Abstract
Recent far-reaching advances in synthetic biology have yielded exciting tools for the creation of new materials. Conversely, advances in the fundamental understanding of soft-condensed matter, polymers, and biomaterials offer new avenues to extend the reach of synthetic biology. The broad and exciting range of possible applications have significant implications to address grand challenges in health, biotechnology, and sustainability. Despite the potentially transformative impact that lies at the interface of synthetic biology and biomaterials, the two fields have to date progressed mostly separately. This perspective article provides a review of recent key advances in these two fields, and a roadmap for collaboration at the interface between these two communities. We highlight the near-term applications of this interface to the development of hierarchically structured biomaterials, from bioinspired building blocks to “living” materials that sense and respond based on the reciprocal interactions between materials and embedded cells.
The field of biomaterials research has expanded significantly in the past few decades, with a growing interest in the concept of ‘programmability’. A key promise of programmable biomaterials is in their use as a synthetic extracellular matrix (ECM) to direct the fate of cells through tunable properties, encoded using chemical approaches. However, in contrast to natural biomaterials like the native ECM—which is highly dynamic, and continuously modified through reciprocal feedback between cells—many current generation biomaterials are static, and transfer information in one direction (i.e. from the material to the cell).
Synthetic biology has long championed programmability as a central feature. Tunability is achieved by introducing genetic modifications in a systematic way via promoters, inducers, and nucleic acid-modifying enzymes. Recent years have also seen an expansion of synthetic biology tools from their origin in bacterial cells to increasing applications in mammalian cell programming. Meanwhile, a new growing front of research is focusing on the construction of artificial / synthetic cells, which can be programmed to have defined input-output relationships, much like a natural cell. Here, we review the state of the field for both synthetic biology and biomaterials, discuss recent work at this interface, and close with an outlook on future opportunities at the intersection of these areas. The idea for this Perspective was triggered by the stimulating discussions held during the two-day National Science Foundation Square-Table workshop in Alexandria, Virginia in October 2019. Nonetheless, there are numerous distinct research groups, clusters, and centers across the world working at the interface between synthetic biology and engineered materials, and this piece aims to provide a broad overview of the recent achievements in this exciting filed.
New frontiers in synthetic biology
Synthetic biology turned 20 years old in 2020 and represents a research topic with immense potential for transforming life in the 21st century1. Remarkable achievements in DNA synthesis and assembly, along with standardization of genetic components and automated sequence designs, have helped biology evolve into an engineering science. From the early days of building genetic circuits in bacteria, synthetic biology approaches have now extended to mammalian systems, and even to engineering completely synthetic cells from the bottom up.
Advances in cell-base technology
Synthetic biology has revolutionized our capability to program how cells perceive, process, and react to external information (Fig. 1a), and cells can now be engineered to perceive specific inputs, such as the concentration or the kinetics of biological, chemical, or physical stimuli2. Of particular interest are optogenetic tools as they allow optical control of molecular reactions with exquisite spatiotemporal precision, dose-dependency, reversibility, and orthogonality3. The information perceived by such input stimuli can subsequently be processed by synthetic networks inspired by fundamental computational operations and algorithms. First pioneered with computing gene networks4, this information-processing capability has recently been transferred to networks of enzymes composed of kinases or proteases2,5. The capability for information processing ranges from Boolean algebra to CRISPR-based networks that record and genetically store sequential information input6, or control-theory-inspired integral feedback controllers that maintain specific biological setpoints in fluctuating environments7. The concept of cellular information processing can be extended to multicellular systems interconnected by ‘chemical wires’ to increase, in a modular fashion, the complexity of computational operations8. Finally, the outcome of such information processing can be translated into desired cellular outputs like cell growth and differentiation, or the synthesis of a product ranging from simple proteins to complex, hitherto unavailable small molecule drugs9.
Figure 1|. Programmability of synthetic biology.
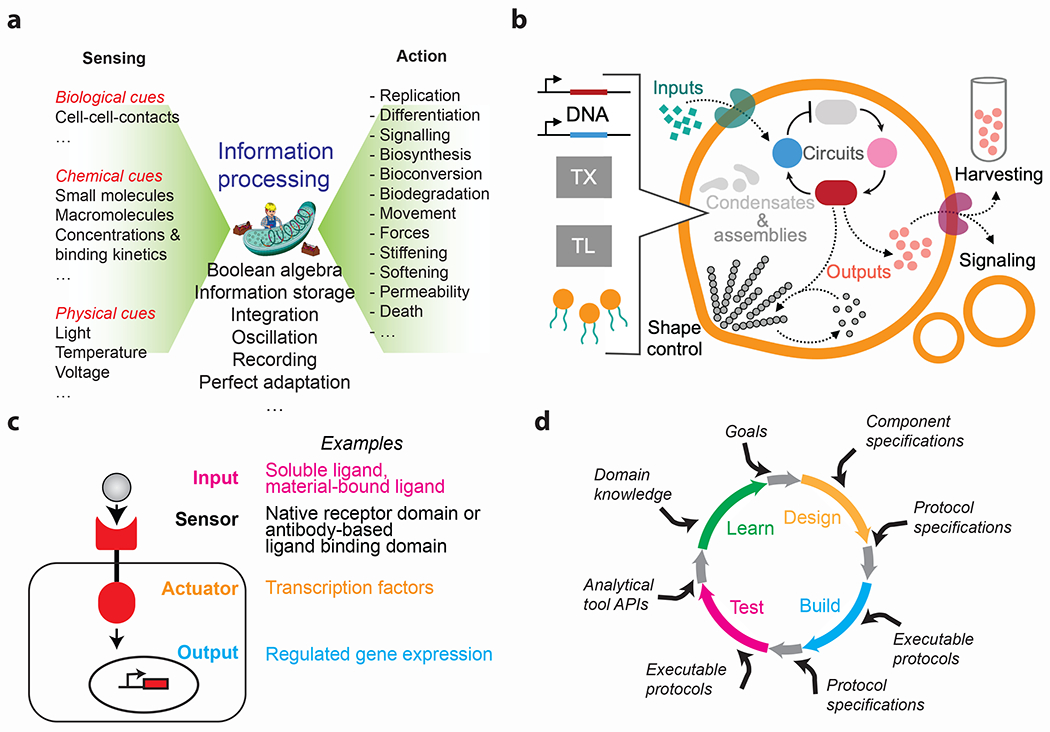
a, The heart of synthetic biology rests on our capability to program cells as information processing units for sensing a variety of inputs and produce discrete actionable outputs. Advances in information processing have been inspired by computational operations and algorithms and more recently propelled by the use of CRISPR-based networks for information recording. b, Artificial / Synthetic cells made from basic biological building blocks and incorporating transcription (TX) and translation (TL) machineries can recapitulate salient features of living cells such as basic chemical signaling, transcriptional dynamics, intracellular organization, and cytoskeleton organization. c, Recent advances in mammalian synthetic biology are focused on engineering receptors to allow customized sensing and response behaviors. This leverages the coupling of a desired input to a transcription factor to alter gene expression. d, Opportunities for automation in the design-build-test-learn cycle of synthetic biology. To date, partial autonomy exists for portions of the cycle while bridging gaps in the connections and curation of machine-interpretable information are emerging. Supplying goals and interpreting the results based on domain knowledge is an important function of the researcher and will be last to be automated. Figure d is adapted from Ref 79.
