Abstract
Observational studies have identified the potential prognostic value for severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) viral load and anti‐SARS‐CoV‐2 antibodies in coronavirus disease 2019 (COVID‐19). However, viral load in nasopharyngeal (NP) swabs produced inconsistent results in prognostic analyses, and the prognostic value of viral load or antibodies has not been confirmed in large clinical trials. COVACTA and REMDACTA were double‐blind, randomized, controlled trials with a combined enrollment of 1078 patients hospitalized with COVID‐19 treated with tocilizumab or placebo in COVACTA or tocilizumab plus remdesivir or placebo plus remdesivir in REMDACTA. We assessed the potential prognostic value of NP and serum SARS‐CoV‐2 viral load and serum anti‐SARS‐CoV‐2 antibodies at baseline as biomarkers for clinical outcomes in patients enrolled in these trials. In adjusted Cox proportional hazard models, serum viral load was a more reliable predictor of clinical outcomes than NP viral load; high serum viral load was associated with higher risk for death and mechanical ventilation/death and lower likelihood of hospital discharge (high vs. negative viral load hazard ratios [95% confidence interval {CI}] were 2.87 [1.57–5.25], 3.86 [2.23–6.68], and 0.23 [0.14–0.36], respectively, in COVACTA and 8.11 [2.95–22.26], 10.29 [4.5–23.55], and 0.21 [0.15–0.29], respectively, in REMDACTA) and high serum viral load correlated with levels of inflammatory cytokines and lung damage biomarkers. High anti‐SARS‐CoV‐2 spike protein antibody (ACOV2S) levels were associated with higher likelihood of hospital discharge (high vs. below the limit of quantification hazard ratios [95% CI] were 2.55 [1.59–4.08] for COVACTA and 1.54 [1.13–2.09] for REMDACTA). These results support the role of baseline SARS‐CoV‐2 serum viral load and ACOV2S antibody titers in predicting clinical outcomes for patients hospitalized with COVID‐19.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Observational studies have identified relationships between severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) viral load, particularly in serum, and humoral responses and severity of outcomes in patients with coronavirus disease 2019 (COVID‐19). However, there are few studies from large clinical trials that explore potential prognostic biomarkers for clinical outcomes in patients hospitalized with COVID‐19.
WHAT QUESTION DID THIS STUDY ADDRESS?
Biomarkers to predict COVID‐19 outcomes are needed to identify high‐risk patients and inform on appropriate treatment approaches.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study of data from two clinical trials of more than 1000 patients across a range of disease severity confirms that serum SARS‐CoV‐2 load has the strongest prognostic value for clinical outcomes and suggests a protective role of anti‐SARS‐CoV‐2 antibodies against viral replication and lung tissue damage with prognostic value for better outcomes.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Biomarkers to identify patients at greatest risk for prolonged hospital stay and progression to mechanical ventilation or death may aid clinical trial design and stratification of treatment approaches in patients hospitalized with COVID‐19.
INTRODUCTION
Most individuals infected with severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) experience mild coronavirus disease 2019 (COVID‐19) symptoms; however, some develop severe disease that necessitates hospitalization and can lead to death. 1 Understanding early risk factors for the development of severe COVID‐19 is essential to identifying appropriate treatments for high‐risk patients to prevent disease progression. Observational studies have identified relationships among SARS‐CoV‐2 viral load, humoral response, and severity of COVID‐19. The value of SARS‐CoV‐2 viral load measured in the nasopharynx for predicting outcomes is uncertain, with some studies showing a relationship between viral load and disease severity 2 , 3 , 4 and others finding no clear association. 5 , 6 SARS‐CoV‐2 viral load in the blood (viremia) is associated with disease severity and mortality and can predict disease outcomes in patients hospitalized with COVID‐19. 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 Furthermore, SARS‐CoV‐2 viremia is associated with upregulation of proteomic signatures indicative of dysregulated inflammatory responses, lung and systemic tissue damage, and tissue fibrosis and repair. 10 , 11 , 12 , 13 , 20 , 21
The humoral immune response to SARS‐CoV‐2 is vital for viral clearance. Antibodies against the SARS‐CoV‐2 spike protein (ACOV2S) are the dominant neutralizing antibody. In hospitalized patients, ACOV2S levels are negatively associated with inflammatory markers in the blood among those who survive to discharge. 22 The time course of antibody generation may be a defining factor between patients with COVID‐19 who survive compared with nonsurvivors who have delayed antibody production despite achieving higher antibody levels later in the disease course. 22 , 23 , 24 Data from large clinical trials with well‐characterized patient populations may advance understanding of the prognostic value of SARS‐CoV‐2 viral load and antibodies on clinical outcomes and inform treatment decisions.
COVACTA and REMDACTA were double‐blind, randomized controlled trials conducted in patients hospitalized with COVID‐19 pneumonia with a combined enrollment of more than 1000 patients. In COVACTA, patients with severe COVID‐19 pneumonia received the anti‐interleukin‐6 receptor alpha antibody, tocilizumab, or placebo and were followed for 60 days. 25 Neither SARS‐CoV‐2 viral clearance measured in nasopharyngeal (NP) swab and serum samples nor the development of anti‐SARS‐CoV‐2 antibodies were affected by tocilizumab treatment in COVACTA. 26 In REMDACTA, patients with severe COVID‐19 pneumonia were treated with tocilizumab plus the antiviral agent, remdesivir, or placebo plus remdesivir. 27 Data collected before study drug administration in these trials provide an opportunity to assess the potential prognostic value of biomarkers, such as SARS‐CoV‐2 titers, and serological responses to SARS‐CoV‐2 in relation to disease outcomes in large, well‐characterized, prospective cohorts of patients hospitalized with severe COVID‐19 pneumonia.
METHODS
Patients
The COVACTA and REMDACTA patient populations and study designs are published. 25 , 27 Briefly, COVACTA (Clinicaltrials.gov NCT04320615) enrolled patients greater than or equal to 18 years of age hospitalized with COVID‐19 pneumonia confirmed by a positive SARS‐CoV‐2 polymerase chain reaction (PCR) test result within 7 days of randomization and evidenced on x‐ray or computed tomography (CT) scans. Eligible patients had blood oxygen saturation less than or equal to 93% or partial pressure of oxygen/fraction of inspired oxygen less than 300 mm Hg. 25 REMDACTA (Clinicaltrials.gov NCT04409262) enrollment criteria included patients greater than or equal to 12 years of age (no patients aged <18 years were enrolled) hospitalized with COVID‐19 pneumonia, confirmed by a positive SARS‐CoV‐2 PCR test result and chest x‐ray or CT scan, and hypoxemia requiring greater than 6 L/min supplemental oxygen. 27 Patients had not received SARS‐CoV‐2 vaccination.
