Abstract
Although the development and clinical application of SARS-CoV-2 vaccines during the COVID-19 pandemic demonstrated unprecedented vaccine success in a short time frame, it also revealed a limitation of current vaccines in their inability to provide broad-spectrum or universal protection against emerging variants. Broad-spectrum vaccines, therefore, remain a dream and challenge for vaccinology. This review will focus on current and future efforts in developing universal vaccines targeting different viruses at the genus and/or family levels, with a special focus on henipaviruses, influenza viruses, and coronaviruses. It is evident that strategies for developing broad-spectrum vaccines will be virus-genus or family specific, and it is almost impossible to adopt a universal approach for different viruses. On the other hand, efforts in developing broad-spectrum neutralizing monoclonal antibodies have been more successful and it is worth considering broad-spectrum antibody-mediated immunization, or “universal antibody vaccine,” as an alternative approach for early intervention for future disease X outbreaks.
Keywords: universal vaccine, broad-spectrum vaccine, passive immunization, ring vaccination, antibody vaccine, disease X
The COVID-19 pandemic demonstrated both the importance and challenges in producing long lasting and broadly protective vaccines. This review discusses the current status and future directions in developing broad-spectrum or universal viral vaccines, focusing on three groups of viruses with the greatest risk of causing the next disease X.
Introduction
The COVID-19 pandemic has heralded a renaissance in vaccinology with unprecedented levels of research activity, from novel antigen design and delivery platforms, preclinical testing in animal models, including non-human primate models and transmission studies, to human clinical trials. There are at least 821 COVID-19 vaccine trials registered in 80 countries, with 92 vaccines in phase 3 efficacy trials and 11 World Health Organization (WHO) emergency use licensed vaccines (as of February 23, 2023, https://covid19.trackvaccines.org/vaccines/). The criteria of COVID-19 vaccine licensure are based on greater than 50% vaccine efficacy against symptomatic infection. This unprecedented research activity and urgency has led to the adoption of new vaccine approaches that may have taken decades, if at all, to reach clinical use, culminating in over 13 billion COVID-19 vaccine doses for 5 billion people being vaccinated at least twice within 3 years of SARS-CoV-2 virus discovery (as of May 7, 2023, https://ourworldindata.org/covid-vaccinations?country=OWID_WRL).
A vaccine is a biological product that can be used to elicit a specific memory immune response mimicking infection that confers protection against infection, disease, or death on subsequent exposure to a pathogen. Vaccines have transformed public health, and second to clean drinking water, they are our greatest intervention against infectious diseases and have been estimated to save millions of lives each year.1 Vaccines provide direct protection of the immunized individual through adaptive memory, establishing B cell and T cell responses. Different vaccine platforms have a distinct fingerprint for triggering different arms of the immune system, with the innate immune response endotype determining antibody titers.2 , 3 A central immune dogma of vaccination is that neutralizing antibodies (nAbs) block infection as a key front-line defense, whereas cytotoxic CD8+ T cells kill infected cells, helper CD4+ T cells coordinate the milieu and formation of memory, whereas non-nAbs engage effector cells; thus, cellular responses and antibody effector functions act after the infection is established.
Despite tremendous progress in vaccine development over the last two centuries, we are still faced with multiple challenges, which have made it difficult or impossible to develop vaccines against some pathogens of significant public health concern. It is harder to vaccinate against pathogens that have antigenic variation, transmitted by vectors or have animal reservoirs, cause chronic or latent infection within the host, employ immune evasion mechanisms, which will require both nAbs and T cells for effective clearance of infection, and due to the presence of multiple serotypes that can lead to antibody-dependent enhancement.
One of the key lessons from COVID-19 vaccine development is the lack of a broad protective “universal” vaccine that is effective against “future” variants. Several strategies are being tried that can induce a broadly reactive immune response by integrating a conserved antigen, such as optimized consensus epitopes,4 sequential immunization of chimeric based antigens,5 or simultaneous priming using mosaic-based vaccines6 , 7 for B cell priming, or using conserved peptides for T cell priming.8 It is difficult, if not impossible, to develop a virus-agnostic strategy for development of universal viral vaccines.
In this review, we will focus our discussion on the development of broad-spectrum vaccines, which target different viruses at the viral genus or family levels, with a special focus on henipaviruses, influenza viruses, and coronaviruses. These viruses are among those with the highest risk of causing diseases and have extensive antigenic variation, animal reservoirs, different host ranges and varying degree of pathogenicity.
Pan-henipavirus universal vaccines
Hendra virus (HeV) was the first bat-borne zoonotic virus to emerge in recent time that caused deadly infections in both human and animals in 1994 in Australia9 and is the prototype virus of the genus Henipavirus family Paramyxoviridae.10 There are four additional members of the genus Henipavirus currently listed by International Committee on Taxonomy of Viruses (ICTV), these are Nipah virus (NiV), Cedar virus (CedV), Ghanaian bat henipavirus (GhV) (also known as Kumasi virus or Ghana virus), and Mojiang virus (MojV). MojV is the only member found in rodents, whereas all other four members use bats as their natural reservoir. HeV and NiV are the only known members of the genus that can cause fatal infections in humans and animals by spillover infection from bats. Recently, a MojV-like virus, the Langya virus (LayV), was found to be associated with human respiratory diseases in China, which most likely originated from shrews.11
Henipaviruses can be divided into two groups. The classical group originates from bats, use ephrin molecules (Eph) as entry receptors, and are best represented by HeV and NiV. The non-classical henipaviruses are from non-bat natural hosts, are unable to use Eph as their entry receptor, and have an ill-defined entry mechanism.12
For HeV and NiV, human or animal sera from natural infections of either virus were shown to be able to efficiently cross-neutralize each other.13 , 14 This laid the foundation for the development of pan-henipavirus vaccines15 and human-neutralizing monoclonal antibodies (mAbs).13 , 16 It remains to be seen whether NiV- or HeV-based vaccines can be effective against other classical henipaviruses, such as CedV and GhV, although the fact that these viruses use similar Eph receptors would suggest a high probability. Although both the glycoprotein (G) and fusion protein (F) have been identified as the target of nAbs, G is the more dominant protective antigen. Multiple vaccine candidates have been developed that target the NiV and HeV G proteins and have been shown to be effective in animal models, which included vaccines based on viral vector delivery, virus-like particle (VLP), messenger RNA (mRNA), and protein subunits (for a detailed review, see Amaya and Broder17). The most advanced vaccine, which has been licensed for use in horses, is a protein subunit vaccine based on the soluble G (sG) protein of HeV (HeVsG).
When vaccines using sG of HeV and NiV were tested in a feline infection model, it was shown that both vaccine candidates were able to provide not only homologous but also heterologous protection against both viruses, with the HeVsG showing equal or better protection against NiV than the NiVsG.18 This initial observation was subsequently confirmed in multiple animal models including non-human primates19 , 20 , 21 , 22 , 23 , 24; thus, HeVsG was chosen to be further developed into a pan-henipavirus vaccine. The HeVsG-based vaccine has since been licensed for immunization of horses, under the trade name Equivac HeV, with great success.23 This represents the first licensed vaccine for any biosafety level 4 agent.
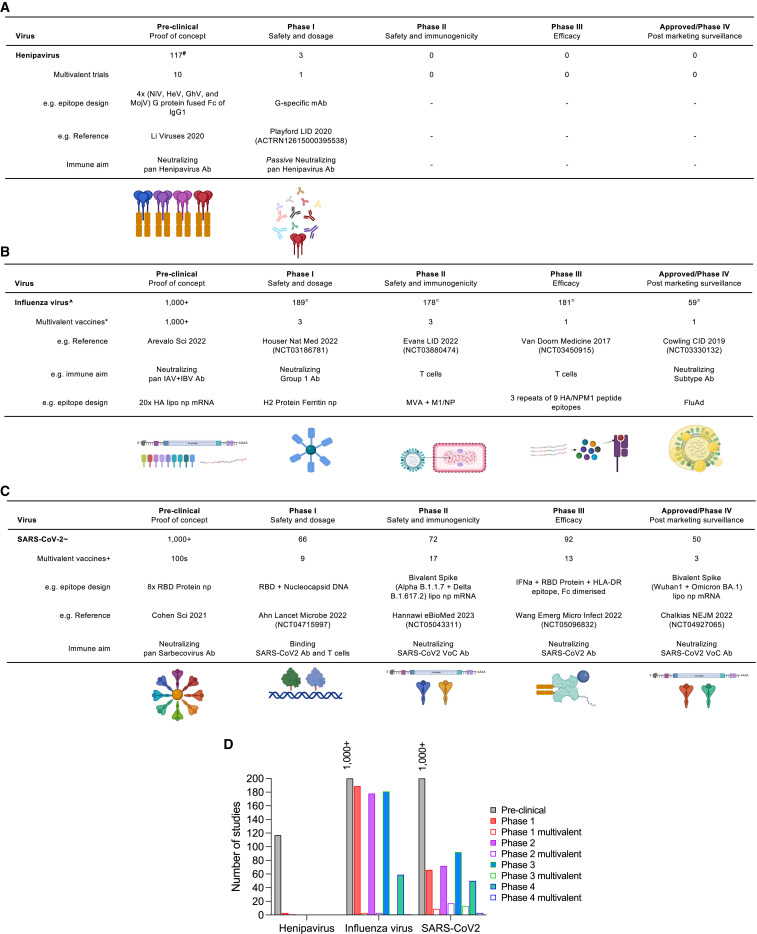
The success of Equivac HeV as a pan-henipavirus vaccine is most likely due to the high structure conservation of the G proteins among different classical henipaviruses and the stringent virus-receptor (EphB2) interaction, which ensures common neutralization targets for universal coverage.12 Currently, there are four NiV vaccines under development with funding from the Coalition for Epidemic Preparedness Innovations (CEPI), which include HeV-sG protein subunit, vesicular stomatitis virus (VSV)-vectored NiV-G, ChAdOx-vectored NiV-G, and measles-virus-vectored NiV-G (https://cepi.net/research_dev/priority-diseases/). Clinical trial of a mRNA NiV vaccine is on-going as well (https://clinicaltrials.gov/ct2/show/NCT05398796). In addition, multivalent henipavirus vaccines are also in pre-clinical development, whereby the G protein of 4 viruses (NiV, HeV, GhV, and MojV) has been fused with Fc region of IgG1 to facilitate presentation and uptake25 to generate neutralizing pan-henipavirus Ab (Figure 1 A). Passive immunization with G-specific mAbs has progressed to phase 1 trials26 (ACTRN12615000395538). Although vaccination against nonclassical henipaviruses will be more difficult until the virus-receptor interaction is known.
Figure 1.
Developmental stages for universal vaccine candidates
(A–C) (A) Clinical development of henipavirus vaccines (A), influenza virus (B), and SARS-CoV-2 (C), (adapted from Biorender).
(A) #Based on PubMed search criteria, “Nipah Hendra or Henipa vaccine, other animals, non-review, cross-reactive.”
(B) ^Based on PubMed search criteria, “universal influenza vaccine human” and filter for clinical trial phase. ∗Based on WHO pandemic influenza vaccine report (October 2022). =Based on Clinicaltrials.gov search criteria, “influenza vaccine.”
