Abstract
Background
Metastatic colorectal cancer (mCRC) patients tend to have modest benefits from molecularly driven therapeutics. Patient-derived tumor organoids (PDTOs) represent an unmatched model to elucidate tumor resistance to therapy, due to their high capacity to resemble tumor characteristics.
Materials and methods
We used viable tumor tissue from two cohorts of patients with mCRC, naïve or refractory to treatment, respectively, for generating PDTOs. The derived models were subjected to a 6-day drug screening assay (DSA) with a comprehensive pipeline of chemotherapy and targeted drugs against almost all the actionable mCRC molecular drivers. For the second cohort DSA data were matched with those from PDTO genotyping.
Results
A total of 40 PDTOs included in the two cohorts were derived from mCRC primary tumors or metastases. The first cohort included 31 PDTOs derived from patients treated in front line. For this cohort, DSA results were matched with patient responses. Moreover, RAS/BRAF mutational status was matched with DSA cetuximab response. Ten out of 12 (83.3%) RAS wild-type PDTOs responded to cetuximab, while all the mutant PDTOs, 8 out of 8 (100%), were resistant. For the second cohort (chemorefractory patients), we used part of tumor tissue for genotyping. Four out of nine DSA/genotyping data resulted applicable in the clinic. Two RAS-mutant mCRC patients have been treated with FOLFOX–bevacizumab and mitomycin–capecitabine in third line, respectively, based on DSA results, obtaining disease control. One patient was treated with nivolumab–second mitochondrial-derived activator of caspases mimetic (phase I trial) due to high tumor mutational burden at genotyping, experiencing stable disease. In one case, the presence of BRCA2 mutation correlated with DSA sensitivity to olaparib; however, the patient could not receive the therapy.
Conclusions
Using CRC as a model, we have designed and validated a clinically applicable methodology to potentially inform clinical decisions with functional data. Undoubtedly, further larger analyses are needed to improve methodology success rates and propose suitable treatment strategies for mCRC patients.
Key words: colorectal cancer, PDTO, resistance, 3D, personalized medicine
Highlights
-
•
CRC is a heterogeneous disease and mechanism of tumor resistance to drugs occurs, impairing treatment success.
-
•
PDTOs resemble three-dimensional tumor characteristics.
-
•
PDTOs provide information for functionally informed therapeutic decisions.
Introduction
The incorporation of complex genomic platforms and circulating tumor DNA (ctDNA) determinations has permitted a more comprehensive biomarker identification and molecular follow-up, which in turn has supported the progressive incorporation of targeted drugs to metastatic colorectal cancer (mCRC) patient treatment pathways.1 Furthermore, gained insights on the complex heterogeneity of cancer are leading to a shift from ‘one drug for every patient’ concept to an individualized patient care, which pursues applying individual therapeutic decisions based on tumor genomic, transcriptomic and immune integrative analysis for every single patient.2 However, despite all these circumstances, mCRC clinical outcomes remain stuck due to the existence of mechanisms of resistance that impair effectiveness of treatment.1
Significant efforts have been made to understand and identify the particular CRC molecular features that lead the disease to resist therapy.3 However, the lack of suitable translational patient-derived tumor models is limiting the progresses achieved. In this regard, two main tumor models have been extensively used for translational studies so far: patient-derived xenografts (PDXs) and immortalized cancer cell lines. PDXs represent established in vivo mouse models that recapitulate tumor heterogeneity and vascularization and consequently they have been explored for patient treatment guidance. In this model, tumor implantation requires a long time, which could lead to delayed clinical decisions, mostly in patients with refractory mCRC who cannot wait long to receive subsequent therapies.4 Moreover, PDXs are expensive and have a low throughput.5 On the other hand, two-dimensional immortalized cell lines have been largely used due to their low cost, simplistic manipulation, infinite growth potential and the possibility of obtaining results in a short time period; however, they fail in recapitulating several tumor features like epithelial polarity or tumor–matrix interaction, impairing their utility to generate informed therapeutic decisions.6
Patient-derived tumor organoids (PDTOs) have been used as a research model in several types of cancer due to their capacity to resemble three-dimensional (3D) tumor characteristics and their ability to recapitulate tumor clinical outcomes and predict drug sensitivity in a timely fashion.7,8 Thus, these models may have the potential to provide information for functionally informed therapeutic decisions of the subsequent treatment lines in a short time frame, thereby improving PDX and cancer primary cell-line performances. In the present work, we present a translational effort to integrate tumor genomics with functional data coming from drug screening assays (DSAs) carried out in mCRC PDTOs, aiming to provide applicable information to guide patient management.
