Abstract
Summary
We report a case of metastatic pancreatic neuroendocrine carcinoma associated with paraneoplastic Cushing’s syndrome, successively treated with five lines of treatment (platin-etoposide, LV5FU2-dacarbazine, FOLFIRINOX, pembrolizumab, and paclitaxel) and anti-secretory treatment. Circulating-free DNA (cfDNA) was analysed at each morphological evaluation starting from the second-line treatment. cfDNA changes were well correlated with the disease course, and cfDNA may be used as a predictive marker and/or as an early marker of response. In addition, the absolute count of atypical cells was elevated upon disease progression.
Learning points
cfDNA changes were well correlated with the Cushing’s syndrome course and with the tumour burden changes assessed by laboratory markers and by RECIST criteria.
cfDNA analysis was used to determine the pharmacogenetic patterns of the present patient.
An elevated number of atypical circulating cells was noticed upon disease progression.
Keywords: cfDNA, neuroendocrine carcinoma, Cushing’s syndrome, immunotherapy
Background
Neuroendocrine carcinomas (NEC) are associated with paraneoplastic syndrome, such as Cushing’s syndrome, in less than 5% of cases. NEC are usually treated with first-line platinum-etoposide chemotherapy and then with FOLFOX, FOLFIRI, or temozolomide/darcarbazine-based chemotherapy with poor effectiveness. However, NEC are heterogeneous and comprise three distinct molecular signatures: the 'small-cell' type characterised by TP53 and RB1 mutations, the 'adenocarcinoma' type characterised by mutations such as KRAS, APC, and/or BRAF, often found in their respective primary tumour location, and the 'well-differentiated neuroendocrine tumour (NET)' type characterised by MEN1, DAXX/ATRX alterations in pancreatic NET for instance. This is interesting especially taking into account that some recent studies have reported Rb loss and KRAS mutation as predictors of the response to platinum-based chemotherapy in pancreatic neuroendocrine neoplasms (Hijioka et al. 2017). Therefore, circulating-free DNA (cfDNA) assessment represents an interesting tool since blood tests are easily accessible, can be regularly performed along the course of the disease, and it may reflect the spatial and temporal heterogeneity of the tumour better than a single localised biopsy (Rizzo & Meyer 2018). Moreover, the response to drugs is also determined by environmental and genetic variants that influence the pharmacokinetics and pharmacodynamics, the latter can be easily identified in the blood by cfDNA assessment (Somogyi et al. 2007). Pharmacokinetics data are reported in pharmvar (https://www.pharmvar.org/) and pharmgkb databases (https://www.pharmgkb.org/).
We reported herein the case of a patient who had metastatic pancreatic NEC associated with Cushing’s syndrome for whom cfDNA analyses were well correlated with the clinical, laboratory, and morphological characteristics of the disease.
Case presentation
In June 2018, a 45-year-old man was diagnosed with liver metastasis of a pancreatic small-cell NEC; the diagnosis was made from liver biopsy, and the Ki67 index was 80%. The NEC was microsatellite stable (MSS), with nuclear expression of TP53 and loss of Rb expression. The physical examination found clinical signs consistent with Cushing’s syndrome, which was confirmed: serum cortisol level was 57.7 µg/dL (normal level 10–20 µg/dL), 24-h urine free cortisol (UFC) measurement was 27,306 µg/24 h (normal level 4.3–176 µg/24 h), and adrenocorticotropic hormone (ACTH) level was 111 pg/mL (normal level 0–46 pg/mL). Brain imaging confirmed the absence of any pituitary abnormality, and the diagnosis of ectopic Cushing’s syndrome was retained.