(C) ∼Based on covid19.trackvaccines.org (December 2022). +Based on Clinicaltrials.gov search criteria, “variant COVID vaccine” and filter for clinical trial phase.
(D) Multivalent vaccines represent a fraction of registered clinical trials (from A–C).
The pathway to a pan-influenza virus vaccine
Diversity of influenza viruses is not addressed by current vaccines
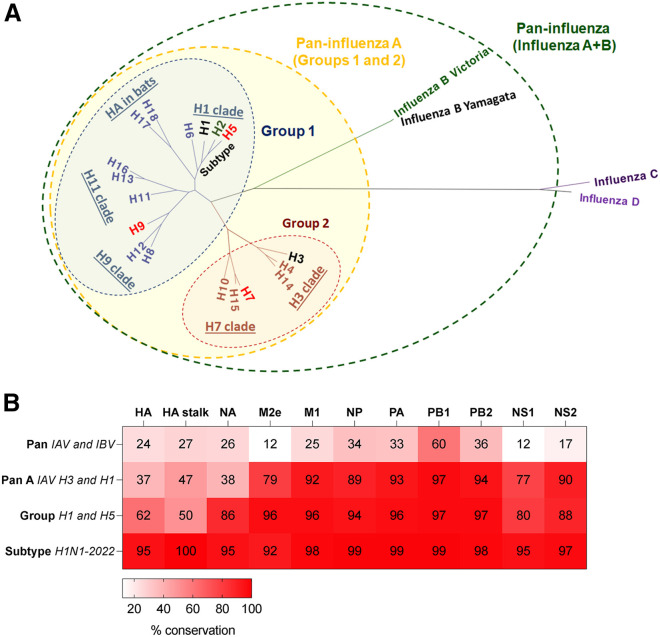
Influenza viruses consist of 4 genera: A, B, C, and D, of which only influenza A and B viruses are endemic in humans (Figure 2 A). Influenza A viruses (IAVs) have the greatest host range, antigenic diversity, and cause seasonal epidemics and pandemics, whereas influenza B viruses (IBVs) cause seasonal epidemics but have limited host range and antigenic diversity and thus reduced pandemic potential. Current inactivated influenza vaccines (IIVs) contain representative IAV and IBV that cause human infections, with limited coverage for zoonotic influenza subtypes.
Figure 2.
Sequence conservation among different influenza virus clades
(A) Definition of influenza breadth based on HA phylogeny. The tree was built with IQ-TREE2 (default parameters with automatic model selection) using HA (HE) protein sequences obtained from the Influenza Virus Database. Human influenza viruses as indicated (black: subtype endemic to humans, green: (H2, B/Yam) previously in humans, red: spill over infections from zoonotic sources).
(B) Amino acid sequence conservation based on sequence alignment (CLUSTALWjp) of proteins (from https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=genomeset) of H1N1 2009 pandemic (A/California/04/2009) versus prototypic virus strains: for Pan (B/Vic, B/Brisbane/60/2008), for Pan A (H3N2, A/Alaska/03/2021), for group 1 (H5N1, A/Indonesia/5/2005), for subtype (H1N1, A/Alabama/01/2020).
IBV is one antigenic subtype with 2 antigenic lineages, Victoria (B/Vic) and Yamagata (B/Yam), which were split from the original B/Lee lineage in the 1980’s, then B/Yam predominated during the 1990’s, B/Vic lineage viruses reappeared in 2001, and they then co-circulated until the COVID-19 pandemic. Due to the public health measures of the COVID-19 pandemic and limited host range of IBV to humans, B/Yam has not been detected since March 2020.27 Elimination of IBV was suggested before the COVID-19 pandemic with the advent of highly effective IBV vaccines due to its limited host range and antigenic diversity. However, B/Yam elimination may be short-lived and continues to be recommended in current quadrivalent vaccines.
The first influenza vaccine was developed in 1936, against IAV, and the standard IIV format in current widespread use remains relatively the same 80 years later,28 but IIV have increased in their valency since. IBV were first isolated in the 1940s and shortly thereafter included in bivalent inactivated influenza vaccination, including one representative IAV and IBV. The IIV strategy was updated again in 1978 after the reintroduction of H1N1 viruses to a trivalent IIV formulation (TIV) containing representative viruses from 2 IAV subtypes, H1N1 and H3N2, and one IBV virus from either B/Vic or B/Yam lineages, and since 2013 quadrivalent (QIV) recommendations have been made to include both representative B/Yam and B/Vic strains. In the past 23 years of WHO vaccine recommendations29 (http://www.influenzacentre.org/Surveillance_Vaccine_Recommendations.html), H3N2 viruses have had 20 strain updates, versus 10 updates for H1N1 viruses, 8 updates for B/Vic, and only 7 strain updates for B/Yam, with the same B/Yam strain used for the past 7 years. Although IBV lineages share 90% hemagglutinin (HA) homology, IBV cross-lineage protection is limited as vaccine efficacy after trivalent vaccination can be just 50% in mismatched seasons.30
Antigenic and genetic characterization of seasonal influenza viruses circulating in humans, with consideration of the latest epidemiological and clinical data from different countries and regions, determine whether there is a need for updating vaccine strain composition. However, because the vaccine composition is decided at least 6 months ahead of influenza epidemic season, there is a risk of mismatch between the vaccine and circulating influenza strains, which occurred recently in 2014/2015 for H3N2 viruses, leading to excess mortality31 and egg adaptations in 2019/2020.32 Greater predictive models from sequence information on immunogenicity, better approaches in capturing latest virus sequence information, and less time-consuming vaccine production pipelines are needed.
Due to the wide variety animal reservoirs from mammals to birds and antigenic variation for IAV, elimination is not possible. IAV has wide antigenic breadth in its surface glycoproteins, resulting in a variety of antigenic subtypes, with at least 18 HA (Figure 2A) and 11 neuraminidase (NA) subtypes. Both HA and NA can be split into distinct phylogenetic groups (e.g., group 1 HA H1/H2/H5/H6/H8/H9/H11/H13/H16/H17/H18 versus group 2 HA H3/H4/H7/H10/H14/H15, and group 1 NA N1/N4/N5/N8, group 2 NA N2/N3/N7/N6/N9, and group 3 NA N10/N11 from bats that lack enzymatic activity) (Figure 2A). Within IAV groups there are further clades, such as the H1 clade, which includes H1/H2/H5/H6, and then each HA subtype, such as H1, which consists of many individual strains. Group 1 and 2 IAV viruses share ∼37% amino acid sequence homology for their HA and NA proteins, and only 24%–27% with IBVs versus group 1 IAV (Figure 2B). Although internal proteins, such as the nucleoprotein (NP), polymerase subunits (PA, PB1, and PB2), and matrix (M), are only 12%–60% conserved between IAV and IBV, they can be 77%–97% conserved across different IAVs of group 1 and 2 (Figure 2B), and within groups and clades homology increases further. Thus, influenza viruses present a challenge, with current IIV only providing short-term, strain-specific protective efficacy that does not even reach different strains within the same subtype.
Heterosubtypic vaccines can be either pan-influenza, covering both IAV and IBV, group, clade, subtype, or strain specific, depending on the breadth of responses elicited and epitope conservation that is targeted by those vaccines (Figure 2A). Indeed, pan-influenza (IAV/IBV) monoclonal nAbs, such as CR9114, which targets the HA-stem, have been identified with protection in lethal heterosubtypic influenza challenge models.33 However, cross-subtype HA-stem antibody responses between H1N1 and H3N2 are also difficult to establish due to differences in HA-stem glycosylation sites, which hide key residues of the H3-stem, and only 47% sequence conservation between these subtypes. Furthermore, pan-influenza (IAV/IBV/ICV)-conserved T cell epitopes, such as HLA-A2-restricted PB1413–421 have been identified; however, these are not immunodominant or protective in mice.34
Although only H1N1 and H3N2 viruses cause seasonal epidemics, spillover IAV infections from avian, swine, equine, and canine sources do occur with limited human to human transmission. H5N1 highly pathogenic avian influenza (HPAI) outbreaks since 1997 instigated greater IAV pandemic preparedness, changes to live animal market practices, and the call for universal influenza vaccines.35 The H7N9 outbreaks in poultry markets of China from 2013 to 2017, were curbed by large-scale poultry bivalent H5/H7 IIV vaccination and, importantly, eliminated human infection with the H7N9 viruses.36 More recently, H5 viruses have reassorted acquiring polymerase genes from H9N2, leading to the greatest number of HPAI ever reported in Europe37 and the USA (https://www.cdc.gov/flu/avianflu/avian-flu-summary.htm) and detections of human cases of H5N6, H9N2, and H3N8 infection (https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20230414.pdf?sfvrsn=5f006f99_113). Infections with IAV from avian origins, H7N9 and H5N1, have followed an epidemiological pattern of age distribution, which has been hypothesized to be attributable to the phenomenon of HA imprinting and conserved epitopes of the HA-stem.38 There is largescale stockpiling of monovalent adjuvanted H5 and H7 vaccines in some countries and preclinical development of broadly reactive vaccines (Figure 1B), as avian IAV remain a very real and imminent threat that we currently have little immune defense against with current seasonal IIV vaccines. Thus, influenza vaccine approaches need to be updated to pre-empt pandemics and be ready for HXNX viruses.
Universal influenza vaccine development
The pathway to a universal influenza vaccine is anticipated to be an iterative process35 and may also require different strategies to immunize individuals with existing immunity compared with naive individuals, such as infants. According to the WHO, there are currently 800 pandemic influenza vaccines in clinical trials, but only 8 are multivalent to provide coverage beyond strain-specific immunity (Figure 1B), and most pandemic preparedness vaccines are taking conservative approaches using a H5 or H7 inactivated monovalent formulation (https://www.who.int/teams/immunization-vaccines-and-biologicals/diseases/seasonal-influenza/tables-on-clinical-evaluation-of-influenza-vaccines).
There are a vast number of universal influenza vaccine strategies being tested in preclinical studies, yet only a handful will reach clinical trials. Many preclinical studies focus on generating broadly neutralizing HA antibodies through mosaic6 , 7 or consensus sequence approaches, such as computationally optimized broadly reactive antigens (COBRAs).4 Recently, a 20-mer HA mRNA lipid nanoparticle vaccine using every IAV and IBV subtype showed protection in mice and ferrets7 via equal breadth of antibodies across all subtypes. It is also possible to use this approach for chimeric HA proteins to be generated, whereby HA trimers may contain subunits of different subtypes, which may augment the ability to generate broadly nAbs.
