Abstract
We report here a Schizosaccharomyces pombe gene (dmc1+) that resembles budding yeast DMC1 in the region immediately upstream of the rad24+ gene. We showed by northern and Southern blot analysis that dmc1+ and rad24+ are co-transcribed as a bicistronic mRNA of 2.8 kb with meiotic specificity, whereas rad24+ itself is constitutively transcribed as a 1.0-kb mRNA species during meiosis. Induction of the bicistronic transcript is under the control of a meiosis-specific transcription factor, Ste11. Disruption of both dmc1+ and rad24+ had no effect on mitosis or spore formation, and dmc1Δ cells displayed no change in sensitivity to UV or γ irradiation relative to the wild type. Tetrad analysis indicated that Dmc1 is involved in meiotic recombination. Analysis of gene conversion frequencies using single and double mutants of dmc1 and rhp51 indicated that both Dmc1 and Rhp51 function in meiotic gene conversion. These observations, together with a high level of sequence identity, indicate that the dmc1+ gene of S.pombe is a structural homolog of budding yeast DMC1, sharing both similar and distinct functions in meiosis.
INTRODUCTION
During meiosis, between DNA replication and prophase of meiosis I, a number of lengthy events which are specific to meiosis, including pairing and recombination of homologous chromosomes, are observed. The budding yeast Saccharomyces cerevisiae has been extensively used to study the molecular mechanisms of genetic recombination during meiosis. Several proteins with sequence similarity to RecA, such as Dmc1 and Rad51, have been isolated and their essential roles in recombination have been characterized (1–4). Dmc1 and Rad51 share significant amino acid sequence homology, particularly within the core region, and similar conserved sequences are also observed in RecA, which plays a pivotal role in homologous recombination in Escherichia coli. Despite this level of similarity, sequence divergence in the N-terminal domain distinguishes the Rad51 from the Dmc1 family. Similar RecA-like proteins have also been isolated from mammals (5,6) and from plants (Lim15, a lily homolog of Dmc1) (7,8), indicating that the structure of factors involved in recombination is conserved among a wide variety of species.
Expression of DMC1, unlike that of RAD51, is meiosis specific. Dmc1 has no role in the repair of double-strand breaks (DSBs) during vegetative growth, but dmc1 mutants are defective in reciprocal meiotic recombination, accumulate DSB recombination intermediates, fail to initiate normal synaptonemal complex (SC) formation, and arrest late in meiotic prophase (1). Both Rad51 and Dmc1 are required for meiotic recombination and promote the synapsis of homologous chromosomes of budding yeast and side-by-side joining of lateral elements in pachytene chromosomes (9). Double mutation of rad51 dmc1 causes more severe meiotic defects (4). Lim15 and Rad51 are also involved in the recombination reaction during meiotic prophase in the lily, and are colocalized at multiple sites on meiotic chromosomes at prophase 1, suggesting that they may be components of early recombination nodules (7,8). Analysis of dmc1 gene knockout mice revealed that both homozygous mutant males and females were sterile, with gametogenesis arrested in meiotic prophase (10,11). This strong similarity in the effects of dmc1 null mutation between yeast and mouse suggests the existence of an evolutionarily conserved mechanism of regulation of meiotic recombination.
Although most organisms form SCs, fission yeast is an intriguing exception in which no SC formation has been reported to date. Thus, it is of interest to investigate whether meiotic recombination regulatory mechanisms are also conserved in fission yeast and, if so, how regulation is achieved without a proteinaceous scaffold such as the SC. Structural and functional analysis of rhp51+, a fission yeast homolog of RAD51, has been carried out (6,12), but no functional analysis of a fission yeast DMC1 homolog has been reported to date. In the course of a systematic search for genes whose transcription is induced in a meiosis-specific manner in fission yeast, we isolated a cDNA clone whose sequence closely resembled that of DMC1. Here we report the structural and functional analysis of this clone. We show that the fission yeast dmc1 gene is transcribed as a bicistronic mRNA in a meiosis-specific manner and carries an additional open reading frame (ORF) downstream of the dmc1-like ORF. The second ORF encodes Rad24, a 14-3-3 protein which is required for a variety of cellular events such as DNA damage checkpoint regulation (13), and which is also known to associate with target proteins via phosphoserine residues (14,15).
MATERIALS AND METHODS
Strains and media
Standard Schizosaccharomyces pombe genetic procedures were conducted as described previously (16,17). Schizosaccharomyces pombe strains used in this study are derivatives of those originally described by Leupold (18), as listed in Table 1. Schizosaccharomyces pombe cells were grown in standard rich media (YPD or YEL) and in synthetic minimal media (EMM2) containing additional leucine (150 mg/ml) or uracil (75 mg/ml) when specified. For the induction of mating and meiosis, cells were cultured in MEA, SPA, SSA, EMM2 or SSL-N medium at 26°C (16,17).
Table 1. List of S.pombe strains.
