Significance
Monoclonal antibody therapy has been sidelined by the rapid evolution of SARS-CoV-2 variants that escape monoclonal antibody neutralization. Receptor decoys are a new approach that avoids this problem. While the viral spike protein may mutate to escape antibody neutralization, the virus needs to conserve affinity for its receptor and therefore cannot easily escape neutralization by a receptor-based decoy. In this report, decoy-expressing viral vectors were found to be effective in mouse models, both to prevent and treat SARS-CoV-2 infection. While the safety profile of lentiviral vectors has not yet been established, adenoassociated virusvectors appear to be safe in humans. The approach could be valuable for immunocompromised individuals and would be useful in the event of zoonosis of a future coronavirus.
Keywords: SARS-CoV-2, AAV vector, lentiviral vector, immunoprophylaxis, ACE2 decoy
Abstract
Vectored immunoprophylaxis was first developed as a means of establishing engineered immunity to HIV using an adenoassociated viral vector expressing a broadly neutralizing antibody. We applied this concept to establish long-term prophylaxis against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in a mouse model using adenoassociated virus and lentiviral vectors expressing a high-affinity angiotensin-converting enzyme 2 (ACE2) decoy. Administration of decoy-expressing (adenoassociated virus) AAV2.retro and AAV6.2 vectors by intranasal instillation or intramuscular injection protected mice against high-titered SARS-CoV-2 infection. AAV and lentiviral vectored immunoprophylaxis was durable and was active against SARS-CoV-2 Omicron subvariants. The AAV vectors were also effective therapeutically when administered postinfection. Vectored immunoprophylaxis could be of value for immunocompromised individuals for whom vaccination is not practical and as a means to rapidly establish protection from infection. Unlike monoclonal antibody therapy, the approach is expected to remain active despite continued evolution viral variants.
Monoclonal antibody therapy had been successful for the treatment of severe COVID-19, decreasing hospitalization and deaths (1) but has since been sidelined as a result of the rapid emergence of viral variants that escape neutralization. The first Omicron variant, BA.1, contained 34 mutations in the spike protein, most of which were within or close to the spike protein receptor-binding domain and allowed for escape from most of the therapeutic monoclonal antibodies. The Regeneron REGN-COV2 cocktail, a cocktail of REGN10933 and REGN10987 monoclonal antibodies, and the Lilly LY-CoV555 potently neutralize the earlier variants of concern (Alpha, Beta, Gamma, and Delta) but their IC50s against the Omicron BA.1 variant was greatly increased (1–10). Vir/GSK VIR-7831 (Sotrovimab) was thought to maintain neutralizing activity against Omicrons BA.1 and BA.2 but was later found to be 10.5- and 340-fold decreased in neutralizing activity against the variants (4–6, 8, 10, 11). Lilly LY-CoV1404 maintained neutralizing titer against BA.1, BA.2 and BA.4/5 (12) but fails to neutralize the more recent, further mutated Omicron variants BQ1.1 and XBB (13). The rapid evolution of the virus is likely to continue over the next several years, imposing a challenge to the development of monoclonal antibodies from which the virus cannot escape. The rapidity of virus evolution is also a challenge for the design of effective vaccines.
A strategy to inhibit virus entry that is less subject to escape by novel variants is that of receptor decoys. The strategy is based on soluble forms of the viral entry receptor fused to the Fc domain of an immunoglobulin heavy chain that serves to increase half-life in vivo and, potentially, to provide antibody effector functions (14). While viruses can mutate epitopes of the spike protein driven by selective pressure to escape neutralization by antibodies elicited from previous infection or vaccination, the spike protein needs to conserve high-affinity binding to its receptor, thereby preserving the neutralizing activity of the receptor decoy. Receptor decoys were first developed as a therapeutic for HIV infection (15, 16). A recombinant protein consisting of the ectodomain of CD4 fused to an immunoglobulin Fc domain was found to bind the viral envelope glycoprotein gp120 with high affinity and potently neutralize the virus in vitro. However, the protein showed no benefit in clinical trials and was not further pursued.
