Comparative analyses of extant microscopic aquatic organisms through laboratory studies and sediment records provide a window into Earth’s history. For example, fossils and modern relatives of aquatic photoautotrophic organisms have been analyzed on an elemental level to estimate the paleo-atmospheric CO2 concentrations, by exploiting distinct changes in the isotopic composition imprinted in the biomass (1–3). As photoautotrophic organisms utilize inorganic carbon to create biomass, enzymatically catalyzed processes discriminate against the heavier 13C, through kinetic isotope fractionation. As a result, the newly built biomass (e.g., carbohydrates) becomes reduced in 13C compared to the source substrate (e.g., CO2). The small changes of the 13C:12C composition of biomass over geological time scales or under different environmental conditions have been used by geochemists and plant biochemists to better understand geological and enzymatic processes (e.g. refs. 4 and 5). The small difference in isotopic composition compared to a standard is defined as δ13C and the difference between the δ13C of a product and a reactant εp. A positive ε value indicates 13C depletion and a negative ε indicates 13C enrichment of the biomass compared to the substrate.
Within this issue of PNAS, Wang et al. (6) draw on and expand the decades-long research goal of explaining signals found in the isotopic carbon record, with both a geochemical and biochemical framework. The authors used numerical modeling to sketch out a new diagrammatic model that overcomes a fundamental limit in which the isotopic fractionation expressed in biomass needs to be smaller (more positive) than the intrinsic fractionation of the main carbon-fixing enzyme RuBisCO (Ribulose Bisphosphate Carboxylase-Oxygenase). In both aquatic and terrestrial plants, RuBisCO is the first step in converting inorganic CO2 to organic carbon. RuBisCO is one of the most abundant and important enzymes on our planet and intrinsically fractionates heavily against the 13C of carbon (intrinsic fractionation of RuBisCO: εf; ~25‰) (7 and references therein) and is thought to be the major driver of the carbon isotope fractionation of biomass (εp). While we have a good understanding of enzymatic processes involved in the acquisition of inorganic carbon (8–10), it is unclear what effect the suite of biochemical transformations of carbon during inorganic carbon acquisition has on the cellular isotope composition. Hence, many questions remain unanswered, e.g., what is the intrinsic fractionation of the different pathways supplying RuBisCO with inorganic carbon? How are these pathways affected by environmental conditions and how would this in turn affect the cellular isotopic signature? Did the evolution of RuBisCO itself and of the C-acquisition pathways change fractionation over Earth’s history? How can we better explain observed changes in the carbon isotope record? These are just a few of the questions that are currently not answered to our satisfaction.
Wang et al. draw on and expand the decades-long research goal of explaining signals found in the isotopic carbon record, with both a geochemical and biochemical framework.
Although the authors do not propose a new model for interpreting the carbon isotope record, or aim to present a novel model to predict atmospheric CO2 concentrations from billions of years ago, their study adds insightful interpretation to studies such as refs. 2, 4 and 11, all of which aimed to rationalize the fossil carbon isotope record. Most notably, Wang et al. constrained their model parameters by using biochemical and physiological measurements. In their methodological approach, they utilized an engineered mutant of the cyanobacterium Synechococcus elongatus PCC7942, which expresses an inferred ancestral form of RuBisCO (Form 1B rubisco dating to >>1 Ga) (12). The authors describe this strain as “a chimeric construct—a modern strain saddled with a predicted Precambrian enzyme.” Based on biomass and RuBisCO fractionation measurements, the ancestral strain displayed much larger cellular fractionation despite its intrinsic RuBisCO fractionation being much weaker compared to the wild type. The authors realized that previous models would not be able to explain the isotopic signature of this ancestral cyanobacterial strain. To resolve this issue, Wang et al. expand on fundamental parametrizations by accounting for a range of cellular processes that can act upon the cellular isotopic signature. Interestingly, the carboxylation rate of the ancestral RuBisCO used for this study was much slower compared to that of the wild type, which by itself likely enhanced the expressed fractionation. Generally, fractionation by enzymes, including RuBisCO, can only be fully expressed if the enzyme can select from an overabundance of a substrate (12C and 13C). If all carbons were utilized (e.g., under low CO2 availability where supply ≤ demand), no fractionation would be expressed by RuBisCO and the organic carbon isotope signature would equal that of the environment plus any processes that might have affected the δ13C pool of the inorganic carbon reaching RuBisCO.
