Abstract
The bacterial trialkyl-substituted aromatic polyketides are structurally featured with the unusual aromatic core in the middle of polyketide chain such as TM-123 (1), veramycin A (2), NFAT-133 (3) and benwamycin I (4), which were discovered from Streptomyces species and demonstrated with antidiabetic and immunosuppressant activities. Though the biosynthetic pathway of 1−3 was reported as a type I polyketide synthase (PKS), the PKS assembly line was interpreted inconsistently, and it remains a mystery how the compound 3 was generated. Herein, the PKS assembly logic of 1−4 was revised by site-mutagenetic analysis of the PKS dehydratase domains. Based on gene deletion and complementation, the putative P450 monooxygenase nftE1 and metallo-beta-lactamase (MBL) fold hydrolase nftF1 were verified as essential genes for the biosynthesis of 1−4. The absence of nftE1 led to abolishment of 1−4 and accumulation of new products (5−8). Structural elucidation reveals 5−8 as the non-aromatic analogs of 1, suggesting the NftE1-catalyzed aromatic core formation. Deletion of nftF1 resulted in disappearance of 3 and 4 with the compounds 1 and 2 unaffected. As a rare MBL-fold hydrolase from type I PKSs, NftF1 potentially generates the compound 3 through two strategies: catalyze premature chain-offloading as a trans-acting thioesterase or hydrolyze the lactone-bond of compound 1 as an esterase.
Keywords: Thioesterase, Metallo hydrolase, Metallo-beta-lactamase, Aromatic polyketide, Polyketide synthase, P450 monooxygenase
1. Introduction
Bacterial aromatic polyketides, mostly isolated from Actinomyces species, represent a rich molecular source for the discovery of novel therapeutic agents [1]. The majority of them are biosynthesized by type II PKSs, which contain the monofunctional proteins of ketoacyl-synthase (KS), chain length factor, and acyl carrier protein (ACP) to carry out repetitive decarboxylative Claisen-condensation of a malonyl-CoA extending unit with an acyl starter unit to yield a reactive poly-ß-ketone backbone followed by aromatic cyclizations [2]. Comparatively, few aromatic polyketides are derived from type I PKSs, which feature a polyketides assembly line of the multifunctional PKS proteins consisted of the core catalytic domains of KS, acyltransferase (AT) and ACP, the accessory domains of ketoreductase (KR), dehydratase (DH) and enoylreductase (ER), and an offloading C-terminal thioesterase (TE) domain [3]. The aromatic polyketides derived from type I PKSs normally contain the terminal benzyl ring derived from the starting building block of PKS assembly [4]. Attractively, there are a group of trialkyl-substituted aromatic polyketides derived from type I PKSs in Streptomyces, structurally featuring with a benzene core in the middle of polyketide chain such as TM-123/veramycin B (1), veramycin A (2), NFAT-133 (3), benwamycin I (4), lorneic acids, and gombapyrones [[5], [6], [7], [8], [9], [10]] (Fig. 1). Recently, the aromatic cores of lorneic acids and gombapyrones were verified to be formed by unusual cytochrome P450 monooxygenases, which form a benzene ring in the linear polyene chain offloaded from type I PKSs [11].
Fig. 1.
Reported trialkyl-substituted aromatic polyketides derived from type I PKSs in bacterium Streptomyces.
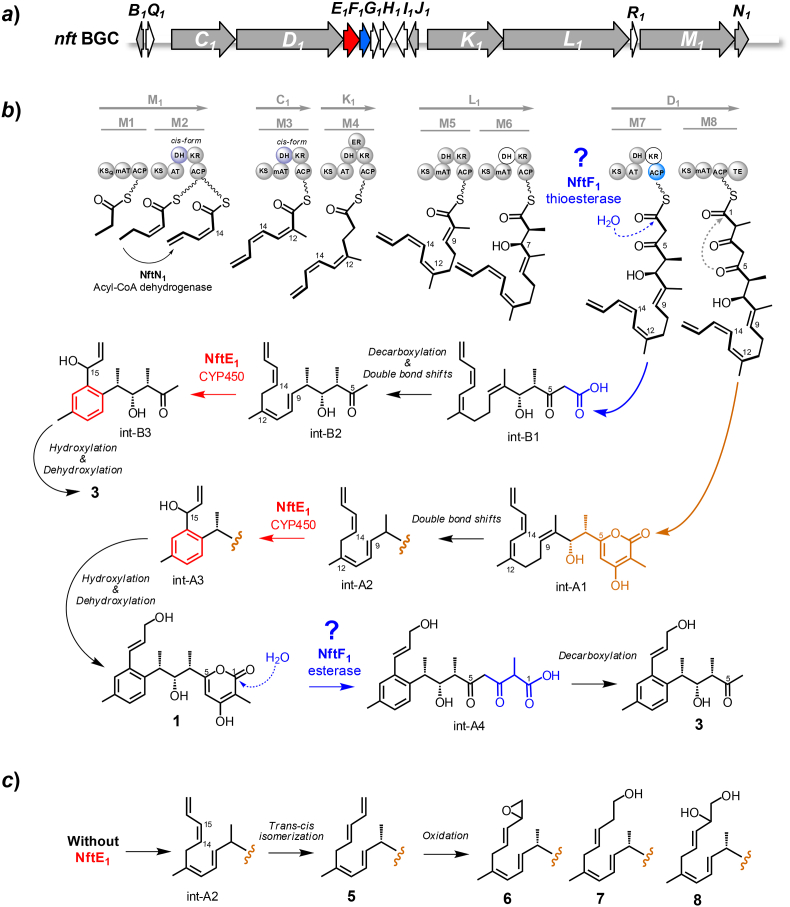
The biosynthetic pathway of 1−3 was investigated through genetic mutation, metabolite characterization and isotope feeding experiments in two Streptomyces species which contain cognate biosynthetic gene cluster of 1−3 (nft BGC) [8,12]. However, the PKS assembly line was interpreted inconsistently, and it remains a mystery how the compound 3 is generated in the biosynthetic pathway. The structure of compound 3 appears as a truncated portion of the compound 1 being lost three carbons in the terminal α-pyrone ring of compound 1 (Fig. 1). This raises a hypothesis that compound 3 could be derived from a chain prematurely offloaded from ACP7 by a trans-acting TE, for instance a MBL-fold hydrolase encoded by nftF1 (Fig. 2). Conventionally, type I PKSs offload chain by an ɑ/ß hydrolase-type TE domain integrated at the C-terminus of PKS assembly line via thioester hydrolysis or O–C macrocyclization [13]. So far, there are not documented MBL-fold TEs from type I PKSs, and only two cases of MBL-fold hydrolases were reported in the type II PKSs of actinorhodin and murayaquinone, in which ActIV and MrqD were characterized as MBL-fold TEs for chain release based on genetic mutation and biochemical analysis with an artificial substrate [14,15]. Interestingly, these cognate genes are discovered frequently in type II PKS BGCs and annotated as ring cyclase/aromatase such as RslC2, AlnR and Ssfy2 from the biosynthetic pathways of rishirilides, alnumycin, and tetracycline SF2575 [[16], [17], [18]].
