Summary
Phages widely exist in numerous environments from wastewater to deep ocean, representing a huge virus diversity, yet remain poorly characterized. Among them, jumbo phages are of particular interests due to their large genome (>200 kb) and unusual biology. To date, only six strains of jumbo phages infecting Klebsiella pneumoniae have been described. Here, we report the isolation and characterization of two jumbo phages from hospital wastewater representing the sixth genus: φKp5130 and φKp9438. Both phages showed lytic activity against broad range of clinical antibiotic-resistant K. pneumoniae strains and distinct physiology including long latent period, small burst size, and high resistance to thermal and pH stress. The treatment of sewage water with the phages cocktail resulted in dramatic decline in K. pneumoniae population. Overall, this study provides detailed molecular and genomics characterization of two novel jumbo phages, expands viral diversity, and provides novel candidate phages to facilitate environmental wastewater treatment.
Subject areas: Applied sciences, Engineering, Water resources engineering
Graphical abstract

Highlights
-
•
Isolation of 2 jumbo phages (∼300 kb, ∼300 nm), representing a novel genus
-
•
Both jumbo phages showed broad lytic activity against K. pneumoniae
-
•
Both jumbo phages exhibited high resistance to thermal and pH stress
-
•
Cocktail of jumbo phages effectively eliminated Klebsiella pneumoniae in sewage water
Applied sciences; Engineering; Water resources engineering
Introduction
Hospitals in developing countries generate 200–400 L per bed per day wastewater whereas for domestic wastewater generation the value is 100–400 L per capita per day. Wastewaters generated from hospitals contain pathogens and other harmful matter compared to domestic wastewater. The misuse of antibiotics with high structural stability leads to their massive release into water1 creating a supportive environment for bacteria to develop antibiotic resistance. And the antibiotic resistance genes (ARGs) in multidrug-resistant bacteria from wastewater treatment plants (WWTPs) can be horizontally transferred among bacterial community via mobile genetic elements (MGEs), thereby inducing the uncontrolled spread of ARGs.2 Several studies in recent years have detected drug-resistant Klebsiella pneumoniae from hospital wastewater.3,4,5 K. pneumoniae is a gram-negative bacterium belonging to the Enterobacteriaceae family, widespread in human and animal host as well as in natural environments including soil, sewage, lake, and ocean. It is an opportunistic pathogen leading to pneumonia, meningitis, septicemia, and urinary tract infection (UTI) and common causative agent of nosocomial and community-acquired infection.6 K. pneumoniae has been recognized by the World Health Organization, European Union, United States, China, and other organizations as a significant threat to global public health due to its high rates of multidrug resistance (MDR). More recently, the convergence of carbapenem-resistant Enterobacteriaceae strains was considered an urgent public health issue as they frequently cause untreatable or hard-to-treat infections in hospitals.7
There is a growing interest in using phages to selectively control bacteria in wastewater and water treatment, inspired by the medicinal use of phage therapy in pathogenic bacterial infections.8 Phages are the most abundant virus in the biosphere and natural predator of bacteria,9,10 targeting and killing bacterial pathogens without causing adverse effects on other organisms in the environment. In the second-stage of effluent disinfection, the number of drug-resistant pathogenic bacteria in the effluent can be minimized by directly using phages to specifically lyse target strains and biofilms or by using the selection pressure formed by phages to select slower-growing phage-resistant bacteria that are more susceptible to biocides or competitive rejection, weakening the health risk posed by drug-resistant pathogens entering the environment.11 For example, Keivan et al. isolated and characterized two novel bacteriophages from Zayandehrood River using Escherichia coli SBSWF27 as host strain and observed a 22-fold decrease of the most probable number (MPN) of the coliform when applied these phages to wastewater treatment for only 2 h.12
In the past two decades, sequencing technology has accelerated the discovery of novel phages and largely expanded the diversity of this being. Among them, jumbo phages are of interests due to their extraordinary genomes (>200 kb) that usually harbor larger virions and unusual aspects of biology, including structural, biochemical, and ecological characteristics. For example, more functional genes in various systems of jumbo phages may help deter or compensate for host immune mechanisms.13,14,15,16,17,18,19,20 To date, the validated genome sequences of K. pneumoniae phages are easily accessible in public database, yet sequenced and characterized jumbo phages infecting K. pneumoniae were still in scarce, with only six jumbo phages reported.14,21,22 To increase the diversity of jumbo phages and make them widely available for combating K. pneumoniae in wastewater, we isolated and characterized two novel jumbo phages against K. pneumoniae and explored the possibility of using them as biocontrol agents in hospital wastewater.
Results
General morphological features
Two phages that infect K. pneumoniae were isolated from sewage samples, namely, vB_KpM_5130 (φKp5130) and vB_KpM_9438 (φKp9438). They belong to the Caudoviricetes class of tailed phages with double-stranded DNA. Electron micrographs show that both phages have long contractile tail and complex tail fiber structure at the baseplate (Figure 1). The morphological features indicate that they belong to the Myovirus morphotype. φKp9438 has slightly longer tail (150 nm) and bigger capsid (140 nm) compared to φKp5130 (tail of 140 nm and capsid of 120 nm). Their contractile tails are shown with extended tail sheath and contracted tail sheath (Figure 1). More than four putative tail fibers and an enveloped head with long contractile tail are seen in φKp5130.
Figure 1.
Transmission electron microscopy (TEM) images of jumbo phages
Phages were stained with 2% phosphortungstic acid and imaged at a final magnification of 16000×. Phage dimensions were measured using image capture software ImageJ. Scale bar = 100 nm.
(A) TEM image of φKp5130.
(B) TEM image of φKp9438.
