Abstract
Recombination is thought to be a mechanism that facilitates cross-species transmission in coronaviruses, thus acting as a driver of coronavirus spillover and emergence. Despite its significance, the mechanism of recombination is poorly understood, limiting our potential to estimate the risk of novel recombinant coronaviruses emerging in the future. As a tool for understanding recombination, here, we outline a framework of the recombination pathway for coronaviruses. We review existing literature on coronavirus recombination, including comparisons of naturally observed recombinant genomes as well as in vitro experiments, and place the findings into the recombination pathway framework. We highlight gaps in our understanding of coronavirus recombination illustrated by the framework and outline how further experimental research is critical for disentangling the molecular mechanism of recombination from external environmental pressures. Finally, we describe how an increased understanding of the mechanism of recombination can inform pandemic predictive intelligence, with a retrospective emphasis on SARS-CoV-2.
Keywords: coronavirus, recombination, pathway, barriers, evolution, cross-species transmission, spillover
In their review, Wells et al. define the coronavirus recombination pathway as a series of environmental and cellular barriers that must be overcome for two coronaviruses to recombine and establish in nature. They review existing coronavirus recombination literature and highlight the specific benefits of experimental and comparative genomics approaches.
Introduction
Despite exponential advances in medical knowledge in the last century, emerging infectious diseases still pose a major threat to global human health.1 , 2 The urgent need to further characterize the drivers of disease emergence has never been understood more clearly than in the last three years since the emergence of SARS-CoV-2 and the onset of the COVID-19 pandemic. Viruses in the family Coronaviridae are of particular importance for disease emergence, not only because of SARS-CoV-2 but also because of the numerous examples of other coronaviruses that infect humans, livestock, and companion animals. Examples in humans include epidemics of SARS-CoV-13 , 4 and MERS-CoV,5 as well as several strains of “common cold” viruses that have fully established within human populations and obtained worldwide distributions (HCoV-229E,6 HCoV-NL63,7 HCoV-OC43,8 HCoV-HKU1,9 and the lesser-known human enteric coronavirus 4408 [HECV-4408]10). Notable pathogenic coronaviruses in domestic animals include transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), swine enteric coronavirus (SECoV), bovine coronavirus (BCoV), and avian infectious bronchitis virus (IBV). Coronaviruses also cause severe disease in the animals we keep as pets, including multiple serotypes of feline coronavirus (FeCoV) in cats, multiple serotypes of canine coronavirus (CaCoV) in dogs, and additionally canine respiratory coronavirus (CRCoV) in dogs. The majority of mammalian coronaviruses are within the Alpha- and Betacoronavirus subgenera, which are mostly evolutionarily associated with bats.11 , 12 , 13 , 14 Conversely, Gamma- and Deltacoronavirus subgenera are largely associated with birds.15 Because closely related bat alpha- and betacoronaviruses have been found for many human and mammalian coronaviruses,16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 a thorough understanding of the pathways of cross-species transmission from bats to other mammals is critical for mitigating the potential for coronavirus outbreaks in humans and non-avian domestic animals in the future.
Host tropism in coronaviruses is primarily mediated by the receptor-binding domain of the spike gene,26 , 27 but other factors, such as host cell proteases and the S2 domain of the spike gene, have been shown to play a role as well.28 , 29 , 30 , 31 , 32 Coronaviruses are hypothesized to expand or alter their range into more distantly related hosts (bat and non-bat) via mechanisms such as receptor-independent entry,33 , 34 adaptation to intermediate hosts,35 , 36 , 37 and genomic recombination38 (Box 1). Signatures of recombination within coronavirus genomes are extremely common, and a single genome can have evidence of mosaic ancestry in multiple genomic regions and from different evolutionary sources for each region.39 , 40 Growing evidence points to the frequent occurrence of recombination between coronaviruses as one of the driving forces of host switching and disease emergence of coronaviruses into novel host species, particularly when it occurs in the spike gene.39 , 40 , 41 Nearly all of the major human coronaviruses have evidence of spike recombination in their evolutionary history (SARS-CoV-1,42 , 43 , 44 MERS-CoV,18 , 45 HCoV-229E,46 HCoV-NL63,46 , 47 HCoV-HKU1,48 and HCoV-OC4349). Many coronaviruses responsible for epidemics in domestic and companion animals also have an evolutionary history of recombination in the spike gene (SECoV,50 CaCoV,51 , 52 and FeCoV53 , 54), strengthening the evidence that recombination may be associated with cross-species coronavirus spillover.
Box 1. Acronyms, terms, and definitions.
gRNA (genomic RNA): a full-length genomic RNA molecule including the 5ʹ and 3ʹ ends of the genome and all existing sequence between.
sgRNA (subgenomic RNA): an RNA transcription molecule where the 5ʹ UTR sequence (the “leader”) of the genome is transcribed until being interrupted at a transcription regulatory sequence, after which transcription continues at a discontinuous downstream gene (the “body”) and ends at the 3ʹ UTR and polyA tail.
dRNA (defective RNA): an RNA molecule including the 5ʹ and 3ʹ ends of the genome but missing internal segments of sequence.
TRS (transcription regulatory sequence): a conserved set of 8 nucleotides which, when encountered during transcription, induce template switching of the polymerase and production of sgRNAs.
Homologous recombination: recombination occurring between two parental viruses such that the crossover occurs at homologous genomic positions between the two viruses.
Breakpoint: the crossover point between two parental genomes in a homologous recombination event.
Non-homologous recombination: recombination occurring between one or two parental viruses such that a crossover occurs at non-homologous genomic positions.
Junction: the crossover point where discontinuous regions of the genome are brought together; may be a leader-body junction in a sgRNA or a deletion junction in a dRNA or in a non-homologous recombinant gRNA.
Intra-molecular recombination: recombination occurring within a single molecule of RNA; typically mediated by secondary structure which is required to bring distant regions of the genome into close enough proximity for RNA-RNA interactions to occur.
Inter-molecular recombination: recombination occurring between two different molecules of RNA; typically mediated by RNA-RNA interactions between two different molecules or by RNA-protein interactions bringing two molecules into close proximity.
Intra-species recombination: recombination occurring between two different strains of virus within the same species.
Inter-species recombination: recombination occurring between two different species of virus.
Copy-choice recombination: a mechanism of recombination where the replicating polymerase undergoes template switching from one RNA molecule to another.
Similarity-essential recombination: a proposed molecular model of copy-choice recombination where the polymerase pauses or dissociates during replication of one parental strand and the nascent strand being synthesized is free to hybridize with a second parental strand in regions of high sequence complementarity; the polymerase then continues replication on the second parental strand.67
Similarity-nonessential recombination: a proposed molecular model of copy-choice recombination where similarity between the parental strands is not required; rather, features such as secondary structure or polymerase binding sites promote dissociation of the polymerase and template switching between parental strands.67
Similarity-assisted recombination: a proposed molecular model of copy-choice recombination where a combination of similarity and other features such as secondary structure interact to promote recombination.67
Although recombination may facilitate spillover, it is not strictly required nor does it universally increase the potential for cross-species transmission to occur. For example, there is disagreement in the literature concerning the role of recombination in the emergence of SARS-CoV-2. Some studies suggest evidence of recombination in the receptor-binding domain of the spike gene of SARS-CoV-2 with a closely related pangolin virus,55 , 56 while others, such as Boni et al., offer evidence that recombination was not a prominent force in shaping the evolutionary history of the SARS-CoV-2 spike.57 Lytras et al. and Temmam et al. have also offered evidence that suggests recombination between SARS-CoV-2 and a pangolin virus may not have actually occurred.58 , 59 Even if recombination did occur with a pangolin virus in the receptor-binding domain of the SARS-CoV-2 progenitor, it likely did not result in a receptor-binding phenotypic change, as the pangolin coronaviruses have also been demonstrated to efficiently utilize ACE2.60 That ACE2 usage is an ancestral trait of SARS-CoV-2-like viruses is also supported by Boni et al.,57 Wells et al.,61 and Starr et al.62 Importantly, while there is no evidence that recombination contributed to the emergence of SARS-CoV-2, this does not mean that recombination has not occurred elsewhere in the genome or at finer phylogenetic scales, such as between different SARS-CoV-2 variants circulating in humans.
Although there is limited evidence that recombination was associated with SARS-CoV-2 spillover, there is compelling evidence supporting a role of recombination in the emergence of SARS-CoV-1 in 2003. Wells et al. suggested that the SARS-CoV-1-like clade of viruses ancestrally did not have the ability to utilize ACE2, instead gaining this trait on more than one occasion via recombination.61 SARS-CoV-1-like viruses that cannot use ACE2 have broadly been shown to have little to no capacity for human infection.63 Thus, the implication is that recombination may have been a critical factor in the eventual emergence of SARS-CoV-1. Similarly, recombination is also thought to have contributed to the emergence of MERS-CoV.18 , 64 , 65 Many MERS-CoV-like viruses, including NeoCoV and PDF-2180, have been shown to have limited capacity for infection of human cells despite their phylogenetic proximity to MERS-CoV.18 , 66 Evidence of recombination in the MERS-CoV spike gene suggests that recombination is responsible for the phenotypic change in receptor binding that conferred the capacity to use human DPP4.18 , 66 With other human coronaviruses, such as 229E and NL63, there is clear evidence that recombination in the spike gene has occurred, but whether recombination resulted in altered receptor-binding phenotypes critical for cross-species transmission has not yet been demonstrated.