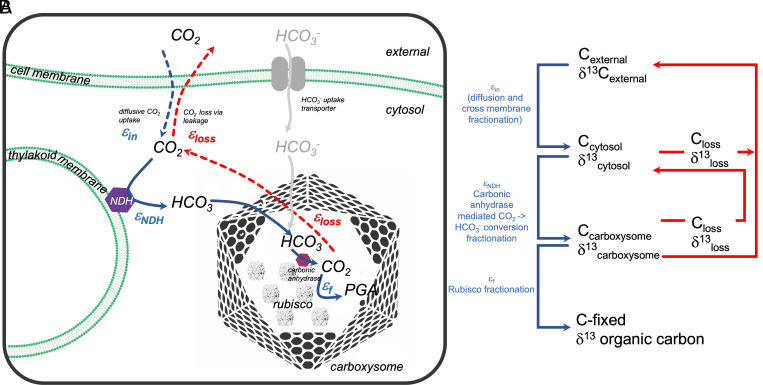
The entirety of processes that allow most aquatic photoautotrophic organisms to fix inorganic carbon efficiently is named the inorganic carbon–concentrating mechanism (CCM). It is comprised of a suite of transmembrane transporters, channels, enzymes, and proteins, all of which function to accumulate inorganic carbon in the close proximity of RuBisCO (8). This suite of mechanisms allows the cells to take up either CO2 and/or HCO3−, convert CO2 internally to HCO3− to facilitate diffusive CO2 uptake into the cell and reduce leakage of inorganic carbon back out, cluster RuBisCO within carboxysomes (in cyanobacteria) or pyrenoids (eukaryotic cells), and increase the inorganic carbon concentration at the site of RuBisCO up to many 100× times (Fig. 1A). Without a CCM, these cells would rely on diffusive CO2 uptake, which would, under current atmospheric CO2 levels, be mostly growth limiting due to the inefficiency of RuBisCO's carboxylase (CO2 utilization) reaction. In order to quantify the impact of diverse CCM components on the fractionation signals, both in eukaryotic and prokaryotic aquatic organisms, Wang et al. build upon findings by previous authors (5, 13–17), some of which emphasized that εp is strongly dependent on nonlinear processes within the cell like the hydration of CO2 to HCO3− through the so-called nicotinamide adenine dinucleotide phosphate dehydrogenase (NDH) complex. Wang et al. focused solely on CO2 as a carbon substrate, which allowed the authors to explain the large εp values measured in the ancestral strain (CO2 is up to ~9‰ enriched in 13C compared to HCO3−). Indeed, if CO2 is the sole carbon source, this CO2 will mostly be hydrated inside the cell by the aforementioned NDH complex, a carbonic anhydrase–like enzyme associated with the photosynthetic electron transport. Both ref. 16 and Wang et al. associate a strong fractionation step to this process, rationalizing that εp > εf. The authors showed an indirect correlation of elevated fractionation with enhanced NDH complex activity under high-growth light (likely high NDH activity). The importance of the NDH complex in affecting fractionation was previously suggested by refs. 18 and 16, who conducted short-term carbon flux experiments. At least two additional processes associated with carbon acquisition might thus cause the variation in εp: 1) the intrinsic fractionation of the NDH complex itself, which will increase the cellular expressed fractionation (as suggested by Wang et al. here), and 2) high CO2 leakage out of the cell, increasing the possibility of near-full fractionation potential by RuBisCO due to an overabundance of inorganic carbon at the site of RuBisCO, which cannot be fixed efficiently due to the slow carboxylation rate of this chimeric enzyme.
Fig. 1.
(A) Schematic pathways of carbon fluxes in cyanobacteria. In this paper, only CO2 fluxes over the cell membrane are considered. (B) Proposed diagrammatic isotopic fractionation processes as described by Wang et al. If supply >> demand, the intrinsic enzymatic fractionations will be fully expressed. Consequently, leakage (loss) of CO2 must be high. If supply < demand, fractionation will be minimal as all carbons will be utilized and the enzymes cannot choose from an overabundance of substrate.
The model and experiments by Wang et al. can explain certain aspects of how cellular processes affect carbon isotope signatures and provide an explanation of εp ≥ εf. Nonetheless, we still need to better understand the extent of fractionation of the different carbon acquisition processes and their regulation. One issue with the Wang et al. study is the simplification of considering CO2 as the sole carbon source. Synechococcus PCC7942 and other cyanobacteria (19) preferentially utilize HCO3−, which is isotopically lighter and hence could not fully explain larger than RuBisCO fractionation values. Nonetheless, Wang et al. elegantly show that the isotopic values measured in biomass strongly rely on distinct internal processes, such as the intrinsic fractionation of RuBisCO itself, the speed of carboxylation, internal carbon acquisition pathways, or others.