Preparation of RNA and northern blot analysis
CD16-1 and CD16-5 cells were shaken at 28°C in PM+N medium (containing nitrogen) until they reached log phase (OD600 = 0.6). Then, the cells were transferred into PM-N medium (without nitrogen) and shaken under the same conditions. Cells were collected at 2-h intervals for 12 h, mixed with 10% SDS, phenol/chloroform and RNA extraction buffer and disrupted with glass beads (φ = 0.5 mm). The samples were centrifuged and the supernatant was treated with phenol/chloroform and then with chloroform before precipitation with ethanol. The precipitate was dissolved in H2O and again precipitated in the presence of 2 M LiCl. Northern blot analysis was carried out according to standard protocols (19). RNA size markers (0.24–9.5 kb) were purchased from Gibco BRL (Rockville, MD). A rad25+-specific probe was constructed by PCR with the oligonucleotide primers 5′-AACCAGTCTGCTAAGGAGGA-3′ (Rad25-M3) and 5′-GGATCCGTTTACTGATAA-3′ (Rad25-M4), using genomic DNA from S.pombe (972) as a template.
Preparation of subtracted cDNA library
We used two diploid strains, CD16-1 and CD16-5 (20; Table 1), for the subtraction process. CD16-1 cells are heterozygous (h+/h–) and initiate meiosis after nitrogen starvation. In contrast, CD16-5 cells are homozygous (h–/h–), and do not proceed to meiosis even after nitrogen starvation. We first constructed a cDNA library in the pAP3neo vector (21) by a linker–primer method, using mRNA from CD16-1 cells which were collected at 1-h intervals for 6 h during incubation in sporulation medium following nitrogen starvation. These cell preparations were combined into a single sample before mRNA preparation. Once constructed, the CD16-1 cDNA library was converted to a single-stranded form using f1 helper phage. In parallel with these procedures, we prepared mRNA from CD16-5 cells which were collected at 1-h intervals for 8 h after nitrogen starvation and mixed before mRNA preparation. This mRNA was labeled with biotin using the photobiotin system (Vector Laboratories Inc., Burlingame, CA), and then mixed in excess in hybridization buffer with the single-stranded form of the CD16-1 cDNA library. Hybrid RNA species were removed using avidin as described previously (21). After two rounds of subtractive hybridization, the single-stranded subtracted cDNA library was converted to the double-stranded form with BcaBEST DNA polymerase (TaKaRa, Japan) and transformed into E.coli (MC1061A strain) by electroporation as described previously (22). Starting from a cDNA library of 1.8 × 106 c.f.u. with an average insert size of 1.5 kb, we constructed a subtracted cDNA library of 1.2 × 104 c.f.u. with an average insert size of 1.45 kb.
Disruption of dmc1+ and rad24+ genes and Southern blot analysis
Using the eta1 cDNA fragment (the definition of which will be given in the Results) as a probe, we cloned the genomic region encompassing the dmc1+ and rad24+ genes from a genomic library which we constructed by inserting partial Sau3AI DNA fragments into the BamHI site of the Bluescript KS(+) vector (Stratagene, La Jolla, CA). For disruption of the dmc1+ gene, a NheI–FbaI fragment carrying the dmc1+ genomic DNA region was subcloned into the Bluescript KS(+) vector, and a 1.8 kb fragment carrying the S.pombe ura4+ gene (23) was then inserted into the BamHI site in the middle of the dmc1+ ORF. For disruption of the rad24+ gene, a ura4+ fragment was inserted into the PstI–FbaI sites of the rad24+ ORF using synthetic linkers. For disruption of both dmc1+ and rad24+ genes, a ura4+ fragment was inserted into the BamHI–FbaI sites of the dmc1+–rad24+ genomic sequence using synthetic linkers. The resulting disrupted genes were introduced into the diploid strain C525 (24) and the Ura+ transformants were screened for disruption of one copy of the targeted genes by Southern blot analysis, which was carried out according to standard protocols (19). Tetrads from these strains were then dissected.
Disruption of rhp51+ gene
For disruption of rhp51+ gene by replacing with ura4+ gene, we performed PCR and obtained DNA fragments carrying the 5′ upstream region or 3′ downstream region of rhp51+ gene, or the coding region of ura4+ gene with genomic DNA of S.pombe as a template. We synthesized the following four oligonucleotides and used them as forward (F) or reverse (R) primers to obtain DNA fragments around the rhp51+ gene: F1, 5′-CCGAATCCCATATCTTCATC-3′; F2, 5′-GGCCTTACGACGTAGTCGACATTATTTTAGTATCGTTTC-3′; R1, 5′-CTAGAAGTTCTCCTCGAGTATAACTTGTTAAGCACG-3′; R2, 5′-GCTTGAACATGAAGTCCTAC-3′. We also synthesized the following set of oligonucleotide primers to obtain a DNA fragment encoding Ura4: 5′-CTCGAGGAGAACTTCTAGAAGCTTAGCTACAAATCCC-3′; 5′-GTCGACTACGTCGTAAGGCCAAGCTTGTGATATTGACG-3′. These DNA fragments were ligated in the order of (5′)rhp51–ura4+–(3′)rhp51 and inserted into a Bluescript KS(+) vector, which was then introduced into the diploid strain C525. Successful disruption of the rhp51+ gene was confirmed by Southern blot analysis on DNA obtained from the dissected tetrads.