COVACTA and REMDACTA were conducted in accordance with Good Clinical Practice guidelines of the International Council for Harmonization E6 and the Declaration of Helsinki or local laws and regulations, whichever afforded greater protection. Informed consent was obtained from the patients or their legally authorized representatives. The studies were approved by institutional review boards or ethics committees at each site.
SARS‐CoV‐2 virology, serology, protein biomarkers, and disease severity
Detailed methods regarding virology, serology, protein biomarker assays, and disease severity categories are described in the Supplementary Material.
Statistical analysis
All patients from the modified intention‐to‐treat (mITT) populations in the study treatment and placebo arms who had at least one virology and/or serology measurement were included in this analysis except for one patient from COVACTA who died on day 1. All analyses were conducted using imputed values for results that were negative or below the limit of quantification (BLOQ). For serum and NP PCR, negative results were imputed as half of the BLOQ threshold (PCR = 60), and BLOQ results were imputed as the BLOQ threshold minus 1 (120–1 = 119). For neutralizing antibody titer results, negative values were imputed as eight, BLOQ as 20, and above upper limit of quantification as 3973 (1 patient). For ACOV2S, BLOQ result (<0.43) was imputed as 0.4, and results reported as greater than 250.0 (6 patients in COVACTA, and 1 in REMDACTA) were assigned missing values. Analyses were performed based on log‐transformed values (continuous virology/log10 for PCR variables, log2 for neutralizing antibody titers, and natural log for ACOV2S), unless otherwise specified. For some analyses, patients were grouped into negative, BLOQ, low, and high groups (viral load variables and neutralizing antibody) or BLOQ, low, and high groups (ACOV2S). Low and high subgroups were defined as below or above the median, respectively, where the median was calculated using the within‐study distribution of nonimputed readings. As a sensitivity analysis, patients were grouped into high and low groups based on medians calculated from combining distributions from both studies. Patterns of missingness were investigated by comparing patients with missing and nonmissing values for virology and serology variables according to baseline demographics and disease characteristics. There was no evidence of informative missingness.
Continuous variables were described using minimum, maximum, mean, standard deviation, and median; percentages were calculated for categorical variables. Spearman correlation coefficients were used to describe the relationship between continuous and/or ordinal scale variables, with correlations zero to less than 0.2 regarded as very weak, 0.2 to less than 0.4 as weak, 0.4 to less than 0.6 as moderate, 0.6 to less than 0.8 as strong, and 0.8 to 1 as very strong. 28 The relationship between continuous and binary variables was tested using the Wilcoxon rank sum test.
Potential prognostic value of NP or serum viral load and antibody levels were investigated by examining associations between baseline viral load or antibody levels and clinical outcomes up to day 28 or day 60 (time to death, time to hospital discharge, time to mechanical ventilation, or death). The prognostic value of biomarkers was tested in Cox proportional hazard models using the same time to event analysis approaches, particularly censoring rules, as those used in COVACTA and REMDACTA. 25 , 27 All models included a continuous biomarker variable (PCR or antibody parameter) and were controlled for treatment arm, age as a continuous variable, stratification factors (geographic region and mechanical ventilation in COVACTA; geographic region and baseline ordinal scale Table S1 in REMDACTA), and, for COVACTA, baseline ordinal scale score of clinical status. Presented p values are for the model with a continuous biomarker term. Hazard ratios and 95% CI estimates comparing each biomarker subgroup with the negative subgroup (BLOQ for ACOV2S) were calculated and plotted as forest plots. In addition, sensitivity analyses were performed to ensure that the prognostic value estimated in the main model was consistent with that of each treatment arm and in patients who had less than or equal to 7 days from symptom onset versus greater than 7 days from symptom onset at baseline.
All analyses were exploratory. Evidence of prognostic value was declared if the continuous biomarker term in the adjusted model had a p value < 0.05, p values were not corrected for multiple comparisons. Analyses were performed using R version 4.0.3.
RESULTS
Patients
In COVACTA, 438 patients were treated and included in the mITT population. In REMDACTA, 640 patients were included in the mITT population. Baseline NP swab samples for virologic testing were available for 389 patients (88.8%) from COVACTA and 605 patients (94.5%) from REMDACTA and serological testing samples were available for 423 (96.6%) and 608 patients (95.0%), respectively.
Demographic characteristics were relatively similar between the two studies—both enrolled patients who were primarily men and white with mean ages of ~60 years (Table S2). A higher proportion of patients in REMDACTA (84.3%) than COVACTA (22.4%) were receiving corticosteroids at baseline. In REMDACTA, 79.8% of patients had a baseline ordinal status score of 4 and 6.6% had a score of 3, whereas in COVACTA, 30.4% of patients had a score of 4 and 27.9% had a score of 3. Approximately 20% of patients in REMDACTA had received one or two doses of remdesivir before baseline. Additional details are published. 25 , 27
At baseline, 349 of 389 patients (89.7%) in COVACTA and 583 of 605 patients (96.4%) in REMDACTA had a positive reverse transcription quantitative PCR (RT‐qPCR) result based on NP samples and 300 of 423 (70.9%) and 532 of 608 patients (87.5%), respectively, had a positive result based on serum samples (Table S2). Antibodies were present in most patients at the time of enrollment; similar proportions of patients in both studies were positive for neutralizing antibodies (COVACTA, 303/344 [88.1%]; REMDACTA, 549/610 [90.0%]) or ACOV2S antibodies (COVACTA, 305/359 [85.0%]; REMDACTA, 504/603 [83.6%]) at baseline.
Characterization of virology and serology
The median time from symptom onset to baseline was 11 days (range: 1–50 days) in COVACTA and 8 days (range: 0–43 days) in REMDACTA. Although there was a wide range in individual viral loads in NP swabs and serum samples, a trend for higher viral loads in NP swabs and serum was observed in baseline samples collected from patients ~5–7 days after symptom onset (Figure 1a,b). Likewise, although individual levels of neutralizing and ACOV2S antibodies at baseline varied, the highest antibody levels were detected in baseline samples collected after ~10 days following symptom onset (Figure 1c,d).
FIGURE 1.