Fig. 2.
The gene organization of nft BGC from S. conglobatus (a), and the proposed biosynthetic logics of compounds 1 and 3 (b) and the mutant compounds 5−8 (c). The question marks are used to indicate the two possible working-mechanisms of NftF1 in producing compound 3.
In this study, the PKS assembly line of 1−3 was revised in Streptomyces conglobatus, based on site-directed mutagenesis of the PKS genes. Structure elucidation of the mutant metabolites suggested an unusual P450 monooxygenase-mediated aromatization occurred in the polyketide biosynthesis of 1−3. Moreover, the production of 3 was identified to be controlled by a MBL-fold hydrolase homolog, which could perform premature chain-offloading as a new-type TE in type I PKSs or hydrolyze the pyrone ring of compound 1 as an esterase.
2. Materials and methods
2.1. Chemical reagents and kits
The primers (Table S1) and Plasmid DNA Extraction Kit were ordered from Shanghai Generay Biotech (China). DNA sequencing was provided by Shanghai Sangon Biotech (China). DNA fragments were assembled by using 2 × Ezmax-Muli CloneMix Plus (Shanghai Tolo Biotech, China). PCR amplifications were carried out by using KOD OnePCR Master Mix (TOYOBO) for gene cloning and by using 2 × Flash PCR MasterMix (CoWin Biotech, China) for colony screening. Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs. Genomic DNA was prepared by using 10% Chelex 100 resin (Bio-Rad) solution. Other chemical reagents used were purchased from Sigma-Aldrich.
2.2. Strains and plasmids
The RJ2 strain was used as starting strain in the study, which was generated by abolishing the conglobatin production from S. conglobatus ATCC 31005 [19]. E. coli DH10B strain was used for plasmid construction. E.coli ET12567 strain containing pUZ8002 plasmid was used for conjugation with Streptomyces. E. coli BL21 (DE3) plysS strain was used for protein expression. Genetic mutations in RJ2 strain were created by using the E. coli−Streptomyces shuttle plasmid pRJ2 [20]. Gene complementation was performed by using the plasmids derived from pRJ5 that contains a phage phiC31 integration element [21]. Plasmid pET-29a was used for protein expression in E. coli. All the plasmids constructed in this study was listed in Table S2.
2.3. Media and culture conditions
TSBY medium (3% tryptone soy broth, 0.5% yeast extract, 10% sucrose, 0.1% antifoam) was used to culture the mycelium of Streptomyces. SFM agar medium (2% soyean flour, 2% D-mannitol, 2% agar, pH7.4) was used for conjugation and mutant screening of Streptomyces. The SGC medium (3% soybean flour, 5% glucose, 0.5% CaCO3, and 0.1% (v/v) antifoam) was used for liquid fermentation. Large-scale fermentation of Streptomyces was carried out in a 500 mL conical flask fitted with a metal spring, with 150 mL of SGC medium inoculated with 1% (v/v) of 3-day TSBY culture, and then incubated at 30 °C, 220 rpm for 5 days. For small-scale fermentation, 250 mL conical flask containing 50 ml medium was used. E. coli strains were grown at 37 °C in the Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or the solid medium (LB plus 1.5% agar) supplemented corresponding antibiotics.
2.4. Isolation of compounds
For isolation of compounds 1–4, a 12-L SGC culture broth of RJ2 strain was extracted five times with an equal volume of ethyl acetate. The combined organic phase was concentrated under vacuum evaporation to yield 12 g syrup, which was subjected to a vacuum liquid chromatography with silica gel (200–300 mesh) in a stepwise elution of CH2Cl2–CH3OH (from 100/1 to 0/1, v/v). Guided by HPLC–MS analysis, the fractions containing target compounds were combined for further separation on an ODS chromatography column (Santai Technologies, Inc., Spherical C18, 31.2 × 257.4 mm, 15 μm, 100 Å) by using MPLC (medium pressure preparative liquid chromatography) under the elution conditions of 20 mL/min and 20–100% CH3CN/H2O (0.1% formic acid) for 6 h. The target fractions were separated by using preparative HPLC fitted with a semipreparative column (Waters Xbridge C18, 10 × 250 mm, 5 μm) with an elution condition of 60% CH3OH/H2O (0.1% formic acid) and 3 mL/min to yield 20.4 mg of 1, 2.5 mg of 2, 29.6 mg of 3, and 5.5 mg of 4.
For isolation of compounds 5–8, a 24-L SGC culture broth of ΔnftE1 strain was extracted with equal volume ethyl acetate for 5 times. The combined extracts were concentrated via vacuum evaporation to yield 18 g syrup, which was separated through a Sephadex LH-20 column chromatography by using the elution of CH2Cl2/CH3OH (1/1, v/v) to give 17 fractions. With the guidance of HPLC–MS analysis, the target products were detected in the fractions 7–10, which were combined and further separated by the MPLC same as above. The mobile phase of CH3CN and H2O (0.1% formic acid, v/v) was eluted at 25 mL/min with a gradient condition of 10–80% CH3CN for 3 h and 80–100% CH3CN for 30 min. According to HPLC–MS analysis, all the 65 fractions were pooled into the 16 fractions. Compound 5 was directly obtained as clearing white crystal (32 mg). Compound 6 (5.9 mg), 7 (19.4 mg), and 8 (37.5 mg) were separated from these fractions by using the same preparative HPLC as above under the condition of 3.0 mL/min and 60% CH3OH/H2O (0.1% FA, v/v).