Materials and methods
We have set up a methodology for generating and storing PDTOs from two cohorts of patients with mCRC at the Department of Precision Medicine of Università della Campania Luigi Vanvitelli, Naples and at the Vall d’Hebron Institute of Oncology (VHIO) Colorectal Cancer Translational Laboratories, Barcelona, respectively. All human samples and biopsies were collected after obtaining a written informed consent from any patient, in accordance with the Declaration of Helsinki. The use of these samples for research purposes was approved by local ethical committees of both institutions.
Generation of PDTO models
Tumor samples were obtained from surgical specimens or biopsies (Figure 1A). Once collected, tumor tissue was transferred into sterile phosphate-buffered saline and processed within 24 h to avoid any type of contamination and guarantee tissue availability. After a 1600 rpm/286 g centrifugation, pellets were collected and transferred to a plate with a sterile scalpel, then transferred into a tube with medium enriched with antibiotics, DNAse and collagenase for tissue dissociation and then incubated for 15' min at 37°C, resuspended and filtered through a 100-μm cell strainer. The resultant cells were cultured in low-attachment wells with medium enriched with specific growth factors and antibiotics to generate PDTOs (Supplementary Materials and Methods, available at https://doi.org/10.1016/j.esmoop.2023.101198). After 3-5 days, clumps were dissociated in TrypLE (Gibco, #12604-013, Grand Island, NY) for 5-10 min, counted and cultured in Matrigel (Matrigel Corning, AZ) to preserve 3D as previously described (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2023.101198).9 Part of all PDTOs was fresh frozen and stored to build a comprehensive living organoid bank to sustain further investigational projects.
Figure 1.
Establishment of PDTOs to recapitulate clinical outcome and provide therapeutic decisions in mCRC. (A) Project overview. (B) Cohort 1 and 2 PDTO diagram. (C) List of drugs used in cohort 1 DSA. (D) List of drugs used in cohort 2 DSA. DSA, drug screening assay; mCRC, metastatic colorectal cancer; PD, progression of disease; PDTO, patient-derived tumor organoid.
Design of drug screening assay
At first, we streamlined the methodology to carry out DSAs using the smallest number of cells to achieve statistically significant results. Initially, to avoid wasting patient tumor samples, we used core needle biopsies from PDX tumor models to calculate the feasibility of the procedure toward obtaining enough tumor viable cells to test our PDTO-deriving methodology and to estimate implantation rates. We estimated a go/no-go threshold of 50% implantation rate to start applying our protocol to patient samples. Once the correct methodology and number of cells to be used were established, we started to apply our mCRC PDTO-generating protocol to patient-derived samples.
For PDTO DSAs, after 3-5 days following organoid generation, clumps were trypsinized with TrypLE and single cells were counted in the presence of trypan blue, using a glass hemocytometer (average cell count from each of the sets of 16 corner squares multiplied by 104). Five hundred cells/well were seeded in 96 multi-wells in 100 μl of medium plus 5% Matrigel, to be exposed to the drug sensitivity screening panel (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2023.101198). We chose 500 cells per well as the minimum cell number that provided statistically significant results in our first PDX drug screening (data not shown). Cells were cultured for 6 days in medium added with chemotherapy and target drugs (Figure 1C and D). Different concentrations of cells were used per well. To conform the panel of drugs, we considered the approved chemotherapy and targeted drugs in mCRC and, as regards the second cohort of the study, we added a subset of drugs investigated in phase I trials conducted in VHIO phase I facility at the time of the project in order to use the intended methodology to inform patient clinical trial recommendation (Figure 1C and D). Experiments were carried out in quadruplicate with drug concentrations based on previous in vitro studies of 3D PDTOs.10, 11, 12, 13
Cell viability was measured at day 0 and day 6. In the first cohort, cell viability was measured with MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay, while, in the second cohort, the CellTiter-Glo® Luminescent Cell Viability Assay (CellTiter Glo, Promega, Madison, WI) was used.14
The MTS assay protocol is based on the reduction of the MTS tetrazolium compound by viable mammalian cells (and cells from other species) to generate a colored formazan dye that is soluble in cell culture media. The formazan dye is quantified by measuring the absorbance at 490-500 nm.
Briefly, cells (500/well) were seeded in a 96-well microtiter plate in a final volume of 100 μl/well and they were treated with anticancer drugs for 6 days. Then, 20 μl/well of MTS reagent was added into each well and incubated for 0.5-4 h at 37°C in standard culture conditions. Finally, the absorbance of treated and untreated cells was measured using a plate reader at absorbance = 490 nm.