Genomic analysis
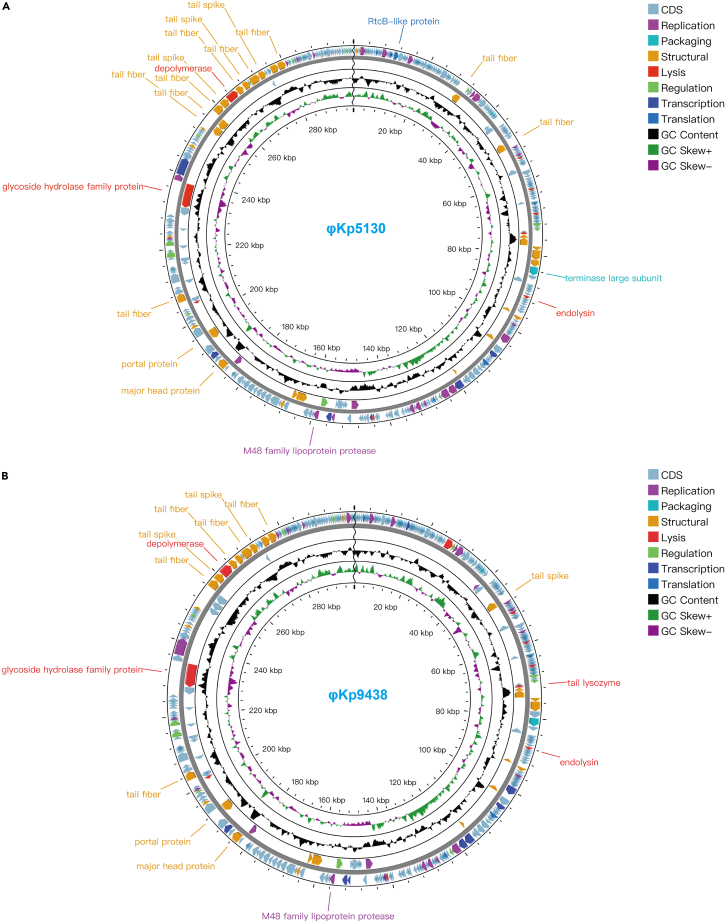
The genomes size of φKp5130 and φKp9438 is 298,475 bp and 299,545 bp, respectively. The large genome (∼300 kb) indicates that they are jumbo bacteriophages. The GC content of φKp5130 and φKp9438 is 45.66% and 45.57% (Table 1), respectively, lower than the median GC content of K. pneumoniae (57.2%). Although two phage genomes share high similarity (93.4% coverage and 98.4% identity), they show very low similarity to any other phage genomes deposited in GenBank, representing a novel phage genus. Genome-based phylogenetic analysis of φKp5130 and φKp9438 with other 46 representative jumbo phages show that the two newly isolated phages are distant from any other phages (Figure 2). These 46 phages have a wide range of genome size (from 196.7 kb to 497.5 kb), GC content (from 27.71% to 58.12%), and tRNA number (from 0 to 32). The phage genus which is relatively close to these two jumbo phages is Moabitevirus whose average genome size is 274.9 kb and average GC content is 46.795% (Figure 2). Both phages did not cluster with any known phage as a subgroup in network reconstructed by vConTACT2 (ProkaryoticViralRefSeq211-Merged). Notably, both phages showed almost no similarity with the giant phages PhiKZ supergroup (<0.03% coverages in genome alignment). The closest genome is a Serratia jumbo phage (GenBank: MK994515.1) of the Moabitevirus genus, sharing less than 2% similarity: 1.3% coverage and 74.3% identity with φKp5130 and 1.3% coverage and 74.8% identity with φKp9438. The GC content and genome size of this Serratia jumbo phage are 46.8% and 273,933 bp, respectively, encoding 337 predicted proteins and 2 tRNA.
Table 1.
Morphological and genomic features of φKp5130 and φKp9438
| φKp5130 | φKp9438 | |
|---|---|---|
| K. pneumoniae host used for isolation | KP5130 | KP9438 |
| Family | Myoviridae | Myoviridae |
| Genome size (bp) | 298,475 | 299,545 |
| Best Blast hit (query coverage, identity) | Serratia phage Moabite (1.3%,68.8%) | Serratia phage_2050HW (0.972%,74.5%) |
| GC content (%) | 45.66% | 45.57% |
| Number of CDS | 354 | 358 |
| Number of hypothetical proteins | 61 | 72 |
| Possible host receptor binding proteins | 17 | 19 |
| Possible depolymerases | 1 | 1 |
| Pectate lyase | 0 | 1 |
| GTPase | 1 | 0 |
| Transglycosylase | 1 | 1 |
| tRNA genes | 9 | 9 |
Figure 2.
Phylogenetic analysis of φKp5130 and φKp9438 with 46 representative jumbo phages of Myovirus and an outgroup phage-based multiple whole-genome sequences alignment
The two new phages are highlighted in purple. The scale bar represents 1 sequence divergence. The green bars represent the average genome size of these jumbo phage families, and the blue and red heatmaps on the periphery represent the average GC content and tRNA numbers of these phages, respectively. The red star represents the outgroup phage Skunavirus bIL170.
Partially due to the novelty of two phages, only 35–39% (138/354 in φKp5130, 126/358 in φKp9438) of the proteins in their genomes can be assigned with known functions, such as structural, replication, translation, and regulation (Figure 3). Genes related to antibiotic resistance, virulence, and lysogenicity encoding were not identified. Notably, both phages contain one DNA polymerase and one multi-subunit RNA polymerase (two alpha, four beta, and one beta-beta subunits), likely to initiate early gene expression before employment of host polymerase.23 Both two phages contain 10 tRNA genes, which is in accordance with previous observation that jumbo phages encode a large number of translation-related genes to increase the translation efficiency.24
Figure 3.
Circular genome maps of jumbo phages
The inner circle represents the GC skew (G − C/G + C). Outwards indicates >0 (green), and inwards indicates <0 (purple). The outermost circle represents CDSs encoded in the genome, clockwise arrow indicates the forward reading frame, and counterclockwise arrow indicates the reverse reading frame. The labeled annotation represents structure-related CDS of phages.
(A) The circular genome map of φKp5130.
(B) The circular genome map of φKp9438.
Host range analysis
Twenty-two K. pneumoniae strains covering 10 capsular types were used for host range determination (Table 2). φKp5130 showed lytic activity against 6 K pneumoniae strains with 4 capsular types, including KL2, KL24, KL38, and KL62. φKp9438 showed much broader host range, which successfully lysed 21 out of 22 (95.45%) tested strains, except KL5 strain.
Table 2.
The host range of two phages to 22 K pneumoniae strains
| K. pneumoniae strains | Capsular locus types | Lytic activity |
|
|---|---|---|---|
| φKp5130 | φKp9438 | ||
| Kp 5137a | KL2 | + | + |
| Kp 5619 | KL2 | + | + |
| Kp 5295 | KL2 | – | + |
| Kp 7338 | KL2 | – | + |
| Kp 9088 | KL5 | + | – |
| Kp 9194 | KL12 | – | + |
| Kp 9385 | KL15 | – | + |
| Kp 9502 | KL19 | – | + |
| Kp 9947 | KL20 | – | + |
| Kp 9841 | KL23 | – | + |
| Kp 9856 | KL23 | – | + |
| Kp 9310a | KL24 | + | + |
| Kp 9349 | KL24 | – | + |
| Kp 9498 | KL24 | – | + |
| Kp 4547 | KL30 | – | + |
| Kp 9773 | KL38 | + | + |
| Kp 8860 | KL47 | – | + |
| Kp 9298 | KL47 | – | + |
| Kp 9816 | KL47 | – | + |
| Kp 5130a | KL62 | + | + |
| Kp 8372 | KL64 | – | + |
| Kp 9628 | KL158 | – | + |
K. pneumoniae strain used for pathogen biocontrol agents test in sewage water.
Interestingly, the two jumbo phages demonstrate distinct host range despite sharing high genome similarity (93.4%). Host range is largely governed by structural proteins of phages targeting bacterial cells, e.g., tail fibers and tail spikes of tailed phages interacting with receptors on bacterial surface.25 A deep survey into the genomes shows that φKp5130 genome encodes 9 tail fiber proteins and 3 tail spikes proteins, while φKp9438 genome encodes 5 tail fiber proteins and 4 tail spikes (Table 3). Notably, 5 tail fiber proteins and 3 tail spikes of φKp9438 are homologous with those of φKp5130, sharing a high identity of 96.77–99.92%. It’s likely that one of the tail spikes of φKp9438 (67.0% coverage and 31.7% identity with GenBank: AUG87748.1) is a key protein for host recognition, which extends the host range of φKp9438 compared with φKp5130.
Table 3.