2.5. Mutant construction and gene complementation
To generate each of the deletion mutations: nftE1−I1, nftC1-DH3 (H972Y), nftK1-DH4 (H969Y), nftD1-DH7 (H964Y), ΔnftE1, ΔnftF1, ΔnftM1, and ΔnftG1, the two DNA fragments used for homologous recombination were prepared through PCR amplification with the genomic DNA of RJ2 as template and the related primers (see primers and descriptions in Table S1). By using Gibson assembly method, the two PCR fragments were assembled with the E. coli–Streptomyces shuttle plasmid pRJ2 linearized by XbaI and EcoRI. The resulting plasmids (Table S2) were individually introduced into the RJ2 strain by conjugation. The target mutant strains were screened by using colony PCR with the related primers (Table S1) from the hygromycin-sensitive colonies prepared after two rounds of propagation on the SFM plates without antibiotics added. The mutant strains were confirmed by sequencing the PCR products.
For mutant complementation, the target genes were amplified by PCR using the genomic DNA of RJ2 as template and the corresponding primers (Table S1). The synthesized fragment of constitutive strong promoter KasO*p [22] and a gene fragment were assembled with an integrative vector pRJ5 (NsiI and EcoRV) by Gibson assembly. The resulting plasmids (Table S2) were transformed into the corresponding mutant strains by conjugation. The exconjugants were selected based on hygromycin-resistance and sequencing of the PCR product.
2.6. Analytical methods
For routine HPLC–MS analysis, 7 mL of 5-d fermentation broth was extracted with an equal volume of ethyl acetate. Then 5 mL of the extract was collected and dried over by nitrogen blowing. The samples were redissolved into 800 μL of CH3OH and centrifuged at 12000 rpm for 6 min before injecting 20 μL supernatant for analysis. The HPLC–MS analysis was performed on a Waters HPLC coupled with a Waters Acquity QDa detector and a Waters Xbridge C18 column (250 mm × 4.6 mm, 5 μm). Samples were eluted with the mobile phases of CH3CN and H2O (0.1% formic acid, v/v) at a flow rate of 0.7 mL/min with a gradient elution of 30–100% CH3CN over 30 min. The mass spectrometer was run in positive ionization mode, scanning from m/z 200 to 1250 with capillary (0.80 kV), cone (15.00 V), and source temperature (120 °C). The HRMS spectra were recorded in a Waters XeVO G2-XS Q TOF mass spectrometer conducted in positive ionization mode, scanning from m/z 100 to 1200. NMR spectra were recorded on Bruker 600 MHz or Bruker NEO 700 MHz instruments. The compounds 2 and 3 were analyzed in CD3CN-d3 and CDCl3, respectively, and the other compounds were detected in DMSO-d6. Chemical shifts (δ) were obtained in reference to tetramethylsilane (TMS) at 0.00 ppm.
2.7. X-ray crystallographic analysis
Crystals data was obtained on Bruker D8 VENTURE diffractometer (Bruker Corp., Karlsruhe, Germany). The crystal structures were solved by direct methods using the SHELXT-2015 software package and refined by the full matrix least-squares method. In the structure refinements, nonhydrogen atoms were refined anisotropically, and the hydrogen atom positions were geometrically idealized and allowed to ride on their parent atoms. The crystallographic data of 5 was deposited in Cambridge Crystallographic Data Center (CCDC number: 2246505).
2.8. NftE1 and NftF1 protein expression and enzymatic assay
For constructing protein expression plasmids, the DNA fragment of nftE1 or nftF1 was amplified from the genomic DNA of RJ2 with the corresponding primers (Table S1). The C-terminal 6 × His tag was introduced by the antisense primer. Each of the DNA fragments was introduced at NdeI and XhoI sites of pET29a through Gibson assembly. The resulting plasmids (Table S2) were transferred into E. coli BL21 (DE3) plysS for protein expression.
For protein expression fermentation, 1-mL overnight culture of the target transformant was inoculated into 500-mL LB medium containing 50 μg/mL kanamycin and incubated at 37 °C and 220 rpm. Protein expression was induced by adding 0.2 mM Isopropyl-β-d-thiogalactopyranoside once the OD600 reached 0.4–0.6. After further cultivation for 15 h at 22 °C, the cells were harvested by centrifugation (11,325×g, 5 min) and resuspended in buffer A (50 mM Tris-HCl, 300 mM NaCl, pH 7.2) for sonication. The cell lysate was centrifuged at 34,925×g for 25 min, and the supernatant was filtrated through a 0.22-μm filter before loaded onto a His-Bind affinity column (1-mL bed volume). The target proteins were separated by stepwise increases of the concentration of imidazole (10–500 mM). Based on SDS-PAGE (12.5%) analysis, the target fractions were concentrated and desalted by using Amicon Ultra-4 concentrators (Millipore, 30-kDa cutoff) and PD10 column (GE Healthcare). Protein concentrations were measured by NanoDrop 1000 spectrophotometer.
The NftE1 enzymatic assay was conducted in a 50 μL reaction system consisted of 1 mM compound 5 or 6 or 7 or 8, 100 μg/mL ferredoxin (Fdx, spinach), 0.2 U/mL ferredoxin-NADP+ reductase (FdR, spinach), 10 U/mL glucose dehydrogenase, 10 mM glucose and 1 mM NADPH in 50 mM Tris-HCl buffer (pH 7.4). The modified NftE1 assay was carried out by using the alternative Fdx and FdR derived from cyanobacterium Synechococcus elongatus [23] with the recipe of 1 mM substrate, 10 μM Fdx, 10 μM FdR, 1 mM NADPH, 10 μM NftE1 in 50 mM PBS Buffer (pH7.5). The assay of NftF1 carried out in a 50 μL reaction system consisted of 1 mM Zn2+ or Mn2+ or Cu2+ or Mg2+, 10 μM NftF1, 0.5 mM compound 1 in 50 mM Tris-HCl buffer (pH6.8). All the reactions were incubated at 30 °C for 5 h, and halted by adding 500 μL of CH3CN before injecting 20 μL of the supernatant for HPLC–MS analysis.
