Abstract
The Pax gene family encodes transcription factors essential for organ and tissue development in higher eukaryotes. Pax proteins are modular with an N-terminal DNA binding domain, a C-terminal transcription activation domain, and a transcription repression domain called the octapeptide. How these domains interact with the cellular machinery remains unclear. In this report, we describe the isolation and characterization of a novel gene and its encoded protein, PTIP, which binds to the activation domain of Pax2 and other Pax proteins. PTIP binds to Pax2 in vitro, in the yeast two-hybrid assay and in tissue culture cells. The binding of PTIP to Pax2 is inhibited by the octapeptide repression domain. The PTIP protein contains five BRCT domains, first identified in BRCA1 and other nuclear proteins involved in DNA repair/recombination or cell cycle control. Pax2 and PTIP co-localize in the cell nucleus to actively expressed chromatin and the nuclear matrix fraction. For the first time, these results point to a link between Pax transcription factors and active chromatin.
INTRODUCTION
The Pax family of genes encodes DNA binding proteins which regulate the development of a variety of tissues in species as diverse as flies, worms and humans (1–3). Pax genes have also been implicated in the initiation and maintenance of the transformed phenotype. Specifically, Pax2 is highly expressed in Wilms’ tumor and renal cell carcinoma and is required for the proliferation of carcinoma-derived cell lines in vitro (4,5). The Pax3 and Pax7 genes are determinants of rhabdomyosarcoma through translocation with transcription factors of the fork-head gene family (6). Despite their critical roles during development and in human disease processes, the biochemical basis of Pax protein function within the cell nucleus and the context of chromatin structure remains obscure.
In mammals, there are nine known Pax genes grouped into four different classes based on the DNA binding paired domain (PD) sequence, gene structure and expression patterns (7). There is an absolute requirement for a functional PD, since the majority of Pax missense mutations occur within this coding region and result in impaired DNA binding activity. Nevertheless, mutations occurring outside the PD, which lead to large C-terminal deletions, indicate that this region is also critical for proper function. Furthermore, mutant phenotypes are dominant, such that heterozygotes with only one normal allele exhibit intermediate phenotypes compared to homozygous null animals. Thus, there is a strict quantitative level of the wild-type gene product required for normal function. For at least one Pax gene, Pax6 and its Drosophila homolog eyeless, it has been proposed that these genes are master regulatory switches, instructing cells to initiate a program of eye-specific development and differentiation (8). In mice and humans, the Pax2 gene is required for specification of the renal epithelium from the intermediate mesoderm (9–11) and also for eye and ear development (12).
Pax proteins have a conserved DNA binding domain, the PD, which spans 128 amino acids near the N-terminus and consists of two helix–turn–helix motifs (13). Sequence conservation among Pax proteins is highest in the PD, but can also extend to a paired-type homeodomain (HD) and a stretch of residues between the PD and HD called the octapeptide. The HD represents a second bona fide DNA binding moiety in some classes of Pax proteins that may enhance DNA target specificity in cooperation with the PD (14–17). However, the Pax2/5/8 subfamily of proteins have only a partial HD which is lacking the third helix and is unlikely to participate in DNA binding (18). Transcription activation and repression activities map to the C-terminus of Pax proteins (19–21). For example in the Pax2 protein, amino acids 197–415 are required for maximal transcription transactivation in cultured mammalian cells (22). This activation domain, generally rich in proline (P), serine (S), threonine (T) and tyrosine (Y), is shared among related Pax proteins. As multiple regions appear to constitute a functional transactivation domain it is not surprising to find that missense mutations have rarely been identified in the C-terminal coding region of Pax genes. Pax2 also contains an octapeptide motif, located between the PD and the downstream activation domain, which is able to inhibit transactivation (22). This octapeptide is conserved in all but the Pax6 class of proteins and is similar to the repressor domains encoded by the engrailed and goosecoid homeobox genes (23,24). In all these proteins, the octapeptide is found N-terminal to the HD, suggesting a potential interaction that has survived evolution.
To understand how these domains regulate transcription, the full-length Pax2 protein was used as a bait in a yeast two-hybrid screen. We have identified a novel nuclear protein, called PTIP, which is able to bind the C-terminus of several Pax proteins. The degree of interaction with the Pax2 C-terminal polypeptides correlates with their transcription transactivation potential and we have therefore designated this factor PTIP for Pax transactivation-domain interacting protein. PTIP contains five copies of the BRCT domain, common to many proteins involved in cell cycle control in response to DNA damage. The PTIP gene is expressed in all tissues and cell lines examined and is associated with active chromatin and the nuclear matrix. These results suggest that PTIP is a common nuclear factor utilized by Pax proteins to affect gene expression.
MATERIALS AND METHODS
Yeast two-hybrid assay
Yeast manipulations (media, culturing and transformations) were carried out as described (25). pPC-Pax2a (see below) was co-transformed with a Gal4 activation domain (GAD):mouse embryonic cDNA fusion library in a 2:1 molar ratio into MaV103 (26) yeast cells and plated on synthetic medium lacking leucine, tryptophan and histidine and containing 35 mM 3-aminotriazole. Approximately 3 × 106 total co-transformants were screened, of which ~300 were judged HIS+ and ~150 lacZ+ when tested by Xgal staining of colony filter lifts. Library plasmids were recovered in Escherichia coli and re-tested for interaction in the two-hybrid system with either the Gal4 DNA binding domain (GDBD) alone, GDBD–Pax2a or unrelated control fusions of DLK (a gift of Dr L. Holzman) and DCC (a gift of Dr E. Fearon). Quantitation of the interaction was assessed by measuring β-galactosidase activity in liquid cultures of MaV103 cells co-transformed with plasmids for the appropriate hybrids. Cultures were grown to mid-log phase and OD600 recorded. Aliquots of cells were removed, pelleted and washed in Z buffer (100 mM NaPO4 pH 7.0, 10 mM KCl, 1 mM MgSO4) and cycled through three rounds of freeze/thaw. ONPG solution (0.67 mg/ml, 28 mM β-mercaptoethanol in Z buffer) was added to the cells and reactions were allowed to proceed at 30°C for 1–3 h. Product formation was monitored by measuring absorbance at 420 nm. Units of β-galactosidase were adjusted for amount of cells used (OD600) and time of incubation. At least two independent colonies were assayed for each co-transformation. Expression of GDBD–Pax2 fusions and GAD–PTIP fusions was confirmed by western blot analysis using antibodies against Pax2, PTIP or GAD (Santa Cruz Biotechnology, Santa Cruz, CA) (data not shown). The positive control fusion proteins, fos/jun or full-length Gal4 were also assayed as described above.