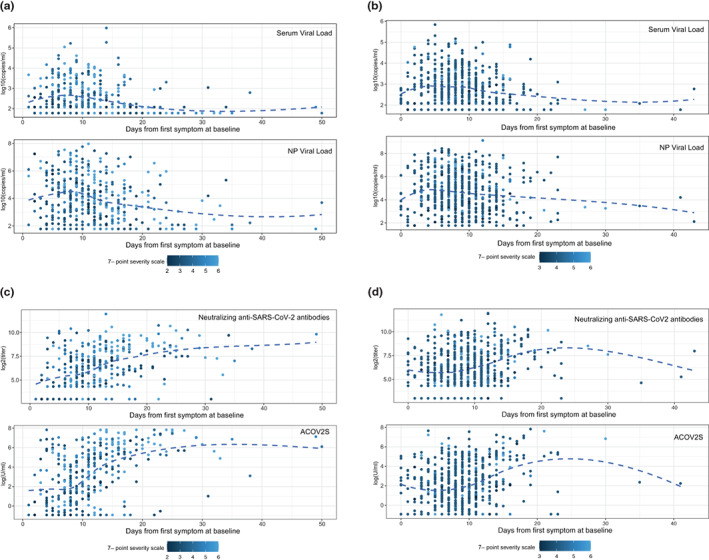
Relationship of viral load and antibodies with time from symptom onset. Baseline SARS‐CoV‐2 viral load was measured by RT‐qPCR from serum samples and NP swabs in (a) COVACTA and (b) REMDACTA. Baseline neutralizing antibodies were measured by PRNT80 and ASCOV2S was measured by immunoassay in (c) COVACTA and (d) REMDACTA. Each dot represents a sample from one patient collected at baseline and is colored according to clinical status on the 7‐point ordinal score at baseline. The dashed line is the locally weighted scatterplot smoothing (LOWESS) curve. Ab, antibodies; ASCOV2S, anti‐SARS‐CoV‐Spike; NP, nasopharyngeal; PRNT, plaque reduction neutralization test; RT‐qPCR, reverse transcriptase‐quantitative polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Weak‐to‐moderate positive correlations were observed between viral loads in NP swabs and serum (Spearman correlations: 0.41 for COVACTA and 0.37 for REMDACTA; both p < 0.01; Figure 2). Strong‐to‐very‐strong positive correlations were observed between neutralizing antibodies and ACOV2S (Spearman correlations: 0.87 for COVACTA and 0.76 for REMDACTA; both p < 0.01), reflecting that ACOV2S correlate with antibodies that are mostly neutralizing. Weak‐to‐moderate negative correlations were observed between viral load (NP swab or serum) and neutralizing or ACOV2S antibodies (Spearman correlations for ACOV2S: −0.53 in NP swab and − 0.36 in serum for COVACTA and − 0.40 in NP swab and − 0.35 in serum for REMDACTA; all p < 0.01).
FIGURE 2.
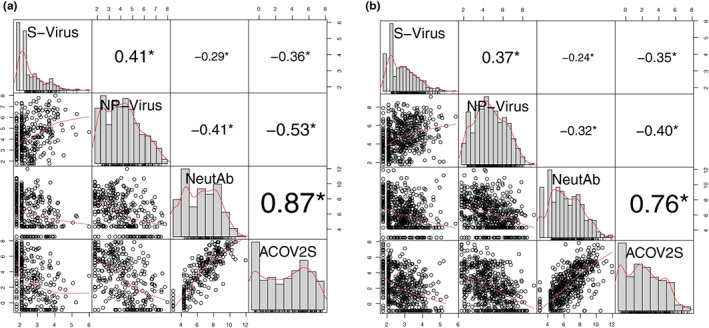
Correlation between viral load and antibodies. (a) COVACTA, (b) REMDACTA. SARS‐CoV‐2 viral load was measured by RT‐qPCR from serum samples and NP swabs. Baseline neutralizing antibodies were measured by PRNT80 and ASCOV2S was measured by immunoassay. Each dot represents a collected sample from 1 patient. Red lines are LOWNESS curves. *p < 0.01. ASCOV2S, anti‐SARS‐CoV‐Spike; NeutAb, neutralizing antibodies; NP‐virus, nasopharyngeal swab viral load; PRNT, plaque reduction neutralization test; RT‐qPCR, reverse transcriptase‐quantitative polymerase chain reaction; S‐virus, serum viral load; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Correlation between virology/serology and patient characteristics
The relative distribution of patients across serum viral load, NP viral load, neutralizing antibodies, and ACOV2S subgroups was evaluated for demographic factors known to be prognostic for COVID‐19 severity (age, sex, race, ethnicity, hypertension, and diabetes) and patient characteristics relevant for the studies included (geographic region, baseline treatment with corticosteroids, baseline treatment with remdesivir, baseline ordinal severity score, and number of days from symptom onset). Across both studies, patients grouped by baseline ordinal severity score, age, and time from symptom onset had significantly different distributions across one or more virology or serology subgroups (Table S3). Although ethnicity was statistically significant across both studies for neutralizing antibodies, the directional trends were opposite.
In REMDACTA, viral loads in serum and NP swabs were statistically significantly higher in patients with worse ordinal status (high indicates worse disease) at baseline according to the seven‐category ordinal scale; this was less pronounced but statistically significant in COVACTA (Figure 3a,b). Serum and NP viral loads appeared to be higher in patients from REMDACTA than those from COVACTA, which may be due to a shorter median time to symptom onset in REMDACTA (Table S2). In both studies, higher titers of neutralizing antibodies and ACOV2S were observed in patients with worse ordinal status at baseline (Figure 3c,d). However, ACOV2S titers were generally higher in patients from COVACTA than REMDACTA across all ordinal scale categories. Subgroup analysis of ACOV2S titers according to clinical status at baseline in patients from COVACTA suggested that lower ACOV2S titers may have been associated with shorter time to death and longer time to hospital discharge in patients with severe disease (ordinal scale score ≥4 [requiring at least noninvasive ventilation or high‐flow oxygen] at baseline; n = 300) but not in patients with more moderate disease (ordinal scale score <4 at baseline; n = 137; Figure S1); however, conclusions from this comparison are limited by the small number of events in moderate disease subgroups. Similar analysis was not performed for REMDACTA because there were only 42 patients with ordinal scale score less than 4 compared to 598 with a score greater than or equal to 4.
FIGURE 3.
