E-02 |
| A~C~B~DRB1~DQA1~DQB1~DPB1 | Control | % | PBC | % | OR (95%CI) | P-value | Pc-value |
| 33:03:01~14:03:01~44:03:01~13:02:01~01:02:01~06:04:01~04:01:01 | 223 | 4.8 | 31 | 0.9 | 0.19 (0.12–0.28) | 2.22E-16 | 1.78E-15 |
| 24:02:01~07:02:01~07:02:01~01:01:01~01:01:01~05:01:01~04:02:01 | 187 | 4.0 | 86 | 2.6 | 0.64 (0.49–0.83) | 6.28E-04 | 5.03E-03 |
| 24:02:01~12:02:02~52:01:01~15:02:01~01:03:01~06:01:01~09:01:01 | 382 | 8.2 | 341 | 10.3 | 1.29 (1.10–1.50) | 1.24E-03 | 9.94E-03 |
OR, odds ratio; 95%CI, 95% confidence interval; Pc-value, multiple testing-corrected p-value; binned, rare HLA alleles with expected count < 5, combined into a common class.
Figure 2.
HLA allele linkage distributions of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, and -DPB1 in Japanese PBC. The heights of the bars show the number of samples carrying the particular HLA alleles, and grey lines connecting orange bars indicate the proportion of linkage between different orange bars. Susceptible and protective HLA haplotypes are highlighted in magenta and yellow, respectively.
Associations of HLA alleles and HLA haplotypes with Japanese PBC-AIH cases
HLA association studies of the 8 HLA genes were undertaken to compare HLA allele frequencies in the 116 PBC-AIH cases as compared to 1,537 non-PBC-AIH cases (Table 4). 7 HLA alleles including HLA-B*59:01:01, HLA-C*15:02:01, HLA-DRB1*04:05:01, HLA-DQB1*04:01:01, HLA-DQB1*06:01:01, HLA-DQA1*01:03:01 and HLA-DQA1*03:03:01 were significantly associated with susceptibility to PBC-AIH after adjusting for multiple corrections. HLA haplotype showed strong DR-DQ associations with HLA-DRB1*04:05:01-HLA-DQB1*04:01:01 and HLA-DRB1*04:05:01-HLA-DQA1*03:03:01-HLA-DQB1*04:01:01 (Table 4).
Table 4.
HLA association analysis and haplotype analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients with AIH and Japanese PBC patients without AIH.
| Locus | Allele | Non-PBC-AIH (2n=3074) | % | PBC-AIH (2n=232) | % | OR (95%CI) | P-value | Pc-value |
|---|---|---|---|---|---|---|---|---|
| B | 59:01:01 | 80 | 2.6 | 14 | 6.0 | 2.40(1.23–4.35) | 2.46E-03 | 3.69E-02 |
| C | 15:02:01 | 92 | 3.0 | 15 | 6.5 | 2.24(1.18–3.97) | 3.95E-03 | 4.35E-02 |
| DRB1 | 04:05:01 | 528 | 17.2 | 68 | 29.6 | 2.02(1.47–2.74) | 2.79E-06 | 3.62E-05 |
| DQB1 | 04:01:01 | 528 | 17.2 | 69 | 29.7 | 2.04(1.49–2.77) | 1.61E-06 | 1.77E-05 |
| DQB1 | 06:01:01 | 873 | 28.4 | 43 | 18.5 | 0.57(0.40–0.81) | 1.21E-03 | 1.33E-02 |
| DQA1 | 01:03:01 | 880 | 28.8 | 44 | 19.0 | 0.58(0.40–0.82) | 1.37E-03 | 1.23E-02 |
| DQA1 | 03:03:01 | 599 | 19.6 | 74 | 31.9 | 1.92(1.42–2.59) | 7.24E-06 | 6.51E-05 |
| Haplotype | Alleles | Non-PBC-AIH (2n=3074) | % | PBC-AIH (2n=232) | % | OR (95%CI) | P-value | Pc-value |
| DRB1~DQB1 | 04:05:01~04:01:01 | 527 | 17.1 | 69 | 29.7 | 2.05(1.5–2.77) | 1.49E-06 | 1.78E-05 |
| DRB1~DQA1~DQB1 | 04:05:01~03:03:01~04:01:01 | 526 | 17.1 | 69 | 29.7 | 2.05(1.5–2.78) | 1.37E-06 | 1.51E-05 |
AIH, autoimmune hepatitis; OR, odds ratio; 95%CI, 95% confidence interval; Pc-value, multiple testing-corrected p-value; NS, not significant; binned, rare HLA alleles with expected count < 5, combined into a common class.
