Significance
Biodegradable polymers are one part of the solution to the global plastics sustainability crisis. Specifically, biodegradable polymers mitigate the challenges of released waste and microplastic accumulation by breaking down in environmental conditions. However, the relationship between the chemical structure of these materials and their biodegradability is poorly understood. Current biodegradation testing methods are time and resource intensive, limiting experimental data from which structure–property relationships can be elucidated. This work introduces high-throughput polymer biodegradation methods applied to a large polymer library from which structure–property relationships are developed and predictive models are built.
Keywords: biodegradation, polymers, high-throughput, structure–property relationships
Abstract
The consistent rise of plastic pollution has stimulated interest in the development of biodegradable plastics. However, the study of polymer biodegradation has historically been limited to a small number of polymers due to costly and slow standard methods for measuring degradation, slowing new material innovation. High-throughput polymer synthesis and a high-throughput polymer biodegradation method are developed and applied to generate a biodegradation dataset for 642 chemically distinct polyesters and polycarbonates. The biodegradation assay was based on the clear-zone technique, using automation to optically observe the degradation of suspended polymer particles under the action of a single Pseudomonas lemoignei bacterial colony. Biodegradability was found to depend strongly on aliphatic repeat unit length, with chains less than 15 carbons and short side chains improving biodegradability. Aromatic backbone groups were generally detrimental to biodegradability; however, ortho- and para-substituted benzene rings in the backbone were more likely to be degradable than metasubstituted rings. Additionally, backbone ether groups improved biodegradability. While other heteroatoms did not show a clear improvement in biodegradability, they did demonstrate increases in biodegradation rates. Machine learning (ML) models were leveraged to predict biodegradability on this large dataset with accuracies over 82% using only chemical structure descriptors.
The applications of polymers in health care, (1) their use for food safety and security, (2) their contribution to energy savings (3) and transportation, (4, 5) and their many additional roles in our economy make them indispensable to modern life. However, they also present challenges related to the use of fossil-based carbon resources and their end-of-life fate (6–8). The lifecycle of polymer products has detrimental effects on the environment; (1) in particular, the fates of plastic as mismanaged waste, (6) incinerated waste, (9) or accumulating landfill waste (10) each present different impacts. To transform into a green economy, it is imperative to develop polymers with a more sustainable life cycle (11). Biodegradable polymers present one solution to the end-of-life fate of plastics, especially mismanaged waste, as they can be industrially degraded or will degrade naturally when there is inevitable leakage of waste into the environment (12). Additionally, biodegradable polymers are able to address the challenges caused by microplastic accumulation as biodegradability enables a polymer to entirely break down in the environment (13).
Although often used as interchangeable descriptors, compostable and biodegradable are separate designations for polymers (14). Here, compostable materials refer to industrially compostable materials which chemically break down in 180 d under conditions specified by American Society for Testing and Materials (ASTM) D6400 and are eligible for certification by the Biodegradable Products Institute in the United States (15). Biodegradable materials are those which break down through ambient microbial action, (14) and there has been no clear biodegradability standard set by ASTM although tests for biodegradation degree and rates in various environments and conditions exist (16–20).
Polyesters are a broadly interesting class of polymers which degrade by well-known hydrolysis mechanisms (21). The most common commercial polyester, poly(ethylene terephthalate), has recently gathered much attention because enzymes capable of its degradation have been engineered, (22–26) but generally it remains nonbiodegradable even under industrial composting conditions. Biodegradable or commercially compostable polyesters, such as poly(hydroxyacids) (PHAs) or poly(lactic acid) (PLA), have been implemented as a sustainable alternative to commodity plastics. However, these materials face significant challenges, with PLA having limited biodegradability, low impact resistance, and toughness, and PHAs showing thermal instability and slow crystallization rates (27). As such, the development of biodegradable polyesters remains a pressing challenge for addressing the polymer sustainability crisis and reducing the environmental burden of the polymer life cycle.
The development of biodegradable polymers is severely limited by current biodegradation testing methods. Biodegradation testing standards include methods for a variety of environments including sewage (ASTM D5210-92), (20) seawater (ASTM D6691 and ISO 19679), (18, 28) industrial compost (ASTM D6400, ASTM D5338, and ISO 14855), (15, 19, 29), home compost or in the environment, (16) and soil (17). However, these tests rely on expensive or challenging analytical methods such as measuring CO2 or CH4 evolution, inorganic carbon analysis, or sample mass loss using quartz crystal balances (30, 31). Additionally, traditional degradation tests often require monitoring between 20 and 130 d, creating a large time burden for running a study on any given polymer (30). Variation in the microbial communities used for testing can cause quantitative differences in test results even when using the same material and testing standard (30). In contrast to the standardized biodegradation methods, clear-zone biodegradation tests, similar to those used for antibiotic or biostatic testing, have been used to determine potential polymer-degrading bacteria across different polymers and have the advantages of being fast and relatively cheap when compared to standard tests; (32, 33) this suggests the potential of the clear-zone method being used for screening new polymer candidates and enabling large dataset construction.
The availability of larger biodegradation sets, combined with modern data-fitting methods, has the potential to yield quantitative structure–property relationships (QSPR) for the prediction of polymer biodegradation based on polymer chemical structure. Structure–property relationships have been developed successfully for a wide variety of polymer properties that depend primarily on chemical structure such as glass transition temperature, molar volume, solubility, and intrinsic viscosity using experimental data (34, 35) and group contribution methods (36). More recently, machine learning (ML) has been leveraged for polymer QSPR; (37) both building predictive models for polymer properties (38–41) and the identification of new, promising polymers from predictive models (42) have been realized. Because it is hypothesized that biodegradation is determined in large part by the chemical structure of a polymer chain, the availability of a sufficiently large dataset should enable the application of ML for QSPR of polymer biodegradability as well.
This work introduces high-throughput polymer synthesis and biodegradation testing methodology to construct a large set of polyester and polycarbonate biodegradation data that can then be applied for the development of biodegradation QSPR. High-throughput polymer synthesis was implemented to develop a chemically diverse polyester and polycarbonate library. To analyze biodegradation in high-throughput, a clear-zone biodegradation assay with digital data extraction was automated and applied across the library of 642 polymers. The resulting biodegradation dataset was used to identify the impact of chemical structures on biodegradation, with features such as ether groups and single carbon side chains being identified as positively impacting biodegradability. Machine learning was then leveraged to develop random forest (RF) and linear regression–based predictive models for biodegradability.
Results and Discussion
Polymer Library Design and Synthesis.
A polymer library comprised of linear polyesters and polycarbonates was synthesized from diols, dicarboxylic acids or corresponding diacid chloride derivatives, dimethyl carbonate, and lactone rings. To make a library for biodegradation testing with maximum chemical diversity, different chemical features were identified as important design variables, including ester group chain length, number of side chains and side chain length, aliphatic and aromatic rings, and heteroatoms. The chemical functionalities included in the library are described in Table 1, along with the number of monomers and polymers that contain them. The inclusion of various side chains was motivated by previous work demonstrating the positive impact of side chains in starch biodegradation (43, 44). Heteroatoms (excluding the ester bond) included in the library were nitrogen, sulfur, and oxygen, and provided additional bonds that could be enzymatically cleaved (45, 46). Nitrogen groups were limited to tertiary nitrogen to avoid amine side reactions during polymerization. Sulfur was included in a diverse set of functionalities including sulfides, disulfides, and sulfones. Oxygen was included as part of ether groups and in the sulfone groups. Hydroxyl, esters outside of the backbone functionality, and carboxylic acid groups were excluded due to effects on polymerization. The impact of ethers was of particular importance due to the prevalence of poly(ethylene glycol)s and interest in their degradation (47).
Table 1.
Summary of the chemical functionalities in the polymer library
| Chemical functionality | Number of monomers | Number of polymers |
|---|---|---|
| All | 94 | 642 |
| Ring (nonaromatic) | 24 | 242 |
| Aromaticity | 8 | 116 |
| Sulfur | 7 | 82 |
| Nitrogen | 2 | 35 |
| Oxygen* | 8 | 106 |
| Double, triple bond | 6 | 46 |
*Oxygen not part of ester or carbonate bonds.