Plasmids and cDNA cloning
The Pax2a open reading frame was fused to the GDBD by inserting a SfiI–BamHI fragment, encoding a flag epitope (DYKDDDDK) preceding amino acids 6–182 of Pax2, into pAS-Cterm2a, generating pAS-PD-Cterm2a. pAS-Cterm2a harbors an EcoRI–BamHI fragment, encoding amino acids 197–415 of Pax2a, in the yeast expression vector pAS2.1 (Clontech, Palo Alto, CA). pAS-PD-Cterm2a lacks amino acids 183–196, which includes the octapeptide motif of Pax2, and the complete coding sequence of Pax2a or Pax2b was generated by swapping in FseI–SalI fragments from pSV-Pax2a and bHA (27). The resultant plasmids were named pAS-Pax2a and pAS-Paxb. pAS-PD was created by deletion of a BamHI fragment from pAS-PD-Cterm2a. GDBD fusions to portions of the Pax2 or Pax6 C-terminus shown in Figure 1 were created by inserting EcoRI–BamHI fragments from fusion constructs previously described (22) into pAS2.1. Pax8 sequences were fused to GDBD using a PvuII–NotI fragment from pCMV-Pax8 inserted into the SmaI and NotI sites of pPC97 (28). This plasmid was then digested with SalI, filled in with Klenow enzyme and re-ligated to preserve the open reading frame. All GDBD–Pax2 fusions in pAS2.1 were subcloned into expression vector pPC97 as HindIII–SpeI fragments. Sequencing confirmed that the open reading frame was maintained across fusion junctions in each clone.
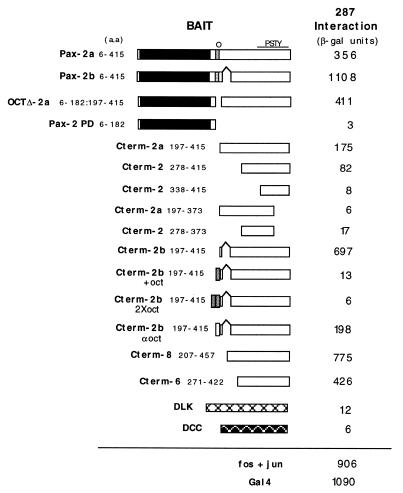
Figure 1.
Interaction between clone 287 (PTIP) and Pax2 in the yeast two-hybrid system. Yeast strain MaV103 was co-transformed with expression plasmids for clone 287 fused to the GAD and Pax2 fused to the GDBD. The portions of Pax2 (or Pax6 and Pax8) encoded by the plasmids are indicated schematically and by amino acid number of the full-length proteins. The paired domain is indicated by a black box, the octapeptide by a shaded box and the region rich in proline, serine, threonine and tyrosine residues by PSTY. Control baits were the mixed lineage kinase DLK and netrin receptor DCC fused to the GDBD. Co-transformants were grown in liquid medium and assayed for activation of the lacZ reporter by measuring β-galactosidase activity. At least two independent clones were tested and an average value is given. The background activity of each GDBD–Pax2 fusion alone is subtracted from this value in every case.
The PTIP cDNA sequence was derived from clone 287 (pPC86-PTIP) from the GAD:cDNA fusion library. This clone contained a 3.5 kb insert in pPC86 (28) with a poly(A) tract and two putative polyadenylation signals at the 3′-end. A 1.0 kb EcoRI fragment from the 5′-end was used to probe a mouse embryonic kidney cDNA library from which a clone was obtained which extended ~300 bp further in the 5′ direction. The complete PTIP sequence is available from GenBank (accession no. AF104261). pMYC-PTIP was made by inserting a SmaI–SpeI fragment from pPC86-PTIP into SmaI and XbaI sites in mammalian expression vector pMYCrk5. pHIS-PTIP was made by inserting an EcoRI–SpeI fragment produced by partial EcoRI digestion of pPC86-PTIP into EcoRI and XbaI sites in pcDNA3.1HisA (Invitrogen, Carlsbad, CA). To create the His-tagged PTIP yeast expression plasmid, a PvuI fragment was swapped between pPC97 and pPC86-PTIP, resulting in pPC86-PTIP-LEU, which carries the LEU2 rather than the TRP1 marker. A HindIII fragment from pHIS-PTIP was then inserted into the corresponding sites in pPC86-PTIP-LEU replacing the N-terminal GAD–PTIP with the HIS–PTIP fusion sequence. A PvuI fragment swap was also performed between pPC86 and pPC97-Pax2a to create pPC97-Pax2a-TRP. Fusion of PTIP to the GDBD was produced by inserting a SalI–NotI fragment from pPC86-PTIP into pPC97. Two HIS-tagged bacterial expression constructs were made by inserting either EcoRI–EcoRI (amino acids 8–316) or EcoRI–HindIII (amino acids 316–591) fragments from pPC86-PTIP into pRSETA and pRSETC (Invitrogen), respectively. To create GST–Pax2 expression plasmids, Pax2b sequences were amplified by PCR with primers corresponding to amino acids 278–373, 197–415 or 160–415. The primers also incorporated BamHI or BglII restriction enzyme sites which were used for insertion into the BamHI site in pGEX-2TK (Pharmacia, Piscataway, NJ).
Northern blot analysis
Tissues from adult and embryonic (gestational day 17) mice were dissected and total RNA was isolated using Trizol reagent (Gibco BRL, Rockville, MD). An aliquot of 10 µg total RNA was electrophoresed in 1% agarose gel containing formaldehyde, blotted to nylon membrane and probed with a 1 kb EcoRI fragment from the PTIP cDNA.
Anti-PTIP antibodies
His-tagged PTIP proteins were expressed in and purified from BL21(DE3) bacterial cells transformed with pRSET/8–316PTIP and pRSET/316–591PTIP according to the manufacturer’s instructions (Invitrogen). Approximately 2.0 mg recombinant protein were isolated from each 500 ml culture using denaturing conditions. Purified proteins were dialyzed into PBS with 0.1% NP-40 and injected into chickens for polyclonal antisera production (Aves Labs, Tigard, OR). Each antigen was injected into two different hens. Purified immunoglobulin fractions of preimmune and immune antisera were obtained and specificity was confirmed by western blot (data not shown). Anti-PTIP/1167 antiserum and anti-PTIP/1169 antiserum recognize epitopes in PTIP polypeptides spanning amino acids 8–316 and 316–591, respectively. Both antisera were purified by affinity chromatography with the corresponding antigen crosslinked to Amino Link Plus beads (Pierce, Rockford, IL). Affinity-purified PTIP antisera were used in each of the applications described below.
In vitro protein interaction
GST–Pax2 proteins were purified from BL21(DE3) bacterial cells transformed with pGEX-2TK, pGEX-2TK:278–373, pGEX-2TK:197–415b or pGEX-2TK:160–415b. At OD600 = 0.6, IPTG (0.4 mM final) was added to cultures to induce fusion protein expression and cultures were harvested 3 h later. Cells were resuspended in PBS, 25% sucrose, 1 mM EDTA, 1 mM DTT and the following protease inhibitor cocktail: 1 mM PMSF, 2 µg/ml aprotinin, 1 µg/ml pepstatin and 1 µg/ml leupeptin. This and subsequent steps were carried out at 4°C, except where noted. EDTA was added to a final concentation of 50 mM and cells lysed by three cycles of freeze/thaw followed by sonication to shear DNA. Triton X-100 was added to a final concentration of 1% and lysates were mixed on ice for 5–10 min. Insoluble material was pelleted by centrifugation at 10 000 g for 30 min. The supernatant was then subjected to affinity chromatography using glutathione–agarose beads (Sigma, St Louis, MO). After binding, beads were washed with 10 vol PBS with 1.0% Triton X-100 and protease inhibitor cocktail and then with an equal volume of the same buffer with 0.5% instant milk and 100 mM ATP at room temperature to elute ~70 kDa contaminating heat shock protein (29). Proteins were eluted with 5 mM glutathione for 10 min at room temperature and dialyzed against PBS with 0.1% NP-40. Protein concentration was judged from Coomassie blue stained gels using BSA as the standard.