The CellTiter-Glo® Luminescent Cell Viability Assay measures the number of viable cells based on the ATP quantitation, an indicator of metabolically active cells. Briefly, after 6 days of treatment, cells were transferred to a transparent 96 multiwell and 100 μl/well of CellTiter-Glo® reagent was added. After 10 min luminescence was measured with the Infinite 200 PRO microplate reader (Tecan Life Sciences, Switzerland).
PDTO molecular analysis
For the second cohort of PDTOs, the results obtained from the drug screening panel were matched with the information from the organoid genotyping to define the exact mechanisms of drug-acquired resistance. Particularly, an amplicon capture-based sequencing panel (VHIO-300) of 431 pan-cancer-related genes was used and, in addition, a NanoString® (Seattle, WA) panel looking for fusions and copy number variations was also used to capture the molecular portrait of the sample (Supplementary Table 1, Supplementary Materials and Methods, available at https://doi.org/10.1016/j.esmoop.2023.101198). The results of the genetic and drug screenings were integrated in order to inform the next most suitable treatment line.
Results
From June 2018 to November 2021, 40 patients were included in the two cohorts of the project, 31 patients in cohort 1 and 9 patients in cohort 2 (Figure 1A and B).
Initially, we pursued to assess the ability of the models generated under our methodology to recapitulate in vitro patient tumor outcomes seen in the clinic. To this end we included a total of 31 PDTOs derived from treatment-naïve patients or after progression to front line in cohort 1. For this cohort, the establishment rate of PDTOs was ∼80% (26/31). DSAs were carried out with drugs that are described in Figure 1C. We carried out MTS viability assays to measure drug activity of the abovementioned drugs.
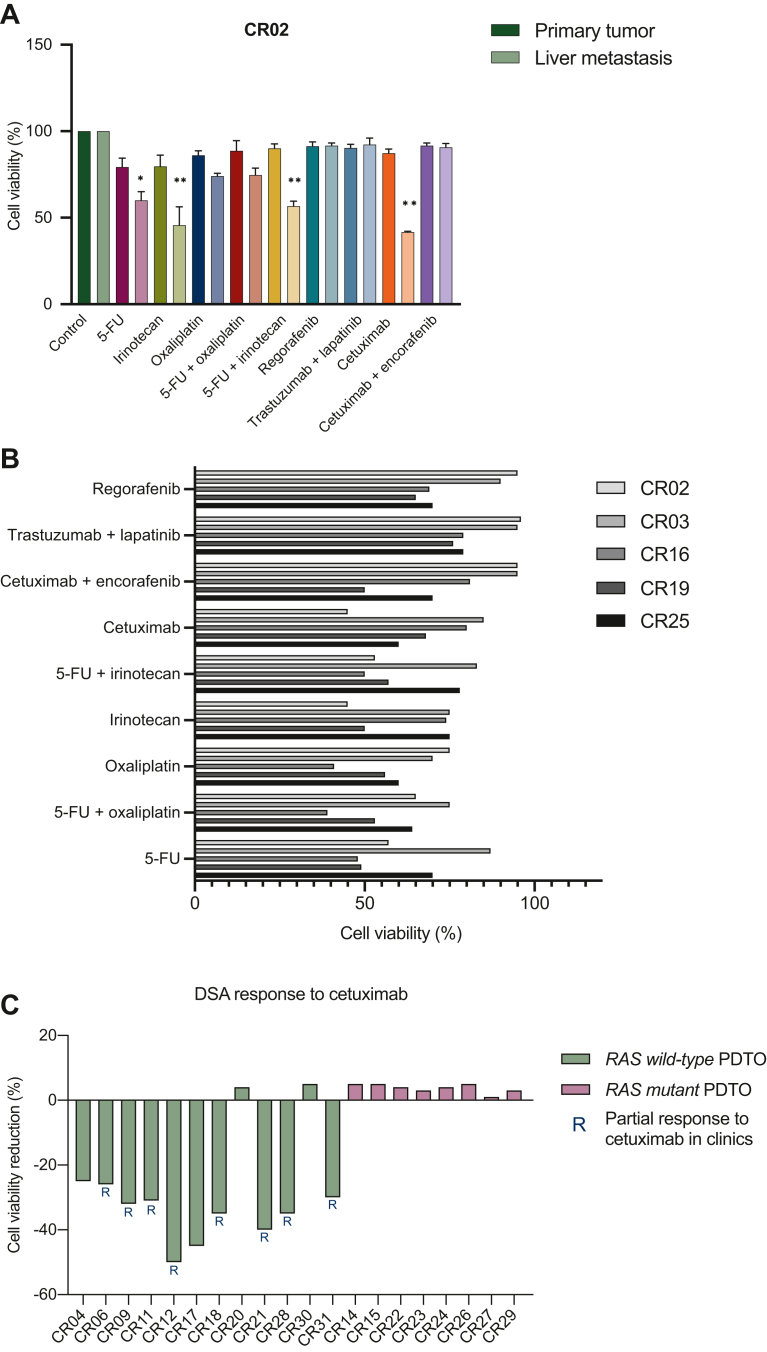
Firstly, we wanted to investigate whether PDTOs derived from metastases could better recapitulate tumor characteristics compared with PDTOs generated from primary tumors. We were able to compare directly the two PDTOs derived from primary tumor and liver metastasis of a patient with synchronic disease (CR02). Interestingly, DSAs of primary tumor PDTO showed no