Cell-free biology and synthetic cells
Cell-free expression (CFE) has emerged as an expedient technology to build de novo synthetic cells from the ground up10. By recapitulating transcription and translation outside living cells, CFE makes it possible to embed selected pathways and functions into cell-sized compartments, to produce desired molecules in situ or to engineer life-like processes or properties (Fig. 1b). First, CFE provides a minimal environment to rapidly characterize and optimize integrated genetic parts producing biological materials, whose properties may depend on a multitude of interdependent parameters inside living cells11,12. The development of synthetic organelles by spontaneous phase separation in CFE is within reach, with synthetic nucleic acids playing a crucial role13,14. Second, CFE makes it easier to engineer the kinetics of reactions and facilitates the control of biomaterial production across many physical scales10, enabling the synthesis of adaptive, self-regulating materials15. Third, CFE is amenable to being embedded into synthetic or composite substrates and biotic-abiotic interfaces16. Altogether, these advantages are spurring the development of CFE-based synthetic cells with the capability for chemical communication that may one day lead to the development of multifunctional biofilms, proto-tissues, and organoids17.
Synthetic receptors with novel functionalities
Biological systems, comprised of cells and materials that interact with each other, give rise to a smart, complex system. Cells read, or sense, their surroundings using membrane-spanning receptors and can modulate their behavior in response to changing environments. Recently, synthetic receptors have been developed for programming this crucial behavior18,19. Both the external information sensed (the ‘input’) and the corresponding change in behavior (the ‘output’) can be user-defined (Fig. 1c). For example, the extracellular domain can be a single-chain antibody that recognizes an exogenous protein, while the intracellular, effector domain can be a synthetic transcription factor. Many of these receptors have been successfully used for improving cell therapy for cancer20 and for programming developmental transitions both in vitro and in vivo8,21. Intriguingly, some of these receptors have proven to be responsive to ligands presented by materials22, paving the way for programmed interaction between cells and materials.
Application of machine learning and lab automation to synthetic biology
Synthetic biology is on the verge of realizing enormous gains in productivity as each step in the “design, build, test, and learn” engineering cycle benefits from automation (Fig. 1d). Abstraction tools, used in electronic circuit design, have also facilitated designs in synthetic biology. For example, Cello was created as a design environment that translates genetic circuits into DNA sequences23. In the lab, mobile robotic workstations can be integrated into workflows with existing equipment. More importantly, in contrast to the overworked (and overcaffeinated) PhD student, the robot can work flawlessly around the clock with perfect reproducibility, freeing the student to focus on more creative work24. Microfluidic approaches (aided by design tools like 3DμF25) provide further gains in efficiency by scaling down the required reagents and experimental footprint. Furthermore, the automated workflow can be harnessed by machine learning algorithms for discovery and optimization, e.g. enabling genotype-to-phenotype predictions for optimizing tryptophan production in Saccharomyces cerevisiae26. While the application of machine learning algorithms and artificial intelligence might sound ‘non-living’, we emphasize the critical role that human/researcher intelligence plays in setting up the parameter space and protocols for such workflows and drawing broader and deeper insights from the results. Lastly, information standards, such graphical notation used in maps of biological processes and machine-readable representations of biochemical models27, facilitate communication between humans and machines, or between independent labs.
New frontiers in biomaterials
Biomaterials include diverse materials, ranging from titanium or silicone implants, metal stents, and plastic drug delivery devices all the way to hydrogels, which are water-swollen polymer networks. In this section, we focus on hydrogels due to their conduciveness to cell-biomaterial interactions in 3D, and the recent explosion of interest in the field. Hydrogels can be formed from fully synthetic polymers, biopolymers derived from natural sources, ECM proteins or other polypeptides, and even nucleic acids. Key traditional applications of biomaterials include drug delivery, tissue engineering and regenerative medicine, and cell culture models that better recapitulate tissue microenvironments. Tremendous effort has gone into designing biomaterials to better capture the complex features of natural ECMs and to produce desired outcomes in these applications, such as: spatiotemporally controlled drug release kinetics, stem cell differentiation, enhanced wound healing, and immune system reprogramming to target cancer28.
Control over intrinsic biomaterial characteristics
At the molecular level, peptide engineering and new chemistries such as bio-orthogonal or photochemical reactions have been applied to achieve more precise control of mechanical and biochemical properties of biomaterials29 (Fig. 2a). Natural ECMs and living tissues are not simple linearly elastic materials, but rather nonlinear viscoelastic systems that undergo irreversible or plastic deformations, motivating the recent development of systems that recapitulate these more complex mechanical behaviours30. For example, viscoelasticity can be achieved by incorporating dynamic or reversible crosslinking interactions such as metal-ligand coordination, host-guest bonds, hydrogen bonds, electrostatic or hydrophobic interactions, and dynamic covalent bonds, in contrast to static covalent bonds that typically result in elastic materials30. These crosslinking methods can be combined with a double network approach to provide additional control of the mechanical behavior of hydrogels, or the signals presented in cellular and biomedical applications31. Changes in stiffness and viscoelasticity impact fundamental cell processes, including cell proliferation, apoptosis, migration, and differentiation, making these parameters critical to control in any application involving cells30.
Figure 2|. The design space for biomaterials.