In addition to heteroatoms, rings and aromatic rings and double and triple bonds were included in the library. These groups were selected to change the backbone rigidity and manipulate the regiochemistry of the polymer chain. Multi-ring structures, bisphenols, and heteroatom-containing rings were all included. The proximity of these groups can affect the accessibility of the ester bonds to enzymes as well as the overall crystallinity of the polymer (34, 36). Specific monomers were selected based on their chemical functionality and commercial availability, with commercial availability being the primary limiting constraint to the library size and diversity. Monomers incorporated into the final library of polymers can be found in SI Appendix, Table S1.
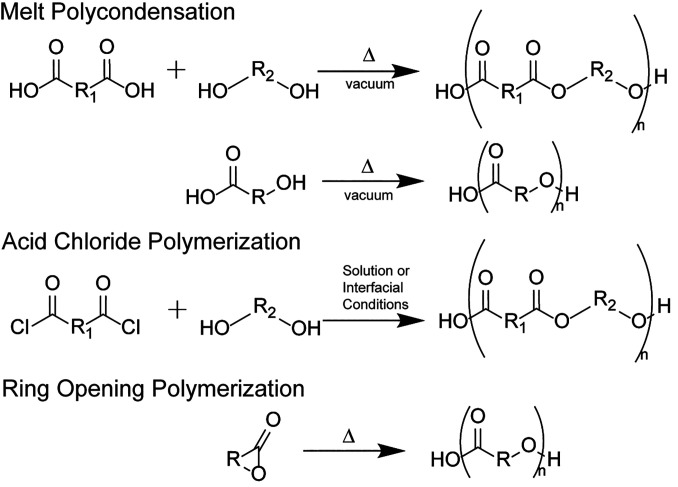
In order to synthesize polymers with the diversity of chemical structures included in the design of the polymer library, several different high-throughput synthetic schemes were developed, each with conditions tunable to the specific subsets of monomers (Fig. 1). The high-throughput nature of the synthesis schemes led to monomers being grouped by similarity in melting points, boiling points, and solubility; conditions best suited to a group of monomers were selected for synthesis. For melt polycondensation (Fig. 1), dicarboxylic acid monomers were crossed with diols, yielding over 350 polymers. Suitable diols typically did not contain aromatic rings and double or triple bonds due to restrictions on thermal stability of unsaturated groups and the need for melting temperatures that are below degradation temperatures. Polycarbonates were also synthesized using melt condensation reactions, with dimethyl carbonate replacing the diacid in the reaction. Acid chloride reactions (Fig. 1) were used for most diols including aromatic rings and double and triple bonds, with interfacial synthesis schemes for polyesters being developed to enable these combinations (48, 49). Despite interfacial reactions being traditionally considered poor for polyesters due to the quenching of the reactive acid chloride, (50) these reactions had a 53% rate of success for all attempted monomer combinations, yielding 166 polymers. Hydroxy-acids and lactone family monomers were synthesized via melt-polymerization methods or ring-opening polymerization methods. Specifically, lactide, ε-caprolactone, and δ-valerolactone were synthesized via ring-opening polymerization, (51) while all other lactones were polymerized via melt ring-opening polymerization. Lactone-family monomers were combined in different ratios to gain insight into the effect of overall polymer composition on polymer structures and were of interest for their often bio-derivable nature (52–54). All reactions performed during library synthesis are summarized in SI Appendix, Fig. S1.
Fig. 1.
Overview of synthesis techniques used for different polymers in the library. Details for individual reactions can be found in Supplementary Dataset S4.
High-Throughput Clear Zone Biodegradation Testing.
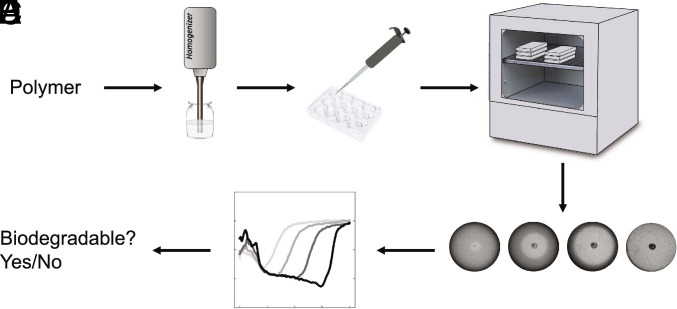
The clear-zone assay is a method for detecting polymer biodegradation and identifying polymer-degrading bacteria; (55–57) here, it is adapted to perform high-throughput biodegradability determination for large polymer libraries. Fig. 2 illustrates the biodegradation test workflow. Polymer was first suspended in agar media above its gelation temperature via precipitation; high shear mixing during the precipitation process is required to form a fine dispersion of small particles. This process requires that the polymer be water insoluble at temperatures around 60 °C, a condition met by the vast majority of polyesters and polycarbonates in the synthetic library. The selected strain was Pseudomonas lemoignei (P. lemoignei) as it has been identified across multiple studies as a PHA-degrading strain (57–59). At least five PHA depolymerase genes have been identified, (60) generating at minimum two types of extracellular enzymes able to degrade short chain length PHA of either native or denatured PHA granules (61). This makes P. lemoignei more likely able to degrade a wide range of polyesters with varying percent crystallinity. Inoculation was done with a single colony in the center of each well once the gel was poured into well plates. Positive controls, negative controls, and six replicates of the test sample were all included on a single plate (See SI Appendix, Fig. S5 for plate layout). The particles of water-insoluble polymer render the gel opaque (Fig. 2D), and their degradation leads to an increase in transmission through the plate which can be monitored using a camera. Because the polymer was finely divided (average particle size of ~15 µm, with a SD of ~10 µm as shown in SI Appendix, Fig. S15), biodegradation was often visible within 48 h, and the assay was terminated after 13 d, making this test much faster than the 180-d testing timeframe typical for standard methods (19, 62). The simplicity of the monitoring allowed for high parallelization with as many as 270 polymers tested in the span of 4 wk using easily accessible equipment and off-the-shelf devices. However, the measured quantity is somewhat different. The clear-zone test measures biofragmentation of the polymer particles and not full biomineralization of the polymer fragments; therefore, there is some difference in the specific physiochemical processes being probed. Given that biofragmentation into bioavailable smaller compounds is typically the rate-limiting step, (63–65) the observed degradation in the plates would reveal what can be expected from these polymers in a natural environment. The test requires a small amount of polymer (40 mg), allowing it to be applied easily to novel chemistries that may be comprised of expensive monomers or screen large numbers of polymers to make in larger quantities (see SI Appendix, Fig. S4 for more details on sample preparation).
Fig. 2.
Clear-zone biodegradation test workflow. (A) Homogenize polymer in agar-growth medium. (B) Add polymer agar mixture to 12-well plate and inoculate with microorganism once gelled. (C) Incubate at 30 °C for up to 13 d. (D) Light transmission pictures are taken every 8 h for the first 3 d, if degradation is detected with the naked eye every 8 h, monitoring continues until day 5, then monitoring is done on the 7th, 9th, 10th, and 13th day. (E) Data processing returns shading curves for each sample, which determine the degradability of the polymer.
After acquisition, data were processed into transmission curves (Fig. 3) that indicated the type of degradation response. First, each photograph was cropped, normalized using the time zero photograph, and converted to optical density (OD) (see SI Appendix, Figs. S17–S22 for details on data processing and analysis). Then, the data were integrated to calculate radial OD curves which can be plotted as a function of time (SI Appendix, Figs. S23 and S24). Four different types of degradation behavior were observed in these curves across all samples, as shown in the four panels of Fig. 3. When there was no degradation, the transmission curves are approximately time independent (panel a), while the other categories (panels b to d) indicate degradation. Classic clear-zone curves (panel b) show a degradation front that progresses to increasing radial distance as a function of time. The edge of the clear zone is very well defined. This type of curve is consistent with diffusion-limited degradation kinetics where a reaction front can be observed, governed by the rate of transport of a species (presumably an enzyme) outward from the central bacterial colony. Distributed degradation curves (panel c) had transmission that increases throughout the sample even from early times. A small degradation front may be present, but it was not a sharp interface in OD like in the classic clear zone. This degradation behavior is consistent with reaction-limited profiles in classic reaction–diffusion systems, and uncatalyzed hydrolysis may also play a role in these samples. Polymers that degraded quickly tended to have classic clear zone profiles, while slow degraders tended to have distributed degradation profiles, consistent with the hypothesized effect of diffusion limitations (SI Appendix, Fig. S16). Finally, degradation of some materials releases strongly absorbing monomers (panel d), resulting in an apparent decrease in transmission due to color interference. The classification of each polymer in the library is shown in SI Appendix, Fig. S7.