HLA-A*26:01:01 association with late-stage PBC (Scheuer 3 and 4)
HLA association studies of the 8 HLA genes were carried out comparing HLA allele frequencies in the 808 patients in the early-stage PBC group (Scheuer 1 and 2) with the 174 patients in the late-stage PBC group (Scheuer 3 and 4) (Supplementary Table 1). HLA-A*26:01:01 and HLA-B*55:02:01 and HLA-C*03:04:01 were significantly associated with susceptibility to the staging of PBC (Supplementary Table 1). HLA haplotype analysis of HLA class I alleles (Supplementary Table 2) revealed significant HLA haplotype associations with HLA-A*26:01:01-HLA-B*40:02:01 and HLA-A*26:01:01- HLA-C*03:04:01-HLA-B*40:02:01.
HLA-DRB1*09:01:02, HLA-DQB1*03:03:02 and HLA-DQA1*03:02:01 associations with symptomatic PBC in the Japanese population
HLA association studies of the 8 HLA genes were carried out comparing HLA allele frequencies in the 892 asymptomatic PBC patients with the 445 symptomatic PBC patients (s0, s1, s2) (Supplementary Table 3). HLA-DRB1*09:01:02, HLA-DQB1*03:03:02 and HLA-DQA1*03:02:01 were significantly associated with susceptibility to symptomatic PBC after adjusting for multiple comparisons (Supplementary Table 3). Strong HLA haplotype of the significant HLA alleles was confirmed (Supplementary Table 4) with HLA-DRB1*09:01:02-HLA-DQB1*03:03:02 and HLA-DRB1*09:01:02-HLA-DQA1*03:02:01-HLA-DQB1*03:03:02.
No HLA association with PBC with HCC
Supplementary Table 5 presents a comparison of demographic and biochemical values between PBC with HCC and those without HCC. Our analysis found no statistically significant differences between the two groups in the variables examined. HLA association studies of the 8 HLA genes were carried out comparing HLA allele frequencies between the 49 PBC with HCC patients and 1,604 PBC without HCC patients (Supplementary Table 5). No HLA allele remained significant after adjusting for multiple comparisons (Supplementary Table 6). However, when we performed age-matched HLA association testing with 49 PBC with HCC patients and 49 PBC without HCC patients (Supplementary Table 7), HLA-DRB1*09:01:02, HLA-DQB1*03:03:02, HLA-DRB1*09:01:02-HLA-DQB1*03:03:02 remained borderline significant after adjusting for multiple comparisons. HLA-DQB1*03:01 and HLA-DQB1*06:02 are reportedly associated with Taiwanese HCV-HCC (28), while HLA-A*33:03 is reportedly associated with Japanese HBV-HCC (29).