Despite the clear role of recombination in the evolution of coronaviruses and the growing evidence of its role as a driver of cross-species transmission, how and why recombination occurs in coronaviruses remains largely undescribed. At the most basic level, the molecular mechanism of recombination in coronaviruses is not definitively known. The leading hypothesis is that of “copy-choice” recombination (Box 1 ), whereby, during replication, the viral RNA-dependent RNA polymerase (RdRp) is dissociated from one RNA template before reassociating and continuing replication on a different template, resulting in a nascent RNA molecule that was replicated from two different templates.67 , 68 , 69 The ways in which ecological and evolutionary forces interact to shape which recombinant viruses might eventually be successful after recombination is also a largely unanswered question. But perhaps an even more interesting question is why coronaviruses recombine at all. Quick and efficient repair of deleterious mutations via recombination with functional genomes is one hypothesis for why this mechanism persists over time.70 But beyond repairing replicative mistakes, recombination also has the potential to act as an evolutionary “fast-forward” by quickly shuffling genetic material between vastly different viruses.68 , 71 For the same result to be produced by mutation alone, long spans of time would be needed for selective forces to shape such extensive nucleotide changes, especially considering the high proofreading capacity of coronaviruses. Given that the shuffling of genetic material has a very high probability of producing genomes that are no longer functional, the selective pressure for the underlying mechanism of recombination to be evolutionarily maintained must presumably be very strong.
The recombination pathway
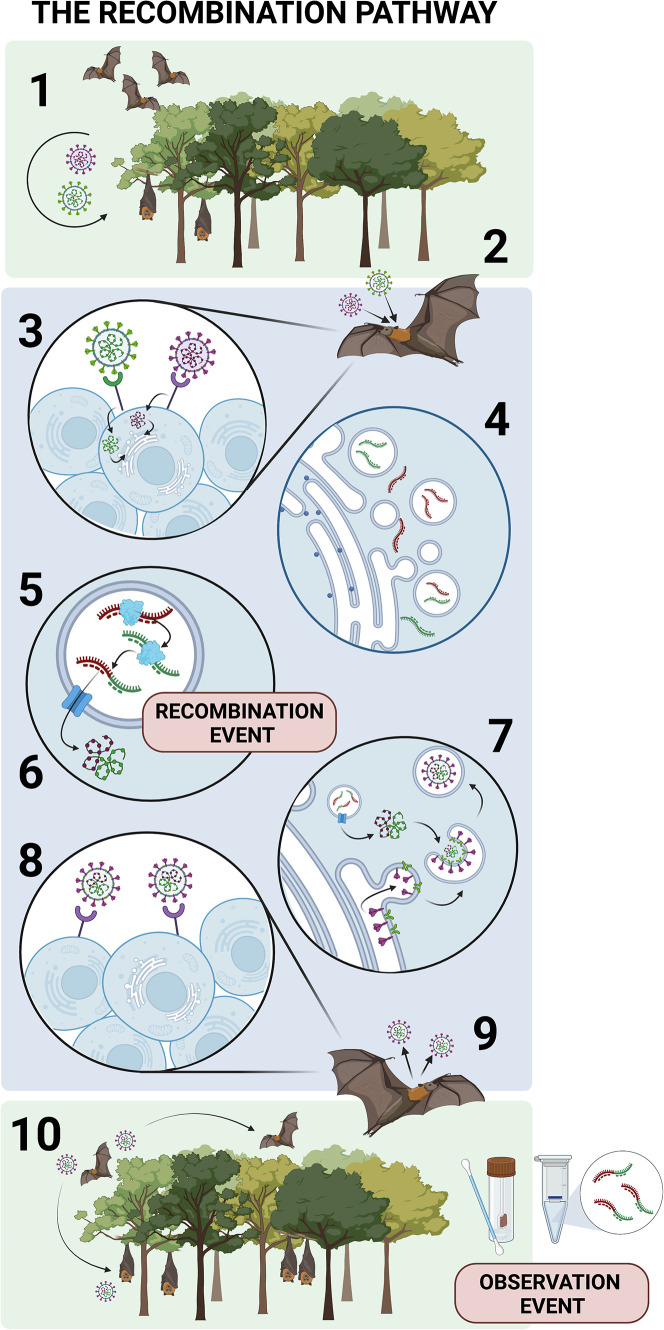
As a hypothetical framework for understanding the various molecular and environmental determinants of coronavirus recombination, the recombination pathway can be considered as a series of barriers that must each be overcome in order for a recombinant virus to be observed in nature. These barriers occur across varying environmental and molecular scales, both before and after the recombination event itself occurs (Figure 1 ). Each step in the pathway differentially impacts the potential for recombination between two viruses to be observed or to even occur in the first place. Broadly, two viruses must first be in the same place at the same time to recombine: in the same host, the same cell, and the same point of replication within the cell. Once barriers to co-occurrence and co-localization have been overcome, two viruses still likely have molecular constraints on being able to recombine. The determinants of such molecular compatibility are currently unknown, but sequence homology or RNA secondary structure are hypothesized to play a role. Once a recombinant RNA genome has been synthesized, it must successfully encapsulate into a virion, escape the cell, and infect enough neighboring cells within the host to amplify to sufficient levels to be shed back into the environment by that host. Finally, the recombinant must infect new host individuals and overcome the forces of natural selection to gain a high enough prevalence in the population such that it has a reasonable probability of being sampled and observed by scientists. Some steps in this pathway are better described than others, but, in general, the factors that distinguish success from failure in all steps of this pathway are poorly understood.
Figure 1.
The hypothetical framework of recombination pathway of coronaviruses
(1) Co-occurrence: two viruses must co-occur in geographic space and share at least one species in their respective host ranges.
(2) Host co-infection: two viruses must co-infect the same individual at the same time.
(3) Cellular co-infection: two viruses must co-infect the same cell at the same time.
(4) Double membrane vesicle (DMV) encapsulation: gRNA from each virus must be encapsulated into the same DMV.
(5) Template switching: the gRNA from each virus must be compatible for RdRp to switch templates during replication. Recombination occurs at step 5.
(6) DMV egress: recombinant gRNA must exit the DMV.
(7) Virion assembly: recombinant gRNA must be encapsulated into a virion.
(8) Cell-to-cell transmission: the recombinant virus must infect neighboring cells.
(9) Viral shedding: the recombinant virus must be shed by the host to infect new individuals.
(10) Host-to-host transmission: the recombinant virus must transmit to additional hosts.
Only once all 10 steps have been passed can a virus have reasonable probability of being observed during wildlife surveillance studies.
Step 1: Co-occurrence in geographic and host space
At the broadest environmental scale, patterns of recombination are shaped simply by which viruses have the opportunity to occur together in space. Viruses which circulate freely in the same host species will have the highest opportunity for co-infection of a single host individual, but viruses with differing host ranges that occur in the same geographic space may still recombine if it is possible for different hosts in that space to share viruses. Studies have shown that coronaviruses have strong co-phylogenetic associations with the genus of their associated chiropteran hosts but that this concordance tends to break down at the host species level.72 , 73 This can clearly be seen in the example of the coronavirus subgenus Sarbecovirus, which is strongly associated with the host genus Rhinolophus but has almost no co-phylogenetic signal for specific species of bats within this genus.61 , 73 This suggests that closely related viruses may be frequently switching between species of bats from the same genus, which would increase opportunities for recombination between them. Frequent host switching of coronaviruses in bats is well-documented14 , 72 , 73; though coronavirus co-occurrence does not have to be exclusively in bats in order for recombination to occur. Multiple types of coronaviruses thought to have evolutionarily originated in bats are known to circulate in several species of domesticated animals (e.g., TGEV, PEDV, and SECoV in pigs), providing novel opportunities for coronavirus co-occurrence outside their bat reservoir hosts.
Steps 2 and 3: Co-infection of a single individual and a single cell
Not only must two viruses be circulating in the same host species but they also must co-infect the same host individual and replicate within the same cell to recombine. There are multiple barriers to recombination in this respect. The probability of co-infection of a single host will depend on the prevalence of each virus within a population, with the greatest probability occurring when prevalence of both viruses is high. This can be shaped by differences in seasonality between viruses, by the size of the host population, or by population-level cross-reactive immunity induced against the second virus after infection with the first.
Furthermore, once the probabilistic barrier of infecting a single host has been overcome, the two viruses must then infect the same cell within that host individual. Tissue tropism is variable within coronaviruses,74 meaning that many pairs of viruses will never have any opportunity for recombination if one replicates exclusively in the respiratory tract and the other in the gastrointestinal tract, for example. The spike proteins of both viruses must be compatible with molecular receptors on the surface of the host cell, which can be complicated when the viruses utilize different receptors altogether. The timing of the two infections may also restrict the potential for recombination, especially if one virus has had the opportunity to establish a robust infection before the second virus is able to infect the same host. Superinfection exclusion, a phenomenon where a virus actively prevents infection by a second virus, is a well-documented but poorly understood mechanism that may further limit the potential for co-infection of a single cell.75 , 76
Step 4: Co-localization in a double membrane vesicle
Like all positive-strand RNA viruses, coronaviruses induce organelle membrane changes in the host to form replication organelles.77 , 78 , 79 Viral proteins nsp3, nsp4, and possibly nsp6 cause convolution of the endoplasmic reticulum membrane and the formation of double membrane vesicles (DMVs),80 which encapsulate viral genomic RNA (gRNA) in the cytoplasm and protect it from host recognition and degradation during replication.81 Thus, if two viruses have managed to infect the same cell at the same time, gRNA from each virus must also co-localize to the same DMV in order for recombination to occur between them during replication or transcription. The mechanism by which gRNA gets inside the DMVs is unknown, and could be either an active (e.g., transport of gRNA from the cytoplasm through a transmembrane pore after DMVs have been formed) or a passive mechanism (such as encapsulation of gRNA in the cytoplasm during the DMV formation process). The timing of co-infection could also significantly impact the potential for co-localization in the same DMV if the first virus is much later in its replication cycle than the second.