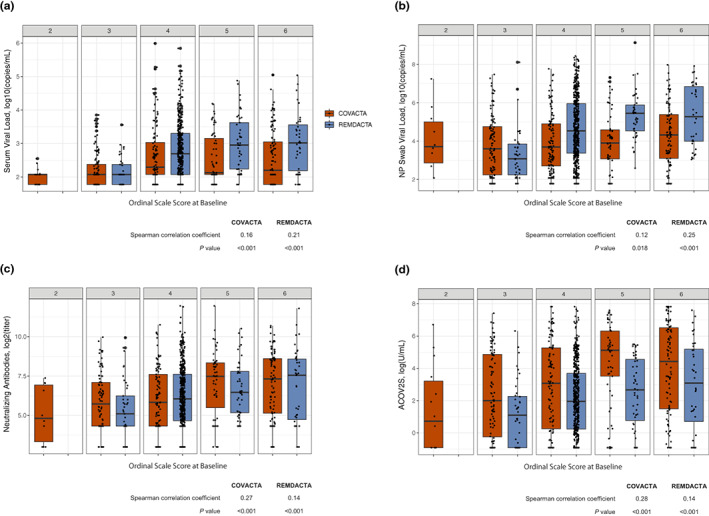
Correlation of viral load and antibodies by clinical status. Correlation of clinical status (assessed by the 7‐category ordinal scale at baseline) with (a) serum viral load, (b) NP swab viral load, (c) neutralizing antibodies, and (d) ASCOV2S. ASCOV2S, anti‐SARS‐CoV‐2‐Spike; NP, nasopharyngeal; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Baseline NP swab viral loads were positively correlated with age group in both studies, with patients aged 65 years or older demonstrating higher NP viral loads than patients aged 18–64 years (Figure S2). In REMDACTA, a similar trend was observed between serum viral load and age. In COVACTA, younger patients had higher ACOV2S titers than older patients. This trend was similar but not significant in REMDACTA.
In REMDACTA, corticosteroid use at baseline was positively correlated with higher viral loads (serum and NP swab) and neutralizing antibody titers; this was not observed in COVACTA, although rates of steroid use in COVACTA were low (~20% at baseline) and dosing was variable because COVACTA enrolled patients before corticosteroid treatment was widely adopted as standard‐of‐care for patients with severe COVID‐19 (Figure S3). In REMDACTA, 123 of 640 patients (19.2%) had already received one or two doses of remdesivir at baseline, and minimal differences in viral loads, neutralizing antibody titers, and ACOV2S were observed in patients who had received remdesivir before randomization compared with those who had not (Figure S4).
Prognostic analyses
Relationships between baseline viral load or antibodies and clinical outcomes of time to mechanical ventilation or death, time to hospital discharge, and time to death were examined. In COVACTA and REMDACTA, higher serum viral load at baseline was strongly prognostic for worse outcome across all end points (Table S4). Higher serum viral load was associated with higher risk for death and the composite outcome of death or mechanical ventilation (Figure 4a,b), and a lower likelihood of hospital discharge (Figure 4c). For all three outcomes, the NP viral load showed a similar directional trend compared to serum viral load but was less robust and had wider CIs for the estimates.
FIGURE 4.
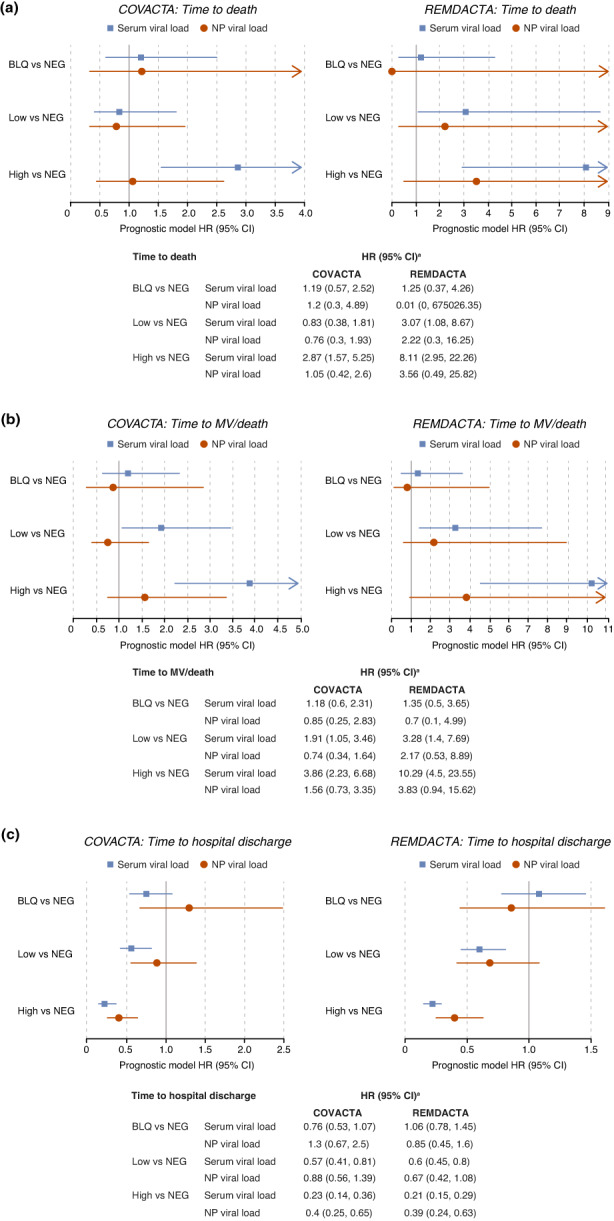
Prognostic value of viral load subgroups for clinical outcomes. Estimates of hazard ratios in BLQ, low, and high versus NEG viral load groups for clinical outcomes of (a) time to death, (b) time to mechanical ventilation/death, and (c) time to hospital discharge. aAll models were controlled for treatment arm, age as a continuous variable, stratification factors (geographic region and mechanical ventilation in COVACTA; geographic region and baseline ordinal scale in REMDACTA), and, for COVACTA, baseline ordinal scale of clinical status. BLQ, below the limit of quantification; CI, confidence interval; HR, hazard ratio; MV, mechanical ventilation; NEG, negative; NP, nasopharyngeal.
Correlations were also observed between ACOV2S titers and clinical outcomes. High levels of ACOV2S were associated with higher likelihood of hospital discharge and, in REMDACTA only, a lower risk for mechanical ventilation or death (Figure 5a). Neutralizing antibodies showed similar directionality but less prognostic value for these outcomes (Figure 5b).
FIGURE 5.