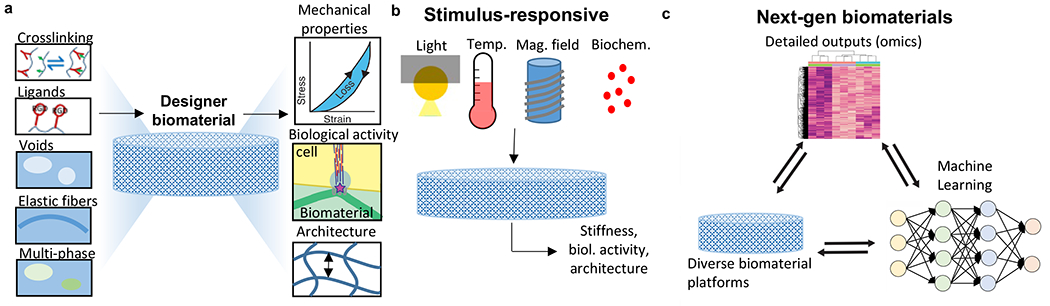
a, Current design paradigm involves selection of biomaterials, crosslinking type, cell-adhesion ligand type and density, specification of void space, inclusion of elastic fibers, and multi-phase materials in order to control mechanical properties (stiffness, viscoelasticity, nonlinear elasticity, plasticity), biological signaling, and micro-scale architecture of the resulting biomaterial. b, Recent advances in biomaterials where stimuli such as light, temperature, magnetic fields, and biomechanical signals can induce changes in the properties of the biomaterial. c, The combination of high-throughput biomaterials production and omics-type measurements of biomaterial properties with the application of machine learning to identify design rules, may pave the way towards next generation biomaterials. Heatmap in Figure 2c is from Ref 80.
Material engineering at the micrometer (or greater) length scale can also impart mechanical properties critical to cell and tissue functions. Micron-scale elastic fibers comprising, or incorporated into, the biomaterial give rise to nonlinear stress-strain responses under tension due to reorientation and alignment32. Complementarily, void spaces in the biomaterial formed through bioprinting, foaming, or microgel annealing provide control over porosity and bulk mechanical properties, which for example might be important for structural stability of the construct, without altering the nanoscale elasticity that cells sense through focal adhesions33. The available palette of tools to control both intrinsic mechanical properties and architectural features will continue to expand with advances in bioprinting multi-component, soft material composites34.
Stimulus-responsive dynamic biomaterials
Over the last decade the state of the art in soft biomaterials transformed from mostly passive, static substrates into dynamic and stimulus-responsive materials that undergo large changes in stiffness, swelling behavior, and 3D structure at the experimentalist’s command, which in turn can elicit specific on-demand responses from embedded cells (Fig. 2b). These systems usually involve the removal or conformational change of crosslinks inside the material, and stimuli include externally applied cues—either physical (e.g. light, temperature, magnetic fields), or chemical (DNA, proteins, small biomolecules)—to provide (spatio)temporal control over the biomaterial properties35. Recent developments highlight hydrogels that show large on-demand stiffness changes36,37 and strategies for the sequential removal and restoration of covalent crosslinks38. Challenges ahead include optimizing response sensitivity (i.e. imparting large changes with small, biologically relevant cues) and the introduction of multiple orthogonal (and reversible) cue responses inside a single biomaterial39. These developments will lay the foundation for incorporating control circuits– used extensively in synthetic biology, but less so in biomaterials– to regulate biomaterial functionality.
Multi-phase biomaterials and their characterization
The human body is full of multi-phase biomaterials, with nearly every tissue containing, at a minimum, cells and a fibrous protein and biopolymer matrix. Such multi-phase biomaterial composites present a challenging target because of their multifunctional and dynamical nature. A recent development toward such systems is the condensation of proteins and other molecules via liquid-liquid phase separation (LLPS)40. LLPS, which has been primarily studied in the context of understanding fundamental cellular organization, has emerged as an intriguing paradigm for creating multi-phase protein biomaterials by tuning their amino acid sequence41. LLPS materials are non-crystalline and stabilized by numerous heterogeneous non-covalent interactions. which can give rise to multiscale structure to the materials. The ability to design protein/polymer systems that selectively co-localize multiple components in a biomolecular condensate is a promising avenue for not only tissue engineering and drug delivery42, but also synthetic biology.
Next generation biomaterials
Further advancement of biomaterials will require both the synthesis of new material compositions and advances in manufacturing, processing, printing, and assembly of material architecture43. 3D bioprinting, multistep lithography and stereolithography, or post-processing for functionalization can each be used to create architectures of responsive or functional material domains. The resulting anisotropic responses of such hierarchical materials will control where the responses occur and can direct complex new responses like shape changes44. A key challenge with some materials, particularly engineered protein-based or DNA-based biomaterials, will be on producing materials at the kilogram scale45, where they can be used broadly. Advances in biomimetic polymers, i.e. protein-like self-assembly46, are a promising alternative for the commercial scale synthesis of shelf-stable materials.
Given the overall complexity of biological systems and the multifarious properties of any given material, the integration of “-omics”-based approaches (e.g. the “materials genome”) with machine learning will help generate materials for diverse applications of interest (Fig. 2c). For example, the combination of high-throughput screening of novel biomaterials with broad yet detailed outputs and machine learning algorithms have enabled the development of antibiofouling biomaterials47 or polymers for pharmaceutical applications48. These approaches can identify materials with unprecedented properties and enable better characterization or engineering of biological systems.
Using synthetic biology to fabricate tailored biomaterials
Although synthetic biology and biomaterials have largely evolved as independent fields, the former has tremendous promise to provide natural or artificially designed modules for the latter. Programmed cells can (1) produce novel building blocks, and/or furnish simplified motifs that can be repurposed by materials scientists to create new biomaterials, or (2) synthesize the material of interest directly.
Innovative bioinspired building blocks can create materials with unique chemical and physical structures to achieve desired functions and properties. For example, coiled-coil peptides, aided by computational design, can be assembled into nanofibers with controllable physical characteristics using click chemistry49. Similarly, DNA nanotechnology has pushed the boundaries of molecularly programmable shape and functionality50. Hybrid systems that combine synthetic DNA with proteins and other biopolymers are enabling building bottom-up hierarchical structures with readily tailored functionalities51.
In a direct application of synthetic biology-based approaches to biomaterials, bacteria can be used to produce engineered variants of natural proteins as components of materials, for applications ranging from engineering of cellular microenvironments to nanowires and self-healing materials52. For example, the SpyTag-SpyCatcher reactive protein partners53 can be incorporated into the bacterial amyloid curli system, leading to tunable functions of the resulting biofilm54. Alternatively, this system can be applied to functionalize the 2D surface-layer proteins of Caulobacter crescentus55, or to engineer the Salmonella microcompartment protein EutM to organize enzymes in a cascade for efficient substrate tunneling56. Engineered proteins can also control biomaterial properties using light-responsive proteins, often directly borrowed from the optogenetic toolbox36, to release tethered enzymes or growth factors from polymeric hydrogels using light57.