statistically significant reduction after treatment of cell viability in DSA, while for liver metastasis PDTO a statistical result was achieved (Figure 2A). Particularly, cells were significantly sensitive to 5-fluorouracil (5-FU), irinotecan and cetuximab, with a cell viability reduction of 40% compared with untreated cells but resulted completely resistant to the combination of cetuximab plus encorafenib, trastuzumab lapatinib and 5-FU plus oxaliplatin. Furthermore, we compared the DSA results with the information coming from the molecular tissue analysis. The patient had an RAS, BRAF, HER2 wild-type (WT) tumor.
Figure 2.
PDTO drug screening assay and correlation with clinical outcomes in Cohort 1. (A) DSA of CR02 PDTO derived from primary tumor and liver metastasis of a patient with a synchronic mCRC. (B) Response to DSA of PDTO derived from patients naïve to treatment for mCRC. (C) Correlation of response/resistance to cetuximab on DSA and patient RAS mutational status. ∗P ≤ 0.033; ∗∗P ≤ 0.001. 5-FU, 5-fluorouracil; DSA, drug screening assay; mCRC, metastatic colorectal cancer; PDTO, patient-derived tumor organoid.
As a further analysis, we split patients into two subgroups according to their treatment naïve or pretreated status. Ten out of 31 PDTOs were derived from patients who were naïve to treatment and underwent surgery of liver metastasis (Table 1 and Figure 1B). Taking into consideration that this subset of PDTOs was derived from patients who did not receive any treatment for mCRC, we carried out a DSA with main approved chemotherapies used in first-line clinical setting and principal targeted agents for mCRC according to patients’ molecular characteristics (RAS, BRAF, HER2 mutational status) (i.e. 5-FU, irinotecan, oxaliplatin, cetuximab, cetuximab plus encorafenib, trastuzumab plus lapatinib, regorafenib) (Figure 1C). DSA showed statistically significant results in terms of cell viability reduction in five PDTOs (CR02, CR03, CR16, CR19, CR25), with different rates of response to different drugs, reflecting in some way an interpatient molecular heterogeneity that is responsible in the clinics of different response rates to chemotherapy plus target agents in the metastatic setting (Figure 2B).
Table 1.