Tail proteins encoded in φKp5130 and φKp9438
| CDS | φKp5130 |
φKp9438 |
Identity (%) | ||
|---|---|---|---|---|---|
| Protein_id | Length (aa) | Protein_id | Length | ||
| Tail fiber | GenBank: YP_003347555.1 | 2324 | GenBank: YP_003347555.1 | 2324 | 96.77% |
| GenBank: QBZ71284.1 | 2621 | GenBank: QBZ71284.1 | 2621 | 99.92% | |
| GenBank: APZ82847.1 | 1802 | GenBank: APZ82847.1 | 1802 | 99.72% | |
| GenBank: QDB71175.1 | 2231 | GenBank: QDB71175.1 | 2231 | 99.60% | |
| GenBank: QDB71241.1 | 2069 | GenBank: QDB71241.1 | 2069 | 99.13% | |
| GenBank: QDB71228.1 | 2522 | N | N | ||
| GenBank: QDB71226.1 | 2210 | N | N | ||
| GenBank: QDB71225.1 | 2117 | N | N | ||
| GenBank: ALT58497.1 | 2015 | N | N | ||
| Tail spike | GenBank: QBQ72011.1 | 2099 | GenBank: QBQ72011.1 | 2099 | 99.90% |
| GenBank: AUG87751.1 | 1781 | GenBank: AUG87751.1 | 1781 | 99.94% | |
| N | GenBank: AUG87748.1 | 2108 | N | ||
| Depolymerase | GenBank: AYP28213.1 | 3059 | GenBank: AYP28213.1 | 3059 | 99.54% |
Plaque assay and one-step growth curves
Plaque assays of φKp5130 and φKp9438 showed that both phages produce large plaques with 1.5 mm diameter surrounded by translucent halos (Figure S1). Halo formation of bacteriophages is associated with depolymerase activity, which facilitates cell wall degradation and DNA ejection.26 Indeed, genomic analysis showed that both φKp5130 and φKp9438 encode a depolymerase containing tail spike probably targeting capsular polysaccharides (CPSs), exopolysaccharides (EPSs), or lipopolysaccharide (LPS) of the host bacteria (Table 1).
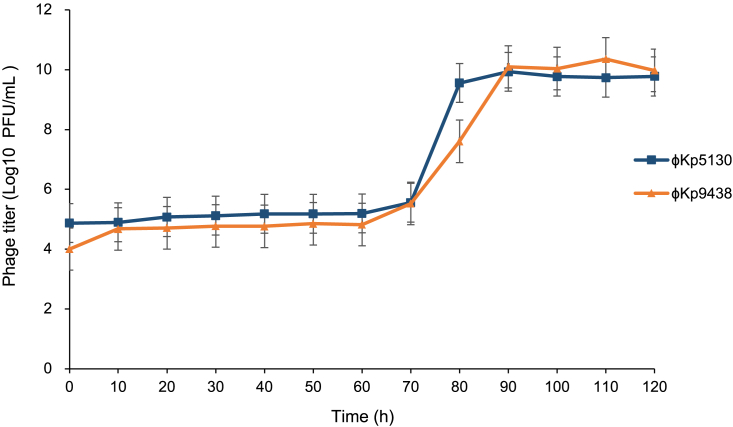
One-step growth analysis of φKp9438 showed a latent period with 60–70 min, a rise period of 20–30 min, and a burst size of 31 plaque-forming unit (PFU)/cell (Figure 4). One-step growth analysis of φKp5130 showed a latent period about 60–70 min, a rise period of 30 min, and a burst size of 57 PFU/cell. Generally, the latent period of the two jumbo phages (60–70 min) is longer than K. pneumoniae phages (10–30 min) with smaller genomes (Table S1). The lytic machinery of bacteriophages usually consists of endolysin and holing. Genomic analysis showed that the two jumbo phages both encode an endolysin of the glycoside hydrolase family, and no holin or holin-like protein was found.
Figure 4.
One-step growth curve of bacteriophage φKp5130 and φKp9438
Phages at different times are shown. Data are represented as mean +/− standard error of mean (SEM).
Thermal and ph stability assay
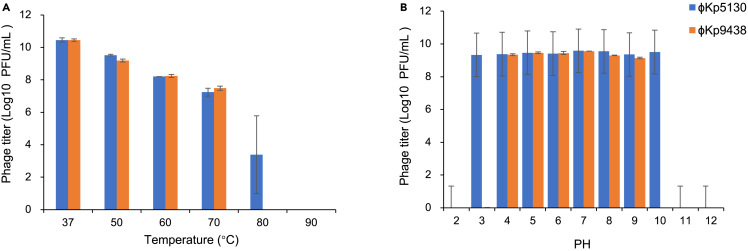
Both jumbo phages presented resistance to thermal and pH stress. In response to heat stress, φKp5130 viability slightly decreased from 37°C to 70°C, showed 5-log reduction when incubated at 80°C, and completely inactivated upon incubation at 90°C for 1 h (Figure 5A). φKp9438 showed similar viability between 37°C and 70°C and drastically decreased upon incubation at 80°C for 1h. Notably, φKp5130 and φKp9438 demonstrated higher tolerance to thermal stress compared to other K. pneumoniae phages with genome less than 200 kb, which cannot survive temperature beyond 60°C (Table S1). Both phages presented a broad pH resistance spectrum, with φKp5130 viable from pH 3 to pH 10 and φKp9438 viable from pH 4 to pH 9 (Figure 5B).
Figure 5.
Viability of jumbo phages in response to stress
Data are represented as mean ± SEM.
(A) Viability of jumbo phages in response to temperature.
(B) Viability of jumbo phages in response to pH.
Utilization of jumbo phages cocktail in sewage water to control potential pathogens
The stability assay indicated that the two jumbo phages maintained stable viability when exposed to a wide range of thermal and pH stress, making them suitable for applications in fluctuating natural environments. To test the biocontrol efficacy of φKp5130 and φKp9438 in controlling antibiotic-resistant pathogens in sewage water, we selected 3 strains of MDR K. pneumoniae isolated from hospital sewage water as target pathogens (Kp5137, Kp5310, Kp9310). All three K. pneumoniae exhibited intermediate to high resistance levels to the antibiotics tested by minimum inhibitory concentration (MIC) and Kirby-Bauer disk diffusion (KB). Kp5137 and Kp9310 were resistant to extended-spectrum beta lactamase, cefazolin, ceftriaxone, ciprofloxacin, and amoxicillin/clavulanic (Tables 4 and 5). Kp5310 was resistant to ciprofloxacin and trimethoprim/sulfamethoxazole (Table 6).
Table 4.
Antibiotics sensitivity testing of Kp5137
| Antibiotics | KB | MIC | Sensitivity |
|---|---|---|---|
| spectrum B-lactamase | Pos | + | |
| Piperacillin/Tazobacta | ≤ 4 | S | |
| Cefoperazone/Sulbactan | 26 | S | |
| Cefazolin | ≥64 | R | |
| Ceftriaxone | 8 | R | |
| Cefepime | ≤ 1 | S | |
| Cefoxitin | ≤ 4 | S | |
| Aztreonam | ≤ 1 | S | |
| Meropenem | 29 | S | |
| Ertapenem | ≤0.5 | S | |
| Imipenem | ≤1 | S | |
| Ciprofloxacin | 1 | R | |
| Levofloxacin | 0. 5 | S | |
| Amikacin | ≤ 2 | S | |
| Tobramycin | 8 | I | |
| Gentamicin | ≥16 | R | |
| Trimethoprim/Sulfameth | ≤1/19 | S | |
| Tigecycline | 2 | S | |
| Amoxicillin/Clavulanic | 16 | I |
R: resistant, S: susceptible, I: intermediate.