2.9. Chemical complementation
Chemical complementation of ΔnftM1 strain was performed by adding 15-μL DMSO solution of compound 1, 5 or 7 at time points of 2 d and 3 d during a 5-d fermentation. The concentration of stock solution was 0.2 M, and the final concentration of each compound added in the culture broth was 0.6 mM. As a negative control, the DMSO solvent was used in same condition. The fermentation was performed with 10-mL SGC medium in a 50-mL conical flask. The broth was extracted for HPLC–MS analysis after the 5-d fermentation.
3. Results and discussions
3.1. Discovery of compounds 1−4 and identification of the nft BGC from S. conglobatus
During screening of new metabolites from the mutant strain RJ2, which was generated by abolishing conglobatin production from S. conglobatus ATCC 31005 [19], we isolated four new components (compounds 1−4) from a 12-liter fermentation extract. The structures of 1−4 were elucidated based on comprehensive analysis with the 1D and 2D NMR (Fig. 1 and Fig. S1). The structures of 1 and 2 bear a 2-pyrone ring and a 1, 2, 4-trisubstituted benzyl moiety, being structurally identical to TM-123/veramycin B (1) and veramycin A (2) that were recently discovered from S. pactum ATCC 27456 and S. sp. ST157608 [8,12] (Tables S3 and 4). The compound 3 is structurally identical to the trialkyl-substituted aromatic polyketide NFAT-133 reported from several Streptomyces species, which possesses antidiabetic and immunosuppressant activities [6,8,24] (Table S5). The compound 4 is the hydroxylated analog of 3, being structurally identical to benwamycin I discovered from S. sp. KIB-H1471 [7] (Table S6).
Given the structural similarity of the trialkyl-substituted aromatic moiety in compounds 1−4 and the reported lorneic acids [25], we speculated a similar mechanism of the intramolecular benzene ring formation occurred in their biosynthesis. A genome mining was then performed toward S. conglobatus using the sequence of P450 monooxygenase homolog Orf1-257, which putatively carries out an oxidative aromatization of the polyolefinic precursor in the lorneic acids biosynthetic pathway [25]. The analysis revealed a BGC of type I PKS from S. conglobatus (GenBank OP948907), which contains a P450 monooxygenase homologous gene nftE1 showing 46% protein sequence identity to Orf1-257. The BGC has the same gene organization to the nft BGC and 77–91% identities of encoded proteins to the ones encoded in the nft BGC (Fig. S2), which was verified to account for the biosynthesis of compounds 1 and 3 in S. pactum [12].
3.2. Revising the PKS assembly line of 1−3 based on site-directed mutagenetic analysis of the DH domains in NftC1, NftK1 and NftD1
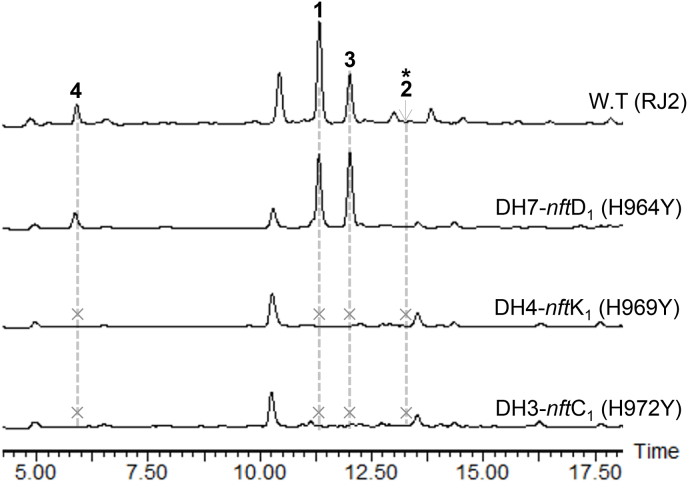
Considering that the PKS assembly logic of NFAT-133 was interpreted inconsistently [8,12], we performed site-directed mutagenetic analysis of the DH domains in NftC1, NftK1 and NftD1, corresponding to the DH3, DH4 and DH7 in the PKS assembly line proposed by in the work (Fig. 2b). By replacing the His residues in their active sites with a Tyr in the conservative motif HX3GX4P of DHs (Fig. S3), the mutant strains DH3-nftC1 (H972Y), DH4-nftK1 (H969Y) and DH7-nftD1 (H964Y) were generated. According to HPLC–MS analysis of the fermentation extracts (Fig. 3), the DH3 and DH4 mutations resulted in complete abolishment of compounds 1−4, while DH7 mutation did not affect the production of 1−4. The results indicate that the DH domain in NftC1 should be active during the PKS assembly. This is contradicted to the previous speculation that the naturally silent DH domain in NftC1 led to the presence of C-7 hydroxyl group in the module-6 of PKS assembly line [8]. Similarly, the results support that the DH domain in NftK1 should be active too, and the NftK1 encoded module could serve as module-4 in the PKS assembly line (Fig. 2b). As expected, the activity of DH7 is not necessary in the PKS owing to that the KR7 domain should be inactive with a Y1532N mutation at the key active site of Tyr (Fig. S4).
Fig. 3.
HPLC analysis of the metabolites of DH domain site-directed mutagenesis strains: DH3-nftC1 (H972Y), DH4-nftK1 (H969Y), and DH7-nftD1 (H964Y). The HPLC is displayed by using 250 nm. The asterisks mean the detected production of compound 2 by target mass extraction (see Fig. S5). The crosses mean that the target compounds were not detected.
Moreover, the double bound geometries are predicted by recognizing the diagnostic motifs of established KR types [26] (Fig. S4), in which KR4, KR5 and KR6 contain the LDD motif of B-type KRs correlating to trans-double bonds. Given that KR2 and KR3 lack the characteristic motifs of B-type KRs, we speculate that the cis-double bonds could be generated by KR2-DH2 and KR3-DH3 to facilitate the downstream aromatization. Taken together, these analyses allowed us to propose a revised PKS assembly line for 1−3 biosynthesis (Fig. 2b).