Novel next-generation vaccine strategies (Figure 1B), such as chimeric HA stem vaccines, NA-based vaccines, peptide mosaic vaccines, and viral-vectored vaccines (such as adenovirus- or vaccinia virus-based vectors), were designed to target conserved IAV regions, such as the NP and M proteins, and generate cross-reactive immune responses by targeting different stages of the virus life cycle. For example, NA antibodies can interfere with virus budding, whereas M2e and NP specific antibodies can interfere with viral fusion and replication. Although T cells are primed after limited replication of the vaccine strains or local antigen presentation. Therefore, T cells and antibodies directed toward the M2e, NA, and NP do not block virus infection but act to reduce the impact of infection and are thus disease mitigating. This non-sterile vaccine-induced immunity may affect virus evolution rates as higher selection pressure is placed on the virus when low-level replication can occur with stringent immune bottlenecks.39
In human clinical influenza vaccine trials (Figure 1B), there are nearly 1,875 active trials (recruiting, not yet recruiting, and completed), whereas 531 trials have been withdrawn, suspended, or terminated (https://clinicaltrials.gov). Various novel strategies are being used in vaccine programs to generate broadly reactive protection against influenza (Figure 1B). For influenza vaccines with multivalent/broad potential, there are 3 recruiting/active phase 1 trials, which include the H2 protein in a ferritin nanoparticle to generate broadly reactive antibodies, especially HA-stem antibodies, as this is a novel subtype in adults under 50 years of age40 (NCT03186781). Among the 3 active phase 2 trials, a T-cell-targeting-based approach uses an MVA vector with NP and M1 proteins to elicit cross-reactive T cells and NP-antibodies; however, this did not show additional protection to seasonal influenza viruses41 (NCT03880474). A phase 3 trial that also aims to elicit cross-reactive T cell responses, using conserved adjuvanted immunogenic peptides (3 repeats of 9 peptides) that bind HLA (human leukocyte antigen) supertypes that recently failed to demonstrate efficacy8 (NCT03450915). There are currently no phase 4 or approved broadly reactive influenza vaccines; however, FluAd, an IIV with MF59 adjuvant, does increase antibody breadth to heterosubtypic influenza viruses (including long drift variants and avian influenza viruses) and provides longer duration antibodies than other available current vaccines.42 , 43 Increased antibody breadth has also been found for other adjuvanted influenza vaccines, such as AS0344 and combinations of TLR agonists.45 Thus, increased use of licensed adjuvanted IIVs may be our current best solution for broader immunity until preclinical universal vaccine candidates are approved.
Vaccines for coronaviruses
Over the last two decades, severe acute respiratory syndrome coronavirus (SARS-CoV, will be labeled following as SARS-CoV-1 to avoid confusion), SARS-CoV-2, and Middle East respiratory syndrome coronavirus (MERS-CoV), have caused epidemics or pandemics in humans, resulting in inordinate human and economic loses worldwide and significantly disrupted global health in an unprecedented manner. The current COVID-19 pandemic had resulted in greater than 765 million confirmed cases and 6.9 million deaths (as of 7 May 2023, https://covid19.who.int/). Clearly, the real number of infection and death is much higher than that because China alone has experienced a massive infection wave from December 2022. Of the seven coronaviruses known to infect humans, five are most likely from bats,46 including SARS-CoV-1, SARS-CoV-2, and MERS-CoV.47 , 48 , 49 , 50 , 51 The unique immune status of bats allows them to harbor a significantly higher proportion of zoonotic viruses than all other mammals.52 , 53 Extensive land-use, intensive farming, and climate changes are key contributing factors for the increasing spillover of zoonotic viruses into the human population54 , 55 , 56; and spillover of bat coronaviruses in Southeast Asia are happening more frequently than previously recognized.57 , 58 The devastating impact of viral pandemics as evidenced by COVID-19 clearly demonstrates that our current preparedness strategy and effort is not sufficient against disease X. Vaccination remains the most cost-effective approach to combat infectious disease outbreaks, but the rapid evolution of SARS-CoV-2 taught us a lesson that the current virus- or strain-specific vaccination strategies are not good enough, and we need to develop more broad-spectrum vaccines preemptively. In this section, we will discuss the past, current, and future coronavirus vaccines and the effort toward making a universal “dream” coronavirus vaccine.
The past: Effort of coronavirus vaccine development before the COVID-19 pandemic
Although seasonal CoVs (especially 229E and OC43) have been associated with human infection for a long time, they do not normally cause severe disease in humans, and the infection is largely self-limiting. On the other hand, human infection with SARS-CoV-1 or MERS-CoV not only results in severe disease or death, but in survivors, it also induces humoral and cellular immunity, lasting for months to decades.59 , 60 , 61 , 62 , 63 The majority of the antibodies elicited during infection target the spike (S) and nucleocapsid proteins, and the nAbs are mainly targeted at the S-receptor-binding domain (RBD) with some minor involvement of epitopes in the N-terminal domain (NTD) and the S2 domains, respectively.64 , 65 , 66
Before the COVID-19 pandemic, no vaccines against coronaviruses had ever been licensed for human use. Research into vaccines against seasonal coronaviruses was considered a low priority, and the circulation of four different viruses also meant the need to develop a quadrivalent vaccine to be truly effective against all four circulating seasonal coronaviruses. In contrast, there were multiple SARS-CoV-1 and MERS-CoV vaccine candidates developed, and some of them went through pre-clinical evaluation, but none reached to the stage of regulatory (e.g., FDA) approval for clinical application.67 Efforts into SARS vaccine development was largely halted after the elimination of human infection in 2003/2004, with no reemergence detected thereafter, despite the fact that multiple SARS-related coronaviruses were detected in bats, and some were shown to be able to use human ACE2 as entry receptor.47 , 48 , 68 Most of the vaccine candidates are based on the S protein in different forms, including trimeric whole S protein, the S1 domain, or the RBD.69 , 70 There are also multiple vaccine delivery platforms used, including protein subunit, VLPs, viral vector, and DNA vaccines. In addition, there were also vaccine candidates based on whole inactivated or live-attenuated SARS-CoV-1, which were experimented in various trials. Most of the vaccine candidates induced high levels of nAbs.71
Although none of the vaccine candidates for SARS-CoV-1 or MERS-CoV reached clinical application, the vast knowledge accumulated from the various research activities built a solid foundation for the development of an effective vaccine for emerging zoonotic coronaviruses. One of the most notable achievements was the demonstration of significant enhancement of immunogenicity by locking the spike protein in a pre-fusion conformation. This was first demonstrated by substituting the residues in the loop between first heptad repeat (HR1) and the central helix with two proline (P) for the MERS-CoV S protein, termed the S-2P construct.72 The same S-2P approach later proved to be a great success in the rapid development of effective SARS-CoV-2 vaccines, including the two most used mRNA COVID-19 vaccines.
The present: Current coronavirus vaccines focusing on SARS-CoV-2
The emergence of SARS-CoV-2 in December 2019 provided new opportunities for demonstration of the importance of rapid vaccine development and deployment. The success has undoubtedly saved millions of lives.73 In this section, we will discuss the vaccine type and protective efficacy of the currently developed SARS-CoV-2 vaccines (summarized in Table 1 ). To date, there are 11 vaccines that have been granted emergency use listing (EUL) by the WHO. It should be emphasized that these vaccines, or the first-generation SARS-CoV-2 vaccines, were all based on the ancestral Wuhan virus strain. Four major vaccine types have been rolled out, including whole inactivated viruses (CoronaVac, Sinovac; Covilo, Sinopharm; and Covaxin, Bharat Biotech), mRNA (Spikevax-mRNA-1273, Moderna; and Comirnaty-BNT162b2, Pfizer-BioNTech), adenovirus vectored (Vaxzevria and Covishield ChAdOx1, AstraZeneca; and Ad26.COV.2, Johnson & Johnson-Janssen), and subunit protein (Nuvaxovid and Covovax NVX-CoV2373, Novavax). Before the emergence of major immune escaping variants and subvariants in the Omicron lineage, randomized, placebo-controlled phase 3 clinical trials showed protective efficacy of 94%–95% against symptomatic COVID-19 infection with two doses of BNT162b2, mRNA-1273, or Ad26.COV.2 vaccine.74 , 75 , 76 , 77 Single dose of adenovirus-vector-based vaccine, Ad26.COV.2, provided 52.9% protective efficacy against moderate to severe-critical COVID-19 infection, and the efficacies varied depending on types of variants.78 Two doses of whole inactivated vaccine, CoronaVac, provided up to 66% protective efficacy against symptomatic infection and about 86% in preventing COVID-19 related deaths.79
Table 1.
SARS-CoV-2 vaccines granted emergency use listing by WHO
| Vaccine | Manufacturer | Platform | Efficacy on preventing symptomatic infection | Efficacy on preventing severe disease | |
|---|---|---|---|---|---|
| 1 | mRNA-127380 | Moderna | mRNA | 93.2% | 98.2% |
| 2 | BNT162b281 | Pfizer/BioNTech | mRNA | 91.3% | 96.7% |
| 3 | Ad26.COV2.S78 | Janssen | viral vector | 52.4% | 74.6% |
| 4 | AZD122282 | AstraZeneca | viral vector | 74.0% | 100% |
| 5 | Covishield | Serum Institute of India | viral vector | NAa | NA |
| 6 | AD5-nCoV83 | CanSino Biologics | viral vector | 57.5% | 91.7% |
| 7 | BBV15284 | Bharat Biotech | inactivated | 77.8% | 93.4% |
| 8 | BBIBP-CorV85 | Sinopharm | inactivated | 78.1% | 100% |
| 9 | CoronaVac86 | Sinovac | inactivated | 50.7% | 100% |
| 10 | NVX-CoV237287 | Novavax | subunit protein | 89.7% | 100% |
| 11 | COVOVAX | Serum Institute of India | subunit protein | NA | NA |
NA, not available.
In November 2021, the SARS-CoV-2 Omicron variant with more than 30 mutations in the S protein was first detected in South Africa and has since spread across the globe.88 In late 2022, a diversity of Omicron subvariants were detected, mostly have convergently evolved from Omicron BA.2 and BA.5 by acquiring amino acid substitutions at a few critical residues in the S protein or by recombination between different Omicron subvariants (e.g., XBB). With the emergence of nAb escaping Omicron variants,89 , 90 , 91 , 92 , 93 , 94 , 95 , 96 , 97 the current monovalent SARS-CoV-2 vaccines are not effective in preventing infection.98 , 99 , 100 Although neutralization antibody levels are highly predictive of immune protection of SARS-CoV-2 symptomatic infection,101 , 102 , 103 , 104 , 105 infection or vaccination-induced CD8+ T cell response provides protection against severe disease and death.106 , 107 , 108 The BNT162b2 and Ad26.COV2.S vaccines provided robust protection against severe disease (∼70%–72%) during Omicron waves despite in the absence of high titers of Omicron-specific nAbs, suggesting that immune protection is supported by cross-reactive non-nAb functions and cellular immunity during Omicron waves.108 , 109 To date, SARS-CoV-2 Omicron subvariant XBB and BQ.1.1 are the most potent in escaping nAbs directed against the ancestral Wuhan strain,97 and these variants most likely emerged under immune selective pressure.93 , 110 After breakthrough infection, there is a significant increase in overall nAb titers and induce mucosal immunity against SARS-CoV-2.94 , 111 , 112 However, nAb titers to Omicron subvariants remain low.94 , 97 Together with waning of SARS-CoV-2-specific nAbs,113 , 114 , 115 individuals might encounter multiple breakthrough infections caused by different SARS-CoV-2 antigenic variants.