Fig. 3.
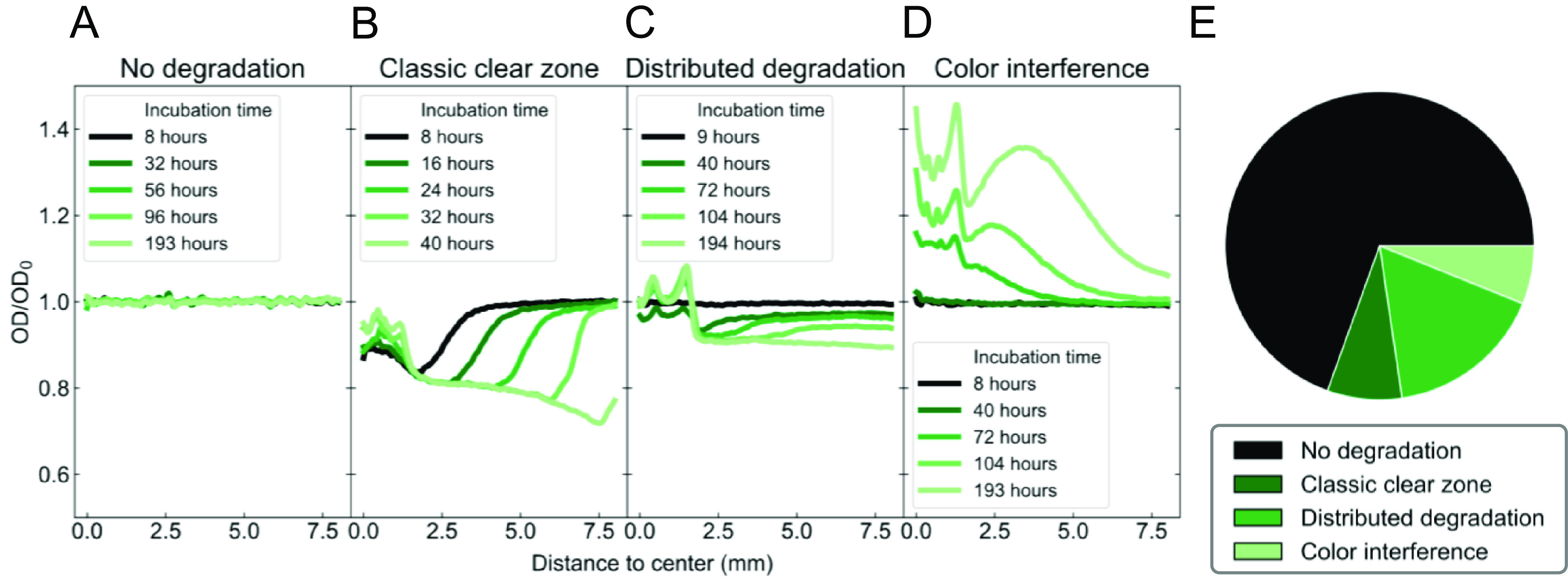
(A–D) Biodegradation behaviors observed from shading curves of clear-zone samples. The different behaviors depend on the enzymatic activity and the rate of biofragmentation. (E) Distribution of all behaviors in the dataset.
Structure–Property Relationships for Biodegradation.
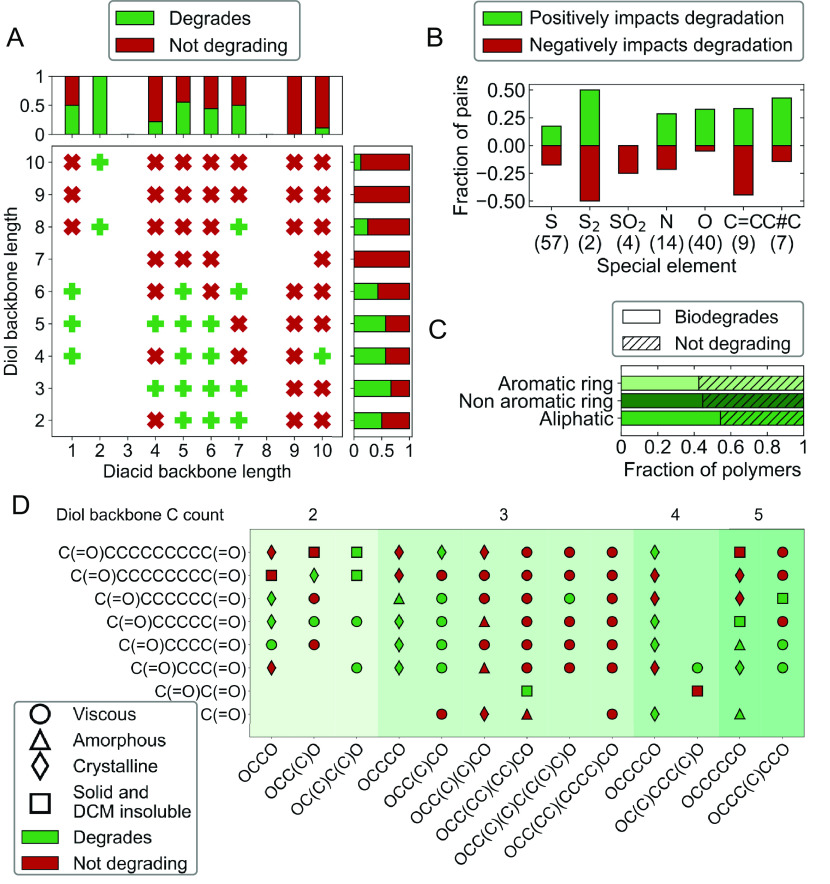
Comparison across the large library of 642 polymers allows the effect of different chemical structures on biodegradation to be elucidated. A simple starting point is systematically examining the monomer backbone length in aliphatic, saturated polyesters (Fig. 4A). There is an obvious trend toward biodegradability for shorter diol and diacid backbone lengths, with most polymers with diacid backbone of 7 or lower and diol of 6 or lower being degradable. Biodegradation was never observed for a total backbone length above 15 carbons (SI Appendix, Fig. S8), strongly suggesting a lower limit of ester functional density that the enzymes of P. lemoignei are able to degrade. Several smaller studies in the literature using different bacterial strains draw contradictory conclusions about whether increases in repeat unit length improve (66) or limit (67) biodegradability. Likely, the optimal chain configurations, hydrophilicity, and crystallinity of the polymer differ between enzymes and among degrading species, leading to this effect. Additionally, differences with previous results, (66) (provided for comparison in SI Appendix, Fig. S36) may be caused by differences in enzymatic and clear-zone biodegradation testing.
Fig. 4.
(A) Biodegradability results for all aliphatic saturated nonsubstituted condensation polymers with no side groups. Marginal histograms show the fraction of polymers degrading for each diol or diacid backbone length. (B) Effect of C or single-bond substitution along the backbone on biodegradation, where a polymer pair identical other than the substitution is compared. The substituted and unsubstituted compared chemistries are listed in SI Appendix, Table S7. (C) Biodegradability of all polymers when separated by presence of rings, distinguishing aromatic and nonaromatic polymers. (D) Biodegradability results for all aliphatic saturated nonsubstituted condensation polymers with and without all carbon side groups. Background color separates the plot by backbone length of the diol repeating units.
As with polyester backbone length, the presence of side groups has a large impact on biodegradability. Examining polyesters composed of the same backbone but different length aliphatic side groups, it can be observed that single-carbon side groups can enhance biodegradability as shown by Fig. 4D. This can be explained by a decrease in crystallinity and packing of the chains, which allows depolymerizing enzymes to attack the cleavable bonds. However, with larger side groups, the opposite effect is observed. The number of biodegradable samples decreases; it is hypothesized that the bulkiness of the side groups prevents enzymes from approaching the polymer backbone. These trends are most discernable for polyesters with diols containing 3 or 4 backbone carbons. Complementary observations were made during enzymatic degradation studies of poly(3-hydroxyalkanoates) where increases in the side-chain length of the monomeric units notably decreased the polymer degradation rate, (67) indicating that long side chains can hinder enzymatically catalyzed degradation.