Receptor decoys for SARS-CoV-2 based on soluble forms of ACE2 have been developed by several groups (17–24). The decoy proteins contain point mutations introduced into the ACE2 spike protein-binding region that increase its affinity for the spike protein resulting in higher potency (18, 20, 22). We previously reported the development of a receptor decoy protein termed an “ACE2 microbody” in which the ACE2 ectodomain is fused to the CH3 domain of a human immunoglobulin IgG1 heavy chain Fc region. The truncated Fc region of the microbody decreased the mass of the protein and prevented binding to Fc receptors, eliminating the possibility of an effect analogous to antibody-dependent enhancement in which suboptimal neutralization increases infectivity through Fc receptor mediated virus uptake (17). Decoy proteins administered by intranasal (i.n.) instillation have been shown in mouse and hamster models to protect from infection when given shortly prior to infection and to therapeutically suppress virus replication when given up to about 12 h postinfection (17–25).
The concept of vectored immunoprophylaxis was first proposed as an approach to establish protection against HIV infection by the viral vectored expression of a broadly neutralizing antibody (26), replacing the need to derive a vaccine immunogen capable of eliciting such antibodies. The approach has since been found to be effective as a therapeutic approach to suppressing virus replication the nonhuman primate simian immunodeficiency virus (SIV) model using adenoassociated virus (AAV) vectors expressing broadly neutralizing antibodies and is currently in clinical trials as a means to suppress HIV replication in infected individuals (27, 28). In addition, Gardner et al. showed that an AAV expressing an enhanced eCD4-Ig protected rhesus macaques from multiple challenges with SIV (28).
Here, we applied vectored immunoprophylaxis to SARS-CoV-2 using AAV and lentiviral vectors expressing a modified high-affinity ACE2 microbody. AAV2.retro and AAV6.2 vectors, administered either i.n. or by intramuscular (i.m.) injection, provided a high degree of protection in ACE2 transgenic and Balb/c mouse models. The protection was long-lasting and effective against recent Omicron variants. The AAV vectors were also effective therapeutically when administered shortly postinfection. The lentiviral vector-based decoy was also effective at suppressing virus replication, providing protection that showed no sign of diminishing two months after i.n. administration. Decoy vectored-immunoprophylaxis could be a useful means to protect immunocompromised individuals for whom vaccination is less effective and could offer a therapy that remains active against new variants as the emerge.