The effects of immune imprinting, or “original antigenic sin,” is observed after Omicron breakthrough infection. Breakthrough infection with Omicron subvariants mainly recalls cross-reactive B cells elicited by ancestral vaccine/infection but rarely produces de novo Omicron-specific B cells or nAbs.97 , 116 , 117 Immune imprinting has caused significant reductions of nAb epitope diversity while causing increased proportion of non-nAbs; thus, it increased the immune pressure on the RBD and promoted convergent RBD evolution.97
The future: Developing a “dream vaccine” for coronaviruses
The current COVID-19 vaccines were proven highly effective in curbing the pandemic in the early phase. However, the emerging SARS-CoV-2 Omicron variants under immune selection has greatly dampened the effectiveness of the current vaccines in preventing infection and/or transmission. This has created a dilemma that, although vaccines are still effective in preventing severe disease, the on-going infection waves raise the possibility of a never-ending cycle of variant emergence, a scenario similar to seasonal influenza viruses. Rapid waning of SARS-CoV-2-specific nAbs also requires frequent boosting, which may or may not be sustainable and may worsen the vaccine uptake in the general population. In this context, agencies such as CEPI and the US National Institute of Allergy and Infectious Disease (NIAID) are calling for increased effort in developing universal vaccine candidates that could induce durable, broad-spectrum immunity that are not only effective for current or future SARS-CoV-2 variants but also for pre-emergent zoonotic sarbecoviruses or coronaviruses known to circulate in wildlife animals.
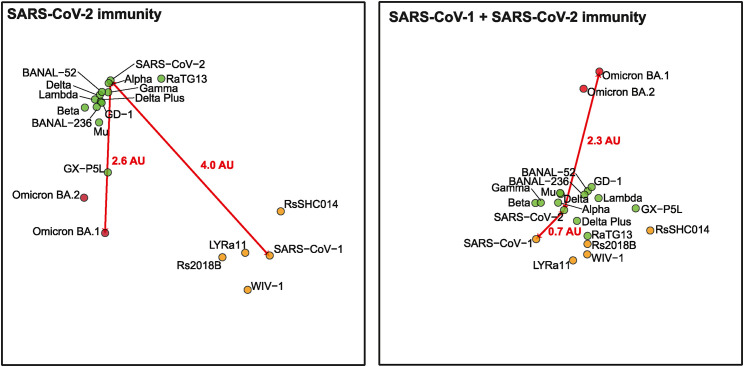
Emergence of nAb escaping variants have prompted the vaccine developers to develop second generation SARS-CoV-2 vaccines (2GCoVax) based on SARS-CoV-2 variants. Both Pfizer and Moderna have rapidly rolled out bivalent vaccines containing ancestral SARS-CoV-2 and Omicron variants. Individuals who had received a bivalent booster vaccine have been shown to have higher nAb titers against Omicron variants.118 , 119 A recent article has demonstrated that BA.5 bivalent vaccine produced comparable nAb titers against Omicron subvariants, including the current circulating subvariant BQ.1.1 and XBB.1.120 However, as with the immune imprinting effect observed from breakthrough infection, the Omicron bivalent vaccines also suffer from the fact that majority of recalled B cells are targeting the ancestral SARS-CoV-2 epitopes, therefore inducing less Omicron-specific nAbs. Further, the antigenic difference among the different Omicron subvariants is significant (Figure 3 ). Naive unvaccinated individuals who had recovered from BA.1 or BA.2 infection failed to produce cross-nAbs against the latest circulating Omicron sub-variants, despite the fact that these Omicron sub-variants are phylogenetically closely related, differing by a few amino acid residues only. Other studies have shown that the bivalent vaccine as a booster might not be able to elicit the desired superior immune responses.121 A recent study based on molecular fate mapping of B cells further demonstrated that responses to sequential homologous boosting derive overwhelmingly from primary cohort B cells, whereas later induction of new antibody responses from naive B cells is strongly suppressed, further highlighting the need to explore other vaccine strategies to combat current and future infections by closely related SARSr-CoVs.122
Figure 3.
Antigenic cartography of SARS-related coronaviruses
Antigenic map was generated by the neutralization titers 50%. The antigenic distance between SARS-CoV-2 and SARS-CoV-1 or Omicron BA.1 in individuals who had been vaccinated against or infected by SARS-CoV-2 (left) and SARS survivors who had received 2 doses of SARS-CoV-2 mRNA vaccines (right). One antigenic unit indicates 2-fold dilution in titer.
As the current data show that 2GCoVax is unlikely to be effective for future variants or novel zoonotic CoVs, efforts are already under the way to develop more broad-spectrum CoV vaccines, including 3rd-generation pan-sarbecovirus vaccine (3GCoVax)—vaccine candidates that can protect us from all the SARS-related coronaviruses, 4th-generation pan-betacoronavirus vaccine (4GCoVax), or the ultimate 5th-generation pan-coronavirus vaccines (5GCoVax).
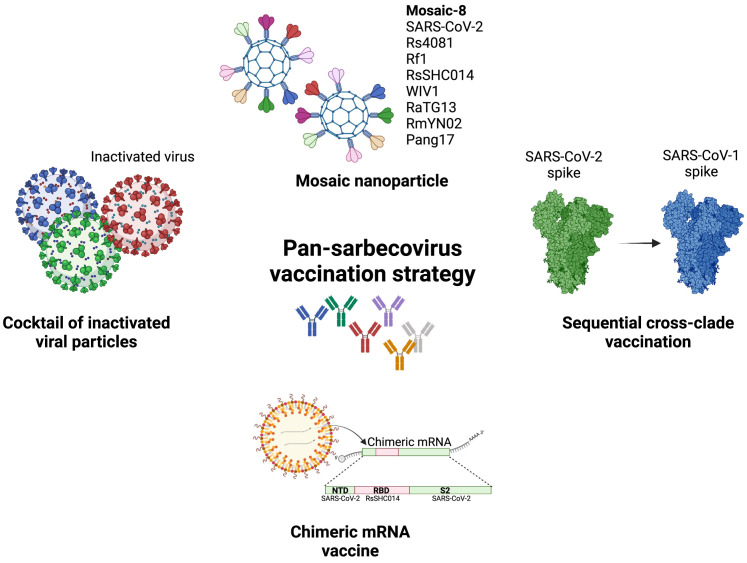
Most of the pan-sarbecovirus vaccine candidates are at preclinical stages, and there are several major strategies toward this aim (Figure 4 ). One of the approaches that showed promising data in preclinical studies used the mosaic nanoparticle platform. The Mosaic-8b developed by a Caltech team is a mosaic nanoparticle containing RBDs from SARS-CoV-2 and seven animal coronaviruses (Bat CoV RaTG13, Bat CoV SHC014, Bat CoV Rs4081, Bat CoV RmYN02, Pangolin CoV Pang17, Bat CoV Rf1, and Bat CoV WIV1). This mosaic vaccine candidate was shown to be capable of inducing broader nAb responses in mice and non-human primates. The immunized animals were protected against the Delta variant and SARS-CoV-1 challenges.123 A similar approach by SK Bioscience, the GBP511 vaccine candidate, contains RBDs from SARS-CoV-2, SARS-CoV-1, Bat CoV RaTG13, and Bat CoV WIV1 and is able to produce broadly protective immune responses against animal sarbecoviruses and protect the vaccinated animals from SARS-CoV-1 infection.124 Chimeric spike mRNA vaccine candidates, containing RBD, NTD, and S2 domains from different SARS-related coronaviruses, have also been shown to be able to induce pan-sarbecovirus immunity in mice and protected the aged mice from SARS-CoV-1, SARS-CoV-2, Beta, Bat CoV RsSHC014, and Bat CoV WIV-1, respectively, in challenge studies.125 Another study, by the Walter Reed Army Medical Center, focused more on the delivery and adjuvant function rather than on antigen design and reported the development of a liposomal adjuvanted SARS-CoV-2 spike protein ferritin nanoparticle that elicits broad-spectrum immunity against SARS-CoV-2 and SARS-CoV-1 in non-human primate studies.126 Similarly, an RBD-sortase-A-conjugated ferritin nanoparticle was reported to offer broadly protective immunity against SARS-CoV-2, variants of concern (VoCs) and Bat CoV RsSHC014 in mice.127 Although encouraging, these studies were mostly conducted in mice with a naive immune background, whereas the vaccine efficacies with pre-existing SARS-CoV-2 immunity remain unknown. Because this will be the predominant immune status in the human population due to vast vaccine efforts globally and widespread breakthrough infections, developing (booster) vaccine strategies targeting a population with pre-existing immunity will be essential and urgently needed.
Figure 4.
Strategies for developing pan-sarbecovirus vaccines
Graphical illustration of the vaccination strategy that elicits pan-sarbecovirus immunity. This includes mosaic nanoparticle, cocktail of inactivated virus particles, chimeric spike mRNA vaccine, and sequential cross-clade boosting.
The clinical trial pipeline for multivalent SARS-CoV-2 vaccines (Figure 1C) includes 189 phase 1 trials, but only 3 are reported as multivalent, including a candidate vaccine for the S RBD and DNA Nucleocapsid128 to stimulate binding antibodies and T cells. Although 17 multivalent SARS-CoV-2 vaccines are in phase 2 trials, some candidates are already outdated, integrating the S of Delta and Beta VoCs129 (NCT05043311). A further 13 multivalent candidates has progressed to phase 3 trials, including a IFNa cytokine-adjuvanted RBD protein with tags for increased antigen presentation, including Fc targeting and CD4+ T cell targeting HLA-DR epitope130 (NCT05096832). This vaccine was assessed in a cohort that had previously been vaccinated with whole inactivated SARS-CoV-2 vaccines; however, it demonstrated an efficacy of only 47.8%. Among the 59 approved vaccines, only one is multivalent: the Ancestral + Omicron bivalent mRNA lipid nanoparticle vaccine, with a 61.8% vaccine efficacy for severe infection compared with ancestral boosters.131 To avoid playing catch up with further VoC and future CoV-X we need development of pan-genus or pan-subgenus vaccines.
A study led by the Duke-NUS Medical School team demonstrated that SARS-CoV-1 survivors who had received BNT162b2 vaccine produced a swathe of pan-sarbecovirus-nAbs,132 reporting the first in-human data supporting the concept of cross-clade boosting to broaden sarbecovirus protective immunity. Preliminary data on Omicron BA.1 and BA.2 showed better protective immunity than individuals who had received two doses of BNT162b293 (Figure 3). Based on this finding, a booster vaccine candidate based on consensus spike protein sequences of all known SARS-CoV-1-related viruses has been shown to elicit broad pan-sarbecovirus immunity in mice that have been primed with SARS-CoV-2 vaccines (W.N. Chia, C.W.T., and L.-F.W., unpublished data).