Measurement of genetic distance in tetrad analysis
Haploid parental strains were grown on a YPD plate at 33°C for 2–3 days. For conjugation and sporulation, two parental strains were picked up from the YPD plate and directly mixed on an ME plate and then incubated at 28°C for 2 days. Ascospores were transferred onto a YPD plate and, after dissection, they were germinated at 28°C for 3 days. Resultant colonies were replicated onto a selective medium plate. The ability of their colony formation on these selective media was inspected by eye after incubation at 33°C for 3–4 days.
Measurement of meiotic gene conversion frequency
For construction of the strain which has ade6-L469 5rpm, GC13 was used in crossing. This strain has a restriction site polymorphism at the ade6 locus for the physical analysis of conversion tracts (25). Yeast cells were grown in YPD medium at 32°C. For conjugation and sporulation, growing cells (mid-log phase) were washed once with water and resuspended in water to be concentrated to 100-fold. Similar numbers of washed cells to be mated were mixed and incubated on an ME plate at 28°C for 1 day. After microscopic analysis for confirming spore formation, spores were collected by scraping from plates and treated with 30% ethanol for 15 min to kill the remaining cells. Spores were spread onto selective or YPD plates with serial concentration of spores, and colonies were counted after incubation for 3–7 days. The gene conversion frequency during meiosis was calculated by counting the ade-dependent and ade-independent colonies. Viability of spores was calculated as the number of ade-dependent colonies in relation to the number of spores on each plate.
Preparation of rad24+- and rad25+-specific probes
A rad24+-specific DNA probe was prepared by digesting the rad24 cDNA with SacI and AccI, generating a 100-bp fragment with a sequence distinct from that of rad25 cDNA. The DNA fragment (120 bp) used as a rad25+-specific probe was prepared by PCR using Rad25-M3 (5′-AACCAGTCTGCTAAGGAGGA-3′) and Rad25-M4 (5′-GGATCCGTTTACTGATAA-3′) as primers and genomic DNA from S.pombe (972) as a template.
RESULTS
Isolation of a meiosis-specific transcript of rad24+
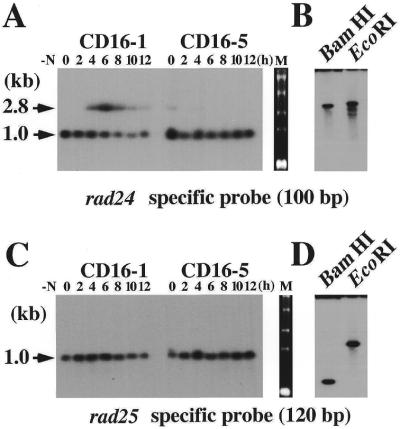
In the fission yeast S.pombe, conjugation of haploid cells of the opposite mating type (h+ and h–) occurs in response to nitrogen starvation and is followed by zygotic meiosis. The heterozygous diploid cells of a distinct mating type (h+/h–) can be induced to initiate meiosis after nitrogen starvation (azygotic meiosis), whereas the homozygous diploid cells with the same mating type (h–/h–) never proceed to meiosis. In order to comprehensively screen for genes that are preferentially expressed during sexual differentiation in S.pombe, we prepared a subtracted cDNA library greatly enriched in mRNA species induced during meiosis. We used two kinds of diploid strains, CD16-1 and CD16-5 (20; Table 1) for subtraction. One of the cDNA clones isolated from this subtracted cDNA library had a 1.0 kb insert but hybridized to an extra band of 2.8 kb in northern analysis. This larger band displayed meiosis-specific induction, with a peak at 4 h only in CD16-1 cells (Fig. 1A). We termed this clone eta (from enhanced transcript after nitrogen starvation).
Figure 1.
rad24+ but not rad25+ displayed a meiosis-specific transcript at 2.8 kb distinct from the 1.0 kb band representing the constitutively transcribed rad24+ transcript. (A and C) Northern blot analyses with rad24- and rad25-specific probes, showing that the 2.8-kb band was derived from the rad24+ but not from the rad25+ gene. CD16-1, a heterozygous (h+/h–) diploid strain initiated meiosis upon nitrogen starvation, whereas CD16-5, a homozygous (h–/h–) diploid strain could not proceed to meiosis. Numbers above the lanes indicate hours after nitrogen starvation. In lane M, RNA size markers that were stained with ethidium bromide before northern blotting are presented. (B and D) Southern blot analyses performed with rad24- and rad25-specific probes (Materials and Methods), showing that these two probes did not cross-hybridize under the conditions used in these experiments.