Discussion
Genome-wide association studies (GWAS) of PBC in populations of different backgrounds, such as Japanese (30, 31) and Caucasian (42, 43), have consistently reported that the strongest SNP associations are located in the HLA class II region. HLA gene-targeted studies have confirmed the strong associations of HLA class II alleles such as HLA-DRB1*08:01 for Caucasian PBC(15); HLA-DRB1*08 and HLA-DRB1*02 for Italian PBC(13); HLA-DRB1*08:03, HLA-DQB1*03:01, HLA-DQB1*04:01 and HLA-DQB1*06:04 for Japanese PBC (12); and HLA-DRB1*08:03, HLA-DQB1*03:01 and HLA-DPB1*17:01 for Chinese PBC (44). In this study, we expanded on previous knowledge from 2-field HLA allele associations (12) to 3-field HLA allele associations incorporating all variants in HLA exonic regions. We have extended the previously known HLA allele associations (12) in Japanese PBC from HLA-DRB1*08:03 to HLA-DRB1*08:03:02, HLA-DQB1*03:01 to HLA-DQB1*03:01:01, HLA-DQB1*04:01 to HLA-DQB1*04:01:01 and HLA-DQB1*06:04 to HLA-DQB1*06:04:01 (Table 2).
In addition, with the increase in detection power after extending the size of Japanese PBC and healthy control samples, besides replicating all 18 significant HLA alleles reported by Yasunami et al., (12) we identified an additional significant novel HLA alleles after adjusting for multiple corrections. The strongest associated novel HLA alleles (Table 2) included 3 novel susceptible HLA-DQA1 alleles, HLA-DQA1*03:03:01, HLA-DQA1*04:01:01, HLA-DQA1*01:04:01, and 1 novel protective HLA-DQA1 allele, HLA-DQA*05:05:01. In addition, HLA-A*33:03:01 and HLA-B*44:03:01 were identified as 2 novel protective HLA class I alleles. Both these protective HLA alleles showed strong linkage disequilibrium (LD) with strong HLA haplotypes of HLA-A*33:03:01-HLA-B*44:03:01 (Table 3). This would represent an extension of associations from HLA class II alleles with HLA-A*33:03:01-HLA-C*14:03:01-HLA-B*44:03:01-HLA-DRB1*13:02:01-HLA-DQA1*01:02:01-HLA-DQB1*06:04:01-HLA-DPB1*04:01:01 (Table 3).
Previous studies focusing on AIH identified susceptible HLA allele associations for HLA-DRB1*04:01 and HLA-DRB1*04:05 in a Japanese population (18), HLA-DRB1*03:01 and HLA-DRB1*04:01 in a European population (45), and HLA-DRB1*04:04, -DRB1*04:05 and -DRB1*13:01 in Latin-American populations (21, 46–48). In a European childhood AIH-1 study (49), HLA-DRB1 alleles carriers are reported to associate with higher onset age, higher IgG levels, median smooth muscle antibodies (SMA) level and anti-nuclear antibodies (ANA). The present study of PBC-AIH confirmed the reported HLA associations of HLA-DRB1*04:01 and HLA-DRB1*04:05 with Japanese AIH (18) (Table 4) suggesting that AIH in PBC carries AIH-associated risk HLA alleles.
We have observed similar HLA allele associations between late-stage PBC and symptomatic PBC. HLA-A*26:01:01 (Supplementary Table 1) was associated with late-stage PBC (Scheuer 3 and 4) and although not significant after adjusting for multiple corrections, the same HLA allele is also associated with a higher risk of symptomatic PBC. On the other hand, HLA-DRB1*09:01:02 and HLA-DQB1*03:03:02 (Supplementary Table 3), which were associated with a higher risk of developing symptomatic PBC, shared a similar tendency of associations with late-stage PBC with HLA-DRB1*09:01:02 and HLA-DQB1*03:03:02 (Supplementary Table 1).
We identified that HLA-DPB1*05:01:01 was associated with PBC with HCC, although this association did not remain significant after adjusting for multiple comparisons (Supplementary Table 6). HLA-DPB1*05:01 is reportedly strongly associated with a higher risk of chronic hepatitis B (CHB) in the Japanese population, in which 15–40% of CHB patients eventually develop cirrhosis and/or HCC (50). Although HCC in PBC is rare, studies have suggested that cirrhotic PBC is a rare precursor of HCC development (23–25) and HLA-DPB1*05:01 would represent a shared-risk HLA allele for HCC development in both CHB and PBC.
