Abstract
During DNA replication, lesion bypass is an important cellular response to unrepaired damage in the genome. In the yeast Saccharomyces cerevisiae, Rad6 and Rad18 are required for both the error-free and error-prone lesion bypass mechanisms. Furthermore, Rad6–Rad18 interaction is thought to be critical at an early step during lesion bypass in yeast. Two closely related human homologs of yeast Rad6 have been identified as HHR6A and HHR6B. Here, we report a full-length cDNA coding for the human homolog of yeast Rad18. The human RAD18 gene codes for a protein of 484 amino acid residues with a calculated molecular weight of 54 804 Da, and the gene is localized to chromosome 3 between reference intervals D3S3591 and D3S1283. Human RAD18 protein (hRAD18) was found to interact with HHR6A and HHR6B. When co-expressed in yeast cells, stable hRAD18–HHR6A and hRAD18–HHR6B protein complexes were identified and purified to near homogeneity. Thus, through interaction and complex formation with HHR6A and HHR6B, RAD18 protein may play an important role in lesion bypass mechanisms in humans. Consistent with its role as a fundamental lesion bypass protein, the RAD18 gene is ubiquitously expressed in various human tissues.
INTRODUCTION
Many endogenous and exogenous agents frequently damage DNA. DNA lesions are subject to removal by DNA repair pathways. However, DNA repair often cannot remove all lesions from the genome due to limited cellular repair capacity, inefficiently repaired regions of the genome or poorly recognized damage. During DNA replication, unrepaired DNA lesions could pose a serious problem for the replication machinery, since many lesions block the replication polymerases. In response, cells have evolved a sophisticated system called lesion bypass to overcome such replication blockage.
In the yeast Saccharomyces cerevisiae, lesion bypass consists of error-free and error-prone mechanisms, both of which require the RAD6 and RAD18 genes (1,2). Rad6 is a ubiquitin-conjugating enzyme (3) and it forms a complex with Rad18 (4–6). Rad18 is a zinc finger protein with single-stranded DNA binding activity (4–6). It has been proposed that Rad18 may act to load Rad6 onto sites of DNA damage where its ubiquitin-conjugating activity could subsequently modify the stalled replicative DNA polymerase complex (4). This model postulates that Rad6 and Rad18 act at an early step of lesion bypass and their interaction is critical for subsequent processes to bypass a lesion. Supporting this hypothesis, the Rad6–Rad18 interaction is indispensable for the function of Rad18 in UV resistance (5).
The damage-induced mutagenesis pathway is a major mechanism of error-prone lesion bypass. This mutagenesis pathway requires at least the following genes: RAD6, RAD18, REV1, REV3, REV6, REV7 and NGM2 (7–13). Rev1 is a dCMP transferase capable of inserting a dCMP opposite a template apurinic/apyrimidinic (AP) site (14) and it likely plays an additional role in error-prone lesion bypass (D.K.Rajpal, X.Wu and Z.Wang, submitted for publication). Rev3 is the catalytic subunit of DNA polymerase ζ (15) and Rev7 is a small subunit of this polymerase (15). REV6 and NGM2 have not yet been cloned.
It has been suggested that yeast error-free lesion bypass is composed of two mechanisms: the Rad5 mechanism (16) and the Rad30 mechanism (17,18). Very little is known about the Rad5 error-free mechanism. Rad5 is a member of the DNA helicase family, and its ATPase activity has been experimentally confirmed (16). Rad30 was recently identified as DNA polymerase η (19). It is capable of error-free translesion synthesis opposite a template TT dimer (19). Thus, Rad30 functions as an error-free lesion bypass mechanism in response to UV radiation. However, recent biochemical studies with purified yeast Rad30 suggest that this polymerase could also function as an error-prone mechanism in response to certain DNA lesions such as AP sites in DNA (20).
Much less is known about lesion bypass in humans. Two closely related homologs of yeast Rad6 have been identified in humans as HHR6A and HHR6B (21). More recently, human REV1 (22), REV3 (23,24) and REV7 (25) genes have been isolated. Thus, a damage-induced mutagenesis pathway similar to that in yeast is probably operational in humans. Additionally, the human counterpart of yeast Rad30 has been identified as the XPV (xeroderma pigmentosum variant) gene product (26,27). These observations strongly suggest that a lesion bypass system may have been well conserved from yeast to humans in response to unrepaired DNA damage during replication. Since Rad18 is indispensable for both error-free and error-prone lesion bypass in yeast, we asked whether humans possess a counterpart to this protein. In this report, we describe the human RAD18 gene, its chromosomal localization and its ubiquitous expression in various tissues. Furthermore, we show that human RAD18 protein interacts and forms stable complexes with HHR6A and HHR6B.