Step 5: Polymerase template switching
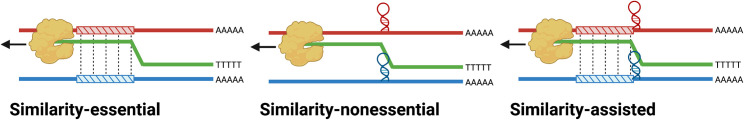
The precise mechanism by which RdRp template switching occurs during coronavirus replication and transcription is currently unknown. There are three hypothetical models for this mechanism: similarity-essential recombination, similarity-nonessential recombination, and similarity-assisted recombination67 (Figure 2 ). In similarity-essential recombination, complementary base pairing or sequence homology is required for polymerase switching between the template and nascent strands. Conversely, similarity-nonessential recombination does not require sequence similarity but instead relies on other signals to trigger template switching, such as RNA secondary structure or signaling sequences. Similarity-assisted recombination can be considered a hybrid model where both sequence similarity and additional triggers, such as secondary structure, play a role in directing recombination. Under all mechanistic hypotheses, a resulting recombinant molecule will be homologous if the polymerase reassociates with the second template in the same genomic position and non-homologous if it does not (Box 1).
Figure 2.
Three proposed molecular mechanisms for RNA recombination
(A) Similarity-essential recombination, where the polymerase dissociates, and the two parental strands hybridize in regions of high homology, allowing the polymerase to switch templates.
(B) Similarity-nonessential recombination, where the polymerase dissociates because of RNA secondary structure.
(C) Similarity-assisted recombination, where both homology and secondary structure promote recombination. Adapted from Nagy and Simon.67.
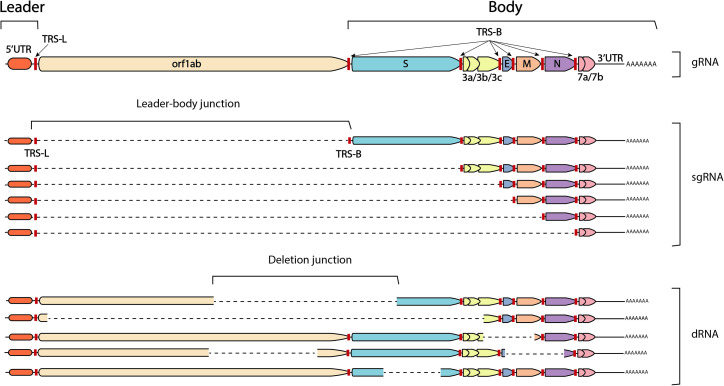
During transcription, coronaviruses generate sub-gRNAs (sgRNA), which are a set of nested co-terminal RNAs encoding structural and accessory proteins82 , 83 (Figure 3 ). Coronaviruses also generate defective RNAs (dRNAs) during replication, also referred to as “defective interfering” RNA (DI-RNA) or “defective viral genomes” (DVGs) in the literature.84 , 85 , 86 It is thought that sgRNAs are generated via a process of intra-molecular non-homologous recombination during negative strand synthesis, whereby the viral polymerase pauses and dissociates from the RNA template once reaching a transcription regulatory sequence (TRS)-B before reassociating downstream at the TRS-L of the same molecule, finishing synthesis at the 5′ leader sequence87 , 88 (Figure 3). Interestingly, sgRNAs can also act as replication templates and can be the source of additional recombinant RNAs, such as smaller sgRNAs or dRNAs.89 dRNAs are also thought to arise by a mechanism of non-homologous recombination,84 , 85 , 86 , 90 but the genomic signal for the dissociation of the RdRp in this case is unknown.91
Figure 3.
The structures of genomic RNA (gRNA), subgenomic RNA (sgRNA), and defective RNA (dRNA)
Colored polygons represent ORFs and are labeled by gene on the gRNA above. Transcription regulatory sequence (TRS) signals are marked with red lines. Dotted lines indicate regions where genomic sequence is missing at either leader-body (sgRNA) or deletion (dRNA) junctions. For sgRNA, junctions are always between the TRS-L and a TRS-B. For dRNA, junctions can be randomly located throughout the genome and vary in size.
During sgRNA formation, complementary base pairing is required to form a structural complex that initiates leader-body joining, forming between an exposed RNA loop containing the TRS-L on the positive-sense gRNA template and the nascent negative-sense TRS-B being synthesized.92 , 93 The sequence similarity of TRS sites and secondary structure required for sgRNA transcription is reminiscent of similarity-assisted recombination,67 where sequence homology in combination with RNA secondary structure direct the RdRp to dissociate from its template. This similarity has led to the tempting hypothesis that the TRS sequences required for the generation of sgRNAs may also play a role in homologous and non-homologous recombination of gRNAs or dRNAs as well.94 , 95 , 96 Despite the clear link between TRS signaling and sgRNA production during transcription, it is not known whether the mechanism resulting in homologous recombinant gRNA occurs via a similar process. It is also possible that similarity-assisted recombination is contingent on complementary sequences other than TRSs.
The mechanism of recombination is also either an intra-molecular mechanism, where a single molecule of RNA folds back on itself to facilitate polymerase template switching, or inter-molecular, where the polymerase separately associates with two entirely different RNA molecules. The production of sgRNAs is known to be an intra-molecular mechanism facilitated by complementary base pairing between TRSs, but it is not known whether non-homologous recombination of dRNAs is occurring via a similar intra-molecular mechanism or via an inter-molecular mechanism requiring two different molecules of RNA. Conversely, by definition, homologous recombinant gRNA requires two different parental viruses and therefore two different RNA molecules. Thus, similarities may exist in the generating mechanisms of sgRNAs, dRNAs, and homologous recombinant gRNAs, but there are parts of these mechanisms that we must infer are different. As homologous gRNA recombination is likely one of the key forces in cross-species transmission, understanding this specific mechanism is of particular importance. The roles of TRS sites, sequence homology, and secondary structure in acting as barriers to homologous gRNA recombination, if any, remain to be determined.
Steps 6 and 7: DMV egress and virion assembly
Replicated gRNA from within DMVs must return to the cytoplasm to be packaged into a forming virion during the process of virus assembly. Transcribed sgRNAs also must be transported to the cytoplasm for translation. In this case, active transport through a transmembrane pore is a requirement to get to the cytoplasm from within the DMV. Viral protein nsp3 is known to form this pore in the walls of DMVs.97 It may be the case that the nsp3 pore will transport any molecule of RNA from the DMV to the cytoplasm, but if the transportation mechanism is partial to particular signals on specific RNA molecules, then the signals of any recombinant RNAs must be compatible with the nsp3 pores in the DMV. If, for example, a recombinant gRNA inherited a transportation signal from the first parental virus but the nsp3 pores on the DMV are formed by proteins from the second, it is possible that this incompatibility will prohibit the recombinant gRNA from exiting the DMV.
In the event that recombinant genomes are able to escape the DMV, additional barriers may then limit packaging of these genomes into new virions. The molecular mechanisms and exact sequence of events involved in coronavirus RNA packaging have yet to be fully determined98; however, it has been established that specific packaging signals present in gRNA interact with the viral nucleoprotein (N) and/or the membrane (M) protein to drive the preferential packaging of gRNA.98 , 99 These packaging signals are not conserved across the coronavirus family and can also vary in their genomic location.98 Limited evidence suggests that closely related viruses (i.e., those within the same subgenus) may have enough conservation in their packaging signals to be functionally interchangeable and allow packaging of heterologous gRNAs,100 while more distantly related viruses are less likely to package gRNA from a heterologous virus.101 In the context of a co-infected cell where two sets of N and M proteins are available, it is conceivable that recombinant genomes would be packaged by whichever N or M is most compatible with the packaging signal present on the recombinant genome. In this case, packaging may not be a significant post-recombination barrier. However, in the event that recombination breakpoints interrupt packaging signals, or if more than one specific packaging signal is required but not contained (in cis) in the same recombinant genome, then functional restrictions imposed by gRNA interaction with structural proteins may prevent packaging into a new virion.
Steps 8 and 9: Cell-to-cell transmission and viral shedding
A virion with a recombinant genome would have the functionality to infect at least one neighboring cell, as the proteins on the surface of the virion were generated during infection of the previous cell by non-recombinant parental RNAs. Once the recombinant gRNA has entered the cell, however, the recombination breakpoint within the genome must be in a position that does not interfere with the function of that genome as it begins a new cycle of replication. Given the many epistatic interactions both within and between genes in the coronavirus genome, a breakpoint that lies between critical interacting regions could easily render a recombinant genome non-functional in this respect. For example, the nonstructural proteins within orf1ab that are first transcribed and translated need to be able to facilitate cellular changes required for replication and transcription of the structural genes, such as the formation of DMVs via interactions between nsp3, nsp4, and nsp6.
Even if replication and transcription are not interrupted, functions which are critical for continued transmission of the recombinant virus may be impaired. For example, interactions between structural proteins required for virion assembly could be affected if structural genes inherited from the opposite parent are not compatible with one another, which has been shown between the M and S proteins.102 It also goes beyond protein-protein interactions to include the conservation of protein-RNA interactions. For example, Kuo et al. showed that by substituting the C-terminal domain of the mouse hepatitis virus (MHV) N gene with that of SARS-CoV-1, the ability of the N protein to interact with gRNA could be abolished.101 Thus, while it is conceivable that a recombinant gRNA with a breakpoint in the N gene could be generated in step 5 and even packaged into virions in steps 6 and 7, its ability to interact with the packaging signals found elsewhere on the recombinant gRNA will not be tested until it has been transmitted to a new cell.
Even still, if the recombinant virus remains functional, it must continue transmitting to additional cells and amplify to sufficient levels to be shed by the host. In this regard, it must compete with both parental viruses for cellular resources, which have already had ample time to establish robust infections and will far outnumber the recombinant. Even the slightest deleterious trait of the recombinant virus could inhibit it from ever being able to exit the host, despite its functionality. Even a recombinant virus of equal fitness to the parental viruses has a steep hill to climb to be shed by its host.