DNA sequencing of the 1.0-kb insert identified it as rad24+, a gene that encodes one of the 14-3-3 proteins of S.pombe (13). Since the other 14-3-3 gene called rad25+ has significant sequence homology to rad24+, we surmized that the upper band at 2.8 kb might be derived from the rad25+ gene. To examine this possibility, we prepared rad24+- and rad25+-specific DNA probes. Northern blot analysis using these probes showed that the rad25+ probe recognized only a 1.0 kb band (Fig. 1C), the expression (northern blot) profile of which during meiosis was distinct from that of rad24+. However, the rad24+ probe recognized bands of 1.0 and 2.8 kb as described above (Fig. 1A), the expression patterns of which resembled those shown in Figure 1A. Genomic Southern blot analyses using these two probes displayed bands of distinct sizes, clearly indicating that the probes did not cross-hybridize (Fig. 1B and D) and that the rad24+ and rad25+ genes map to distinct loci in the genome. These results indicate that the 2.8-kb meiosis-specific band derived from a genomic locus close to the rad24+ gene.
eta1 carries two ORFs
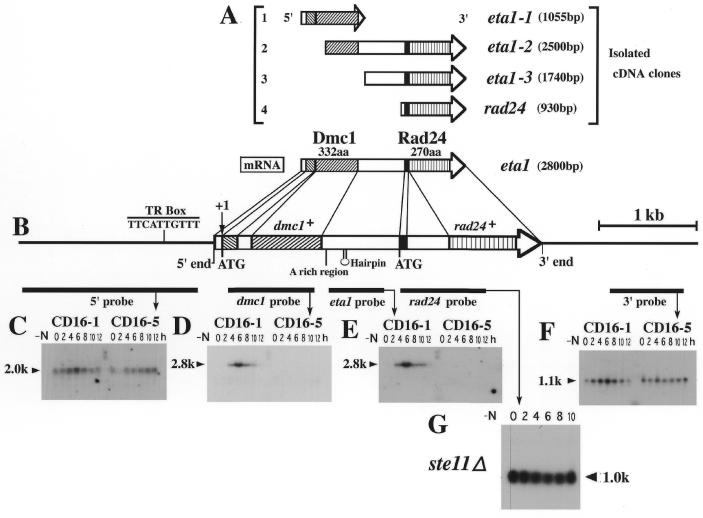
We then screened the original CD16-1 cDNA library to isolate a cDNA clone corresponding to the 2.8 kb band, using the rad24+ cDNA insert as a probe. We isolated three cDNAs other than rad24+, which we named eta1-2 and -3 (Fig. 2A). Then, with eta1-2 cDNA insert as a probe, we cloned eta1-1 cDNA. DNA sequencing showed that eta1-2 (2.5 kb) and -3 (1.74 kb) were partial cDNA clones derived from the 2.8 kb mRNA and contained 5′ sequence extensions relative to rad24+. eta1-1 (1055 bp) contained the missing 5′ portion of eta1-2. We also screened an S.pombe (h– 972) genomic library with the same probe, cloned a DNA fragment including the surrounding regions of the rad24+ gene, and determined the DNA sequence.
Figure 2.
Northern blot analyses showing that the dmc1+ and rad24+ genes are positioned in the order dmc1+ rad24+, and that the meiosis-specific eta1 mRNA is bicistronic, including both dmc1+ and rad24+ ORFs. (A) Structure of cloned eta1 and rad24 cDNA fragments. The rad24 cDNA was isolated from a subtracted cDNA library (CD16-1 minus CD16-5). Using this rad24 cDNA fragment as a probe, eta1-1, -2 and -3 cDNAs were isolated from a CD16-1 cDNA library by colony hybridization. (B) Schematic representation of the dmc1+ and rad24+ genes and the bicistronic eta1 mRNA. (C–F) Northern blot analyses of the dmc1+ and rad24+ genes; the locations of the five different DNA fragments used as probes are indicated by thick horizontal bars. The 1.0-kb band was observed only when the rad24+ probe was used, whereas the 2.8-kb band was observed only when the eta1 cDNA region was included in the probe. (G) The 2.8-kb band disappeared in northern analysis using an ste11+ null mutant (ste11Δ), indicating that meiosis-specific transcription of dmc1+ takes place downstream of the Ste11 transcriptional cascade.
A comparison of the genomic DNA and cDNA sequences revealed that the eta1-1, -2, -3 and rad24+ cDNAs were derived from the same genomic region, as illustrated in Figure 2B. We also concluded that the eta1-1 cDNA was an artifact generated by in vitro initiation of the cDNA synthesis reaction from the poly(A)-rich region of the gene, since the poly(A) sequence at the 3′ end of the eta1-1 cDNA contained a cytosine, and northern blot analysis using the eta1 probe showed no band at ∼1.0 kb (Fig. 2E). Nonetheless, this clone was useful, since it contained the missing portion of the eta1-2 and -3 clones.
The second ORF in eta1 cDNA resembles Dmc1 of S.cerevisiae
Analysis of the DNA sequence corresponding to the 2.8-kb mRNA showed that it contained an additional ORF encoding 332 amino acids upstream of the rad24+ ORF. The genomic DNA sequence showed that this additional ORF is split by a single intron (Fig. 2B). Homology searching showed that the predicted amino acid sequence of this ORF had significant homology to Dmc1 of S.cerevisiae (1). Optimized alignment revealed an overall sequence identity of 72% (data not shown). We therefore refer to this sequence as the dmc1+ gene of S.pombe. The sequences of the S.pombe rad24+ and dmc1+ genes have been deposited in the DDBJ/EMBL/GenBank under the combined accession number AB008545. Optimized alignment also showed that the gene product of dmc1+ (SpDmc1) is not only homologous to human Dmc1 (5) but also to Rad51 of S.pombe (12), and its S.cerevisiae (2) and human (6) counterparts (data not shown). Motifs A and B, which are consensus sequences for ATP-binding enzymes, are also conserved in SpDmc1 (26).