Table 5.
Antibiotics sensitivity testing of Kp9310
| Antibiotics | KB | MIC | Sensitivity |
|---|---|---|---|
| spectrum B-lactamase | Pos | + | |
| Piperacillin/Tazobacta | 8 | S | |
| Cefoperazone/Sulbactan | 21 | S | |
| Cefazolin | ≥64 | R | |
| Ceftriaxone | ≥64 | R | |
| Cefepime | ≥64 | R | |
| Cefoxitin | ≤4 | S | |
| Aztreonam | ≥64 | R | |
| Meropenem | 28 | S | |
| Ertapenem | ≤0. 5 | S | |
| Imipenem | ≤1 | S | |
| Ciprofloxacin | ≥4 | R | |
| Levofloxacin | ≥8 | R | |
| Amikacin | ≤2 | S | |
| Tobramycin | 8 | I | |
| Gentamicin | ≤1 | S | |
| Trimethoprim/Sulfameth | ≥16/3 | R | |
| Tigecycline | 2 | S | |
| Amoxicillin/Clavulanic | ≥32 | R |
R: resistant, S: susceptible, I: intermediate.
Table 6.
Antibiotics sensitivity testing of Kp5310
| Antibiotics | KB | MIC | Sensitivity |
|---|---|---|---|
| spectrum B-lactamase | Neg | – | |
| Piperacillin/Tazobacta | ≤ 4 | S | |
| Cefoperazone/Sulbactan | 28 | S | |
| Cefazolin | ≤2 | S | |
| Ceftriaxone | ≤ 1 | S | |
| Cefepime | ≤1 | S | |
| Cefoxitin | ≤ 4 | S | |
| Aztreonam | ≤ 1 | S | |
| Meropenem | 29 | S | |
| Ertapenem | ≤0.5 | S | |
| Imipenem | ≤ 1 | S | |
| Ciprofloxacin | 1 | R | |
| Levofloxacin | 0.5 | S | |
| Amikacin | ≤ 2 | S | |
| Tobramycin | ≤1 | S | |
| Gentamicin | ≤ 1 | S | |
| Trimethoprim/Sulfameth | ≥16/3 | R | |
| Tigecycline | 2 | S | |
| Amoxicillin/Clavulanic | ≤2 | S |
R: resistant, S: susceptible, I: intermediate.
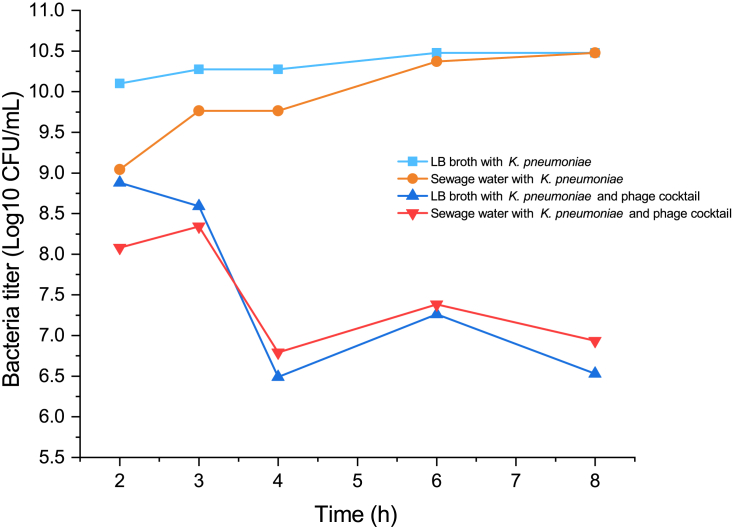
Two isolated jumbo phages φKp5130 and φKp9438 were mixed to make a phage cocktail and applied to biocontrol of aforementioned 3 MDR K. pneumoniae target strains. To ensure the success of phage treatment, the appropriate phage should be isolated and enriched to produce sufficient numbers. It is recommended phages number to be inoculated should be 3–10 times greater than bacteria.27 Therefore, the phage cocktail was incubated with K. pneumoniae strains at an MOI of 5. Two incubation conditions were tested: sterilized wastewater to mimic the real environment and Luria-Bertani (LB) medium as a control. As showed in Figure 6, the bacteria population increased steadily in phage-free groups (G1 and G2) and showed a dramatic decline in the phage treatment group, no matter in the environment of LB broth or wastewater. There was an increase in K. pneumoniae population but not as much as in phage-treated group from 2 h, possibly due to phage particles adsorbing to the samples and altering their metabolic rate. At subsequent time points, the target population steadily increased in uninoculated groups whereas that in the phage-inoculated group was more significantly inhibited. Thus, there was a reduction in target population as a result of the phage. According to the single-step growth experiment, the phage population reached its peak in 2–3 h. So, we maintained the incubation time for 8 h in this experiment. As showed in Figure 6, the bacteria population increased steadily in phage-free groups (G1 and G2) and showed a dramatic decline in the phage-treatment group, no matter in the environment of LB or wastewater. These results showed that the jumbo phages cocktail effectively eliminated target K. pneumoniae population in sewage water (Figure 6).
Figure 6.
Cell count of bacteria K. pneumoniae (CFU/mL) in sewage water at different time point 2–8 h after G1-G4 treatments.
Discussion
In this study, we isolated two jumbo phages that infect K. pneumoniae from hospital sewage. Genomic analysis showed a large genome (∼300 kb) for both phages and indicated that they are jumbo bacteriophage of the Myovirus. Both phages showed almost no similarity with the existing giant phages supergroup. Due to low similarity to any other phage genomes and the fact that only about 35% of the proteins in their genomes can be assigned with known functions, these two phages are considered a novel phage genus. The phage tail plays the most critical role for host receptor recognition, attachment, and DNA ejection. Both phages we isolated have long contractile tail and complex tail fiber structure at the baseplate. A contractile tail sheath surrounding the tail tube facilitates the puncture of the bacterial cell envelope. Host range is largely governed by tail fibers and tail spikes of phages interacting with receptors on bacterial cells. According to host range determination, φKp9438 successfully lysed 21 out of 22 K pneumoniae strains while φKp5130 showed lytic activity against 6 K pneumoniae strains. It’s suggested that tail spike GenBank: AUG87748.1 of φKp9438 is a key protein for host recognition, which extends the host range of φKp9438 compared with φKp5130 despite their high genome similarity. This finding will allow us to take next step in creating phage cocktails to broaden the host range to target a wide range of drug-resistant bacteria in sewage water.