Results
Decoy-Expressing AAV Vectors Inhibit SARS-CoV-2 Infection.
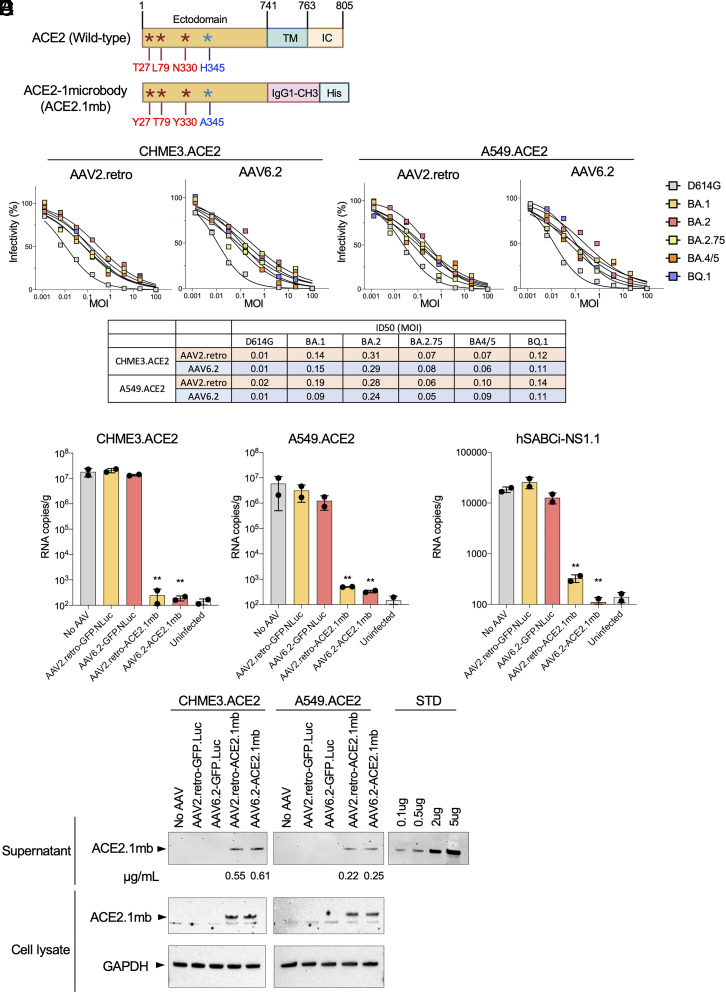
To determine the feasibility of vectored prophylaxis for SARS-CoV-2, we constructed AAV vectors expressing the ACE2 receptor decoy. The decoy, termed ACE2.1mb, is similar to the ACE2 microbody we previously reported (17) that consists of the ACE2 ectodomain fused to a single CH3 domain of an IgG1 heavy chain Fc domain (Fig. 1A). The protein has been modified by the introduction of point mutations in the ACE2 spike protein-binding region reported by Chen et al. to increase affinity for the spike protein (18) and by the introduction of an H345A point mutation that inactivates its catalytic activity (29). The coding sequence was cloned into an AAV vector containing a the cytomegalovirus immediate‑early enhancer/chicken β‑actin (CAG) promoter and virus stock was produced with AAV2.retro and AAV6.2 capsids. AAV2 and AAV6 are reported to have tropism for mouse and human lung and airway cells (30–32). AAV2.retro is an AAV2 variant that was selected for increased central nervous system (CNS) tropism and retrograde transport in axons (33, 34). It has not been reported to transduce lung cells but in pilot experiments, we found that it worked surprisingly well. AAV6.2 is an AAV6 variant containing a single F129L mutation that was found to increase the efficiency of mouse and human airway cell transduction (35). The ability of the vectors to protect cells from SARS-CoV-2 infection was tested in the lung cell-line A549.ACE2 and microglial cell-line CHME3.ACE2. The cells were transduced with serial dilutions of the decoy vectors and then challenged 5 d later with D614G, BA.1, BA.2, BA.2.75, BA.4/5, and BQ.1 spike protein-pseudotyped lentiviruses carrying a luciferase reporter genome. At 2 days postinfection (dpi), luciferase activity in the cultures was measured. The results showed that both decoy-expressing AAVs protected A549.ACE2 and CHME3.ACE2 cells from infection (Fig. 1B). Virus with the D614G spike was most potently neutralized, while BA.2 was the most resistant, with a 20-33-fold higher ID50 [defined as the multiplicity of infection (MOI) resulting in a 50% decrease in luciferase activity]. The ability of the vector to protect cells in the culture at low MOI indicated that the decoy was active on bystander cells. The inhibition curves are shallower than those generated by the addition of recombinant decoy protein (17, 25) to cultures most likely as a result of less efficient transduction at high MOI.
Fig. 1.
