Abstract
Multiple drug resistance poses a significant threat to public health worldwide, with a substantial increase in morbidity and mortality rates. Consequently, searching for novel strategies to control microbial pathogenicity is necessary. With the aid of auto-inducers (AIs), quorum sensing (QS) regulates bacterial virulence factors through cell-to-cell signaling networks. AIs are small signaling molecules produced during the stationary phase. When bacterial cultures reach a certain level of growth, these molecules regulate the expression of the bound genes by acting as mirrors that reflect the inoculum density.
Gram-positive bacteria use the peptide derivatives of these signaling molecules, whereas Gram-negative bacteria use the fatty acid derivatives, and the majority of bacteria can use both types to modulate the expression of the target gene. Numerous natural and synthetic QS inhibitors (QSIs) have been developed to reduce microbial pathogenesis. Applications of QSI are vital to human health, as well as fisheries and aquaculture, agriculture, and water treatment.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-023-01154-9.
Keywords: Virulence factors, Antibiotics, Quorum sensing, Autoinducers, Multiple drug resistance
Introduction
The discovery of antibiotics by Sir Alexander Fleming in 1945 put an end to infectious diseases. However, in recent decades, the efficacy of these extraordinary drugs has diminished. Microorganisms must adapt to survive, which could be accomplished by the evolution of antimicrobial resistance. The development of resistance by microorganisms was not surprising, as it is a typical survival mechanism. According to the World Health Organization (WHO), multiple drug resistance (MDR) is one of the top ten severe threats to public health, with high morbidity and mortality rates worldwide. The emergence of antibiotic resistance in pathogenic Gram-positive and Gram-negative bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Staphylococcus aureus (VRSA), vancomycin-resistant Enterococci (VRE), and carbapenem-resistant Klebsiella pneumoniae (CRKP) poses a threat to humans. The world has experienced seven epidemics caused by infectious organisms, with Vibrio cholerae ranking first [1]. Pseudomonas aeruginosa, S. aureus, Escherichia coli, and Candida albicans are the leading causes of the increasing number of deaths in hospitals and intensive care units [2].
MDR is a complicated issue that has recently been exacerbated by multiple factors. Continuous exposure to some antibiotics certain antibiotics has enabled the development of multidrug and extensive drug resistance [3]. In Gram-negative bacteria, plasmid-mediated resistance to quinolones has been reported [4]. Due to the production of the β-lactamase enzyme, other bacteria can develop resistance to most β-lactam antibiotics. In addition, MDR can also be acquired through genetic mutation and horizontal gene transfer [5].
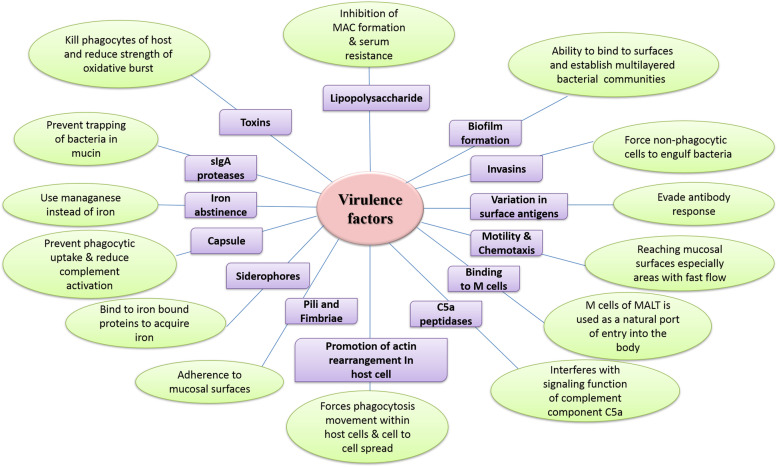
Furthermore, social factors, such as the absence of government policies and the self-medication of consumers who are not certified health professionals, have contributed to the development of MDR. For instance, antimicrobial drugs can be purchased over the counter in developing nations. Lack of medical knowledge has led to inappropriate prescriptions, such as prescribing antibiotics for influenza and inadequate doses. The misuse and overuse of antibiotics diminished their efficacy against infectious diseases and contributed to the emergence of microbial resistance [6]. External factors, such as the inappropriate use of antibiotics in poultry, aquaculture, and agriculture, have contributed to MDR. Infection with drug-resistant bacteria is now significantly more prevalent than infection with drug-susceptible organisms. Antibiotics are released into the environment through domestic use, hospitals, pharmaceutical companies, and research facilities contaminating drinking water similarly to other drugs. The Food and Drug Administration (FDA) and the Environmental Protection Agency (EPA) have not yet developed legislation and regulations, resulting in a significant problem for all forms of life, including aquatic ecosystems [7]. The spread of MDR has challenged researchers to develop new methods in order to overcome this problem. One of these methods is to control bacteria’s virulence and pathogenic factors. Many virulence factors are controlled by cell-to-cell signaling pathways communicative network known as QS [8] (Fig. 1). QS is a ubiquitous phenomenon in the bacterial world that plays a significant role in controlling myriad activities and facilitating survival in hostile environments [9]. For instance, it causes some changes in gene expression resulting in phenotypic modulation in bacteria [10]. As a result, processes like biofilm formation, development of genetic competence, transfer of conjugative plasmid, regulation of virulence, sporulation, symbiosis, and the production of antimicrobial peptides contribute to bacterial adaptation to stress during growth and harsh environmental conditions [11] and antibiotic action [12].
Fig. 1.
Various virulence factors of pathogenic bacteria regulated by quorum sensing
Auto-inducers
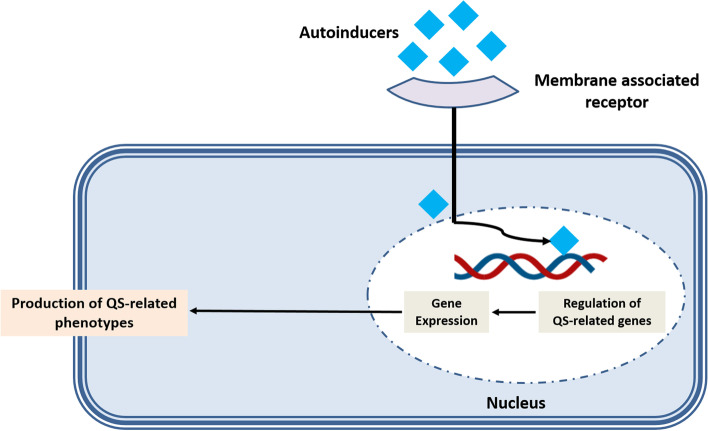
QS promotes communication among bacterial cells by sensing and releasing AIs. AIs are small signaling molecules produced in the stationary phase at the basal level [13]. These molecules act as mirrors that reflect the density of the inoculum. Once the threshold of the growth is reached, they regulate the expression of related genes [14, 15] (Fig. 2). Gram-positive bacteria use peptide derivatives as signaling molecules, whereas Gram-negative bacteria use fatty acid derivatives. Most bacteria can use both AI types of AIs to modulate the target gene expression [16].
Fig. 2.
Mechanism of action of auto-inducers
Classes of auto-inducers
Acyl homoserine lactone
Acyl homoserine lactone (AHL) consists of a lactone ring and a side chain of carbon atoms ranging from C8 to C14 in length (Fig. 3). They occur primarily in Gram-negative bacteria and are involved in intraspecies communication. Moreover, they are produced by a group of homologous LuxI (AHL synthase) proteins that use S-adenosyl methionine as a building block, providing the homoserine lactone moiety. At low cell density, low concentrations of LuxI are produced, followed by the production of AHLs at low concentrations that can diffuse freely through the cell membrane. AHLs accumulate with the bacterial growth until the threshold level at which the transcriptional activating protein LuxR (AHL receptor) binds to the AHL molecules [16]. The AHL-LuxR complex forms dimers or multimers then binds to its cognate promotor and activates QS-related bacterial gene expression [14, 17–19]. Most AHL biosensors have a variety of similar topologies for detecting QS gene networks: (a) a QS transcription activator expressed by an induced or constitutive promoter: and (b) a reporter gene expressed by the LuxR’s cognate promoter homolog.
Fig. 3.
Acyl homoserine lactone
Auto-inducing peptides
As QS molecules, Gram-positive bacteria produce auto-inducing peptides (AIPs). AIPs are small, heterogeneous oligopeptides with a linear or cyclic structure [20]. At high doses, AIP binds to histidine kinase (HK) receptors. The autophosphorylation of the HK receptor activates the cytoplasmic regulator, which then stimulates the transcription of the QS-related genes. After being released from the HK receptor, the AIPs are recirculated within the cell cytoplasm, where they regulate the activity of transcription factors [17].