The production of useful inorganic biomaterials using synthetic biology is in its infancy but represents an area with great potential, particularly in the context of sustainable building materials. The development of bio-concrete—by precipitating calcium carbonate through a biomineralization process that involves the urease enzyme—is a prime example of using programmed cells to produce a material in situ. Microorganisms genetically engineered for slightly lower urease activity produced larger calcite crystals with high moduli58, demonstrating that the morphology and material properties of biogenic calcite can be tailored by using a synthetic biology approach. Again by employing microbially induced CaCO3 precipitation, it was shown that photosynthetic cyanobacteria could biomineralize inert sand-gelatin scaffolds and significantly toughen the hydrogel matrix59; regulating the cell metabolic activity that in turn allowed for multiple ‘regenerations’ using temperature and humidity switches. Mirroring the rewiring of bacterial metabolic networks for enhanced production of foods, fuels, and other chemicals60, synthetic biology-based rewiring of cellular circuits can be harnessed to further amplify natural biomineralization processes, or potentially generate novel, bio-orthogonal pathways for inorganic materials production.
Many natural biological materials result from the cooperation of multiple cell types. Secreted glucan chains, which are produced by various species of Gram-negative acetic acid bacteria, can become bundled into cellulose fibrils and form a floating mat around the embedded cells. Recently, an approach to fabricate functional bacterial cellulose using a stable co-culture of yeast and cellulose-producing bacteria was demonstrated61. Enzymes secreted by yeast can modify bacterial cellulose, generating autonomously grown catalytic materials with enzymatic functions, illustrating that complex biofabricated materials can be achieved by programming multi-cellular consortia.
The studies highlighted above represent the beginning of the convergence between synthetic biology and biomaterials research. Computational approaches are enabling the design of new building blocks inspired by (but not directly derived from) nature. Engineered peptides and proteins are essential building blocks for modular, bottom-up design of innovative materials with programmable and dynamic functions and structures. Controlling the stability, size, and spatial display of chemical functionalities on these molecules will enable well-defined hierarchical structures that span the nano- and micro- to macro-scales. Applying the concepts of parametric chemistry and tunable hierarchical structuring, a multi-material, 3D printing platform for additive manufacturing of bio-cement has been developed62.
Beyond proteins, polysaccharides or hybrid systems that combine proteins, polysaccharides, DNA, and engineered cells enable the bottom-up assembly of multi-component and hierarchically structured materials systems (Fig 3a). For example, plant-derived starches are already used for the production of biodegradable bioplastics, yet their widespread adoption is limited by a combination of high costs and narrow range of material properties. Since metabolic pathways for many polysaccharides are well characterized, the prospect of controlling features like molecular weight distribution, sugar composition, and branching could lead to bioderived-materials with a wider range of material properties and replace existing petrochemical-derived materials. Polysaccharides can also be produced in much higher yields (>10 g/L) relative to proteins and nucleic acids, by using engineered cells or enzymes. Structural RNA self-assembly has been investigated as a scaffold for intracellular enzyme display63, raising intriguing questions about whether oligonucleotide nanomaterials could be secreted extracellularly by cellular systems. Finally, the growing interest in engineering non-living artificial cells (i.e. protocells) from the bottom-up provides yet another opportunity for creating novel biomaterials and/or providing unique sense-response capabilities.
Figure 3|. Using synthetic biology to fabricate biomaterials with tailored properties toward ‘living’ materials systems.
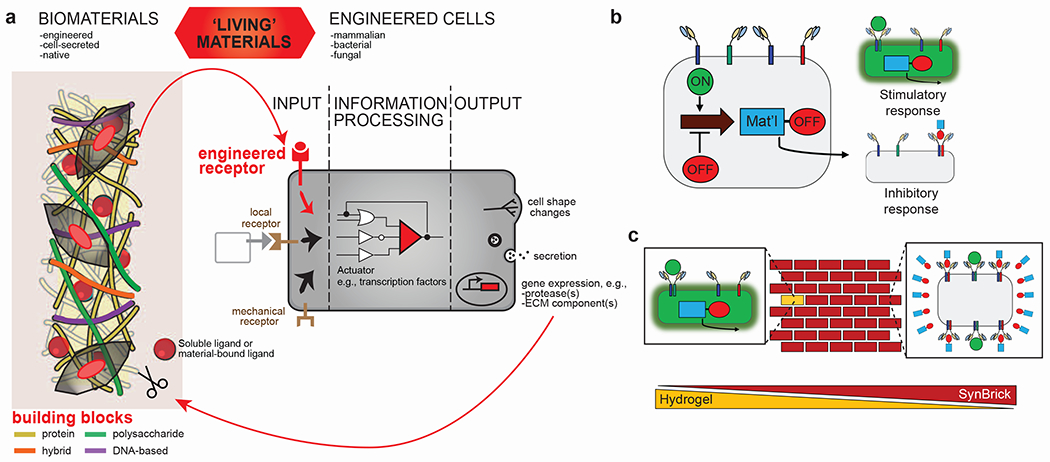
a, Biomaterials inspired, derived, or produced by natural systems, including proteins, polysaccharides, and DNA, are being used in a variety of applications. In particular, cellular systems are being engineered for the production of materials. An exciting direction for merging synthetic biology and biomaterials is to create ‘living’ materials that not only instruct the cells, but where the cells in turn modulate the material properties. b, Schematic of cells programmed with biological circuitry. Stimulatory molecules (green circles) induce stimulatory receptor dimerization which causes the cell to perform a specific task (e.g., fluoresce green and secrete material with an inhibitory molecule). Similarly, inhibitory molecules (synthesized material with a red circle) cause inhibitory receptors to dimerize, signaling the cell to stop fluorescing and producing material. c, Schematic of SynBricks. Cells programmed with stimulatory and inhibitory biological feedback can be encapsulated in sacrificial hydrogel scaffolds with stimulatory molecules, causing resident cells to glow green and replace the hydrogel with calcium carbonate (raw material of bricks) tagged with inhibitory molecules. Once the inhibitory molecule concentration surpasses a threshold, cells will stop fluorescing and producing SynBrick material.