MATERIALS AND METHODS
Materials
Human EST clones EST182472 (GenBank accession no. AA311754), yd06a07.r1 (GenBank accession no. T80555) and nm33a08.s1 (GenBank accession no. AA572955) were purchased from Research Genetics (Huntsville, AL). Yeast strains used for the expression of human proteins were W303-1B (28) and FF18739 (MATa leu2,3-112 trp1-289 ura3-52 his7-2 lys1-1 rad18::Leu2). The yeast strain used for the two-hybrid assays was Y190 (29). Protease inhibitors, alkaline phosphatase-conjugated anti-mouse IgG, alkaline phosphatase-conjugated anti-rat IgG, 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium were obtained from Sigma (St Louis, MO). Protein G–agarose beads and restriction endonucleases were purchased from Gibco BRL (Gaithersburg, MD). A rat monoclonal anti-HA antibody was from Boehringer Mannheim (Indianapolis, IN). A mouse monoclonal anti-His antibody was from Qiagen (Valencia, CA).
Protease inhibitors
The final concentrations of protease inhibitors used were 1 mM PMSF, 300 µg/ml benzamidine and 1 µg/ml each of antipain, chymostatin, leupeptin and pepstatin. These protease inhibitors were prepared as a 100× mixture in ethanol and stored at –20°C. Benzamidine and PMSF were weighed and dissolved directly in ethanol. Antipain, chymostatin, leupeptin and pepstatin were prepared as 10 mg/ml stock solutions in H2O, DMSO, H2O and methanol, respectively. When needed, protease inhibitors were added to buffers just before use.
Plasmid constructions
To construct plasmids for human RAD18 expression in yeast, the human cDNA clone EST182472 was amplified by PCR with Pfu DNA polymerase (Stratagene, La Jolla, CA) using the primers 5′-CGCGGATCCTCCATGGACTCCCTGGCC-3′ and 5′-GCTCATGACCATGGAAGAGAAGGAGAATGGC-3′. The resulting 1.9 kb human RAD18 cDNA was cleaved with BamHI and XbaI restriction endonucleases and cloned into the corresponding sites of the yeast expression vector pEGLha, generating pEGLha-hRAD18. Expression of pEGLha-hRAD18 yielded human RAD18 protein containing the HA tag at its N-terminus. To construct plasmids for yeast two-hybrid assays, the human RAD18 cDNA was removed from the EST182472 clone by NcoI restriction digestion and then cloned into the NcoI site of the yeast two-hybrid vectors pAS2 and pACT2 (29), generating pAS2-hRAD18 and pACT2-hRAD18, respectively.
To construct a plasmid for HHR6A expression in yeast, the human EST clone yd06a07.r1 was amplified by PCR with Pfu DNA polymerase using the primers 5′-GAAGATCTCCCATGGACATGTCCACCCCGGCTC-3′ and 5′-GAAGATCTAAGCTTCTTTAAACTGTACCCGGGG-3′. The resulting human HHR6A cDNA was cleaved with HindIII and BglII restriction endonucleases and cloned into the HindIII and BamHI sites of the yeast expression vector pEGUh6, generating pEGUh6-HHR6A. Expression of this plasmid yielded 6×His-tagged human HHR6A at its N-terminus. To construct HHR6A plasmids for yeast two-hybrid assays, the amplified HHR6A DNA fragment was cleaved with BglII and NcoI restriction endonucleases and cloned into the BamHI and NcoI sites of the yeast two-hybrid vectors pAS2 and pACT2 (29), generating pAS2-HHR6A and pACT2-HHR6A, respectively. To construct HHR6B plasmids for yeast two-hybrid assays, the human EST clone nm33a08.s1 was amplified by PCR with Pfu DNA polymerase using the primers 5′-CATGCCATGGGATCCATGCATGTCGACCCCGGCC-3′ and 5′-GAAGATCTAAGCTTTGCAGAATCCTTAAAACCTGTGGC-3′. The resulting human HHR6B cDNA was cleaved with NcoI and BglII restriction endonucleases and cloned into the BamHI and NcoI sites of the yeast two-hybrid vectors pAS2 and pACT2 (29), generating pAS2-HHR6B and pACT2-HHR6B, respectively.
Detection of RAD18 gene expression in human tissues
Expression of the RAD18 gene in various human tissues was detected by RT–PCR. Human multiple tissue cDNA panels were purchased from Clontech Laboratories (Palo Alto, CA) and used for PCR. These cDNAs were derived from poly(A)+ mRNA samples from various human tissues with reverse transcriptase and normalized against glyceraldehyde-3-phospate dehydrogenase cDNA by the manufacturer. Two PCR primers, CCACTCTCGACATCCACTTTG and CCTTTTCTGTTTGGTCCTTTG, were used to amplify a 433 bp region of human RAD18 cDNA. PCR reactions (20 µl) contained 0.4 ng cDNA, 5 pmol each of primers, 2 mM MgCl2, 20 mM Tris–HCl, pH 8.0, 50 mM KCl, 200 µM each dATP, dCTP, dGTP and dTTP and 1 U Taq DNA polymerase. After heating at 94°C for 30 s, 35 cycles of amplification were performed according to the following conditions: 20 s denaturation at 94°C, 30 s annealing at 55°C and 45 s extension at 68°C. Reaction products were separated by electrophoresis on a 1% agarose gel containing 0.5 µg/ml ethidium bromide.