Step 10: Host-to-host transmission and population spread
Finally, a functional recombinant virus has emerged from its host, but more barriers await to inhibit its success. Although recombination may not have hindered cell-to-cell transmission, additional traits required for environmental transmission must also remain intact in the recombinant for continued spread. The environmental conditions in which the recombinant virus emerges will also have a significant impact on its continued transmission. For example, unless recent host switching into a naive population is involved, the recombinant virus is likely emerging into a population with some degree of widespread immunity against one or both parental viruses, which have already reached sufficient levels of prevalence to co-infect the same individual. Unless the recombination event has resulted in a large enough antigenic shift away from the parental viruses, such a landscape will result in strong competition for immunologically naive hosts. Even in the absence of competition, it is also quite possible that a recombinant virus in this situation could simply be lost to genetic drift because its population size is so small. Recombinant viruses that are most likely to succeed under competitive circumstances with small population sizes are those for which the recombination event has conferred a strong selective advantage over both parental viruses.
Existing research on coronavirus recombination
Comparative genomics
Studies comparing genomic sequences to detect recombination between coronaviruses can be found in the literature spanning back to the 1990s.53 , 103 , 104 , 105 Once the recombinant origin of SARS-CoV-1 had been established,106 , 107 however, focus began to shift toward detecting recombination at a much broader scale, between coronavirus genomes that were not of the same species.43 , 108 The consensus that bats were the evolutionary origin of alpha- and betacoronaviruses also solidified in the late 2000s,11 , 13 spurring an increase in bat virus surveillance and the generation of novel genomes and more detections of recombination.42 , 109 , 110 Similarly, the emergence of MERS-CoV in 2012 also brought increased attention to coronaviruses, recombination, and the utility of comparative genomics,18 and small-scale comparative genomics studies that identified specific recombination events within single genomes continued to be relatively frequently published in the following years.46 , 111 , 112 , 113 , 114
It was not until the emergence of SARS-CoV-2 that comparative genomics studies began to examine patterns of recombination at an even broader scale. By compiling recombination data across entire classes of coronaviruses, these studies intended to illuminate broad, emergent patterns of recombination. One of the first genus-wide comparative genomics studies was published by Bobay et al. in 2020.115 Rather than using typical similarity-based recombination detection methods, they use a previously unpublished method that tests observed shared polymorphisms against a null model where sequence similarity can only arise via convergent mutations to determine where recombination likely occurred. Using this strategy, they found that recombination is probably occurring exclusively between viruses in the same betacoronavirus subgenus. They also identified that recombination is strongly overrepresented in the spike gene within the Sarbecovirus subgenus. A finding of increased recombination signals in the spike gene of sarbecoviruses was similarly demonstrated by Lytras et al. using the RDP5 program for recombination detection.58 Forni et al. also examined natural patterns of recombination on a set of complete betacoronavirus genomes using the 3SEQ detection method,116 and they also found an increase in recombination in the spike gene. Additionally, they show that breakpoints tend to occur as pairs within genomes, such that an entire section of a genome is recombined in a modular manner. Forni and coauthors conclude that, generally, the majority of recombination events across coronavirus genomes are shaped by the viability of the recombinant genome and natural selection rather than in a mechanistic manner, which is similar to conclusions raised by Bobay et al. Yang et al. also performed comparative genomics using a dataset of alpha- and betacoronavirus coronavirus genomes using RDP4. The authors had the specific intention of examining the relationship between TRS sequences and recombination.96 They found a statistical association between the position of the TRSs and the distribution of breakpoints over what would be expected by chance, but not all breakpoints could be explained by a proximally positioned TRS.
In perhaps the most comprehensive comparative genomics study to date, de Klerk et al. also identified recombination events in a large dataset of alpha- and betacoronavirus genomes using RDP5.117 They found variable rates of recombination across the genome for all coronavirus subgenera they examined; in particular, they show that the S gene appears more recombinogenic than other regions of the genome. Similar to the findings of Yang et al., the authors found a statistical association between TRSs and breakpoints compared with what would be expected by chance. However, they also found a significant increase in the presence of breakpoints immediately upstream of genes. As TRSs also lie immediately upstream of genes, whether the TRS is involved or whether natural selection prefers recombination events that do not interrupt genes is unclear. In addition, there are a vast number of recombination events that do not occur in proximity to any TRS, such as at the S1/S2 junction, which was also identified as a potential “hotspot” of recombination by de Klerk and coauthors. In this study, the authors also investigated the association between sequence similarity and breakpoint positions, but they were unable to identify a significant relationship. They did, however, identify a hotspot cluster at the 5′ end of the genome where secondary structure is highly conserved, which perhaps offers some support to a similarity-nonessential model of recombination.
Together, these studies highlight strong evidence for increased rates of recombination in the spike gene and at positions where gene function is least likely to be interrupted. It is critical, however, that findings of comparative genomics studies are interpreted in light of the multiple environmental and molecular factors involved in shaping the naturally observed viruses that are used for these studies. Inferring that any patterns identified are attributable to the molecular mechanism of recombination is highly confounded by the fact that the environment also plays a role in shaping which viruses have the opportunity for recombination and which resulting recombinants will successfully amplify in a population and eventually be observed, which each group of authors acknowledge in their findings. For example, it is unclear whether a hotspot of recombination in the spike gene is the result of a mechanistic predisposition (step 5 in the pathway) or of natural selection favoring spike recombinants (steps 8–10). Likewise, the absence of observed recombinants across certain taxonomic scales cannot be conclusively attributed to a limitation of the molecular mechanism of recombination (step 5), as a lack of co-occurrence between two viruses in the environment before recombination could occur (step 1) and would result in the same pattern (this limitation is also raised in the discussion by Bobay et al.). Thus, comparative genomics studies can be valuable for highlighting hotspots of recombination in the coronavirus genome or for defining taxonomic boundaries beyond which recombination has never been observed, but, ultimately, the distinction between a mechanistic origin versus environmental shaping of patterns of recombination cannot be resolved through comparative genomics approaches alone.
Experimental studies
Despite the important role recombination plays in coronavirus evolution and host switching, recombination between two different coronaviruses has not been studied experimentally in decades. In contrast to comparative genomics, which are essentially investigating patterns produced by the recombination pathway at large, experimental studies of recombination benefit from the ability to isolate the molecular steps in the recombination pathway from environmental ones. By co-infecting viruses in vitro, the requirements for co-occurrence in geographic space, co-infection of the same host individual, and to some extent co-infection of the same cell are circumvented. In this manner, patterns emerging from experimental studies of recombination will much more closely represent the actual molecular mechanism of recombination and not the predispositions of the environment or of post-recombination influences of natural selection.
The majority of the existing literature on in vitro coronavirus recombination is from a series of experiments throughout the 1980s and 1990s that used different strains of MHV (genus Betacoronavirus, subgenus Embecovirus) to investigate patterns of intra-species recombination.88 , 118 , 119 , 120 , 121 , 122 , 123 , 124 , 125 , 126 , 127 , 128 The earliest study of in vitro coronavirus recombination by Makino et al. in 1986118 showed that a temperature-sensitive strain of MHV could be restored during mixed strain infection with temperature-permissive strains and that this event occurred at least five independent times during their experiments. in 1988, Keck et al. followed these experiments with an in vivo study of MHV infection in mouse brain tissue and demonstrated that recombination also occurs not only in culture but also under more natural circumstances.122 Several additional studies of coronavirus recombination using MHV were also performed around this time, and recombination events observed in orf1ab,120 spike (S),124 envelope (E),123 nucleocapsid (N),123 and in the 3′ UTR121 led to the conclusion that recombination was possible anywhere along the genome.126 Recombination was also observed more than once in a single molecule120 and at a rate of up to 25% at high multiplicity of infection (MOI).125 , 128 This rate was soon shown to vary in different regions of the genome, particularly the S gene, where one study by Fu and Baric in 1992 recorded three times the rate of recombination in S compared with orf1ab.124 , 127
The observation of varying rates of recombination along the genome led to the development of a hotspot model of recombination, where the presence of intrinsic molecular signals in the genome increased the odds of recombination at specific sites124; however, the identification of the mechanism responsible for the generation of hotspots has remained elusive. Hypothetical models of recombination proposed by Nagy and Simon in 1997 suggested that either nucleotide homology, RNA secondary structure, or both could potentially act as a molecular signal for recombination.67 Some lines of evidence during this time supported the similarity-nonessential model, where RNA secondary structure but not nucleotide homology define potential sites of recombination.90 , 129 A significant amount of evidence showing the importance of RNA secondary structure for genomic replication and transcription also arose during this time.130 , 131 , 132 Banner and Lai in 1991 went as far as to suggest that recombination was instead an entirely random mechanism at the genomic level and that hotspots were simply an emergent property of the purging of defective recombinants via natural selection.126
In the late 1990s and early 2000s, studies of coronavirus recombination spread beyond studies of MHV, eventually being observed in additional in vitro experiments between two different strains of avian IBV (genus Gammacoronavirus) and in experiments between two different strains of porcine TGEV (genus Alphacoronavirus). Some of the first evidence of recombination between viruses that were more distantly related was the discovery of a naturally circulating recombinant coronavirus in 1998, when FeCoV type II was shown to be the result of recombination between FeCoV type I and CaCoVtype II.53 More evidence that recombination could occur across larger taxonomic scales began to emerge from studies of 5′ and 3′ replication signals, which were shown to be incompatible across certain taxonomic boundaries. Hsue and Masters showed in 1997 that the 3′ UTR of MHV could be replaced through targeted recombination by that of BCoV (also genus Betacoronavirus, subgenus Embecovirus), which shared the same secondary structure but not the same nucleotide sequence, with no loss of viability.131 Wu et al. in 2003 replicated these studies for the 5′ UTR by demonstrating that a BCoV helper coronavirus could successfully replicate MHV.133 Goebel et al. took this one step further in 2003, showing that the SARS-CoV-1 3′ UTR (genus Betacoronavirus, subgenus Sarbecovirus) can also functionally replace that of MHV, but the 3′ UTR of either TGEV (Alphacoronavirus) or IBV (Gammacoronavirus) cannot.134
Taken together, these results demonstrate a clear taxonomic boundary for recombination between coronaviruses of different genera, and it has been suggested that any co-infecting coronaviruses from within the same genus should, in theory, be able to successfully recombine.133 , 134 Though theoretically possible given the studies by Goebel et al., inter-species homologous recombination between betacoronavirus subgenera has not yet been observed in nature,96 although, interestingly, inter-species recombination has been shown to occur across subgenera in alphacoronaviruses.24 In contrast with the Alphacoronavirus subgenera, which each have the same arrangements of accessory proteins, each Betacoronavirus subgenus has a different genomic structure and accessory proteins,135 which perhaps has some bearing on their potential for recombination. Incompatibilities between 5′ and 3′ UTRs may act as a barrier for recombination between genera, while incompatibilities mediated by genome structure or accessory proteins may introduce an additional barrier to recombination for the betacoronaviruses.