Towards ‘living’ materials
A second exciting direction for merging synthetic biology and biomaterials is to create ‘living’ materials that not only instruct the cells but can in turn be modulated by the cells, to together provide functionality to the material. For example, all living organisms move in some way; in mammals, myocytes within muscle convert chemical energy into reversible mechanical work, in response to electrical and chemical stimuli. Designing living actuators driven by myocyte activity requires materials that define cell shape and orientation, cytoskeletal assembly and organization, and enable communication with excitatory inputs. Beyond repairing or replacing injured muscle tissue, such materials could create soft, biologic robotics. For example, recent work patterned genetically engineered rat cardiomyocytes that contract in response to light to create a biohybrid laser-guided stingray64.
Living organisms also sense and respond to physiological cues, and cells programmed with stimulatory transmembrane receptors can induce numerous responses upon activation, including differentiation and immunomodulation. For example, researchers engineered synthetic Notch receptors that cause cells to undergo an epithelial to mesenchymal transition upon contact from specific sender cell ligands65. In addition, T cells can be engineered with transmembrane receptors that dimerize in the presence of vascular endothelial growth factor and respond by producing a cytokine (IL-2)66. In these systems, ligands only induce a positive response, but cells programmed with stimulatory and inhibitory transmembrane receptors would provide biological feedback useful for engineering living materials. For example, cells embedded in a temporary scaffold with a stimulatory ligand could secrete a matrix that replaces the scaffold with nascent material containing an inhibitory ligand to regulate production (Fig 3b). It was recently shown that cells secrete matrix in biomaterials that they then respond to67,68, and can remodel certain biomaterials through protease activity and mechanical force30, but control over these dynamic interactions is missing. This paradigm has wide applicability ranging from creating regenerative tissues that otherwise have limited healing capacity, to adding lifelike properties to inanimate materials, like bricks that repair themselves (Fig 3c).
Another hallmark of living systems is the processing of raw materials into useful products. Cells within exocrine systems like the pancreas act as factories that assemble individual proteins into large complexes that are excreted from the cell. This natural process could be adapted for bioremediation, where dispersed pollutants are converted into environmentally inert materials. Recently, an implantable bioactive material was developed that degrades into succinate, which can enter the tricarboxylic acid cycle in mitochondria to accelerate bone regeneration69. In the future, cells programmed with engineered metabolic pathways could serve as central processors that support homeostasis and growth of living materials.
An alternate conception of ‘living materials’ is systems that can evolve like a living system, with successive rounds of selection, ‘mutation’ (of material properties), and amplification. Such efforts require high-throughput synthesis and characterization/screening, with machine learning approaches to identify the best candidates for subsequent rounds of ‘evolution.’ Advances in robotic-enabled chemical synthesis and separation provide unprecedented opportunities to rapidly generate polymeric materials. These solution chemistry approaches are complemented by emerging advances in bioprinting (which provide spatial control of material composition70) as well as efforts in bio-templated synthesis71 and engineered evolution, selection, and amplification72. However, the development of property libraries linked to formulation and composition remains a significant challenge73, due to the vast property space of biomaterials, which includes (time-dependent) physical, structural, and biochemical parameters. There is also a need for clever screening/selection schemes (devised by talented, though probably still caffeinated, PhD students freshly liberated by automatic workflows to pursue creative work) that can maintain physical connections between functional material properties and specific genetic variants. There is often a tradeoff between accuracy and speed, requiring a detailed formulation-property analysis on a select subset of systems.
Despite these challenges, several promising platforms have emerged for high throughput analysis, using volumes in the pico- to nanoliter range and measuring changes optically. Polymer microarrays, with the content of each reaction spot encoded by its spatial coordinates, enable screening hundreds or thousands of substrates for supporting the growth of different cell types, such as stem cells or islet cells, or for those minimizing cell adhesion for anti-fouling applications74. Alternatively, cells and other materials can be encapsulated in aqueous droplets suspended in an immiscible oil using microfluidics, where each droplet serves as an independent reactor. Coupling continuous flow capabilities to a high droplet generation rate (>10 kHz) enables, for example, the directed evolution of enzymes from a library of >107 enzyme variants within 10 hours75. Finally, for materials synthesized directly by cells, flow cytometers or fluorescence-activated cell or droplet sorting76 provide powerful interrogation methods. Extending these approaches to measure a wider range of material types and properties is a priority for biomaterials discovery.
High throughput synthesis and emerging advances in characterization provide important inputs to establishing design, build, test, and learn cycles, but advances in data science and high-performance computing infrastructure are also critically important. Standardization of data collection and management are needed, including the wide adoption of uniform naming formats for polymers and composite formulations, and standardizing experimental devices and procedures. Standardized formats are particularly critical for machine learning and algorithmic searching of data sets across numerous laboratories. These are concepts at the heart of the Materials Genome Initiative, launched across multiple federal agencies in the US aimed at “discovering, manufacturing, and deploying advanced materials twice as fast and at a fraction of the cost compared to traditional methods”77. It is also likely that granting and regulatory agencies will play a key role in defining and enforcing these metrics within the field. We further highlight the emergence of various interdisciplinary consortiums and research clusters throughout the world that are interested in similar questions. International cooperation can likely make these developments more rapid and efficient.
Outlook
Synthetic biology and biomaterials research will each continue to flourish as independent fields, but the intersection between the two is poised to become a major focus of research efforts over the near- to mid-term, with tremendous potential for pressing materials challenges. Bridging the gap between laboratory demonstrations and fabrication techniques that can be implemented on larger scales will be increasingly important. Some of the concepts of synthetic biopolymeric materials and cell-based fabrication have already penetrated the commercial arena78, including the production of textiles from microbially-produced recombinant spider silk proteins, tissue-engineered leather and meat, colorless polyimides for electronics, and rigid materials based on fungal mycelium. Continued innovation in this area will enable the large-scale production of synthetic biological materials that rival or surpass existing materials, from plastics to concrete.
We have identified three major areas where we envision that this ‘living interface’ can have the greatest societal impact: medicine, biotechnology, and sustainability. In medicine, near-term goals include the ability for designer cells to serve as drug delivery vehicles with closed-loop control; long-term goals include moonshots like building artificial organs by hierarchical assembly of engineered cells. In biotechnology, we anticipate an impact in accelerated vaccine production and delivery. An even longer-term vision is the non-biologic evolution of materials, especially in conjunction with dynamic, stimulus-responsive, and ‘computational’ biomaterials. Finally, in sustainability, we envision many possibilities ranging from cell-based or synthetic cell-based decomposable materials to completely living building materials that can sense, respond, and regenerate autonomously. Given that nature has an ecosystem with balanced biotic and abiotic factors, the living interface of synthetic biology and biomaterials may accelerate how nature comes up with new biomaterials. We recognize that engineering control/feedback of ‘living’ systems is inherently difficult and will require continuous innovation. We have no doubt that by imparting life-like properties into materials—both in conjunction with cells or mimicking their key properties—scientists will come with many advances that we cannot foresee today, but that will fundamentally transform both basic and applied research across many fields of science.