AAV-ACE2.1mb prevents SARS-CoV-2 infection in cell culture. (A) The domain structure of full-length ACE2 is shown above with the ectodomain, transmembrane (TM) and intracellular domain (IC). The structure of the decoy consisting of the ACE2 ectodomain, human IgG1-CH3, and carboxy-terminal His tag. The ACE2 domain contains three high-affinity mutations as described by Chan et al. and a H345A mutation in the ACE2 peptidase catalytic site. (B) A549.ACE2 and CHME3.ACE2 cells were transduced with a fivefold serial dilution of AAV2.retro and AAV6.2 decoy vectors and then challenged with ancestral D614G, Omicron spike protein-pseudotyped lentiviral vectors. Luciferase activity was measured 2-dpi. The curves shown above indicate the infectivity based on luciferase activity normalized to mock vector-transduced cells. Each measurement is shown as the average of duplicates. The table below shows that ID50 calculated from the curves shown above. (C) CHME3.ACE2 and A549.ACE2 cells were transduced with AAV2.retro-GFP.nLuc (GFP.Luc), AAV6.2-GFP.nLuc, AAV2.retro-ACE2.1mb, AAV6.2-ACE2.1mb at MOI = 0.5. 2 d post transduction, the cells were infected with SARS-CoV-2 WA1/2020 (MOI = 0.01). The cultures were lysed 2-dpi and RNA was prepared. Viral RNA copy numbers were determined by RT-qPCR. (D) hSABCi-NS1.1 cells were transduced with AAV2.retro-GFP.nLuc, AAV6.2-GFP.nLuc, AAV2.retro-ACE2.1mb, AAV6.2-ACE2.1mb at MOI = 0.5. Two days post transduction, the cells were infected with SARS-CoV-2 WA1/2020 (MOI = 0.01). The cultures were lysed 2-dpi, and RNA was prepared. Viral RNA copy numbers were determined by RT-qPCR. Confidence intervals are shown as mean ± SD. **P ≤ 0.01. (E) CHME3.ACE2 and A549.ACE2 cells were transduced with AAV2.retro or AAV6.2-ACE2.1mb at MOI = 0.5. 3-dpi, secreted decoy protein in the supernatant was pulled-down on NTA beads and bead-bound decoy was detected on an immunoblot probed with His-tag antibody. Pure recombinant decoy protein is shown at right as a standard and was used to determine the amount of protein pulled-down, which is shown below each lane as micrograms decoy pulled-down from 1 mL culture supernatant. At right is shown decoy protein in the cell lysates is shown below with GAPDH as a loading control.
The ability of decoy-expressing vectors to inhibit live SARS-CoV-2 replication was tested on A549.ACE2, CHME3.ACE2, and hSABCi-NS1.1 cells. hSABCi-NS1.1 is a human small airway basal cell-line grown, which in air–liquid interface culture conditions differentiates into mature airway epithelium cell types that model the respiratory tract. The cells were transduced with decoy-expressing or control GFP.nLuc AAV2.retro and AAV6.2 vectors and challenged a day later with SARS-CoV-2 WA1/2020. Virus replication was measured by RT-qPCR quantification of cell-associated viral RNA copies. The 3 cell lines supported high levels of SARS-CoV-2 replication (Fig. 1C). Transduction of the CHME3.ACE2 cells with either of the AAV vectors resulted in a 4–5 log decrease in viral RNA, a level that was not significantly higher than uninfected cells (Fig. 1 C, Left). Transduction of the cells by the control AAV had no effect on SARS-CoV-2 replication. The results in the A549.ACE2 cells were similar (Fig. 1 C, Right). The vectors were also effective in the hSABCi-NS1.1 human small airway basal culture although the decrease was less pronounced (50- to 100-fold), most likely because the cells did not support virus replication to the level of the other cell lines. The AAV6.2 vector was significantly more effective than the AAV2.retro vector at decreasing virus replication in the airway cell cultures (Fig. 1D). Production of the decoy protein by the transduced CHME3.ACE2 and A549.ACE2 cells was confirmed by immunoblot analysis of the protein pulled-down from the culture supernatant on anti-His tag-coated magnetic beads (Fig. 1E). The CHME3.ACE2 cells were found to produce about twofold more decoy than A549.ACE2 which may have contributed to the greater extent of protection in these cells. The concentration of the decoy protein in the culture medium was 0.2 to 0.6 μg/mL, a concentration greater than the IC50 0.15 μg/mL (17).
Vectored Immunoprophylaxis by Decoy-Expressing AAV Vectors.