Auto-inducers-2
Auto-inducers-2 (AI-2) consist of furanosyl borate diester and 4, 5-dihydroxy- 2,3-pentane dione (DPD) derivatives. It is found in Gram-negative and Gram-positive bacteria and is produced by intraspecies. In addition, it is regarded as the most prevalent signaling molecule [18]. The mechanism of action of AI-2 is still uknown, but it occurs by activation of the Lsr transport system by the phosphoenolpyruvate phosphotransferase system [21]. AI-2 was originally identified in the bioluminescence system of the marine bacterium Vibrio harveyi and is synthesized by two complex components, one of which is catalyzed by luxS gene locus and related homologs and the other is catalyzed by the S-adenosyl homocysteine nucleosidase enzyme [17]. AI-2 regulates different activities in many bacterial species, including biofilm formation in V. cholerae, Streptococcus mutans, and Salmonella Typhimurium [22]. Moreover, they modulate motility in E. coli and Campylobacter jejuni [23, 24]. The conjugation of AI-2 with AIPs and AHLs can regulate a wide variety of bacterial properties, such as the growth of Bacillus anthracis, pathogenicity of V. cholerae, and bioluminescence of V. harveyi [25].
Methods to screen AI producers
QS molecules can regulate an excessive number of physiological processes that directly affect human and plant life, so it is crucial to study the properties of these ubiquitous small molecules. Quantitative and qualitative methods to characterize and detect AIs have been developed recently. The optimal method is determined by the nature of the study and the type of autoinducer manufactured [26]
T-Streak plate method
Auto-inducer molecules can be detected by QS biosensor strains, and recently, numerous biosensors have been developed to detect them and detect either AI-1 or AI-2. Both detecting the presence of AIs in bacteria and screening for QS inhibitors are effective [27]. The bacterial biosensor is a genetically modified bacterium with a remarkable QS gene circuit and a reporter gene circuit that is simple to measure and detect. The reporter gene generates a variety of observable outputs, including luminescence, fluorescence, and color pigments. It is the most commonly used technique due to being fast, inexpensive, and easy to apply for qualitatively screening AIs [26]. The tester strain is streaked on agar, producing a distinguished visible output such as Chromobacterium violaceum, the large motile Gram-negative cosmopolitan β-proteobacteria that produce a violet pigment termed “violacein” [28].
C. violaceum is the most frequently used biosensor. Its QS signaling system utilizes N-hexanoyl and N-decanoyl-L-homoserine lactone mediated by CviI and CviR (LuxI/LuxR) homolog genes. CviI AHL-synthase is responsible for C10-HSL synthesis, and CviR is the transcriptional regulatory protein that binds to AHL molecules on reaching the threshold level. This results in the formation of a complex that activates C. violaceum QS-regulated genes such as violacein pigment production, biofilm formation, chitinase, lipase, exopolysaccharide (EPS), and flagellar proteins [29–32]. Other types of reporter strains, such as E. coli JM109 ( [33], S. aureus 8325–4 [34], and Acinetobacter baumannii ATCC 19,606 and ATCC 17,978 can be utilized [35].
Thin-Layer Chromatography
The thin-layer chromatography (TLC) method is more accurate and sensitive than the streak plate method because it provides information on both the type and size of AIs in the tester strain. AIs are organic compounds that must be separated using organic solvents [26]. The supernatants are placed on a TLC plate, dried to allow separation, and then covered with agarose containing the biosensor strain. On the TLC plate, a tear or circular shape is created for a specific type of visible spot when a biosensor and AI are combined. This shape can predict the presence of a specific AI in the tester strain, but mass spectrometry is required for confirmation [36].
Calorimetric assay
The calorimetric assay is an accurate method that is used for quantitative and qualitative detections of AHLs. However, it does not provide any information regarding the size or type of AHLs. The biosensor should be grown with a test strain and O-nitrophenyl-beta-D-galactopyranoside (ONPG). The galactosidase enzyme is produced as a result of converting ONPG to ortho-nitrophenol and galactose. It is a consistent method for biosensors that use the lacZ reporter gene. In this case, ortho-nitrophenol can be quantified by the Miller test method using a spectrophotometer at 420 nm. The quantity of AHL in the test strain may also be helpful in determining the AHL standard curve [26, 37].
Luminescence assay
The luminescence test provides a qualitative determination of AIs, and it can also give a quantitative determination by drawing a standard curve. This test is carried out using liquid test strain extracts mixed with a biosensor strain. This method is ideal, especially for biosensors that utilize luxCDABE reporter luminescence genes, and is comparable to test [38]. In addition, it is highly accurate but provides no information regarding the nature or size of the AHL. It is detected using a luminometer, and the assay is performed on an extract of the test and biosensor strains [39].
Strategies for disrupting the signaling systems
There are at least two significant areas of research that focus on finding antibiotic alternatives. The first is antimicrobial peptide application (AMP), which involves the use of small, effective molecules against resistant pathogens [40]. The second approach involves searching for natural and synthetic substances with QSI activity that targets cell–cell communication and thus control pathogenesis [41].
Targeting the biosynthesis of AIs
Minimizing cell-to-cell communication and hindering their formation plays a vital role in QS inhibition and limits the emergence of pathogenic symptoms. This strategy inhibits the production of AHLs and AI-2, which are responsible for interspecies and intraspecies communication. This method is rarely employed for deactivation and signal degradation.
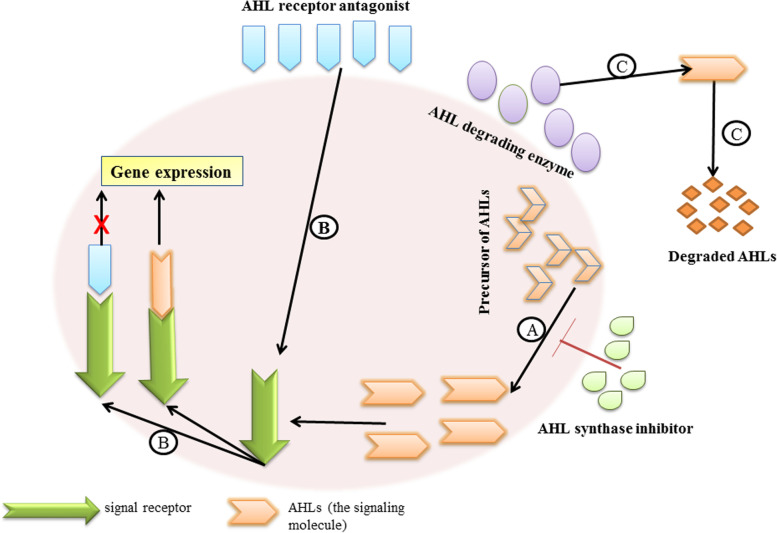
Blocking of AHLs production in Gram-Negative Bacteria
The three enzymatic systems that produce AHL synthase enzyme are HdtS, LuxI, and LuxM; however, the most frequent target of these enzymes is Lux I-type synthase [42] (Fig. 4A). The most studied AHL synthase inhibitors are structural analogs of S-adenosylmethionine (SAM). It has been reported that compounds such as sinefungin, L/D-S-adenosyl homocysteine, and butyryl-SAM inhibited the first step in QS signaling and suppressed AHL synthesis in pathogenic P. aeruginosa in vitro [43]. Notably, SAM is a fundamental component of other enzymes in biological systems, and its analogs may inhibit AHL production [44, 45]. For instance, when E. coli was cultivated in a methionine-free environment, all elements of the bacterial methyl cycle were reduced [46]. The lack of methyl cycle components affects the production of the AHL signal because it is composed of carbon, which is derived from the fatty acid biosynthesis intermediates of the host [47].
Fig. 4.
Mechanisms of quorum sensing inhibition
Blocking of AIPs production in Gram-positive bacteria
There is a paucity of studies on blocking AIPs because AIPs are synthesized by ribosomes, and peptidase enzymes are also included in bacteria growth and survival. Consequently, their inhibition is bactericidal and contributes to the emergence of bacterial resistance [41]. Nevertheless, numerous studies have been conducted to comprehend the mechanism of signal mechanisms, which will eventually lead to the development of signal inhibition methods[45, 48].