Yeast two-hybrid assays for protein interactions
Yeast Y190 cells were transformed with plasmids pAS2-hRAD18, pAS2-HHR6A, pAS2-HHR6B, pACT2-hRAD18, pACT2-HHR6A and pACT2-HHR6B, individually or in combination: pAS2-hRAD18 + pACT2-HHR6A, pAS2-hRAD18 + pACT2-HHR6B, pAS2-HHR6A + pACT2-hRAD18 and pAS2-HHR6B + pACT2-hRAD18. Transformed cells were grown on minimum plates for 4–6 days at 30°C. Colonies from each plate were transferred to a nitrocellulose filter and permeablized by freezing the filters in liquid nitrogen for 10 s followed by thawing at room temperature. The filters with the colony side up were placed on Whatman no. 1 paper presoaked with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl and 1 mM MgSO4) containing 38.6 mM β-mercaptoethanol and 0.5 mg/ml X-Gal. Following incubation at 30°C, cellular β-galactosidase activity was detected by the appearance of blue color in colonies derived from the colorless X-Gal.
Co-immunoprecipitation
Yeast cells containing pEGLha-hRAD18, pEGUh6-HHR6A or pEGUh6-HHR6B individually or pEGLha-hRAD18 + pEGUh6-HHR6A or pEGLha-hRAD18 + pEGUh6-HHR6B were grown in minimum medium containing 2% sucrose at 30°C for 1.5 days. Expression of the human proteins was induced by diluting the culture 10-fold in YPG medium (2% Bacto-peptone, 1% yeast extract and 2% galactose) supplemented with 0.5% sucrose and growing at 30°C for 12 h. Cells were collected by centrifugation and then homogenized with Zirconium beads in a Bead-beater essentially as described before (30) in HNT buffer containing 20 mM HEPES, pH 7.9, 0.1% Triton X-100, 50 mM NaCl, 5 mM EDTA, 10 mM DTT, 10 mM MgSO4 and protease inhibitors. Cell-free extracts were obtained following centrifugation to remove cell debris.
Protein G–agarose beads (10 µl) were washed with 1 ml of HNT buffer three times and then incubated with 200 ng of a rat monoclonal anti-HA antibody at 4°C for 3 h. After washing with 1 ml of HNT buffer four times to remove free antibody, cell-free extracts (1–1.5 mg) containing the human proteins were added to the protein G–antibody complex and incubated at 4°C for 3 h. Proteins bound to the protein G–agarose–anti-HA antibody complex were collected by centrifugation and washed thoroughly with 300 µl HNT buffer six to eight times. Immunoprecipitated proteins were resuspended in buffer (25 mM Tris–HCl, pH 6.8, 0.05% SDS, 71.5 mM β-mercaptoethanol, 5% glycerol and 0.02% bromophenol blue) and separated by electrophoresis on a 15% SDS–polyacrylamide gel (31). After transfer to a nitrocellulose membrane, human RAD18, HHR6A and HHR6B proteins were identified by western blotting (31) using two sequential detection steps. First, the membrane was incubated with a mouse monoclonal anti-His antibody, followed by incubation with alkaline phosphatase-conjugated anti-mouse IgG. His-tagged human HHR6A or HHR6B was visualized after color development by incubating the membrane with 5-bromo-4-chloro-3-indolyl phosphate (0.3 mM) and nitroblue tetrazolium (0.4 mM) in buffer containing 5 mM Tris–HCl, pH 7.5, 17 mM NaCl and 0.05% Tween 20. Then, HA-tagged human RAD18 was detected by incubating the same membrane with a rat monoclonal anti-HA antibody, followed by secondary antibody incubation and color development as before.
Purification of hRAD18–HHR6A and hRAD18–HHR6B protein complexes
Yeast cells harboring pEGLha-hRAD18 and pEGUh6-HHR6A or pEGLha-hRAD18 and pEGUh6-HHR6B were grown in minimum medium containing 2% sucrose for 2 days. Expression of the human proteins was induced by diluting the culture 10-fold in 16 l of YPG medium supplemented with 0.5% sucrose and growing for 15 h at 30°C. Cells were collected by centrifugation and washed with water. After resuspending in an extraction buffer containing 50 mM Tris–HCl, pH 7.5, 600 mM KCl, 10% sucrose, 5 mM β-mercaptoethanol and protease inhibitors, cells were homogenized on ice with Zirconium beads in a Bead-beater for 15 pulses of 30 s each. The clarified extract (∼100 ml) was loaded onto a HiTrap chelating column charged with NiSO4 (2 × 5 ml; Amersham Pharmacia Biotech, Piscataway, NJ). The column was washed with 240 ml of Ni buffer A (50 mM Tris–HCl, pH 7.5, 0.5 M NaCl, 10% glycerol, 5 mM β-mercaptoethanol and protease inhibitors) containing 10 mM imidazole, followed by a wash with 240 ml of Ni buffer A containing 35 mM imidazole. Bound proteins were eluted with a linear gradient (300 ml) of 35–250 mM imidazole in Ni buffer A. The His-tagged HHR6A or HHR6B protein was identified by western blotting using a mouse monoclonal anti-His antibody. The HA-tagged human RAD18 was detected by western blotting using a rat monoclonal anti-HA antibody. The pooled sample was concentrated with PEG 10,000 and desalted through 5 × 5 ml Sephadex G-25 columns in buffer A (50 mM Tris–HCl, pH 7.5, 1 mM EDTA, 10% glycerol, 5 mM β-mercaptoethanol and protease inhibitors) containing 50 mM KCl.