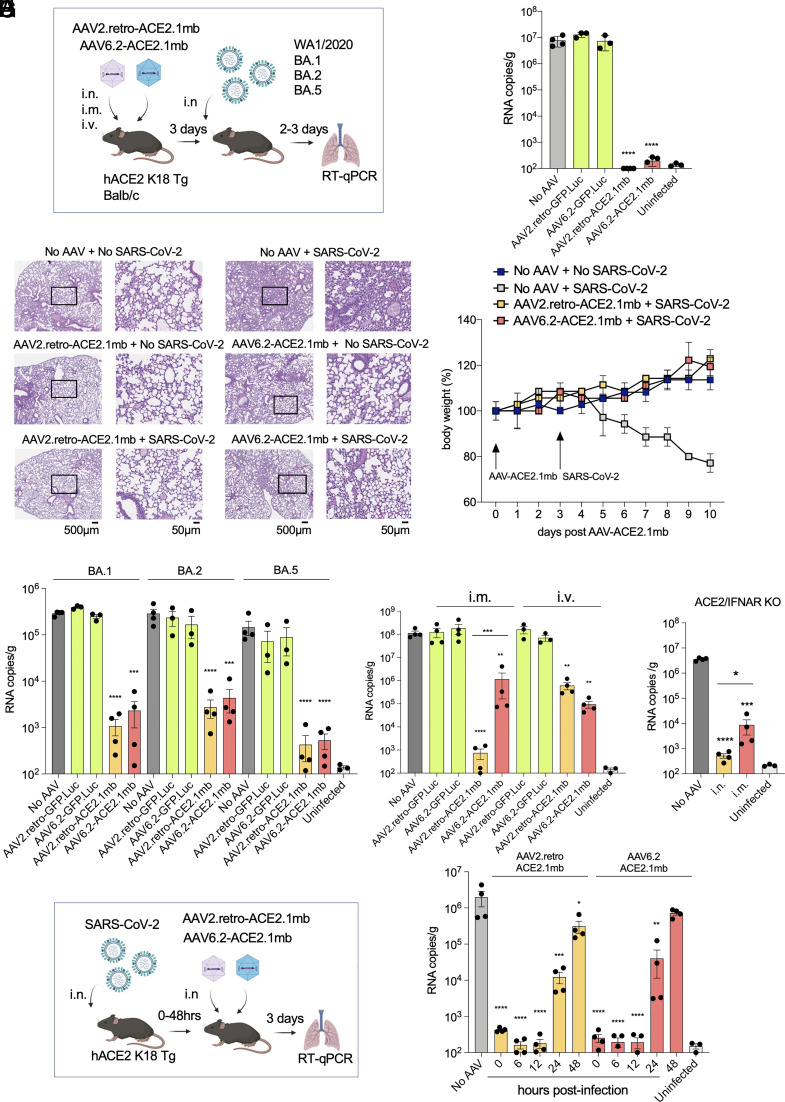
The feasibility of vectored immunoprophylaxis for SARS-CoV-2 with the decoy-expressing AAV vectors was tested in transgenic and nontransgenic mouse models. Decoy-expressing and control GFP AAV2.retro and AAV6.2 vectors were administered to human ACE2 K18 transgenic mice (hACE2 K18 Tg) i.n., i.v. or i.m. After 3 d, the mice were challenged with the WA1/2020 SARS-CoV-2 isolate and virus loads in the lung were measured 3-dpi (Fig. 2A). The results showed that administration of the vector i.n. strongly suppressed virus replication in the mice, decreasing the virus load by 5-logs, a level that was indistinguishable from uninfected mice (Fig. 2B). The control vectors had no effect on virus loads. Histology showed prominent signs of interstitial pneumonia with thickened alveolar septa and inflammatory cell infiltration in the lungs of infected untreated mice while the lungs of decoy-expressing AAV vector-treated mice showed no signs of pneumonia and were free of infiltrating inflammatory cells (Fig. 2C). The lungs of mice treated with the decoy-expressing AAV vectors alone in the absence of SARS-CoV-2 infection were clear, indicating that the decoy vectors themselves did not cause pulmonary inflammation (Fig. 2C). Treatment with the decoy vectors prevented the characteristic loss of body mass associated with untreated SARS-CoV-2 infection (Fig. 2D). Analysis of proinflammatory and antiinflammatory cytokine levels (IFNγ, IL-10, TNFa, IL12-p70, IL-6 and MCP-1) in the lungs of vector-treated, uninfected mice showed no alteration in cytokine levels, suggesting that the decoy-expressing AAV vector treatment itself was not inflammatory (SI Appendix, Fig. S1).