Although the efforts, as discussed in the preceding section, point to the feasibility of developing successful pan-sarbecovirus vaccines (or 3GCoVax), pan-betacoronavirus (4GCoVax), or pan-coronavirus vaccine (5GCoVax) remain as dream vaccine candidates, the path to such vaccine development is less clear or convincing at the present time. Another approach to producing broad-spectrum coronavirus vaccines is to develop hybrid or multi-valent vaccines just focusing on known zoonotic viruses or animal viruses with zoonotic potential, such as SARSr-CoV plus MERS-CoV and SARS-CoV. DIOSynVax, a Cambridge startup, is partnering with CEPI to establish broadly protective betacoronavirus vaccine using mRNA platform. The antigen design was conducted using the combination of protein structure, computational biology, and immune-optimization to maximize the protection of different viruses (https://cepi.net/news_cepi/cepi-and-diosynvax-partner-in-quest-to-develop-broadly-protective-betacoronavirus-vaccine/). A study by a group in Beijing has used structural-guided universal design of Beta coronavirus vaccines using dimeric RBD, which yielded promising mouse protection data against MERS-CoV and SARS-CoV-2.133 It is clear that more studies are required to find the best path to producing a truly effective broad-spectrum vaccine(s), whether it is at family, genus, or sub-genus level. It is also too early to determine whether the multi-subunit display approach, such as the 8-RBD mosaic nanoparticle vaccine candidate,123 or the sequential immunization approach will be more productive. It is thus interesting to note that a recent study has conducted such a comparison using LNP-mRNA coding the spike protein of SARS-CoV-2, SARS-CoV-1, and MERS-CoV. The data indicated that the sequential immunization approach produced significantly stronger species-specific nAbs against all three viruses than the simultaneous vaccination approach using a mixture of the three mRNA vaccine candidates.134
No pan-coronavirus candidate (5GCoVax) has been reported to date. As a first step toward such a goal, Moderna announced an expansion of its mRNA pipeline to develop a vaccine candidate (mRNA-1287) against four seasonal coronaviruses (Human Coronavirus 229E, NL63, OC43, HKU1).
Universal antibody vaccines
In contrast to the limited success in developing pan-virus vaccines, multiple broad-spectrum human-neutralizing mAbs have been isolated and characterized for our focused viruses. It is time to consider stockpiling broad-spectrum mAbs for antibody-medicated immunization or ring vaccination as an effective early intervention.
The first pan-henipavirus mAb (m102.4) was isolated from a naive human Fab phage display library.13 , 135 Structural studies revealed that key binding residues of the viral G proteins overlap with the key contact points between the G proteins and their main cellular receptor, EphB2.136 This ensures structural conservation and stability among different viruses, making it a true pan-henipavirus-neutralizing mAb. This mAb has proven to be efficacious in both therapeutic and prophylactic applications in animal infection models137 , 138 , 139 and passed phase-1 human clinical trials.26 Since then, there have been other pan-henipavirus mAbs that were developed and shown to be effective, including those targeting the F protein.140
For influenza viruses, there are broadly reactive mAbs against HA, NA, and M2 viral proteins. These antibodies target conserved epitopes shared by different subtypes or even types. This review mainly focuses on the HA. For those that are related to NA and M2, please refer to a comprehensive review contributed by Sun et al.141 There are many mAbs targeting the HA head and HA stem domains.141 These HA mAbs can inhibit virus replication via different mechanisms, such as inhibiting sialic acid receptor binding, virus-host membrane fusion, virus budding, HA0 cleavage and HA trimer formation. However, clinical trials examining some of these antibodies, mainly the stem antibodies, have reported mixed results because some studies showed potential clinical benefits of these antibodies,142 whereas some did not.143 , 144 Thus, the potential use of these mAbs for therapeutic and prophylactic purpose requires further investigation. Furthermore, some of these broad-spectrum mAbs can play roles in mediating other immune responses, such as antibody-dependent cellular cytotoxicity, antibody-dependent cellular phagocytosis, and complement-dependent cytotoxicity. This indicates that the Fc receptor (FcR)-dependent effector functions of these antibodies might have key roles in mediating their protective effect against influenza virus infections. Thus, systematic analysis of broad-spectrum HA mAbs using different Fc and Fab combinations might further enhance the potency of these mAbs.
Within a relatively short time, many pan-variant-, pan-sarbecovirus-, or pan-betacoronavirus-neutralizing mAbs have been identified and characterized. Although many target the RBD region, there are also those targeting NTD, S1, S2, or fusion peptide regions.145 , 146 , 147 , 148 , 149 The most potent and broad-spectrum mAbs were isolated from SARS-CoV-1 survivors who have been subsequently exposed to SARS-CoV-2 through vaccination or breakthrough infections.89 , 97 For a more detailed review, please refer to Cox et al.150
Although the cost of mAbs in passive immunization or ring vaccination application remains a major stumbling block, it might still be cost effective to stockpile universal neutralizing mAbs against high-pandemic-potential viruses, including those discussed in this review, for early containment of future disease X outbreaks caused by the known-unknown viruses, such as a new sarbecovirus. In this context, the recent development in administration route and half-life extension provides promises to a wider use of mAbs as vaccines. First, it is now possible to provide mAbs intramuscularly rather than the traditional intravenous route.151 , 152 Second, it has been shown that mAbs given intramuscularly can last 6–12 months systemically. Last, there are also promising trials in delivering potent mAbs using nasal spray and some with effective half-life of 6 h or more.153
In addition to cost, there are other potential limitations or challenges for prophylactic use of mAbs. Widespread use of mAbs at the beginning of a pandemic may preclude immune imprinting and herd immunity to be generated in the population. Another potential downside is the hard-to-predict virus-specific immunological consequences (e.g., potential for enhancement), which will be difficult to assess at the early stage of an outbreak. Another challenge is the dosing and pharmacodynamics of administering mAbs in a population-wide basis is poorly understood.
Concluding remarks
Unlike the pathogens that are eradicated or near eradication, diverse animal reservoir-harbored influenza viruses, coronaviruses, and henipaviruses make eradication of these pathogens not possible with our current vaccines. The lessons learned from our past efforts in developing universal vaccines are many.
First, the efficacy of the vaccine platform varies on the basis of the pathogen, presence of reservoirs, extent of antigenic variation, and level of immunity needed to maintain protection. Unlike influenza and COVID-19, despite global high-level uptake for decades of measles virus (MeV) vaccines, there has not been the selection of antigenic drift capable of evading vaccine antibodies. This has been attributed to the existence of numerous co-dominant MeV antigenic sites in both the haemagglutinin and fusion surface proteins with no substantial advantage of any mutants in evasion of nAbs without combined mutations and fitness costs.154 Unlike MeV, the surface HA and NA proteins of influenza viruses so far have not formed a similar dual target. Although the genetic and antigenic difference between SARS-CoV-1 and SARS-CoV-2 is very similar to those between HeV and NiV, the effectiveness of cross protection offered by their respective vaccines is vastly different.
Second, successful development of any vaccine depends on the knowledge accumulated from decades of multidisciplinary basic research. The rapid deployment of successful mRNA vaccines against SARS-CoV-2 was made possible by multiple basic research efforts over the years, building on respiratory syncytial virus vaccine development for the prefusion conformation. Among them, the mRNA platform and the pre-fusion structure and immunogenicity of the spike proteins play a pivotal role for the success.
Third, success in production of universal nAbs have not yielded a direct path to development of universal vaccines. For all three groups of viruses, there are multiple reports of successful isolation and characterization of broad-spectrum human-nAbs. Yet, the detailed definition and structure characterization of their corresponding epitopes has not directly translated to or facilitated the production of vaccines targeting such epitopes. Effective vaccines designed by structure-based reverse vaccinology remain to enter the clinic.
Lastly, clinical application of mAbs as therapy has received mixed outcomes. With the improvement of half-life and using the cocktail approach, it is worthy to explore that concept of antibody vaccine stockpiling for early intervention of future outbreaks, as the generation of pan-genus vaccines remains in development.
Acknowledgments
C.W.T.’s work is partially supported by OF-YIRG grant (MOH-000535/MOH-OFYIRG19nov-0002) from National Medical Research Council, Singapore. S.A.V. is supported by an NHMRC EL2 fellowship and the Theme-based Research Scheme of the Research Grants Council of the Hong Kong Special Administrative Region, China (LLMP: T11-705/21-N). L.L.M.P.’s work is supported by Collaborative Research Fund, RGC (C7145-20G) and HMRF commissioned fund (CID-HKU2). Research in L.-F.W.’s team is supported by grants from the Singapore National Research Foundation (NRF2016NRF-NSFC002-013 and NRF2018NRF-NSFC003SB-002) and National Medical Research Council (STPRG-FY19-001, COVID19RF-003, COVID19RF-006, COVID19RF-0014, and OFLCG19May-0034).
Declaration of interests
C.W.T. and L.-F.W. are co-inventors of multiple patent applications on development of pan-sarbecovirus vaccines and human-nAbs.