For further purification of the hRAD18–HHR6B complex, the desalted Ni column sample (∼20 ml) was loaded onto a FPLC Mono S HR5/5 column. After washing the column with 10 ml of buffer A, bound proteins were eluted with a 30 ml linear gradient of 50–400 mM KCl in buffer A. The hRAD18–HHR6B protein complex eluted at ∼140 mM KCl. The Mono S sample was desalted as before and loaded onto a FPLC Mono Q HR5/5 column. After washing the column with 10 ml of buffer A, bound proteins were eluted with a 30 ml linear gradient of 50–500 mM KCl in buffer A. The hRAD18–HHR6B protein complex was eluted at ∼350 mM KCl. In addition to western blots, silver staining (31) of SDS–polyacrylamide gels was also used to visualize proteins during purification. The Silver Stain Plus kit from Bio-Rad (Hercules, CA) was used according to the manufacturer’s recommended conditions.
For further purification of the hRAD18–HHR6A complex, the desalted Ni column sample (∼20 ml) was loaded onto a FPLC Mono Q HR5/5 column and eluted as described above. Fractions containing the hRAD18–HHR6A complex (3 ml) were concentrated with PEG 10,000 to 250 µl and loaded onto a FPLC Superdex 200 HR 10/30 gel filtration column (Amersham Pharmacia Biotech) that had been equilibrated in buffer A containing 150 mM KCl. The hRAD18–HHR6A complex was then eluted in the same buffer.
RESULTS
Human RAD18 gene
Using the yeast Rad18 protein sequence, we searched the non-redundant GenBank CDS database for its human homolog. A human EST sequence (GenBank accession no. AA311754) was identified. We then determined the entire nucleotide sequence of this clone. It contains an open reading frame of 1455 bp, a 70 bp 5′ non-coding region and a 1524 bp 3′ non-coding region. An in-frame stop codon lies 24 bp upstream of the putative ATG start codon. The sequence context of this ATG (CGACCATGG) matches well with the Kozak consensus sequence (CCACCATGG) that is commonly found surrounding the mammalian ATG initiator codon (32). Therefore, this clone contains a full-length cDNA coding for a human protein of 484 amino acid residues with a calculated molecular weight of 54 804 Da. Upon searching GenBank with the putative protein sequence, we identified yeast Rad18 as its homolog (PN = 1 × e–13). Hence, we conclude that this cDNA clone codes for a human homolog of the yeast lesion bypass protein Rad18. Accordingly, this gene is referred to as human RAD18 (GenBank accession no. AF169796).
Comparison of the RAD18 proteins between human and yeast showed significant sequence conservation, with 26% identity and 59% similarity (Fig. 1). Furthermore, several structural features of yeast Rad18 are also present in the human protein, including an N-terminal C3HC4 zinc finger motif, also known as the RING finger motif (33), a C2HC zinc finger motif, a basic region near the C-terminus and a C-terminal acidic region (Fig. 1).
Figure 1.
Conserved sequence features of RAD18 from yeast to humans. The human RAD18 (hRAD18, top) and its yeast counterpart (yRAD18, bottom) were aligned. Identical amino acids are indicated by two dots and similar amino acids are indicated by a single dot. The two zinc finger motifs are shown in red and the putative nucleotide binding motif in yeast Rad18 is shown in blue. A positively charged segment and a negatively charged segment in the C-terminal region of human RAD18 are highlighted in green and purple, respectively.
Chromosomal localization of the human RAD18 gene
We searched the non-redundant database of GenBank STS division with the human RAD18 cDNA sequence and identified a human STS (WI-13593). The location of this STS was assigned to 55.8 cR from the top of the chromosome 3 linkage group by Radiation Hybrid Mapping (Whitehead Institute/MIT Center for Genome Research, Cambridge, MA). On GeneMap’98 it was mapped to a physical position between reference intervals D3S3591 and D3S1283. Therefore, we conclude that the human RAD18 gene is located on chromosome 3 between D3S3591 and D3S1283.
Expression of the RAD18 gene in various human tissues
Apparently, lesion bypass mechanisms exist in humans (22–24,26,27). However, it is not known whether hRAD18 functions in various human tissues. As an indication of the importance of the human RAD18 gene in response to DNA damage, we examined its expression in different human tissues. As shown in Figure 2, RAD18 expression was detected by RT–PCR in all of the 16 human tissues examined. Therefore, we conclude that the RAD18 gene is ubiquitously expressed in humans.