Acknowledgements
The authors wish to thank all the participants of the second Square Table workshop where the ideas in this perspective article originated from. The workshop was funded by the National Science Foundation (NSF) grant BMAT-1939310. We especially acknowledge Germano Iannacchione for his stewardship of the Square Table workshops. The authors also acknowledge support from the National Institutes of Health grants R01 EB030031 (A.P.L), R35 GM138256 (L.M.), R21 CA232244 (K.A.H.), NSF grants CMMI 1846367 (O.C.), DMR-BMAT CAREER 1753387 (N.S.), EF-1934496 (V.N.), DMR-2004875 (N.S.J.), DMR-2004937 (M.V.), CBET-2033654 (B.M.B), DMR-2037055 (S.L.V.), MCB-2033387 (S.T.), DMR-2004796 (J.M.), DMR-2011824 (A.M.K.), the Human Frontiers in Science Program RGP0045/2018 (S.P.), Department of Energy grants DOE BES DE-SC-0010595 (E.F. and R.S.), DGF grant DFG-EXC-2189 (W.W.), Dutch Ministry of Education, Culture and Science-Gravitation 024.001.035 (P.H.J.K.), and the DARPA Engineered Living Materials Program (P.D.A.).
Footnotes
Competing interests
The authors declare no competing interests
References
- 1.Meng F & Ellis T The second decade of synthetic biology: 2010–2020. Nat. Commun 11, 5174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sedlmayer F, Aubel D & Fussenegger M Synthetic gene circuits for the detection, elimination and prevention of disease. Nat. Biomed. Eng 2, 399–415 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Kolar K, Knobloch C, Stork H, Žnidarič M & Weber W OptoBase: A Web Platform for Molecular Optogenetics. ACS Synth. Biol 7, 1825–1828 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Moon TS, Lou C, Tamsir A, Stanton BC & Voigt CA Genetic programs constructed from layered logic gates in single cells. Nature 491, 249–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao XJ, Chong LS, Kim MS & Elowitz MB Programmable protein circuits in living cells. Science (80-.). 361, 1252–1258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang W & Liu DR Rewritable multi-event analog recording in bacterial and mammalian cells. Science (80-.). 360, eaap8992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aoki SK, Lillacci G, Gupta A, Baumschlager A, Schweingruber D & Khammash M A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 570, 533–537 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Toda S, Blauch LR, Tang SKY, Morsut L & Lim WA Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science (80-.). (2018) doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smanski MJ, Zhou H, Claesen J, Shen B, Fischbach MA & Voigt CA Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol 14, 135–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noireaux V & Liu AP The New Age of Cell-Free Biology. Annu. Rev. Biomed. Eng 22, 51–77 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Godino E, López JN, Zarguit I, Doerr A, Jimenez M, Rivas G & Danelon C Cell-free biogenesis of bacterial division proto-rings that can constrict liposomes. Commun. Biol 3, 539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garenne D, Libchaber A & Noireaux V Membrane molecular crowding enhances MreB polymerization to shape synthetic cells from spheres to rods. Proc. Natl. Acad. Sci. U. S. A 117, 1902–1909 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleh OA, Jeon BJ & Liedl T Enzymatic degradation of liquid droplets of DNA is modulated near the phase boundary. Proc. Natl. Acad. Sci. U. S. A (2020) doi: 10.1073/pnas.2001654117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokolova E, Spruijt E, Hansen MMK, Dubuc E, Groen J, Chokkalingam V, Piruska A, Heus HA & Huck WTS Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc. Natl. Acad. Sci. U. S. A 110, 11692–11697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green LN, Subramanian HKK, Mardanlou V, Kim J, Hariadi RF & Franco E Autonomous dynamic control of DNA nanostructure self-assembly. Nat. Chem 11, 510–520 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Efrat Y, Tayar AM, Daube SS, Levy M & Bar-Ziv RH Electric-Field Manipulation of a Compartmentalized Cell-Free Gene Expression Reaction. ACS Synth. Biol 7, 1829–1833 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Dupin A & Simmel FC Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits. Nat. Chem (2019) doi: 10.1038/s41557-018-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santorelli M, Lam C & Morsut L Synthetic development: building mammalian multicellular structures with artificial genetic programs. Curr. Opin. Biotechnol 59, 130–140 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheller L, Strittmatter T, Fuchs D, Bojar D & Fussenegger M Generalized extracellular molecule sensor platform for programming cellular behavior article. Nat. Chem. Biol 14, 723–729 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Rivière I & Sadelain M Chimeric Antigen Receptors: A Cell and Gene Therapy Perspective. Mol. Ther 25, 1117–1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stapornwongkul KS, de Gennes M, Cocconi L, Salbreux G & Vincent JP Patterning and growth control in vivo by an engineered GFP gradient. Science (80-.). 370, 321–327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Williams JZ, Chang R, Li Z, Burnett CE, Hernandez-Lopez R, Setiady I, Gai E, Patterson DM, Yu W, Roybal KT, Lim WA & Desai TA DNA scaffolds enable efficient and tunable functionalization of biomaterials for immune cell modulation. Nat. Nanotechnol Online ahead of print (2020) doi: 10.1038/s41565-020-00813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen AAK, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D & Voigt CA Genetic circuit design automation. Science (80-.). 352, aac7341 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Burger B, Maffettone PM, Gusev VV, Aitchison CM, Bai Y, Wang X, Li X, Alston BM, Li B, Clowes R, Rankin N, Harris B, Sprick RS & Cooper AI A mobile robotic chemist. Nature 583, 237–241 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Sanka R, Lippai J, Samarasekera D, Nemsick S & Densmore D 3DμF - Interactive Design Environment for Continuous Flow Microfluidic Devices. Sci. Rep 9, 9166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Petersen SD, Radivojevic T, Ramirez A, Pérez-Manríquez A, Abeliuk E, Sánchez BJ, Costello Z, Chen Y, Fero MJ, Martin HG, Nielsen J, Keasling JD & Jensen MK Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism. Nat. Commun. 