Significance
Tuberculosis (TB) has been a mortal enemy to humanity for thousands of years. Despite the introduction of chemotherapeutic drug treatments dating back to the 1950s, TB today remains the leading cause of death by infectious diseases. Mycobacteria tuberculosis (Mtb) is the causative agent for TB. The highly complex and unique cell wall is one of the crucial factors to Mtb pathogenicity. In order to explore unique anti-TB drug target candidates, we sought to determine the structure of the arabinosyltransferase AftA from Mtb, an essential membrane-bound glycosyltransferase-C (GT-C) enzyme of cell wall arabinogalactan biosynthesis which primes the first arabinofuranosyl residue to the matured galactan chain. The structural and functional data will open avenues for anti-TB drug development.
Keywords: Mycobacterium tuberculosis, arabinosyltransferase, AftA, structure
Abstract
Arabinogalactan (AG) is an essential cell wall component in mycobacterial species, including the deadly human pathogen Mycobacterium tuberculosis. It plays a pivotal role in forming the rigid mycolyl–AG–peptidoglycan core for in vitro growth. AftA is a membrane-bound arabinosyltransferase and a key enzyme involved in AG biosynthesis which bridges the assembly of the arabinan chain to the galactan chain. It is known that AftA catalyzes the transfer of the first arabinofuranosyl residue from the donor decaprenyl-monophosphoryl-arabinose to the mature galactan chain (i.e., priming); however, the priming mechanism remains elusive. Herein, we report the cryo-EM structure of Mtb AftA. The detergent-embedded AftA assembles as a dimer with an interface maintained by both the transmembrane domain (TMD) and the soluble C-terminal domain (CTD) in the periplasm. The structure shows a conserved glycosyltransferase-C fold and two cavities converging at the active site. A metal ion participates in the interaction of TMD and CTD of each AftA molecule. Structural analyses combined with functional mutagenesis suggests a priming mechanism catalyzed by AftA in Mtb AG biosynthesis. Our data further provide a unique perspective into anti-TB drug discovery.
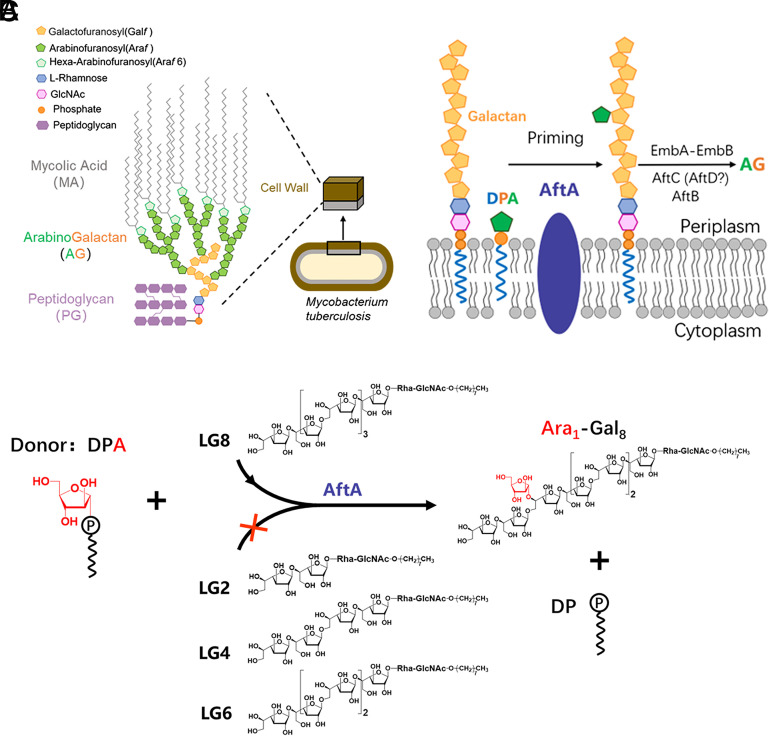
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is one of the leading causes of human death globally (1) only second to the COVID-pandemic during recent times. Of great concern is the continuing development of multidrug-resistant and extensively drug-resistant TB clinical isolates which have further exacerbated the global TB burden (2). This calls for an urgent need for the search for new and effective anti-TB targets and lead molecules for clinical development. The arabinogalactan (AG) layer within the core mycoyl-arabinogalactan-peptidoglycan (mAGP) structure of Mtb cell wall plays a central role in maintaining the complex structure by covalently linking the mycolic acid layer on the outer membrane to the inner peptidoglycan (PG) framework (3–5) (Fig. 1A). This highly unique and complex cell wall is key to the pathogenicity of Mtb. In addition, targeting the essential enzymes involved in cell wall biosynthesis is a successful approach to the development of new anti-TB lead molecules, as shown by the use of two of the four first-line drugs ethambutol and isoniazid (6–11) during directly observed therapy, and inhibitors currently in clinical trials, such as SQ109 and BTZs (12–14). Recent studies have demonstrated that AG serves as an important virulence factor by interaction with a novel receptor, galectin-9, which activates the TAK1–ERK–MMP signaling axis (15). The synthesis of AG requires three biological events in Mtb, first the synthesis of the lipid-linked linear galactan domain in the cytoplasm, followed by the crossmembrane transport of the matured galactan domain to the periplasm, and finally the assembly of the heavily branched arabinan domains onto the galactan in the periplasm (4, 16–18) (Fig. 1B). The arabinan polymerization is completed by at least six membrane-bound arabinosyltransferases in a stepwise manner initiated by AftA transferring the first arabinofuranose (Araf) residue to the sixth Galf residue in Mycobacterium smegmatis and the 8th, 10th, and 12th Galf residues in Corynebacterium glutamicum (19–25) (Fig. 1B). However, the precise priming model for AG formation in the pathogenic Mtb remains elusive.
Fig. 1.