Genomic structure in the vicinity of dmc1+
The above results suggest that the eta1 mRNA contains two ORFs. Since it is believed that eukaryotic mRNA is essentially monocistronic, we wished to confirm that the unusual structure of eta1 cDNA was not an artifact generated by fusion of two distinct cDNAs. For that purpose, we performed a series of northern blots using distinct probes specific for the 5′ upstream and 3′ downstream regions, dmc1+, eta1+ and rad24+. As shown in Figures 1A and 2C–F, the rad24+ probe recognized two mRNAs, one at 2.8 kb and the other at 1.0 kb (Fig. 1A), as described above, whereas the dmc1+ and eta1+ probes recognized only the 2.8-kb band (Fig. 2C–F). In contrast, the 5′ upstream and 3′ downstream probes recognized bands of 2.0 and 1.1 kb, respectively. These results indicate that both the eta1+ (2.8 kb) and rad24+ (1.0 kb) mRNAs are bona fide transcripts from the eta1 genomic region.
Expression of dmc1+ is controlled by ste11+
A variety of meiosis-related genes whose transcription is induced during nitrogen starvation and conjugation are known to be directly or indirectly regulated by the ste11+ gene product, which encodes a high-mobility-group (HMG) box transcription factor (27). Therefore, in order to determine whether induction of eta1 transcription is also regulated by ste11+, we carried out northern analysis using RNA obtained from an ste11 null mutant. As shown in Figure 2G, the 2.8-kb eta1+ band, but not the 1.0-kb rad24+ band, was completely eliminated in the ste11 null mutant during the time course of meiosis indicating that the induction of dmc1+ transcription was dependent on ste11+. The presence of a short sequence (TTCATTGTTT), which resembles the Ste11-responsive element known as a ‘TR box’ 261 bp upstream of the eta1 ORF (Fig. 2B), further suggests that transcription of eta1+ is under the direct control of the Ste11 transcription factor (27).
Dmc1 is not essential for growth or meiosis
To investigate the physiological role of dmc1+ in relation to rad24+ and the biological significance of the meiosis-specific bicistronic transcription of dmc1+ and rad24+, we constructed three types of mutants with disrupted genes by one-step gene replacement (Table 1). We disrupted dmc1+ alone (D10, D16), rad24+ alone (R10, R16) or both dmc1+ and rad24+ (DR10, DR16). Disruption of the targeted genes was confirmed by screening for the uracil auxotrophic marker and by Southern blotting (data not shown). Diploid cells with one of the targeted genes replaced by ura4+ sporulated and germinated with a segregation ratio of 1:1 to the wild type. All of the mutants produced viable spores, indicating that the dmc1+ and rad24+ genes are not essential for growth (data not shown), and that the disruption of both genes also has no effect on cell growth. The growth properties and cell morphology of gene-disrupted cells were also indistinguishable from those of wild-type cells except that rad24Δ and dmc1Δ rad24Δ cells displayed semi-wee phenotype (data not shown) as reported previously (28,29). Moreover, all of the gene-disrupted cells underwent normal meiosis when subjected to nitrogen starvation (data not shown). The dispensability of dmc1+ in meiosis is in contrast to its homolog, the DMC1 gene of S.cerevisiae, which has been shown by mutational analysis to be essential for meiosis (1). Since dmc1+ mRNA in the rad24Δ strain has no rad24+ ORF at its 3′ end, our results also suggest that the downstream rad24+ ORF in dmc1 mRNA plays no essential role in the progression of meiosis.
The frequency of crossing over is reduced in dmc1 mutants
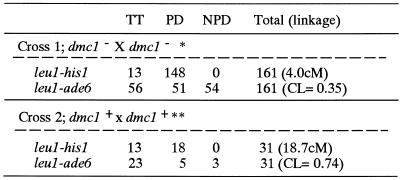
To determine the effect of dmc1 mutation on the frequency of recombination, we performed tetrad analysis and determined the genetic distance between his2 and leu1 on chromosome II. As shown in Table 2, the genetic distance was 18.7 cM in a control mating (dmc1+ × dmc1+), whereas in crosses between two dmc1Δ mutant strains, it was decreased to 4.0 cM, ∼21% of the control distance. The result indicates that dmc1Δ mutant is deficient in meiotic crossing over and Dmc1 plays a pivotal role in meiotic recombination.
Table 2. The frequency of crossing-over is reduced in dmc1 mutants.
Strains used: *NP16-6A × NP16-6D; **NP16-6B × NP16-6C. TT, tetratype; PD, parental ditype; NPD, non-parental ditype; cM, centiMorgan = (TT + 6NPD)/2(TT + PD + NPD); CL, centromere linkage = TT/(TT + PD + NPD).