Fig. 2.
Vectored immunoprophylaxis by decoy-expressing AAV decoy vector and therapeutic use. (A) The experimental scheme for testing decoy prophylaxis is diagrammed. hACE2 K18 Tg and Balb/c mice were treated by i.v. injection or i.n. instillation with decoy-expressing AAV vector or control GFP.nLuc-expressing AAV vector (1 × 1012 vg). Three days posttreatment, the hACE2 K18 Tg mice were challenged with SARS-CoV-2 WA1/2020 and the Balb/c mice were challenged with Omicron variants (2 × 104 PFU). Viral RNA copies were quantified 2 d (Omicron) or 3 d (SARS-CoV-2 WA1/2020) postinfection. (B) Mice (n = 3 to 4) were treated with decoy-expressing or control GFP-expressing AAV vectors and challenged with SARS-CoV-2 WA1/2020. Viral RNA copies in the lungs were quantified 3-dpi. (C) H&E-stained lung sections from control and decoy-expressing AAV vectors and SARS-CoV-2-infected mice are shown on the left (2×, scale bars 500 μm) and with the boxed area enlarged on the right (20×, scale bars 50 μm). (D) Mice (n = 3) were treated with decoy-expressing AAV vectors on day 0 and challenged with SARS-CoV-2 WA1/2020 on day 3. Body weight was measured daily. (E) Balb/c mice (n = 3 to 4) were treated with decoy-expressing or GFP-expressing control AAV vectors and then infected 3 d later with Omicron BA.1, BA.2, or BA.5. Viral RNA copies in the lungs were quantified 2-dpi. (F) Mice (n = 4) were administered decoy-expressing AAV vectors i.m. or i.v. and challenged 3-dpi with SARS-CoV-2 WA1/2020. (G) hACE2/IFNAR KO mice (n = 4) were administered decoy-expressing AAV2.retro vector i.m. or i.n. and then infected 3 d later with SARS-CoV-2 WA1/2020. Viral RNA copies in the lungs were quantified 3-dpi. (H) Therapeutic use of the decoy-expressing AAV vectors was tested as diagrammed (Left). hACE2 K18 Tg (n = 4) were infected with SARS-CoV-2 WA1/2020 (2 × 104 PFU) and then treated with decoy-expressing AAVs at time-points up to 48 h postinfection. Viral RNA in the lung was quantified 3-dpi (Right). CIs are shown as mean ± SD. ***P ≤ 0.001, ****P ≤ 0.0001.
To test the effectiveness of the decoy-expressing AAV vectors in protecting against the Omicron variants, Balb/c mice, which support the replication of SARS-CoV-2 Omicron variants through the endogenous murine ACE2 (36, 37), were administered decoy-expressing or control AAV2.retro or AAV6.2 vector i.n. and then challenged with Omicrons BA.1, BA.2 or BA.5. The results showed that the decoy-expressing AAV vectors caused a marked decrease in virus load as compared to the control vectors (Fig. 2E). The decoy-expressing vectors were most effective against the BA.5 variant, decreasing the virus load 300-fold. They were least effective against BA.2, decreasing the virus load 100-fold (Fig. 2E). The AAV2.retro vector appeared to suppress the replication of all three Omicron variants slightly better. This difference did not reach statistical significance but was confirmed in a dose–response analysis, which showed that the AAV2.retro vector was about 10-fold more effective. A 20,000-fold decrease in virus load by AAV2.retro required 1 × 1010 vector genomes (vg); the same degree of suppression by AAV6.2 required 1 × 1011 vg (SI Appendix, Fig. S2).