11, 4880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waltemath D, Golebiewski M, Blinov ML, Gleeson P, Hermjakob H, Hucka M, Inau ET, Keating SM, König M, Krebs O, Malik-Sheriff RS, Nickerson D, Oberortner E, Sauro HM, Schreiber F, Smith L, Stefan MI, Wittig U & Myers CJ The first 10 years of the international coordination network for standards in systems and synthetic biology (COMBINE). J. Integr. Bioinform 17, 20200005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huebsch N & Mooney DJ Inspiration and application in the evolution of biomaterials. Nature 462, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimarco RL & Heilshorn SC Multifunctional materials through modular protein engineering. Adv. Mater 24, 3923–3940 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Chaudhuri O, Cooper-White J, Janmey PA, Mooney DJ & Shenoy VB Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 584, 535–546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodell CB, MacArthur JW, Dorsey SM, Wade RJ, Wang LL, Woo YJ & Burdick JA Shear-thinning supramolecular hydrogels with secondary autonomous covalent crosslinking to modulate viscoelastic properties in vivo. Adv. Funct. Mater 25, 636–644 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, Shenoy VB, Burdick JA & Chen CS Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat. Mater 14, 1262–1268 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin DR, Weaver WM, Scumpia PO, Di Carlo D & Segura T Accelerated wound healing by injectable microporous gel scaffolds assembled from annealed building blocks. Nat. Mater 14, 737–744 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skylar-Scott MA, Mueller J, Visser CW & Lewis JA Voxelated soft matter via multimaterial multinozzle 3D printing. Nature 575, 330–335 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Guo Z, Liu H, Dai W & Lei Y Responsive principles and applications of smart materials in biosensing. Smart Mater. Med 1, 54–65 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hörner M, Raute K, Hummel B, Madl J, Creusen G, Thomas OS, Christen EH, Hotz N, Gübeli RJ, Engesser R, Rebmann B, Lauer J, Rolauffs B, Timmer J, Schamel WWA, Pruszak J, Rümer W, Zurbriggen MD, Friedrich C, Walther A, Minguet S, Sawarkar R & Weber W Phytochrome-Based Extracellular Matrix with Reversibly Tunable Mechanical Properties. Adv. Mater 31, e1806727 (2019). [DOI] [PubMed] [Google Scholar]
- 37.de Almeida P, Jaspers M, Vaessen S, Tagit O, Portale G, Rowan AE & Kouwer PHJ Cytoskeletal stiffening in synthetic hydrogels. Nat. Commun 10, 609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosales AM, Vega SL, DelRio FW, Burdick JA & Anseth KS Hydrogels with Reversible Mechanics to Probe Dynamic Cell Microenvironments. Angew. Chemie - Int. Ed 56, 12132–12136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badeau BA & Deforest CA Programming Stimuli-Responsive Behavior into Biomaterials. Annu. Rev. Biomed. Eng 21, 241–265 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Chao Y & Shum HC Emerging aqueous two-phase systems: From fundamentals of interfaces to biomedical applications. Chem. Soc. Rev 49, 114–142 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Schuster BS, Dignon GL, Tang WS, Kelley FM, Ranganath AK, Jahnke CN, Simpkins AG, Regy RM, Hammer DA, Good MC & Mittal J Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. U. S. A 117, 11421–11431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein IA, Boija A, Afeyan LK, Hawken SW, Fan M, Dall’Agnese A, Oksuz O, Henninger JE, Shrinivas K, Sabari BR, Sagi I, Clark VE, Platt JM, Kar M, McCall PM, Zamudio AV, Manteiga JC, Coffey EL, Li CH, Hannett NM, Guo YE, Decker TM, Lee TI, Zhang T, Weng JK, Taatjes DJ, Chakraborty A, Sharp PA, Chang YT, Hyman AA, Gray NS & Young RA Partitioning of cancer therapeutics in nuclear condensates. Science (80-.). 368, 1386–1392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Champeau M, Heinze DA, Viana TN, de Souza ER, Chinellato AC & Titotto S 4D Printing of Hydrogels: A Review. Adv. Funct. Mater 30, 1910606 (2020). [Google Scholar]
- 44.Cangialosi A, Yoon CK, Liu J, Huang Q, Guo J, Nguyen TD, Gracias DH & Schulman R DNA sequence–directed shape change of photopatterned hydrogels via high-degree swelling. Science (80-.). 357, 1126–1130 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D & Dietz H Biotechnological mass production of DNA origami. Nature 552, (2017). [DOI] [PubMed] [Google Scholar]
- 46.Barbee MH, Wright ZM, Allen BP, Taylor HF, Patteson EF & Knight AS Protein-Mimetic Self-Assembly with Synthetic Macromolecules. Macromolecules 54, (2021). [Google Scholar]
- 47.Chan D, CHien J-C, Axpe E, Blankemeier L, Baker SW, Swaminathan S, Piunova VA, Zubarev DY, Soh HT & Appel EA Combinatorial Polyacrylamide Hydrogels for Preventing Biofouling on Implantable Biosensors. bioRxiv (2020) doi: 10.1101/2020.05.25.115675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Upadhya R, Kosuri S, Tamasi M, Meyer TA, Atta S, Webb MA & Gormley AJ Automation and data-driven design of polymer therapeutics. Adv. Drug Deliv. Rev 171, 1–28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu D, Sinha N, Lee J, Sutherland BP, Halaszynski NI, Tian Y, Caplan J, Zhang HV, Saven JG, Kloxin CJ & Pochan DJ Polymers with controlled assembly and rigidity made with click-functional peptide bundles. Nature 574, 658–662 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Seeman NC & Sleiman HF DNA nanotechnology. Nat. Rev. Mater 3, 17068 (2017). [Google Scholar]
- 51.Buchberger A, Simmons CR, Fahmi NE, Freeman R & Stephanopoulos N Hierarchical Assembly of Nucleic Acid/Coiled-Coil Peptide Nanostructures. J. Am. Chem. Soc 142, 1406–1416 (2020). [DOI] [PubMed] [Google Scholar]
- 52.An B, Wang Y, Jiang X, Ma C, Mimee M, Moser F, Li K, Wang X, Tang TC, Huang Y, Liu Y, Lu TK & Zhong C Programming Living Glue Systems to Perform Autonomous Mechanical Repairs. Matter 3, 2080–2092 (2020). [Google Scholar]