The inclusion of aromatic and aliphatic rings along the polymer backbone, commonly incorporated to control thermomechanical properties, (68, 69) creates an overall decrease in biodegradability (Fig. 4C). The library is composed of 320 aliphatic polymers, 226 aromatic ring-containing polymers, and 94 nonaromatic ring–containing polymers. The effect of rings in decreasing biodegradability aligns with prior results in the literature on studies of copolyesters with mixed aromatic and aliphatic diacids (70, 71). Interestingly, having the ester groups as meta-substituents on the aromatic ring yields fewer biodegrading polymers than either the ortho- or para- substituted rings, indicating that the ring substitution may be just as important in determining biodegradability as the presence of the aromatic groups (SI Appendix, Fig. S9).
Heteroatom substitutions can affect polarity, mobility, crystallinity, and mechanical properties of polymers; on average, the addition of polar atoms enhances biodegradation, either through biodegradability or biodegradation rate. Heteroatom effects were analyzed by comparing pairs of polymers where one had a carbon and the other had a heteroatom or functionality, classifying the effect of the substitution as making a nondegradable polymer degradable, creating no change, or making a degradable polymer nondegradable (Fig. 4D). Most substitutions had a balanced effect, with nearly equal number of pairs showing increased and decreased degradability. However, three clear trends are visible. First, of the 40 pairs of polymers with oxygen substitution in the form of an ether group, over 31% showed improved degradability while under 6% showed decreased degradability, which supports previous hypotheses that a balance between hydrophobic and hydrophilic elements in a polymer backbone improves biodegradability by facilitating enzymatic attack (72). The current study demonstrates the applicability of heteroatom substitution on a much wider range of chemistries. In the dataset presented here, this balance of hydrophobic and hydrophilic elements improving degradation is readily observable in polymers including dipropylene glycol and tripropylene glycol (SI Appendix, Fig. S10). Although there are substantially fewer polymer pairs in the library for probing triple-bond substitutions, these substitutions also show a positive effect on biodegradation. Finally, sulfonates have a negative effect on degradation, although the number of pairs in the library is too small to draw firm conclusions. From the full set of tested polymers (Supplementary Dataset S4), 47 of them displayed a classic clear zone behavior, which was associated with faster degradation (SI Appendix, Fig. S16). Of these 47 faster degrading samples, 26 of them had an oxygen in the backbone and 16 had a sulfur atom (one had both a sulfur and an oxygen). This demonstrates that the presence of heteroatoms along the backbone of a polyester can improve biodegradability and also enhance biodegradation rates.
To further understand the biodegradation results, the physical state of each polymer at room temperature was determined. Polymers were classified in one of three categories: fluid, glassy, or crystalline. Overall, it was observed that crystallinity lowers biodegradability, consistent with previous literature findings (72, 73). The fraction of biodegradable polymers decreases as the physical state changes from fluid to solid amorphous to solid crystalline as shown in SI Appendix, Fig. S11.
Machine Learning Models.
Machine learning classification models were applied to the large biodegradation dataset to predict biodegradability based on chemical structure. To featurize the polymers, each polymer dataset was written as a Simplified Molecular-Input Line-Entry System (SMILES) string (74) of eight repeat units with the first and last atoms bonded to form a closed ring structure, approximating infinite molar mass chains (75). Morgan fingerprints, RDKit hashing fingerprints, and RDKit descriptors (RDS) were generated for each polymer and used as the chemical descriptors (35), where features that were constant for all samples were removed to reduce dimensionality. Chemical descriptors were augmented with polymer molecular weight data obtained by gel permeation chromatography (GPC) measurements and crystallinity obtained by polarized optical microscopy. These last two features were only available for a subset of polymers as they required solubility of the polymer under relevant conditions. Each dataset was split into six subsets where care was taken to have a close to even distribution of different chemistries in each split (SI Appendix, Figs. S26 and S27). Five splits were used for training and hyperparameter optimization in a five-fold crossvalidation 80/20 data split, with the sixth subset used for validation. Validation results are shown in Fig. 5A for both logistic regression and RF models. Classification of the full dataset using only chemical descriptor results has a maximum prediction accuracy of 82% for the RF model using RDKit fingerprinting (RFP), which is comparable to other works using machine learning for polymer property prediction (41, 76). A recent report of small-molecule biodegradability classification using RF models yielded higher prediction accuracy on a fourfold larger dataset, (77) suggesting that the data size may still be a limiting factor for accuracy even for the large dataset presented here.
Fig. 5.
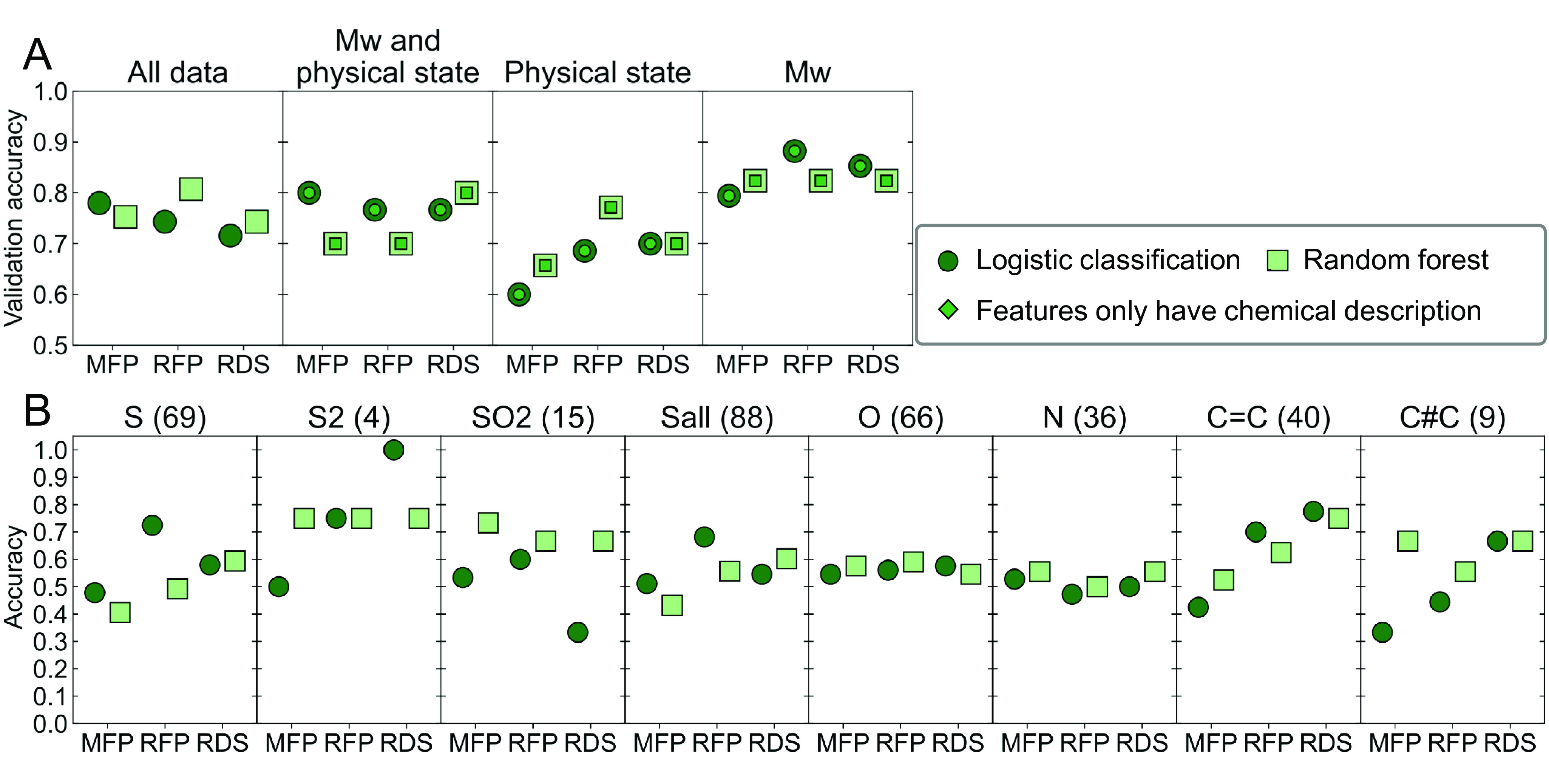
(A) Biodegradability prediction validation accuracy after a hyperparameter optimization of a fivefold crossvalidation on 83% of the data, and final accuracy determined on the 17% validation data. Polymers are vectorized using three chemical descriptions: Morgan fingerprinting (MFP), RDKit fingerprinting (RFP), and RDKit descriptors (RDS). For polymers that could have physical state or molecular weight values determined, the feature vectors were completed with this information, as indicated by the title of each quadrant. (B) Extrapolation prediction accuracy for the full dataset using only chemical description for the feature vectors. The functionalities studied were substitutions along the backbone of sulfides, disulfides, sulfur oxide, all sulfurs, oxygen, tertiary amine, double-bonded carbons, and triple-bonded carbons (from Left to Right, excluding atoms and bonds of the ester group). In each case, the testing set constituted of all the polymers in the dataset that contained the substitution of interest.