Among the 140 PBC patients who underwent liver transplantation or died of hepatic failure (end-stage PBC), 92.3% were positive for AMA, while the AMA positivity was 88.2% in the group of 1392 PBC patients who did not progress to end-stage PBC, indicating that AMA positivity is not significantly associated with PBC progression to end-stage. HLA association analysis in PBC patients with end-stage and those without progression to end-stage did not reveal any significant HLA allele associations after multiple corrections. Nevertheless, HLA-DPB1*05:01:01 was marginally associated with an increased risk of liver transplantation or death from liver failure (Supplementary Table 7).
1- and 2-field HLA alleles associations with PBC have been confirmed in multiple population backgrounds. Our results confirmed and extended the current understanding of HLA allele associations by investigating 3-field HLA alleles associations with predisposition risk, staging, symptomatic status, AIH and HCC events for PBC patients in the Japanese population.
Data availability statement
The GWAS data summary statistics are deposited in the National Bioscience Database Center (NBDC): https://humandbs.biosciencedbc.jp/hum0261-v2.
Ethics statement
The studies involving human participants were reviewed and approved by Japanese National Hospital Organization Study Group for Liver Disease in Japan (NHOSLJ). The patients/participants provided their written informed consent to participate in this study.
Author contributions
S-SK, MiN, KT conceived and planned the experiments. YA and MiN contributed to sample preparation. S-SK, KU, NN, MK, YK, YH and MaN contributed to interpretation of the results. S-SK took the lead in writing the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
This work was supported by the Japan Agency for Medical Research and Development under grant number JP22fk0210111 and 20K08370, Grants-in-Aid for Scientific Research from the Japan Society for Promotion of Science (JSPS) to MN (#20590800, #23591006, #26293181), a Grant-in-Aid for Clinical Research from the National Hospital Organization (NHO) to MN. Part of the figure generations was created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1151502/full#supplementary-material
Association analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients with Scheuer Stage 0–2 and Japanese PBC patients with Scheuer Stage 3–4. Abbreviations: OR = odd ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class.
HLA haplotype analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients with Scheuer Stage 0–2 and Japanese PBC patients with Scheuer Stage 3–4. Abbreviations: OR = odds ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class.
Association analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in asymptomatic Japanese PBC patients and s0, s1 and s2 Japanese PBC patients. Abbreviations: OR = odds ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class; aPBC = asymptomatic PBC (i.e., without itching, without varices, without ascites, without encephalopathy, without jaundice); s0 = positive symptoms of portal hypertension such as esophageal varices; s1 = positive itching; s2 = positive jaundice (T.bil. > 2.5 mg/dl).
HLA haplotype analysis of HLA-DRB1, -DQA1 and -DQB1 in asymptomatic Japanese PBC patients and s0, s1 and s2 Japanese PBC patients. Abbreviations: OR = odds ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class.