References
- 1.Boylston A. The origins of inoculation. J. R. Soc. Med. 2012;105:309–313. doi: 10.1258/jrsm.2012.12k044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fourati S., Tomalin L.E., Mulè M.P., Chawla D.G., Gerritsen B., Rychkov D., Henrich E., Miller H.E.R., Hagan T., Diray-Arce J., et al. Pan-vaccine analysis reveals innate immune endotypes predictive of antibody responses to vaccination. Nat. Immunol. 2022;23:1777–1787. doi: 10.1038/s41590-022-01329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulendran B. Systems vaccinology: probing humanity's diverse immune systems with vaccines. Proc. Natl. Acad. Sci. USA. 2014;111:12300–12306. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crevar C.J., Carter D.M., Lee K.Y., Ross T.M. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccin. Immunother. 2015;11:572–583. doi: 10.1080/21645515.2015.1012013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nachbagauer R., Feser J., Naficy A., Bernstein D.I., Guptill J., Walter E.B., Berlanda-Scorza F., Stadlbauer D., Wilson P.C., Aydillo T., et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021;27:106–114. doi: 10.1038/s41591-020-1118-7. [DOI] [PubMed] [Google Scholar]
- 6.Kanekiyo M., Joyce M.G., Gillespie R.A., Gallagher J.R., Andrews S.F., Yassine H.M., Wheatley A.K., Fisher B.E., Ambrozak D.R., Creanga A., et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019;20:362–372. doi: 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arevalo C.P., Bolton M.J., Le Sage V., Ye N., Furey C., Muramatsu H., Alameh M.G., Pardi N., Drapeau E.M., Parkhouse K., et al. A multivalent nucleoside-modified mRNA vaccine against all known influenza virus subtypes. Science. 2022;378:899–904. doi: 10.1126/science.abm0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Doorn E., Pleguezuelos O., Liu H., Fernandez A., Bannister R., Stoloff G., Oftung F., Norley S., Huckriede A., Frijlink H.W., Hak E. Evaluation of the immunogenicity and safety of different doses and formulations of a broad spectrum influenza vaccine (FLU-v) developed by SEEK: study protocol for a single-center, randomized, double-blind and placebo-controlled clinical phase IIb trial. BMC Infect. Dis. 2017;17:241. doi: 10.1186/s12879-017-2341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray K., Selleck P., Hooper P., Hyatt A., Gould A., Gleeson L., Westbury H., Hiley L., Selvey L., Rodwell B. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 10.Lee B., Broder C.C., Wang L.F. In: Fields Virology. Howley P.M., Knipe D., Whalen C., editors. Lippincott; 2020. Henipaviruses; pp. 559–595. [Google Scholar]
- 11.Zhang X.A., Li H., Jiang F.C., Zhu F., Zhang Y.F., Chen J.J., Tan C.W., Anderson D.E., Fan H., Dong L.Y., et al. A zoonotic Henipavirus in febrile patients in China. N. Engl. J. Med. 2022;387:470–472. doi: 10.1056/NEJMc2202705. [DOI] [PubMed] [Google Scholar]
- 12.Zeltina A., Bowden T.A., Lee B. Emerging paramyxoviruses: receptor tropism and zoonotic potential. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z., Bossart K.N., Bishop K.A., Crameri G., Dimitrov A.S., McEachern J.A., Feng Y., Middleton D., Wang L.F., Broder C.C., Dimitrov D.S. Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J. Infect. Dis. 2008;197:846–853. doi: 10.1086/528801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossart K.N., Crameri G., Dimitrov A.S., Mungall B.A., Feng Y.R., Patch J.R., Choudhary A., Wang L.F., Eaton B.T., Broder C.C. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 2005;79:6690–6702. doi: 10.1128/JVI.79.11.6690-6702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broder C.C., Geisbert T.W., Xu K., Nikolov D.B., Wang L.F., Middleton D., Pallister J., Bossart K.N. Immunization strategies against henipaviruses. Curr. Top. Microbiol. Immunol. 2012;359:197–223. doi: 10.1007/82_2012_213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang H.V., Cross R.W., Borisevich V., Bornholdt Z.A., West B.R., Chan Y.P., Mire C.E., Da Silva S.C., Dimitrov A.S., Yan L., et al. Broadly neutralizing antibody cocktails targeting Nipah virus and Hendra virus fusion glycoproteins. Nat. Struct. Mol. Biol. 2021;28:426–434. doi: 10.1038/s41594-021-00584-8. [DOI] [PubMed] [Google Scholar]
- 17.Amaya M., Broder C.C. Vaccines to emerging viruses: nipah and Hendra. Annu. Rev. Virol. 2020;7:447–473. doi: 10.1146/annurev-virology-021920-113833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mungall B.A., Middleton D., Crameri G., Bingham J., Halpin K., Russell G., Green D., McEachern J., Pritchard L.I., Eaton B.T., et al. Feline model of acute Nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J. Virol. 2006;80:12293–12302. doi: 10.1128/JVI.01619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pallister J., Middleton D., Wang L.F., Klein R., Haining J., Robinson R., Yamada M., White J., Payne J., Feng Y.R., et al. A recombinant Hendra virus G glycoprotein-based subunit vaccine protects ferrets from lethal Hendra virus challenge. Vaccine. 2011;29:5623–5630. doi: 10.1016/j.vaccine.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pallister J.A., Klein R., Arkinstall R., Haining J., Long F., White J.R., Payne J., Feng Y.R., Wang L.F., Broder C.C., Middleton D. Vaccination of ferrets with a recombinant G glycoprotein subunit vaccine provides protection against Nipah virus disease for over 12 months. Virol. J. 2013;10:237. doi: 10.1186/1743-422X-10-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bossart K.N., Rockx B., Feldmann F., Brining D., Scott D., LaCasse R., Geisbert J.B., Feng Y.R., Chan Y.P., Hickey A.C., et al. A Hendra virus G glycoprotein subunit vaccine protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 2012;4:146ra107. doi: 10.1126/scitranslmed.3004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mire C.E., Geisbert J.B., Agans K.N., Feng Y.R., Fenton K.A., Bossart K.N., Yan L., Chan Y.P., Broder C.C., Geisbert T.W. A recombinant Hendra virus G glycoprotein subunit vaccine protects nonhuman primates against Hendra virus challenge. J. Virol. 2014;88:4624–4631. doi: 10.1128/JVI.00005-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middleton D., Pallister J., Klein R., Feng Y.R., Haining J., Arkinstall R., Frazer L., Huang J.A., Edwards N., Wareing M., et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis. 2014;20:372–379. doi: 10.3201/eid2003.131159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickering B.S., Hardham J.M., Smith G., Weingartl E.T., Dominowski P.J., Foss D.L., Mwangi D., Broder C.C., Roth J.A., Weingartl H.M. Protection against henipaviruses in swine requires both, cell-mediated and humoral immune response. Vaccine. 2016;34:4777–4786. doi: 10.1016/j.vaccine.2016.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Li R., Wang M., Liu Y., Yin Y., Zai X., Song X., Chen Y., Xu J., Chen W. Fc-based recombinant Henipavirus vaccines elicit broad neutralizing antibody responses in mice. Viruses. 2020;12 doi: 10.3390/v12040480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Playford E.G., Munro T., Mahler S.M., Elliott S., Gerometta M., Hoger K.L., Jones M.L., Griffin P., Lynch K.D., Carroll H., et al. Safety, tolerability, pharmacokinetics, and immunogenicity of a human monoclonal antibody targeting the G glycoprotein of henipaviruses in healthy adults: a first-in-human, randomised, controlled, phase 1 study. Lancet Infect. Dis. 2020;20:445–454. doi: 10.1016/S1473-3099(19)30634-6. [DOI] [PubMed] [Google Scholar]
- 27.Dhanasekaran V., Sullivan S., Edwards K.M., Xie R., Khvorov A., Valkenburg S.A., Cowling B.J., Barr I.G. Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat. Commun. 2022;13:1721. doi: 10.1038/s41467-022-29402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plotkin S. History of vaccination. Proc. Natl. Acad. Sci. USA. 2014;111:12283–12287. doi: 10.1073/pnas.1400472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C., Thompson M.G., Cowling B.J. Influenza vaccination in tropical and subtropical areas. Lancet Respir. Med. 2017;5:920–922. doi: 10.1016/S2213-2600(17)30377-6. [DOI] [PubMed] [Google Scholar]
- 30.Skowronski D.M., Chambers C., De Serres G., Sabaiduc S., Winter A.L., Dickinson J.A., Gubbay J.B., Drews S.J., Fonseca K., Charest H., et al. Vaccine Effectiveness against Lineage-matched and -mismatched Influenza B Viruses Across 8 Seasons in Canada, 2010–2011 to 2017–2018. Clin. Infect. Dis. 2019;68:1754–1757. doi: 10.1093/cid/ciy876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambers B.S., Parkhouse K., Ross T.M., Alby K., Hensley S.E. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep. 2015;12:1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouma S., Weirick M., Hensley S.E. Potential antigenic mismatch of the H3N2 component of the 2019 southern hemisphere influenza vaccine. Clin. Infect. Dis. 2020;70:2432–2434. doi: 10.1093/cid/ciz723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreyfus C., Laursen N.S., Kwaks T., Zuijdgeest D., Khayat R., Ekiert D.C., Lee J.H., Metlagel Z., Bujny M.V., Jongeneelen M., et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koutsakos M., Illing P.T., Nguyen T.H.O., Mifsud N.A., Crawford J.C., Rizzetto S., Eltahla A.A., Clemens E.B., Sant S., Chua B.Y., et al. Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat. Immunol. 2019;20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
- 35.Erbelding E.J., Post D.J., Stemmy E.J., Roberts P.C., Augustine A.D., Ferguson S., Paules C.I., Graham B.S., Fauci A.S. A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J. Infect. Dis. 2018;218:347–354. doi: 10.1093/infdis/jiy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J., Deng G., Ma S., Zeng X., Yin X., Li M., Zhang B., Cui P., Chen Y., Yang H., et al. Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe. 2018;24:558–568.e7. doi: 10.1016/j.chom.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Food Safety Authority. European Centre for Disease Prevention and Control. European Union Reference Laboratory for Avian Influenza. Cornelia Adlhoch. Alice Fusaro. José L Gonzales. Thijs Kuiken. Stefano Marangon. Éric Niqueux. Christoph Staubach. Calogero Terregino. Inma Aznar Avian influenza overview March - June 2022. EFSA J. 2022;20 doi: 10.2903/j.efsa.2022.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gostic K.M., Ambrose M., Worobey M., Lloyd-Smith J.O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bull M.B., Gu H., Ma F.N.L., Perera L.P., Poon L.L.M., Valkenburg S.A. Next-generation T cell-activating vaccination increases influenza virus mutation prevalence. Sci. Adv. 2022;8:eabl5209. doi: 10.1126/sciadv.abl5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Houser K.V., Chen G.L., Carter C., Crank M.C., Nguyen T.A., Burgos Florez M.C., Berkowitz N.M., Mendoza F., Hendel C.S., Gordon I.J., et al. Safety and immunogenicity of a ferritin nanoparticle H2 influenza vaccine in healthy adults: a phase 1 trial. Nat. Med. 2022;28:383–391. doi: 10.1038/s41591-021-01660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans T.G., Bussey L., Eagling-Vose E., Rutkowski K., Ellis C., Argent C., Griffin P., Kim J., Thackwray S., Shakib S., et al. Efficacy and safety of a universal influenza A vaccine (MVA-NP+M1) in adults when given after seasonal quadrivalent influenza vaccine immunisation (FLU009): a phase 2b, randomised, double-blind trial. Lancet Infect. Dis. 2022;22:857–866. doi: 10.1016/S1473-3099(21)00702-7. [DOI] [PubMed] [Google Scholar]