3.3. Identification of the essential genes from the nftE1−I1 gene cassette at the center of nft BGC
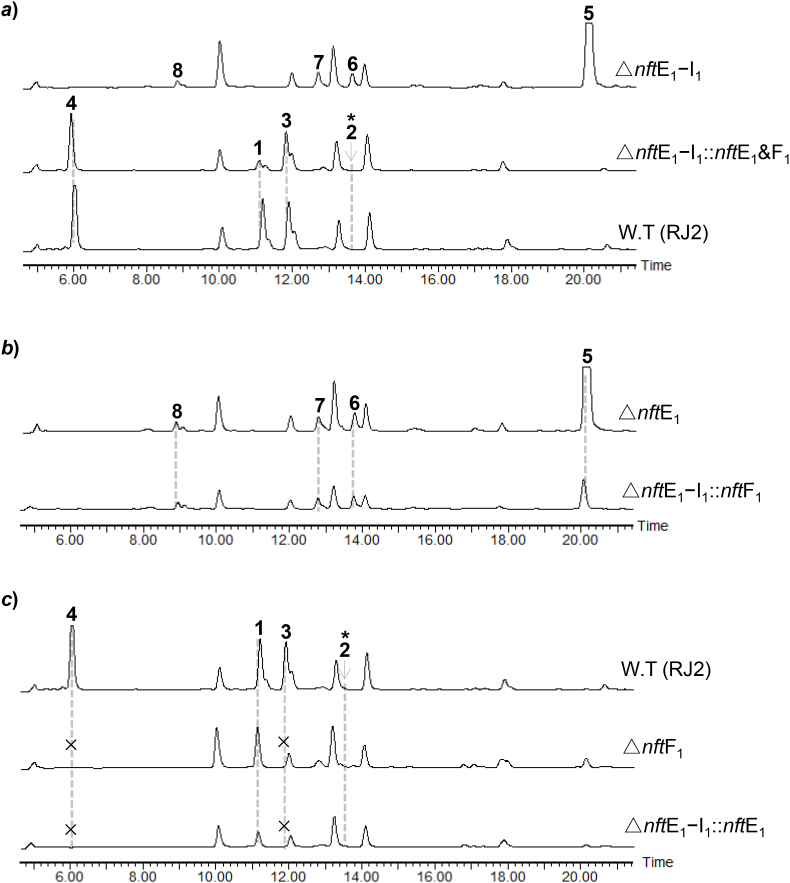
To functionally identify the five non-PKS genes nftE1−I1 at the center of nft BGC (Fig. 2a), we first constructed a mutant by deleting the whole genes cassette in the staring strain RJ2. HPLC–MS analysis of the fermentation culture revealed that the resulting mutant ΔnftE1−I1 completely lost the capacity of producing 1−4, and meanwhile four new components (compounds 5−8) were detected (Fig. 4a). Next, to discriminate the redundant genes, complementation of the ΔnftE1−I1 mutant was performed. Surprisingly, the introduction of nftE1 and nftF1 genes was able to restore the production of 1−4 in ΔnftE1−I1 strain along with the disappearance of 5−8 (Fig. 4a), revealing the redundant genes of nftG1, nftH1 and nftI1. We then performed individual gene deletion towards nftE1 and nftF1. Interestingly, the resulting ΔnftE1 mutant produced the same metabolite profiles of ΔnftE1−I1 strains (Fig. 4b). The results indicate that the P450 monooxygenase-encoding gene nftE1 should involve in a common step for the biosynthesis of 1−4, very likely the aromatic cyclization of a linear polyene chain offloaded from PKS. The proposal was supported by the recent report about the unusual cytochrome P450 monooxygenases LonP and GbnP, showing 46/91 and 46/97 identity%/coverage% to NftE1, which were verified to carry out the benzene ring formation in the biosynthesis of lorneic acids and gombapyrones [11]. Furthermore, the ΔnftF1 mutant lost the capacity of producing compounds 3 and 4 and still kept the production of 1 and 2 (Fig. 4c), indicating that nftF1 should specifically serve for generating compounds 3 and 4.
Fig. 4.
HPLC analysis of the metabolites from the mutant strains ΔnftE1−I1 (a), ΔnftE1 (b) and ΔnftF1 (c), and the complementary strains. The HPLC is displayed by using 250 nm. The asterisks mean the detected production of compound 2 by target mass extraction (see Fig. S5). The crosses mean that the target compounds were not detected.
The functions of nftE1 and nftF1 were further confirmed by individual gene complementation of the ΔnftE1−I1 mutant. As expected, introduction of nftF1 gene did not change the metabolic profile of the ΔnftE1−I1 strain (Fig. 4b), and introduction of nftE1 restored the production of 1 and 2 along with the absence of 3 and 4 in the culture of ΔnftE1−I1 :: nftE1 strain (Fig. 4c). Additionally, Zhou et al. recently reported that deletion of the putative decarboxylase gene nftG resulted in significantly reduced amount of compound 3 [27]. However, in-frame deletion of the cognate gene nftG1 in our strain showed no influence to the production of 1−4 (Fig. S6), suggesting that nftG1 should be redundant or an alternative factor encoded by the gene outside of nft BGC could bear the same activity of NftG1 in the RJ2 strain.
3.4. Structural characterization of the compounds 5−8 produced by ΔnftE1 mutant strain
To characterize the structures of compounds 5−8 by spectral analysis, a 12-liter liquid fermentation of ΔnftE1 strain was performed to isolate 32 mg of 5 (white crystal), 5.9 mg of 6 (yellow amorphous powder), 19.4 mg of 7 (pale yellow amorphous powder), and 37.5 mg of 8 (white amorphous powder) though column chromatography and preparative HPLC.