We next examined the centromere linkage of two relevant loci, namely ade6 on chromosome III and leu1 on chromosome II. As shown in Table 2, in a control mating (dmc1+ × dmc1+), centromere linkage (ade6–leu1) was 0.74. We thus observed no centromere linkage between ade6 and leu1 markers in a wild-type background, which is as expected since the genetic distances between ade6 and leu1 and the respective centromeres are ∼15 and >100 cM, respectively (30). In contrast, in the dmc1Δ mutant strains, centromere linkage was reduced to 0.35 (Table 2), indicating that the recombination frequency in centromere-proximal regions is also reduced in dmc1Δ mutants.
Frequency of meiotic gene conversion was reduced in dmc1 mutants
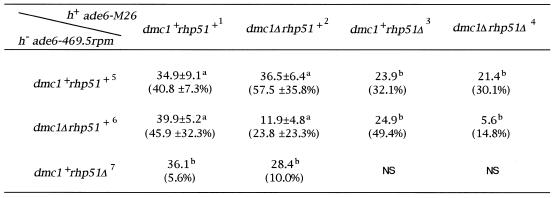
We next analyzed the frequency of meiotic gene conversion by crossing ade6-M26 and ade6-469 mutants, and counting the ade-independent and ade-dependent colonies generated from ethanol-resistant spores of mating mixtures (23). As shown in Table 3, disruption of the dmc1+ gene caused a reduction of meiotic gene conversion at the ade6 locus, and rhp51Δ mutants were completely deficient for sporulation. The proportion of ade-independent colonies from dmc1Δ × dmc1Δ crosses (0.12%) was reduced to one-third of that observed in wt × wt (0.35%) or wt × dmc1Δ (0.37%) crosses, although sporulation frequency was indistinguishable between these matings. In contrast to dmc1Δ mutant cells, rhp51Δ mutant cells could not form spores. In a cross heterozygous for rhp51 (wt × rhp51Δ crosses), cells proceeded to partial meiosis with a reduced meiotic gene conversion rate (0.36 or 0.24%). The reduction in the gene conversion rate in dmc1+ × rhp51Δ crosses was not significant when dmc1+ gene was eliminated (0.28 or 0.21%). A mutation heterozygous for dmc1 did not affect the gene conversion rate in any crosses (0.37, 0.40 and 0.21%), whereas a mutation homozygous for dmc1 resulted in remarkable reductions in the gene conversion rate (0.12 and 0.056%). Spore viability was not affected in crosses involving dmc1Δ mutants, but was severely reduced in dmc1Δrhp51+ × dmc1Δrhp51Δ crosses (10% of wt crosses). No spore was formed in rhp51Δ × rhp51Δ crosses. These results indicated that mutation of the dmc1+ gene did not affect meiosis but caused defects in the meiotic gene conversion process, and that, in contrast to dmc1+, rhp51+ was essential for meiosis. Because a mutation heterozygous for rhp51 led to reduction of meiotic gene conversion, and more severe defects were observed both in gene conversion and in spore viability when the dmc1+ gene was eliminated from the crossing strain, it appears likely that Rhp51 also functions in the process of meiotic gene conversion, and that the Rhp51 function may be indispensable for meiosis.
Table 3. Meiotic gene conversion frequencies in dmc1Δ and rhp51Δ strains.
NS, spore formation is too poor to analyze statistically. Strains along the top row were of the genotype h– ade6-M26. Strains on the left column were of the genotype h+ ade6-469.5rpm. The full genotype of these strains is described in Table 1. In each cross, zygotic spores produced on ME medium were treated with 30% ethanol and plated on EMM2 medium containing appropriate supplements in several dilution series (Materials and Methods). More than 50 Ade+ recombinations were scored for each recombination frequency determination. More than 50 (and generally more than 100 colonies) were counted to determine total viable spores. Strain name: 1, K37; 2, NP28-3D; 3, YSM6-6; 4, YSM6-12; 5, YSM2-16; 6, YSM1-50; 7, YSM8-24.
aThe average of recombination frequency (×10–4) and viability (below the frequency) in three independent assays.
bRecombination frequency (×10–4) and viability (below the frequency) from a single assay.
DISCUSSION
SpDmc1 is a fission yeast homolog of budding yeast Dmc1
In the present study, we isolated the dmc1+ gene from the fission yeast S.pombe in the course of screening for genes whose expression is specifically induced during meiosis. The deduced amino acid sequence of the dmc1+ gene strongly resembled that of S.cerevisiae DMC1, which plays an important role in meiosis-specific recombination and synaptonemal complex formation (1). Dmc1 is also structurally homologous to budding yeast Rad51 and to Rhp51, a fission yeast homolog of Rad51 (12), both of which are believed to be involved in mitotic recombination and repair of DNA damage (2). The timing of dmc1+ expression during meiosis (Fig. 1) was very similar to that of S.cerevisiae DMC1 (1). These levels of structural similarity, and its meiosis-specific expression with a similar timing, strongly suggest that dmc1+ is a fission yeast homolog of DMC1.