- 42.Kavian N., Hachim A., Li A.P., Cohen C.A., Chin A.W., Poon L.L., Fang V.J., Leung N.H., Cowling B.J., Valkenburg S.A. Assessment of enhanced influenza vaccination finds that FluAd conveys an advantage in mice and older adults. Clin. Transl. Immunology. 2020;9 doi: 10.1002/cti2.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li A.P.Y., Cohen C.A., Leung N.H.L., Fang V.J., Gangappa S., Sambhara S., Levine M.Z., Iuliano A.D., Perera R.A.P.M., Ip D.K.M., et al. Immunogenicity of standard, high-dose, MF59-adjuvanted, and recombinant-HA seasonal influenza vaccination in older adults. npj Vaccines. 2021;6:25. doi: 10.1038/s41541-021-00289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goll J.B., Jain A., Jensen T.L., Assis R., Nakajima R., Jasinskas A., Coughlan L., Cherikh S.R., Gelber C.E., Khan S., et al. The antibody landscapes following AS03 and MF59 adjuvanted H5N1 vaccination. NPJ Vaccines. 2022;7:103. doi: 10.1038/s41541-022-00524-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez-Davies J.E., Dollinger E.P., Pone E.J., Felgner J., Liang L., Strohmeier S., Jan S., Albin T.J., Jain A., Nakajima R., et al. Magnitude and breadth of antibody cross-reactivity induced by recombinant influenza hemagglutinin trimer vaccine is enhanced by combination adjuvants. Sci. Rep. 2022;12:9198. doi: 10.1038/s41598-022-12727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 49.Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q., Qi J., Yuan Y., Xuan Y., Han P., Wan Y., Ji W., Li Y., Wu Y., Wang J., et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olival K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., Daszak P. Host and viral traits predict zoonotic spillover from mammals. Nature. 2017;546:646–650. doi: 10.1038/nature22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irving A.T., Ahn M., Goh G., Anderson D.E., Wang L.F. Lessons from the host defences of bats, a unique viral reservoir. Nature. 2021;589:363–370. doi: 10.1038/s41586-020-03128-0. [DOI] [PubMed] [Google Scholar]
- 54.Eby P., Peel A.J., Hoegh A., Madden W., Giles J.R., Hudson P.J., Plowright R.K. Pathogen spillover driven by rapid changes in bat ecology. Nature. 2023;613:340–344. doi: 10.1038/s41586-022-05506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mora C., McKenzie T., Gaw I.M., Dean J.M., von Hammerstein H., Knudson T.A., Setter R.O., Smith C.Z., Webster K.M., Patz J.A., et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat. Clim. Chang. 2022;12:869–875. doi: 10.1038/s41558-022-01426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plowright R.K., Reaser J.K., Locke H., Woodley S.J., Patz J.A., Becker D.J., Oppler G., Hudson P.J., Tabor G.M. Land use-induced spillover: a call to action to safeguard environmental, animal, and human health. Lancet Planet Health. 2021;5:e237–e245. doi: 10.1016/S2542-5196(21)00031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sánchez C.A., Li H., Phelps K.L., Zambrana-Torrelio C., Wang L.F., Zhou P., Shi Z.L., Olival K.J., Daszak P. A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia. Nat. Commun. 2022;13:4380. doi: 10.1038/s41467-022-31860-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans T.S., Tan C.W., Aung O., Phyu S., Lin H., Coffey L.L., Toe A.T., Aung P., Aung T.H., Aung N.T., et al. Exposure to diverse sarbecoviruses indicates frequent zoonotic spillover in human communities interacting with wildlife. Int. J. Infect. Dis. 2023;131:57–64. doi: 10.1016/j.ijid.2023.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mo H.Y., Xu J., Ren X.L., Zeng G.Q., Tan Y.X., Chen R.C., Chan-Yeung M., Zhong N.S. Evaluation by indirect immunofluorescent assay and enzyme linked immunosorbent assay of the dynamic changes of serum antibody responses against severe acute respiratory syndrome coronavirus. Chin. Med. J. (Engl) 2005;118:446–450. [PubMed] [Google Scholar]
- 60.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I., Tiu C., Hu Z., Chen V.C., Young B.E., Sia W.R., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 61.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 62.Xu X., Gao X. Immunological responses against SARS-coronavirus infection in humans. Cell. Mol. Immunol. 2004;1:119–122. [PubMed] [Google Scholar]
- 63.Zhong X., Yang H., Guo Z.F., Sin W.Y., Chen W., Xu J., Fu L., Wu J., Mak C.K., Cheng C.S., et al. B-cell responses in patients who have recovered from severe acute respiratory syndrome target a dominant site in the S2 domain of the surface spike glycoprotein. J. Virol. 2005;79:3401–3408. doi: 10.1128/JVI.79.6.3401-3408.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou H., Chen Y., Zhang S., Niu P., Qin K., Jia W., Huang B., Zhang S., Lan J., Zhang L., et al. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat. Commun. 2019;10:3068. doi: 10.1038/s41467-019-10897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang X.C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S., Sheehan J., Zhu Q., et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corti D., Zhao J., Pedotti M., Simonelli L., Agnihothram S., Fett C., Fernandez-Rodriguez B., Foglierini M., Agatic G., Vanzetta F., et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2015;112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y.D., Chi W.Y., Su J.H., Ferrall L., Hung C.F., Wu T.C. Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 2020;27:104. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1781. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Folegatti P.M., Bittaye M., Flaxman A., Lopez F.R., Bellamy D., Kupke A., Mair C., Makinson R., Sheridan J., Rohde C., et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect. Dis. 2020;20:816–826. doi: 10.1016/S1473-3099(20)30160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watson O.J., Barnsley G., Toor J., Hogan A.B., Winskill P., Ghani A.C. Global impact of the first year of COVID-19 vaccination: a mathematical modelling study. Lancet Infect. Dis. 2022;22:1293–1302. doi: 10.1016/S1473-3099(22)00320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hardt K., Vandebosch A., Sadoff J., Le Gars M., Truyers C., Lowson D., Van Dromme I., Vingerhoets J., Kamphuis T., Scheper G., et al. Efficacy, safety, and immunogenicity of a booster regimen of Ad26.COV2.S vaccine against COVID-19 (ENSEMBLE2): results of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Infect. Dis. 2022;22:1703–1715. doi: 10.1016/S1473-3099(22)00506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Van Dromme I., Spiessens B., et al. Final analysis of efficacy and safety of single-dose Ad26.COV2.S. N. Engl. J. Med. 2022;386:847–860. doi: 10.1056/NEJMoa2117608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jara A., Undurraga E.A., González C., Paredes F., Fontecilla T., Jara G., Pizarro A., Acevedo J., Leo K., Leon F., et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N. Engl. J. Med. 2021;385:875–884. doi: 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El Sahly H.M., Baden L.R., Essink B., Doblecki-Lewis S., Martin J.M., Anderson E.J., Campbell T.B., Clark J., Jackson L.A., Fichtenbaum C.J., et al. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N. Engl. J. Med. 2021;385:1774–1785. doi: 10.1056/NEJMoa2113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomas S.J., Moreira E.D., Jr., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Polack F.P., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N. Engl. J. Med. 2021;385:1761–1773. doi: 10.1056/NEJMoa2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Falsey A.R., Sobieszczyk M.E., Hirsch I., Sproule S., Robb M.L., Corey L., Neuzil K.M., Hahn W., Hunt J., Mulligan M.J., et al. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N. Engl. J. Med. 2021;385:2348–2360. doi: 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halperin S.A., Ye L., MacKinnon-Cameron D., Smith B., Cahn P.E., Ruiz-Palacios G.M., Ikram A., Lanas F., Lourdes Guerrero M., Muñoz Navarro S.R., et al. Final efficacy analysis, interim safety analysis, and immunogenicity of a single dose of recombinant novel coronavirus vaccine (adenovirus type 5 vector) in adults 18 years and older: an international, multicentre, randomised, double-blinded, placebo-controlled phase 3 trial. Lancet. 2022;399:237–248. doi: 10.1016/S0140-6736(21)02753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ella R., Reddy S., Blackwelder W., Potdar V., Yadav P., Sarangi V., Aileni V.K., Kanungo S., Rai S., Reddy P., et al. Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): interim results of a randomised, double-blind, controlled, phase 3 trial. Lancet. 2021;398:2173–2184. doi: 10.1016/S0140-6736(21)02000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Al Kaabi N., Zhang Y., Xia S., Yang Y., Al Qahtani M.M., Abdulrazzaq N., Al Nusair M., Hassany M., Jawad J.S., Abdalla J., et al. Effect of 2 inactivated SARS-CoV-2 vaccines on symptomatic COVID-19 infection in adults: A randomized clinical trial. JAMA. 2021;326:35–45. doi: 10.1001/jama.2021.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jin L., Li Z., Zhang X., Li J., Zhu F. CoronaVac: a review of efficacy, safety, and immunogenicity of the inactivated vaccine against SARS-CoV-2. Hum. Vaccin. Immunother. 2022;18:2096970. doi: 10.1080/21645515.2022.2096970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heath P.T., Galiza E.P., Baxter D.N., Boffito M., Browne D., Burns F., Chadwick D.R., Clark R., Cosgrove C., Galloway J., et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N. Engl. J. Med. 2021;385:1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 91.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., Liu X., Lambe T., Crook D., Stuart D.I., et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2022;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan C.W., Chia W.N., Zhu F., Young B.E., Chantasrisawad N., Hwa S.H., Yeoh A.Y., Lim B.L., Yap W.C., Pada S.K.M.S., et al. SARS-CoV-2 Omicron variant emerged under immune selection. Nat. Microbiol. 2022;7:1756–1761. doi: 10.1038/s41564-022-01246-1. [DOI] [PubMed] [Google Scholar]
- 94.Tan C.W., Lim B.L., Young B.E., Yeoh A.Y., Yung C.F., Yap W.C., Althaus T., Chia W.N., Zhu F., Lye D.C., et al. Comparative neutralisation profile of SARS-CoV-2 omicron subvariants BA.2.75 and BA.5. Lancet Microbe. 2022;3:e898. doi: 10.1016/S2666-5247(22)00220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hachmann N.P., Miller J., Collier A.Y., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariant BA.4.6. N. Engl. J. Med. 2022;387:1904–1906. doi: 10.1056/NEJMc2212117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsang N.N.Y., So H.C., Cowling B.J., Leung G.M., Ip D.K.M. Effectiveness of BNT162b2 and CoronaVac COVID-19 vaccination against asymptomatic and symptomatic infection of SARS-CoV-2 omicron BA.2 in Hong Kong: a prospective cohort study. Lancet Infect. Dis. 2023;23:421–434. doi: 10.1016/S1473-3099(22)00732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang B., Wong I.O.L., Xiao J., Tsang T.K., Liao Q., Cowling B.J. Effectiveness of CoronaVac and BNT162b2 vaccines against severe acute respiratory syndrome coronavirus 2 omicron BA.2 Infections in Hong Kong. J. Infect. Dis. 2022;226:1382–1384. doi: 10.1093/infdis/jiac360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., et al. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022;387:21–34. doi: 10.1056/NEJMoa2203965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 102.Gilbert P.B., Donis R.O., Koup R.A., Fong Y., Plotkin S.A., Follmann D. A Covid-19 milestone attained - A correlate of protection for vaccines. N. Engl. J. Med. 2022;387:2203–2206. doi: 10.1056/NEJMp2211314. [DOI] [PubMed] [Google Scholar]