Radiolabeled PTIP polypeptides were synthesized by coupled in vitro transcription and translation reactions from pMYC-PTIP or pHIS-PTIP templates with SP6 or T7 polymerase, respectively, in the presence of Trans-35S-label (ICN, Costa Mesa, CA) according to the manufacturer’s instructions (Promega, Madison, WI). Incorporation of [35S]amino acids was confirmed by SDS–PAGE and autoradiography. For binding experiments, GST–Pax2 proteins were bound to glutathione–agarose beads and preblocked by 1 h incubation in NP-40 buffer (150 mM NaCl, 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 1 mM DTT plus protease inhibitor cocktail) with 12.5% normal rabbit serum. PTIP polypeptides synthesized in vitro were precleared by incubating with 100 ml of a 50% slurry of glutathione–agarose beads in NP-40 buffer. Equal amounts of precleared PTIP polypeptide were incubated with 1–2 µg GST fusion protein on beads and incubated for 2–3 h, after which the beads were washed three times with 1 ml of NP-40 buffer. Retained protein complexes were boiled for 5 min in Laemmli sample buffer, separated by SDS–PAGE and subjected to autoradiography.
Mammalian cell expression and co-immunoprecipitation
NIH 3T3 cells were transiently transfected with pMYC-PTIP and/or p1-415bHA (22) using lipofectamine reagent as per the manufacturer’s instructions (Gibco BRL). Samples of 1 × 106 cells were transfected with pMYC-PTIP, p1-415bHA or pCB6+ (empty CMV expression plasmid) using a total of 15 µg plasmid DNA. Cells were harvested 48 h later and lysed in NP-40 buffer. Cell lysis was performed on ice and all subsequent steps were carried out at 4°C. Lysates were clarified by incubating on ice for 15 min and centrifuged for 15 min at 10 000 g. Supernatants were precleared for 1 h by incubating with 100 µl of a 50% slurry of protein G–Sepharose (Pharmacia), preimmune chicken antisera, mouse IgG2b and IgG1 (10 µg each; Sigma). Precleared extracts were immunoprecipitated with 2–5 µg appropiate antibody and incubated for 3–12 h before addition of 40 µl of protein G–Sepharose slurry. Antibodies used were anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim Biochemical, Indianapolis, IN), anti-myc monoclonal antibody 9E10 (Calbiochem, San Diego, CA) and polyclonal anti-PTIP chicken antisera 1167 and 1169. For anti-PTIP immunoprecipitations, fixed anti-chicken IgY (Promega) was used to preclear extracts and collect immune complexes rather than protein G–Sepharose. Reactions were incubated for another 30 min and then beads were washed three times with 1 ml of NP-40 buffer. Protein complexes were boiled for 5 min in Laemmli sample buffer, separated by SDS–PAGE and electroblotted to PVDF membranes. In some cases two membranes were used per gel and western blots were performed using anti-PTIP 1167 antiserum (1:5000 dilution) on the first membrane and anti-Pax2 antiserum (1:2000 dilution) on the second. Horseradish peroxidase-conjugated secondary antibodies, anti-chicken IgY (Promega) or anti-rabbit IgG (Bio-Rad, Hercules, CA) were used at a 1:5000 dilution and antigen–antibody complexes visualized with ECL reagents (DuPont-NEN).
Indirect immunofluorescence
NIH 3T3 cells grown in glass chamber slides were washed in PBS and fixed for 5 min in 4% paraformaldehyde/PBS. Cells were washed again in PBS, permeablized with 0.1% Triton X-100/PBS for 10 min and then washed twice in 0.1% Tween 20/PBS. Cells were incubated for 60–90 min with a 1:1000 dilution of anti-PTIP antiserum (1167 or 1169) or preimmune antiserum. Alternatively, cells were incubated with PTIP antiserum which had been preincubated with a molar excess of purified antigen. All antibodies were diluted in 0.1% Tween 20/PBS with 2% normal goat serum. Cells were washed twice with 0.1% Tween 20/PBS to remove excess primary antibodies and then incubated with FITC-conjugated goat anti-chicken secondary antibodies (1:300 dilution; Sigma) for 30 min. Cells were washed and then mounted on coverslips with Gelvatol containing 2.5% DABCO.
Chromatin fractionation
Nuclei were prepared by centrifugation through sucrose step gradients as described (27). The micrococcal nuclease digestion procedure was as described by Rose and Garrard (30) with the modifications of Reyes et al. (31). Briefly, 107 nuclei were resuspended in 200 µl of nuclear buffer (20 mM Tris pH 7.6, 70 mM NaCl, 20 mM KCl, 5 mM MgCl2, 3 mM CaCl2 and protease inhibitors). The nuclei suspension was digested with 3 U microccocal nuclease for 1, 2 or 5 min at 37°C. Digestions were stopped with EDTA and EGTA, added to 5 mM. After 5 min at maximum speed in an Eppendorf centrifuge, the supernatant was designated the S1 fraction. The pellet was lysed in 2 mM EDTA at 4°C, with slow rotation. After centrifugation, the sample was split into the supernatant, or S2 fraction, and the pellet fraction. Proteins were analyzed by adding 2× SDS–PAGE loading buffer to the S1 and S2 fractions and 1× SDS–PAGE to the pellet and running an equal number of cell equivalents on acrylamide SDS–PAGE gels. DNA was isolated by protease K digestion and phenol/chloroform extraction.
Nuclear matrix isolation
The cells were washed in PBS and extracted with 0.5 ml (per 100 mm dish) of CSK buffer (10 mM PIPES pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mm EGTA, 0.5% Triton X-100, 1 mM DTT and protease inhibitors) according to the method of He et al. (32). After 5 min at 4°C the monolayer was scraped off and centrifuged at 5000 g for 2 min. The soluble cytoplasmic fraction was removed and the pellets resuspended in 200 µl of CSK buffer with protease inhibitors and 100 U RNase-free DNase I. After 15 min at 37°C, ammonium sulfate was added from a 4 M stock to a final concentration of 0.25 M. The samples were rotated at 4°C for 5 min and centrifuged. The supernatant, or chromatin fraction, was removed and the pellets extracted in CSK buffer with 2 M NaCl. After further centrifugation, the 2 M NaCl fraction was removed and the pellets lysed directly in 1× SDS–PAGE. For western blot analyses, equal cell equivalents were loaded from each fraction and probed with the various antibodies. Anti-BRM was from Transduction Laboratories, (Lexington, KY). Anti-glucocorticoid receptor was a gift of D. Robins.