Function of Mtb AftA. (A) Schematic representation of the components and assembly of the Mtb cell wall. (B) Biosynthesis of Mtb AG. AftA catalyzes the transfer of the first arabinosyl residue from donor DPA to the galactan chain, followed by elongation and branching steps catalyzed by EmbA–EmbB and AftC, AftB, and possibly AftD. (C) Cell-free-based arabinosyltransferase activity of purified Mtb AftA, using DPA and synthetic galactan acceptor analogs ( LG2–LG8). The enzymatic activity was monitored by the transfer of arabinofuranose to the C-5 position of the sixth galactofuranosyl residue of LG8 as reported previously (19, 20). The arabinofuranosyl residue is colored in red.
Results
To better understand the priming mechanism catalyzed by AftA, we determined the cryo-EM structure of the full-length AftA from Mtb. The AftA structure revealed a dimeric assembly and a conserved glycosyltransferase-C (GT-C) fold (26–28). More broadly, our structural and functional study provides the molecular insight into how this essential priming enzyme catalyzes the first Araf transfer in AG biosynthesis.
Enzyme Purification, Characterization, and Structure Determination.
To gain insights into the structure and function of AftA from Mtb, the N terminal His-tagged full-length Mtb AftA protein was recombinantly expressed in Msm. The n-dodecyl-β-D-maltopyranoside (DDM)-purified AftA protein was subsequently used in all cell-free arabinosyltransferase activity assays as previously established for AftA identification (19, 20). As the physiologically matured AftA acceptor substrate is chemically complex, different lengths of linear galactan acceptors decorated with the native linker unit Rha-GlcNAc of AG were prepared as previously reported (20) (Fig. 1C). Only the linear octyl-galactoside (LG8) was shown to be a functional AftA acceptor in our cell-free reaction and is consistent with previous reports using Msm membrane extracts (20) (Fig. 1C), indicating a precise acceptor selectivity of AftA. The reaction product analyses from these previous studies have shown the priming activity of AftA on the C-5 position of the sixth galactofuranosyl residue of LG8 (Fig. 1 B and C). For cryo-EM studies, the Mtb AftA protein was purified in glyco-diosgenin (GDN) and appeared homogeneous in negative-staining EM (SI Appendix, Fig. S1 A–C). Single-particle cryo-EM was used to determine the Mtb AftA structure at an overall resolution at 3.1 Å from 6,113 micrographs (Fig. 2A and SI Appendix, Fig. S2 and Table S1). The cryo-EM map revealed a dimeric assembly of AftA (SI Appendix, Fig. S2). The map is of good quality which allowed us to build a near-atomic model of AftA that included most of the residues (SI Appendix, Table S2). It is worth noting that, since no symmetry was imposed during three-dimensional (3D) reconstitution, the two AftA protomers were individually built and shown to be nearly identical upon superimposing to each other (rmsd of all Cα atoms is 0.642 Å) except for amino acids from 340 to 350, and from 599 to 608, which were missing in one protomer (Fig. 2A and SI Appendix, Fig. S3 and Table S2). In the following description of the AftA structure, we focus on the protomer built with the higher map quality unless otherwise stated.
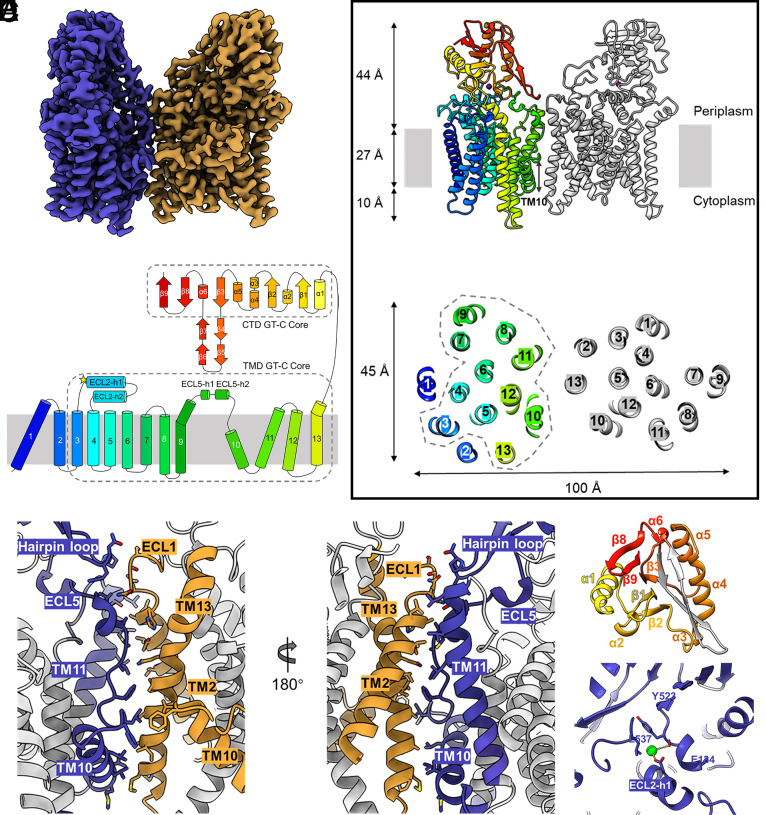
Fig. 2.
AftA structure and conserved GT-C fold. (A) Cryo-EM map of the dimeric AftA, colored by protomer. (B) Top: Cartoon representation of the dimeric AftA, with one protomer color-coded from N- to C-terminus. Bottom: The GT-C core arrangement of the TMD comprised of TM3-13 is enclosed in dash line. (C) Topology diagram of an AftA protomer using the same color as B. Both TMD and CTD GT-C cores are enclosed in dash lines. (D) An asymmetric dimer interface of the two AftA protomers colored as in A. Contact residues are shown as sticks. (E) The GT-C core of the CTD comprised of five α helix and five β strands are shown as colored cartoon. (F) Close-up view of the bound Ca2+.
Overall Structure of Mtb AftA.
Detergent-purified AftA was determined as a dimer. To validate the oligomeric status of AftA, a structural guided disulfide crossbridge was introduced in the native mycobacterial lipid membranes as described (29). The results show that AftA can form a dimer on the Msm membrane, consistent with the EM results (SI Appendix, Fig. S1 D–E).