Targeting the AI-2 synthases
Targeting the LuxS enzyme splits S-ribosyl-L-homocysteine (SHR) to produce 4,5 dihydroxy-2,3- pentanedione (DPD) and L-homocysteine could disrupt AI-2 and mitigate microbial pathogenesis. S-homoribosyl-L-cysteine and L-homocysteine were approved because they could inhibit LuxS synthases and SHR hydrolysis across a wide spectrum of bacteria [49]. n addition, numerous naturally occurring brominated furanone have been demonstrated to inhibit the LuxS enzyme in a concentration-dependent manner [50]. Numerous combinations of brominated furanone have been synthesized and evaluated for their ability to inhibit QS processes in the V. harveyi reporter strain [51]. V. harveyi can only respond to cross-bacterial communication using the AI-2 signal because it does not have the AHL signal receptor. The presence of furanone derivatives has been shown to reduce the bioluminescence in V. harveyi, indicating their potential role in disrupting bacterial communication. Additionally, furanone reduced the biofilm formation of S. epidermidis by 57% [51]. Natural compounds like surfactin that have been isolated from Bacillus subtilis have the potential to target the LuxS/AI-2 QS system and inhibit biofilm formation by 70% at 1/2 MIC (16 μg/mL) [52].
Targeting AI receptors
Numerous quorum sensing inhibitors (QSIs) inhibit bacterial signal receptors, resulting in an inactive receptor–signal complex that inhibits cell-to-cell communication and limits the pathogenesis and virulence of infectious bacteria. Targeting AI signal receptors is the most obvious strategy, but not all of the listed QSIs have been identified as AI receptor-disrupting agents [44].
Targeting receptors of AHL in Gram-negative bacteria
LuxR-AHL is the most widespread IA receptor protein found in Gram-negative bacteria. Consequently, the targeting of this complex by certain QSIs is an effective alternative strategy for controlling pathogenesis (Fig. 4B). Strategies to disrupt the bond between the signal receptor protein and AHLs are based on the synthesis and design of inhibitors like AHL analogs, structurally independent AHLs, and naturally occurring QS inhibitors.
AHL analogs
Cell-to-cell communication via AHL signals is primarily determined by their chemical structure (lactone ring and acyl side chain). Since any change in their structure, such as the incorporation of any functional group in the acyl side chain, changes in chirality and geometry block the interaction between the receptor and signal. For instance, when the active methylene group was incorporated into the AHL, the protein-signal binding of the receptor decreased by 50%, and when an additional methylene group was added, the activity was reduced by 90%. Therefore, the AHL modifications represent a highly effective strategy for controlling processes with QS signals [33]. Several bulky groups that could inhibit the LasR, TraR, and LuxR receptors have been added to the acyl side chain to create certain AHL analogs in P. aeruginosa, Agrobacterium tumefaciens, and Vibrio fischeri, respectively [53].
Structural unrelated AHLs
Although a 90% loss of binding capacity was achieved in-vitro, actual application in-vivo is still very limited, and more structurally unrelated AHL signals as alternative compounds need to be discovered. It has been demonstrated that certain antibiotics in sub-minimum inhibitory concentration (MIC) doses, such as ciprofloxacin, ceftazidime, azithromycin, cefoperazone, and various synthetic furanone, can reduce virulence factors like motility and biofilm formation in some Gram-negative bacteria and inhibit QS signaling [54]. In addition, the efficacy of nonsteroidal anti-inflammatory drugs (NSAIDs) such as piroxicam and meloxicam was approved for efficacy as QSIs [55]. Similarly, nanoparticles (NPs) could attenuate the pathogenesis of some pathogenic bacteria. For instance, copper ion NPs inhibited the Qs signaling system of P. aeruginosa [56], gold NPs in V.cholerae [57], and silver NPs against P. aeruginosa [58].
Natural QS inhibitors analogs
Several natural compounds have the potential to inhibit the QS signaling systems of certain pathogens. For example, the limonene compound extracted from Citrus reticulate inhibited biofilm formation by 41% at 0.1 mg/mL and AHL signaling production by 33% in Pseudomonas aeruginosa [59]. Moreover, the same compound was isolated from Eucalyptus radiate and could inhibit QS-regulated pyomelanin pigment production in A. baumannii [60]. Similarly, the phenolic extract of Rubus rosifolius inhibited swarming motility and biofilm formation in Serratia marcescens [61].
Pyocyanin production, biofilm formation, swarming motility, elastolytic, and proteolytic activities in P. aeruginosa PAO1 were inhibited in a concentration-dependent manner by a flavonoid-rich fraction of Centella Asiatica [62]. The methoxyisoflavan compound isolated from Trigonella stellate reduced violacein production in C. violaceum and inhibited pyocyanin, protease production, hemolysin activity, and biofilm formation in P. aeruginosa [63]. The biofilm formation of Escherichia coli and Pseudomonas aeruginosa was inhibited by the methanolic extract of Buchanania lanzana Spreng [64]. The biofilms of foodborne pathogens, S. typhimurium, S. aureus, and P. aeruginosa, were reduced by 51.96%, 47.06%, and 45.28%, respectively, using the ethanol extract of Amomum tsaoko (Zingiberaceae) [65]. Garlic extract could inhibit the signaling system in P. aeruginosa infections and attenuate their biofilm formation [66]. Similarly, water-soluble cranberry extracts inhibited the biofilm formation of V. cholerae [67]. It also increased susceptibility toward tobramycin and azithromycin antibiotics and toward phagocytosis mediated by neutrophils [68].
Targeting histidine kinase receptors in Gram-positive bacteria
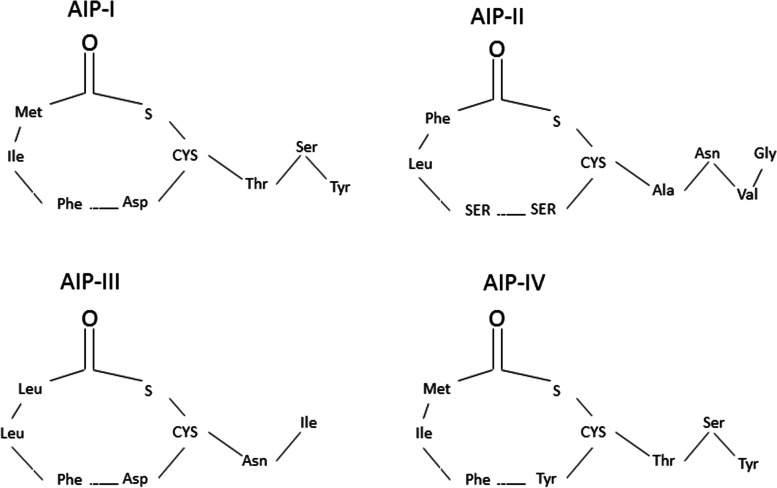
Two QS systems are regulated in Gram-positive bacteria a reactive transcriptional regulator and a histidine kinase receptor attached to the membrane [69]. Now, targeting these receptors with specific AIP antagonists can inhibit the action of the Gram-positive bacterial receptor and pathogenesis [41]. For example, the S. aureus agr system, which is an AIP-mediated QS system, uses four distinct thiolactone peptides (AIP I–IV) to control bacterial behavior [12, 44] (Fig. 5).
Fig. 5.
Chemical structure of autoinducer peptides (AIPs) in S. aureus
Targeting LuxP receptors
The signaling of AI-2 is mediated by three protein receptors that have been described so far. The first is the LuxP gene in V. harveyi, the second is the LsrB gene in S. typhimurium, and the third is RbsB in Aggregatibacter actinomycetemcomitans [70]. The emergence of compounds capable of targeting these receptors is a promising strategy for silencing processes mediated by the AI-2 QS system [41, 71]. For instance, the sulphone compound showed an antagonistic effect on V. harveyi LuxP receptors [72]. Similarly, bioluminescence production by V. harveyi was inhibited by certain aromatic groups, such as polyols and phenylboronics, at sub-MIC concentrations [73]. Furthermore, the nucleoside analog LMC-21 decreased biofilm formation in V. cholerae, V. vulnificus, and V. anguillarum and reduced pigment and protease production in V. anguillarum [74].