Compound 5 was determined with the molecular formula C21H28O4 by HR-ESIMS (m/z 367.1883 [M + Na]+, calcd for C21H28O4Na, 367.1880, error 0.8 ppm), displaying 8° of unsaturation. The 13C NMR and DEPT spectra of 5 revealed 21 carbon resonances, including five quaternary carbons (δC 165.4, 165.2, 164.9, 134.3 and 96.7), ten methine carbons (δC 137.1, 135.4, 132.1, 131.7, 126.2, 125.8, 99.2, 74.5, 40.9, 40.8), two methylene carbons (δC 115.7, 35.0), and four methyl carbons (δC 23.5, 17.1, 10.8, 8.4) (Table S7). The 1H–1H COSY of 5 reveals four spin systems (Fig. 5). The presence of α-pyrone ring in 5 is confirmed through the HMBC correlations between H-14 and C-16 as well as between H-21 and C-15/C-17 (Table S7, Fig. 5). A long aliphatic chain of 15 carbons is established by the HMBC correlations from H-3 to C-1, from H-4 to C-2, from H-18 to C-5/C-7, from H-7 to C-5, from H-9 to C-7/C-19, from H-19 to C-11, and from H-20 to C-11/C-13 (Table S7, Fig. 5). The positions of substituted methyl and aliphatic chain at the α-pyrone ring were confirmed based on the HMBC correlations between H-20 and C-13 as well as between H-21 and C-15/C-17 (Table S7, Fig. 5). Consequently, compound 5 was established as a methyl substituted α-pyrone ring linking with a 15-carbons polyene chain, which is identical to the planar structure of TM-125 discovered during our investigation by Zhou et al. from another ΔnftE mutant generated in S. pactum [12]. Moreover, the absolute structure of 5 was first determined in the work by using single-crystal X-ray diffraction (Fig. 5).
Fig. 5.
ORTEP drawing of 5, and the 1H–1H COSY and key HMBC correlations of 5–8.
The compounds 6−8 were determined with the molecular formulas C21H28O5, C21H30O5, and C21H30O6, corresponding to the unsaturation degrees of 8, 7 and 7, respectively, based on the HR-ESIMS analysis: 6 (m/z 383.1841 [M + Na]+, calcd for C21H28O5Na, 383.1829, error 3.1 ppm), 7 (m/z 385.1983 [M + Na]+, calcd for C21H30O5Na, 385.1985, error −0.5 ppm), and 8 (m/z 401.1928 [M + Na]+, calcd for C21H30O6Na, 401.1935, error 1.5 ppm). The 1D and 2D NMR data suggests that compounds 6−8 should be the structural derivatives of 5, differing by the terminal epoxide ring in 6, the C-1 hydroxyl in 7, and the 1, 2-ortho hydroxyls in 8 (Fig. 5). Compared to 5, the upfield shifts of C-1 (Δδ −51.2) and of C-2 (Δδ −56.8) were observed in 6 (Table S8), and one more oxygen should be possessed by 6 according to the determined molecular formula, suggesting a C-1−C-2 epoxy ring in 6. The C-1 hydroxyl in 7 could be deduced from the downfield shift resonance of C-1 (δC 60.9) and C-2 (δC 35.9) (Table S9). Compared to 7, the downfield shift of C-2 (Δδ +35.1) along with one more oxygen in the determined molecular formula suggest the 1, 2-ortho hydroxyls in 8 (Table S10). These elucidations were further confirmed by 2D NMR analysis of 6−8 (Fig. 5, Tables S7–10).
3.5. Exploration of the NftE1-mediate polyketide aromatic cyclization
The non-aromatic structure of compounds 5−8 produced by the ΔnftE1 mutant suggests that the P450 monooxygenase homologous gene nftE1 indeed involved in the aromatization process during the 1−4 biosynthesis (Fig. 2b). To investigate the in vitro activity of NftE1, we overexpressed the soluble protein of C-His6-fused NftE1 in E. coli BL21 (DE3) and purified the homogeneous protein by using Ni-NTA chromatography (Fig. S7). However, no new product was generated from the assay consisting of 10 μM NftE1, 1 mM compound 5 or 6 or 7 or 8, 100 μg/mL ferredoxin (Fdx, spinach), 0.2 U/mL ferredoxin-NADP+ reductase (FdR, spinach), 10 U/mL glucose dehydrogenase, 10 mM glucose, and 1 mM NADPH in 50 mM Tris.HCl Buffer (pH 7.4) at 30 °C for 5 h. During our investigation, Yang et al. confirmed that the cytochrome P450 LonP/Orf1-257 in lorneic acids biosynthesis catalyzes the biotransformation of an acyclic polyene substrate to the benzene ring [11]. As such, we re-investigated the activity of NftE1 by using the alternative Fdx and FdR derived from cyanobacterium Synechococcus elongatus [23]. However, the modified assay still did not provide desirable products. We speculate that the authentic substrate for NftE1 could be the intermediate int-A2 (Fig. 2b), which could be converted into compound 5 upon the absence of NftE1 by receiving a trans-cis isomerization at C14–C15 double (Fig. 2c). The proposal may explain the unable restoration of compound 1 through chemical complementation of the ΔnftM1 strain with each of compounds 5 and 7 (data not shown).
3.6. Functional identification of nftF1 gene in producing 3 and 4
The nftF1 gene, showing homologous to MBL-fold hydrolase, was originally annotated as the conventional resistance gene of secondary metabolite BGCs since that metallo-hydrolases usually confer host resistance by hydrolyzing β-lactam antibiotics [28]. Surprisingly, gene deletion mutation and complementation revealed that nftF1 gene was essential for the production of compounds 3 and 4, and uncorrelated to compounds 1 and 2 (Fig. 4c). Owing to the truncated structure feature of 3 compared to 1, we hypothesized that compound 3 could be derived from either a heptaketide chain prematurely offloaded from ACP7 or an NftF1-hydrolyzed product of compound 1 (Fig. 2b). With the recombinant protein of C-His6-fused NftF1 expressed in E. coli BL21 (DE3) (Fig. S7), we first tested the hydrolysis ability of NftF1 towards the lactone ring of 1. However, no new product was detected in the assay consisting of 10 μM NftF1, 0.5 mM compound 1, 50 mM Tris.HCl buffer (pH6.8), and 1 mM divalent metal ions of Zn2+ or Mn2+ or Cu2+ or Mg2+. Moreover, we evaluated the hydrolytic activity of NftF1 towards compound 1 by feeding, with 1, the fermentation of the ΔnftM1 strain which lost the production of 1−4 due to deletion of the PKS gene nftM1. However, no restoration of compound 3 was detected in the fermentation fed with compound 1 (Fig. S8). As such, it is rather likely that NftF1 contains TE activity to specifically offload a nascent chain from ACP7 for producing the compound 3 (Fig. 2b).