Geochemical-relevant processes are strongly dependent on the specifics of the underlying biological mechanisms.
Acknowledgments
Author contributions
S.A.K. wrote the paper.
Competing interests
The author declares no competing interest.
Footnotes
See companion article, “Carbon isotope fractionation by an ancestral RuBisCO suggests that biological proxies for CO2 through geologic time should be reevaluated,” 10.1073/pnas.2300466120.
References
- 1.Schidlowski M., A 3,800-Million-year isotopic record of life from carbon in sedimentary-rocks. Nature 333, 313–318 (1988). [Google Scholar]
- 2.Hayes J. M., Strauss H., Kaufman A. J., The abundance of C-13 in marine organic matter and isotopic fractionation in the global biogeochemical cycle of carbon during the past 800 Ma. Chem. Geol. 161, 103–125 (1999). [Google Scholar]
- 3.Francois R., et al. , Changes in the Delta-C-13 of surface-water particulate organic-matter across the subtropical convergence in the Sw Indian-Ocean. Global Biogeochem. Cy 7, 627–644 (1993). [Google Scholar]
- 4.Farquhar G. D., Oleary M. H., Berry J. A., On the relationship between carbon isotope discrimination and the inter-cellular carbon-dioxide concentration in leaves. Aust. J. Plant Physiol. 9, 121–137 (1982). [Google Scholar]
- 5.Sharkey T. D., Berry J. A., "Carbon isotope fractionation of algae as influenced by an inducible CO2 concentrating mechanism" in Inorganic Carbon Uptake by Aquatic Photosynthetic Organisms, Lucas W. J., Berry J. A., Eds. (American Society of Plant Physiologists, 1985). [Google Scholar]
- 6.Wang R. Z., et al. , Carbon isotope fractionation by an ancestral RuBisCO suggests biological proxies for CO2 through geologic time should be re-evaluated. Proc. Natl. Acad. Sci. U.S.A. 120, e2300466120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kedzior M., et al. , Resurrected RuBisCO suggests uniform carbon isotope signatures over geologic time. Cell Rep. 39, 110726 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Giordano M., Beardall J., Raven J. A., CO2 concentrating mechanisms in algae: Mechanisms, environmental modulation, and evolution. Annu. Rev. Plant Biol. 56, 99–131 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Badger M. R., Price G. D., CO2 concentrating mechanisms in cyanobacteria: Molecular components, their diversity and evolution. J. Exp. Bot. 54, 609–622 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Kaplan A., Reinhold L., CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Phys. 50, 539-+ (1999). [DOI] [PubMed] [Google Scholar]
- 11.Park R., Epstein S., Carbon isotope fractionation during photosynthesis. Geochim. Cosmochim. Ac 21, 110–126 (1960). [Google Scholar]
- 12.Shih P. M., et al. , Biochemical characterization of predicted Precambrian RuBisCO. Nat. Commun. 7, 10382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popp B. N., et al. , Effect of phytoplankton cell geometry on carbon isotopic fractionation. Geochim. Cosmochim. Ac. 62, 69–77 (1998). [Google Scholar]
- 14.Laws E. A., Popp B. N., Bidigare R. R., Kennicutt M. C., Macko S. A., Dependence of phytoplankton carbon isotopic composition on growth-rate and [CO2](Aq) - Theoretical considerations and experimental results. Geochim. Cosmochim. Ac. 59, 1131–1138 (1995). [Google Scholar]
- 15.Popp B. N., Takigiku R., Hayes J. M., Louda J. W., Baker E. W., The post-paleozoic chronology and mechanism of C-13 depletion in primary marine organic-matter. Am. J. Sci. 289, 436–454 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Eichner M., Thoms S., Kranz S. A., Rost B., Cellular inorganic carbon fluxes in Trichodesmium: A combined approach using measurements and modelling. J. Exp. Bot. 66, 749–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkes E. B., Pearson A., A general model for carbon isotopes in red-lineage phytoplankton: Interplay between unidirectional processes and fractionation by RuBisCO. Geochim. Cosmochim. Ac. 265, 163–181 (2019). [Google Scholar]
- 18.Kranz S. A., et al. , Combined effects of CO2 and light on the N2-fixing cyanobacterium Trichodesmium IMS101: Physiological responses. Plant Physiol. 154, 334–345 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J. W., Price G. D., Badger M. R., Characterization of CO2 and HCO3- uptake during steady-state photosynthesis in the Cyanobacterium Synechococcus-PCC7942. Aust. J. Plant Physiol. 21, 185–195 (1994). [Google Scholar]