Demographic and biochemical values of PBC with HCC versus PBC without HCC. Abbreviations: ALT: alanine transaminase, ALP: alkaline phosphatase
References
- 1. Poupon R. Primary biliary cirrhosis: a 2010 update. J Hepatol (2010) 52(5):745–58. doi: 10.1016/j.jhep.2009.11.027 [DOI] [PubMed] [Google Scholar]
- 2. Lleo A, Wang GQ, Gershwin ME, Hirschfield GM. Primary biliary cholangitis. Lancet (2020) 396(10266):1915–26. doi: 10.1016/S0140-6736(20)31607-X [DOI] [PubMed] [Google Scholar]
- 3. Huet PM, Deslauriers J, Tran A, Faucher C, Charbonneau J. Impact of fatigue on the quality of life of patients with primary biliary cirrhosis. Am J Gastroenterol (2000) 95(3):760–7. doi: 10.1111/j.1572-0241.2000.01857.x [DOI] [PubMed] [Google Scholar]
- 4. Selmi C, Bowlus CL, Gershwin ME, Coppel RL. Primary biliary cirrhosis. Lancet (2011) 377(9777):1600–9. doi: 10.1016/S0140-6736(10)61965-4 [DOI] [PubMed] [Google Scholar]
- 5. Sherlock S, Dooley J. Diseases of the liver and biliary system. John Wiley & Sons; (2008). [Google Scholar]
- 6. Reshetnyak VI. Primary biliary cirrhosis: clinical and laboratory criteria for its diagnosis. World J Gastroenterol (2015) 21(25):7683–708. doi: 10.3748/wjg.v21.i25.7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Invernizzi P, Selmi C, Gershwin ME. Update on primary biliary cirrhosis. Dig Liver Dis (2010) 42(6):401–8. doi: 10.1016/j.dld.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cordell HJ, Fryett JJ, Ueno K, Darlay R, Aiba Y, Hitomi Y, et al. An international genome-wide meta-analysis of primary biliary cholangitis: novel risk loci and candidate drugs. J Hepatol (2021) 75(3):572–81. doi: 10.1016/j.jhep.2021.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cordell HJ, Fryett JJ, Ueno K, Darlay R, Aiba Y, Hitomi Y, et al. Corrigendum to 'An international genome-wide meta-analysis of primary biliary cholangitis: novel risk loci and candidate drugs' [J hepatol 2021;75(3):572-581]. J Hepatol (2022) 76(2):489. doi: 10.1016/j.jhep.2021.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Onishi S, Sakamaki T, Maeda T, Iwamura S, Tomita A, Saibara T, et al. DNA Typing of HLA class II genes; DRB1*0803 increases the susceptibility of Japanese to primary biliary cirrhosis. J Hepatol (1994) 21(6):1053–60. doi: 10.1016/s0168-8278(05)80617-8 [DOI] [PubMed] [Google Scholar]
- 11. Nakamura M, Yasunami M, Kondo H, Horie H, Aiba Y, Komori A, et al. Analysis of HLA-DRB1 polymorphisms in Japanese patients with primary biliary cirrhosis (PBC): the HLA-DRB1polymorphism determines the relative risk of antinuclear antibodies for disease progression in PBC. Hepatol Res (2010) 40(5):494–504. doi: 10.1111/j.1872-034X.2010.00631.x [DOI] [PubMed] [Google Scholar]
- 12. Yasunami M, Nakamura H, Tokunaga K, Kawashima M, Nishida N, Hitomi Y, et al. Principal contribution of HLA-DQ alleles, DQB1*06:04 and DQB1*03:01, to disease resistance against primary biliary cholangitis in a Japanese population. Sci Rep (2017) 7(1):11093. doi: 10.1038/s41598-017-11148-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Invernizzi P, Selmi C, Poli F, Frison S, Floreani A, Alvaro D, et al. Human leukocyte antigen polymorphisms in Italian primary biliary cirrhosis: a multicenter study of 664 patients and 1992 healthy controls. Hepatology (2008) 48(6):1906–12. doi: 10.1002/hep.22567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Begovich AB, Klitz W, Moonsamy PV, Van de Water J, Peltz G, Gershwin ME. Genes within the HLA class II region confer both predisposition and resistance to primary biliary cirrhosis. Tissue Antigens (1994) 43(2):71–7. doi: 10.1111/j.1399-0039.1994.tb02303.x [DOI] [PubMed] [Google Scholar]