Administration of the vectors i.n. delivered the vectors to the relevant tissues but it was possible that alternative routes of administration would also be effective given that the decoy protein is stable in vivo and free to diffuse upon secretion from transduced cells (25). I.m. administration of an antibody-expressing AAV to SIV-infected nonhuman primates resulted in long-term suppression of virus loads suggesting that this route can provide durable expression in myocytes (38). In addition, i.v. administration has also been found to be effective for certain AAV serotypes (39, 40). We therefore tested the effectiveness of i.v. and i.m. administration of the decoy-expressing AAV vectors (Fig. 2A). The results showed that i.m. administration of the decoy-expressing AAV2.retro vector was highly effective, decreasing the virus load by 5-logs as compared to control vector, a level that was indistinguishable from uninfected mice (Fig. 2F). The AAV6.2 vector administered i.m. was less effective, decreasing the virus load 2-logs. Administration of the vectors i.v. was only moderately effective, with AAV2.retro decreasing the virus load 200-fold and AAV6.2, 1,000-fold. To determine the concentration of the decoy protein in the sera of mice that had been administered the AAV2.retro, antiviral activity in the sera of treated mice was measured in the pseudotyped lentivirus assay and then calibrated with a standard curve generated with recombinant decoy protein diluted in normal mouse serum. The results showed concentrations of 0.09, 0.10, and 0.12 μg/mL at 1, 2, and 4 wk postadministration, respectively, demonstrating stable expression by the vector over the time period.
The innate immune response rapidly responds to SARS-CoV-2 infection, limiting viral pathogenicity (41, 42). It was therefore possible that the innate immune response was required to enforce the suppressive effect of the decoy on virus replication. To determine whether this was the case, we tested the effectiveness of the decoy-expressing vectors in ACE2-expressing IFNα/β receptor knockout mice (hACE2/IFNAR KO). The mice are deficient in innate immune responses and upon SARS-CoV-2 infection are subject to increased virus replication and more severe pathogenesis (43, 44). The results showed that the i.n. administered vector maintained a high degree of viral load suppression; i.m. administration was somewhat less effective, resulting in a 3-log (rather than 4-log) suppression of virus load (Fig. 2G). The findings suggested that in the vectors are effective in the absence of an innate immune response.
Therapeutic Use of Vectored Immunoprophylaxis for SARS-CoV-2.
The studies described above tested the prophylactic effect of the decoy-expressing AAVs administered prior to SARS-CoV-2 infection but it was possible that the approach might also be effective therapeutically postinfection. The effectiveness of postinfection administration would depend both on how soon postinfection the vector is administered and the time it takes for the vector to transduce cells and establish an inhibitory concentration of decoy in the respiratory tract. To determine the therapeutic effectiveness of the AAV vectors, we infected hACE2 K18 Tg mice with SARS-CoV-2 and then administered decoy-expressing AAV2.retro and AAV6.2 vectors at increasing times postinfection. The results showed that the decoy-expressing vectors were effective when administered concomitant with SARS-CoV-2 and up to 12 h postinfection (Fig. 2H); the treatment was partially effective at 24 h and lost efficacy at 48 h. The results demonstrate remarkably rapid transduction and biosynthesis of the decoy protein by the AAV vectors. While this time course would appear to be too short to be of therapeutic use, the kinetics closely mirror what is seen in monoclonal antibody therapy of SARS-CoV-2 in mouse models (7, 8, 11) suggesting that in humans, where the time-course of disease is slower, the AAV vectors might act with the kinetics similar to that of effective monoclonal antibodies.
Durable Immunoprophylaxis with a Decoy-Expressing AAV.