After the mid-2000s and the emergence of SARS-CoV-1, studies examining undirected in vitro recombination between two different coronaviruses did not appear in the literature again. Recently, a study by Gribble et al. examined recombination via the production of dRNAs; however, their study did not assess recombination between two co-infecting viruses.95 Instead, they individually cultured SARS-CoV-2, MERS-CoV, and MHV and examined the resulting dRNAs (i.e., non-homologous recombination events). They found some evidence of “microhomology,” where the ∼2–7 bp flanking deletion junctions showed higher than expected sequence similarity. They also showed that, statistically, there is evidence that non-homologous recombination near TRSs is more frequent than would be expected by chance; however, TRSs or complementarity alone could not explain all non-homologous recombination. It is not known whether the non-homologous, single-virus recombination studied by Gribble et al. and the homologous recombination that occurs between two co-infecting viruses are occurring via the same molecular mechanism. Thus, the roles of homology, secondary structure, and TRSs in homologous inter-species coronavirus recombination still remain unclear.
The future of coronavirus recombination research
The limitations of comparative genomics support a clear need for additional experimental evidence of recombination to resolve the confounded relationship between molecular mechanism and environmental pressure in shaping patterns of coronavirus recombination. Nearly all existing experimental studies, whether recent or not, are limited strictly to the study of intra-species recombination. Those involving more than one species have used sequences that were generated with targeted recombination,131 , 133 , 134 , 136 , 137 , 138 where recombinants can be purposefully designed to have genetic components such as the 3′ or 5′ UTR from heterologous viruses and their functionality can then be assessed. Undirected recombination, where two species of coronaviruses are allowed to freely recombine, has not been performed. We know, however, that inter-species recombination is quite common within the coronavirus family due to the vast number of recombinant virus genomes that have been found naturally.18 , 21 , 42 , 43 , 45 , 46 , 48 , 49 , 50 , 51 , 52 , 54 , 61 , 139 , 140 , 141 , 142 , 143 The uncertainty of the molecular boundaries of recombination, such as incompatibility between different taxonomic groups or requirements for specific genomic sequence similarities, can ultimately only be ascertained experimentally, and thus this direction of further research promises to provide a wealth of important knowledge.
Early experimental evidence also has relied completely on PCR (which has low sensitivity for detecting recombination unless primers are designed to specifically span a recombination breakpoint) for the identification of recombination events. There is much to be gained from more thorough analyses with next-generation sequencing, where current technology allows sequencing of single RNA molecules without amplification and even sequencing of RNA molecules from within a single cell. This vastly expands the types of recombination events we have the ability to observe in vitro and the patterns that might be possible to identify.
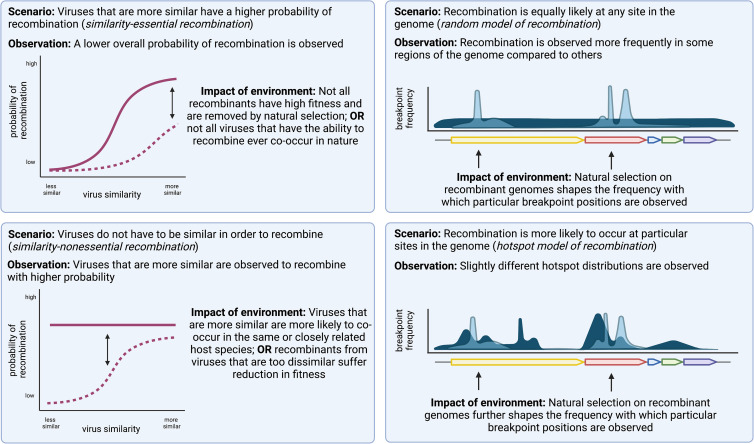
Perhaps paradoxically, experimental patterns of recombination in vitro also allow for insights into the environmental pressures shaping recombination patterns in nature, even though recombinant RNAs detected in laboratory experiments essentially represent patterns generated only by steps 3–5 of the recombination pathway. A recombinant RNA presumably does not have to successfully exit a DMV, assemble into a virion, or be functional for replication to be observed, particularly when single-molecule sequencing is employed. Essentially, this kind of experimental design isolates almost as closely as possible the “true” molecular potential of recombination between two viruses, without any post-recombination barriers filtering what is ultimately observed. Using this framework, by effectively subtracting patterns observed in laboratory settings from recombination patterns in nature, we can attribute the differences directly to environmental circumstances or post-recombination natural selection (Figure 4 ).
Figure 4.
Hypothetical scenarios of coronavirus recombination and possible observations in nature associated with each
Left: observations in nature of the probabilities of recombination between two viruses are impacted by natural selection and environmental co-occurrence in a similarity-essential model of recombination scenario (top) and a similarity-nonessential model of recombination scenario (bottom).
Right: observations of recombination breakpoints across genomic positions are impacted by natural selection in a random model of recombination scenario (top) and a hotspot model of recombination (bottom).
For example, in a scenario where recombination occurs via a similarity-essential mechanism, we would expect to see higher rates or probabilities of recombination between viruses that are genetically more similar in laboratory experiments. In nature, we would expect to see much the same pattern, but greatly reduced in strength because of the barriers that exist for recombinants to be successful after recombination occurred. Conversely, in a similarity-nonessential mechanism, we might expect to see a relatively uniform probability of recombination, regardless of genetic similarity between the two viruses being tested. If in nature, we instead observe a pattern such that closely related viruses appear to recombine more frequently than distantly related ones, it can be inferred that the driver of this pattern exists outside of steps 3–5. It may be such that closely related viruses tend to occur in closely related species and have higher chances of crossing species barriers and co-infecting the same host, making recombination between them appear more frequent. Alternatively, recombinants between viruses that are quite distantly related may be suffering strong fitness deficits and being filtered out post-recombination by natural selection. Critically, further ecological experiments have the potential to answer whether closely related coronaviruses are more likely to co-occur, which would further whittle down whether the patterns we observe are driven by natural selection or not.
The same theoretical framework applies for investigation of a hotspot model of recombination versus a random model of recombination. If recombination is indeed random, experimental evidence will show relatively equal frequencies of recombination breakpoints across the entire genome. Thus, hotspots of recombination observed in naturally occurring viruses can be directly attributed to the effects of post-recombination natural selection and provide valuable insight into the types of recombination events that confer selective advantages. An observed increase in recombination in the spike gene, for example, would be clear evidence that this type of event has strong evolutionary implications. One can also imagine that, even if experimental evidence supports a hotspot model of recombination where events are directed to particular regions of the genome via some type of signaling, there will still likely be differences in hotspot distributions in the lab compared with those observed in natural viruses and still much to be inferred about post-recombination selective pressure. Taken together, this framework supports that a range of experimental evidence of recombination in coronaviruses in vitro in combination with continued broad comparative genomics studies will be immensely valuable in illuminating barriers of recombination across multiple steps of the recombination pathway.
In light of the COVID-19 pandemic and increased scrutiny on coronavirus research, we emphasize here that coronavirus recombination research can be safely undertaken and remain firmly outside of gain-of-function territory. Careful consideration should be given to which pairs of viruses are chosen for study as well as to any necessary and appropriate safety precautions. For viruses that have not been assessed for the potential for human infection (i.e., uncharacterized bat coronaviruses), we recommend higher biosafety precautions, such as performing work under Biosafety Level 3 (BSL-3) containment. Perhaps more importantly, cultures in which viruses have been co-infected to allow for recombination can be chemically inactivated, thus eliminating the potential for any novel recombinant coronaviruses to remain infectious. This of course does not allow for investigation of many of the molecular post-recombination barriers, such as the functionality of the recombinant genome, which we suggest could be more safely evaluated using other methods.
Informing pandemic predictive intelligence
Genomic recombination facilitates rapid, large-scale shuffling of genotypes in coronaviruses, often resulting in viruses with novel combinations of phenotypic traits. When it comes to predicting spillover risk, recombination poses the particular challenge that a viral community at one time point may be entirely different at another, with evolution occurring in multiple, nonlinear directions in a short period of time. Thus, even if surveillance were to capture all viruses within a community at a given time and characterize the risk of each one causing a pandemic in humans, the true overall risk of a virus with pandemic potential occurring in that population would not be fully realized because of the potential for large genotypic shifts to generate such a virus at any time in the future. Assessing risk on any scale beyond that of a single virus must consider the additional evolutionary dimension that recombination allows within a group of co-circulating viruses. The ability to predict the potential for recombination between any two given viruses will be crucial for evaluating the total risk at larger scales.
Prediction of recombination potential between any two viruses requires a much more complete understanding of the recombination pathway. Perhaps most importantly, the molecular mechanism by which recombination occurs must be conclusively elucidated. With a more accurate picture of how recombination is occurring at the molecular level, we can begin to build statistical models predicting the probability of recombination between any two viruses. This is particularly important when coronaviruses with known potential for human infection overlap in geographic space and host range with coronaviruses that do not—any co-occurring coronavirus could presumably gain this ability nearly immediately through recombination with a human-infecting virus. The ability to predict the potential for recombination between co-occurring viruses would thus be invaluable in targeting surveillance efforts to geographic hotspots of recombination where novel human-infecting recombinant viruses are most likely to arise. This of course requires an expansive knowledge of where coronaviruses are currently circulating, underscoring the need for continued broad-scale wildlife coronavirus surveillance.