Figure 2.
Expression of the RAD18 gene in various human tissues. First strand cDNA was synthesized from poly(A)+ mRNA of various human tissues as indicated and normalized against the constitutive gene glyceraldehyde-3-phosphate dehydrogenase. A 433 bp DNA fragment corresponding to the human RAD18 cDNA was amplified by 35 cycles of PCR. DNA products were separated by electrophoresis on a 1% agarose gel and visualized by ethidium bromide staining. Size markers in bp are shown on the right.
Interaction of human RAD18 protein with HHR6A and HHR6B
Based on the yeast Rad6–Rad18 interaction (4–6), we asked whether human RAD18 protein interacts with the human Rad6 counterparts, HHR6A and HHR6B. This question was initially addressed using the yeast two-hybrid assay. Human RAD18 (hRAD18) was cloned into the vector pAS2, yielding pAS2-hRAD18. HHR6A and HHR6B were cloned into the vector pACT2, yielding pACT2-HHR6A and pACT2-HHR6B, respectively. Expression of pAS2-hRAD18 in yeast cells produced a RAD18 fusion protein containing the Gal4 DNA binding domain. Expression of pACT2-HHR6A and pACT2-HHR6B produced the respective fusion proteins containing the Gal4 activation domain. Interactions between human RAD18 and HHR6A or HHR6B would physically bring the Gal4 DNA binding and activation domains close together to activate Gal4-dependent promoters. One Gal4 target is the β-galactosidase gene, whose activation in yeast is indicated by blue colonies in the presence of X-Gal (29). As shown in Figure 3G and H, yeast cells containing both pAS2-hRAD18 and pACT2-HHR6A or both pAS2-hRAD18 and pACT2-HHR6B activated the β-galactosidase gene as evidenced by the appearance of blue colonies following β-galactosidase filter lift assays. In control experiments using yeast cells transformed with pAS2-hRAD18, pACT2-HHR6A or pACT2-HHR6B alone, the β-galactosidase gene was not activated (Fig. 3A–C).
Figure 3.
Interactions between human RAD18 and HHR6A or HHR6B determined by the yeast two-hybrid assay. The two-hybrid vectors pAS2 and pACT2 contained the GAL4 DNA binding domain and the GAL4 activation domain, respectively. Yeast Y190 cells were transformed with one plasmid or two plasmids as follows: (A) pAS2-hRAD18; (B) pAS2-HHR6A; (C) pAS2-HHR6B; (D) pACT2-hRAD18; (E) pACT2-HHR6A; (F) pACT2-HHR6B; (G) pAS2-hRAD18 + pACT2-HHR6A; (H) pAS2-hRAD18 + pACT2-HHR6B; (I) pAS2-HHR6A + pACT2-hRAD18; (J) pAS2-HHR6B + pACT2-hRAD18. The expression of the two-hybrid target gene, β-galactosidase, was monitored by filter assays using X-Gal as the enzyme substrate. Expression of β-galactosidase turned yeast colonies blue in the presence of X-Gal, indicating physical interactions between the two proteins tested.
Similarly, when human RAD18 was cloned into the pACT2 vector (pACT2-hRAD18) and HHR6A or HHR6B were cloned into the pAS2 vector (pAS2-HHR6A and pAS2-HHR6B, respectively), these plasmids alone did not activate the β-galactosidase gene in yeast cells (Fig. 3D–F). In contrast, plasmids pACT2-hRAD18 and pAS2-HHR6A together or pACT2-hRAD18 and pAS2-HHR6B together activated the β-galactosidase gene (Fig. 3I and J). These results suggest that human RAD18 protein interacts with both HHR6A and HHR6B.
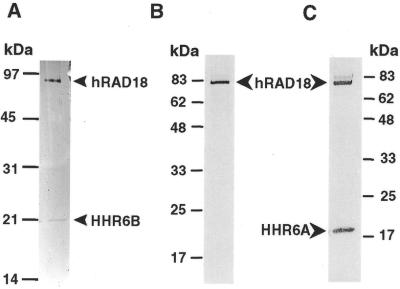
To further test the proposed hRAD18–HHR6B interaction, we performed an immunoprecipitation experiment. First, we tagged human RAD18 with the HA epitope (HA–hRAD18) and HHR6B with 6×His (His–HHR6B) at their N-termini using yeast expression vectors. Then, the tagged human RAD18 and HHR6B were expressed individually or together in yeast cells, which was confirmed by western blot analysis using antibodies specific to the HA or the 6×His tag (Fig. 4, lanes 1–3). Cell-free extracts containing both human RAD18 and HHR6B were subsequently prepared for in vitro immunoprecipitation. As shown in Figure 4 (lane 5), the monoclonal antibody against the HA tag precipitated HA–hRAD18 from the cell extract, as expected. With human RAD18 precipitation, the HA antibody additionally co-precipitated the His-tagged HHR6B (Fig. 4, lane 5). Without HA–hRAD18 in the cell extract, His–HHR6B was not detected following immunoprecipitation with the HA antibody (Fig. 4, lane 4). These results indicate that human RAD18 and HHR6B form a protein complex when co-expressed.