First cohort PDTOs
| PDTO ID | Primary tumor or liver met | RAS mutational status | Treatment received before surgery |
|---|---|---|---|
| CR01 | Primary tumor | Mutant | FOLFOX plus bevacizumab |
| CR02 | Primary tumor/liver metastasis | Wild type | No treatment |
| CR03 | Liver metastasis | Mutant | No treatment |
| CR04 | Liver metastasis | Wild type | FOLFOX |
| CR05 | Primary tumor | Unknown | No treatment |
| CR06 | Liver metastasis | Wild type | FOLFOX plus panitumumab |
| CR07 | Liver metastasis | Wild type | No treatment |
| CR08 | Primary tumor | Wild type | No treatment |
| CR09 | Liver metastasis | Wild type | FOLFOX plus cetuximab |
| CR10 | Liver metastasis | Unknown | No treatment |
| CR11 | Liver metastasis | Wild type | FOLFOX plus cetuximab |
| CR12 | Liver metastasis | Wild type | FOLFIRI plus panitumumab |
| CR13 | Liver metastasis | Mutant | No treatment |
| CR14 | Liver metastasis | Mutant | FOLFOX |
| CR15 | Liver metastasis | Mutant | FOLFOX plus bevacizumab |
| CR16 | Primary tumor | Unknown | No treatment |
| CR17 | Liver metastasis | Wild type | FOLFOX |
| CR18 | Liver metastasis | Wild type | FOLFIRI plus cetuximab |
| CR19 | Liver metastasis | Wild type | No treatment |
| CR20 | Liver metastasis | Wild type | FOLFOX plus bevacizumab |
| CR21 | Liver metastasis | Wild type | FOLFIRI plus panitumumab |
| CR22 | Liver metastasis | Mutant | XELOX plus bevacizumab |
| CR23 | Liver metastasis | Mutant | FOLFOXIRI plus bevacizumab |
| CR24 | Liver metastasis | Mutant | FOLFOX |
| CR25 | Liver metastasis | Wild type | No treatment |
| CR26 | Liver metastasis | Mutant | FOLFOX plus bevacizumab |
| CR27 | Liver metastasis | Mutant | XELOX plus bevacizumab |
| CR28 | Liver metastasis | Wild type | FOLFIRI plus panitumumab |
| CR29 | Liver metastasis | Mutant | FOLFOX plus bevacizumab |
| CR30 | Liver metastasis | Wild type | Adjuvant XELOX |
| CR31 | Liver metastasis | Wild type | FOLFIRI plus panitumumab |
FOLFOX, 5-fluorouracil plus oxaliplatin; FOLFIRI, 5-fluorouracil plus irinotecan; FOLFOXIRI, 5-fluorouracilplus oxaliplatin plus irinotecan; PDTO, patient-derived tumor organoid.
We complemented the cohort with patients with CRC liver metastases, already treated in first-line setting for mCRC, who obtained a partial response and underwent liver resection, with the aim of demonstrating the predictive value of PDTOs by a direct comparison of the effectiveness of treatment in the clinics with DSA cell viability. We believed this analysis was important to assess the possible impact of the growth factors used to derive the organoids on the drug sensitivity profiles of primary patient tumor cells. DSAs for CR04, CR06, CR09, CR11, CR12, CR14, CR15, CR17, CR18, CR20, CR21, CR22, CR23, CR24, CR26, CR27, CR28, CR29, CR30 and CR31 PDTOs were carried out with a large panel of drugs (Figure 1C). The RAS, BRAF, microsatellite instable status was available for all the patients. All the patients had a microsatellite stable status. Twelve patients had a RAS, BRAF WT mCRC while eight patients had an RAS-mutant tumor. Patients with RAS/BRAF WT tumor (PDTOs: CR04, CR06, CR09, CR11, CR12, CR17, CR18, CR20, CR21, CR28, CR30 and CR31) received as first-line regimen: chemotherapy alone, chemotherapy plus anti-epidermal growth factor receptor (EGFR) and chemotherapy plus anti-vascular endothelial growth factor (VEGF) (Table 1). Patients with RAS/BRAF-mutant tumor (PDTOs: CR14, CR15, CR22, CR23, CR24, CR26, CR27, CR29) received chemotherapy alone or chemotherapy plus anti-VEGF. All the PDTOs derived from patients who were treated with anti-EGFR, achieving a partial response, presented response to anti-EGFR at DSA level too. Of note, while none of the PDTOs derived from patients with RAS/BRAF-mutant tumor showed response to cetuximab treatment in the DSA, 10 out of 12 RAS/BRAF WT PDTOs showed a significant reduction of cell viability at the DSA assay with cetuximab (Fisher’s exact test: P = 0.001) (Figure 2C). These data confirm previous findings of concordance between treatment activity in patients and in PDTO models, demonstrating how PDTOs maintain tumor molecular characteristics and could therefore represent a useful model to predict response to treatment. We also investigated the correlation between clinical and DSA response to the combination of chemotherapies used in first-line treatment, alone or in combination with anti-EGFR or anti-VEGF monoclonal antibodies (Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2023.101198).