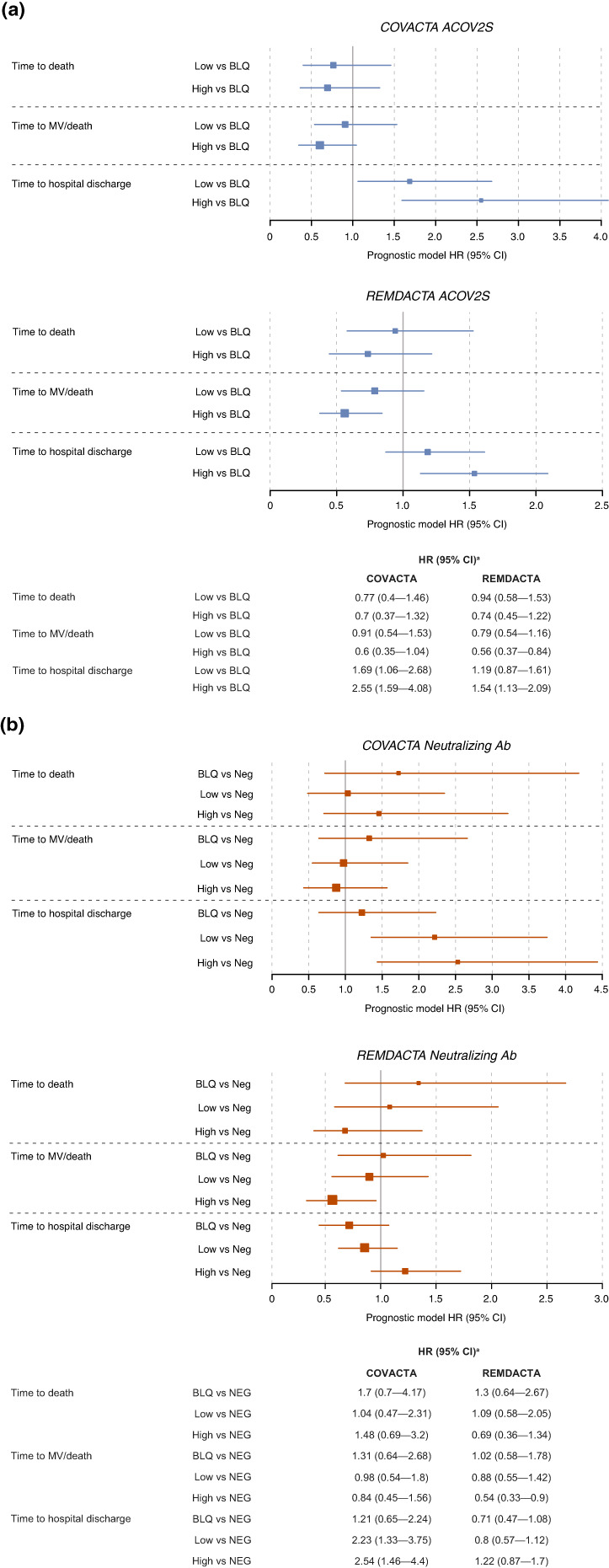
Prognostic value of antibody subgroups for clinical outcomes. Estimates of HR in (a) low and high versus BLQ ACOV2S or (b) BLQ, low, and high versus NEG neutralizing antibodies for clinical outcomes of time to death, time to mechanical ventilation/death, and time to hospital discharge. aAll models were controlled for treatment arm, age as a continuous variable, stratification factors (geographic region and mechanical ventilation in COVACTA; geographic region and baseline ordinal scale in REMDACTA), and, for COVACTA, baseline ordinal scale of clinical status. Ab, antibodies; ACOV2S, anti‐SARS‐CoV‐2 spike protein antibody; BLQ, below the limit of quantification; CI, confidence interval; HR, hazard ratio; MV, mechanical ventilation; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Relationship of virology/serology to biomarkers of inflammation and lung damage
In COVACTA and REMDACTA, laboratory‐measured markers for worse outcome in COVID‐19, including C‐reactive protein (CRP), interleukin‐6 (IL‐6), ferritin, blood neutrophils, and lymphopenia were weakly correlated with serum viral load, and less correlated with NP swab viral load or antibodies. Cytokines such as C‐X‐C motif chemokine ligand 10 (CXCL10) and IL‐10 were positively correlated with serum and NP swab viral load, more strongly in serum, and negatively correlated with antibodies (Figure 6).
FIGURE 6.
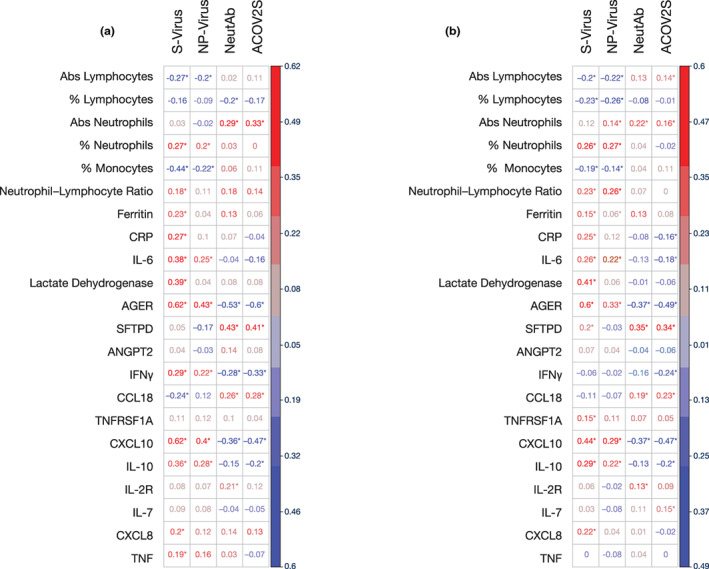
Correlation of serum viral load and antibodies with biomarkers in (a) COVACTA and (b) REMDACTA. Heatmaps indicated strength of correlation between biomarker and viral load or antibody levels where * indicates statistically significant difference of P < 0.05 based on correlation tests. Abs, absolute; ACOV2S, anti‐SARS‐CoV‐2‐Spike; AGER, advanced glycation end product‐specific receptor; ANGPT2, angiopoietin‐2 CCL18, chemokine ligand 18; CRP, C‐reactive protein; CXCL, C‐X‐C motif chemokine ligand; IFN, interferon; IL, interleukin; NeutAb, neutralizing antibodies; NP‐virus, nasopharyngeal viral load; SFTPD, surfactant protein‐D; S‐virus, serum viral load; TNF, tumor necrosis factor; TNFRSF1A, tumor necrosis factor receptor superfamily member 1A.
We hypothesized that injury to the lung barrier from viral or inflammatory‐mediated damage may result in serum viremia and, therefore, we assessed correlations of biomarkers of lung epithelial and endothelial injury with virology and serology. The soluble form of advanced glycation end product‐specific receptor (AGER) in serum is a marker of alveolar epithelial cell injury damage that can predict severe clinical outcomes in acute lung injury and acute respiratory distress syndrome (ARDS). 29 , 30 , 31 In COVACTA and REMDACTA (Figure 6a,b), the levels of AGER were positively correlated with viral load, more strongly in serum, and negatively correlated with antibody titer. Likewise, in both studies, correlations were observed between the general cell damage biomarker lactate dehydrogenase (LDH) and serum viral load, but not with NP swab viral load. Another lung tissue biomarker, surfactant protein‐D (SFTPD), had modest correlation with serum viral load that was only significant in REMDACTA, but positively correlated with neutralizing and ACOV2S antibodies in COVACTA and REMDACTA. Serum angiopoietin‐2 (ANGPT2), a biomarker of endothelial damage associated with severe clinical outcomes in acute lung injury and ARDS, 32 , 33 had no correlation with viral load or antibodies.