RESULTS
Identification of a Pax2 interacting clone in the yeast two-hybrid system
In order to identify potential cofactors that mediate Pax2-dependent transactivation, a yeast two-hybrid screen was undertaken using either the whole Pax2 coding region or the previously defined C-terminal Pax2 transactivation domain fused in-frame to the GDBD. More than 3 × 106 clones of an embryonic mouse cDNA library were screened and 300 surviving co-transformants were picked for the β-galactosidase assay. Plasmid DNA was isolated from the 150 lacZ+ co-transformants and retransformed into yeast with the Pax2 bait plasmid. Only a single clone (287) was able to activate the HIS and LacZ reporter genes upon retransformation. Clone 287 interacted with both the full-length Pax2 bait and the C-terminal activation domain, but not with unfused or control baits (Fig. 1). Deletions made in the C-terminal fragment, containing the partial HD and PSTY-rich domain, greatly diminished the interaction with clone 287 in the yeast assays. The presence or absence of the Pax2 octapeptide motif also affects interaction in yeast. In comparison to full-length Pax2a, an octapeptide deletion form was able to interact with clone 287 at a modestly higher level. Pax2b C-terminal baits (amino acids 197–415) containing one or two copies of the octapeptide element failed to interact with clone 287. An unrelated peptide inserted at the octapeptide position reduced, but clearly did not abolish, interaction with clone 287. C-terminal fragments from both Pax6 and Pax8 were also capable of interacting with clone 287 (Fig. 1), suggesting that it may interact with all members of the Pax family. As negative control baits, the kinase DLK and the netrin receptor DCC did not show any interaction with 287. Clone 287 interacts with both splice forms of Pax2, although higher activation is observed with Pax2b. This may be a result of the increased level of Pax2b relative to Pax2a when the proteins are expressed exogenously in yeast or mammalian cells (22).
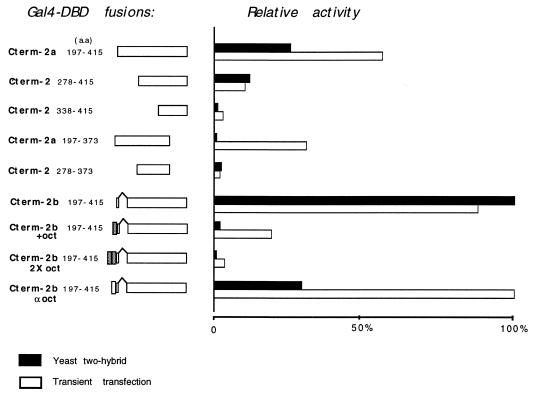
Deletion mutants with reduced potential to activate transcription in transfected cells also bound poorly to clone 287 in the yeast two-hybrid assay (Fig. 2). The GDBD–Pax2 fusion constructs were tested for transcription activation by transient transfection in NIH 3T3 cells using a CAT reporter plasmid with five copies of the Gal4 upstream activating sequence. The same Gal4–Pax2 fusion proteins were tested for their ability to bind PTIP in yeast. Although the absolute values are not comparable between the two assays, the relative activity of each GDBD–Pax2 chimera is similar within the assays. This relationship suggests that the product of clone 287 may play a functional role in Pax2-dependent transcriptional regulation. Based on these observations we have designated it PTIP, for Pax transactivation domain interacting protein.
Figure 2.
Relative transcription activation activity of the Pax2 C-terminal domains and their affinitites for PTIP in the yeast two-hybrid assay. GDBD–Pax2 fusions were tested for activation of a CAT reporter plasmid containing multiple copies of the Gal4 upstream activation sequence (open bars). The same fusion proteins were also analyzed for interaction with PTIP in the two hybrid system (solid bars). The bars indicate the relative average activity for each fusion in the two assays.
Cloning and sequence characterization of PTIP
A full-length PTIP cDNA was obtained by screening additional murine cDNA libraries with a probe from yeast two-hybrid cDNA clone 287. The predicted primary sequence from the complete PTIP cDNA (GenBank accession no. AF104261) is shown in Figure 3. The open reading frame from the PTIP cDNA encodes a polypeptide of 1056 residues with a predicted molecular weight of 119.3 kDa and a pI of 6.8. The sequence near the putative initiating methionine is preceded by two in-frame stop codons and generates a 271 nt 5′ untranslated region. Although the sequence surrounding this methionine codon departs from the consensus Kozak sequence (33), no other ATG codons were present in the 5′ sequence which were in-frame with the open reading frame predicted from yeast two-hybrid cDNA clone 287.
Figure 3.
Primary amino acid sequence from the conceptual translation of the PTIP cDNA. The polypeptide sequence was deduced from the full-length cDNA which was generated from overlapping yeast and phage clones. The five BRCT domains are underlined and a central glutamine-rich region spans approximately residues 396–577. The complete PTIP sequence is available under GenBank accession no. AF104261.
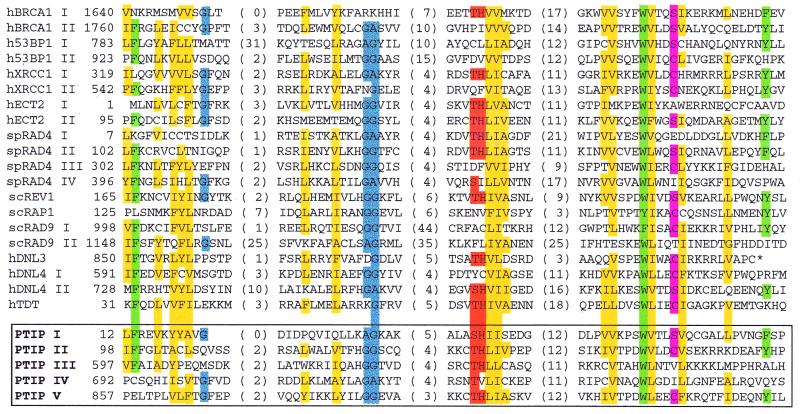
Initial database searches failed to show strong similarity to any known proteins and no obvious structural motifs were found from analysis of the PTIP primary sequence. However, the yeast rad4/cut5 protein has several domains with 30–35% similarity to regions of PTIP. This similarity to rad4/cut5 was limited to the BRCT domains, a protein module identified in BRCA1 using profile and motif search methods (34,35) and hydrophobic cluster analysis (36). The BRCT domain is roughly 100 amino acids in size and is present in numerous nuclear proteins which are involved in DNA repair, recombination or cell cycle checkpoint control (34,36). Based on the defining criteria, we have assigned five likely BRCT domains in the PTIP protein, two at the N-terminus and three at the C-terminus of PTIP (Fig. 4). Between the BRCT domains there is a glutamine (Q)-rich region, with 95 glutamines spread over 181 residues including a homopolymeric stretch of 13 glutamine residues.
Figure 4.
Alignment of amino acid residues conserved among BRCT domains. Alignment is based upon BLAST analysis of the non-redundant protein sequence database and previously described hydrophobic cluster analysis (36). The first amino acid position of each domain is given as well as the number of intervening residues between conserved blocks. An asterisk indicates a stop codon. Yellow boxes, A, F, G, I, L, V, Y; blue boxes, G, A; green boxes, F, W, Y; pink boxes, S, C; orange boxes, H, S, T. h designates human protein sequences, sc, Saccharomyces cerevisiae and sp, Schizosaccharomyces pombe. The five BRCT domains (I–V) of murine PTIP are outlined by the open box.
A partial human cDNA sequence has been described (37) (GenBank accession no. U80735) whose conceptual protein is >90% identical to murine PTIP from amino acid 650 to 1056. Using a radiation hybrid panel, this human sequence was mapped to chromosome 7q36. We have also identified human genomic clones using mouse PTIP as probe and mapped these clones to 7q36 by FISH analysis, confirming the genetic mapping (data not shown).