- 15. Donaldson PT, Baragiotta A, Heneghan MA, Floreani A, Venturi C, Underhill JA, et al. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: a large-scale study. Hepatology (2006) 44(3):667–74. doi: 10.1002/hep.21316 [DOI] [PubMed] [Google Scholar]
- 16. Heneghan MA, Yeoman AD, Verma S, Smith AD, Longhi MS. Autoimmune hepatitis. Lancet (2013) 382(9902):1433–44. doi: 10.1016/S0140-6736(12)62163-1 [DOI] [PubMed] [Google Scholar]
- 17. Tanaka A, Mori M, Matsumoto K, Ohira H, Tazuma S, Takikawa H. Increase trend in the prevalence and male-to-female ratio of primary biliary cholangitis, autoimmune hepatitis, and primary sclerosing cholangitis in Japan. Hepatol Res (2019) 49(8):881–9. doi: 10.1111/hepr.13342 [DOI] [PubMed] [Google Scholar]
- 18. Oka S, Furukawa H, Yasunami M, Kawasaki A, Nakamura H, Nakamura M, et al. HLA-DRB1 and DQB1 alleles in Japanese type 1 autoimmune hepatitis: the predisposing role of the DR4/DR8 heterozygous genotype. PloS One (2017) 12(10):e0187325. doi: 10.1371/journal.pone.0187325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Umemura T, Katsuyama Y, Yoshizawa K, Kimura T, Joshita S, Komatsu M, et al. Human leukocyte antigen class II haplotypes affect clinical characteristics and progression of type 1 autoimmune hepatitis in Japan. PloS One (2014) 9(6):e100565. doi: 10.1371/journal.pone.0100565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lim YS, Oh HB, Choi SE, Kwon OJ, Heo YS, Lee HC, et al. Susceptibility to type 1 autoimmune hepatitis is associated with shared amino acid sequences at positions 70-74 of the HLA-DRB1 molecule. J Hepatol (2008) 48(1):133–9. doi: 10.1016/j.jhep.2007.08.019 [DOI] [PubMed] [Google Scholar]
- 21. Duarte-Rey C, Pardo AL, Rodriguez-Velosa Y, Mantilla RD, Anaya JM, Rojas-Villarraga A. HLA class II association with autoimmune hepatitis in Latin America: a meta-analysis. Autoimmun Rev (2009) 8(4):325–31. doi: 10.1016/j.autrev.2008.11.005 [DOI] [PubMed] [Google Scholar]
- 22. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers (2021) 7(1):6. doi: 10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 23. Villa E, Baldini GM, Pasquinelli C, Melegari M, Cariani E, Di Chirico G, et al. Risk factors for hepatocellular carcinoma in italy. Male sex, hepatitis b virus, non-a non-b infection, and alcohol. Cancer (1988) 62(3):611–5. doi: [DOI] [PubMed] [Google Scholar]
- 24. De Bac C, Stroffolini T, Gaeta GB, Taliani G, Giusti G. Pathogenic factors in cirrhosis with and without hepatocellular carcinoma: a multicenter Italian study. Hepatology (1994) 20(5):1225–30. doi: 10.1016/0270-9139(94)90761-7 [DOI] [PubMed] [Google Scholar]
- 25. Kaczynski J, Hansson G, Wallerstedt S. Incidence of primary liver cancer and aetiological aspects: a study of a defined population from a low-endemicity area. Br J Cancer (1996) 73(1):128–32. doi: 10.1038/bjc.1996.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones DE, Metcalf JV, Collier JD, Bassendine MF, James OF. Hepatocellular carcinoma in primary biliary cirrhosis and its impact on outcomes. Hepatology (1997) 26(5):1138–42. doi: 10.1002/hep.510260508 [DOI] [PubMed] [Google Scholar]
- 27. Lin ZH, Xin YN, Dong QJ, Wang Q, Jiang XJ, Zhan SH, et al. Association between HLA-DRB1 alleles polymorphism and hepatocellular carcinoma: a meta-analysis. BMC Gastroenterol (2010) 10:145. doi: 10.1186/1471-230X-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee MH, Huang YH, Chen HY, Khor CS, Chang YH, Lin YJ, et al. Human leukocyte antigen variants and risk of hepatocellular carcinoma modified by HCV genotypes: a genome-wide association study. Hepatology (2018) 67(2):651–61. doi: 10.1002/hep.29531 [DOI] [PubMed] [Google Scholar]