Figure 4.
Co-immunoprecipitation of human RAD18 with HHR6B. Human RAD18 and HHR6B were tagged at their N-termini with HA (HA–hRAD18) and 6×His (His–HHR6B), respectively, and expressed in yeast cells. Yeast cell-free extracts (40 µg) containing HA–hRAD18 (lane 1), His–HHR6B (lane 2) or both (lane 3) were detected by western blot analysis for the presence of these proteins. Extracts (1 mg) containing His–HHR6B (lane 4) or both His–HHR6B and HA–hRAD18 (lane 5) were precipitated with a monoclonal anti-HA antibody as described in Materials and Methods. Protein pellets (anti-HA IP, +) were analyzed by western blot. During western blot analyses, His–HHR6B protein bands on the nitrocellulose membrane were detected first by a monoclonal anti-His antibody. Then, the same membrane was probed again with the anti-HA antibody to detect HA–hRAD18 protein bands. Protein markers are indicated on the left.
The immunoprecipitation experiment was extended to test the proposed hRAD18–HHR6A interaction. HA–hRAD18 and the His-tagged HHR6A (His–HHR6A) were co-expressed in yeast and cell-free extracts were then prepared for immunoprecipitation. As shown in Figure 5 (lane 2), HA–hRAD18 protein was precipitated by the anti-HA monoclonal antibody from extracts containing HA–hRAD18 as the only human protein. Without HA–hRAD18 in the cell extract, His–HHR6A was not detected following immunoprecipitation with the HA antibody (Fig. 5, lane 3). However, in the presence of human RAD18, the HA antibody precipitated both HA–RAD18 and His–HHR6A from the cell extracts (Fig. 5, lane 4). These results indicate that human RAD18 and HHR6A form a protein complex when co-expressed.
Figure 5.

Co-immunoprecipitation of human RAD18 with HHR6A. Human RAD18 and HHR6A were tagged at their N-termini with HA (HA–hRAD18) and 6×His (His–HHR6A), respectively, and expressed in yeast cells. Extracts (1.5 mg) containing HA–hRAD18 (lane 2), His–HHR6A (lane 3) or both His–HHR6A and HA–hRAD18 (lane 4) were precipitated with a monoclonal anti-HA antibody as described in Materials and Methods. Protein pellets were analyzed by western blot. Lane 1 is the immunoprecipitation control without any cell extracts. During western blot analyses, His–HHR6A protein bands on the nitrocellulose membrane were detected first by a monoclonal anti-His antibody. Then, the same membrane was probed again with the anti-HA antibody to detect HA–hRAD18 protein bands. Protein markers are indicated on the left.
Stable protein complexes of hRAD18–HHR6A and hRAD18–HHR6B
To examine whether human RAD18 interactions with HHR6A and HHR6B result in stable protein complexes that can be isolated and studied, we fractionated yeast cell extracts containing the co-expressed human proteins by liquid chromatography. Cell-free extracts containing hRAD18 and HHR6B were sequentially fractionated on Ni–Sepharose, FPLC Mono S and FPLC Mono Q columns. Human RAD18 was co-purified with HHR6B to near homogeneity (Fig. 6A). Cell-free extracts containing hRAD18 and HHR6A were then sequentially fractionated on Ni–Sepharose and FPLC Mono Q columns. During purification we noticed that HHR6A was poorly stained by silver. As a result, the HHR6A protein band was barely detectable in the most pure Mono Q fraction following silver staining (Fig. 6B). The presence of the His-tagged HHR6A in this fraction, however, was clearly revealed by western blot analysis using the specific anti-His antibody (Fig. 6C). Thus, human RAD18 was also co-purified with HHR6A to near homogeneity (Fig. 6B and C). To provide further evidence that the purified hRAD18 and HHR6A were in a complex, we loaded this Mono Q fraction onto a FPLC Superdex 200 gel filtration column. Human RAD18 and HHR6A were identified by western blotting. Again, human RAD18 was co-purified with HHR6A and eluted at a position corresponding to 86–105 kDa. In contrast, pure HHR6A alone eluted at a position corresponding to ∼14 kDa from this gel filtration column under identical chromatographic conditions. Due to the small size of HHR6A and the possibility of non-globular hRAD18 and/or HHR6A, we were unable to estimate a stoichiometry between these two proteins in the complex. These results show that human RAD18 forms a stable protein complex with HHR6A and another stable protein complex with HHR6B.
Figure 6.