- 103.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34:108728. doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moss P. The T cell immune response against SARS-CoV-2. Nat. Immunol. 2022;23:186–193. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 108.Collie S., Champion J., Moultrie H., Bekker L.G., Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N. Engl. J. Med. 2022;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gray G., Collie S., Goga A., Garrett N., Champion J., Seocharan I., Bamford L., Moultrie H., Bekker L.G. Effectiveness of Ad26.COV2.S and BNT162b2 Vaccines against Omicron Variant in South Africa. N. Engl. J. Med. 2022;386:2243–2245. doi: 10.1056/NEJMc2202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lim J.M.E., Tan A.T., Le Bert N., Hang S.K., Low J.G.H., Bertoletti A. SARS-CoV-2 breakthrough infection in vaccinees induces virus-specific nasal-resident CD8+ and CD4+ T cells of broad specificity. J. Exp. Med. 2022;219 doi: 10.1084/jem.20220780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wright P.F., Prevost-Reilly A.C., Natarajan H., Brickley E.B., Connor R.I., Wieland-Alter W.F., Miele A.S., Weiner J.A., Nerenz R.D., Ackerman M.E. Longitudinal systemic and mucosal immune responses to SARS-CoV-2 infection. J. Infect. Dis. 2022;226:1204–1214. doi: 10.1093/infdis/jiac065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chia W.N., Zhu F., Ong S.W.X., Young B.E., Fong S.W., Le Bert N., Tan C.W., Tiu C., Zhang J., Tan S.Y., et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. 2021;2:e240–e249. doi: 10.1016/S2666-5247(21)00025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marot S., Malet I., Leducq V., Zafilaza K., Sterlin D., Planas D., Gothland A., Jary A., Dorgham K., Bruel T., et al. Rapid decline of neutralizing antibodies against SARS-CoV-2 among infected healthcare workers. Nat. Commun. 2021;12:844. doi: 10.1038/s41467-021-21111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reynolds C.J., Pade C., Gibbons J.M., Otter A.D., Lin K.M., Muñoz Sandoval D., Pieper F.P., Butler D.K., Liu S., Joy G., et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science. 2022;377:eabq1841. doi: 10.1126/science.abq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.K., Adams-Quack P., Bacher M., Finlayson A., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022;7:eabq2427. doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chalkias S., Harper C., Vrbicky K., Walsh S.R., Essink B., Brosz A., McGhee N., Tomassini J.E., Chen X., Chang Y., et al. A bivalent omicron-containing booster vaccine against Covid-19. N. Engl. J. Med. 2022;387:1279–1291. doi: 10.1056/NEJMoa2208343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis-Gardner M.E., Lai L., Wali B., Samaha H., Solis D., Lee M., Porter-Morrison A., Hentenaar I.T., Yamamoto F., Godbole S., et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA Bivalent Booster. N. Engl. J. Med. 2023;388:183–185. doi: 10.1056/NEJMc2214293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kurhade C., Zou J., Xia H., Liu M., Chang H.C., Ren P., Xie X., Shi P.Y. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med. 2023;29:344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 121.Offit P.A. Bivalent Covid-19 vaccines - A cautionary tale. N. Engl. J. Med. 2023;388:481–483. doi: 10.1056/NEJMp2215780. [DOI] [PubMed] [Google Scholar]
- 122.Schiepers A., van 't Wout M.F.L., Greaney A.J., Zang T., Muramatsu H., Lin P.J.C., Tam Y.K., Mesin L., Starr T.N., Bieniasz P.D., et al. Molecular fate-mapping of serum antibody responses to repeat immunization. Nature. 2023;615:482–489. doi: 10.1038/s41586-023-05715-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cohen A.A., van Doremalen N., Greaney A.J., Andersen H., Sharma A., Starr T.N., Keeffe J.R., Fan C., Schulz J.E., Gnanapragasam P.N.P., et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science. 2022;377:eabq0839. doi: 10.1126/science.abq0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Walls A.C., Miranda M.C., Schäfer A., Pham M.N., Greaney A., Arunachalam P.S., Navarro M.J., Tortorici M.A., Rogers K., O'Connor M.A., et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447.e16. doi: 10.1016/j.cell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martinez D.R., Schäfer A., Leist S.R., De la Cruz G., West A., Atochina-Vasserman E.N., Lindesmith L.C., Pardi N., Parks R., Barr M., et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Joyce M.G., Chen W.H., Sankhala R.S., Hajduczki A., Thomas P.V., Choe M., Martinez E.J., Chang W.C., Peterson C.E., Morrison E.B., et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021;37:110143. doi: 10.1016/j.celrep.2021.110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li D., Martinez D.R., Schäfer A., Chen H., Barr M., Sutherland L.L., Lee E., Parks R., Mielke D., Edwards W., et al. Breadth of SARS-CoV-2 neutralization and protection induced by a nanoparticle vaccine. Nat. Commun. 2022;13:6309. doi: 10.1038/s41467-022-33985-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ahn J.Y., Lee J., Suh Y.S., Song Y.G., Choi Y.J., Lee K.H., Seo S.H., Song M., Oh J.W., Kim M., et al. Safety and immunogenicity of two recombinant DNA COVID-19 vaccines containing the coding regions of the spike or spike and nucleocapsid proteins: an interim analysis of two open-label, non-randomised, phase 1 trials in healthy adults. Lancet Microbe. 2022;3:e173–e183. doi: 10.1016/S2666-5247(21)00358-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hannawi S., Saifeldin L., Abuquta A., Alamadi A., Mahmoud S.A., Hassan A., Liu D., Yan L., Xie L. Safety and immunogenicity of a bivalent SARS-CoV-2 protein booster vaccine, SCTV01C, in adults previously vaccinated with mRNA vaccine: a randomized, double-blind, placebo-controlled phase 1/2 clinical trial. EBiomedicine. 2023;87:104386. doi: 10.1016/j.ebiom.2022.104386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang X.Y., Mahmood S.F., Jin F., Cheah W.K., Ahmad M., Sohail M.A., Ahmad W., Suppan V.K., Sayeed M.A., Luxmi S., et al. Efficacy of heterologous boosting against SARS-CoV-2 using a recombinant interferon-armed fusion protein vaccine (V-01): a randomized, double-blind and placebo-controlled phase III trial. Emerg. Microbes Infect. 2022;11:1910–1919. doi: 10.1080/22221751.2022.2088406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin D.Y., Xu Y., Gu Y., Zeng D., Wheeler B., Young H., Sunny S.K., Moore Z. Effectiveness of bivalent boosters against severe omicron infection. N. Engl. J. Med. 2023;388:764–766. doi: 10.1056/NEJMc2215471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tan C.W., Chia W.N., Young B.E., Zhu F., Lim B.L., Sia W.R., Thein T.L., Chen M.I., Leo Y.S., Lye D.C., Wang L.F. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., et al. A universal design of Betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733.e11. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peng L., Fang Z., Renauer P.A., McNamara A., Park J.J., Lin Q., Zhou X., Dong M.B., Zhu B., Zhao H., et al. Multiplexed LNP-mRNA vaccination against pathogenic coronavirus species. Cell Rep. 2022;40:111160. doi: 10.1016/j.celrep.2022.111160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhu Z., Dimitrov A.S., Bossart K.N., Crameri G., Bishop K.A., Choudhry V., Mungall B.A., Feng Y.R., Choudhary A., Zhang M.Y., et al. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 2006;80:891–899. doi: 10.1128/JVI.80.2.891-899.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu K., Rockx B., Xie Y., DeBuysscher B.L., Fusco D.L., Zhu Z., Chan Y.P., Xu Y., Luu T., Cer R.Z., et al. Crystal structure of the Hendra virus attachment G glycoprotein bound to a potent cross-reactive neutralizing human monoclonal antibody. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bossart K.N., Zhu Z., Middleton D., Klippel J., Crameri G., Bingham J., McEachern J.A., Green D., Hancock T.J., Chan Y.P., et al. A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute Nipah virus infection. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bossart K.N., Geisbert T.W., Feldmann H., Zhu Z., Feldmann F., Geisbert J.B., Yan L., Feng Y.R., Brining D., Scott D., et al. A neutralizing human monoclonal antibody protects african green monkeys from Hendra virus challenge. Sci. Transl. Med. 2011;3:105ra103. doi: 10.1126/scitranslmed.3002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Geisbert T.W., Mire C.E., Geisbert J.B., Chan Y.P., Agans K.N., Feldmann F., Fenton K.A., Zhu Z., Dimitrov D.S., Scott D.P., et al. Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci. Transl. Med. 2014;6:242ra82. doi: 10.1126/scitranslmed.3008929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dang H.V., Chan Y.P., Park Y.J., Snijder J., Da Silva S.C., Vu B., Yan L., Feng Y.R., Rockx B., Geisbert T.W., et al. An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections. Nat. Struct. Mol. Biol. 2019;26:980–987. doi: 10.1038/s41594-019-0308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun X., Ling Z., Yang Z., Sun B. Broad neutralizing antibody-based strategies to tackle influenza. Curr. Opin. Virol. 2022;53:101207. doi: 10.1016/j.coviro.2022.101207. [DOI] [PubMed] [Google Scholar]
- 142.McBride J.M., Lim J.J., Burgess T., Deng R., Derby M.A., Maia M., Horn P., Siddiqui O., Sheinson D., Chen-Harris H., et al. Phase 2 randomized trial of the safety and efficacy of MHAA4549A, a broadly neutralizing monoclonal antibody, in a human influenza A virus challenge model. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01154-17. e01154–e01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Han A., Czajkowski L., Rosas L.A., Cervantes-Medina A., Xiao Y., Gouzoulis M., Lumbard K., Hunsberger S., Reed S., Athota R., et al. Safety and efficacy of CR6261 in an influenza A H1N1 healthy human challenge model. Clin. Infect. Dis. 2021;73:e4260–e4268. doi: 10.1093/cid/ciaa1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lim J.J., Nilsson A.C., Silverman M., Assy N., Kulkarni P., McBride J.M., Deng R., Li C., Yang X., Nguyen A., et al. A Phase 2 randomized, double-blind, placebo-controlled trial of MHAA4549A, a monoclonal antibody, plus oseltamivir in patients hospitalized with severe influenza A virus infection. Antimicrob. Agents Chemother. 2020;64:2845–2847. doi: 10.1128/AAC.00352-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pinto D., Sauer M.M., Czudnochowski N., Low J.S., Tortorici M.A., Housley M.P., Noack J., Walls A.C., Bowen J.E., Guarino B., et al. Broad betacoronavirusBetacoronavirus neutralization by a stem helix-specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Low J.S., Jerak J., Tortorici M.A., McCallum M., Pinto D., Cassotta A., Foglierini M., Mele F., Abdelnabi R., Weynand B., et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science. 2022;377:735–742. doi: 10.1126/science.abq2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 149.Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., Pegu A., Oloniniyi O.K., Henry A.R., Darko S., et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373 doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cox M., Peacock T.P., Harvey W.T., Hughes J., Wright D.W., COVID-19 Genomics UK (COG-UK) Consortium. Willett B.J., Thomson E., Gupta R.K., Peacock S.J., et al. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2023;21:112–124. doi: 10.1038/s41579-022-00809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lanini S., Milleri S., Andreano E., Nosari S., Paciello I., Piccini G., Gentili A., Phogat A., Hyseni I., Leonardi M., et al. Safety and serum distribution of anti-SARS-CoV-2 monoclonal antibody MAD0004J08 after intramuscular injection. Nat. Commun. 2022;13:2263. doi: 10.1038/s41467-022-29909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levin M.J., Ustianowski A., De Wit S., Launay O., Avila M., Templeton A., Yuan Y., Seegobin S., Ellery A., Levinson D.J., et al. Intramuscular AZD7442 (Tixagevimab-Cilgavimab) for prevention of Covid-19. N. Engl. J. Med. 2022;386:2188–2200. doi: 10.1056/NEJMoa2116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin Y., Yue S., Yang Y., Yang S., Pan Z., Yang X., Gao L., Zhou J., Li Z., Hu L., et al. Nasal spray of neutralizing monoclonal antibody 35B5 confers potential prophylaxis against severe acute respiratory syndrome coronavirus 2 variants of concern: A small-scale clinical trial. Clin. Infect. Dis. 2023;76:e336–e341. doi: 10.1093/cid/ciac448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Muñoz-Alía M.Á., Nace R.A., Zhang L., Russell S.J. Serotypic evolution of measles virus is constrained by multiple co-dominant B cell epitopes on its surface glycoproteins. Cell Rep. Med. 2021;2:100225. doi: 10.1016/j.xcrm.2021.100225. [DOI] [PMC free article] [PubMed] [Google Scholar]