The main disinfection process in the current wastewater treatment uses chlorination which is able to remove most of the pathogenic bacteria in the water.28 However, this process has some defects such as being prone to residues, high cost, and easy-to-produce by-products such as chloroform. At the same time, the extensive use of antibiotics has left large amounts of antibiotics in the water and environment and has induced the large numbers of antibiotic-resistant bacteria. Due to their high specificity, phages display more advantages than antibiotics in treating bacterial infections and can be an excellent potential antimicrobial, with capability of killing target pathogens without causing adverse effects on other organisms in the environment. Here we demonstrated the utility of jumbo phage cocktail in biocontrol of MDR K. pneumoniae populations in both laboratory environment (LB medium) and real environment (wastewater). Water chemistry and environmental factors could affect phage-specific interactions and compromise phage absorption and infectivity. The stable viability of these two jumbo phages when exposed to thermal and pH stress can make them suitable for use in sterilization in unpredictable natural environments. Moreover, we have carried out detailed analysis and comparison of the physiological properties and genomic information of the phages. These works have laid the foundation for the next step of phage synthesis and modification for specific antibiotic-resistant bacteria for medical and sewage water treatment.
Limitations of the study
Our study describes the isolation of two jumbo phages infecting K. pneumoniae and provides a comprehensive molecular and genomic characterization. These two jumbo phages represent a novel genus and show remote phylogenetic relation with other jumbo phages. However, the number of reported jumbo phages is still limited, and further studies are needed to better understand the biology and potential applications of these phages. Future studies could focus on elucidating the mechanisms by which jumbo phages infect and lyse bacteria, as well as exploring how they interact with other microorganisms in various environmental contexts. Additionally, the potential applications of jumbo phages could be further investigated, such as their use as control agents in wastewater treatment and in food or water safety or as a potential alternative to conventional antibiotics.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| K. pneumoniae strains Kp9438 | This paper | Kp9438 |
| K. pneumoniae strains Kp5130 | This paper | Kp5130 |
| Phage φKp5130 | This paper | φKp5130 |
| Phage φKp9438 | This paper | φKp9438 |
| K. pneumoniae strains Kp 5137 | This paper | Kp 5137 |
| K. pneumoniae strains Kp 5619 | This paper | Kp 5619 |
| K. pneumoniae strains Kp 5295 | This paper | Kp 5295 |
| K. pneumoniae strains Kp 7338 | This paper | Kp 7338 |
| K. pneumoniae strains Kp 9088 | This paper | Kp 9088 |
| K. pneumoniae strains Kp 9194 | This paper | Kp 9194 |
| K. pneumoniae strains Kp 9385 | This paper | Kp 9385 |
| K. pneumoniae strains Kp 9502 | This paper | Kp 9502 |
| K. pneumoniae strains Kp 9947 | This paper | Kp 9947 |
| K. pneumoniae strains Kp 9841 | This paper | Kp 9841 |
| K. pneumoniae strains Kp 9856 | This paper | Kp 9856 |
| K. pneumoniae strains Kp 9310 | This paper | Kp 9310 |
| K. pneumoniae strains Kp 9349 | This paper | Kp 9349 |
| K. pneumoniae strains Kp 9498 | This paper | Kp 9498 |
| K. pneumoniae strains Kp 4547 | This paper | Kp 4547 |
| K. pneumoniae strains Kp 9773 | This paper | Kp 9773 |
| K. pneumoniae strains Kp 8860 | This paper | Kp 8860 |
| K. pneumoniae strains Kp 9298 | This paper | Kp 9298 |
| K. pneumoniae strains Kp 9816 | This paper | Kp 9816 |
| K. pneumoniae strains Kp 8372 | This paper | Kp 8372 |
| K. pneumoniae strains Kp 9628 | This paper | Kp 9628 |
| Chemicals, peptides, and recombinant proteins | ||
| PEG8000 | SIGMA | P5413-2KG |
| DNase I | SIGMA | Cat # DN25 |
| RNase A | Invitrogen | Cat # 12091021 |
| Proteinase K | NEB | P8107S |
| LB Broth (Lennox) | Hopebio | HB0128 |
| Agar | Sangon Biotech | A505255-0250 |
| Sodium chloride (NaCl) | Sangon Biotech | A610476-0001 |
| Magnesium Sulfate Heptahydrate (MgSO4 · 7H2O) | HUSHI | 10013018 |
| Trizma® Hydrochloride Solution (Tris-HCl) | SIGMA | T2694-1L |
| Gelatin | BBI | G9764-500g |
| Meropenem | Thermo Scientific | Cat#CT0774B |
| Cefoperazone | Thermo Scientific | Cat#CT0249B |
| Critical commercial assays | ||
| Norgen Phage DNA Isolation Kit | BIOTEK | Cat # 46800 |
| Qubit™ dsDNA HS Assay Kit | INVITROGEN | Q32854 |
| VITEK® 2 Gram Negative ID card (VITEK2 AST-GN13) | BioMérieux | Cat#22095 |
| Deposited data | ||
| Assembly and annotation data of phages | China National GeneBank DataBase (CNGBdb) | https://ftp.cngb.org/pub/CNSA/data3/CNP0001966/ |
| Raw data of phages | China National GeneBank DataBase (CNGBdb) | https://db.cngb.org/search/project/CNP0001966/ |
| Software and algorithms | ||
| Fastp | Chen et al.29 | https://github.com/OpenGene/fastp |
| SOAPnuke | Chen et al.30 | https://github.com/BGI-flexlab/SOAPnuke |
| SPAdes | Bankevich et al.31 | https://github.com/ablab/spades |
| prodigal | Hyatt et al.32 | https://github.com/hyattpd/Prodigal |
| BLASTp | Camacho et al.33 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| hmmscan | Johnson et al.34 | https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan |
| tRNAscan-SE | Chan et al.35 | http://trna.ucsc.edu/tRNAscan-SE/ |
| CGView | Stothard et al.36 | https://cgview.ca/ |
| mafft | Katoh et al.37 | https://mafft.cbrc.jp/alignment/software/ |
| IQ-TREE | Minh et al.38 | https://github.com/iqtree/iqtree2 |
| iTOL | Letunic et al.39 | https://itol.embl.de/ |
| vConTACT2 | Bin Jang et al.40 | https://bitbucket.org/MAVERICLab/vcontact2/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ziqing Deng: dengziqing@genomics.cn.
Materials availability
This study did not generate new unique reagents. Bacterial isolates and isolated phages are available by request from the lead contact under the conditions of a material transfer agreement.
Experimental model and subject details
K. pneumoniae bacterial isolates in this study were isolated from clinical blood culture samples and cryopreserved in Luria-Bertani (LB) media with 20% glycerol and stored at -80°C. All the phages were in dependently isolated by our team and stored at 4°C in SM buffer for optimal storage conditions. All bacteria and phages are available from the lead contact upon request.
Method details
Phage isolation and purification
K. pneumoniae strains Kp9438 and Kp5130 were isolated from clinical blood culture samples and used for phage isolation and propagation. The bacteria species were first identified using the VITEK 2 system (bioMérieux). Phage stocks in this study were isolated from sewage samples at approximately 112.95⁰ Latitude and 28.23⁰ Longitude. The supernatant of sewage water was passed through a 0.45 μm membrane filter (VWR) after centrifuged at 5,000 ×g for 20 minutes at 4°C. Kp9438 and Kp5130 was verified by 16S rRNA sequencing before laboratory subculture. The single colony was inoculated into 3 mL LB broth at 37°C, 220 rpm for overnight. And then 500 μL of overnight bacterial culture was co-incubated with 40 mL of filtered sewage in a 250 mL flask containing 10 mL 5× LB broth for overnight. After centrifuged at 10,000 ×g for 20 minutes and filter-sterilized (0.2 μm VWR), 200 μL filtrate was mixed with 100 μL early log-phase cultures (OD600 ∼0.6) and incubated at 37°C for 5 min. The mixture was mixed well with top agar and overlaid on top of LB medium using double-overlay agar assays. Plates were incubated at 37°C, 220 rpm for overnight and single plaques were purified three times until all plaques were uniform.43 Phages culture was produced in LB broth with their respective host, centrifuged, filter-sterilized and stored as phage lysates (>108 PFU/mL) at 4°C in SM buffer (100 mM NaCl; 10 mM MgSO4·7H2O; 50 mM Tris-HCl; 0.1 g/L gelatin; pH 7.5), or at -80°C with 50 % (v/v) glycerol.