- 29. Sawai H, Nishida N, Khor SS, Honda M, Sugiyama M, Baba N, et al. Genome-wide association study identified new susceptible genetic variants in HLA class I region for hepatitis b virus-related hepatocellular carcinoma. Sci Rep (2018) 8(1):7958. doi: 10.1038/s41598-018-26217-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet (2012) 91(4):721–8. doi: 10.1016/j.ajhg.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawashima M, Hitomi Y, Aiba Y, Nishida N, Kojima K, Kawai Y, et al. Genome-wide association studies identify PRKCB as a novel genetic susceptibility locus for primary biliary cholangitis in the Japanese population. Hum Mol Genet (2017) 26(3):650–9. doi: 10.1093/hmg/ddw406 [DOI] [PubMed] [Google Scholar]
- 32. Kawai Y, Mimori T, Kojima K, Nariai N, Danjoh I, Saito R, et al. Japonica array: improved genotype imputation by designing a population-specific SNP array with 1070 Japanese individuals. J Hum Genet (2015) 60(10):581–7. doi: 10.1038/jhg.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hitomi Y, Ueno K, Kawai Y, Nishida N, Kojima K, Kawashima M, et al. POGLUT1, the putative effector gene driven by rs2293370 in primary biliary cholangitis susceptibility locus chromosome 3q13.33. Sci Rep (2019) 9(1):102. doi: 10.1038/s41598-018-36490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet (2007) 81(3):559–75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scheuer PJ, Standish RA, Dhillon AP. Scoring of chronic hepatitis. Clin Liver Dis (2002) 6(2):335–347, v-vi. doi: 10.1016/s1089-3261(02)00009-0 [DOI] [PubMed] [Google Scholar]
- 36. Poupon R. Autoimmune overlapping syndromes. Clin Liver Dis (2003) 7(4):865–78. doi: 10.1016/s1089-3261(03)00092-8 [DOI] [PubMed] [Google Scholar]
- 37. Nakamura M, Kondo H, Tanaka A, Komori A, Ito M, Yamamoto K, et al. Autoantibody status and histological variables influence biochemical response to treatment and long-term outcomes in Japanese patients with primary biliary cirrhosis. Hepatol Res (2015) 45(8):846–55. doi: 10.1111/hepr.12423 [DOI] [PubMed] [Google Scholar]
- 38. Nakamura M, Kondo H, Mori T, Komori A, Matsuyama M, Ito M, et al. Anti-gp210 and anti-centromere antibodies are different risk factors for the progression of primary biliary cirrhosis. Hepatology (2007) 45(1):118–27. doi: 10.1002/hep.21472 [DOI] [PubMed] [Google Scholar]
- 39. Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, et al. HIBAG–HLA genotype imputation with attribute bagging. Pharmacogenomics J (2014) 14(2):192–200. doi: 10.1038/tpj.2013.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khor SS, Yang W, Kawashima M, Kamitsuji S, Zheng X, Nishida N, et al. High-accuracy imputation for HLA class I and II genes based on high-resolution SNP data of population-specific references. Pharmacogenomics J (2015) 15(6):530–7. doi: 10.1038/tpj.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pappas DJ, Marin W, Hollenbach JA, Mack SJ. Bridging ImmunoGenomic data analysis workflow gaps (BIGDAWG): an integrated case-control analysis pipeline. Hum Immunol (2016) 77(3):283–7. doi: 10.1016/j.humimm.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med (2009) 360(24):2544–55. doi: 10.1056/NEJMoa0810440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet (2011) 43(4):329–32. doi: 10.1038/ng.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang C, Zheng X, Tang R, Han C, Jiang Y, Wu J, et al. Fine mapping of the MHC region identifies major independent variants associated with han Chinese primary biliary cholangitis. J Autoimmun (2020) 107:102372. doi: 10.1016/j.jaut.2019.102372 [DOI] [PubMed] [Google Scholar]