Pax2 and PTIP physically associate
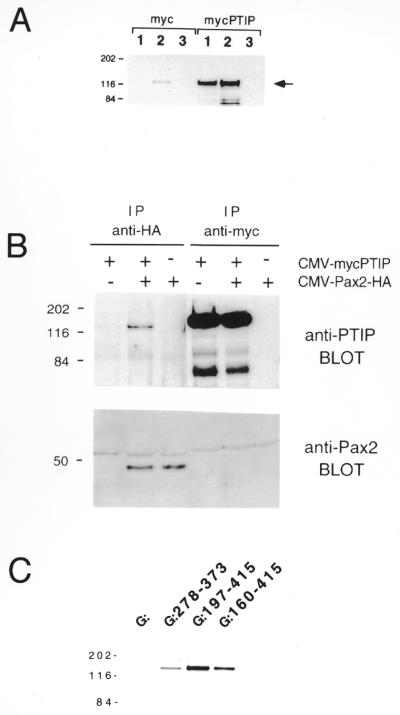
In order to examine the PTIP protein in eukaryotic cells, antibodies were generated against two different bacterial fusion proteins, 1167 and 1169, which correspond to amino acids 8–316 and 316–591, respectively. Endogenous PTIP was compared to PTIP generated with a myc-tagged expression vector (Fig. 5A). Immunoprecipitation with 1167 antiserum followed by western blotting analysis with 1169 antiserum detected a 130 kDa protein from cells transfected with a control empty expression plasmid (Fig. 5A, lane 2, left). A similar sized protein was detected in mycPTIP transfected cells by immunoprecipitation with anti-myc (Fig. 5A, lane 1, right) or anti-PTIP-1167 (Fig. 5A, lane 2, right). These data demonstrate that endogenous PTIP is an ∼130 kDa nuclear protein and that our cDNA sequence most probably covers the complete coding region.
Figure 5.
Pax2 and PTIP interactions. (A) Specificity of anti-PTIP antibodies. Extracts were prepared from cells transiently transfected with control expression vector (pMYC) or pMYC-PTIP and immunoprecipitated with affinity purified anti-PTIP-1167 (lane 2) or anti-myc monoclonal antibody (lane 1). As a negative control, a chicken anti-mTcf4 antibody was used (lane 3). Immunocomplexes were collected with anti-chicken IgY affinity resin or protein G–Sepharose, washed and separated by SDS–PAGE. Western blots were probed with anti-PTIP-1169. Molecular weight standards are shown to the left (kDa) and the ~130 kDa PTIP protein is indicated by an arrow. Note the endogenous PTIP protein in lane 2, myc panel, and the transfected PTIP protein in lanes 1 and 2, mycPTIP panel. (B) Co-immunoprecipitation of Pax2 and PTIP. NIH 3T3 cells were transiently transfected with expression vectors for epitope-tagged forms of Pax2 and PTIP or a control expression vector (indicated above lanes by + or –). Cell extracts were immunoprecipitated with anti-HA or anti-myc monoclonal antibodies and the immune complexes collected with protein G–Sepharose, washed and separated by SDS–PAGE. Proteins were analyzed by western blotting with polyclonal antisera against either Pax2 or PTIP. Co-immunoprecipitation of PTIP with Pax2 was observed (second lane). Molecular weight standards are shown to the left (kDa). (C) [35S]PTIP from in vitro transcription/translation reactions was incubated with GST–Pax2 fusion proteins. The regions of Pax2b fused to GST are indicated by the amino acids above each lane. After incubation, glutathione–agarose beads were washed and the protein complexes separated by SDS–PAGE. Gels were stained with Coomassie blue to visualize integrity and loading of GST–Pax2 proteins, amplified for fluorography and exposed to film. Experiments for each binding reaction were repeated at least twice and typical results for full-length PTIP are shown.
Evidence that PTIP and Pax2 proteins form a complex in eukaryotic cells was obtained by co-immunoprecipitation experiments (Fig. 5B). NIH 3T3 cells were transiently transfected singly or in combination with expression vectors encoding epitope-tagged forms of PTIP and Pax2. Extracts from these cells were immunoprecipitated with monoclonal antibodies recognizing the epitope tags and then the bound proteins were subjected to western blot analysis after separation by SDS–PAGE. Western blots developed with polyclonal anti-PTIP antiserum showed an ∼130 kDa protein immunoprecipitated by the monoclonal antibody to the epitope-tagged PTIP (Fig. 5B). PTIP co-immunoprecipitated with HA-tagged Pax2 using a monoclonal anti-HA antibody. The 130 kDa PTIP protein was not co-immunoprecipitated in cells which did not express Pax2, indicating that co-immunoprecipitation is not a result of non-specific binding. However, the reciprocal experiment using an anti-myc monoclonal antibody to immunoprecipitate PTIP did not pull down detectable amounts of Pax2. This may be due to the lower affinity of the anti-myc antibody, the position of the epitope tag in PTIP, or other structural constraints in the anti-myc/PTIP complex.
To confirm that PTIP and Pax2 proteins physically associate, recombinant GST–Pax2 fusion proteins were purified from bacteria and used in binding reactions with PTIP produced by in vitro translation. Results of the binding reactions show that full-length PTIP is specifically bound by GST–Pax2 fusions and not by the unfused GST protein (Fig. 5C). While moderate binding of PTIP to amino acids 278–373 was found, the highest level observed was with amino acids 197–415 of Pax2. A larger C-terminal fragment of Pax2 (amino acids 160–415), which includes the octapeptide motif, bound PTIP less efficiently. These data are consistent with the level of interaction determined by the yeast two-hybrid system and indicate that the octapeptide motif of Pax proteins may reduce the affinity for PTIP.
Expression and localization of PTIP
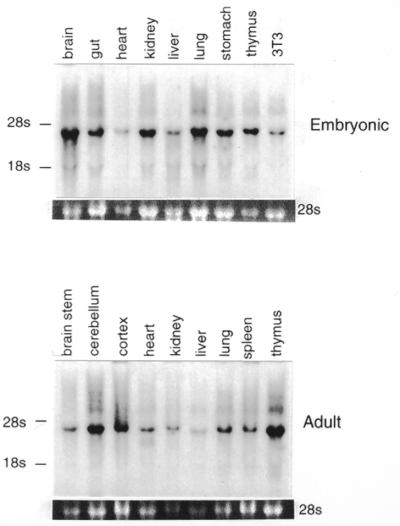
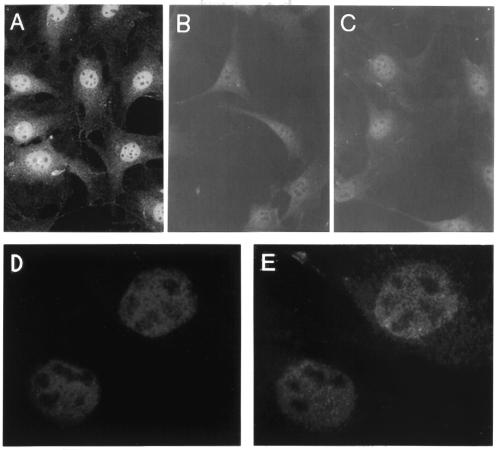
PTIP mRNA is expressed in both adult and embryonic tissues (Fig. 6). By northern blotting, a single transcript of ∼4 kb, closely matching the size of the full-length PTIP cDNA, was detected in all tissues examined. Although the levels are variable, particularly high levels are seen in the embryonic kidney and brain, which also express high levels of Pax2. In situ hybridization also detects widespread expression of PTIP during several stages of embryonic mouse development (data not shown). In addition, PTIP mRNA is present in all human and mouse cell lines we have examined (data not shown). In NIH 3T3 cells, strong nuclear staining is detected in all cells using anti-PTIP 1167 antiserum, which was generated against amino acids 8–361 (Fig. 7A). The specificity of this signal was verified by the lack of staining observed with either preimmune antiserum (Fig. 7B) or by preincubating the antiserum with antigen (Fig. 7C). Double labeling of Pax2 transfected NIH 3T3-derived stable cell lines with anti-PTIP and anti-Pax2 antibodies showed an overlapping pattern of nuclear staining (Fig. 7D and E). Pax2 and PTIP are detected throughout the nucleus but not in the nucleoli. Staining is diffuse but granular. Overall, the data indicate that the PTIP protein is nuclear and co-localizes, at the gross level, with Pax2.