Once a direct correlation of drug sensitivity between patients’ treatment and PDTO DSA was established, we speculated if PDTOs could produce clear data to guide novel strategies in the case of patients with chemorefractory mCRC, which we speculated may be the main applicability of our approach. To this end, we recruited in the second cohort of our study patients with refractory mCRC who had received at least two lines of treatment. A total of nine PDTOs were generated. Seven out of nine PDTOs were derived from liver metastasis biopsies, one out of nine from a lateral cervical lymph node biopsy and one out of nine from a rectal local recurrence biopsy (Table 2 and Figure 1B). DSAs with drugs shown in Figure 1D were conducted for all the nine PDTOs. The PDTO establishment rate of this cohort was ∼60% (5/9). For five out of nine patients an additional biopsy core was used for the genotyping. In four cases, DSA/next-generation sequencing (NGS) data resulted applicable in the clinic. Particularly, two patients with RAS-mutant refractory mCRC were treated with FOLFOX–bevacizumab ‘rechallenge’ and with mitomycin–capecitabine in third line, respectively, based on DSA results, obtaining a stabilization of disease (SD) lasting 6 and 5 months (Figure 3). One patient was treated in a phase I trial with nivolumab plus second mitochondrial-derived activator of caspases mimetic due to the presence of high tumor mutational burden at NGS, experiencing SD for 6 months; unfortunately, we could not predict this outcome in the DSA since the recapitulation of the immune microenvironment was out of reach for our methodology. For one patient the presence of a tumor BRCA2 mutation correlated with DSA sensitivity to olaparib in the PDTO model (Table 2 and Figure 3). Since olaparib was not available as a therapeutic option for the patient, we decided to begin a third-line treatment with mitomycin C plus 5-FU combination based on the mutagenic mechanism of action of mitomycin C. The patient achieved a remarkable disease control of 7 months with SD as best response. Unfortunately, after progression of disease, the patient’s clinical conditions worsened and treatment with an off-label use of olaparib was not possible.
Table 2.
Second cohort PDTOs
| PDTO ID | Primary tumor or liver metastasis | RAS mutational status | Genomic profile (VHIO panel 300) | DSA/NGS result-based treatment decision |
|---|---|---|---|---|
| CR32 | Liver metastasis | Mutant | Not carried out | NA |
| CR33 | Primary tumor | Mutant | KRAS EXON 2 G13D, AMER1, PMS1, SOX9 | Mitomycin C plus 5-FU |
| CR34 | Liver metastasis | Mutant | Not carried out | NA |
| CR35 | Liver metastasis | Wild type | APC, ARID1B exon 16 AND ARID1B exon 20, TP53, EGFR, mTOR, BRCA1, CHD4, IRF2, TAF1, PI3KC2G, DICER1 | Nivolumab + SMAC mimetic (phase I trial) |
| CR36 | Liver metastasis | Not carried out | NA | |
| CR37 | Node metastasis | Wild type | APC, ATRX, HGF,BRCA2, NOTCH3, ROS1, TP53 | Olaparib off label |
| CR38 | Liver metastasis | Mutant | APC, ARIDB1, KRAS EXON 2 G12V, TP53 | FOLFOX plus bevacizumab |
| CR39 | Liver metastasis | Wild type | APC, TP53 | NA |
| CR40 | Liver metastasis | Mutant | Not carried out | NA |
5-FU, 5-fluorouracil; DSA, drug screening assay; NA, not available; NGS, next-generation sequencing; PDTO, patient-derived tumor organoid; SMAC, second mitochondrial-derived activator of caspases.
Figure 3.
Correlation of drug screening luminescence assay and tumor genomic profile of PDTO CR33, CR37 and CR38. 5-FU, 5-fluorouracil; PDTO, patient-derived tumor organoid.
Discussion
Clinical outcomes of chemorefractory mCRC are poor due to the fast development of therapy resistance. Organoids have been investigated due to their capacity to form mini-organs that could recapitulate the tissue of origin. A large body of literature has been dedicated to the use of these experimental models to better study several diseases, including solid tumors.7,15,16 PDTOs derived from a biopsy or a resected tissue and cultured in vitro represent an unmatched model to better assess drug response and its predictive therapeutic potential represents an innovative manner to guide personalized treatment in cancer.9 We succeeded to establish a fast patient-oriented ‘from bedside to bench and back to the bedside’ methodology that uses PDTOs to inform functional data-based treatment decisions on a patient-by-patient basis. We followed a step-by-step approach aimed at maximizing the reliability of the results and feasibility of the whole working flow by optimizing tumor biopsy and PDTO derivation protocols using PDX samples. Moreover, we were able to obtain the drug screening results in a patient-affordable timely fashion. In fact, between patient biopsy and analysis of data only 3 weeks intercurred, a very short period if we consider that the generation of PDX models that could give specific molecular information needs at least 3 months. This particularly relevant since chemorefractory mCRC conform a heavy pretreated population carrying relatively high tumor burdens, and therefore long decision-making windows have the risk of facilitating patient deterioration. Finally, we designed an affordable fit-for-purpose mCRC drug sensitivity panel streamlined in order to be able to run with the number of cells that can be obtained from a patient biopsy.