Enzymatic inactivation of auto-inducers
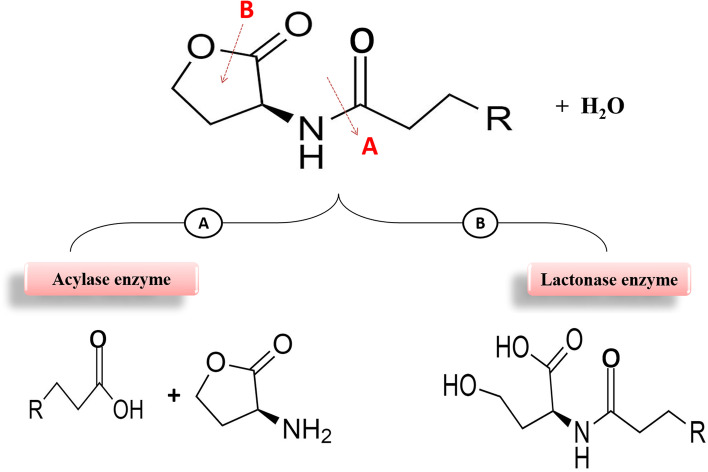
Enzymatic inactivation or degradation of extracellular antimicrobial peptides is a promising strategy because it can significantly reduce microbial resistance without placing stress on the bacterial cell. Certain enzymes, such as lactonase and acylase, can degrade AHL in Gram-negative bacteria by inactivating the lactone ring [75]. Oxidoreductases are a different class of enzymes that may interfere with bacterial communication [76] (Fig. 4C).
Lactonase enzyme
The lactonase enzyme acts by hydrolyzing the ester bond of the homoserine lactone ring (Fig. 6A), and the first microorganism to produce the lactonase enzyme was the Bacillus species that were isolated from soil [77]. Similarly, Agrobacterium tumefaciens [78, 79], Rhizobium sp. [80], Chryseobacterium sp. [81], Mycobacterium avium [82], Microbacterium testaceum [83], V. cholerae [84], Brucella melitensis [85], Arthrobacter sp. IBN110 and Klebsiella pneumonia [86]. These enzymes degrade all signals and have the broadest spectrum of AHL specificity regardless of acyl-side chain length or substitutions [87].
Fig. 6.
Enzymatic inactivation action of auto-inducers by acylase enzyme (A) and lactonase enzyme (B)
The lactonase enzyme has also been identified in mammalian tissues in the form of paraoxonases (PONs), which belong to the three categories PON1, PON2, and PON3. PON1 and PON3 genes have been detected in the kidneys and liver, and their amino acid products are detected in the blood circulation associated with high-density lipoprotein (HDL). In contrast, PON2 showed the best lactonase activity and was present in a variety of tissues [88].
Acylase enzyme
The acylase enzyme breaks down the amide bond between the fatty acid side chain and lactone ring (Fig. 6B), as it can use AHL molecules as the sole energy source and nitrogen [89]. Variovorax paradoxus is the first microorganism reported to produce the acylase enzyme. Similarly, the acylase enzyme was produced by Ralstonia strain XJ12B [90], Pseudomonas strain PAI-A, P. aeruginosa PAO1 [91], and the kidney in Porcine [92]. The acylase enzyme has more substrate selectivity than lactonase as it can identify the acyl chain, such as the acylase PvdQ of Pseudomonas [93].
Oxidoreductase enzyme
The oxidoreductase enzyme reduces or oxidizes the acyl chain of AHLs rather than breaking them down, as in acylase and lactonase enzymes. This inhibits AHL receptor binding and further gene expression controlled by QS [76]. For instance, the oxidoreductase enzyme produced by Burkholderia sp. GG4 inhibited the virulence factors of Erwinia carotovora by the modification of the 3-oxo-C6-HSL signaling molecule [94].
Antibodies
It has also been reported that antibodies play a role in the inhibition of QS signaling molecules. [33]. Some organisms have receptors that inhibit QS compounds, and adaptive mammalian immune systems produce antibodies in response to antigen exposure. Signals from artificial intelligence are not expected to stimulate the human immune system because they lack protein and have a low molecular weight. However, it has been found that AHL bacterial molecules act as small molecule toxins in mammalian cells, inducing apoptosis and modulating NF-kB activity [33, 95]. For instance, the XYD-11G2 antibody prevented P. aeruginosa from producing pyocyanin and neutralized the 3-oxo-C12-HSL signal [96]. Monoclonal antibodies have also been shown to neutralize AIPs and disrupt QS pathways, though less attention has been paid to them. The S. aureus agr pathogen and E. faecalis agr-like systems were studied as AIP-mediated QS systems in Gram-positive bacteria [97]. For example, the AP4-24 H11 antibody could act as a sequester of the AIP-4 produced by S. aureus and inhibit QS pathogenicity in vivo and in vitro [98].
Active uptake of AI signaling molecules by beneficial bacteria
As competing bacteria, certain other bacteria, such as those in the family Enterobacteriaceae, can sequester and disrupt cell communication. These pathogens include B. anthracis, E. coli O157, commensal E. coli K12, Salmonella typhimurium, and Salmonella meliloti [99]. Other members will no longer be able to use AI-2 signals to control their behavior because it takes AI-2 out of the environment. It was reported that when E. coli was cultured with V. harveyi, the bioluminescence mediated by QS signals was decreased by 18%. In contrast, using a mutant E. coli strain that contains a constitutively inhibited LsrK and decreased the bioluminescence by 90% [99], as the mutant E.coli disrupted the QS signaling system of V. harveyi.
Application of molecularly imprinted polymers
The application of molecularly imprinted polymers has been proposed as an innovative approach. The authors have developed various polymers based on computational modeling to have the ability to form complex compounds that act as sequesters for the signals and eliminate the extracellular signals from the environment. The first generation of these polymers could target the signals of V. fischeri and inhibit biofilm formation and bioluminescence production. Some other polymers were designed to target AHLs signals in P. aeruginosa and inhibited 80% of QS-regulated biofilm [100]. For instance, the application of salicylate-based polymers exhibited slower adhesion rates, and the development of P. aeruginosa biofilms was significantly decreased compared to that of a control polymer [101]
Targeting the efflux pumps
Efflux pumps play a significant role in the MDR of some antimicrobial agents as they have transport proteins that prevent these antimicrobial agents from reaching the target sites [102]. Recent research has demonstrated that QS is required for the development of efflux pumps in bacteria [103, 104]. For instance, it was found that P. aeruginosa’s active efflux pump and QS were related, and the addition of the C4-HSL signaling molecule enhanced the expression of the MexABOprM efflux pump in P. aeruginosa [105, 106]. So, inhibiting QS mechanisms play a significant role in the MDR of some antimicrobial agents [102]. Additionally, some substances such as phenothiazines and trifluoromethyl ketones (TFs) prevented bacteria’s QS systems from functioning properly, resulting in reduced production of efflux pumps and QS-regulated virulence factors [107]. Similarly, TFs approved efficacy in the inhibition of QS and efflux pump of E. coli and C. violaceum CV026 [108]. Moreover, phenylalanine arginyl b-naphthylamide efflux pump inhibitor significantly eliminated the virulence factors of P. aeruginosa as protease, elastase, pyocyanin, and motility and suppressed the QS cascade [109].
Applications of QSI in different fields
Health
The human body and bacteria are closely associated with each other. Recent studies have shown that the human body has 10 trillion bacterial cells on the skin, known for being the largest organ as it is full of bacteria and dominated by Staphylococcus, Proteobacteria, Corynebacteria, and Propionibacterium [110]. The gut contains approximately 160 species of bacteria [111] that contribute to the human body's health and fitness, especially those belonging to Firmicutes and Bacteroidetes [112]. The body appears healthy when Firmicutes and Bacteroidetes make up 44% to 48% of the gut. However, when more carbohydrates are consumed than the body requires, the Bacteroidetes grow on these carbohydrates, causing obesity because they make up 82% to 86% of humans [113]. Firmicutes and Bacteroidetes work silently in our bodies; we do not always feel them in silence or feel it. However, another group of bacteria forces our body to listen because they are pathogenic and cause many diseases and symptoms that can lead to death if it is not controlled, and in some cases, antibiotic resistance [114]. QS signals are the key regulator of pathogenicity and virulence factors. Consequently, controlling these signals with the aforementioned strategies will significantly impact bacterial resistance and human health [115–117] by targeting the efflux pump gene expression and inhibiting biofilm formation. Conversely, these signals inhibited human breast cancer cell lines [118]. Therefore, the application of genetically modified bacteria with QS signals is an effective method for targeting cancerous cells (Choudhary and Dannert 2010). The use of QSI-coated plastics and polyurethanes on medical implants and catheters can be a practical strategy for medical applications. For example, dispersin B could inhibit Staphylococcus epidermidis from forming a biofilm. A combination of dispersin B with certain antibiotics such as triclosan demonstrated a synergistic effect and inhibited the formation of QS and biofilms. of E. coli, S. aureus, and S. epidermidis ( [119, 120].