With the recent emergence of SARS-CoV-2, we have learned that the sarbecoviruses are a perfect example of why this type of knowledge and surveillance is so important. SARS-CoV-2 and the discovery of other SARS-CoV-2-like viruses in bats and pangolins highlighted that ACE2 usage is an ancestral trait in the SARS-CoV-2 clade57 , 61 , 62 and that a virus from this clade likely transferred the ACE2 usage trait to SARS-CoV-1-like viruses via recombination,61 leading to the emergence of SARS-CoV-1 in 2003. Notably, all closest known non-ACE2-using relatives of SARS-CoV-1 and many of the ACE2-using relatives of SARS-CoV-2 have all been found in or near Yunnan Province in China,43 , 61 , 111 , 112 , 144 demonstrating that viruses that use ACE2 indeed overlap with viruses that do not in geographic space. The majority of sarbecoviruses are also found in closely related rhinolophid hosts,42 , 73 , 145 demonstrating a strong potential for overlapping host ranges as well. Given that the recombinogenicity of sarbecoviruses is already well established,57 , 61 , 96 , 117 , 146 there is extremely strong evidence supporting that continued sarbecovirus surveillance in and around this region of Southeast Asia is critical. Once broad-scale surveillance can provide a more detailed picture of the sarbecoviruses that currently exist, where they circulate, and in which hosts, we can begin to identify interfaces that are at the highest risk of producing recombinant viruses, and specifically those that may have the capacity to use ACE2 and infect humans. Identifying potential hotspots of recombination at such a fine scale can vastly increase the effectiveness of aggressive surveillance efforts.
Predictive intelligence about coronavirus recombination will also be valuable for designing surveillance strategies for coronaviruses in domesticated animals. In pigs, for example, SECoV is known to have arisen from a recombination event between TGEV and PEDV.50 The knowledge that multiple coronaviruses were circulating in pigs has been known for some time, but the potential for recombination between them was unrealized until it had already occurred. Similar circumstances exist surrounding feline and canine alphacoronaviruses co-circulating in cats and dogs, which have recombined to form new serotypes multiple times.51 , 53 , 54 Recombination is also thought to have contributed to the emergence of MERS-CoV,18 , 65 , 66 potentially facilitated by co-occurrence of different coronaviruses in camelids.45 , 147 Monitoring for co-occurring coronaviruses in domesticated animals will be a far easier task than surveillance in wildlife, and preventing coronavirus recombination in domesticated animals has strong implications for the health of both humans and animals.
Surveillance and prediction of recombination in coronaviruses will be a dynamic and continuously evolving endeavor. This is highlighted by the broad host range of SARS-CoV-2, which has been shown to infect a multitude of wildlife and domesticated animal species.148 , 149 , 150 The spread of SARS-CoV-2 across the globe has given a sarbecovirus unprecedented access to novel host species outside of the Rhinolophus genus, such as white-tailed deer151 , 152 and mink.153 , 154 Many of these hosts harbor their own species of coronaviruses, such as mink coronavirus (genus Alphacoronavirus, subgenus Minacovirus)155 and strains of BCoV (genus Betacoronavirus, subgenus Embeccovirus) in white-tailed deer,156 , 157 each of which has unknown recombinogenic potential with SARS-CoV-2. Mink coronavirus is an alphacoronavirus and BCoV is in the betacoronavirus subgenus Embecovirus, and given what we currently know about the taxonomic boundaries of recombination, neither are likely to recombine with SARS-CoV-2, which is a sarbecovirus. However, this example highlights how quickly the host range of a virus can change and massively impact the ecological circumstances of co-circulating viruses, especially when anthropogenic forces are at play. If and when new coronaviruses emerge in humans, the astronomical increase in access to novel hosts facilitated by pandemic spread will be an important factor to monitor to keep track of the potential for recombination with other existing coronavirus species.
Ultimately, surveillance alone will be insufficient in measuring the true risk of the pandemic potential of a community of co-occurring viruses because there will always be continued generation of novel combinations of genotypes via recombination. Only in combination with predictive recombination intelligence can we begin to fully explore the evolutionary dimension of spillover risk. But before the potential for recombination between two co-occurring coronaviruses can be accurately predicted, the molecular mechanism of recombination must be more definitively known. Some of the biggest remaining unanswered questions are (1) whether sequence homology is indeed required for recombination, (2) whether it occurs randomly across the genome or not, and (3) where epistatic incompatibilities are most likely to render recombinants non-functional. Comparative genomics as a method of study for coronavirus recombination offers a quick, inexpensive, and comprehensive solution toward that goal, but ultimately will never be a tool that can definitively separate the molecular mechanism from ecology and natural selection. The barriers to performing comparative genomics studies are much lower than those of performing experimental recombination, but experimental studies of recombination will be pivotal in disentangling the molecular mechanism of recombination from the rest of the recombination pathway. Answering these critical remaining questions, among others, is essential for modeling and predicting potential recombination events in the future.
Acknowledgments
The research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI149693 (Principle Investigator S.J.A.). Partial support was also provided by the National Science Foundation as part of the Predictive Intelligence for Pandemic Prevention (PIPP) program award number 2200221 (co-Principle Investigator S.J.A.).
Author contributions
Conceptualization, H.L.W. and S.J.A.; writing – original draft, H.L.W. and S.J.A.; writing – review & editing, H.L.W., C.M.B., B.V., A.L.R., and S.J.A.; administrative support, I.N.-M.; funding acquisition, S.J.A. and H.L.W.
Declaration of interests
The authors declare no competing interests.
References
- 1.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 4.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 7.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C., Wertheim-van Dillen P.M., Kaandorp J., Spaargaren J., Berkhout B. Identification of a new human coronavirus. Nat. Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y., Li C., Chen L., Xu B., Zhou Y., Cao L., Shang Y., Fu Z., Chen A., Deng L., et al. A novel human coronavirus OC43 genotype detected in mainland China. Emerg. Microbes Infect. 2018;7:173. doi: 10.1038/s41426-018-0171-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo P.C.Y., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H., Poon R.W., Cai J.J., Luk W.K., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X.M., Herbst W., Kousoulas K.G., Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Aravena M., McKee C., Gamble A., Lunn T., Morris A., Snedden C.E., Yinda C.K., Port J.R., Buchholz D.W., Yeo Y.Y., et al. Ecology, evolution and spillover of coronaviruses from bats. Nat. Rev. Microbiol. 2022;20:299–314. doi: 10.1038/s41579-021-00652-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijaykrishna D., Smith G.J., Zhang J.X., Peiris J.S., Chen H., Guan Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007;81:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anthony S.J., Johnson C.K., Greig D.J., Kramer S., Che X., Wells H., Hicks A.L., Joly D.O., Wolfe N.D., Daszak P., et al. Global patterns in coronavirus diversity. Virus Evol. 2017;3 doi: 10.1093/ve/vex012. vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo P.C.Y., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau S.K.P., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 18.Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R., Navarrete-Macias I., Liang E., Wells H., Hicks A., et al. Further evidence for bats as the evolutionary source of middle east respiratory syndrome coronavirus. mBio. 2017;8 doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P.C., Lau S.K., Li K.S., Tsang A.K., Yuen K.Y. Genetic relatedness of the novel human group C Betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg. Microbes Infect. 2012;1 doi: 10.1038/emi.2012.45. e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ithete N.L., Stoffberg S., Corman V.M., Cottontail V.M., Richards L.R., Schoeman M.C., Drosten C., Drexler J.F., Preiser W. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corman V.M., Baldwin H.J., Tateno A.F., Zerbinati R.M., Annan A., Owusu M., Nkrumah E.E., Maganga G.D., Oppong S., Adu-Sarkodie Y., et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfefferle S., Oppong S., Drexler J.F., Gloza-Rausch F., Ipsen A., Seebens A., Müller M.A., Annan A., Vallo P., Adu-Sarkodie Y., et al. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg. Infect. Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y.W., Dickerman A.W., Piñeyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4 doi: 10.1128/mBio.00737-13. e00737–e00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacroix A., Duong V., Hul V., San S., Davun H., Omaliss K., Chea S., Hassanin A., Theppangna W., Silithammavong S., et al. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y., Shang J., Yang Y., Liu C., Wan Y., Geng Q., Wang M., Baric R., Li F. Lysosomal proteases are a determinant of coronavirus tropism. J. Virol. 2018;92 doi: 10.1128/JVI.01504-18. e01504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Haan C.A.M., Te Lintelo E., Li Z., Raaben M., Wurdinger T., Bosch B.J., Rottier P.J. Cooperative involvement of the S1 and S2 subunits of the murine coronavirus spike protein in receptor binding and extended host range. J. Virol. 2006;80:10909–10918. doi: 10.1128/JVI.00950-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Promkuntod N., Wickramasinghe I.N.A., de Vrieze G., Gröne A., Verheije M.H. Contributions of the S2 spike ectodomain to attachment and host range of infectious bronchitis virus. Virus Res. 2013;177:127–137. doi: 10.1016/j.virusres.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McRoy W.C., Baric R.S. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J. Virol. 2008;82:1414–1424. doi: 10.1128/JVI.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher T.M., Buchmeier M.J., Perlman S. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology. 1992;191:517–522. doi: 10.1016/0042-6822(92)90223-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montoya V., McLaughlin A., Mordecai G.J., Miller R.L., Joy J.B. Variable routes to genomic and host adaptation among coronaviruses. J. Evol. Biol. 2021;34:924–936. doi: 10.1111/jeb.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kan B., Wang M., Jing H., Xu H., Jiang X., Yan M., Liang W., Zheng H., Wan K., Liu Q., et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J. Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peck K.M., Burch C.L., Heise M.T., Baric R.S. Coronavirus host range expansion and middle east respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu. Rev. Virol. 2015;2:95–117. doi: 10.1146/annurev-virology-100114-055029. [DOI] [PubMed] [Google Scholar]