DISCUSSION
These results from two large clinical trials of more than 1000 hospitalized patients across a range of disease severity confirm that baseline SARS‐CoV‐2 viral load, especially in serum, predicts the risk for death and is associated with markers of lung tissue damage and elevated inflammation. Our findings support the prognostic value of higher anti‐SARS‐CoV‐2 antibodies for better clinical outcomes, which, based on inverse relationships with markers of lung tissue damage and viral load, may be associated with a protective role against viral replication and lung tissue damage. In both studies, baseline disease severity was positively associated with viral load (serum and NP swab) and antibodies (neutralizing and ACOV2S), and age was positively associated with NP swab viral load; however, the prognostic models demonstrate an association of viral load and anti‐SARS‐CoV‐2 antibodies with worse clinical outcomes independent of age or baseline disease severity. We previously identified several biomarkers prognostic for clinical outcomes in COVACTA, including IL‐6, CRP, ferritin, lymphocytes, monocytes, neutrophils, platelets, and white blood cells. 34 Results of the current analysis suggest that serum viral load is among the strongest prognostic biomarkers for clinical outcomes.
Although most studies of SARS‐CoV‐2 infection have quantified virus in the nasopharynx, our results confirm previous reports showing that viremia (serum viral RNA) is prevalent in hospitalized patients with COVID‐19. 7 , 8 , 9 , 11 , 12 , 14 , 15 , 16 , 20 , 35 SARS‐CoV‐2 viral load at baseline in NP swabs and serum was highly variable; however, the highest viral load in serum and NP samples was generally observed in patients enrolled ~5–7 days after symptom onset, which is consistent with previous reports. 36 , 37 , 38 , 39 Serum viral load had greater prognostic value and was more strongly correlated with inflammatory cytokines and tissue damage biomarkers than NP swab viral load. These findings imply that serum viral load is a better biomarker of more severe disease with organ damage; however, it is also possible that weak correlation of NP viral load with organ damage biomarkers could be related to greater variability in the NP viral load introduced by repeat swabbing and use of supplemental oxygen.
Most patients enrolled in COVACTA and REMDACTA had detectable ACOV2S and neutralizing anti‐SARS‐CoV‐2 antibodies at baseline and levels increased over time with all patients having detectable antibodies by day 28, as reported previously for COVACTA. 26 The prognostic relationship of low antibody levels at baseline with worse clinical outcomes may seem contradictory to the higher baseline antibody levels observed in patients with worse ordinal scale at baseline; however, subgroup analysis in COVACTA showed the prognostic value of low antibody titers for shorter time to death and longer time to hospital discharge was stronger for patients with severe disease (ordinal scale score ≥4 [requiring at least noninvasive ventilation or high‐flow oxygen] at baseline). One potential explanation for this is that timing of the antibody response may be an important determinant of the protective effect, with greater protection afforded in patients with earlier antibody responses. 22 , 23 Patients with higher antibody levels tended to have longer duration from the time of symptom onset (Figure 1), likely reflecting further progression to the adaptive phase of immunity. However, patients with delayed antibody onset or persistently low antibody levels may have progressed to a more severe phase of disease with greater lung damage compared to patients with lower antibody levels who may have received intervention earlier during the course of infection before extensive lung damage occurred. Indeed, ACOV2S and neutralizing antibodies were negatively correlated with AGER, a biomarker of lung epithelial damage. As most patients were positive for anti‐SARS‐CoV‐2 antibodies at the time of enrollment in COVACTA and REMDACTA and samples were not available from patients before enrollment, the relative influence of time to seropositivity on clinical outcomes could not be assessed in our study.
Serum viral load correlated more strongly with circulating levels of AGER compared with other biomarkers measured across both studies. This extends findings from studies of fewer patients and biomarkers. 20 , 21 AGER is expressed by alveolar epithelial cells, and elevated serum levels are interpreted as a biomarker of epithelial injury and loss of barrier integrity. Serum viral load also correlated with elevated serum LDH, an intracellular protein released by damaged cells and used as a biomarker of cell injury. These results imply that the viral RNA detected in serum may result from the loss of lung barrier integrity in severe disease. This loss of pulmonary barrier integrity may allow replication‐competent virus to establish extrapulmonary infection reservoirs that could also contribute viral RNA to serum. Conversely, the association between high antibody levels and low AGER levels supports a protective effect of antibodies against lung damage and virus leakage into the blood, possibly by controlling infection before damage occurs. SFTPD, previously identified as a biomarker of alveolar‐capillary damage in severe infections, such as influenza, was positively correlated with antibodies at baseline. Another study reported that SFTPD was elevated in patients with severe influenza infection but not severe COVID‐19, suggesting there may be a unique effect of SARS‐CoV‐2 infection on expression of SFTPD. 40
Higher NP swab and serum viral loads were correlated with increased blood interferon (IFN)γ and CXCL10 levels in our study, consistent with reports that have demonstrated an association between high SARS‐CoV‐2 viral loads from NP swabs and autopsied lungs with elevated expression of interferon‐stimulated genes. 41 , 42 Our results expand upon this to show that, similar to findings in the airway, viral load in serum is associated with elevated IFN pathway biomarkers in blood. Because serum includes proteins in the blood produced by a mixture of cells, identifying the cellular source of these cytokines will require quantification using flow cytometry or transcriptional analyses of single cells to account for cell composition.