Agriculture
It is well known that bacteria and plants have mutually beneficial relationships. For instance, epiphytes such as Pantoea, Pseudomonas, and Erwinia support Nicotiana (tobacco) plants by influencing the behavior of plant pathogens and assisting the host in triggering its defense mechanisms against pathogenic attacks [121]. Furthermore, the expression of the lactonase enzyme that inhibits AHL may protect transgenic plants like potatoes, cabbage, and tobacco from infection caused by Erwinia carotovora. The application of these inhibiting enzymes is effective in plant protection and controlling the virulence factors and pathogenesis of infectious organisms [83]. The release of certain chemical compounds during bacterial infection is another method to protect some plants, such as Pisum sativum and Medicago truncatula, and increase crop yield from bacterial infection. These compounds act as QS mimics, significantly controlling bacterial pathogens' virulence factors [41].
Water treatment
Recent studies have demonstrated that biofilm is associated with the vast majority of bacterial infections in humans [122]. Biofilm development is crucial in treating bacterial infections and MDR because it exhibits new biological characteristics and enhanced environmental adaptability [123, 124]. Furthermore, bioreactors used for desalination and reclamation of seawater as well as on any artificial objects submerged in marine water, cause biofouling phenomena [125]. This phenomenon begins with the formation of biofilm, which is composed of microorganisms and microalgae, followed by the settlement of invertebrate larvae and algal spores. This has serious implications and increases the costs of fuel shipping and treatment [126]. The most prevalent bacteria are Pseudomonas putida and Aeromonas [127]. There are several strategies to control biofouling, (i) physical methods such as surface modification, but their application on a wide scale is not easy as more studies are needed to understand the adhesion mechanisms of organisms. (ii) Chemical methods, such as the use of antifouling paints such as tributyltin, are very toxic for marine organisms [128]. (iii) The application of biological strategies involves eco-friendly systems such as coatings with QSIs, using genetically modified marine organisms, or immobilizing QQ enzymes. [129]. Due to the short lifespan of enzymes, this highly effective method cannot be implemented on a large scale [130, 131]. Using a filtration membrane coated with the acylase enzyme, for instance, inhibited biofilm formation, exopolysaccharide production, and an anti-biofouling activity [130].
Aquaculture and fisheries
Aquaculture and fisheries departments are a highly viable food source, especially for those living in coastal regions [132]. Nevertheless, pathogenic bacteria cause significant harm to aquatic organisms and substantial economic loss. The application of antibiotics to protect aquatic organisms is a simple option, but it can lead to MDR, particularly when they are used in sub-MIC concentrations [1]. Consequently, QSIs must be utilized to control the pathogenicity and virulence factors of these pathogens [133]. Some micro- and macroalgae produce QSIs, like Chlorella saccharophila CCAP211, which decreases the bioluminescence of V. harveyi and reduces the violacein production in C. violaceum CV026 without any effect on growth [134]. Moreover, Chlamydomonas reinhardtii microalga inhibited some QS-mediated phenotypes, such as luminescence [135]. Similarly, the red macroalga Delisea pulchra was reported to produce halogenated furanone that inhibited QS-regulated phenotypes [136]. Some sponges, such as Haliclona megastoma, Aphrocallistes bocagei, and Clathria atrasanguinea approved for their efficacy against violacein pigment production in C. violaceum CV026 and ATCC 12,472. They also inhibited biofilm formation, protease, and hemolysin production of S. marcescens [137].
Development of resistance against QSIs
QS inhibition has been considered a potential alternative antipathogenic therapy. It has been demonstrated to be an efficient anti-infective technique in several host-microbe systems [45]. Prior studies reported that pathogens are unlikely to develop resistance to the QSI tactics because they pose no selective pressure on bacteria [5, 12, 138]. However, some studies showed the possibility of development of resistance against QSI [139]. Due to nalC and mexR mutations, P. aeruginosa developed resistance to the QSI; brominated furanone C-30 [140]. Similarly, García-Contreras et al. [141] reported that P. aeruginosa developed resistance against 5-fluorouracil QSI in addition to brominated furanone C-30 for at least one QS-regulated phenotype.
Conclusion
The emergence of resistance is the result of a natural evolutionary process that encourages the development of resistant strains through selection pressure. Furthermore, the formation of bacterial biofilms has been identified as one of the most challenging aspects of treating clinical bacterial infections. Additionally, the QS system has been shown to regulate bacterial efflux pumps and biofilm formation in multidrug-resistant bacteria. Consequently, QQ strategies could offer a novel approach to combating microbial resistance by regulating the expression of efflux pump genes and inhibiting bacterial invasiveness and infectiousness. Antimicrobial resistance regulation through control of QS signaling molecules in clinical isolates requires additional research. Consequently, innovation and advancement in research on QQ molecules and their effects on regulatory systems will result in the introduction of new QQ molecules to combat microbial resistance. The discovery of QQ molecules is expected to increase significantly over the upcoming decades, and they could be used as coating agents in medical devices, replacing bacteriostatic and bactericidal antibiotics in-vivo.
Acknowledgements
The authors would like to thank the Egyptian knowledge bank.
Authors’ contributions
Nourhan G. Naga and Mona I. Shaaban wrote the main manuscript text. Nourhan G. Naga prepared the figures. Dalia E. El-Badan and Khaled M. Ghanem reviewed the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The original contributions presented in the review are included in the article material. Further inquiries can be directed to the corresponding authors.
Declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Haidari RA, Shaaban MI, Ibrahim SRM, Mohamed GA. Anti-quorum sensing activity of some medicinal plants. African J Tradit Complement Altern Med. 2016;13:67–71. doi: 10.21010/ajtcam.v13i5.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes AH, Moore LSP, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 4.Robicsek A, Strahilevitz J, Jacoby GA, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12:83–88. doi: 10.1038/nm1347. [DOI] [PubMed] [Google Scholar]
- 5.Chadha J, Harjai K, Chhibber S. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ Microbiol. 2022;24:2630–2656. doi: 10.1111/1462-2920.15784. [DOI] [PubMed] [Google Scholar]
- 6.Rather IA, Kim B-C, Bajpai VK, Park Y-H. Self-medication and antibiotic resistance: Crisis, current challenges, and prevention. Saudi J Biol Sci. 2017;24:808–812. doi: 10.1016/j.sjbs.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borges A, Simões M, Todorović TR, et al. Cobalt Complex with Thiazole-Based Ligand as New Pseudomonas aeruginosa Quorum Quencher. Biofilm Inhibitor and Virulence Attenuator Molecules. 2018;23:1385. doi: 10.3390/molecules23061385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silpe JE, Duddy OP, Bassler BL. Natural and synthetic inhibitors of a phage-encoded quorum-sensing receptor affect phage–host dynamics in mixed bacterial communities. Proc Natl Acad Sci. 2022;119:e2217813119. doi: 10.1073/pnas.2217813119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prescott RD, Decho AW. Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 2020;28:436–444. doi: 10.1016/j.tim.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monterrosa MA, Galindo JF, Lorduy JV, et al. The role of LasR active site amino acids in the interaction with the acyl homoserine lactones (AHLs) analogues: a computational study. J Mol Graph Model. 2019;86:113–124. doi: 10.1016/j.jmgm.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Azimi S, Klementiev AD, Whiteley M, Diggle SP. Bacterial quorum sensing during infection. Annu Rev Microbiol. 2020;74:201–219. doi: 10.1146/annurev-micro-032020-093845. [DOI] [PubMed] [Google Scholar]
- 13.Czajkowski R, Jafra S. Quenching of acyl-homoserine lactonedependent quorum sensing by enzymatic disruption of signal molecules. Acta Biochim Pol. 2009;56(1):1–16. doi: 10.18388/abp.2009_2512. [DOI] [PubMed] [Google Scholar]
- 14.Boo A, Amaro RL, Stan G-B. Quorum sensing in synthetic biology: A review. Curr Opin Syst Biol. 2021;28:100378. doi: 10.1016/j.coisb.2021.100378. [DOI] [Google Scholar]
- 15.Elgaml A, Higaki K, Miyoshi S. Effects of temperature, growth phase and luxO-disruption on regulation systems of toxin production in Vibrio vulnificus strain L-180, a human clinical isolate. World J Microbiol Biotechnol. 2014;30:681–691. doi: 10.1007/s11274-013-1501-3. [DOI] [PubMed] [Google Scholar]
- 16.Prazdnova EV, Gorovtsov AV, Vasilchenko NG, et al. Quorum-sensing inhibition by Gram-positive bacteria Microorganisms. 2022;10:350. doi: 10.3390/microorganisms10020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scutera S, Zucca M, Savoia D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert Opin Drug Discov. 2014;9:353–366. doi: 10.1517/17460441.2014.894974. [DOI] [PubMed] [Google Scholar]
- 19.Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 20.Okada M, Sato I, Cho SJ, et al. Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat Chem Biol. 2005;1:23–24. doi: 10.1038/nchembio709. [DOI] [PubMed] [Google Scholar]
- 21.Pereira CS, Santos AJM, Bejerano-Sagie M, et al. Phosphoenolpyruvate phosphotransferase system regulates detection and processing of the quorum sensing signal autoinducer-2. Mol Microbiol. 2012;84:93–104. doi: 10.1111/j.1365-2958.2012.08010.x. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida A, Ansai T, Takehara T, Kuramitsu HK. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl Environ Microbiol. 2005;71:2372–2380. doi: 10.1128/AEM.71.5.2372-2380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girón JA, Torres AG, Freer E, Kaper JB. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol. 2002;44:361–379. doi: 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 24.Sperandio V, Torres AG, Girón JA, Kaper JB. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157: H7. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zohar BA, Kolodkin-Gal I. Quorum sensing in Escherichia coli: Interkingdom, inter- and intraspecies dialogues, and a suicide inducing peptide. In: Quorum Sensing vs. Quorum Quenching: a Battle with No End in Sight. New Delhi: Springer; 2015. p. 85–99.