For many polymer prediction tasks, the inclusion of physical property data improves model performance (78, 79). For biodegradation, measurements of molar mass and a one-hot encoding for polymer crystallinity were added as key material properties thought to be important for biodegradation. Due to physical property limitations, both measurements could only be performed on subsets of the full dataset. Modeling the subset that includes the crystallinity classifier yields lower accuracy, while when molecular weight data are included, performance improves to 88% accuracy. However, in both cases, when the subset is modeled using only chemical descriptors, the model performs nearly identically to those with added features. Because crystallinity and molar mass screening methods require dissolving the polymers in specific solvents, it is hypothesized that the measured subsets have higher chemical similarity which yields the differing model results. Pair-wise Tanimoto scores were obtained for each polymer pair of the different datasets; the average score of the full dataset was 0.22, while that of all other datasets was between 0.28 and 0.31 (SI Appendix, Fig. S35), consistent with this hypothesis.
To explore the robustness of the classification models, extrapolation studies were performed by specifically excluding chemical functionalities from the training datasets (Fig. 5B). The chemistries studied were atom and bond substitutions along the backbone: sulfides, disulfides, sulfur dioxide, all sulfurs, oxygen atoms, tertiary amines, double-bonded carbons, and triple-bonded carbons. Overall, extrapolations to polymers with backbone functionalities not included in the training data performed only modestly better than random. No model systematically extrapolated to new chemistries better, and some chemistries, such as the oxygen on nitrogen atoms along the backbone, were very hard to predict for all models. This further reinforces the need for a chemically diverse training set.
Conclusions
As concern for the accumulation of polymer waste in the environment rises, the need to design functional biodegradable polymers becomes urgent. However, the design of biodegradable polymers faces several large hurdles—expensive or time-consuming biodegradation tests and small datasets for biodegradable polymers cumulating in a lack of design principles or structure—property relationships for biodegradable chemistries. Here, each of these challenges is addressed. High-throughput synthesis was applied to generate a large, chemically diverse library of 642 unique polyesters and polycarbonates. Challenges for biodegradation testing were addressed by developing a high-throughput and automated clear-zone assay–based biodegradation test using P. lemoignei. The high-throughput nature of the polymer synthesis and biodegradation testing generated a dataset which revealed insights into structure–property relationships for biodegradation and was large enough to leverage machine learning as a predictive tool for biodegradability.
Shorter backbone lengths, short side chains, ortho- and para-substituted rings, and oxygen substitution were correlated with improved biodegradability. Saturated, aliphatic polyesters demonstrated that polymers with diacids of seven or less carbons or diols with six or less carbons were largely biodegradable, while polymers with a combined backbone repeat unit of greater than 15 carbons were ubiquitously non-biodegradable, indicating that there exists a minimum ester density necessary for degradation by this bacterial species. Short single-carbon side chains were beneficial for biodegradability; however, an opposite effect was observed for large side chains which are hypothesized to block accessibility of the cleavable ester bonds. The inclusion of aromatic or aliphatic backbone rings decreased biodegradability overall. Substitution of the ring was relevant to biodegradability, with ortho- or para- substitution on the ring yielding more biodegradable samples. Oxygen substitution as backbone ethers had a positive effect, increasing biodegradability for 31% of the samples; sulfur substitutions did not clearly positively or negatively impact biodegradability but were correlated with faster degradation. The size of the dataset enabled the development of simple classification models for biodegradability, with accuracies up to 82%. Machine learning analysis revealed that chemical structure was a sufficient descriptor for biodegradability prediction, when compared to using chemical structure, molecular weight, and crystallinity.
These findings demonstrate the power of high-throughput experimentation and testing in accelerating the search for biodegradable plastic alternatives. The tools developed here offer an accelerated screening approach with environmental assessment as a primary screening characteristic rather than as an afterthought. Resulting models can be used to guide new monomer discovery, particularly from biosourced targets, helping to reduce the risk and cost in commercializing new polymers by improving the success rate of early-stage candidates with QSPR property prediction that applies not only to thermomechanical properties but also to environmental fate.
Materials and Methods
Polymers were synthesized using standard polycondensation and ring-opening polymerization techniques. Details of polymerization methods for each specific polymer are provided in SI Appendix, Table S3 and specifics on resulting polymers is available in Supplementary Dataset S4. Sample reaction procedures for a melt polycondensation reaction, interfacial polycondensation reaction, and ring-opening polymerization are provided below. Polymerization reaction products were screened for polymer products by GPC when soluble in either tetrahydrofuran (THF) or dimethylformamide (DMF). The resulting determined molecular weight and polydispersity information can be found in Supplementary Dataset S4. For remaining polymers, those soluble in chloroform, dimethylsulfoxide (DMSO), or 1,1,1,3,3,3-hexafluoroisopropanol were screened via ester-adjacent H1 NMR peak shifts for polymer products. Materials insoluble in the previously listed solvents were rinsed with methanol to remove starting reagents and treated as polymeric given that all monomers were soluble in one or more of these solvents.
Melt Polycondensation Procedure (Polymer 345).
To a 20-mL vial with a Teflon stir bar, 2,2,4-Trimethyl-1,3-pentanediol (1.05 equivalents) and 1,4-Cyclohexanedicarboxylic acid (1 equivalent) were added. The vial was then fitted with a septum cap and purged with nitrogen. The vial was heated to 150 °C for 1 h under 1 atm of nitrogen, followed by heating to 175 °C for 1 h before being placed under vacuum (~10 torr) and cooled to 150 °C for an additional 2 h. Following that, the vial was refilled with nitrogen (1 atm), and titanium (IV) isopropoxide (0.02 equivalents) was added. The reaction was allowed to stir for 30 min before being placed back under vacuum for 12 h at 180 °C. The polymer was used without further processing.
Interfacial Polycondensation (Polymer 1).
In a 20-mL vial, Bisphenol A (1 equivalent) was dissolved in water (7.5 mL) with sodium hydroxide base (1.3 equivalent). A SpinPlus stir bar was added to enable emulsification of the two solvents during reaction. In a second 20-mL vial, malonyl chloride (1.1 equivalents) was added to diethyl ether (7.5 mL). After dissolution of the acid chloride, the solution was added dropwise to the stirring (600 rpm) diol solution. The resulting interfacial reaction was left stirring overnight, and the resulting polymer was precipitated into methanol (200 mL) and dried before use.
Ring-Opening Polymerization in a Melt (Polymer 610).
To a 20-mL vial with a Teflon octagonal stir bar, lactide and ε-decalactone (1,000 and 500 equivalents, respectively), triphenyl bismuth (30 equiv), and tin (II) octoate (10 equiv) were added. The reaction mixture was stirred at 110 °C for 20 h, and the resulting polymer was dissolved in dichloromethane, filtered over aluminum oxide, and precipitated into cold hexanes (−20 °C) before being dried.
General Characterization.
Polymer identity and composition was confirmed using a Bruker Neo500 or Neo600 nuclear magnetic resonance spectrometer for polymers soluble in chloroform, dimethyl sulfoxide, or hexafluoroisopropanol. Polymer’s molecular weight was determined using size-exclusion chromatography on an Agilent 1260 Infinity liquid chromatography system with a Wyatt Technology Optilab rEX Refractive Index Detector or using a Waters liquid chromatography system with a Wyatt Technology Optilab T-rEX Refractive index detector and miniDAWN TREOS light scattering detector for polymers soluble in tetrahydrofuran or dimethylformamide, respectively. Data for monomer ratios and molecular weight are summarized in Supplementary Dataset S4.