While other studies have used a similar methodology to carry out PDTOs,17, 18, 19 the innovative aspect of our project is the potential clinical applicability of the whole methodology. We developed a screening assay composed of a large panel of drugs concomitantly used in the clinics and investigated in ongoing clinical trials that harbored the potential to guide patient treatment decisions on a real basis. Moreover, when NGS analysis was feasible, matched information between drug screening and molecular profile was able to provide more specific results. We investigated the applicability of the methodology at first on PDTOs derived from patients with mCRC naïve for treatment in the metastatic setting, and then we focused our experiment on patients with liver metastases who received a first-line treatment and then, after obtaining a partial response, underwent surgery. For these patients, we correlated the response obtained in the clinics with DSA results (Figure 2C and Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2023.101198). The discrepancy observed between primary tumor and CRC metastasis DSAs could reflect the biological and molecular intrapatient spatial heterogeneity between primary tumor and distant metastases due to variations in stroma, local tumor cells and microenvironmental factors, as previously reported by different studies.20
Then, we wanted to apply this methodology to those patients affected by refractory mCRC who received at least two treatment lines in the metastatic setting but developed resistance. The DSA results obtained in cohort 1 were reproducible also in cohort 2, even if experiments were carried out in a different laboratory with diverse readout assays for DSAs. Also, in this validation cohort, we were able to match the pharmacological information obtained from DSA with the genotyping of patients’ tissue, to provide more specific information to patients.
However, our study retains some limitations to be considered: (i) the modest number of PDTOs collected in the study and the low derivation rate in cohort 2 of our study (60%). We believe this can be in relation to the heavy pretreated nature of the cohort and the use of tumor biopsies for tissue collection. In contrast, the success rate for cohort 1, when patients were less pretreated and surgical specimens were used, was slightly higher (80%). Unfortunately, in the metastatic setting, few patients can undergo surgery and tissue biopsies represent the unique possibility to generate PDTO models. (ii) The existence of intratumor heterogeneity, which could impair the drug screening results’ applicability. A way to diminish the impact of tumor heterogeneity, at least in relation to tumor genotyping, can be by the incorporation of ctDNA to the tumor genomic screening in our workflow.
In conclusion, even with all the technical limitations, our study demonstrated that PDTOs are easily applicable models to quickly obtain significant data on drug sensitivity and could be a way to maximize clinical outcomes in the refractory setting of selected CRC patients. When integrated with genotyping findings, functional data coming from PDTOs have the potential to elucidate the concrete patterns of drug sensitivity in any single patient and shift CRC treatment to a more integrative vision that involves preclinical and translational studies, to finally achieve a better personalized treatment.
Funding
This Translational Research Fellowship Project was supported by the ESMO with the aid of a grant from Amgen, by the Accelerator (ACRCelerator) [grant number A26825] and Ayuda a médicos jóvenes investigadores from Fundacion Científica—Asociacion Española Contra el Cancer (FC-AECC)/Associazione Italiana per la Ricerca sul Cancro (AIRC)/Cancer Research United Kingdom (CRUK) and by Familia Armangué. Any views, opinions, findings, conclusions or recommendations expressed in this material are those solely of the author(s) and do not necessarily reflect those of ESMO or Amgen. We thank Regione Campania (I-Cure Research Project) [grant number: Cup 21C17000030007], ESMO Translational Research Fellowship Program.
Disclosure
GM: honoraria from Servier and Incyte. DC: travel support from Sanofi and Bristol Myers Squibb (BMS). AA: advisory and speaker for Amgen and Servier. SN: honoraria from Bristol Myers Squibb and Novartis. TT: advisor and speaker for Roche, Merck-Serono, Sanofi, Servier, Novartis, Bayer. CDC: travel grants from Amgen, personal fee for conference/advisory boards Merck, AZ, Roche. EE: honoraria for an advisory role, travel grants and research grants (past 5 years) from Hoffmann-La Roche, Bristol-Myers Squibb, Servier, Amgen, Merck Serono, Array Biopharma, Sanofi and Bayer. JT reports having a personal financial interest in the form of a scientific consultancy role for Array Biopharma, AstraZeneca, Avvinity, Bayer, Boehringer Ingelheim, Chugai, Daiichi Sankyo, F. Hoffmann-La Roche, Genentech, HalioDX, Hutchison MediPharma International, Ikena Oncology, Inspirna, IQVIA, Lilly, Menarini, Merck Serono, Merus, MSD, Mirati, Neophore, Novartis, Ona Therapeutics, Orion Biotechnology, Peptomyc, Pfizer, Pierre Fabre, Samsung Bioepis, Sanofi, Seattle Genetics, Scandion Oncology, Servier, Sotio Biotech, Taiho, Tessa Therapeutics and TheraMyc, as well as educational collaboration with Imedex, Medscape Education, MJH Life Sciences, PeerView Institute for Medical Education and Physicians Education Resource (PER). RD: advisory role for Roche and Boehringer Ingelheim and speaker’s fee from Roche, Boehringer Ingelheim, Ipsen, Amgen, Servier, Sanofi, Libbs and Merck Sharp & Dohme and research grants from Merck and Pierre Fabre. JR: honoraria from Sanofi and travel and accommodation expenses from Amgen, Merck and Sanofi. AV: personal fees from Bayer, Bristol Meyers Squibb, Guardant Health, Merck, Novartis, Roche, Incyte. EM has served as advisor and speaker for AstraZeneca, Amgen, Bayer, Merck Serono, Roche, Sanofi, Servier, Pierre Fabre. FC: institutional research grants from Amgen, Merck KGaA, Merck Sharp & Dohme, Pfizer, Pierre Fabre, Roche and Servier; and service on advisory boards for Bayer, Merck KGaA, Merck Sharp & Dohme, Pierre Fabre, Roche and Servier outside the submitted work. GA: advisory honoraria from Gadetta BV. Amgen, Bayer and Servier. All other authors have declared no conflicts of interest.