- 26.Rai N, Rai R, Venkatesh KV. Quorum sensing biosensors. In: Kalia V, editor. Quorum sensing vs quorum quenching: a battle with no end in sight. New Delhi: Springer; 2015. pp. 173–183. [Google Scholar]
- 27.Tello E, Castellanos L, Duque C. Synthesis of cembranoid analogues and evaluation of their potential as quorum sensing inhibitors. Bioorg Med Chem. 2013;21:242–256. doi: 10.1016/j.bmc.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Howard AJ, Ison CA. Haemophilus, Gardnerella and other bacilli. Mackie McCartney Pract Med Microbiol. 1996;14:329–341. [Google Scholar]
- 29.Brumbach KC, Eason BD, Anderson LK. The Serratia-type hemolysin of Chromobacterium violaceum. FEMS Microbiol Lett. 2007;267:243–250. doi: 10.1111/j.1574-6968.2006.00566.x. [DOI] [PubMed] [Google Scholar]
- 30.McClean KH, Winson MK, Fish L, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 31.Stauff DL, Bassler BL. Quorum sensing in Chromobacterium violaceum: DNA recognition and gene regulation by the CviR receptor. J Bacteriol. 2011;193:3871–3878. doi: 10.1128/JB.05125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatramanan M, Sankar Ganesh P, Senthil R, et al. Inhibition of quorum sensing and biofilm formation in Chromobacterium violaceum by fruit extracts of Passiflora edulis. ACS Omega. 2020;5:25605–25616. doi: 10.1021/acsomega.0c02483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31:224–245. doi: 10.1016/j.biotechadv.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Singh RP, Nakayama J. Development of quorum-sensing inhibitors targeting the fsr system of Enterococcus faecalis. In: Kalia VC, editor. Quorum Sensing vs Quorum Quenching: a Battle with No End in Sight. New Delhi: Springer; 2015. pp. 319–24. [Google Scholar]
- 35.Bouvet PJM, Grimont PAD. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus a. Int J Syst Evol Microbiol. 1986;36:228–240. [Google Scholar]
- 36.Anbazhagan D, Mansor M, Yan GOS, et al. Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS ONE. 2012;7:e36696. doi: 10.1371/journal.pone.0036696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson JP, Gray KM, Passador L, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simanek KA, Taylor IR, Richael EK, et al. The PqsE-RhlR interaction regulates RhlR DNA binding to control virulence factor production in Pseudomonas aeruginosa. Microbiol Spectr. 2022;10:e02108–e2121. doi: 10.1128/spectrum.02108-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massai F, Imperi F, Quattrucci S, et al. A multitask biosensor for micro-volumetric detection of N-3-oxo-dodecanoyl-homoserine lactone quorum sensing signal. Biosens Bioelectron. 2011;26:3444–3449. doi: 10.1016/j.bios.2011.01.022. [DOI] [PubMed] [Google Scholar]
- 40.Brogden NK, Brogden KA. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int J Antimicrob Agents. 2011;38:217–225. doi: 10.1016/j.ijantimicag.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Churchill MEA, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem Rev. 2011;111:68–85. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hentzer M, Givskov M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Invest. 2003;112:1300–1307. doi: 10.1172/JCI20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni N, Li M, Wang J, Wang B. Inhibitors and antagonists of bacterial quorum sensing. Med Res Rev. 2009;29:65–124. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Luo Y. Bacterial quorum-sensing systems and their role in intestinal bacteria-host crosstalk. Front Microbiol. 2021;12:611413. doi: 10.3389/fmicb.2021.611413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin C-CJ, Wang MC. Microbial metabolites regulate host lipid metabolism through NR5A–Hedgehog signalling. Nat Cell Biol. 2017;19:550–557. doi: 10.1038/ncb3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papenfort K, Bassler BL. Quorum sensing signal–response systems in Gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Loughlin CT, Miller LC, Siryaporn A, et al. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X, Yu Z, Ding T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020;8:425. doi: 10.3390/microorganisms8030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zang T, Lee BWK, Cannon LM, et al. A naturally occurring brominated furanone covalently modifies and inactivates LuxS. Bioorg Med Chem Lett. 2009;19:6200–6204. doi: 10.1016/j.bmcl.2009.08.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benneche T, Hussain Z, Scheie AA, Lönn-Stensrud J. Synthesis of 5-(bromomethylene) furan-2 (5 H)-ones and 3-(bromomethylene) isobenzofuran-1 (3 H)-ones as inhibitors of microbial quorum sensing. New J Chem. 2008;32:1567–1572. doi: 10.1039/b803926g. [DOI] [Google Scholar]
- 52.Liu J, Li W, Zhu X, et al. Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces. Appl Microbiol Biotechnol. 2019;103:4565–4574. doi: 10.1007/s00253-019-09808-w. [DOI] [PubMed] [Google Scholar]
- 53.Geske GD, Mattmann ME, Blackwell HE. Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators. Bioorg Med Chem Lett. 2008;18:5978–5981. doi: 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naga NG, El-Badan DE-S, Rateb HS, et al. Quorum sensing inhibiting activity of cefoperazone and its metallic derivatives on pseudomonas aeruginosa. Front Cell Infect Microbiol. 2021;11:716789. [DOI] [PMC free article] [PubMed]
- 55.Soheili V, Bazzaz BSF, Abdollahpour N, Hadizadeh F. Investigation of Pseudomonas aeruginosa quorum-sensing signaling system for identifying multiple inhibitors using molecular docking and structural analysis methodology. Microb Pathog. 2015;89:73–78. doi: 10.1016/j.micpath.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 56.Mishra A, Mishra N. Antiquorum Sensing Activity of Copper Nanoparticle in Pseudomonas Aeruginosa: An in Silico Approach. Proc Natl Acad Sci India Sect B Biol Sci. 2021;91:29–36. doi: 10.1007/s40011-020-01193-z. [DOI] [Google Scholar]
- 57.Chatterjee T, Saha T, Sarkar P, et al. The gold nanoparticle reduces Vibrio cholerae pathogenesis by inhibition of biofilm formation and disruption of the production and structure of cholera toxin. Colloids Surfaces B Biointerfaces. 2021;204:111811. doi: 10.1016/j.colsurfb.2021.111811. [DOI] [PubMed] [Google Scholar]
- 58.Ali SG, Ansari MA, Jamal S, et al. Antiquorum sensing activity of silver nanoparticles in P. aeruginosa: an in silico study. Silico Pharmacol. 2017;5:1–7. doi: 10.1007/s40203-017-0031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luciardi MC, Blázquez MA, Cartagena E, et al. Mandarin essential oils inhibit quorum sensing and virulence factors of Pseudomonas aeruginosa. LWT-Food Sci Technol. 2016;68:373–380. doi: 10.1016/j.lwt.2015.12.056. [DOI] [PubMed] [Google Scholar]
- 60.Luís Â, Duarte A, Gominho J, et al. Chemical composition, antioxidant, antibacterial and anti-quorum sensing activities of Eucalyptus globulus and Eucalyptus radiata essential oils. Ind Crops Prod. 2016;79:274–282. doi: 10.1016/j.indcrop.2015.10.055. [DOI] [Google Scholar]
- 61.Oliveira BD, Rodrigues AC, Cardoso BMI, et al. Antioxidant, antimicrobial and anti-quorum sensing activities of Rubus rosaefolius phenolic extract. Ind Crops Prod. 2016;84:59–66. doi: 10.1016/j.indcrop.2016.01.037. [DOI] [Google Scholar]