Dichloromethane Solubility and Polymer Physical State.
Polymers were first classified as fluid or solid by placing every sample vial on its side for 3 d. Any vial where polymer sample had flowed to the side was marked as fluid; others were marked as solid. To distinguish glassy and crystalline samples, the solid samples were further examined using polarized optical microscopy. Polymer samples were dissolved in DCM at a concentration of 40 mg/mL and left mixing on a plate shaker for at least 30 min (solubility data for DCM are shown in SI Appendix, Fig. S15). Samples with leftover visible polymer pieces were marked as insoluble in DCM. A drop of DCM with dissolved sample was placed on a microscope slide and left to dry in a vented hood. Once dried, the slide was observed under polarized light with a ×20 magnification, and the presence or absence of crystals determined the physical state of the polymer. Only polymers that were DCM soluble were classified as glassy or crystalline.
Growth Medium.
Double-concentrated growth medium was prepared following the America Type Culture Collection (ATCC) 179 Pseudomonas medium and was composed of a basal medium, solution A, and solution B. The basal medium was prepared by adding 2.56 g K2HPO4, 2.08 g KH2PO4, 1.0 g NH4Cl, and 0.247 g anhydrous MgSO4 in 500 mL MilliQ water. The solution’s pH was adjusted to 6.8 using NaOH and then filter-sterilized. Solution A was prepared by adding 1.0 g of ferric ammonium citrate and 0.1 g CaCl2 to 100 mL MilliQ water and then filter-sterilized. Solution B was a 1.0 M succinic acid solution adjusted to pH 6 with NaOH and filter-sterilized. The microorganism growth medium was composed of 50 µL of solution A and 150 µL of solution B for 5 mL of basal medium, adding 5 mL of sterile water to achieve normal concentrations.
Microorganism.
P. lemoignei (80) Jendrossek German Collection of Microorganisms and Cell Cultures (DSMZ) 7445 [LMG 2207, NCTC 10937] was purchased from ATCC. The strain was grown in single concentrated growth medium cryofrozen in a 30% glycerol solution. All experiments were conducted with the same source of microorganism.
Clear-Zone Plate Preparation.
Forty milligrams of polymer was added to a 5.54-mL (1 dram) vial with 0.8 mL of DCM. The vial was left on a shaker for at least 45 min before making the plate samples. Double-concentrated agar solution was prepared by adding 40 g of agar to 500 mL MilliQ and autoclaving. For each polymer, an 80-mL sterile autoclave vial was filled with 16 mL of the double-concentrated growth basal medium and heated to 60 °C in a water bath. Twenty milliliters of double-concentrated agar at 60 °C was added to the bottle. Four milliliters of double-concentrated basal medium was added to the polymer vial. The vial was vigorously shaken to mix the aqueous solution and DCM, and the content was quickly poured into the autoclave bottle set at 60 °C. After waiting for 30 s allowing most of the DCM to bubble off to prevent overflowing during homogenizing, the solution was mixed using a Scilogex D500 homogenizer with a S20F/ER20 probe head for 15 s at 10,000 rpm. After 15 s, the solution was mixed again for another 15 s. Ten milliliters of this mixed solution was added to a sterile conical tube containing 50 µL of solution A and 150 µL of solution B. The conical tube was then turned upside down a few times to gently mix the solution without generating any bubbles. Finally, 0.8 mL was sterilely pipetted into 8 wells of a sterile 12-well plate. The remaining 4 wells were filled with the same solution not containing any polymer as controls. A plate layout is shown in SI Appendix, Fig. S5.
Microorganism Growth and Plate Inoculation.
The bacteria were cultured in growth medium for about 30 h in an incubator shaker set to 30 °C and 180 rpm, reaching an OD between 0.5 and 0.8. Wells in rows A and C of the well plate were inoculated with 1 µL of the grown bacteria. The drop was deposited at the center of the well. Plates were incubated for a total of 13 d at 30 °C with a humidity source within the chamber to prevent the agar from drying.
Biodegradation Monitoring.
The transmission of each well was monitored by imaging the wells at regular time intervals using an Opentrons 1st-generation robot with a 2.8 to 12 mm Varifocal Universal Serial Bus (USB) Webcam Mini Camera with adjustable focus and an 8 × 11 in Kaiser Slimlite Plano light tablet. Before each plate measurement, the lid of the 12-well plate was passed over a flame in a sterile environment to remove any water condensation which would interfere with imaging. For the first 72 h, all samples were monitored every 8 h. For the following 3 d (until the 6th day after inoculation), all samples were monitored every 24 h, with the exception of samples where a clear zone could be detected with the naked eye, in which cases monitoring every 8 h was maintained. After that the 6th day, all samples were monitored on days 8, 10, and 13.
Biodegradation Data Extraction.
As the polymer degrades in the plate, the agar’s transmission increases, and more light can pass through to reach the camera. To extract OD curves, after pinpointing the center of the colony, the average pixel color on a black-and-white scale as a distance from the center is computed. These shading curves can be plotted for each measurement for each well. The full process is highly automated and is illustrated and thoroughly explained in SI Appendix, Figs. S17–S25.
Biodegradation of Commercial Polymers.
This high-throughput biodegradation test was applied to 6 commercially available polymers: poly(3-hydroxybutyrate), polybutylene adipate, polybutylene adipate terephthalate, polycaprolactone (two different molecular weights), and polylactic acid. The results are described in SI Appendix, Table S5. The average molecular weight (when given by the supplier) and the biodegradation behavior observed are specified. Of particular note, the commercial polybutylene adipate and polymer 326, also polybutylene adipate, both showed classic clear-zone degradation behavior.
Biodegradation Modeling.
Octamer rings written as Simplified Molecular-Input Line-Entry System (SMILES) strings were vectorized using the Rational Discovery kit (RDKit) python library. Three vectorizations were tested: RDS, RFP, and Morgan fingerprinting. Classification was performed using the sci-kit learn python library (81), and both logistic and RF models were explored to determine whether a linear or nonlinear model would perform best for this property prediction. Four different datasets were studied: the full set of tested polymer with only the chemical vectorization as input, a subdataset of all the tested polymers with molecular weight data where the input vector was a combination of the chemical vectorization and the molecular weight data, a subdataset of all the tested polymers with reliable physical state data where the input vector is chemical vectorization and physical state encoded as a one-hot vector, and a subdataset of the polymers that have both physical state and molecular weight determined where the input vector is a combination of the chemical vectorization and the property-specific encoding as previously described. Each feature was divided by the largest occurring value, so each input was between −1 and 1 (or 0 and 1 for strictly positive values). For training and validation, each dataset was split in 6 equal parts with caution to distribute different chemistries and physical states (when applicable) in each split (as shown in SI Appendix, Fig. S26). One split was used for validation, while the other 5 were used for training and testing as a fivefold hyperparameter optimization. Optimization for vector size for both models, number or trees, minimum number of samples to split, minimum number of samples at a leaf node, maximum depth, and maximum number of leaf nodes specifically for the RF model was performed. The results of the optimization runs are shown in SI Appendix, Figs. S28–S33. Extrapolation to new chemistries was tested only on the full dataset by making the testing set be all the polymers with the chemistry to be extrapolated to and the training set the other polymers of the dataset, using the optimized hyperparameters for the RF and the logistic classification models. Software and data are available at Zenodo (82).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (CSV)
Dataset S02 (CSV)
Dataset S03 (CSV)
Dataset S04 (PDF)
Acknowledgments
This work was supported by Abdul Latif Jameel Water and Food Systems Seed Grants and DIC Corporation. K.A.F. was supported in part by the NSF Graduate Research Fellowship under Grant No. 2141064. We would like to thank the MIT Department of Chemistry Instrumentation Facility for use of their NMR Instruments and Dr. Walter Massefski for his assistance in obtaining NMR data.
Author contributions
K.A.F., S.H.M.A., T.R.B., and B.D.O. designed research; K.A.F., S.H.M.A., T.R.B., D.J.W., D.T.R., and L.V.N. performed research; K.A.F. and S.H.M.A. contributed new reagents/analytic tools; K.A.F., S.H.M.A., T.R.B., D.T.R., L.V.N., and B.D.O. analyzed data; and K.A.F., S.H.M.A., and B.D.O. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Results data are summarized in Datasets S1–S3. Code was used as made available in High-Throughput Clear-Zone on Zenodo Repository (10.5281/zenodo.7348718) (82).