Figure 6.
Northern blot analysis of PTIP mRNA expression. An aliquot of 10 µg of total RNA from various tissues of embryonic and adult mice was separated by denaturing agarose gel electrophoresis, blotted to nylon membrane and probed with a 1 kb EcoRI fragment from the murine PTIP cDNA. 3T3 designates RNA isolated from cultured NIH 3T3 cells. A single PTIP transcript is found with an estimated size of 4000 nt. The position of the 28S and 18S rRNAs are indicated. Variation in RNA loading is shown by the relative intensities of the 28s ribosomal band.
Figure 7.
Endogenous PTIP is localized to the nucleus. Affinity purified chicken antisera to murine PTIP were used for indirect immunofluorescent staining of NIH 3T3 cells. (A) Anti-PTIP 1167 antiserum. (B) Preimmune 1167 antiserum. (C) Anti-PTIP 1167 antiserum preincubated with immunizing antigen. (D) Pax2 transformed NIH 3T3 cells stained with anti-Pax2 antibodies. (E) Same cells as (D) stained with anti-PTIP.
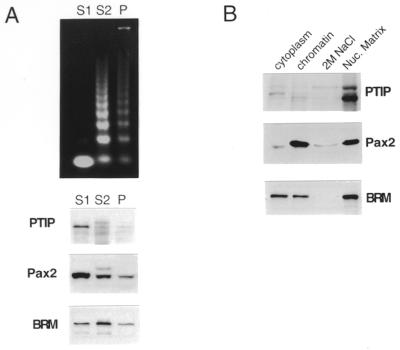
To refine the localization of both Pax2 and PTIP, nuclei from transformed embryonic kidney cells and Pax2 transformed NIH 3T3 cells were subject to limited micrococcal nuclease digestion. After 2 min digestion, the soluble chromatin (S1 fraction), the bulk chromatin (S2 fraction) and the insoluble pellets were assayed for DNA and protein (Fig. 8A). Both Pax2 and PTIP were found predominantly in the soluble S1 fraction, representing >80% of the total Pax2 and PTIP in the cell. In contrast, the mouse BRM1 protein is found in all fractions, with much of the total BRM1 in the bulk chromatin or S2 fraction. In addition, the nuclei were extracted with 2 M NaCl after DNase I digestion to enrich for nuclear matrix-associated proteins (Fig. 8B). Under these conditions, PTIP co-purifies predominantly with the nuclear matrix fraction, whereas Pax2 is found in the soluble chromatin fraction and the nuclear matrix fraction (Fig. 8B). Thus, ∼50% of the Pax2 protein is in the matrix fraction where it may interact with PTIP. As a control for the purification procedure, the glucocorticoid receptor was assayed in normal and dexamethasone-treated cells (data not shown). The glucocorticoid receptor is cytoplasmic in the absence of dexamethasone but becomes associated with the nuclear matrix upon stimulation. These data suggest that the Pax2–PTIP interaction occurs within the context of the nuclear matrix and is associated with active, micrococcal nuclease-sensitive chromatin.
Figure 8.
Association of PTIP with active chromatin and the nuclear matrix. (A) Micrococcal nuclease digestion of nuclei. (Top) An aliquot of 1 µg DNA extracted from fractions S1, S2 and pellet. Western blots of protein fractions shown below with antibodies against the indicated proteins. (B) Extraction of nuclear matrix. Cells were separated into cytoplasmic fractions, whole chromatin, 2 M NaCl extracted chromatin and insoluble nuclear matrix. Proteins from each fraction were analyzed by western blotting with the indicated antibodies.
DISCUSSION
PTIP is a novel protein that interacts with the C-terminal transactivation domain of Pax2 and is associated with actively expressed chromatin. Although initially identified in a yeast two-hybrid screen, interaction between Pax2 and PTIP was verified through in vitro biochemical association assays and co-immunoprecipitation from tissue culture cells. Furthermore, C-terminal deletions of Pax2 that are inefficient transactivators in tissue culture cells do not interact with PTIP in yeast, whereas Pax2 constructs that bound PTIP strongly, in yeast and in vitro, also exhibited the highest amount of transactivation. These are consistent with a role for PTIP in mediating Pax-dependent transactivation. Significantly, both Pax2 and PTIP can be found associated with the nuclear matrix and with active chromatin.
PTIP contains five copies of the BRCT domain, as identified by hydrophobic cluster analysis. These domains were first identified at the C-terminal end of the BRCA1 protein and are essential for BRCA1 tumor suppressor function (38). The two BRCT domains from BRCA1 can also act as transcription activator motifs when fused to a DNA binding domain (39,40) and mediate the association with RNA polymerase II holoenzyme (41). It has been proposed that the BRCT domain acts as a protein–protein binding domain, as has been demonstrated between the BRCT domain proteins BRCA1 and BARD (42), rad4/cut5 and Crb2 (43) or XRCC1 and human DNA ligase III (44). Thus, the BRCT domains may enable the assembly of multiprotein complexes with various enzymatic activities. For example, BRCT domains are found in proteins with DNA ligase, DNA polymerase, DNA binding and guanyl nucleotide exchange activities (34,36). Where known, the majority of proteins in this group function in DNA recombination or repair and cell cycle control. For example, rad4/cut5 is essential for DNA replication and sensitivity to UV irradiation (45,46) and XRCC1 is involved in repairing DNA after treatment with alkylating agents or ionizing radiation (47).
A second notable feature of the PTIP protein is the glutamine-rich domain spanning amino acids 403–577. This region is >53% glutamines with several long polyglutamine stretches. Such glutamine-rich domains are known to function as activators and are found in a number of eukaryotic transcription factors (48,49). Although PTIP can activate transcription in yeast when fused to the GDBD (data not shown), we have not observed this effect in transfected tissue culture cells.