- 62.Vasavi HS, Arun AB, Rekha PD. Anti-quorum sensing activity of flavonoid-rich fraction from Centella asiatica L. against Pseudomonas aeruginosa PAO1. J Microbiol Immunol Infect. 2016;49:8–15. doi: 10.1016/j.jmii.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 63.Naga NG, Zaki AA, El-Badan DE, et al. Methoxyisoflavan derivative from Trigonella stellata inhibited quorum sensing and virulence factors of Pseudomonas aeruginosa. World J Microbiol Biotechnol. 2022;38:1–13. doi: 10.1007/s11274-022-03337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pattnaik A, Sarkar R, Sharma A, et al. Pharmacological studies on Buchanania lanzan Spreng.-A focus on wound healing with particular reference to anti-biofilm properties. Asian Pac J Trop Biomed. 2013;3:967–974. doi: 10.1016/S2221-1691(13)60187-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahman MRT, Lou Z, Yu F, et al. Anti-quorum sensing and anti-biofilm activity of Amomum tsaoko (Amommum tsao-ko Crevost et Lemarie) on foodborne pathogens. Saudi J Biol Sci. 2017;24:324–330. doi: 10.1016/j.sjbs.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jakobsen TH, van Gennip M, Phipps RK, et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother. 2012;56:2314–2325. doi: 10.1128/AAC.05919-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pederson DB, Dong Y, Blue LB, et al. Water-soluble cranberry extract inhibits Vibrio cholerae biofilm formation possibly through modulating the second messenger 3’, 5’-Cyclic diguanylate level. PLoS ONE. 2018;13:e0207056. doi: 10.1371/journal.pone.0207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gui Z, Wang H, Ding T, et al. Azithromycin reduces the production of α-hemolysin and biofilm formation in Staphylococcus aureus. Indian J Microbiol. 2014;54:114–117. doi: 10.1007/s12088-013-0438-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Gestel J, Bareia T, Tenennbaum B, et al. Short-range quorum sensing controls horizontal gene transfer at micron scale in bacterial communities. Nat Commun. 2021;12:2324. doi: 10.1038/s41467-021-22649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 71.Guo M, Gamby S, Zheng Y, Sintim HO. Small molecule inhibitors of AI-2 signaling in bacteria: state-of-the-art and future perspectives for anti-quorum sensing agents. Int J Mol Sci. 2013;14:17694–17728. doi: 10.3390/ijms140917694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng H, Cheng Y, Ni N, et al. Synthesis and evaluation of new antagonists of bacterial quorum sensing in Vibrio harveyi. ChemMedChem Chem Enabling Drug Discov. 2009;4:1457–1468. doi: 10.1002/cmdc.200900180. [DOI] [PubMed] [Google Scholar]
- 73.Ni N, Choudhary G, Li M, Wang B. Pyrogallol and its analogs can antagonize bacterial quorum sensing in Vibrio harveyi. Bioorg Med Chem Lett. 2008;18:1567–1572. doi: 10.1016/j.bmcl.2008.01.081. [DOI] [PubMed] [Google Scholar]
- 74.Brackman G, Celen S, Baruah SK, et al. AI-2 quorum sensing inhibitors affect the starvation response and reduce virulence in several Vibrio species, most likely by interfering with LuxPQ. MICROBIOLOGY-SGM. 2009;155:4114–4122. doi: 10.1099/mic.0.032474-0. [DOI] [PubMed] [Google Scholar]
- 75.Whitehead NA, Barnard AML, Slater H, et al. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 76.Hong K-W, Koh C-L, Sam C-K, et al. Quorum quenching revisited—from signal decays to signalling confusion. Sensors. 2012;12:4661–4696. doi: 10.3390/s120404661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong Y-H, Wang L-H, Xu J-L, et al. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature. 2001;411:813–817. doi: 10.1038/35081101. [DOI] [PubMed] [Google Scholar]
- 78.Carlier A, Uroz S, Smadja B, et al. The Ti plasmid of Agrobacterium tumefaciens harbors an attM-paralogous gene, aiiB, also encoding N-acyl homoserine lactonase activity. Appl Environ Microbiol. 2003;69:4989–4993. doi: 10.1128/AEM.69.8.4989-4993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H-B, Wang L-H, Zhang L-H. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krysciak D, Schmeisser C, Preuss S, et al. Involvement of multiple loci in quorum quenching of autoinducer I molecules in the nitrogen-fixing symbiont Rhizobium (Sinorhizobium) sp. strain NGR234. Appl Environ Microbiol. 2011;77:5089–5099. doi: 10.1128/AEM.00112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang W-Z, Morohoshi T, Someya N, Ikeda T. AidC, a novel N-acylhomoserine lactonase from the potato root-associated cytophaga-flavobacteria-bacteroides (CFB) group bacterium Chryseobacterium sp. strain StRB126. Appl Environ Microbiol. 2012;78:7985–7992. doi: 10.1128/AEM.02188-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chow JY, Wu L, Yew WS. Directed evolution of a quorum-quenching lactonase from Mycobacterium avium subsp. paratuberculosis K-10 in the amidohydrolase superfamily. Biochemistry. 2009;48:4344–4353. doi: 10.1021/bi9004045. [DOI] [PubMed] [Google Scholar]
- 83.Wang W-Z, Morohoshi T, Ikenoya M, et al. AiiM, a novel class of N-acylhomoserine lactonase from the leaf-associated bacterium Microbacterium testaceum. Appl Environ Microbiol. 2010;76:2524–2530. doi: 10.1128/AEM.02738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shevate SN, Shinde SS, Bankar AV, Patil NP. Identification of Quorum Quenching N-Acyl Homoserine Lactonases from Priestia aryabhattai J1D and Bacillus cereus G Isolated from the Rhizosphere. Curr Microbiol. 2023;80:86. doi: 10.1007/s00284-023-03186-3. [DOI] [PubMed] [Google Scholar]
- 85.Terwagne M, Mirabella A, Lemaire J, et al. Quorum sensing and self-quorum quenching in the intracellular pathogen Brucellamelitensis. PLoS ONE. 2013;8:e82514. doi: 10.1371/journal.pone.0082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park S-Y, Lee SJ, Oh T-K, et al. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology. 2003;149:1541–1550. doi: 10.1099/mic.0.26269-0. [DOI] [PubMed] [Google Scholar]
- 87.Liu D, Momb J, Thomas PW, et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures Biochemistry. 2008;47:7706–7714. doi: 10.1021/bi800368y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Camps J, Pujol I, Ballester F, et al. Paraoxonases as potential antibiofilm agents: their relationship with quorum-sensing signals in Gram-negative bacteria. Antimicrob Agents Chemother. 2011;55:1325–1331. doi: 10.1128/AAC.01502-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leadbetter JR, Greenberg EP. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J Bacteriol. 2000;182:6921–6926. doi: 10.1128/JB.182.24.6921-6926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin Y, Xu J, Hu J, et al. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003;47:849–860. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- 91.Huang JJ, Han J-I, Zhang L-H, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69:5941–5949. doi: 10.1128/AEM.69.10.5941-5949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu F, Byun T, Dussen H-J, Duke KR. Degradation of N-acylhomoserine lactones, the bacterial quorum-sensing molecules, by acylase. J Biotechnol. 2003;101:89–96. doi: 10.1016/S0168-1656(02)00305-X. [DOI] [PubMed] [Google Scholar]
- 93.Bokhove M, Jimenez PN, Quax WJ, Dijkstra BW. The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket. Proc Natl Acad Sci. 2010;107:686–691. doi: 10.1073/pnas.0911839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan K-G, Atkinson S, Mathee K, et al. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:1–14. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pustelny C, Albers A, Büldt-Karentzopoulos K, et al. Dioxygenase-mediated quenching of quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem Biol. 2009;16:1259–1267. doi: 10.1016/j.chembiol.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 96.Marin SDL, Xu Y, Meijler MM, Janda KD. Antibody catalyzed hydrolysis of a quorum sensing signal found in Gram-negative bacteria. Bioorg Med Chem Lett. 2007;17:1549–1552. doi: 10.1016/j.bmcl.2006.12.118. [DOI] [PubMed] [Google Scholar]
- 97.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 98.Park J, Jagasia R, Kaufmann GF, et al. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem Biol. 2007;14:1119–1127. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell–cell communication. Nature. 2005;437:750–753. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Piletska EV, Stavroulakis G, Larcombe LD, et al. Passive control of quorum sensing: prevention of Pseudomonas aeruginosa biofilm formation by imprinted polymers. Biomacromol. 2011;12:1067–1071. doi: 10.1021/bm101410q. [DOI] [PubMed] [Google Scholar]