Supporting Information
References
- 1.Shekhar A. R., Kumar A., Syamsai R., Cai X., Pol V. G., Is the plastic pandemic a greater threat to humankind than COVID-19? ACS Sustainable Chem. Eng. 10, 3150–3154 (2022). [DOI] [PubMed] [Google Scholar]
- 2.López-Rubio A., et al. , Overview of active polymer-based packaging technologies for food applications. Food Rev. Int. 20, 357–387 (2004). [Google Scholar]
- 3.Thompson R. C., Moore C. J., vom Saal F. S., Swan S. H., Plastics, the environment and human health: Current consensus and future trends. Philos. Trans. R. Soc. B Biol. Sci. 364, 2153–2166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao L., et al. , Synergistic reinforcement of silanized silica-graphene oxide hybrid in natural rubber for tire-tread fabrication: A latex based facile approach. Compos. B Eng. 161, 667–676 (2019). [Google Scholar]
- 5.Safri S. N. A., Sultan M. T. H., Jawaid M., Jayakrishna K., Impact behaviour of hybrid composites for structural applications: A review. Compos. B Eng. 133, 112–121 (2018). [Google Scholar]
- 6.Nielsen T. D., Hasselbalch J., Holmberg K., Stripple J., Politics and the plastic crisis: A review throughout the plastic life cycle. WIREs Energy Environ. 9, e360 (2020). [Google Scholar]
- 7.Mulder K. F., Sustainable consumption and production of plastics? Technol. Forecast. Soc. Change 58, 105–124 (1998). [Google Scholar]
- 8.Satti S. M., Shah A. A., Polyester-based biodegradable plastics: An approach towards sustainable development. Lett. Appl. Microbiol. 70, 413–430 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Geyer R., Jambeck J. R., Law K. L., Production, use, and fate of all plastics ever made. Sci. Adv. 3, e1700782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geyer R., “Chapter 2–Production, use, and fate of synthetic polymers” in Plastic Waste and Recycling, Letcher T. M., Ed. (Academic Press, 2020), pp. 13–32. [Google Scholar]
- 11.Haque F. M., et al. , Defining the macromolecules of tomorrow through synergistic sustainable polymer research. Chem. Rev. 122, 6322–6373 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Agarwal S., Biodegradable polymers: Present opportunities and challenges in providing a microplastic-free environment. Macromol. Chem. Phys. 221, 2000017 (2020). [Google Scholar]
- 13.Wayman C., Niemann H., The fate of plastic in the ocean environment–A minireview. Environ. Sci. Processes Impacts 23, 198–212 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Goel V., Luthra P., Kapur G. S., Ramakumar S. S. V., Biodegradable/bio-plastics: Myths and realities. J. Polymers Environ. 29, 3079–3104 (2021). [Google Scholar]
- 15.ASTM, Standard Specification for Labeling of Plastics Designed to be Aerobically Composted in Municipal or Industrial Facilities (Version D6400-22, ASTM, West Conshohocken, PA, 2022). [Google Scholar]
- 16.ASTM, Standard Guide for Exposing and Testing Plastics that Degrade in the Environment by a Combination of Oxidation and Biodegradation (Version D6954-18, ASTM, West Conshohocken, PA, 2018). [Google Scholar]
- 17.ASTM, Standard Test Method for Determining Aerobic Biodegradation of Plastic Materials in Soil (Version D5988-18, ASTM, West Conshohocken, PA, 2018). [Google Scholar]
- 18.ASTM, Standard Test Method for Determining Aerobic Biodegradation of Plastic Materials in the Marine Environment by a Defined Microbial Consortium or Natural Sea Water Inoculum (Version D6691-17, ASTM, West Conshohocken, PA, 2018). [Google Scholar]
- 19.ASTM, Standard Test Method for Determining Aerobic Biodegradation of Plastic Materials Under Controlled Composting Conditions, Incorporating Thermophilic Temperatures (Version D5338-15, ASTM, West Conshohocken, PA, 2021). [Google Scholar]
- 20.ASTM, Standard Test Method for Determining Anaerobic Biodegradation of Plastic Materials Under High-Solids Anaerobic-Digestion Conditions (Version D5511-18, ASTM, West Conshohocken, PA, 2018). [Google Scholar]
- 21.Zhu Y., Romain C., Williams C. K., Sustainable polymers from renewable resources. Nature 540, 354–362 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Chen X.-Q., et al. , Directional-path modification strategy enhances PET hydrolase catalysis of plastic degradation. J. Hazard. Mater. 433, 128816 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Brott S., et al. , Engineering and evaluation of thermostable IsPETase variants for PET degradation. Eng. Life Sci. 22, 192–203 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu H., et al. , Machine learning-aided engineering of hydrolases for PET depolymerization. Nature 604, 662–667 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Wei R., et al. , Mechanism-based design of efficient PET hydrolases. ACS Catal. 12, 3382–3396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao R., Pan H., Kai L., Han K., Lian J., Microbial degradation and valorization of poly (ethylene terephthalate)(PET) monomers. World J. Microbiol. Biotechnol. 38, 1–14 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Naser A. Z., Deiab I., Defersha F., Yang S., Expanding poly(lactic acid) (PLA) and polyhydroxyalkanoates (PHAs) applications: A review on modifications and effects. Polymers (Basel). 13, 4271 (2021), 10.3390/polym13234271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ISO, “Plastics—Determination of aerobic biodegradation of non-floating plastic materials in a seawater/sediment interface—Method by analysis of evolved carbon dioxide” (ISO 19679:2020, ISO, Geneva, Switzerland, 2020). [Google Scholar]
- 29.ISO, “Determination of the ultimate aerobic biodegradability of plastic materials under controlled composting conditions—Method by analysis of evolved carbon dioxide—Part 2: Gravimetric measurement of carbon dioxide evolved in a laboratory-scale test” (ISO 14855-2:2018, ISO, Geneva, Switzerland, 2018). [Google Scholar]
- 30.Albright V. C., Chai Y., Knowledge gaps in polymer biodegradation research. Environ. Sci. Technol. 55, 11476–11488 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Ruggero F., Gori R., Lubello C., Methodologies to assess biodegradation of bioplastics during aerobic composting and anaerobic digestion: A review. Waste Manag. Res. 37, 959–975 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Augusta J., Müller R.-J., Widdecke H., A rapid evaluation plate-test for the biodegradability of plastics. Appl. Microbiol. Biotechnol. 39, 673–678 (1993). [Google Scholar]
- 33.Tachibana Y., Yamahata M., Kimura S., Kasuya K.-I., Synthesis, physical properties, and biodegradability of biobased poly(butylene succinate-co-butylene oxabicyclate). ACS Sustainable Chem. Eng. 6, 10806–10814 (2018). [Google Scholar]
- 34.D. W. van Krevelen†, Properties of Polymers (Elsevier Science, 2012). [Google Scholar]
- 35.Bicerano J., Prediction of Polymer Properties (cRc Press, 2002). [Google Scholar]
- 36.Bicerano J., Prediction of the properties of polymers from their structures. J. Macromol. Sci. Part C 36, 161–196 (1996). [Google Scholar]
- 37.Gormley A. J., Webb M. A., Machine learning in combinatorial polymer chemistry. Nat. Rev. Mater. 6, 642–644 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miccio L. A., Schwartz G. A., Mapping chemical structure-glass transition temperature relationship through artificial intelligence. Macromolecules 54, 1811–1817 (2021). [Google Scholar]
- 39.Nazarova A. L., et al. , Dielectric polymer property prediction using recurrent neural networks with optimizations. J. Chem. Inf. Model. 61, 2175–2186 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Sahu H., et al. , An informatics approach for designing conducting polymers. ACS Appl. Mater. Interfaces 13, 53314–53322 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Barnett J. W., et al. , Designing exceptional gas-separation polymer membranes using machine learning. Sci. Adv. 6, eaaz4301 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim C., Batra R., Chen L., Tran H., Ramprasad R., Polymer design using genetic algorithm and machine learning. Comput Mater. Sci. 186, 110067 (2021). [Google Scholar]