Transmission electron microscopy (TEM)
The morphological characteristics of isolated phages were determined as previously described.44 High titer phage sample (108 PFU/mL) was prepared after purification, placed on carbon-coated copper grids for 15 minutes for adsorption. After further drying, samples were stained with 2% uranyl acetate and examined in Tecnai G2 Spirit BioTWIN 120-kV transmission electron microscope (FEI Company, Hillsboro, USA) with magnification of 12000-16000 ×. Phage dimensions were measured using Image capture software ImageJ.
Host range assay
To further investigate the physiology of isolated phages, 22 strains of K. pneumoniae, representing 10 capsular locus types as listed in Table 2, were used. The spotting assay methods as previously describe45 were used to determine the host range. The phage lysates with a concentration of 108 PFU/mL were spotted onto LB double layer agar plates of the 22 bacterium strains, each with three replicates. The plates were then incubated overnight at 37°C to observe the formation of phage plaques.
One-step growth assays
A one-step growth curve was performed as previously described17 with slight modifications. 50 mL exponential-phase culture (108 CFU/mL) of Kp9438 and Kp5130 were infected with 500 μL phages supernatant (109 PFU/mL). The final multiplicity of infection (MOI) of the mixture was 0.1. The mixture was incubated 15 minutes for adsorption at 37°C and centrifuged at 12,000 ×g for 5 minutes to remove the phage that no adsorbed in the supernatant. Then the two samples resuspended in 50 mL LB broth were incubated for 120 minutes at 37°C, 220 rpm. Subsequently, 400 μL aliquots were taken from the culture every 10 minutes and centrifuged at 12,000 ×g for 30 seconds. The supernatants were collected and serially diluted and enumerated by the spot assay method immediately after incubation overnight at 37°C. All experiments were performed in triplicate.
Thermostability and pH sensitivity assay
For thermostability assay, a filter-sterilized bacteriophage solution of 109 PFU/mL was prepared and incubated at 37°C, 50°C, 60°C, 70°C, 80°C and 90°C for 1 hour. For pH Sensitivity assay, phages preparation (109 PFU/mL) was incubated at pH 2-12 with 1 M NaOH or 1 M HCl for 1 hour at 37°C.46 The bacteriophage titer was then assessed by double-agar layer method.47,48 All experiments were performed in triplicate. Three plates were counted, and the results were taken as mean values.
DNA extraction
50ml Phage lysates were centrifuged at 8,000 ×g for 5 minutes and then filtered through 0.22 μm filters (VWR) to remove cell debris. Add 10 μg/mL DNase and RNase, and incubate at 37°C for 1 h. Then the final lysate treated at a rate of 1:2=PEG-8000:lysate (10% PEG-8000,1 M NaCl final), mix gently by inversion and incubate at 4°C overnight. Samples were centrifuged at 10,000 ×g for 20 minutes at 4°C. Phage pellets were suspended in 200 μl TE buffer (0.5 M EDTA pH8, 0.1 M Tris·HCl pH7.4, 0.5% SDS). Then, we added 10 μl Proteinase K(20 mg/mL) in 56 ºC for 2 h. The DNA extraction protocol with Norgen Phage DNA Isolation KIT (Cat. 46800) was used for phage DNA isolation.
Genome assembly and annotation
Phage genome analysis refers to a previous article.49 The genomic DNA of the bacteriophages were ultrasonic fragmented before subjected to circularization. DNA nanoball (DNB)-based libraries were constructed by rolling circle replication (RCR) and then sequenced on the MGISEQ-2000 platform (BGI-Shenzhen, China) with paired-end 100 nt strategy, generating 5.1Gb sequencing data for φKp9438 and 10.8 Gb sequencing data for φKp5130. The sequencing depth of each sample was >10000×. Poor quality reads were filtered out with SOAP nuke (https://github.com/BGI-flexlab/SOAPnuke) and Fastp.29,30 Cleaned reads were assembled with SPAdes v3.13.031. The assembly result produced a single contig. Functional annotation of representative genes on jumbo phage genomes were done using prodigal,32 BLASTp33 searches against NCBI nr database (snapshot of 2020-08-17) and further searched by hmmscan34 against UniProt/Swiss-Prot database (snapshot of 2020-08-17). Final function annotation results were then performed by manual investigation of the amino acid homology search results. tRNA scan-SE (version 2.0.5)35 was used to search for tRNA genes on phage genome. Lysogenic genes were checked for the following proteins: integrases, Cro/CI repressor proteins, immunity repressors, DNA partitioning protein A (ParA), and anti-repressor proteins. Virulence factors and drug resistance genes were identified through comparison with databases such as the Virulence Factors Database (VFDB, http://www.mgc.ac.cn/VFs/), The Comprehensive Antibiotic Resistance Database (CARD, https://card.mcmaster.ca/) and ResFinder Database (https://cge.cbs.dtu.dk/services/ResFinder/). Schematics of phage genomes were built with CGView Server online.36 vConTACT240 was used for taxonomic assignment of phages. Closest homology known phage search by BLAST, we used total number of matching bases over the whole genome size as phage’s similarity.
Phylogenetic analysis
For phylogenetic analysis in Myovirus morphotype, 46 isolates whole genome sequences17 from http://warwick.s3.climb.ac.uk/inphared/ICTV_genera.tar.gz in different genus were chosen to make multiple sequence alignment by mafft 7.453.37 Skunavirus bIL170 of the Siphoviridae family was used as an outgroup. Phylogenetic tree was generated using the Maximum Likelihood method and the best-fit nucleotide substitution models were determined using IQ-TREE Model Finder.38 All Maximum Likelihood (ML) phylogenies for jumbo phage sequences were constructed with 1,000 ultrafast bootstrap pseudo-replications50 and tree was embellished by iTOL.39
Utilization of phages cocktail in sewage water to control potential pathogens
Hospital wastewater samples used for the study were collected from secondary settling tank of Qingdao University Affiliated Hospital sewage treatment plant. All samples were taken before the addition of chemical bactericides. After the sewage water was settled, the supernatant was taken and aliquoted. To avoid the influence of other native organisms during phage treatment, sewage water was sterilized before introducing the target organism. Then each sample was inoculated with 3 strains of K. pneumoniae (Kp5130, Kp5137, Kp9310) listed in Table 2 at 104 CFU/ml51 The VITEK 2 system (bioMérieux) was used for bacterial identification. Minimum inhibitory concentration (MIC) method and Kirby-Bauer disk diffusion method (K-B methods) were used to determine the antimicrobial susceptibility testing of these strains. We followed these sensitivity standards which established by the Clinical and Laboratory Standards Institute (CLSI). MIC test was conducted according to a previously study.52 Briefly, antibiotic powder was dissolved and diluted by the two-fold method using sterile distilled water, and the MIC of antibiotics was determined by comparing the growth of bacteria under different antibiotic concentrations. Disk-diffusion method was conducted according to a previously protocol.53 Briefly, the disks containing different antibiotics were placed on the medium inoculated with bacteria and incubated overnight at 37°C. The sensitivity of bacteria to antibiotics were determined according to the diameter of the inhibition zone.