Protein BLAST analysis of NftF1 reveals a widespread MBL-fold hydrolase homolog from the BGCs of type II PKS, which were generally annotated as aromatic ring cyclases due to their non-similarities to any typical TE. Protein sequence alignment of the selected homologs from the documented type II PKS BGCs enables us to recognize the conserved THxHxDH motif and metal-binding sites from MBL family (Fig. S9). Of them, two MBL-fold hydrolases were functionally characterized with TE activities in type II PKSs, including ActIV (GenBank: CAC44204.1) and MrqD (GenBank: AQW35062.1) which show 38% identity (90% query-cover) and 31% identity (95% query-cover) to NftF1, respectively. ActIV was characterized as a bifunctional cyclase-thioesterase via in vitro reconstitution of actinorhodin biosynthesis pathway, and it was also demonstrated with TE activity towards a model substrate of type II PKS, anthraquinone-2-carboxylic acid-N-acetylcysteamine [14]. MrqD was suggested to function as a dedicated product releasing TE in murayaquinone biosynthesis based on interpretations of the gene deletion and site-directed mutagenetic analysis [15]. Thus, these bioinfomatic informations imply the TE activity of NftF1. However, more experimental evidences are required to exclude another possible mechanism, in which, with yet unidentified cofactor, NftF1 hydrolyzes the lactone-bond of 1 to yield compound 3 (Fig. 2b). Further investigation will be performed to determine the exact mechanism of NftF1 in producing the compound 3 such as chemically synthesis of the mimic substrate of N-acetylcysteaminyl thioester for analyzing the TE activity of NftF1 or screening of the cofactors of NftF1 in hydrolytic degradation of compound 1.
4. Conclusion
The PKS assembly line of trialkyl-substituted aromatic polyketides 1−4 is revised based on site-directed mutagenetic analysis of the three DH domains. The nftE1 and nftF1 genes were identified to be essential for 1−4 biosynthesis through gene deletion and complementation analysis of the nftE1−I1 gene cassette at the center of nft BGC. Structure elucidation of the new products 5−8 accumulated in the ΔnftE1 mutant suggests that the unusual benzene core formation in 1−4 should be controlled by the P450 monooxygenase NftE1. Gene deletion and complementation reveal that nftF1 gene is essential to produce 3 and 4. Protein BLAST analysis reveals that NftF1 belongs to MBL-fold hydrolase family. The homologs of NftF1 are widespread in type II PKSs, in which two homologs were demonstrated with TE activities. We speculate that NftF1 could serve as a trans-acting TE to mediate a premature chain-offloading from ACP7 yielding the precursor chain of compound 3. Alternatively, with yet unidentified cofactors, NftF1 is capable to hydrolyze the lactone-bond of 1 for yielding the compound 3.
Funding
This work received financial support from the National Natural Science Foundation of China (Nos. 32070070, 32211530074 and 31929001), and the innovative research team of high-level local universities in Shanghai. H. D. thanks Royal Society-NSFC international exchange grant (IEC\NSFC\211349).
CRediT authorship contribution statement
Ming Yang: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. Wanlu Li: Data curation, Formal analysis, Investigation, Visualization. Lin Zhou: Investigation. Xiao Lin: Investigation. Wenyu Zhang: Writing – review & editing. Yaoyao Shen: Investigation. Hai Deng: Writing – review & editing. Hou-wen Lin: Funding acquisition, Resources, Supervision. Yongjun Zhou: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
Authors declare that they have no conflict of interest.
Acknowledgements
Dr. Yongjun Zhou gratefully acknowledges Prof. Peter F. Leadlay from University of Cambridge for kindly providing the S. conglobatus strain. We thank Dr. Simon R. Williams from Department of Chemistry, University of Cambridge for the help to elucidate the strucure of compound 1 during Dr. Yongjun Zhou's postdoctoral research (2012–2015) in Prof. Peter F. Leadlay's group.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2023.05.003.
Contributor Information
Hou-wen Lin, Email: franklin67@126.com.
Yongjun Zhou, Email: zhouyongjun66@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Das A., Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc Chem Res. 2009;42(5):631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J., Zhang R., Chen X., Sun X., Yan Y., Shen X., et al. Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb Cell Factories. 2020;19(1):110. doi: 10.1186/s12934-020-01367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nivina A., Yuet K.P., Hsu J., Khosla C. Evolution and diversity of assembly-line polyketide synthases. Chem Rev. 2019;119(24):12524–12547. doi: 10.1021/acs.chemrev.9b00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floss H.G., Yu T.W., Arakawa K. The biosynthesis of 3-amino-5-hydroxybenzoic acid (AHBA), the precursor of mC7N units in ansamycin and mitomycin antibiotics: a review. J Antibiot (Tokyo) 2011;64(1):35–44. doi: 10.1038/ja.2010.139. [DOI] [PubMed] [Google Scholar]
- 5.Zhou W., Posri P., Abugrain M.E., Weisberg A.J., Chang J.H., Mahmud T. Biosynthesis of the nuclear factor of activated T cells inhibitor NFAT-133 in Streptomyces pactum. ACS Chem Biol. 2020;15(12):3217–3226. doi: 10.1021/acschembio.0c00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Y., Yu L., Komaki H., Oku N., Igarashi Y. Absolute configuration of NFAT-133, an aromatic polyketide with immunosuppressive and antidiabetic activity from actinomycetes. J Antibiot (Tokyo) 2016;69(1):69–71. doi: 10.1038/ja.2015.80. [DOI] [PubMed] [Google Scholar]
- 7.Yang F.X., Huang J.P., Liu Z., Wang Z., Yang J., Tang J., et al. Benwamycins A-G, trialkyl-substituted benzene derivatives from a soil-derived Streptomyces. J Nat Prod. 2020;83(1):111–117. doi: 10.1021/acs.jnatprod.9b00903. [DOI] [PubMed] [Google Scholar]