The structure of the Mtb AftA shows a unique dimer assembly not observed in any other GT-C superfamily, such as the dimeric Emb protein complexes from mycobacteria (10, 30). Specifically, one AftA protomer rotates roughly 77° relatively to the other along the axis perpendicular to the putative membrane plane. This rotation leads to a more open dimer assembly compared to the more closely packed Emb dimers and the yeast Pmt1–Pmt2 complex (31) (Fig. 2 A and B and SI Appendix, Fig. S4). Each AftA protomer contains an N-terminal transmembrane domain (TMD) comprised of 13 TM helices and a soluble C-terminal domain (CTD) in the periplasm. TM helices vary in length within the membrane, particularly TM10 which forms the shortest TM helix and crosses only a half of the membrane as indicated by the detergent micelles. Two long loops commonly observed in these membrane-bound enzymes were characterized as extracellular loop 2 ECL2 and ECL 5 (Fig. 2C).
An asymmetric dimer interface is maintained by TM10, TM11, and ECL5 from one protomer and TM2, TM10, and TM13 from the other on the TMD. On the CTD, notably, a β hairpin loop protrudes into the dimer interface and interacts with ECL1 of the other (Fig. 2D). On the other protomer, however, the amino acids corresponding to this β hairpin loop are far away from the dimer interface and disordered, most possibly due to the instability caused by the protomer rotation (SI Appendix, Table S2). The CTD displays a mixed α and β structure that covers the TMD in the periplasm (Fig. 2 B, C, and E). A bound cation was observed above the TMD, clamped by ECL2-h1 and CTD (Fig. 2 B and F). The cation is modeled as calcium as it was shown in high abundance in the purified AftA solution in the ICP-MS study. The Ca2+ binding mode in AftA is quite different from the Ca2+ bound in the CTD far away from the TMD in the EmbA/B/C complexes, the Mn2+ observed near the catalytic site of PglB or the Zn2+ between JM1 and PL4 in the apo-state of ArnT (10, 30, 32–34) (SI Appendix, Fig. S5). It appears shielded from the solvent and forms an electrostatic interaction with the carboxyl group of the side chain of Glu134 and the main chain of Tyr523 (Fig. 2F). Substitution of Glu134 by Ala had little effect on the activity of AftA, suggesting that the interaction of Ca2+ with the ECL2 is not essential for catalysis (Fig. 3F). Hence, it is proposed that this metal ion may be responsible for the structural integrity of the enzyme.
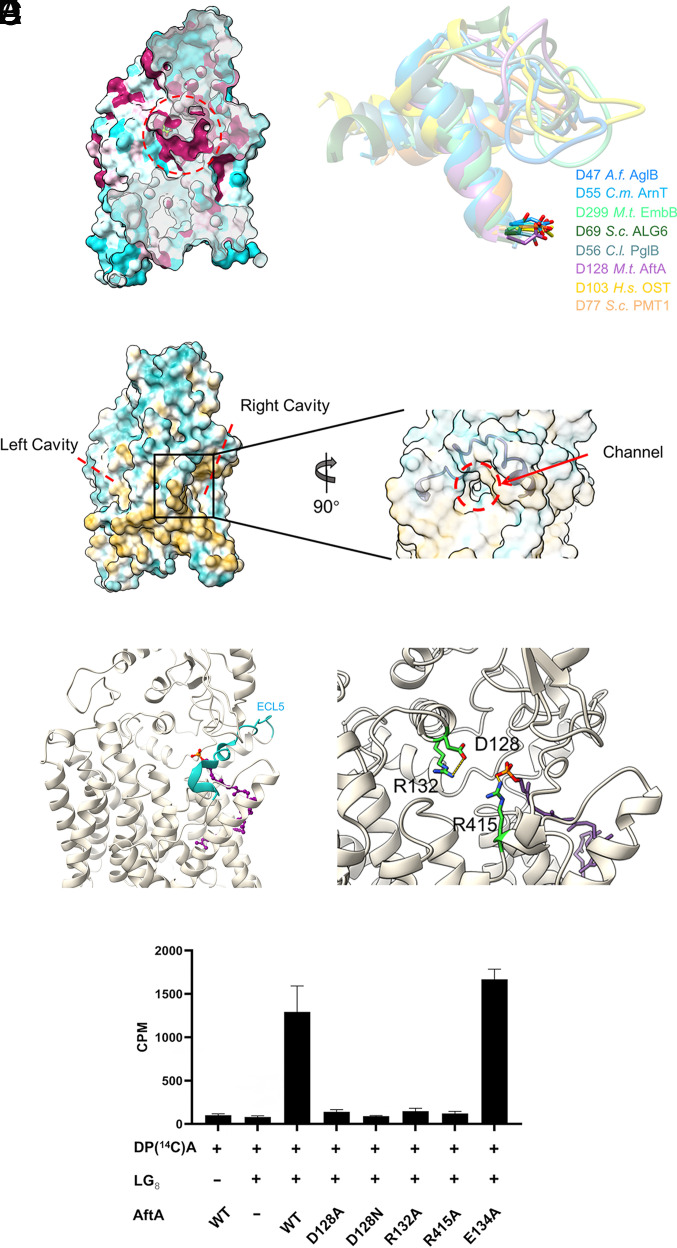
Fig. 3.
Active site and donor-binding cavity. (A) Left: Conservation mapping on the Mtb AftA structure. Sequence conservation is colored on one protomer with a gradient from magenta (absolutely conserved) to cyan (least conserved) based on 10 AftA sequences (SI Appendix, Fig. S7). Right: Superimposing of the conserved ECL-h1, ECL-h2, and the connecting loop of GT-C enzymes where the catalytic role of the conserved Asp shown in stick representation was experimentally demonstrated. (B) Left: Two cavities in the AftA structure which is shown as hydrophobicity surface. Right: Surface representation of AftA in semi-transparent mode shows a channel close to the active site in the putative donor cavity which is covered by ECL5 shown as cartoon representation. (C) Ribbon representation of the DP in AftA structure upon superimposing the TMD GT-C core to the structure of the UndP-bound ArnT (PDB: 5F15, SI Appendix, Fig. S6) (D) Close-up view of the interactions of the catalytic residue D128 with R132, and the phosphate group of the DP with R415. (E) Enzymatic activity of WT Mtb AftA and mutants.