The C-terminal transactivation domain of Pax proteins are generally rich in P, S and T residues, although there are no clearly defined consensus sequences which might facilitate transactivation. Small deletions or mutations in the C-terminus do not appear to eliminate Pax function as the majority of naturally occurring mutations fall within the PD or generate frameshifts and truncations which remove most of the C-terminus (50). Alternatively, missense mutations in the transactivation domain could reduce function but might not fall below a threshold of activity that enables the mutations to be detected phenotypically, as null alleles are routinely detected in heterozygotes. Likewise, alternatively spliced forms of human Pax8 which generate different P-rich reading frames in the C-terminus do not activate transcription (21). Thus, gross alterations in the sequence and structure of the Pax C-terminal transactivation domain are necessary to completely abolish function. These large deletions within the Pax2 transactivation domain that abolish transactivation also reduce or eliminate the interaction with PTIP.
A second modulator of the Pax2–PTIP interaction is the octapeptide sequence. The Pax2 octapeptide motif has homology to the engrailed suppresser (24) and can suppress the interaction between PTIP and the activation domain of Pax2. In transfection assays, a single octapeptide sequence reduces the transactivation potential of Pax2 by 3- to 4-fold, relative to octapeptide deletion constructs, and two copies of the octapeptide totally abolish all transactivation potential (22). This inhibition of transactivation by the octapeptide is not due to decreased protein stability or reduction in DNA binding activity. Similarly, the presence of the single octapeptide motif in the full-length Pax2 protein does not prevent PTIP interaction, it merely reduces the magnitude relative to the octapeptide deletion. However, when two copies of the octapeptide are inserted upstream of the Pax2 transactivation domain, PTIP binding is completely eliminated. Thus, it appears that the octapeptide motif may regulate downstream interactions, perhaps by binding a repressor protein or imposing structural constraints on Pax2 that inhibit its interaction with PTIP. In general, repressor activity with full-length Pax2 has not been observed with the synthetic reporters used in transfection assays. However, Pax2 and Pax5 can repress expression of the p53 gene and p53-mediated gene activation, although it is not known whether this repression requires the octapeptide (51). In Drosophila, groucho binds to and mediates some of the repressor function of the engrailed octapeptide (52). Whether mammalian groucho homologs can bind the Pax octapeptide remains to be determined.
The nuclear matrix by definition is the set of proteins remaining after nuclease digestion and salt extraction of nuclei. Although many transcription factors and matrix-associated DNA sequences from actively expressed genes are found in this fraction (for a review see 53), the structural determinants and significance of the matrix remain somewhat controversial (54). Visualization of a chromatin-depleted scaffolding was recently achieved by crosslinking and DNase I digestion (55). The matrix attachment regions are DNA sequences thought to bind this scaffolding such that actively expressed chromatin could loop out (56). Yet, looping out of expressed regions from the axial core is also observed in extracted metaphase chromosomes (57), presumably in the absence of any extrachromosomal scaffolding. Our experiments show that PTIP fractionates with the insoluble nuclear matrix, together with ∼50% of the total Pax2 protein. Yet, PTIP is easily solubilized upon limited digestion of nuclei with micrococcal nuclease, which digests the linker region between nucleosomes. Actively transcribed DNA sequences are found in the most soluble S1 fraction and in the insoluble pellet (30,58), presumably because this DNA corresponds to the loops and the matrix attachment regions. Pax2 is found mostly in the S1 fraction, with significant amounts also in S2 and the pellet. Taken together, the data are consistent with the idea that PTIP associates with that fraction of Pax2 bound to actively expressed genes at, or near, the nuclear matrix attachment regions.
Whether PTIP can alter Pax2 DNA binding activity or transcription activation of a synthetic reporter construct has been examined in transfected tissue culture cells. Despite its ability to activate transcription in yeast, we have not observed a significant change in Pax2 function when PTIP is overexpressed in transiently transfected cells, although our results indicate that Pax2 and PTIP are co-localized in these cells and can be isolated in a complex by immunoprecipitation. One possible explanation for this apparent lack of an effect may be the presence of endogenous PTIP. In this case, the activity of Pax2 is already determined by endogenous factors and additional PTIP does not significantly alter the activity. In order to see an effect upon Pax2 it may be necessary to eliminate or reduce PTIP levels. All cell lines we have examined express PTIP and it is not clear if a loss of PTIP function can be tolerated. A more important caveat may be that transfection assays do not reflect a true in vivo state, since much of the transfected plasmid DNA may not exhibit higher order chromatin structure. The association of PTIP and Pax2 with the nuclear matrix may be essential for proper Pax2-mediated regulation of gene expression.
Pax genes encode essential developmental regulators that control the morphogenesis of complex tissues such as the eye, the vertebral column, the CNS and the kidney. Yet, despite a wealth of genetic data in flies, mouse and humans, the biochemical basis of Pax protein function remains obscure. This paper presents the first defined interaction between Pax2 and a novel, ubiquitous nuclear factor, PTIP. Clearly, it will be essential to determine whether PTIP is a link between Pax proteins and cell cycle checkpoints, chromatin remodeling complexes or transcription activators and repressors to fully understand the significance of this interaction.
Acknowledgments
ACKNOWLEDGEMENTS
We thank L. Holzman for the DLK bait plasmid and E. Fearon for the yeast two-hybrid vectors, DCC bait plasmid and the embryonic cDNA library. This work was supported in part by NIH grants DK51043 and DK54740 to G.R.D.