Antibody levels and viral loads (NP swab and serum) were weakly inversely correlated in our study, consistent with other studies that have shown no or a weak relationship between viral load and antibodies. 20 , 22 These findings could indicate that other contributors to viral clearance, such as T cell‐mediated adaptive immunity, were dysregulated in these patients, although T cell function was not measured in COVACTA or REMDACTA. The weak correlation between antibody levels and viral load might also reflect a delay in RNA clearance after antibody neutralization because SARS‐CoV‐2 RNA can be detected in the airway for months after the onset of symptoms. 43
In conclusion, SARS‐CoV‐2 viremia is associated with systemic inflammation and lung tissue damage, whereas antibodies to SARS‐CoV‐2 may be protective against lung tissue damage. The findings of this study confirm the utility of viral load and antibody titers in predicting clinical outcomes in patients with COVID‐19 in two clinical trials encompassing more than 1000 hospitalized patients. There are several important limitations of this work. This post hoc analysis is observational and, therefore, the results are limited to hypothesis generation. Mechanistic studies are required to confirm whether the observed associations of viral load with lung damage and inflammation biomarkers reflect uncontrolled SARS‐CoV‐2 replication as a driver or consequence of COVID‐19 pathogenesis. Because COVACTA and REMDACTA enrolled patients in 2020 to early 2021, before the emergence of Omicron strains and availability of vaccines, clinical utility of SARS‐CoV‐2 antibody titers and viral load requires confirmation in the current context of widespread use of vaccines, variants with differing virulence, and high rates of prior SARS‐CoV‐2 infection. Although PCR viral load assessment is widely available, serum PCR testing is not frequently performed outside of clinical research in hospital settings and point‐of‐care devices that rapidly quantify viral load would be needed for clinical utility in the context of fast‐progressing disease. Last, further analyses are required to define and confirm quantitative assay cutoff points for clinical utility. Notably, although we previously reported that treatment response to tocilizumab did not vary with respect to baseline NP swab or serum viral load levels in the COVACTA study, 26 clinical utility of antibody levels is supported by studies of the anti‐SARS‐CoV‐2 monoclonal antibody cocktail casirivimab/imdevimab, in which greater efficacy was observed in patients who were seronegative for antibodies to SARS‐CoV‐2. 44 , 45 Viral load may be particularly relevant to guide treatment with antivirals, but this requires further assessment. Despite these limitations, our findings support consideration of viral load, especially in the serum, for identification of patients at greatest risk for the most adverse outcomes and underscore the relevance of treatments aimed at controlling viral replication or boosting humoral response to SARS‐CoV‐2 in patients hospitalized with COVID‐19.
AUTHOR CONTRIBUTIONS
R.N.B., A.T., C.M.R., F.C., O.G., I.T.L., J.J.W., D.P., K.J., G.C., and S.W. wrote the manuscript. R.N.B., J.M., C.M.R., F.C., M.B., L.T., O.G., I.T.L., J.J.W., D.P., and S.W. designed the research. R.N.B., L.T., I.T.L., I.O.R., and S.W. performed the research. A.T., H.S., J.M., M.B., O.G., I.T.L., and K.J. analyzed the data.
FUNDING INFORMATION
F. Hoffmann‐La Roche Ltd and the US Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, under OT number HHSO100201800036C.
CONFLICT OF INTEREST STATEMENT
R.N.B. is an employee of Roche/Genentech and holds stock in Roche. A.T. is an employee of Roche. H.S. is an employee of Roche/Genentech and holds stock in Roche. J.M. is an employee of Roche/Genentech and holds stock in Roche. C.M.R. is an employee of Roche/Genentech and holds stock/stock options in Roche, received travel support from Genentech, and has a patent filed with Genentech unrelated to the current study. F.C. is a former employee of Roche/Genentech. M.B. is an employee of Roche/Genentech and holds stock/stock options in Roche, received funding from the Biomedical Advanced Research and Development Authority related to the current work, and is an author of an unpublished pending patent “Method for Treating Pneumonia, Including COVID‐19 Pneumonia, with an IL‐6 Antagonist.” L.T. is an employee of Roche/Genentech and holds stock/stock options in Roche, received funding from the Biomedical Advanced Research and Development Authority related to the current work, and is an author of an unpublished pending patent “Method for Treating Pneumonia, Including COVID‐19 Pneumonia, with an IL‐6 Antagonist.” O.G. is an employee of Roche and shareholder of Roche Holding AG. I.T.L. is a former clinical research fellow of Genentech/Roche, and received funding from Genentech/Roche during the conduct of the study. J.J.W. is an employee of and shareholder in Gilead Sciences. D.P. is an employee of and shareholder in Gilead Sciences. K.J. is a former employee of and shareholder in Gilead Sciences, and is an employee of Corcept Therapeutics. G.C. is a former employee of and shareholder in Gilead Sciences, and is an employee of Vir Biotechnology. I.O.R. received grants/contracts from Roche/Genentech and Boehringer, and consulting fees from Roche/Genentech, Boehringer, Bristol Myers Squibb, and Immunomet. S.W. is an employee of Roche and holds stock in Roche.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Table S1
Table S2
Table S3
Table S4
Appendix S1
ACKNOWLEDGMENTS
Medical writing support was provided by Stacie Dilks, PhD, and Sara Duggan, PhD, of ApotheCom and funded by F. Hoffmann‐La Roche Ltd.
Bauer RN, Teterina A, Shivram H, et al. Prognostic value of severe acute respiratory syndrome coronavirus‐2 viral load and antibodies in patients hospitalized with COVID‐19. Clin Transl Sci. 2023;16:1049‐1062. doi: 10.1111/cts.13511
DATA AVAILABILITY STATEMENT
Qualified researchers may request access to individual patient‐level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available here (https://vivli.org/members/ourmembers/). For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here (https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm).