Purified hRAD18–HHR6A and hRAD18–HHR6B protein complexes. (A) HHR6B and hRAD18 tagged with 6×His and HA, respectively, were co-expressed in yeast cells (W303-1B) and purified through Ni–Sepharose, FPLC Mono S and FPLC Mono Q columns. A sample (10 µl) from the most pure Mono Q fraction was separated by electrophoresis on a 15% SDS–polyacrylamide gel and visualized by silver staining of the gel. The identities of HHR6B and hRAD18 on the gel (indicated by arrowheads) were confirmed by western blot analysis. (B) HHR6A and hRAD18 tagged with 6×His and HA, respectively, were co-expressed in yeast cells (FF18739) and purified through Ni–Sepharose and FPLC Mono Q columns. A sample (10 µl) from the most pure Mono Q fraction was separated by electrophoresis on a 15% SDS–polyacrylamide gel and visualized by silver staining of the gel. (C) The same Mono Q sample (10 µl) as in (B) was analyzed by western blot using both anti-HA and anti-His monoclonal antibodies. Protein markers are indicated on the sides.
DISCUSSION
We have identified and sequenced a human EST clone coding for full-length RAD18. In addition to sequence homology, several structural features of yeast Rad18 are also conserved in the human protein. However, the Walker type A nucleotide binding motif (GKS) found in yeast Rad18 (8) is not present in the human protein. As a result, we were unable to detect an ATPase activity of the purified human RAD18–HHR6B protein complex. RAD18 homologs have been identified in Schizosaccharomyces pombe (SPBC1734.06, pombe genome sequencing project), Emericella nidulans (uvsH) (34), Neurospora crassa (uvs-2) (35) and mouse (Van der Laan et al., GenBank Accession no. AF205278). We examined protein sequences of these RAD18 homologs and found that none of them contains the GKS Walker type A nucleotide binding motif. Thus, this nucleotide binding motif and weak ATPase activity are most likely unique features restricted to yeast Rad18 protein. This raised the possibility that the ATPase activity of yeast Rad18 may not be important to its lesion bypass function.
Genetic analyses in yeast have indicated the importance of lesion bypass in responding to DNA damage. As demonstrated by yeast rad6 mutants, complete loss of this lesion bypass function results in extreme sensitivity of cells to many DNA damaging agents, such as UV radiation. The effect on cell survival is as dramatic as the loss of nucleotide excision repair (11). Since yeast Rad6 and Rad18 counterparts exist in humans, we strongly believe that lesion bypass mechanisms dependent on RAD18, HHR6A and/or HHR6B are operational in humans. Consistent with the notion that lesion bypass is also a fundamental response to DNA damage in mammals, we observed ubiquitous expression of the RAD18 gene in various human tissues. Different mRNA levels were apparent in some tissue samples as revealed by RT–PCR (Fig. 2). However, due to the commercial nature of these samples, we were unable to rigorously control the mRNA preparations. This limitation prevented us from conducting a more quantitative comparison of the hRAD18 expression levels in different tissues.
In yeast, complex formation between Rad18 and Rad6 is thought to be important during lesion bypass, as mutations that abolish such an interaction lead to a severe cellular sensitivity to UV radiation (5). This feature is likely of functional importance in humans as well. Supporting this notion, we observed interactions between human RAD18 and the two human homologs of Rad6, HHR6A and HHR6B. The interactions result in the formation of stable protein complexes, hRAD18–HHR6A and hRAD18–HHR6B. Since Rad18 and Rad6 are required for both error-free and error-prone lesion bypass in yeast (1,2), it is likely that hRAD18–HHR6A and hRAD18–HHR6B may also participate in both modes of lesion bypass. At present, however, it is not known how these two complexes participate in different lesion bypass mechanisms.
The importance of DNA lesion bypass in human physiology is underscored by the recent discovery that XPV disease is caused by inactivation of the lesion bypass DNA polymerase η (26,27). Clinically, XPV is characterized by photosensitivity and a predisposition to skin cancer (36). Since damage-induced mutagenesis is an important factor in carcinogenesis, the REV3 mutagenesis pathway is also expected to play an important role in human health. Our results in yeast demonstrate that high levels of DNA polymerase ζ encoded by the REV3 and REV7 genes lead to enhanced mutagenesis following UV radiation (D.K.Rajpal, X.Wu and Z.Wang, submitted for publication). Thus, mutations that activate human DNA polymerase ζ are expected to stimulate damage-induced mutagenesis and may consequently lead to a higher risk of human cancers. Based on our understanding of the yeast model system, it is predicted that hRAD18, HHR6A and HHR6B likely play important roles at an early step in the REV3 mutagenesis pathway and the polymerase η lesion bypass mechanism. Isolation of the human RAD18 gene and purification of the hRAD18–HHR6A and hRAD18–HHR6B protein complexes should facilitate future studies on both error-free and error-prone lesion bypass in humans.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Errol Friedberg and I. Fabre for providing us with yeast strains Y190 and FF18739, respectively. We thank Deepak Rajpal for assistance in the yeast two-hybrid assays. We thank Elena Braithwaite, Elizabeth Lin and Deepak Rajpal for critical review of this manuscript. These studies were supported by a THRI grant from the Tobacco and Health Research Institute of the University of Kentucky and a New Investigator Award in Toxicology from the Burroughs Wellcome Fund.