- 45. Strettell MD, Donaldson PT, Thomson LJ, Santrach PJ, Moore SB, Czaja AJ, et al. Allelic basis for HLA-encoded susceptibility to type 1 autoimmune hepatitis. Gastroenterology (1997) 112(6):2028–35. doi: 10.1053/gast.1997.v112.pm9178696 [DOI] [PubMed] [Google Scholar]
- 46. Vazquez-Garcia MN, Alaez C, Olivo A, Debaz H, Perez-Luque E, Burguete A, et al. MHC class II sequences of susceptibility and protection in mexicans with autoimmune hepatitis. J Hepatol (1998) 28(6):985–90. doi: 10.1016/s0168-8278(98)80347-4 [DOI] [PubMed] [Google Scholar]
- 47. Goldberg AC, Bittencourt PL, Mougin B, Cancado EL, Porta G, Carrilho F, et al. Analysis of HLA haplotypes in autoimmune hepatitis type 1: identifying the major susceptibility locus. Hum Immunol (2001) 62(2):165–9. doi: 10.1016/s0198-8859(00)00234-2 [DOI] [PubMed] [Google Scholar]
- 48. Fortes Mdel P, Machado IV, Gil G, Fernandez-Mestre M, Dagher L, Leon RV, et al. Genetic contribution of major histocompatibility complex class II region to type 1 autoimmune hepatitis susceptibility in Venezuela. Liver Int (2007) 27(10):1409–16. doi: 10.1111/j.1478-3231.2007.01581.x [DOI] [PubMed] [Google Scholar]
- 49. Ma Y, Su H, Yuksel M, Longhi MS, McPhail MJ, Wang P, et al. Human leukocyte antigen profile predicts severity of autoimmune liver disease in children of European ancestry. Hepatology (2021) 74(4):2032–46. doi: 10.1002/hep.31893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lai CL, Yuen MF. The natural history and treatment of chronic hepatitis b: a critical evaluation of standard treatment criteria and end points. Ann Intern Med (2007) 147(1):58–61. doi: 10.7326/0003-4819-147-1-200707030-00010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Association analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients with Scheuer Stage 0–2 and Japanese PBC patients with Scheuer Stage 3–4. Abbreviations: OR = odd ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class.
HLA haplotype analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in Japanese PBC patients with Scheuer Stage 0–2 and Japanese PBC patients with Scheuer Stage 3–4. Abbreviations: OR = odds ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class.
Association analysis of HLA-A, -C, -B, -DRB1, -DQA1, -DQB1, -DPA1 and -DPB1 in asymptomatic Japanese PBC patients and s0, s1 and s2 Japanese PBC patients. Abbreviations: OR = odds ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class; aPBC = asymptomatic PBC (i.e., without itching, without varices, without ascites, without encephalopathy, without jaundice); s0 = positive symptoms of portal hypertension such as esophageal varices; s1 = positive itching; s2 = positive jaundice (T.bil. > 2.5 mg/dl).
HLA haplotype analysis of HLA-DRB1, -DQA1 and -DQB1 in asymptomatic Japanese PBC patients and s0, s1 and s2 Japanese PBC patients. Abbreviations: OR = odds ratio; 95%CI = 95% confidence interval; Pc-value = multiple testing-corrected p-value; binned = rare HLA alleles with expected count < 5, combined into a common class.
Demographic and biochemical values of PBC with HCC versus PBC without HCC. Abbreviations: ALT: alanine transaminase, ALP: alkaline phosphatase
Data Availability Statement
The GWAS data summary statistics are deposited in the National Bioscience Database Center (NBDC): https://humandbs.biosciencedbc.jp/hum0261-v2.