- 53.Keeble AH & Howarth M Power to the protein: Enhancing and combining activities using the Spy toolbox. Chem. Sci 11, 7281–7291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen PQ, Botyanszki Z, Tay PKR & Joshi NS Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun 5, 4945 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Charrier M, Li D, Mann VR, Yun L, Jani S, Rad B, Cohen BE, Ashby PD, Ryan KR & Ajo-Franklin CM Engineering the S-Layer of Caulobacter crescentus as a Foundation for Stable, High-Density, 2D Living Materials. ACS Synth. Biol 8, 181–190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang G, Johnston T, Quin MB & Schmidt-Dannert C Developing a Protein Scaffolding System for Rapid Enzyme Immobilization and Optimization of Enzyme Functions for Biocatalysis. ACS Synth. Biol 8, 1867–1876 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Shadish JA, Strange AC & Deforest CA Genetically Encoded Photocleavable Linkers for Patterned Protein Release from Biomaterials. J. Am. Chem. Soc 141, 15619–15625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heveran CM, Liang L, Nagarajan A, Hubler MH, Gill R, Cameron JC, Cook SM & Srubar WV Engineered Ureolytic Microorganisms Can Tailor the Morphology and Nanomechanical Properties of Microbial-Precipitated Calcium Carbonate. Sci. Rep 9, 14721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heveran CM, Williams SL, Qiu J, Artier J, Hubler MH, Cook SM, Cameron JC & Srubar WV Biomineralization and Successive Regeneration of Engineered Living Building Materials. Matter 2, 481–494 (2020). [Google Scholar]
- 60.Nielsen J & Keasling JD Engineering Cellular Metabolism. Cell 164, 1185–1197 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Gilbert C, Tang TC, Ott W, Dorr BA, Shaw WM, Sun GL, Lu TK & Ellis T Living materials with programmable functionalities grown from engineered microbial co-cultures. Nat. Mater (2021) doi: 10.1038/s41563-020-00857-5. [DOI] [PubMed] [Google Scholar]
- 62.Duro-Royo J, Van Zak J, Tai YJ, Ling AS & Oxman N Parametric chemistry reverse engineering biomaterial composites for additive manufacturing of bio-cement structures across scales, in Challenges for Technology Innovation: An Agenda for the Future (CRC Press, 2017). doi: 10.1201/9781315198101-39. [DOI] [Google Scholar]
- 63.Sachdeva G, Garg A, Godding D, Way JC & Silver PA In vivo co-localization of enzymes on RNA scaffolds increases metabolic production in a geometrically dependent manner. Nucleic Acids Res. 42, 9493–9503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park SJ, Gazzola M, Park KS, Park S, Di Santo V, Blevins EL, Lind JU, Campbell PH, Dauth S, Capulli AK, Pasqualini FS, Ahn S, Cho A, Yuan H, Maoz BM, Vijaykumar R, Choi JW, Deisseroth K, Lauder GV, Mahadevan L & Parker KK Phototactic guidance of a tissue-engineered soft-robotic ray. Science (80-.). 353, 158–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morsut L, Roybal KT, Xiong X, Gordley RM, Coyle SM, Thomson M & Lim WA Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 164, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarz KA, Daringer NM, Dolberg TB & Leonard JN Rewiring human cellular input-output using modular extracellular sensors. Nat. Chem. Biol 13, 202–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loebel C, Mauck RL & Burdick JA Local nascent protein deposition and remodelling guide mesenchymal stromal cell mechanosensing and fate in three-dimensional hydrogels. Nat. Mater 18, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferreira SA, Motwani MS, Faull PA, Seymour AJ, Yu TTL, Enayati M, Taheem DK, Salzlechner C, Haghighi T, Kania EM, Oommen OP, Ahmed T, Loaiza S, Parzych K, Dazzi F, Varghese OP, Festy F, Grigoriadis AE, Auner HW, Snijders AP, Bozec L & Gentleman E Bi-directional cell-pericellular matrix interactions direct stem cell fate. Nat. Commun 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu H, Du Y, St-Pierre JP, Bergholt MS, Autefage H, Wang J, Cai M, Yang G, Stevens MM & Zhang S Bioenergetic-active materials enhance tissue regeneration by modulating cellular metabolic state. Sci. Adv 6, 32232154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li YC, Zhang YS, Akpek A, Shin SR & Khademhosseini A 4D bioprinting: The next-generation technology for biofabrication enabled by stimuli-responsive materials. Biofabrication 9, 012001 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Nam KT, Kim DW, Yoo PJ, Chiang CY, Meethong N, Hammond PT, Chiang YM & Belcher AM Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science (80-.). 312, 885–888 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Kan A & Joshi NS Towards the directed evolution of protein materials. MRS Commun. 9, 441–455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Green ML, Choi CL, Hattrick-Simpers JR, Joshi AM, Takeuchi I, Barron SC, Campo E, Chiang T, Empedocles S, Gregoire JM, Kusne AG, Martin J, Mehta A, Persson K, Trautt Z, Van Duren J & Zakutayev A Fulfilling the promise of the materials genome initiative with high-throughput experimental methodologies. Appl. Phys. Rev 4, 011105 (2017). [Google Scholar]
- 74.Algahtani MS, Scurr DJ, Hook AL, Anderson DG, Langer RS, Burley JC, Alexander MR & Davies MC High throughput screening for biomaterials discovery. J. Control. Release 190, 115–126 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Agresti JJ, Antipov E, Abate AR, Ahn K, Rowat AC, Baret JC, Marquez M, Klibanov AM, Griffiths AD & Weitz DA Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. U. S. A 107, 4004–4009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma F, Chung MT, Yao Y, Nidetz R, Lee LM, Liu AP, Feng Y, Kurabayashi K & Yang GY Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nat. Commun 9, 1030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Niu C, Wang Z, Gan Y, Zhu Y, Sun S & Shen T Machine learning in materials genome initiative: A review. J. Mater. Sci. Technol 57, 113–122 (2020). [Google Scholar]
- 78.Voigt CA Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun 11, 6379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beal J & Rogers M Levels of autonomy in synthetic biology engineering. Mol. Syst. Biol 16, e10019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stowers RS, Shcherbina A, Israeli J, Gruber JJ, Chang J, Nam S, Rabiee A, Teruel MN, Snyder MP, Kundaje A & Chaudhuri O Matrix stiffness induces a tumorigenic phenotype in mammary epithelium through changes in chromatin accessibility. Nat. Biomed. Eng 3, 1009–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