DDBJ/EMBL/GenBank accession no. AF104261
REFERENCES
- 1.Dahl E., Koseki,H. and Balling,R. (1997) Bioessays, 19, 755–765. [DOI] [PubMed] [Google Scholar]
- 2.Noll M. (1993) Curr. Opin. Genet. Dev., 3, 595–605. [DOI] [PubMed] [Google Scholar]
- 3.Stuart E.T., Kioussi,C. and Gruss,P. (1993) Annu. Rev. Genet., 27, 219–236. [DOI] [PubMed] [Google Scholar]
- 4.Dressler G.R. and Douglass,E.C. (1992) Proc. Natl Acad. Sci. USA, 89, 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnarra J.R. and Dressler,G.R. (1995) Cancer Res., 55, 4092–4098. [PubMed] [Google Scholar]
- 6.Barr F.G. (1997) Curr. Top. Microbiol. Immunol., 220, 113–129. [DOI] [PubMed] [Google Scholar]
- 7.Walther C., Guenet,J.L., Simon,D., Deutsch,U., Jostes,B., Goulding,M., Plachov,D., Balling,R. and Gruss,P. (1991) Genomics, 11, 424–434. [DOI] [PubMed] [Google Scholar]
- 8.Halder G., Callaerts,P. and Gehring,W.J. (1995) Science, 267, 1788–1792. [DOI] [PubMed] [Google Scholar]
- 9.Rothenpieler U.W. and Dressler,G.R. (1993) Development, 119, 711–720. [DOI] [PubMed] [Google Scholar]
- 10.Sanyanusin P., Schimmenti,L.A., McNoe,L.A., Ward,T.A., Pierpont,M.E.M., Sullivan,M.J., Dobyns,W.B. and Eccles,M.R. (1995) Nature Genet., 9, 358–364. [DOI] [PubMed] [Google Scholar]
- 11.Torres M., Gomez-Pardo,E., Dressler,G.R. and Gruss,P. (1995) Development, 121, 4057–4065. [DOI] [PubMed] [Google Scholar]
- 12.Torres M., Gomez-Pardo,E. and Gruss,P. (1996) Development, 122, 3381–3391. [DOI] [PubMed] [Google Scholar]
- 13.Xu W., Rould,M.A., Jun,S., Desplan,C. and Pabo,C.O. (1995) Cell, 80, 639–650. [DOI] [PubMed] [Google Scholar]
- 14.Bertuccioli C., Fasano,L., Jun,S., Wang,S., Sheng,G. and Desplan,C. (1996) Development, 122, 2673–2685. [DOI] [PubMed] [Google Scholar]
- 15.Jun S. and Desplan,C. (1996) Development, 122, 2639–2650. [DOI] [PubMed] [Google Scholar]
- 16.Miskiewicz P., Morrissey,D., Lan,Y., Raj,L., Kessler,S., Fujioka,M., Goto,T. and Weir,M. (1996) Development, 122, 2709–2718. [DOI] [PubMed] [Google Scholar]
- 17.Fujioka M., Miskiewicz,P., Raj,L., Gulledge,A.A., Weir,M. and Goto,T. (1996) Development, 122, 2697–2707. [DOI] [PubMed] [Google Scholar]
- 18.Czerny T., Schaffner,G. and Busslinger,M. (1993) Genes Dev., 7, 2048–2061. [DOI] [PubMed] [Google Scholar]
- 19.Chalepakis G., Jones,F.S., Edelman,G.M. and Gruss,P. (1994) Proc. Natl Acad. Sci. USA, 91, 12745–12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorfler P. and Busslinger,M. (1996) EMBO J., 15, 1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 21.Kozmik Z., Kurzbauer,R., Doerfler,P. and Busslinger,M. (1993) Mol. Cell. Biol., 13, 6024–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lechner M.S. and Dressler,G.R. (1996) J. Biol. Chem., 271, 21088–21093. [DOI] [PubMed] [Google Scholar]
- 23.Mailhos C., Andre,S., Mollereau,B., Goriely,A., Hemmati-Brivanlou,A. and Desplan,C. (1998) Development, 125, 937–947. [DOI] [PubMed] [Google Scholar]
- 24.Smith S.T. and Jaynes,J.B. (1996) Development, 122, 3141–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundblad V. (1995) In Ausubel,F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds), Current Protocols in Molecular Biology, 2nd Edn. John Wiley & Sons, New York, NY, pp. 13.1–13.13.7.
- 26.Vidal M., Brachmann,R.K., Fattaey,A., Harlow,E. and Boeke,J.D. (1996) Proc. Natl Acad. Sci. USA, 93, 10315–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phelps D.E. and Dressler,G.R. (1996) J. Biol. Chem., 271, 7978–7985. [DOI] [PubMed] [Google Scholar]
- 28.Chevray P.M. and Nathans,D. (1992) Proc. Natl Acad. Sci. USA, 89, 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman M. y. and Goldberg,A. (1991) J. Bacteriol., 173, 7249–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose S.M. and Garrard,W.T. (1984) J. Biol. Chem., 259, 8534–8544. [PubMed] [Google Scholar]
- 31. Reyes J.C., Muchardt,C. and Yaniv,M. (1997) J. Cell Biol., 137, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He D.C., Nickerson,J.A. and Penman,S. (1990) J. Cell Biol., 110, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. (1996) Mamm. Genome, 7, 563–574. [DOI] [PubMed] [Google Scholar]
- 34.Bork P., Hofmann,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- 35.Koonin E.V., Altschul,S.F. and Bork,P. (1996) Nature Genet., 13, 266–267. [DOI] [PubMed] [Google Scholar]
- 36.Callebaut I. and Mornon,J.-P. (1997) FEBS Lett., 400, 25–30. [DOI] [PubMed] [Google Scholar]
- 37.Margolis R.L., Abraham,M.R., Gatchell,S.B., Li,S.-H., Kidwai,A.S., Breschel,T.S., Stine,O.C., Callahan,C., McInnis,M.G. and Ross,C.A. (1997) Hum. Genet., 100, 114–122. [DOI] [PubMed] [Google Scholar]
- 38.Couch F.J. and Weber,B.L. (1996) Hum. Mutat., 8, 8–18. [DOI] [PubMed] [Google Scholar]
- 39.Chapman M.S. and Verma,I.M. (1996) Nature, 382, 678–679. [DOI] [PubMed] [Google Scholar]
- 40.Monteiro A.N., August,A. and Hanafusa,H. (1996) Proc. Natl Acad. Sci. USA, 93, 13595–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scully R., Anderson,S.F., Chao,D.M., Wei,W., Ye,L., Young,R.A., Livingston,D.M. and Parvin,J.D. (1997) Proc. Natl Acad. Sci. USA, 94, 5605–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L.C., Wang,Z.W., Tsan,J.T., Spillman,M.A., Phung,A., Xu,X.L., Yang,M.C., Hwang,L.Y., Bowcock,A.M. and Baer,R. (1996) Nature Genet., 14, 430–440. [DOI] [PubMed] [Google Scholar]
- 43.Saka Y., Esashi,F., Matsusaka,T., Mochida,S. and Yanagida,M. (1997) Genes Dev., 11, 3387–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 45.Saka Y. and Yanagida,M. (1993) Cell, 74, 383–393. [DOI] [PubMed] [Google Scholar]
- 46.Saka Y., Fantes,P., Sutani,T., McInerny,C., Creanor,J. and Yanagida,M. (1994) EMBO J., 13, 5319–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson L.H., Brookman,K.W., Jones,J.J., Allen,S.A. and Carrano,A.V. (1990) Mol. Cell. Biol., 10, 6160–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka M. and Herr,W. (1994) Mol. Cell. Biol., 14, 6056–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Courey A.J. and Tjian,R. (1988) Cell, 55, 887–898. [DOI] [PubMed] [Google Scholar]
- 50.Brown A., McKie,M., van Heyningen,V. and Prosser,J. (1998) Nucleic Acids Res., 26, 259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuart E.T., Haffner,R., Oren,M. and Gruss,P. (1995) EMBO J., 14, 5638–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jimenez G., Paroush,Z. and Ish-Horowicz,D. (1997) Genes Dev., 11, 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrett T.J. and Spelsberg,T.C. (1999) Vitam. Horm., 55, 127–163. [DOI] [PubMed] [Google Scholar]
- 54.Singer R.H. and Green,M.R. (1997) Cell, 91, 291–294. [DOI] [PubMed] [Google Scholar]
- 55.Nickerson J.A., Krockmalnic,G., Wan,K.M. and Penman,S. (1997) Proc. Natl Acad. Sci. USA, 94, 4446–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laemmli U.K., Kas,E., Poljak,L. and Adachi,Y. (1992) Curr. Opin. Genet. Dev., 2, 275–285. [DOI] [PubMed] [Google Scholar]
- 57.Bickmore W.A. and Oghene,K. (1996) Cell, 84, 95–104. [DOI] [PubMed] [Google Scholar]
- 58.Xu M., Barnard,M.B., Rose,S.M., Cockerill,P.N., Huang,S.Y. and Garrard,W.T. (1986) J. Biol. Chem., 261, 3838–3845. [PubMed] [Google Scholar]