Conserved GT-C Superfamily Topology and the Active Site.
AftA belongs to the GT85 subfamily according to the CAZy database (35). Using the Dali server (36), we sought to find models with structural similarities to AftA. The top hits of structural homologs to TMD of AftA retrieved by DALI correspond to the GT-C superfamily members deposited in the database: bacterial aminoarabinose-transferase ArnT (34), human oligosaccharyltransferases OST-A and OST-B (37), mycobacterial arabinosyltransferases EmbA/B/C (10, 30) and AftD (38), bacterial and archaeal oligosaccharyltransferases PglB (32, 33) and AglB (39), yeast glucosyltransferase ALG6 (40), and mannosyltransferases Pmt1 and Pmt2 (31). Generally, the TMD of AftA follows a conserved GT-C fold as described in previous studies by showing a GT-C core comprising TM3 to 13 (Fig. 2 B and C and SI Appendix, Fig. S6). The structural homologs of the GT-C core not only refer to i) the TM helix arrangement, but also extend to ii) the catalytically essential aspartate residue Asp128 at the first long ECL (ECL2 in AftA) formed by two crossed helices (see below), and iii) two distal TMs (TM9/10 in AftA) connected by the long ECL5 which has been regarded as the donor entrance gate and harbors at least one helix parallel to the membrane plane (SI Appendix, Fig. S6). The additional TM1-2 which does not belong to the conserved GT-C core, in AftA, participates in forming the dimer interface via ECL1 and is suggested to play a role as a scaffold on the outside of the GT-C core (Fig. 2 B and C). On the other hand, the CTD of AftA also contains a soluble core formed by an α5β5-sandwich, which covers the catalytic sites as also observed in the periplasmic domain of prokaryotic GT-C enzymes including mycobacterial AftD (38), bacterial PglB (32, 33), ArnT (34), archaeal AglB (39), and the luminal domain of eukaryotic GT-C enzymes involved in N- and C- glycosylation (Fig. 2E and SI Appendix, Fig. S6).
To gain further functional insight, we mapped the sequence conservation onto the AftA structure using unique AftA sequences across the Mycobacterium and Corynebacterium genera. We found a highly conserved region harboring Asp128 above the membrane (Fig. 3A). Asp128 is stabilized by a conserved Arg132 on the helix of ECL2 (Fig. 3D). Mutation of Asp128 to either Ala or Asn abolished AftA function, suggesting that the acidic residue is essential for catalysis. Additionally, an R132A mutant also abolished the activity of AftA (Fig. 3E). We conclude that the highly conserved Asp128 plays as a catalytic base in AftA-mediated arabinose transfer reaction. All GT-C enzymes require two substrates for catalysis: a similar phospholipid-activated sugar donor and an otherwise dissimilar peptide/carbohydrate acceptor. The two substrates get access to the active site marked by the conserved aspartic residue on ECL1 of most enzymes and ECL2 of the EmbA/B/C. AftA forms two cavities that converge at the active site (Fig. 3B). The right-side cavity forms a hydrophobic groove below the membrane boundary and a narrow channel above the membrane boundary that connects the left-side cavity. Moreover, the former cavity is on the right side under ECL5 which generally serves as the donor entrance gate (Fig. 3B). Hence, this cavity probably serves as the donor decaprenol-phospho-arabinose (DPA)–binding cavity. Indeed, when superimposing on the structure of the UndP-bound aminoarabinose transferase ArnT around the GT-C core, the phospholipid moiety of the sugar donor lies within the proposed donor cavity (Fig. 3C and SI Appendix, Fig. S6B). In the donor-bound structure of ArnT, PglB, EmbA/B/C, the N-terminal segment of ECL5 (ECL4 in ArnT) forms a helix upon donor substrate binding. In the structure of AftA, this part forms two short helices and a less ordered loop, probably due to the absence of DPA (Fig. 3C). The phosphate group reaches the putative active site and is 5.5 Å to the side chain of Asp128, a distance allowing an arabinose residue to bind. In the proposed SN2 mechanism reaction, the release of the leaving group of the substitution is stabilized by positively charged molecules, such as positively charged residues in the high-resolution donor-bound structure of EmbA, ArnT, CeDPY19, and the metal ion in the LLO-bound PglB (10, 34, 41). In the AftA structure, Arg415 on TM12 is close to the phosphate group (Fig. 3D). Given the high conservation and its essential role in the enzymatic activity of AftA (Fig. 3E and SI Appendix, Fig. S7), it is proposed to interact with the donor DPA. It seems that the activity of AftA is not strictly metal ion dependent, as the cell-free enzymatic activity performed in the reaction buffer containing Mg2+ is not entirely abolished in the presence of the metal ion chelator EDTA at 10 mM, a concentration that has little effect on the thermostability of the purified AftA (SI Appendix, Fig. S8).
Putative Galactan Acceptor–Binding Site and Catalytic Model.
Among all GT-C enzymes of Mtb, AftA is unique in catalyzing the first arabinosyl residue to the galactan domain in a highly selective manner (Fig. 1A). To reveal the selection mechanism of AftA, we performed molecular docking for one sugar acceptor of AftA, octyl-galactoside, with the AftA protomer (Fig. 4A). The modeled complex shows that the octyl-galactoside could fit into a cleft flanked by TM5, TM12, TM13, ECL2, ECL5, and one β-hairpin region on CTD. Further sequence alignment analyses indicate that the acceptor-binding sites are not well conserved across mycobacteria species, suggesting that the priming selection in AG biosynthesis might differ across mycobacterial species (Fig. 4B and SI Appendix, Fig. S7). Particularly, the sixth Gal could insert deep into the AftA active site by forming one hydrogen bond with the Asp128 side chain via its O5 atom (Fig. 4B), poised for the following catalysis.