- 39.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baric R.S., Fu K., Chen W., Yount B. High recombination and mutation rates in mouse hepatitis virus suggest that coronaviruses may be potentially important emerging viruses. Adv. Exp. Med. Biol. 1995;380:571–576. doi: 10.1007/978-1-4615-1899-0_91. [DOI] [PubMed] [Google Scholar]
- 42.Lau S.K.P., Li K.S., Huang Y., Shek C.T., Tse H., Wang M., Choi G.K., Xu H., Lam C.S., Guo R., et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus Bat coronavirus in China Reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., Yip C.W., Zeng F., Lam P.Y., Leung F.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X.W., Yap Y.L., Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2005;150:1–20. doi: 10.1007/s00705-004-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabir J.S.M., Lam T.T., Ahmed M.M., Li L., Shen Y., Abo-Aba S.E., Qureshi M.I., Abu-Zeid M., Zhang Y., Khiyami M.A., et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 46.Tao Y., Shi M., Chommanard C., Queen K., Zhang J., Markotter W., Kuzmin I.V., Holmes E.C., Tong S. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J. Virol. 2017;91 doi: 10.1128/JVI.01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pyrc K., Dijkman R., Deng L., Jebbink M.F., Ross H.A., Berkhout B., van der Hoek L. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J. Mol. Biol. 2006;364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woo P.C.Y., Lau S.K., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch. Virol. 2005;150:2299–2311. doi: 10.1007/s00705-005-0573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau S.K.P., Lee P., Tsang A.K., Yip C.C., Tse H., Lee R.A., So L.Y., Lau Y.L., Chan K.H., Woo P.C., et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J. Virol. 2011;85:11325–11337. doi: 10.1128/JVI.05512-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C., Faccini S., Bonilauri P., Cordioli P., Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg. Infect. Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regan A.D., Millet J.K., Tse L.P., Chillag Z., Rinaldi V.D., Licitra B.N., Dubovi E.J., Town C.D., Whittaker G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology. 2012;430:90–99. doi: 10.1016/j.virol.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Licitra B.N., Duhamel G.E., Whittaker G.R. Canine enteric coronaviruses: emerging viral pathogens with distinct recombinant spike proteins. Viruses. 2014;6:3363–3376. doi: 10.3390/v6083363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., de Groot R.J. Feline coronavirus type II strains 79–1683 and 79–1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terada Y., Matsui N., Noguchi K., Kuwata R., Shimoda H., Soma T., Mochizuki M., Maeda K. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106534. e106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X., Giorgi E.E., Marichannegowda M.H., Foley B., Xiao C., Kong X.P., Chen Y., Gnanakaran S., Korber B., Gao F. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb9153. eabb9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makarenkov V., Mazoure B., Rabusseau G., Legendre P. Horizontal gene transfer and recombination analysis of SARS-CoV-2 genes helps discover its close relatives and shed light on its origin. BMC Ecol. Evol. 2021;21:5. doi: 10.1186/s12862-020-01732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 58.Lytras S., Hughes J., Martin D., Swanepoel P., de Klerk A., Lourens R., Kosakovsky Pond S.L., Xia W., Jiang X., Robertson D.L. Exploring the natural origins of SARS-CoV-2 in the light of recombination. Genome Biol. Evol. 2022;14 doi: 10.1093/gbe/evac018. evac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Temmam S., Vongphayloth K., Baquero E., Munier S., Bonomi M., Regnault B., Douangboubpha B., Karami Y., Chrétien D., Sanamxay D., et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330–336. doi: 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 60.Wrobel A.G., Benton D.J., Xu P., Calder L.J., Borg A., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2. Nat. Commun. 2021;12:837. doi: 10.1038/s41467-021-21006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells H.L., Letko M., Lasso G., Ssebide B., Nziza J., Byarugaba D.K., Navarrete-Macias I., Liang E., Cranfield M., Han B.A., et al. The evolutionary history of ACE2 usage within the coronavirus subgenus Sarbecovirus. Virus Evol. 2021;7 doi: 10.1093/ve/veab007. veab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starr T.N., Zepeda S.K., Walls A.C., Greaney A.J., Alkhovsky S., Veesler D., Bloom J.D. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature. 2022;603:913–918. doi: 10.1038/s41586-022-04464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y., Liu D., Shi W., Lu R., Wang W., Zhao Y., Deng Y., Zhou W., Ren H., Wu J., et al. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. mBio. 2015;6 doi: 10.1128/mBio.01280-15. e01280–e01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corman V.M., Ithete N.L., Richards L.R., Schoeman M.C., Preiser W., Drosten C., Drexler J.F. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiong Q., Cao L., Ma C., Tortorici M.A., Liu C., Si J., Liu P., Gu M., Walls A.C., Wang C., et al. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. Nature. 2022;612:748–757. doi: 10.1038/s41586-022-05513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 68.Worobey M., Holmes E.C. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 69.Kim M.J., Kao C. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. USA. 2001;98:4972–4977. doi: 10.1073/pnas.081077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muller H.J. The relation of recombination to mutational advance. Mutat. Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 71.Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011;9:617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leopardi S., Holmes E.C., Gastaldelli M., Tassoni L., Priori P., Scaravelli D., Zamperin G., De Benedictis P. Interplay between co-divergence and cross-species transmission in the evolutionary history of bat coronaviruses. Infect. Genet. Evol. 2018;58:279–289. doi: 10.1016/j.meegid.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Latinne A., Hu B., Olival K.J., Zhu G., Zhang L., Li H., Chmura A.A., Field H.E., Zambrana-Torrelio C., Epstein J.H., et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020;11:4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Everest H., Stevenson-Leggett P., Bailey D., Bickerton E., Keep S. Known cellular and receptor interactions of animal and human coronaviruses: a review. Viruses. 2022;14:351. doi: 10.3390/v14020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Folimonova S.Y. Superinfection exclusion is an active virus-controlled function that requires a specific viral protein. J. Virol. 2012;86:5554–5561. doi: 10.1128/JVI.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunter M., Fusco D. Superinfection exclusion: a viral strategy with short-term benefits and long-term drawbacks. PLOS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1010125. e1010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. e00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blanchard E., Roingeard P. Virus-induced double-membrane vesicles. Cell. Microbiol. 2015;17:45–50. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hagemeijer M.C., Monastyrska I., Griffith J., van der Sluijs P., Voortman J., van Bergen en Henegouwen P.M., Vonk A.M., Rottier P.J., Reggiori F., de Haan C.A. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y., Grunewald M., Perlman S. Coronaviruses: an updated overview of their replication and pathogenesis. Methods Mol. Biol. 2020;2203:1–29. doi: 10.1007/978-1-0716-0900-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J. Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Makino S., Shieh C.K., Soe L.H., Baker S.C., Lai M.M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988;166:550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brian D.A., Spaan W.J.M. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sawicki S.G., Sawicki D.L. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 88.Lai M.M.C., Baric R.S., Makino S., Keck J.G., Egbert J., Leibowitz J.L., Stohlman S.A. Recombination between nonsegmented RNA genomes of murine coronaviruses. J. Virol. 1985;56:449–456. doi: 10.1128/jvi.56.2.449-456.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu H.Y., Brian D.A. Subgenomic messenger RNA amplification in coronaviruses. Proc. Natl. Acad. Sci. USA. 2010;107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rowe C.L., Baker S.C., Nathan M.J., Fleming J.O. Evolution of mouse hepatitis virus: detection and characterization of spike deletion variants during persistent infection. J. Virol. 1997;71:2959–2969. doi: 10.1128/jvi.71.4.2959-2969.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu H.Y., Ozdarendeli A., Brian D.A. Bovine coronavirus 5′-proximal genomic acceptor hotspot for discontinuous transcription is 65 nucleotides wide. J. Virol. 2006;80:2183–2193. doi: 10.1128/JVI.80.5.2183-2193.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yount B., Roberts R.S., Lindesmith L., Baric R.S. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc. Natl. Acad. Sci. USA. 2006;103:12546–12551. doi: 10.1073/pnas.0605438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gribble J., Stevens L.J., Agostini M.L., Anderson-Daniels J., Chappell J.D., Lu X., Pruijssers A.J., Routh A.L., Denison M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009226. e1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Y., Yan W., Hall A.B., Jiang X. Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination. Mol. Biol. Evol. 2021;38:1241–1248. doi: 10.1093/molbev/msaa281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wolff G., Limpens R.W.A.L., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W.M., Koning R.I., Agard D.A., Grünewald K., Koster A.J., et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morales L., Mateos-Gomez P.A., Capiscol C., del Palacio L., Enjuanes L., Sola I. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5′ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J. Virol. 2013;87:11579–11590. doi: 10.1128/JVI.01836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cologna R., Hogue B.G. Identification of a bovine coronavirus packaging signal. J. Virol. 2000;74:580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kuo L., Koetzner C.A., Hurst K.R., Masters P.S. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J. Virol. 2014;88:4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bosch B.J., de Haan C.A.M., Smits S.L., Rottier P.J.M. Spike protein assembly into the coronavirion: exploring the limits of its sequence requirements. Virology. 2005;334:306–318. doi: 10.1016/j.virol.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuan S., Nelsen C.J., Murtaugh M.P., Schmitt B.J., Faaberg K.S. Recombination between North American strains of porcine reproductive and respiratory syndrome virus. Virus Res. 1999;61:87–98. doi: 10.1016/S0168-1702(99)00029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kusters J.G., Niesters H.G., Lenstra J.A., Horzinek M.C., van der Zeijst B.A. Phylogeny of antigenic variants of avian coronavirus IBV. Virology. 1989;169:217–221. doi: 10.1016/0042-6822(89)90058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wesley R.D. The S gene of canine coronavirus, strain UCD-1, is more closely related to the S gene of transmissible gastroenteritis virus than to that of feline infectious peritonitis virus. Virus Res. 1999;61:145–152. doi: 10.1016/S0168-1702(99)00032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stanhope M.J., Brown J.R., Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect. Genet. Evol. 2004;4:15–19. doi: 10.1016/j.meegid.2003.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Y.Y., Lu C.P. Analysis of putative recombination hot sites in the S gene of canine coronaviruses. Acta Virol. 2009;53:111–120. doi: 10.4149/av_2009_02_111. [DOI] [PubMed] [Google Scholar]