Treatment
The patient was initially treated with cisplatin-etoposide (three cycles): the initial laboratory response (alkaline phosphatase-ALP, ACTH) was associated with stable disease (SD) according to RECIST criteria and worse symptoms. Ketoconazole was not administered because of interference with chemotherapy (enzymatic induction). Thus, metyrapone (1000 mg p.o. every 8 h) and cabergoline (0.5 mg daily) treatments were initiated. A month later, a new CT scan revealed a RECIST progressive disease (PD) and LV5FU2-dacarbazine treatment was initiated: the Cushing’s syndrome symptoms temporarily improved (metyrapone was then reduced at 750 mg p.o. every 8 h), partial response was obtained after three cycles according to RECIST criteria, but PD occurred after six cycles. Then, third-line chemotherapy was initiated: FOLFIRINOX (eight cycles) followed by six cycles of FOLFIRI (oxaliplatin was discontinued because of grade 2 neurotoxicity); a dramatic clinical, laboratory, and morphological improvement was observed after four cycles. In April 2019, metyrapone was no longer available in France, it was, therefore, substituted with osilodrostat (2 mg p.o. twice daily), and the dosage was progressively increased (up to 20 mg p.o. twice daily) because of refractory hypercortisolaemia. PD occurred 8 months after the initiation of FOLFIRINOX treatment. Pembrolizumab, as single-agent immunotherapy, was initiated but failed to control the disease progression. In November 2019, the patient was administered cisplatin-etoposide as a fourth-line therapy and PD occurred after three cycles. Paclitaxel was initiated at the end of January 2020 and discontinued after one cycle because of major asthenia with hepatalgia relieved by morphine. The patient died in February 2020.
Investigation
cfDNA levels were monitored at each morphological evaluation starting from the second-line treatment. cfDNA analysis was used to determine the pharmacogenetic patterns of the present patient for chemotherapy and supportive treatments. Absolute count of circulating atypical cells was carried out at the latest disease stage (Supplementary material, see section on supplementary materials given at the end of this article).
Outcome
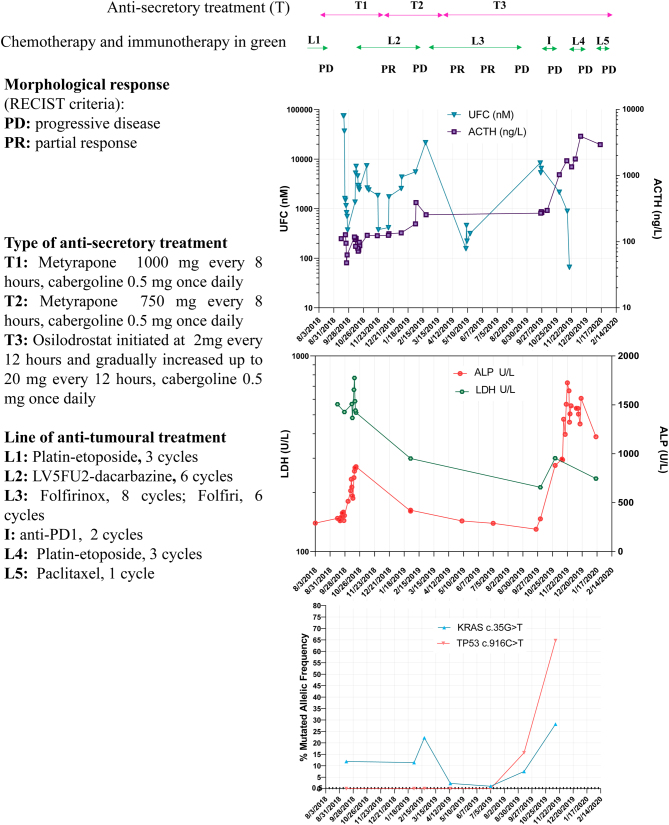
Based on cfDNA analysis, a KRAS c.35G>T and then a TP53 c.916C>T mutation were found, but no RB1 or BRAF mutation nor mutation on mismatch repair (MMR) genes were detected. The mutated allelic frequency (MAF) of KRAS mutation significantly and dramatically decreased during FOLFIRINOX treatment, so did the paraneoplastic secretion of ACTH while a biochemical improvement (ALP/LDH) was observed; all these parameters increased upon disease progression, in addition, a predominant tumoural clone with a TP53 mutation was newly detected (Fig. 1).
Figure 1.
Course of the Cushing’s syndrome (A, UFC and ACTH) and neuroendocrine carcinoma along the treatment according to the morphology (response assessed by RECIST criteria), the laboratory analysis (B, LDH and ALP), and the mutated allelic frequency in cfDNA (C). UFC, urine free cortisol; ACTH, adrenocorticotropic hormone; LDH, lactate deshydrogenase; ALP, alkaline phosphatase; cfDNA, circulating-free DNA.