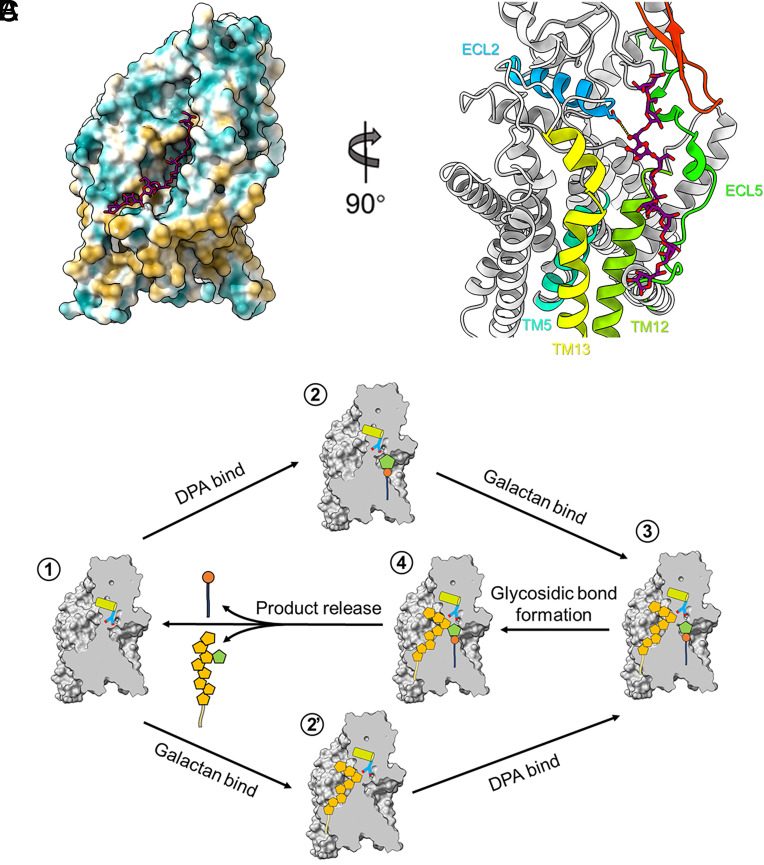
Fig. 4.
Putative acceptor-binding site and working model of AftA. (A) Molecular docking of an octyl-galactoside in the AftA structure, the ligand is shown as stick and AftA as hydrophobicity surface representation. (B) Contacts to the docked LG8 including ECL2, ECL5, TM5, TM12, and TM13 in the TMD, and the β hairpin (red) in the CTD. (C) Proposed priming mechanism catalyzed by AftA. See text for description. ECL2-h1 is indicated as cylinder, the catalytic base D128 is represented as stick. Arabinosyl and galactosyl residues are shown as pentagons and colored in orange and green, respectively.
On the basis of the structural and functional data, a priming model is now proposed for AftA (Fig. 4C). Upon either a DPA or a galactan chain binding first to the active site through the right-side cavity or the left-side cavity, the N- and C-ECL5 are stabilized by the bound substrates, respectively, by forming more-ordered helices. The carboxyl group of Asp128 orientates and activates the O5 of the sixth Gal for a nucleophilic attack on the C1 carbon of the arabinose ring of DPA. A new glycosidic bond is formed, while the leaving group DP is stabilized by R415 and possibly an additional metal ion.
Discussion
In this study, we present the first structure of a highly purified and enzymatically active AftA from Mtb. The structure of AftA reveals a conserved GT-C fold in both TMD and CTD by comprehensive structural comparison with the reported GT-C enzymes. Enzymatic assays, conservation analysis, and mutagenesis studies have probed the active site for catalysis and the DPA-binding site of AftA. Experimental structures of the substrate-bound AftA are required to complete the more precise catalytic mechanism. Although belonging to the same GT-C superfamily as the EmbA/B/C enzymes, AftA is not inhibited by ethambutol (SI Appendix, Fig. S8). Our previous work has shown that ethambutol binds to the active sites of both EmbB and EmbC in a conserved binding mode. Structures of the active sites of Mtb AftA and Mtb EmbB revealed that EmbB has a more negatively charged pocket with an appropriate size, which favors the binding of the positively charged imino groups of ethambutol (SI Appendix, Fig. S9). Together with its essential role in AG synthesis, AftA is regarded as an attractive new drug target for the generation of new anti-TB drugs by inhibiting Mtb cell wall formation, therefore overcoming drug resistance of one the current front-line agents, ethambutol. Our structure provides a solid foundation for future drug discovery efforts.
In all previously reported mycobacterial GT-C structures including EmbA/B/C and AftD, an acyl carrier protein AcpM either from Msm (in the EmbA/B/C structure) or Escherichia coli (in the AftD structure) was found associated with each GT-C enzyme at the cytoplasmic side. The association is tight as the AcpM subunit was copurified endogenously from the expressing host strain, and mainly mediated by electrostatic interaction via the positively charged region of GT-C enzymes and the negatively charged AcpM (10, 30, 38, 42) (SI Appendix, Fig. S10). It is worth noting that all resolved AcpMs are modified at the conserved Ser with a well-defined lipid 4′-Phosphopantetheine (Ppant) which extends into its membrane protein partner. We did not observe any signal corresponding to AcpM on the cytoplasmic side of our AftA structure during the whole data-processing procedure. Purified AftA protein without AcpM or similar protein appears active in our biochemical assay, suggesting that AcpM is not required for all arabinosyltransferases in mycobacteria. Structural analysis reveals two key factors on the GT-C enzymes required for AcpM binding: i) a highly positively charged surface on the cytoplasmic side and ii) a relatively hydrophobic tunnel formed by the TMHs near the cytoplasm where the 4′-Ppant can be buried. In the structure of EmbA/B/C, this tunnel is formed by TMH2, 6, 7 and a long ICL1, whereas in AftD, it is formed by TMH13-16 which has been shown to be important for AftD function. Both TMH2, ICL1 in EmbA/B/C and TMH13-16 in AftD are non-GT-C core elements, which indicates that some mycobacterial GT-C enzymes have gained additional features to cofunction with the AcpM partner during evolution. In the AftA structure, however, despite a mostly positively charged surface in the cytoplasm which abides by the principle “positive inside” for most membrane proteins, no notable tunnel with appropriate depth is found to favor the binding of 4′-Ppant (SI Appendix, Fig. S10C). Hence, we predict that a tunnel formed by the non-GT-C-core elements near the cytoplasm is a prerequisite to the association for AcpM.