To increase the host range, we mixed two isolated jumbo phages φKp5130 and φKp9438 into a phage cocktail, and the initial concentration of the phage cocktail was 107 PFU/mL. The phage cocktail was mixed with K. pneumoniae culture and added to 10 mL of LB broth (G3) and sterile sewage (G4), respectively. The multiplicity of infection (MOI) was 5. As a control, K. pneumoniae culture was added alone to 10 mL LB broth (G1) and sterilized sewage water (G2). All the mixtures were incubated in a room with fluctuating temperatures of 22ºC ± 5ºC to simulate natural environments and the K. pneumoniae survival was evaluated for up to 8 hours. After inoculation serial dilutions were carried out and 0.1 mL cultures were added to LB plates at 37°C for 24 hours. The number of bacterial colonies was measured by standard plate count (SPC).
Quantification and statistical analysis
Phage titers were defined as phage plaque forming units per mL. All phage titers were log10 transformed and presented as the mean with error bars representing the standard deviation of triplicate measures.
Acknowledgments
This work is supported by National Key R&D Program of China under Grant (2018YFA0903100 and 2020YFA0908700) and the Natural Science Foundation of Shandong Province (ZR2021MH177). Genome sequencing is supported by China National GeneBank and the Global Phage Hub initiated by BGI-Shenzhen.
Author contributions
H.M., D.Z.Q., and X.M.F. conceived and designed the project. Y.M.H., L.Z., and H.R. performed experimental analysis. X.B. and H.R. performed genomic analysis. D.K. and Z.C.Z. performed electron microscopy analysis. P.H.Z and G.H. performed antibiotic-resistant test. H.X., Z.Q, L.W.J., and H.T. provided the clinical isolates. Y.M.H., J.S.Y, H.M., and D.Z.Q. wrote the manuscript. H.M., X.B., L.J.H, S.W.C., D.Z.Q., and X.M.F. reviewed and edited the manuscript with input from all authors. All authors approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106947.
Contributor Information
Ziqing Deng, Email: dengziqing@genomics.cn.
Minfeng Xiao, Email: xiaominfeng@genomics.cn.
Supplemental information
Data and code availability
-
•
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA)41 of China National GeneBank DataBase (CNGBdb)42 with accession number CNP0001966.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Che Y., Xia Y., Liu L., Li A.D., Yang Y., Zhang T. Mobile antibiotic resistome in wastewater treatment plants revealed by Nanopore metagenomic sequencing. Microbiome. 2019;7:44. doi: 10.1186/s40168-019-0663-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galarde-López M., Velazquez-Meza M.E., Bobadilla-Del-Valle M., Carrillo-Quiroz B.A., Cornejo-Juárez P., Ponce-de-León A., Sassoé-González A., Alpuche-Aranda C.M. Surveillance of antimicrobial resistance in hospital wastewater: identification of carbapenemase-producing Klebsiella spp. Antibiotics. 2022;11:288. doi: 10.3390/antibiotics11030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins W., Cino J., Lenzi M.H., Sands K., Portal E., Hassan B., Dantas P.P., Migliavacca R., Medeiros E.A., Gales A.C., Toleman M.A. Diversity of lytic bacteriophages against XDR Klebsiella pneumoniae sequence type 16 recovered from sewage samples in different parts of the world. Sci. Total Environ. 2022;839:156074. doi: 10.1016/j.scitotenv.2022.156074. [DOI] [PubMed] [Google Scholar]
- 4.Tiwari A., Paakkanen J., sterblad M., Kirveskari J., Hendriksen R.S., Heikinheimo A. Wastewater surveillance detected carbapenemase enzymes in clinically relevant gram-negative bacteria in helsinki, Finland. Front. Microbiol. 2022;13:2011–2012. doi: 10.3389/fmicb.2022.887888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mutuku C., Melegh S., Kovacs K., Urban P., Virág E., Heninger R., Herczeg R., Sonnevend Á., Gyenesei A., Fekete C., Gazdag Z. Characterization of beta-lactamases and multidrug resistance mechanisms in enterobacterales from hospital effluents and wastewater treatment plant. Antibiotics. 2022;11:776. doi: 10.3390/antibiotics11060776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyres K.L., Holt K.E. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Gu D., Dong N., Zheng Z., Lin D., Huang M., Wang L., Chan E.W.C., Shu L., Yu J., Zhang R., Chen S. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 2018;18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 8.Levin B.R., Bull J.J. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2004;2:166–173. doi: 10.1038/nrmicro822. [DOI] [PubMed] [Google Scholar]
- 9.Jamal M., Bukhari S., Andleeb S., Ali M., Raza S., Nawaz M.A., Hussain T., Rahman S.U., Shah S.S.A. Bacteriophages: an overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2019;59:123–133. doi: 10.1002/jobm.201800412. [DOI] [PubMed] [Google Scholar]
- 10.Wintachai P., Naknaen A., Thammaphet J., Pomwised R., Phaonakrop N., Roytrakul S., Smith D.R. Characterization of extended-spectrum-β-lactamase producing Klebsiella pneumoniae phage KP1801 and evaluation of therapeutic efficacy in vitro and in vivo. Sci. Rep. 2020;10:11803. doi: 10.1038/s41598-020-68702-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieu J., Yu P., Zuo P., Da Silva M.L.B., Alvarez P.J.J. Going viral: emerging opportunities for phage-based bacterial control in water treatment and reuse. Acc. Chem. Res. 2019;52:849–857. doi: 10.1021/acs.accounts.8b00576. [DOI] [PubMed] [Google Scholar]
- 12.Beheshti Maal K., Soleimani Delfan A., Salmanizadeh S. Isolation and identification of two novel Escherichia coli bacteriophages and their application in wastewater treatment and coliform's phage therapy. Jundishapur J. Microbiol. 2015;8:e14945. doi: 10.5812/jjm.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imam M., Alrashid B., Patel F., Dowah A.S.A., Brown N., Millard A., Clokie M.R.J., Galyov E.E. vB_PaeM_MIJ3, a novel jumbo phage infecting Pseudomonas aeruginosa, possesses unusual genomic features. Front. Microbiol. 2019;10:2772. doi: 10.3389/fmicb.2019.02772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y.J., Lin T.L., Chen C.C., Tsai Y.T., Cheng Y.H., Chen Y.Y., Hsieh P.F., Lin Y.T., Wang J.T. Klebsiella phage φk64-1 encodes multiple depolymerases for multiple host capsular types. J. Virol. 2017;91 doi: 10.1128/jvi.02457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempf M.J., McBride M.J. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 2000;182:1671–1679. doi: 10.1128/jb.182.6.1671-1679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone L.M., Warring S.L., Jackson S.A., Warnecke C., Gardner P.P., Gumy L.F., Fineran P.C. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 2020;5:48–55. doi: 10.1038/s41564-019-0612-5. [DOI] [PubMed] [Google Scholar]