DDBJ/EMBL/GenBank accession no. AF169796
REFERENCES
- 1.Torres-Ramos C.A., Yoder,B.L., Burgers,P.M., Prakash,S. and Prakash,L. (1996) Proc. Natl Acad. Sci. USA, 93, 9676–9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassier-Chauvat C. and Fabre,F. (1991) Mutat. Res., 254, 247–253. [DOI] [PubMed] [Google Scholar]
- 3.Jentsch S., McGrath,J.P. and Varshavsky,A. (1987) Nature, 329, 131–134. [DOI] [PubMed] [Google Scholar]
- 4.Bailly V., Lamb,J., Sung,P., Prakash,S. and Prakash,L. (1994) Genes Dev., 8, 811–820. [DOI] [PubMed] [Google Scholar]
- 5.Bailly V., Prakash,S. and Prakash,L. (1997) Mol. Cell. Biol., 17, 4536–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailly V., Lauder,S., Prakash,S. and Prakash,L. (1997) J. Biol. Chem., 272, 23360–23365. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds P., Weber,S. and Prakash,L. (1985) Proc. Natl Acad. Sci. USA, 82, 168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones J.S., Weber,S. and Prakash,L. (1988) Nucleic Acids Res., 16, 7119–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence C.W., Krauss,B.R. and Christensen,R.B. (1985) Mutat. Res., 150, 211–216. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence C.W., Das,G. and Christensen,R.B. (1985) Mol. Gen. Genet., 200, 80–85. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence C. (1994) Bioessays, 16, 253–258. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence C.W. and Hinkle,D.C. (1996) Cancer Surv., 28, 21–31. [PubMed] [Google Scholar]
- 13.Morrison A., Christensen,R.B., Alley,J., Beck,A.K., Bernstine,E.G., Lemontt,J.F. and Lawrence,C.W. (1989) J. Bacteriol., 171, 5659–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Nature, 382, 729–731. [DOI] [PubMed] [Google Scholar]
- 15.Nelson J.R., Lawrence,C.W. and Hinkle,D.C. (1996) Science, 272, 1646–1649. [DOI] [PubMed] [Google Scholar]
- 16.Johnson R.E., Prakash,S. and Prakash,L. (1994) J. Biol. Chem., 269, 28259–28262. [PubMed] [Google Scholar]
- 17.Roush A.A., Suarez,M., Friedberg,E.C., Radman,M. and Siede,W. (1998) Mol. Gen. Genet., 257, 686–692. [DOI] [PubMed] [Google Scholar]
- 18.McDonald J.P., Levine,A.S. and Woodgate,R. (1997) Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson R.E., Prakash,S. and Prakash,L. (1999) Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 20.Yuan F., Zhang,Y., Rajpal,D.K., Wu,X., Guo,D., Wang,M., Taylor,J.-S. and Wang,Z. (2000) J. Biol. Chem., 275, 8233–8239. [DOI] [PubMed] [Google Scholar]
- 21.Koken M.H.M., Reynolds,P., Jaspers-Dekker,I., Prakash,L., Prakash,S., Bootsma,D. and Hoeijmakers,J.H.J. (1991) Proc. Natl Acad. Sci. USA, 88, 8865–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W., Xin,H., Zhang,Y., Wu,X., Yuan,F. and Wang,Z. (1999) Nucleic Acids Res., 27, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W., Wu,X. and Wang,Z. (1999) Mutat. Res., 433, 89–98. [DOI] [PubMed] [Google Scholar]
- 24.Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakumo Y., Roth,T., Ishii,H., Rasio,D., Numata,S., Croce,C.M. and Fishel,R. (2000) J. Biol. Chem., 275, 4391–4397. [DOI] [PubMed] [Google Scholar]
- 26.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 27.Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999) Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z., Wei,S., Reed,S.H., Wu,X., Svejstrup,J.Q., Feaver,W.J., Kornberg,R.D. and Friedberg,E.C. (1997) Mol. Cell. Biol., 17, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai C. and Elledge,S.J. (1996) Methods Enzymol., 273, 331–347. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z., Wu,X. and Friedberg,E.C. (1995) Methods, 7, 177–186. [Google Scholar]
- 31.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Kozak M. (1987) Nucleic Acids Res., 15, 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saurin A.J., Borden,K.L., Boddy,M.N. and Freemont,P.S. (1996) Trends Biochem. Sci., 21, 208–214. [PubMed] [Google Scholar]
- 34.Yoon J.H., Lee,B.J. and Kang,H.S. (1995) Mol. Gen. Genet., 248, 174–181. [DOI] [PubMed] [Google Scholar]
- 35.Tomita H., Soshi,T. and Inoue,H. (1993) Mol. Gen. Genet., 238, 225–233. [DOI] [PubMed] [Google Scholar]
- 36.Cleaver J.E. and Kraemer,K.H. (1989) In Scriver,C.R., Beaudet,A.L., Sly,W.S. and Valle,D. (eds), The Metabolic Basis of Inherited Disease, 6th Edn. McGraw-Hill, New York, NY.