Materials and Methods
Protein Expression and Membrane Preparation.
The Mtb AftA protein was overexpressed in Msm mc2155 from an engineered pMV261 vector containing the full-length AftA gene from genomic Mtb H37Rv DNA fused with an N-terminal 10×His tag, under the control of an acetamide promoter. Recombinant plasmid was introduced into Msm mc2155 competent cells by electroporation. For large-scale production, cells were cultured in 1 L Luria broth medium supplemented with 50 μg/mL kanamycin, 20 μg/mL carbenicillin, and 0.1% (v/v) Tween 80 (to avoid cell aggregation) at 37 °C with shaking at 220 rpm until the OD600 reached 1.0. Four days after induction with 0.2% (w/v) acetamide at 16 °C, the cells were collected in Buffer A containing 20 mM HEPES, 150 mM NaCl, pH 8.0. The cells were lysed by French Press at 1,200 bar and 4 °C. Cell debris was cleared by centrifugation at 12,500 rpm for 15 min at 4 °C. The membrane pellet was collected by ultracentrifugation (36,500 rpm, 1.5 h) at 4 °C and then resuspended in Buffer A stored at −80 °C until use. All mutants were expressed using the same protocol as the wild-type (WT) protein.
Protein Purification.
Thawed membrane fractions were solubilized with 1% (w/v) DDM (Anatrace) by gently agitating for 1.5 h at 4 °C. Detergent-insoluble material was removed by ultracentrifugation (18,500 rpm for 30 min). The supernatant was purified by nickel affinity resin (Qiagen), followed by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) preequilibrated with Buffer B containing 20 mM HEPES, 150 mM NaCl, pH 8.0, and 0.04% (w/v) GDN (Anatrace). The peak fraction corresponding to the AftA dimer was concentrated to 6.5 mg/mL for cryo-grid preparation. Protein for activity assays was purified using the same protocol except the gel filtration buffer which was exchanged to an assay buffer (Buffer C) containing 50 mM MOPS, 10 mM MgCl2, pH 7.9, 5 mM β-mercaptoethanol, 5% (v/v) glycerol, and DDM at twice the critical micelle concentration.
Grid Preparation and Data Collection.
Aliquots of the freshly purified sample were applied to a glow-discharged holey carbon grid (Quantifoil Cu R0.6/1.0, Solarus Gatan Plasma System H2/O2 for 30 s). The grids were blotted for 3.5 s, flash-frozen in liquid ethane, and cooled in liquid nitrogen using an FEI Mark IV Vitrobot (humidity 100%, temperature 281 K, blotting paper TED PELLA 595 filter paper). Images were taken using an FEI Titan Krios electron microscope operating at 300 kV with a Gatan K3 Summit direct electron detector at a nominal magnification of 105,000. Images were recorded in superresolution mode and binned to a pixel size of 0.832 Å. Automated single-particle data acquisition was performed with Serial EM data collection software (43). Defocus values varied from 1.2 to 2.0 μm. Each stack was exposed for 2.4 s with a total dose of 60 e−/Å2, with 40 frames per stack. The details of electron microscopy data collection parameters are listed in SI Appendix, Table S1.
EM Image Processing.
All dose-fractioned images were motion-corrected and dose-weighted by MotionCorr2 software (44) and their contrast transfer functions were estimated by Gctf (resolution range: 4 to 25 Å; search defocus: 0.1 to 4 μm). A total of 2,752,481 particles were picked automatically from 6,113 images (particle diameter: 200 Å; minimum separation distance: 110 Å) and extracted with a box size of 384 pixels using cryoSPARC (45). The subsequent two-dimensional (2D) and 3D classifications and refinement steps were all performed in cryoSPARC. A total of 1,109,935 particles were selected after two rounds of 2D classification (number of 2D classes: 100; window inner radius: 0.85; maximum resolution: 6 Å; iterations: 20). A total of 451,978 particles were used to do ab-initio reconstruction in four classes (classes: 4; maximum resolution: 12 Å; initial resolution: 35 Å; class similarity: 0.1) which were used as 3D volume templates for heterogeneous refinement with all the selected particles (refinement box size: 128 pixels). After heterogeneous refinement, 147,896 particles were converged into one class, yielding a 5.11 Å initial map. After nonuniform refinement, the final resolution reached 3.14 Å (SI Appendix, Fig. S4 and Table S1).
Model Building and Refinement.
The 3.14 Å cryo-EM map was used for the model building of AftA. The Alpha fold (46) model was first manually fitted into the 3.14 Å resolution cryo-EM map using UCSF Chimera (47). Manual adjustment of the complete model was first performed in COOT 0.8.8 (48), followed by iterative rounds of real-space refinement in PHENIX 1.12 (49) and manual adjustment in COOT. Refinement strategies used include “minimization_global,” “local_grid_search,” and “atomic displacement parameters (ADP).” Restraints included rotamer restraints, Ramachandran restraints, and NCS (noncrystallographic symmetry) constraints.
Arabinosyltransferase Activity Assays.
AftA arabinosyltransferase assays were essentially performed as described (19, 20) using LG8 (500 µM in water), DP[14C]A (50,000 cpm, stored in 1% Ige-Pal), 1 mM ATP, 1 mM NADPH, AftA proteins (4 µM), and in some cases ethambutol (6 µg per assay) and divalent cations (2 mM), with the appropriate amount of buffer (50 mM MOPS pH 7.9) and made up to a final volume of 120 µL. Samples were incubated at 37 °C for 2 h, quenched by the addition of ethanol (120 µL), and loaded onto a 1-mL Supelclean LC-SAX SPE cartridge (Supleco) to retain unreacted DP[14C]A. The SAX cartridge was washed with 2 mL ethanol and the eluate-containing LG8-[14C]Ara was collected and dried. The sample was resuspended in water (0.5 mL) and applied onto a Bio-Gel P-2 Gel filtration column (20 cm × 1.0 cm; Bio-Rad) to separate the product LG8-[14C]Ara from any degraded [14C]Ara from the assay using water at a flow rate of 0.1 mL/min. Fractions (0.5 mL) were collected and subsequently mixed with 10 mL of scintillant cocktail (Ultima-flo AF, Perkin Elmer) and radioactivity was measured using a Tri-Carb 2810 TR scintillation counter (Perkin Elmer). The LG8-[14C]Ara product was observed at the void volume of the Bio-Gel P-2 column and each assay was performed in triplicate.