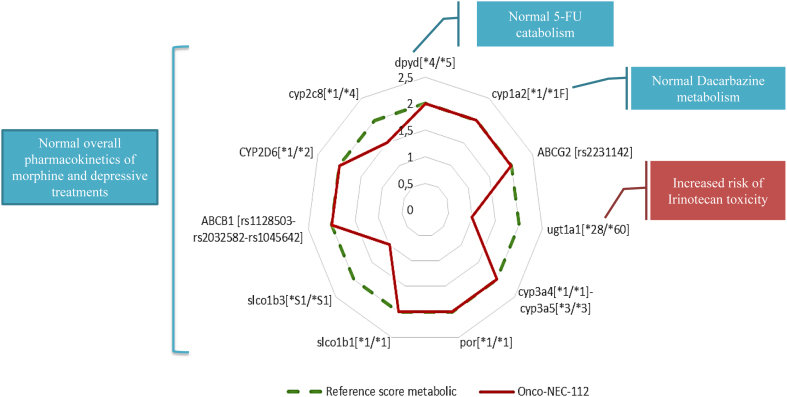
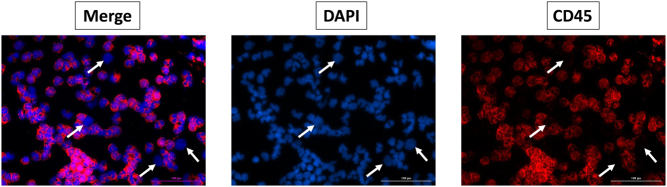
The analysis of cfDNA did not predict any modification in terms of pharmacokinetics and pharmacodynamics regarding 5FU catabolism (DYPD gene) and dacarbazine metabolism (CYP1A2 gene). The predicted enzymatic activity of UGT1A was decreased, which usually increases the risk of digestive and haematological toxicity of irinotecan treatment, although the standard dose of irinotecan was well-tolerated in the present case. The pharmacogenetics analysis did not predict any modification in terms of pharmacokinetics for analgesic and neuroleptic treatments (Fig. 2), suggesting no need for dose adaptation (based on https://www.pharmvar.org/ and pharmgkb https://www.pharmgkb.org/ data). Moreover, numerous atypical cells (CD45 negative) were observed in the whole blood after microfluidic enrichment using ClearCell® FX1. At the latest disease stage, the atypical cell count was high (71.4 atypical cells per mL of whole blood; Fig. 3).
Figure 2.
Pharmacogenetic profile.
Figure 3.
Atypical cell analysis after enrichment (534/7.5 mL of whole blood) by ClearCell® FX1 and immunostaining using DAPI (blue) and anti-CD45 antibody (red). Example of four atypical cells CD45− (white arrow). Image scale 100 µm.
Discussion
To our knowledge, the present study is the first report of a pancreatic NEC case for which repeated cfDNA analyses were performed. The molecular changes were well correlated with the Cushing’s syndrome course (paraneoplastic secretion of ACTH) and with the tumour burden changes assessed by laboratory markers (ALP/LDH) and by RECIST criteria, as already reported in other cancers. The pathological diagnosis, performed in an outside laboratory, was reviewed, but unfortunately, there was not enough material to perform a genomic comparison between tumour tissue and cfDNA. This case highlights several points. NEC cases seem to be good candidates for cfDNA assessment because the amount of tissue available for genomic analysis is often low, they often are associated with liver metastases, a high proliferative index with apoptosis/necrosis, and high mutational tumour load, which are factors associated with high MAFs. Close monitoring of cfDNA changes could forecast the course of the disease under treatment; an increase in cfDNA levels detected disease progression 5 months prior to radiological evidence in 89% of patients with metastatic breast cancer (Dawson et al. 2013). The most interesting is probably the interest of cfDNA to detect predictive factors of response in NEC. Hijioka et al. have reported that loss of Rb expression was significantly associated with a higher response rate under platinum-based chemotherapy in 49 patients with pancreatic NEC. This association was not observed with KRAS mutation in NEC, by contrast with panNEN G3 (Hijioka et al. 2017). This could at least partially explain the poor response to first-line cisplatin-based chemotherapy in the patient presented herein. In contrast, FOLFIRINOX had an impressive effectiveness in the present case, which may be explained by the molecular 'adenocarcinoma' signature, probably more responsive to the chemotherapy regimen used in adenocarcinomas than those usually used in 'pure' NEC (Gerard et al. 2020). This will be evaluated in the FOLFIRINEC study (NCT04325425) comparing FOLFIRINOX and platinum-etoposide in gastroenteropancreatic NEC and establishing a mutational molecular profile of tumour tissue and cfDNA. If the results are convincing, cfDNA genotyping results could thus represent an easy tool to orientate patients to a specific treatment. After FOLFIRINOX treatment, we were not able to propose BRAF/MEK inhibitors because there was no BRAF mutation in the present cfDNA. At the end of the disease course, the patient was administered pembrolizumab, which had low effectiveness. The treatment options were limited at that time, and the disappointed results of studies evaluating immunotherapy in NEC had not been reported yet. In addition to the MSS status, high cortisol levels, secondary to the Cushing syndrome and impacting T cell function, could explain the little clinical benefit of pembrolizumab in the present case. cfDNA could probably also help to detect the few patients with good predictive factors of response to immunotherapy by identifying high tumour mutational burden and/or mutation on MMR/POLE gene.