Tetrad analysis to examine the genetic distance between his2 and leu1 on chromosome II indicated that Dmc1 plays a role in recombination (Table 2). Examination of the centromere linkage between ade6 on chromosome III and leu1 on chromosome II indicated that the recombination frequency in such centromere-proximal regions is also reduced in dmc1 mutants (Table 2). Analysis of the gene conversion frequency by crossing ade6-M26 and ade6-469 mutants showed that gene conversion at the ade6 gene was reduced although sporulation was normal in the dmc1Δ cells (Table 3). These results further indicate that the dmc1+ gene of S.pombe is a functional homolog of budding yeast DMC1, although their functions are somewhat different.
Relationship between Dmc1 and Rhp51
In S.cerevisiae, a dmc1 mutation displayed more severe defects in crossover recombination, DSB formation and meiotic progression, but less severe defects in viability and intrachromosomal recombination during return to growth than does a rad51 mutation (2). A dmc1 rad51 double mutant is more defective than a dmc1 mutant in inter-homolog and inter-chromosomal recombination during return to growth. Thus, Dmc1 and Rad51 seemed to have both distinct and overlapping roles in meiotic recombination.
On a structural basis, Dmc1 and Rhp51 of S.pombe are closely related to one another, each of the two fission yeast proteins being more similar to its orthologs than they are to one another. In the rhp51Δ mutant, the spore viability is only 1.7%, suggesting an essential role for Rhp51 in meiosis. The rhp51Δ mutant showed a 15-fold reduction in mitotic homologous recombination compared with the wild-type strain, and also showed an increase in the percentages of gene conversions relative to homologous integrations (12). In budding yeast, Dmc1 and Rad51 (a homolog of Rhp51) have been shown to make unique contributions to meiotic recombination, sharing at least one overlapping function but otherwise playing distinct roles (4).
As shown in this report (Table 3), a mutant heterozygous for dmc1 did not affect the gene conversion rate, while a mutation homozygous for dmc1 caused remarkable reductions in the gene conversion rate, indicating that Dmc1 plays an important role in meiotic recombination. A mutation heterozygous for rhp51 caused a reduced meiotic gene conversion rate, which was further reduced by cumulative elimination of dmc1+. A mutant homozygous for rhp51 is inviable since rhp51+ is required for vegetative growth. Furthermore, a mutant homozygous for dmc1 and heterozygous for rad51 showed a greater defect in the formation of inter-homolog recombinants during meiosis than the mutant heterozygous for both dmc1 and rhp51. Disruption of dmc1+ resulted in no apparent defect in meiotic progression. This is in contrast to budding yeast DMC1, which is essential for meiosis and mutation of which causes cell cycle arrest late in meiotic prophase (1). On the other hand, rhp51+ was essential for meiosis and rhp51 mutants were deficient for sporulation. It is of note that, unlike rhp51+ mutants (12), the dmc1Δ cells were not sensitive to γ or UV radiation (data not shown). These results suggest that both Dmc1 and Rhp51 play important roles in meiotic recombination, sharing distinct but overlapping functions during meiosis in a manner similar to budding yeast Dmc1 and Rad51.
The rhp51Δ mutant cells are extremely sensitive to X-rays, indicating that the rhp51+ gene is involved in the repair of X-ray damage (12). Similarly, cells with disrupted rad24 gene are sensitive to γ and UV radiation (13). In contrast, dmc1Δ cells are sensitive neither to γ irradiation nor to UV radiation (data not shown). Moreover, when we compared the γ or UV sensitivity of wild-type (C525a), rad24Δ, dmc1Δ or rad24Δdmc1Δ cells, we detected no effect relative to rad24Δ cells (data not shown). Thus, the dmc1+ gene itself does not contribute to UV and γ radiation tolerance, nor is it directly involved in the DNA repair mechanism mediated by the product of the rad24+ gene.
The dmc1+ mRNA contains two ORFs
We report here that Rad24 was translated from eta1 mRNA during meiosis. However, the level of endogenous 1.0 kb transcript derived from the rad24+ gene driven by its own promoter was equal to that of eta1 (Fig. 1A). The large amount of Rad24 protein translated from this 1.0-kb mRNA may overshadow the small amount of Rad24 protein expressed from eta1 mRNA and preclude any necessity for additional Rad24 expression. Indeed, we detected no physiological effect on meiotic progression even when the rad24+ region of eta1 was deleted (data not shown). Gene products derived from polycistronic mRNAs in bacteria generally have related functions. In the present case, however, no relationship between the physiological functions of Dmc1 and Rad24 has been detected (data not shown). Lack of a binding motif for 14-3-3 proteins (31) in Dmc1 may exclude the possibility of direct binding of Rad24 to Dmc1. In fact, we examined the in vivo interaction between Dmc1 and Rad24 by immunoprecipitation, and could not detect any stable association between them (data not shown). The bicistronic mode of eta1 mRNA transcription therefore appears to have little physiological significance, suggesting that it may be a run-off resulting from the absence of an appropriate termination signal in the dmc1+ gene.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Dr C. Shimoda for gifts of CD16-1, CD16-5 and mei4Δ strains, Dr J. Kohli for gifts of strains to perform experiments on meiotic recombination, and Dr M. Yamamoto and Dr Y. Iino for a gift of the ste11Δ strain. We thank Ms Y. A. Kishi and Mr W. Sahara for technical assistance during the preparation of S.pombe RNA. We also thank Dr P. Hughes for critical reading of the manuscript. This work was supported by a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan, and grants from The Naito Foundation and Uehara Foundation.