- 17.Kwon J., Kim S.G., Kim H.J., Giri S.S., Kim S.W., Lee S.B., Park S.C. Isolation and characterization of Salmonella jumbo-phage pSal-SNUABM-04. Viruses. 2020;13 doi: 10.3390/v13010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawato Y., Istiqomah I., Gaafar A.Y., Hanaoka M., Ishimaru K., Yasuike M., Nishiki I., Nakamura Y., Fujiwara A., Nakai T. A novel jumbo Tenacibaculum maritimum lytic phage with head-fiber-like appendages. Arch. Virol. 2020;165:303–311. doi: 10.1007/s00705-019-04485-6. [DOI] [PubMed] [Google Scholar]
- 19.Guan J., Bondy-Denomy J. Intracellular organization by jumbo bacteriophages. J. Bacteriol. 2020;203 doi: 10.1128/jb.00362-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M Iyer L., Anantharaman V., Krishnan A., Burroughs A.M., Aravind L., Aravind L. Jumbo phages: a comparative genomic overview of core functions and adaptions for biological conflicts. Viruses. 2021;13:63. doi: 10.3390/v13010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis R., Clooney A.G., Stockdale S.R., Buttimer C., Draper L.A., Ross R.P., Hill C. Isolation of a novel jumbo bacteriophage effective against Klebsiella aerogenes. Front. Med. 2020;7:67. doi: 10.3389/fmed.2020.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simoliūnas E., Kaliniene L., Truncaitė L., Zajančkauskaitė A., Staniulis J., Kaupinis A., Ger M., Valius M., Meškys R. Klebsiella phage vB_KleM-RaK2 - a giant singleton virus of the family Myoviridae. PLoS One. 2013;8:e60717. doi: 10.1371/journal.pone.0060717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokolova M.L., Misovetc I., K V.S. Multisubunit RNA polymerases of jumbo bacteriophages. Viruses. 2020;12 doi: 10.3390/v12101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y., Gao M. Jumbo bacteriophages: an overview. Front. Microbiol. 2017;8:403. doi: 10.3389/fmicb.2017.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fokine A., Rossmann M.G. Molecular architecture of tailed double-stranded DNA phages. Bacteriophage. 2014;4:e28281. doi: 10.4161/bact.28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leiman P.G., Molineux I.J. Evolution of a new enzyme activity from the same motif fold. Mol. Microbiol. 2008;69:287–290. doi: 10.1111/j.1365-2958.2008.06241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennes K.P., Simon M. Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl. Environ. Microbiol. 1995;61:333–340. doi: 10.1128/aem.61.1.333-340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shomar B., Al-Darwish K., Vincent A. Optimization of wastewater treatment processes using molecular bacteriology. J. Water Process Eng. 2020;33:101030. doi: 10.1016/j.jwpe.2019.101030. [DOI] [Google Scholar]
- 29.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., Li Y., Ye J., Yu C., Li Z., et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7:1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson L.S., Eddy S.R., Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinf. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan P.P., Lin B.Y., Mak A.J., Lowe T.M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stothard P., Grant J.R., Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Briefings Bioinf. 2019;20:1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bin Jang H., Bolduc B., Zablocki O., Kuhn J.H., Roux S., Adriaenssens E.M., Brister J.R., Kropinski A.M., Krupovic M., Lavigne R., et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019;37:632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 41.Guo X., Chen F., Gao F., Li L., Liu K., You L., Hua C., Yang F., Liu W., Peng C., et al. CNSA: a data repository for archiving omics data. Database. 2020 doi: 10.1093/database/baaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen F.Z., You L.J., Yang F., Wang L.N., Guo X.Q., Gao F., Hua C., Tan C., Fang L., Shan R.Q., et al. CNGBdb: China national GeneBank DataBase. Yi Chuan. 2020;42:799–809. doi: 10.16288/j.yczz.20-080. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y., Sun E., Song J., Yang L., Wu B. Complete genome sequence of a novel T7-like bacteriophage from a pasteurella multocida capsular type A isolate. Curr. Microbiol. 2018;75:574–579. doi: 10.1007/s00284-017-1419-3. [DOI] [PubMed] [Google Scholar]
- 44.Du K., Yang F., Zhang J.T., Yu R.C., Deng Z., Li W.F., Chen Y., Li Q., Zhou C.Z. Comparative genomic analysis of five freshwater cyanophages and reference-guided metagenomic data mining. Microbiome. 2022;10:128. doi: 10.1186/s40168-022-01324-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutter E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009;501:141–149. doi: 10.1007/978-1-60327-164-6_14. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Zheng K., Liu B., Liang Y., You S., Zhang W., Zhang X., Jie Y., Shao H., Jiang Y., et al. Characterization and genomic analysis of marinobacter phage vB_MalS-PS3, representing a new lambda-like temperate siphoviral genus infecting algae-associated bacteria. Front. Microbiol. 2021;12:726074. doi: 10.3389/fmicb.2021.726074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao M., Wang C., Qiang X., Liu H., Li P., Pei G., Zhang X., Mi Z., Huang Y., Tong Y., Bai C. Isolation and characterization of a novel bacteriophage infecting carbapenem-resistant Klebsiella pneumoniae. Curr. Microbiol. 2020;77:722–729. doi: 10.1007/s00284-019-01849-8. [DOI] [PubMed] [Google Scholar]
- 48.D'Andrea M.M., Marmo P., Henrici De Angelis L., Palmieri M., Ciacci N., Di Lallo G., Demattè E., Vannuccini E., Lupetti P., Rossolini G.M., Thaller M.C. phiBO1E, a newly discovered lytic bacteriophage targeting carbapenemase-producing Klebsiella pneumoniae of the pandemic Clonal Group 258 clade II lineage. Sci. Rep. 2017;7:2614. doi: 10.1038/s41598-017-02788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng L., Deng Z., Tao H., Song W., Xing B., Liu W., Kong L., Yuan S., Ma Y., Wu Y., et al. Harnessing stepping-stone hosts to engineer, select, and reboot synthetic bacteriophages in one pot. Cell Rep. Methods. 2022;2:100217. doi: 10.1016/j.crmeth.2022.100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoang H.A., Nhung N.T.T. Development of a bacteriophage-based method for detection of Escherichia coli O157:H7 in fresh vegetables. Food Saf. 2018;6:143–150. doi: 10.14252/foodsafetyfscj.2018010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiggins B.A., Alexander M. Minimum bacterial density for bacteriophage replication: implications for significance of bacteriophages in natural ecosystems. Appl. Environ. Microbiol. 1985;49:19–23. doi: 10.1128/aem.49.1.19-23.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiegand I., Hilpert K., Hancock R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 53.Liguori K., Keenum I., Davis B.C., Calarco J., Milligan E., Harwood V.J., Pruden A. Antimicrobial resistance monitoring of water environments: a framework for standardized methods and quality control. Environ. Sci. Technol. 2022;56:9149–9160. doi: 10.1021/acs.est.1c08918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA)41 of China National GeneBank DataBase (CNGBdb)42 with accession number CNP0001966.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.