- 43.Rivard C., Moens L., Roberts K., Brigham J., Kelley S., Starch esters as biodegradable plastics: Effects of ester group chain length and degree of substitution on anaerobic biodegradation. Enzyme Microbial. Technol. 17, 848–852 (1995). [Google Scholar]
- 44.Aburto J., et al. , Synthesis, characterization, and biodegradability of fatty-acid esters of amylose and starch. J. Appl. Polymer Sci. 74, 1440–1451 (1999). [Google Scholar]
- 45.Kawai F., Microbial degradation of polyethers. Appl. Microbiol. Biotechnol. 58, 30–38 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Ye J., Singh A., Ward O. P., Biodegradation of nitroaromatics and other nitrogen-containing xenobiotics. World J. Microbiol. Biotechnol. 20, 117–135 (2004). [Google Scholar]
- 47.Kawai F., Biodegradation of polyethers (polyethylene glycol, polypropylene glycol, polytetramethylene glycol, and others), in Biopolymers Online, Steinbüchel A., Ed. (Wiley-VCH, 2003), vol. 9, p. 272–281. [Google Scholar]
- 48.P. W. Morgan, “Interfacial polymerization” in Encyclopedia of Polymer Science and Technology (Wiley, 2011). [Google Scholar]
- 49.Nakamura H., Imanishi S., Sanui K., Ogata N., Synthesis of aromatic polyesters by interfacial polycondensation using immiscible binary solvents. Polymer J. 11, 661–664 (1979). [Google Scholar]
- 50.Odian G., “Step polymerization” in Principles of Polymerization (Wiley, 2004), pp. 39–197. [Google Scholar]
- 51.Binczak J., Dziuba K., Chrobok A., Recent developments in lactone monomers and polymer synthesis and application. Materials (Basel) 14, 2881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rydz J., Sikorska W., Kyulavska M., Christova D., Polyester-based (bio)degradable polymers as environmentally friendly materials for sustainable development. Int. J. Mol. Sci. 16, 546–596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rai P., Mehrotra S., Priya S., Gnansounou E., Sharma S. K., Recent advances in the sustainable design and applications of biodegradable polymers. Bioresour. Technol. 325, 124739 (2021). [DOI] [PubMed] [Google Scholar]
- 54.De Hoe G. X., et al. , Sustainable polyester elastomers from lactones: Synthesis, properties, and enzymatic hydrolyzability. J Am. Chem. Soc. 140, 963–973 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Augusta J., Miiller R.-J., Widdecke H., A rapid evaluation plate-test for the biodegradability of plastics. Appl. Microbiol. Biotechnol. 39, 673–678 (1993). [Google Scholar]
- 56.Pranamuda H., Tokiwa Y., Tanaka H., Polylactide degradation by an Amycolatopsis sp. Appl. Environ. Microbiol. 63, 1637–1640 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elbanna K., Lütke-Eversloh T., Jendrossek D., Luftmann H., Steinbüchel A., Studies on the biodegradability of polythioester copolymers and homopolymers by polyhydroxyalkanoate (PHA)-degrading bacteria and PHA depolymerases. Arch. Microbiol. 182, 212–225 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Numata K., Abe H., Iwata T., Biodegradability of poly(hydroxyalkanoate) materials. Materials 2, 1104–1126 (2009). [Google Scholar]
- 59.Shah A. A., Hasan F., Hameed A., Ahmed S., Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 26, 246–265 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Briese B. H., Schmidt B., Jendrossek D., Pseudomonas lemoignei has five poly(hydroxyalkanoic acid) (PHA) depolymerase genes: A comparative study of bacterial and eukaryotic PHA depolymerases. J. Environ. Polymer Degrad. 2, 75–87 (1994). [Google Scholar]
- 61.Knoll M., et al. , The PHA depolymerase engineering database: A systematic analysis tool for the diverse family of polyhydroxyalkanoate (PHA) depolymerases. BMC Bioinformatics 10, 89 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.ASTM, Standard Specification for Labeling of End Items that Incorporate Plastics and Polymers as Coatings or Additives with Paper and Other Substrates Designed to be Aerobically Composted in Municipal or Industrial Facilities (Version D6868-21, ASTM, West Conshohocken, PA, 2021). [Google Scholar]
- 63.Kawai F., et al. , Biodegradability of scott-gelead photodegradable polyethylene and polyethylene wax by microorganisms. Macromol. Symp. 144, 73–84 (1999). [Google Scholar]
- 64.Albertsson A.-C., Barenstedt C., Karlsson S., Lindberg T., Degradation product pattern and morphology changes as means to differentiate abiotically and biotically aged degradable polyethylene. Polymer 36, 3075–3083 (1995). [Google Scholar]
- 65.Kawai F., et al. , Comparative study on biodegradability of polyethylene wax by bacteria and fungi. Polymer Degrad. Stab. 86, 105–114 (2004). [Google Scholar]
- 66.Herzog K., Müller R. J., Deckwer W. D., Mechanism and kinetics of the enzymatic hydrolysis of polyester nanoparticles by lipases. Polymer Degrad. Stab. 91, 2486–2498 (2006). [Google Scholar]
- 67.Kanesawa Y., Tanahashi N., Doi Y., Saito T., Enzymatic degradation of microbial poly(3-hydroxyalkanoates). Polymer Degrad. Stab. 45, 179–185 (1994). [Google Scholar]
- 68.Cai X., Yang X., Zhang H., Wang G., Aliphatic-aromatic poly(carbonate-co-ester)s containing biobased furan monomer: Synthesis and thermo-mechanical properties. Polymer 134, 63–70 (2018). [Google Scholar]
- 69.Ali M. A., Kaneko T., Syntheses of aromatic/heterocyclic derived bioplastics with high thermal/mechanical performance. Ind. Eng. Chem. Res. 58, 15958–15974 (2019). [Google Scholar]
- 70.Witt U., Müller R.-J., Deckwer W.-D., Biodegradation behavior and material properties of aliphatic/aromatic polyesters of commercial importance. J. Environ. Polymer Degrad. 5, 81–89 (1997). [Google Scholar]
- 71.Kleeberg I., Hetz C., Kroppenstedt R. M., Müller R. J., Deckwer W. D., Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates. Appl. Environ. Microbiol. 64, 1731–1735 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larrañaga A., Lizundia E., A review on the thermomechanical properties and biodegradation behaviour of polyesters. Eur. Polym. J. 121, 109296 (2019). [Google Scholar]
- 73.Huang Y., et al. , Effect of hydrophobic fluoropolymer and crystallinity on the hydrolytic degradation of poly(lactic acid). Eur. Polym. J. 97, 308–318 (2017). [Google Scholar]
- 74.Weininger D., SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988). [Google Scholar]
- 75.Arora A., et al. , Random forest predictor for diblock copolymer phase behavior. ACS Macro Lett. 10, 1339–1345 (2021). [DOI] [PubMed] [Google Scholar]
- 76.Park J., et al. , Prediction and interpretation of polymer properties using the graph convolutional network. ACS Polym. Au 2, 213–222 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee M., Min K., A comparative study of the performance for predicting biodegradability classification: The quantitative structure-activity relationship model vs the graph convolutional network. ACS Omega 7, 3649–3655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jenkins M. J., Harrison K. L., The effect of crystalline morphology on the degradation of polycaprolactone in a solution of phosphate buffer and lipase. Polym. Adv. Technol. 19, 1901–1906 (2008). [Google Scholar]
- 79.Hu H., et al. , Recent advances in rational design of polymer nanocomposite dielectrics for energy storage. Nano Energy 74, 104844 (2020). [Google Scholar]
- 80.Delafield F. P., Doudoroff M., Palleroni N. J., Lusty C. J., Contopoulos R., Decomposition of poly-beta-hydroxybutyrate by pseudomonads. J. Bacteriol. 90, 1455–1466 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pedregosa F. V., et al. , Scikit-Learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 82.Av-Ron S., High-throughput clear-zone (Zenodo, 2022) 10.5281/zenodo.7348718. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (CSV)
Dataset S02 (CSV)
Dataset S03 (CSV)
Dataset S04 (PDF)
Data Availability Statement
Results data are summarized in Datasets S1–S3. Code was used as made available in High-Throughput Clear-Zone on Zenodo Repository (10.5281/zenodo.7348718) (82).