Supplementary data
References
- 1.Bertotti A., Papp E., Jones S., et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciardiello F., Ciardiello D., Martini G., et al. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin. 2022;72:372–401. doi: 10.3322/caac.21728. [DOI] [PubMed] [Google Scholar]
- 3.Guinney J., Dienstmann R., Wang X., et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivera M., Fichtner I., Wulf-Goldenberg A., et al. Patient-derived xenograft (PDX) models of colorectal carcinoma (CRC) as a platform for chemosensitivity and biomarker analysis in personalized medicine. Neoplasia. 2021;23:21–35. doi: 10.1016/j.neo.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachs N., Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Junttila M.R., De Sauvage F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 7.Sato T., Stange D.E., Ferrante M., et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 8.Buzzelli J.N., Ouaret D., Brown G., Allen P.D., Muschel R.J. Colorectal cancer liver metastases organoids retain characteristics of original tumor and acquire chemotherapy resistance. Stem Cell Res. 2018;27:109–120. doi: 10.1016/j.scr.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Della Corte C.M., Barra G., Ciaramella V., et al. Antitumor activity of dual blockade of PD-L1 and MEK in NSCLC patients derived three-dimensional spheroid cultures. J Exp Clin Cancer Res. 2019;38:253. doi: 10.1186/s13046-019-1257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van De Wetering M., Francies H.E., Francis J.M., et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roerink S.F., Sasaki N., Lee-Six H., et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556:457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 12.Vlachogiannis G., Hedayat S., Vatsiou A., et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orditura M., Della Corte C.M., Diana A., et al. Three dimensional primary cultures for selecting human breast cancers that are sensitive to the anti-tumor activity of ipatasertib or taselisib in combination with anti-microtubule cytotoxic drugs. Breast. 2018;41:165–171. doi: 10.1016/j.breast.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Riss T.L., Moravec R.A., Niles A.L., et al. Cell viability assays. Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda, MD: 2013. [PubMed] [Google Scholar]
- 15.Sato T., Vries R.G., Snippert H.J., et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 16.Qu J., Kalyani F.S., Liu L., Cheng T., Chen L. Tumor organoids: synergistic applications, current challenges, and future prospects in cancer therapy. Cancer Commun. 2021;41:1331–1353. doi: 10.1002/cac2.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho Y.W., Min D.W., Kim H.P., et al. Patient-derived organoids as a preclinical platform for precision medicine in colorectal cancer. Mol Oncol. 2022;16:2396–2412. doi: 10.1002/1878-0261.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mauri G., Durinikova E., Amatu A., et al. Empowering clinical decision making in oligometastatic colorectal cancer: the potential role of drug screening of patient-derived organoids. JCO Precis Oncol. 2021;5 doi: 10.1200/PO.21.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrino B., Herencia-Ropero A., Llop-Guevara A., et al. Preclinical in vivo validation of the RAD51 test for identification of homologous recombination deficient tumors and patient stratification. Cancer Res. 2022;15:1646–1657. doi: 10.1158/0008-5472.CAN-21-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan D.K.H., Buczacki S.J.A. Tumour heterogeneity and evolutionary dynamics in colorectal cancer. Oncogenesis. 2021;10:53. doi: 10.1038/s41389-021-00342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.