Cross-Linking Experiments.
To examine structure-based disulfide bridge studies of AftA on Msm, cell membranes of Msm overexpressing WT AftA and the F360C mutant were isolated. Protein loading buffer containing SDS was subsequently added to the membranes to a final SDS concentration of 2% (w/w) for solubilization. The oligomeric status of AftA was identified by western blot using anti-His antibodies. Reducing agent DTT was added at RT as a control.
Molecular Docking.
To reveal the substrate-binding site in AftA, molecular docking for one AftA substrate LG8 was performed using the Rosetta software suite. The chemical formula and 3-dimensional structure of LG8 were obtained by ChemBioDraw Ultra 14.0 and ChemBio3D Ultra 14.0, respectively. We generated 2,000 conformers of LG8 using RDKit (https://www.rdkit.org) and created corresponding parameter and Protein Data Bank (PDB) files required by Rosetta. For the metal ion, we constrained the coordinate bonds between Mg2+ and surrounding atoms in desired distance. The molecular docking in Rosetta consists of three steps: low-resolution docking, high-resolution docking, and final energy minimization. For the low-resolution docking, the transform module performs a grid-based Monte Carlo search of the ligand-binding site, which iterates through cycles of random translation, rotation, and conformation sampling. Then, at the stage of high-resolution docking, the High Res Docker module carries out cycles of rotamer trials (sampling of side chain rotamers, one side chain at a time) or repacking (simultaneous sampling of rotamers for multiple side chains), coupled with small movements of ligand. After each cycle, energy minimization and Monte Carlo sampling were performed, with a Boltzmann criterion to determine whether to accept or reject the new structure. Finally, the Final Minimizer module executes a gradient-based energy minimization of the final docking complex, including side-chain torsion angle sampling and backbone torsion angle minimization with constraints on the Cα atoms. At the end, a total of 10,000 docking structures were generated, from which the desired docking complex was selected based on two criteria: 1) one hydrogen bond could be formed between the C5-linked hydroxyl group on the sixth Gal and AftA-D128 (necessary for the catalysis) and 2) the Rosetta score. The chosen AftA-LG8 docking structure was employed for further construction of AftA–LG8–DPA ternary complex.
Creation of Figures and Data Analysis.
Figures of molecular structures were generated using PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC.) (50) and UCSF ChimeraX (47). Protein sequence alignments were performed using Clustal Omega (51) and ESPript (52).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We would like to thank the Bio-Electron Microscopy Facility of ShanghaiTech University for their support and assistance with microscopy data collection. We thank Z. Wang for assisting with functional experiments. We also thank Dr. M.F. Wang for protein expression screening. This work was supported by Shanghai Frontiers Science Center for Biomacromolecules and Precision Medicine, ShanghaiTech University, the Lingang Laboratory (Grant No. LG 202101-01-08) to L.Z. and Medical Research Council, UK (MR/S000542/1 and MR/R001154/1), to G.S.B.
Author contributions
G.S.B., Z.R., and L.Z. designed research; Y.G., C. Wei, J.W., N.M., Q.L., C. Wu, N.Y., H.Y., Y.Z., X.Y., S.S.G., N.V., S.M.B., Z.H., L.D., G.S.B., and L.Z. performed research; Y.G., C. Wei, J.W., N.M., Q.L., C. Wu, S.S.G., N.V., S.M.B., Z.H., L.D., G.S.B., Z.R., and L.Z. analyzed data; and Y.G., J.W., L.D., G.S.B., Z.R., and L.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Gurdyal S. Besra, Email: g.besra@bham.ac.uk.
Lu Zhang, Email: zhanglu1@shanghaitech.edu.cn.
Data, Materials, and Software Availability
Cryo-EM data from this study have been deposited in the Electron Microscopy Data Bank (EMDB) with the entry ID EMD-35410 (53). Coordinates for the atomic model have been deposited in the Protein Data Bank (PDB) with the entry ID 8IF8 (53). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Lawn S. D., Zumla A. I., Tuberculosis. Lancet 378, 57–72 (2011), 10.1016/s0140-6736(10)62173-3. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Global tuberculosis report 2022 (2022).