To go further, the same blood samples used for cfDNA analysis can also provide information on the pharmacogenetic patterns of patients to anticipate possible toxicities or optimise chemotherapy doses and supportive treatments (Somogyi et al. 2007). The UGT1A1*28 allelic variation of UGT1A1 has been associated with an increased risk of severe neutropenia and may justify a lower dose of irinotecan. In contrast, patients non-homozygous for UGT1A1*28 with metastatic colorectal cancer may benefit from high doses of irinotecan (260–300 mg/m2) with an improved overall response rate (Páez et al. 2019). We can speculate that the patient presented herein had, due to his mutation, a higher blood concentration of irinotecan, which contributed to the high response to FOLFIRINOX he displayed.
Some obstacles still limit the implementation of cfDNA in routine clinical practise. cfDNA can be released in the bloodstream by primary tumour cell necrosis, apoptosis, and/or active secretion. The quantity of cfDNA released is variable, and its determination requires highly specific and sensitive methods of detection. Strict pre-analytical conditions ensured by specialised laboratories centralising the samples or trained local biology laboratories are required to prevent cfDNA contamination with genomic DNA from cell lysis. EDTA tubes could facilitate cfDNA analysis in classical pre-analytical routine workflow up to 24 h after blood sampling, as no release of genomic DNA was observed at 24 h following shipping by car at room temperature (Garcia et al. 2017). After this time period, tubes containing nucleated-cell stabilisers are preferred but are more expensive and are not validated in some countries for routine diagnosis. Besides, cfDNA analysis is still expensive, but its costs are gradually coming down. In addition, the cfDNA use could improve NEC management in several ways but further studies are warranted to formally validate cfDNA as a biomarker that can be incorporated into routine clinical care: (i) first, to choose the best treatment according to one predictive factor of response, as already demonstrated in non-small cell lung cancer (NSCLC) with third generation of EGFR inhibitor (osimertinib) according to EGFR mutation detected in cfDNA; this has been reported in cases of colon NEC successfully treated by a BRAF inhibitor after the detection of V600E-BRAF mutation on cfDNA (Dizdar et al. 2019); (ii) secondly, cfDNA could be used as an early marker of response (and/or therapeutic failure), but prospective studies are warranted to prove that an early rise in cfDNA levels or the detection of new oncogenic mutations is indeed associated with shorter survival and, more importantly, if an early treatment change does translate in survival gain (Gerard et al. 2020); (iii) thirdly, cfDNA could be used as an early marker of recurrence or a marker of persistent disease after a curative treatment (surgery or chemoradiotherapy) of localised NEC; this is currently investigated not only in rectal adenocarcinoma but also in NEC by the NEONEC phase II study (NCT04268121).
Finally, many studies have shown that the presence of circulating tumour cells in NETs was correlated with the tumour burden and disease progression (Rizzo & Meyer 2018). Herein, we noticed an elevated number of atypical cells corresponding to DAPI+ CD45− labelling late in the course of the disease. Atypical cell enumeration could represent another tool to monitor the course of the disease and to identify other IHC predictive factors of response, such as PDL1 or SSTR expression (Garcia et al. 2019); additional studies are warranted to evaluate its interest in NEC.