REFERENCES
- 1. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsukagoshi H, Shinoda D, Saito M, et al. Relationships between viral load and the clinical course of COVID‐19. Viruses. 2021;13(2):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mancini N, Clementi N, Ferrarese R, et al. Very high SARS‐CoV‐2 load at the emergency department presentation strongly predicts the risk of admission to the intensive care unit and death. Clin Chem Lab Med. 2021;59:e247‐e250. [DOI] [PubMed] [Google Scholar]
- 4. Tanner AR, Phan H, Brendish NJ, et al. SARS‐CoV‐2 viral load at presentation to hospital is independently associated with the risk of death. J Infect. 2021;83:458‐466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Le Borgne P, Solis M, Severac F, et al. SARS‐CoV‐2 viral load in nasopharyngeal swabs in the emergency department does not predict COVID‐19 severity and mortality. Acad Emerg Med. 2021;28:306‐313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salto‐Alejandre S, Berastegui‐Cabrera J, Camacho‐Martínez P, et al. SARS‐CoV‐2 viral load in nasopharyngeal swabs is not an independent predictor of unfavorable outcome. Sci Rep. 2021;11:12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fajnzylber J, Regan J, Coxen K, et al. SARS‐CoV‐2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang K, Wu L, Luo Y, Gong B. Quantitative assessment of SARS‐CoV‐2 RNAemia and outcome in patients with coronavirus disease 2019. J Med Virol. 2021;93:3165‐3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacobs JL, Bain W, Naqvi A, et al. Severe acute respiratory syndrome coronavirus 2 viremia is associated with coronavirus disease 2019 severity and predicts clinical outcomes. Clin Infect Dis. 2022;74:1525‐1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miki S, Sasaki H, Horiuchi H, et al. On‐admission SARS‐CoV‐2 RNAemia as a single potent predictive marker of critical condition development and mortality in COVID‐19. PLoS One. 2021;16:e0254640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bermejo‐Martin JF, González‐Rivera M, Almansa R, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID‐19. Crit Care. 2020;24:691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gutmann C, Takov K, Burnap SA, et al. SARS‐CoV‐2 RNAemia and proteomic trajectories inform prognostication in COVID‐19 patients admitted to intensive care. Nat Commun. 2021;12:3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu D, Zhou F, Sun W, et al. Relationship between serum severe acute respiratory syndrome coronavirus 2 nucleic acid and organ damage in coronavirus 2019 patients: a cohort study. Clin Infect Dis. 2021;73:68‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prebensen C, Myhre PL, Jonassen C, et al. Severe acute respiratory syndrome coronavirus 2 RNA in plasma is associated with intensive care unit admission and mortality in patients hospitalized with coronavirus disease 2019. Clin Infect Dis. 2021;73:e799‐e802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hogan CA, Stevens BA, Sahoo MK, et al. High frequency of SARS‐CoV‐2 RNAemia and association with severe disease. Clin Infect Dis. 2021;72:e291‐e295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monchi M, Bruneau T, Jochmans S, et al. Association of high SARS‐CoV‐2 RNAemia with diabetes and mortality in critically ill COVID‐19 patients. iScience. 2022;25:104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ram‐Mohan N, Kim D, Zudock EJ, et al. SARS‐CoV‐2 RNAemia predicts clinical deterioration and extrapulmonary complications from COVID‐19. Clin Infect Dis. 2022;74:218‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mertz C, Glowinski R, Cohen SH, et al. Severe acute respiratory syndrome coronavirus 2 RNAemia and clinical outcomes in children with coronavirus disease 2019. J Infect Dis. 2022;225:208‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solis M, Gallais F, Garnier‐Kepka S, et al. Combining predictive markers for severe COVID‐19: Torquetenovirus DNA load and SARS‐CoV‐2 RNAemia. J Clin Virol. 2022;148:105120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Schneider AM, Mehta A, et al. SARS‐CoV‐2 viremia is associated with distinct proteomic pathways and predicts COVID‐19 outcomes. J Clin Invest. 2021;131(13):e148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wick KD, Siegel L, Neaton JD, et al. RAGE has potential pathogenetic and prognostic value in nonintubated hospitalized patients with COVID‐19. JCI Insight. 2022;7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lucas C, Klein J, Sundaram ME, et al. Delayed production of neutralizing antibodies correlates with fatal COVID‐19. Nat Med. 2021;27:1178‐1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zohar T, Loos C, Fischinger S, et al. Compromised humoral functional evolution tracks with SARS‐CoV‐2 mortality. Cell. 2020;183:1508‐1519.e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Röltgen K, Powell AE, Wirz OF , et al. Defining the features and duration of antibody responses to SARS‐CoV‐2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54):eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosas IO, Bräu N, Waters M, et al. Tocilizumab in hospitalized patients with severe covid‐19 pneumonia. N Engl J Med. 2021;384:1503‐1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosas IO, Bräu N, Waters M, et al. Tocilizumab in patients hospitalised with COVID‐19 pneumonia: efficacy, safety, viral clearance, and antibody response from a randomised controlled trial (COVACTA). EClinicalMedicine. 2022;47:101409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rosas IO, Diaz G, Gottlieb RL, et al. Tocilizumab and remdesivir in hospitalized patients with severe COVID‐19 pneumonia: a randomized clinical trial. Intensive Care Med. 2021;47(11):1258‐1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Swinscow TDV. Chapter 11–Correlation and Regression. In: Statistics at Square One. 9th ed. BMJ Publishing Group; 1997. [Google Scholar]
- 29. Erusalimsky JD. The use of the soluble receptor for advanced glycation‐end products (sRAGE) as a potential biomarker of disease risk and adverse outcomes. Redox Biol. 2021;42:101958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calfee CS, Ware LB, Eisner MD, et al. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax. 2008;63:1083‐1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oczypok EA, Perkins TN, Oury TD. All the "RAGE" in lung disease: the receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr Respir Rev. 2017;23:40‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA. Plasma angiopoietin‐2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med. 2012;40:1731‐1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parikh SM, Mammoto T, Schultz A, et al. Excess circulating angiopoietin‐2 may contribute to pulmonary vascular leak in sepsis in humans. PLoS Med. 2006;3:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tom J, Bao M, Tsai L, et al. Prognostic and predictive biomarkers in patients with coronavirus disease 2019 treated with tocilizumab in a randomized controlled trial. Crit Care Med. 2021;50:398‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X, Zhao B, Qu Y, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:1937‐1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS‐CoV‐2 in clinical samples. Lancet Infect Dis. 2020;20:411‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Kampen JJA, van de Vijver D, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease‐2019 (COVID‐19). Nat Commun. 2021;12:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cevik M, Kuppalli K, Kindrachuk J, Peiris M. Virology, transmission, and pathogenesis of SARS‐CoV‐2. BMJ. 2020;371:m3862. [DOI] [PubMed] [Google Scholar]
- 40. Choreño‐Parra JA, Jiménez‐Álvarez LA, Ramírez‐Martínez G, et al. Expression of surfactant protein D distinguishes severe pandemic influenza a(H1N1) from coronavirus disease 2019. J Infect Dis. 2021;224:21‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lieberman NAP, Peddu V, Xie H, et al. In vivo antiviral host transcriptional response to SARS‐CoV‐2 by viral load, sex, and age. PLoS Biol. 2020;18:e3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID‐19. Nat Commun. 2020;11:5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS‐CoV‐2, SARS‐CoV‐1 and MERS‐CoV viral load dynamics, duration of viral shedding 2 and infectiousness – a living systematic review and meta‐analysis. Lancet Microbe. 2021;2:e13‐e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN‐COV2, a neutralizing antibody cocktail, in outpatients with COVID‐19. N Engl J Med. 2021;384:238‐251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. RECOVERY Collaborative Group . Casirivimab and imdevimab in patients admitted to hospital with COVID‐19 (RECOVERY): a randomised, controlled, open‐label, platform trial. Lancet. 2022;399(10325):665‐676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Table S1
Table S2
Table S3
Table S4
Appendix S1
Data Availability Statement
Qualified researchers may request access to individual patient‐level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available here (https://vivli.org/members/ourmembers/). For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here (https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm).