- 3.Grzegorzewicz A. E., et al. , Assembling of the My cobacterium tuberculosis cell wall core*. J. Biol. Chem. 291, 18867–18879 (2016), 10.1074/jbc.M116.739227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankute M., Cox J. A. G., Harrison J., Besra G. S., Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423 (2015), 10.1146/annurev-micro-091014-104121. [DOI] [PubMed] [Google Scholar]
- 5.Alderwick L. J., Harrison J., Lloyd G. S., Birch H. L., The mycobacterial cell wall-peptidoglycan and arabinogalactan. Cold Spring Harb. Perspect. Med. 5, 15 (2015), 10.1101/cshperspect.a021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batt S. M., Burke C. E., Moorey A. R., Besra G. S., Antibiotics and resistance: The two-sided coin of the mycobacterial cell wall. Cell Surface 6, 100044 (2020), 10.1016/j.tcsw.2020.100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belanger A. E., et al. , The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. U.S.A. 93, 11919–11924 (1996), 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goude R., Amin A. G., Chatterjee D., Parish T., The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 53, 4138–4146 (2009), 10.1128/AAC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee A., et al. , inhA, a dene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230 (1994), 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., et al. , Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol. Science 368, 1211+ (2020), 10.1126/science.aba9102. [DOI] [PubMed] [Google Scholar]
- 11.Bansal R., Sharma D., Singh R., Tuberculosis and its treatment: An overview. Mini-Rev. Med. Chem. 18, 58–71 (2018), 10.2174/1389557516666160823160010. [DOI] [PubMed] [Google Scholar]
- 12.Sacksteder K. A., Protopopova M., Barry C. E., Andries K., Nacy C. A., Discovery and development of SQ109: A new antitubercular drug with a novel mechanism of action. Future Microbiol. 7, 823–837 (2012), 10.2217/fmb.12.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B., et al. , Crystal structures of membrane transporter MmpL3, an anti-TB drug target. Cell 176, 636–648.e613 (2019), 10.1016/j.cell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Makarov V., et al. , Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324, 801–804 (2009), 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., et al. , Sensing of mycobacterial arabinogalactan by galectin-9 exacerbates mycobacterial infection. EMBO Rep. 22, e51678 (2021), 10.15252/embr.202051678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crick D. C., Mahapatra S., Brennan P. J., Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11, 107R–118R (2001), 10.1093/glycob/11.9.107R. [DOI] [PubMed] [Google Scholar]
- 17.Holzheimer M., Buter J., Minnaard A. J., Chemical synthesis of cell wall constituents of Mycobacterium tuberculosis. Chem. Rev. 121, 9554–9643 (2021), 10.1021/acs.chemrev.1c00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison J., et al. , Lcp1 is a phosphotransferase responsible for ligating arabinogalactan to peptidoglycan in mycobacterium tuberculosis. mBio 7, e00972–00916 (2016), 10.1128/mBio.00972-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alderwick L. J., Seidel M., Sahm H., Besra G. S., Eggeling L., Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall Arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 281, 15653–15661 (2006), 10.1074/jbc.M600045200. [DOI] [PubMed] [Google Scholar]
- 20.Angala S. K., et al. , Use of synthetic glycolipids to probe the number and position of arabinan chains on Mycobacterial arabinogalactan. ACS Chem. Biol. 16, 20–26 (2021), 10.1021/acschembio.0c00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi L. B., et al. , Transfer of the first arabinofuranose residue to galactan is essential for Mycobacterium smegmatis viability. J. Bacteriol. 190, 5248–5255 (2008), 10.1128/jb.00028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alderwick L. J., et al. , Deletion of Cg-emb in corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an Arabinan-deficient mutant with a cell wall galactan core. J. Biol. Chem. 280, 32362–32371 (2005), 10.1074/jbc.M506339200. [DOI] [PubMed] [Google Scholar]
- 23.Seidel M., et al. , Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall Arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis*. J. Biol. Chem. 282, 14729–14740 (2007), 10.1074/jbc.M700271200. [DOI] [PubMed] [Google Scholar]
- 24.Birch H. L., et al. , Biosynthesis of mycobacterial arabinogalactan: Identification of a novel α(1→3) arabinofuranosyltransferase. Mol. Microbiol. 69, 1191–1206 (2008), 10.1111/j.1365-2958.2008.06354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Škovierová H., et al. , AftD, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 19, 1235–1247 (2009), 10.1093/glycob/cwp116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albuquerque-Wendt A., Hutte H. J., Buettner F. F. R., Routier F. H., Bakker H., Membrane topological model of glycosyltransferases of the GT-C superfamily. Int. J. Mol. Sci. 20, 16 (2019), 10.3390/ijms20194842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lairson L. L., Henrissat B., Davies G. J., Withers S. G., Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008), 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 28.Liu J., Mushegian A., Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12, 1418–1431 (2003), 10.1110/ps.0302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung B. C., et al. , Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341, 1012–1016 (2013), 10.1126/science.1236501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., et al. , Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2. Protein Cell 11, 505–517 (2020), 10.1007/s13238-020-00726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai L., Kovach A., You Q. L., Kenny A., Li H. L., Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex. Nat. Struct. Mol. Biol. 26, 704−+ (2019), 10.1038/s41594-019-0262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Napiorkowska M., et al. , Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase. Nat. Struct. Mol. Biol. 24, 1100−+ (2017), 10.1038/nsmb.3491. [DOI] [PubMed] [Google Scholar]
- 33.Lizak C., Gerber S., Numao S., Aebi M., Locher K. P., X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355 (2011), 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 34.Petrou V. I., et al. , Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science 351, 608–612 (2016), 10.1126/science.aad1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drula E., et al. , The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 50, D571–D577 (2021), 10.1093/nar/gkab1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L., Dali server: Structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022), 10.1093/nar/gkac387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez A. S., Kowal J., Locher K. P., Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B. Science 366, 1372-+ (2019), 10.1126/science.aaz3505. [DOI] [PubMed] [Google Scholar]
- 38.Tan Y. Z., et al. , Cryo-EM structures and regulation of arabinofuranosyltransferase AftD from mycobacteria. Mol. Cell 78, 683–699.e611 (2020), 10.1016/j.molcel.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto S., et al. , Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc. Natl. Acad. Sci. U.S.A. 110, 17868–17873 (2013), 10.1073/pnas.1309777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloch J. S., et al. , Structure and mechanism of the ER-based glucosyltransferase ALG6. Nature 579, 443−+ (2020), 10.1038/s41586-020-2044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bloch F. N., et al. , Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase. Nat. Chem Biol. 19, 575–584 (2023), 10.1038/s41589-022-01219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong H. C., Liu G. H., Zhang Y. M., Rock C. O., Zheng J., The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277, 15874–15880 (2002), 10.1074/jbc.M112300200. [DOI] [PubMed] [Google Scholar]
- 43.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017), 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017), 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 46.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021), 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021), 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. Section D 60, 2126–2132 (2004), 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Section D 75, 861–877 (2019), 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delano W. L., The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA, 2002). [Google Scholar]
- 51.Sievers F., Higgins D. G., Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018), 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robert X., Gouet P., Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014), 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong Y.C., Rao Z.H., Zhang L., PDB entry 8IF8, Arabinosyltransferase AftA. RCSB Protein Data Bank. https://www.rcsb.org/structure/unreleased/8IF8. Deposited 17 February 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Cryo-EM data from this study have been deposited in the Electron Microscopy Data Bank (EMDB) with the entry ID EMD-35410 (53). Coordinates for the atomic model have been deposited in the Protein Data Bank (PDB) with the entry ID 8IF8 (53). All other data are included in the manuscript and/or SI Appendix.