In conclusion, cfDNA burden was well correlated to the course of the disease. Interestingly, this illustrates that cfDNA could be a useful tool to better identify subgroups of NEC patients and propose more personalised treatments.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this case report.
Funding
The analyses were supported by a grant from the Comité de Savoie de la Ligue Contre le Cancer and the Comité Départemental de l’Allier de la Ligue Contre le Cancer.
Acknowledgements
The authors thank Hélène Boyer (DRCI, Hospices Civils de Lyon) for her help in manuscript editing and also thank Dr Marie-Sophie Soubeyrand and Dr Jérôme Desramé who had initially made the diagnosis and followed the patient.
References
- Dawson SJ, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo Bet al. 2013. Analysis of circulating tumor DNA to monitor metastatic breast cancer. New England Journal of Medicine 368 1199–1209. ( 10.1056/NEJMoa1213261) [DOI] [PubMed] [Google Scholar]
- Dizdar L, Werner TA, Drusenheimer JC, Möhlendick B, Raba K, Boeck I, Anlauf M, Schott M, Göring W, Esposito Iet al. 2019. BRAFV600E mutation: a promising target in colorectal neuroendocrine carcinoma. International Journal of Cancer 144 1379–1390. ( 10.1002/ijc.31828) [DOI] [PubMed] [Google Scholar]
- Garcia J, Dusserre E, Cheynet V, Bringuier PP, Brengle-Pesce K, Wozny AS, Rodriguez-Lafrasse C, Freyer G, Brevet M, Payen Let al. 2017. Evaluation of pre-analytical conditions and comparison of the performance of several digital PCR assays for the detection of major EGFR mutations in circulating DNA from non-small cell lung cancers: the CIRCAN_0 study. Oncotarget 8 87980–87996. ( 10.18632/oncotarget.21256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Barthelemy D, Geiguer F, Ballandier J, Li KW, Aurel JP, Le Breton F, Rodriguez-Lafrasse C, Manship B, Couraud Set al. 2019. Semi-automatic PD-L1 characterization and enumeration of circulating tumor cells from non-small cell lung cancer patients by immunofluorescence. Journal of Visualized Experiments 150 e59873. ( 10.3791/59873) [DOI] [PubMed] [Google Scholar]
- Gerard L, Garcia J, Gauthier A, Lopez J, Durand A, Hervieu V, Lemelin A, Chardon L, Landel V, Gibert Bet al. 2020. ctDNA in neuroendocrine carcinoma of gastroenteropancreatic origin or of unknown primary: the CIRCAN-NEC pilot study. Neuroendocrinology [epub]. ( 10.1159/000512502) [DOI] [PubMed] [Google Scholar]
- Hijioka S, Hosoda W, Matsuo K, Ueno M, Furukawa M, Yoshitomi H, Kobayashi N, Ikeda M, Ito T, Nakamori Set al. 2017. Rb loss and KRAS mutation are predictors of the response to platinum-based chemotherapy in pancreatic neuroendocrine neoplasm with Grade 3: a Japanese multicenter pancreatic NEN-G3 study. Clinical Cancer Research 23 4625–4632. ( 10.1158/1078-0432.CCR-16-3135) [DOI] [PubMed] [Google Scholar]
- Páez D, Tobeña M, Fernández-Plana J, Sebio A, Virgili AC, Cirera L, Barnadas A, Riera P, Sullivan I, Salazar J. 2019. Pharmacogenetic clinical randomised phase II trial to evaluate the efficacy and safety of FOLFIRI with high-dose irinotecan (HD-FOLFIRI) in metastatic colorectal cancer patients according to their UGT1A 1 genotype. British Journal of Cancer 120 190–195. ( 10.1038/s41416-018-0348-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo FM, Meyer T. 2018. Liquid biopsies for neuroendocrine tumors: circulating tumor cells, DNA, and microRNAs. Endocrinology and Metabolism Clinics of North America 47 471–483. ( 10.1016/j.ecl.2018.04.002) [DOI] [PubMed] [Google Scholar]
- Somogyi AA, Barratt DT, Coller JK. 2007. Pharmacogenetics of opioids. Clinical Pharmacology and Therapeutics 81 429–444. ( 10.1038/sj.clpt.6100095) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a