DDBJ/EMBL/GenBank accession no. AB008545
REFERENCES
- 1.Bishop D.K., Park,D., Xu,L. and Kleckner,N. (1992) Cell, 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 2.Shinohara A., Ogawa,H. and Ogawa,T. (1992) Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- 3.Bishop D.K. (1994) Cell, 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- 4.Shinohara A., Gasior,S., Ogawa,T., Kleckner,N. and Bishop,D. (1997) Gene Cell, 2, 615–629. [DOI] [PubMed] [Google Scholar]
- 5.Habu T., Taki,T., West,A., Nishimune,Y. and Morita.T. (1996) Nucleic Acids Res., 24, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 7.Terasawa M., Shinohara,A., Hotta,Y., Ogawa,H. and Ogawa,T. (1995) Genes Dev., 9, 925–934. [DOI] [PubMed] [Google Scholar]
- 8.Ikeya T., Shinohara,A., Sato,S., Tabata,S. and Ogawa,T. (1996) Gene Cell, 1, 379–389. [DOI] [PubMed] [Google Scholar]
- 9.Rockmill B., Sym,M., Scherthan,H. and Roeder,G.S. (1995) Genes Dev., 9, 2684–2695. [DOI] [PubMed] [Google Scholar]
- 10.Pittman D.L., Cobb,J., Schimenti,K.J., Wilson,L.A., Cooper,D.M., Brignull,E., Handel,M.A. and Schimenti,J.C. (1998) Mol. Cell., 1, 697–705. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida K., Kondoh,G., Matsuda,Y., Habu,T., Nishimune,Y. and Morita,T. (1998) Mol. Cell, 1, 707–718. [DOI] [PubMed] [Google Scholar]
- 12.Muris D.F.R., Vreeken,K., Carr,A.M., Broughton,B.C., Lehmann,A.R., Lohman,P.H.M. and Pastink,A. (1993) Nucleic Acids Res., 21, 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford J.C., Al-Khodairy,F., Fotou,E., Sheldrick,K.S., Griffiths,D.J. and Carr,A.M. (1994) Science, 265, 533–535. [DOI] [PubMed] [Google Scholar]
- 14.Aitken A. (1996) Trends Cell Biol., 6, 341–347. [DOI] [PubMed] [Google Scholar]
- 15.Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- 16.Gutz H., Heslot,H., Leupold,U. and Loprieno,N. (1974) In King,R.C. (ed.), Handbook of Genetics. Plenum Press, NY, Vol. I, pp. 395–446.
- 17.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments with Fission Yeast. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Leupold U. (1970) Methods Cell Physiol., 4, 169–177. [Google Scholar]
- 19.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Kitamura K. and Shimoda,C. (1991) EMBO J., 10, 3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobori M., Ikeda,Y., Nara,H., Kumegawa,M., Nojima,H. and Kawashima,H. (1998) Gene Cell, 3, 459–475. [DOI] [PubMed] [Google Scholar]
- 22.Kobori M. and Nojima,H. (1993) Nucleic Acids Res., 21, 2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grimm C., Kohli,J., Murray,J. and Maundrell,K. (1988) Mol. Gen. Genet., 215, 81–86. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa H., Nakagawa,T., Uno,Y., Kitamura,K. and Shimoda,C. (1994) Mol. Gen. Genet., 244, 456–464. [DOI] [PubMed] [Google Scholar]
- 25.Grimm C., Bahler,J. and Kohli,J. (1994) Genetics, 136, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker J.M., Saraste,M., Runwick,M. and Gay,N. (1982) EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugimoto A., Iino,Y., Maeda,T., Watanabe,Y. and Yamamoto,M. (1991) Genes Dev., 5, 1990–1999. [DOI] [PubMed] [Google Scholar]
- 28.Al-Khodairy F., Fotou,E., Sheldrick,K.S., Griffiths,D.J., Lehmann,A.R. and Carr,A.M. (1994) Mol. Biol. Cell. 5, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford J.C., Al-Khodairy,F., Fotou,E., Sheldrick,K.S., Griffiths,D.J. and Carr,A.M. (1994) Science 265, 533–535. [DOI] [PubMed] [Google Scholar]
- 30.Münz P., Wolf,K., Kohli,J. and Leupold,U. (1989) In Nasim,A., Young,P. and Johnson,B.F. (eds), Molecular Biology of the Fission Yeast. Academic Press, San Diego, CA, pp. 1–30.
- 31.Yaffe M.B., Rittinger,K., Volinia,S., Caron,P.R., Aitken,A., Leffers,H., Gamblin,S.J., Smerdon,S.J. and Cantley,L.C. (1997) Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]