- 101.Bryers JD, Jarvis RA, Lebo J, et al. Biodegradation of poly (anhydride-esters) into non-steroidal anti-inflammatory drugs and their effect on Pseudomonas aeruginosa biofilms in vitro and on the foreign-body response in vivo. Biomaterials. 2006;27:5039–5048. doi: 10.1016/j.biomaterials.2006.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Blázquez J. Antibiotics and antibiotic resistance: a bitter fight against evolution. Int J Med Microbiol. 2013;303:293–297. doi: 10.1016/j.ijmm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 103.Subhadra B, Oh MH, Choi CH. RND efflux pump systems in Acinetobacter, with special emphasis on their role in quorum sensing. J Bacteriol Virol. 2019;49:1–11. doi: 10.4167/jbv.2019.49.1.1. [DOI] [Google Scholar]
- 104.Wang Y, Liu B, Grenier D, Yi L. Regulatory mechanisms of the LuxS/AI-2 system and bacterial resistance. Antimicrob Agents Chemother. 2019;63:e01186–e1219. doi: 10.1128/AAC.01186-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pang Z, Raudonis R, Glick BR, et al. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37:177–192. doi: 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 106.Sawada I, Maseda H, Nakae T, et al. A quorum-sensing autoinducer enhances the mexAB-oprM efflux-pump expression without the MexR-mediated regulation in Pseudomonas aeruginosa. Microbiol Immunol. 2004;48:435–439. doi: 10.1111/j.1348-0421.2004.tb03533.x. [DOI] [PubMed] [Google Scholar]
- 107.Amaral L, Spengler G, Martins A, et al. Inhibitors of bacterial efflux pumps that also inhibit efflux pumps of cancer cells. Anticancer Res. 2012;32:2947–2957. [PubMed] [Google Scholar]
- 108.Varga ZG, Armada A, Cerca P, et al. Inhibition of quorum sensing and efflux pump system by trifluoromethyl ketone proton pump inhibitors. In Vivo (Brooklyn) 2012;26:277–285. [PubMed] [Google Scholar]
- 109.El-Shaer S, Shaaban M, Barwa R, Hassan R. Control of quorum sensing and virulence factors of Pseudomonas aeruginosa using phenylalanine arginyl β-naphthylamide. J Med Microbiol. 2016;65:1194–1204. doi: 10.1099/jmm.0.000327. [DOI] [PubMed] [Google Scholar]
- 110.Peterson RM, Huang T, Rudolf JD, et al. Mechanisms of self-resistance in the platensimycin-and platencin-producing Streptomyces platensis MA7327 and MA7339 strains. Chem Biol. 2014;21:389–397. doi: 10.1016/j.chembiol.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.White DG, Zhao S, Simjee S, et al. Antimicrobial resistance of foodborne pathogens. Microbes Infect. 2002;4:405–412. doi: 10.1016/S1286-4579(02)01554-X. [DOI] [PubMed] [Google Scholar]
- 115.Haque S, Ahmad F, Dar SA, et al. Developments in strategies for Quorum Sensing virulence factor inhibition to combat bacterial drug resistance. Microb Pathog. 2018;121:293–302. doi: 10.1016/j.micpath.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 116.Jiang Q, Chen J, Yang C, et al. Quorum sensing: a prospective therapeutic target for bacterial diseases. Biomed Res Int. 2019;2019:2015978. [DOI] [PMC free article] [PubMed]
- 117.Zhao X, Zhong J, Wei C, et al. Current perspectives on viable but non-culturable state in foodborne pathogens. Front Microbiol. 2017;8:580. doi: 10.3389/fmicb.2017.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li L, Hooi D, Chhabra SR, et al. Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene. 2004;23:4894–4902. doi: 10.1038/sj.onc.1207612. [DOI] [PubMed] [Google Scholar]
- 119.Arciola CR, Montanaro L, Costerton JW. New trends in diagnosis and control strategies for implant infections. Int J Artif Organs. 2011;34:727–736. doi: 10.5301/IJAO.2011.8784. [DOI] [PubMed] [Google Scholar]
- 120.Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dulla GFJ, Lindow SE. Acyl-homoserine lactone-mediated cross talk among epiphytic bacteria modulates behavior of Pseudomonas syringae on leaves. ISME J. 2009;3:825–834. doi: 10.1038/ismej.2009.30. [DOI] [PubMed] [Google Scholar]
- 122.Fei C, Mao S, Yan J, et al. Nonuniform growth and surface friction determine bacterial biofilm morphology on soft substrates. Proc Natl Acad Sci. 2020;117:7622–7632. doi: 10.1073/pnas.1919607117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.France MT, Cornea A, Kehlet-Delgado H, Forney LJ. Spatial structure facilitates the accumulation and persistence of antibiotic-resistant mutants in biofilms. Evol Appl. 2019;12:498–507. doi: 10.1111/eva.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kalia VC, Wood TK, Kumar P. Evolution of resistance to quorum-sensing inhibitors. Microb Ecol. 2014;68:13–23. doi: 10.1007/s00248-013-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Azizi S, Valipour A, Sithebe T. Evaluation of different wastewater treatment processes and development of a modified attached growth bioreactor as a decentralized approach for small communities. Sci World J. 2013;2013:156870. [DOI] [PMC free article] [PubMed]
- 126.Satheesh S, Ba-akdah MA, Al-Sofyani AA. Natural antifouling compound production by microbes associated with marine macroorganisms—A review. Electron J Biotechnol. 2016;21:26–35. doi: 10.1016/j.ejbt.2016.02.002. [DOI] [Google Scholar]
- 127.Zhang Y, Zhang Y, Yang Y, et al. Identification of a Pseudomonas sp. that inhibits RHL system of quorum sensing. Indian J Microbiol. 2013;53:28–35. doi: 10.1007/s12088-012-0340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qian P-Y, Xu Y, Fusetani N. Natural products as antifouling compounds: recent progress and future perspectives. Biofouling. 2009;26:223–234. doi: 10.1080/08927010903470815. [DOI] [PubMed] [Google Scholar]
- 129.Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol. 2010;86:1267–1279. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 130.Kim J-H, Choi D-C, Yeon K-M, et al. Enzyme-immobilized nanofiltration membrane to mitigate biofouling based on quorum quenching. Environ Sci Technol. 2011;45:1601–1607. doi: 10.1021/es103483j. [DOI] [PubMed] [Google Scholar]
- 131.Yeon K-M, Cheong W-S, Oh H-S, et al. Quorum sensing: a new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ Sci Technol. 2009;43:380–385. doi: 10.1021/es8019275. [DOI] [PubMed] [Google Scholar]
- 132.Smith MD, Roheim CA, Crowder LB, et al. Sustainability and global seafood. Science. 2010;327:784–786. doi: 10.1126/science.1185345. [DOI] [PubMed] [Google Scholar]
- 133.Defoirdt T, Sorgeloos P, Bossier P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr Opin Microbiol. 2011;14:251–258. doi: 10.1016/j.mib.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 134.Natrah FMI, Defoirdt T, Sorgeloos P, Bossier P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar Biotechnol. 2011;13:109–126. doi: 10.1007/s10126-010-9346-3. [DOI] [PubMed] [Google Scholar]
- 135.Teplitski M, Chen H, Rajamani S, et al. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 2004;134:137–146. doi: 10.1104/pp.103.029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Manefield M, Harris L, Rice SA, et al. Inhibition of luminescence and virulence in the black tiger prawn (Penaeus monodon) pathogen Vibrio harveyi by intercellular signal antagonists. Appl Environ Microbiol. 2000;66:2079–2084. doi: 10.1128/AEM.66.5.2079-2084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Annapoorani A, Parameswari R, Pandian SK, Ravi AV. Methods to determine antipathogenic potential of phenolic and flavonoid compounds against urinary pathogen Serratia marcescens. J Microbiol Methods. 2012;91:208–211. doi: 10.1016/j.mimet.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 138.Defoirdt T, Boon N, Bossier P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010;6:e1000989. doi: 10.1371/journal.ppat.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.García-Contreras R, Maeda T, Wood TK. Resistance to quorum-quenching compounds. Appl Environ Microbiol. 2013;79:6840–6846. doi: 10.1128/AEM.02378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maeda T, García-Contreras R, Pu M, et al. Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 2012;6:493–501. doi: 10.1038/ismej.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.García-Contreras R, Martínez-Vázquez M, Velázquez Guadarrama N, et al. Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Pathog Dis. 2013;68:8–11. doi: 10.1111/2049-632X.12039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the review are included in the article material. Further inquiries can be directed to the corresponding authors.