- 109.He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y., et al. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014;88:7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luk H.K.H., Li X., Fung J., Lau S.K.P., Woo P.C.Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect. Genet. Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu B., Zeng L.P., Yang X.L., Ge X.Y., Zhang W., Li B., Xie J.Z., Shen X.R., Zhang Y.Z., Wang N., et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006698. e1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Han Y., Du J., Su H., Zhang J., Zhu G., Zhang S., Wu Z., Jin Q. Identification of diverse bat alphacoronaviruses and betacoronaviruses in China provides new insights into the evolution and origin of coronavirus-related diseases. Front. Microbiol. 2019;10:1900. doi: 10.3389/fmicb.2019.01900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., et al. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 2015;474:19–27. doi: 10.1016/j.virol.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bobay L.M., O’Donnell A.C., Ochman H. Recombination events are concentrated in the spike protein region of betacoronaviruses. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009272. e1009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Forni D., Cagliani R., Sironi M. Recombination and positive selection differentially shaped the diversity of betacoronavirus subgenera. Viruses. 2020;12:1313. doi: 10.3390/v12111313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Klerk A., Swanepoel P., Lourens R., Zondo M., Abodunran I., Lytras S., MacLean O.A., Robertson D., Kosakovsky Pond S.L., Zehr J.D., et al. Conserved recombination patterns across coronavirus subgenera. Virus Evol. 2022;8 doi: 10.1093/ve/veac054. veac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Makino S., Keck J.G., Stohlman S.A., Lai M.M. High-frequency RNA recombination of murine coronaviruses. J. Virol. 1986;57:729–737. doi: 10.1128/jvi.57.3.729-737.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baric R.S., Shieh C.K., Stohlman S.A., Lai M.M.C. Analysis of intracellular small RNAs of mouse hepatitis virus: evidence for discontinuous transcription. Virology. 1987;156:342–354. doi: 10.1016/0042-6822(87)90414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keck J.G., Stohlman S.A., Soe L.H., Makino S., Lai M.M.C. Multiple recombination sites at the 5′-end of murine coronavirus RNA. Virology. 1987;156:331–341. doi: 10.1016/0042-6822(87)90413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keck J.G., Makino S., Soe L.H., Fleming J.O., Stohlman S.A., Lai M.M. RNA recombination of coronavirus. Adv. Exp. Med. Biol. 1987;218:99–107. doi: 10.1007/978-1-4684-1280-2_11. [DOI] [PubMed] [Google Scholar]
- 122.Keck J.G., Matsushima G.K., Makino S., Fleming J.O., Vannier D.M., Stohlman S.A., Lai M.M. In vivo RNA-RNA recombination of coronavirus in mouse brain. J. Virol. 1988;62:1810–1813. doi: 10.1128/jvi.62.5.1810-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Keck J.G., Soe L.H., Makino S., Stohlman S.A., Lai M.M. RNA recombination of murine coronaviruses: recombination between fusion-positive mouse hepatitis virus A59 and fusion-negative mouse hepatitis virus 2. J. Virol. 1988;62:1989–1998. doi: 10.1128/jvi.62.6.1989-1998.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Banner L.R., Keck J.G., Lai M.M.C. A clustering of rna recombination sites adjacent to a hypervariable region of the peplomer gene of murine coronavirus. Virology. 1990;175:548–555. doi: 10.1016/0042-6822(90)90439-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baric R.S., Fu K., Schaad M.C., Stohlman S.A. Establishing a genetic recombination map for murine coronavirus strain A59 complementation groups. Virology. 1990;177:646–656. doi: 10.1016/0042-6822(90)90530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Banner L.R., Lai M.M. Random nature of coronavirus RNA recombination in the absence of selection pressure. Virology. 1991;185:441–445. doi: 10.1016/0042-6822(91)90795-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fu K., Baric R.S. Evidence for variable rates of recombination in the MHV genome. Virology. 1992;189:88–102. doi: 10.1016/0042-6822(92)90684-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fu K., Baric R.S. Map locations of mouse hepatitis virus temperature-sensitive mutants: confirmation of variable rates of recombination. J. Virol. 1994;68:7458–7466. doi: 10.1128/jvi.68.11.7458-7466.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang X., Lai M.M. Unusual heterogeneity of leader-mRNA fusion in a murine coronavirus: implications for the mechanism of RNA transcription and recombination. J. Virol. 1994;68:6626–6633. doi: 10.1128/jvi.68.10.6626-6633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hsue B., Hartshorne T., Masters P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2000;74:6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hsue B., Masters P.S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Williams G.D., Chang R.Y., Brian D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu H.Y., Guy J.S., Yoo D., Vlasak R., Urbach E., Brian D.A. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavius molecules. Virology. 2003;315:174–183. doi: 10.1016/S0042-6822(03)00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Goebel S.J., Taylor J., Masters P.S. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 2004;78:7846–7851. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.de Haan C.A.M., Haijema B.J., Masters P.S., Rottier P.J.M. Manipulation of the coronavirus genome using targeted RNA recombination with interspecies chimeric coronaviruses. Methods Mol. Biol. 2008;454:229–236. doi: 10.1007/978-1-59745-181-9_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koetzner C.A., Parker M.M., Ricard C.S., Sturman L.S., Masters P.S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sánchez C.M., Izeta A., Sánchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Durán J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang X., Zhu Y., Zhu X., Shi H., Chen J., Shi D., Yuan J., Cao L., Liu J., Dong H., et al. Identification of a natural recombinant transmissible gastroenteritis virus between Purdue and Miller clusters in China. Emerg. Microbes Infect. 2017;6 doi: 10.1038/emi.2017.62. e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ji W., Wang W., Zhao X., Zai J., Li X. Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross-species transmission from snake to human. J. Med. Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Woo P.C.Y., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou P., Fan H., Lan T., Yang X.L., Shi W.F., Zhang W., Zhu Y., Zhang Y.W., Xie Q.M., Mani S., et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tian P.F., Jin Y.L., Xing G., Qv L.L., Huang Y.W., Zhou J.Y. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg. Infect. Dis. 2014;20:1735–1738. doi: 10.3201/eid2010.140338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yu P., Hu B., Shi Z.L., Cui J. Geographical structure of bat SARS-related coronaviruses. Infect. Genet. Evol. 2019;69:224–229. doi: 10.1016/j.meegid.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yuan J., Hon C.C., Li Y., Wang D., Xu G., Zhang H., Zhou P., Poon L.L., Lam T.T., Leung F.C., et al. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J. Gen. Virol. 2010;91:1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
- 146.Goldstein S.A., Brown J., Pedersen B.S., Quinlan A.R., Elde N.C. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. Genome Biol. Evol. 2022;14 doi: 10.1093/gbe/evac161. evac161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dudas G., Carvalho L.M., Rambaut A., Bedford T. MERS-CoV spillover at the camel-human interface. eLife. 2018;7 doi: 10.7554/eLife.31257. e31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cui S., Liu Y., Zhao J., Peng X., Lu G., Shi W., Pan Y., Zhang D., Yang P., Wang Q. An updated review on SARS-CoV-2 infection in animals. Viruses. 2022;14:1527. doi: 10.3390/v14071527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.P., Pfenning A.R., Zhao H., et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hossain Md.G., Javed A., Akter S., Saha S. SARS-CoV-2 host diversity: an update of natural infections and experimental evidence. J. Microbiol. Immunol. Infect. 2021;54:175–181. doi: 10.1016/j.jmii.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Palmer M.V., Martins M., Falkenberg S., Buckley A., Caserta L.C., Mitchell P.K., Cassmann E.D., Rollins A., Zylich N.C., Renshaw R.W., et al. Susceptibility of White-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 2021;95 doi: 10.1128/JVI.00083-21. e00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pickering B., Lung O., Maguire F., Kruczkiewicz P., Kotwa J.D., Buchanan T., Gagnier M., Guthrie J.L., Jardine C.M., Marchand-Austin A., et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat. Microbiol. 2022;7:2011–2024. doi: 10.1038/s41564-022-01268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lu L., Sikkema R.S., Velkers F.C., Nieuwenhuijse D.F., Fischer E.A.J., Meijer P.A., Bouwmeester-Vincken N., Rietveld A., Wegdam-Blans M.C.A., Tolsma P., et al. Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands. Nat. Commun. 2021;12:6802. doi: 10.1038/s41467-021-27096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Have P., Moving V., Svansson V., Uttenthal A., Bloch B. Coronavirus infection in mink (Mustela vison). Serological evidence of infection with a coronavirus related to transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Vet. Microbiol. 1992;31:1–10. doi: 10.1016/0378-1135(92)90135-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tsunemitsu H., El-Kanawati Z.R., Smith D.R., Reed H.H., Saif L.J. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J. Clin. Microbiol. 1995;33:3264–3269. doi: 10.1128/jcm.33.12.3264-3269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Alekseev K.P., Vlasova A.N., Jung K., Hasoksuz M., Zhang X., Halpin R., Wang S., Ghedin E., Spiro D., Saif L.J. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J. Virol. 2008;82:12422–12431. doi: 10.1128/JVI.01586-08. [DOI] [PMC free article] [PubMed] [Google Scholar]