- 8.Dardić D., Böhringer N., Plaza A., Zubeil F., Pohl J., Sommer S., et al. Antidiabetic profiling of veramycins, polyketides accessible by biosynthesis, chemical synthesis and precursor-directed modification. Org Chem Front. 2022;9(6):1604–1615. [Google Scholar]
- 9.Raju R., Gromyko O., Fedorenko V., Luzhetskyy A., Müller R. Lorneic acids C and D, new trialkyl-substituted aromatic acids isolated from a terrestrial Streptomyces sp. J Antibiot. 2013;66(6):347–349. doi: 10.1038/ja.2013.18. [DOI] [PubMed] [Google Scholar]
- 10.Helaly S., Schneider K., Nachtigall J., Vikineswary S., Tan G.Y.A., Zinecker H., et al. Gombapyrones, new α-pyrone metabolites produced by Streptomyces griseoruber Acta 3662. J Antibiot. 2009;62(8):445–452. doi: 10.1038/ja.2009.70. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y.M., Zhao E.J., Wei W., Xu Z.F., Shi J., Wu X., et al. Cytochrome P450 catalyzes benzene ring formation in the biosynthesis of trialkyl-substituted aromatic polyketides. Angew Chem Int Ed. 2023;62(5) doi: 10.1002/anie.202214026. [DOI] [PubMed] [Google Scholar]
- 12.Zhou W., Posri P., Abugrain M.E., Weisberg A.J., Chang J.H., Mahmud T. Biosynthesis of the nuclear factor of activated T cells inhibitor NFAT-133 in Streptomyces pactum. ACS Chem Biol. 2020;15(12):3217–3226. doi: 10.1021/acschembio.0c00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Little R.F., Hertweck C. Chain release mechanisms in polyketide and non-ribosomal peptide biosynthesis. Nat Prod Rep. 2022;39(1):163–205. doi: 10.1039/d1np00035g. [DOI] [PubMed] [Google Scholar]
- 14.Taguchi T., Awakawa T., Nishihara Y., Kawamura M., Ohnishi Y., Ichinose K. Bifunctionality of ActIV as a cyclase-thioesterase revealed by in vitro reconstitution of actinorhodin biosynthesis in Streptomyces coelicolor A3(2) Chembiochem. 2017;18(3):316–323. doi: 10.1002/cbic.201600589. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y., Zhao Y., Zhou J., Yang M., Lin L., Wang W., et al. Unexpected role of a short-chain dehydrogenase/reductase family protein in type II polyketide biosynthesis. Angew Chem Int Ed Engl. 2022;61(7) doi: 10.1002/anie.202110445. [DOI] [PubMed] [Google Scholar]
- 16.Alali A., Zhang L., Li J., Zuo C., Wassouf D., Yan X., et al. Biosynthesis of the tricyclic aromatic type II polyketide rishirilide: new potential third ring oxygenation after three cyclization steps. Mol Biotechnol. 2021;63(6):502–514. doi: 10.1007/s12033-021-00314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oja T., Palmu K., Lehmussola H., Lepparanta O., Hannikainen K., Niemi J., et al. Characterization of the alnumycin gene cluster reveals unusual gene products for pyran ring formation and dioxan biosynthesis. Chem Biol. 2008;15(10):1046–1057. doi: 10.1016/j.chembiol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Pickens L.B., Kim W., Wang P., Zhou H., Watanabe K., Gomi S., et al. Biochemical analysis of the biosynthetic pathway of an anticancer tetracycline SF2575. J Am Chem Soc. 2009;131(48):17677–17689. doi: 10.1021/ja907852c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen Y., Sun F., Zhang L., Cheng Y., Zhu H., Wang S.P., et al. Biosynthesis of depsipeptides with a 3-hydroxybenzoate moiety and selective anticancer activities involves a chorismatase. J Biol Chem. 2020;295(16):5509–5518. doi: 10.1074/jbc.RA119.010922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Lin X., Williams S.R., Liu L., Shen Y., Wang S.P., et al. Directed accumulation of anticancer depsipeptides by characterization of neoantimycins biosynthetic pathway and an NADPH-dependent reductase. ACS Chem Biol. 2018;13(8):2153–2160. doi: 10.1021/acschembio.8b00325. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L., Shen Y., Chen N., Li W., Lin H.-w., Zhou Y. Targeted accumulation of selective anticancer depsipeptides by reconstructing the precursor supply in the neoantimycin biosynthetic pathway. Bioresources and Bioprocessing. 2021;8(1):43. doi: 10.1186/s40643-021-00397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W., Li X., Wang J., Xiang S., Feng X., Yang K. An engineered strong promoter for streptomycetes. Appl Environ Microbiol. 2013;79(14):4484–4492. doi: 10.1128/AEM.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma L., Du L., Chen H., Sun Y., Huang S., Zheng X., et al. Reconstitution of the in vitro activity of the cyclosporine-specific P450 hydroxylase from sebekia benihana and development of a heterologous whole-cell biotransformation system. Appl Environ Microbiol. 2015;81(18):6268–6275. doi: 10.1128/AEM.01353-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakkar C.S., Kate A.S., Desai D.C., Ghosh A.R., Kulkarni-Almeida A.A. NFAT-133 increases glucose uptake in L6 myotubes by activating AMPK pathway. Eur J Pharmacol. 2015;769:117–126. doi: 10.1016/j.ejphar.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Zhou T., Komaki H., Ichikawa N., Hosoyama A., Sato S., Igarashi Y. Biosynthesis of akaeolide and lorneic acids and annotation of type I polyketide synthase gene clusters in the genome of Streptomyces sp. NPS554. Mar Drugs. 2015;13(1):581–596. doi: 10.3390/md13010581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissman K.J. Polyketide stereocontrol: a study in chemical biology. Beilstein J Org Chem. 2017;13:348–371. doi: 10.3762/bjoc.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou W., Alharbi H.A., Hummingbird E., Keatinge-Clay A.T., Mahmud T. Functional studies and revision of the NFAT-133/TM-123 biosynthetic pathway in Streptomyces pactum. ACS Chem Biol. 2022;17(8):2039–2045. doi: 10.1021/acschembio.2c00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawara H., Kawamura N., Kudo T., Suzuki K.I., Nakase T. Distribution of beta-lactamases in actinomycetes. Antimicrob Agents Chemother. 1999;43(12):3014–3017. doi: 10.1128/aac.43.12.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.