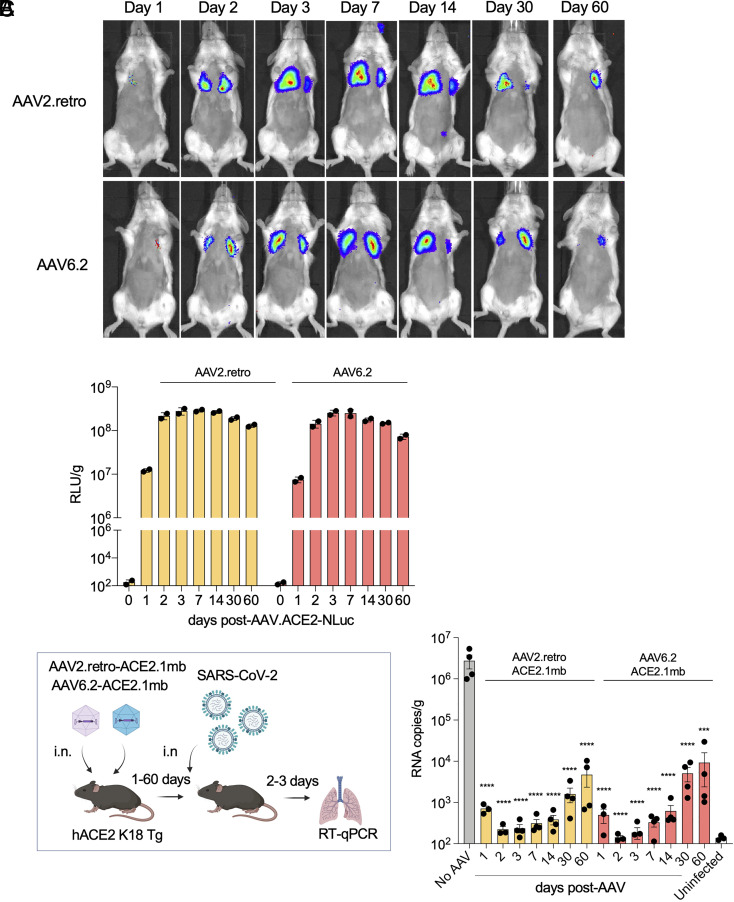
Although AAV does not integrate at a significant frequency into the host cell genome, the genome can remain stable in host cells for extended periods of time as demonstrated in nonhuman primate and clinical trials in which AAV vectors were found to maintain long-term transgene expression (45). To test the durability of the decoy-expressing AAV vectors, we constructed AAV2.retro and AAV6.2 vectors that expressed decoy–luciferase fusion proteins. The vectors were administered i.n. to mice, which were then live-imaged over 60 d. Expression by both vectors in the lungs was first detected 24 h posttreatment and then increased to maximal by day 3 (Fig. 3A). Expression levels remained stable through day 14 and by day 60 had only slightly decreased. Measurement of luciferase activity in tissue homogenates further demonstrated durable expression by the vectors (Fig. 3B). The decoy proteins were readily detectable on day 1 and the following day, expression increased 25-fold. To determine the durability of viral load suppression, hACE2 K18 Tg mice were administered the decoy-expressing vectors i.n. and then challenged with SARS-CoV-2 over a 60-d period. The results showed that the decoy-expressing vectors strongly suppressed the virus loads in mice that had been infected up to 60 d posttreatment (Fig. 3C). The protection began to wane 60 d posttreatment but still suppressed the virus load 300-to 600-fold. The results were consistent with the small decrease in expression in vivo of the decoy–luciferase fusion protein.
Fig. 3.
Durable vectored immunoprophylaxis by decoy-expressing AAV vectors. (A) Balb/c mice (n = 3) were injected i.n. with decoy-luciferase fusion protein-expressing AAV vectors (1 × 1012 vg). Luciferase activity was visualized by live imaging over 60 d at the indicated time points. Representative images of a mouse from each group are shown. (B) Luciferase activity in lung tissue homogenates from mice treated with the decoy-luciferase-expressing AAV vectors (n = 2) was measured over the time course. (C) The experimental scheme to test the durability of AAV vectored immunoprophylaxis is diagrammed (Left). hACE2 K18 Tg (n = 4) were injected with AAV decoy. At 1-, 2-, 3-dpi, the mice were challenged with SARS-CoV-2 (2 × 104 PFU) and viral RNA in the lungs was quantified. The results are shown as a histogram (Right). SARS-CoV-2-infected/AAV-untreated (No AAV) and AAV-untreated/SARS-CoV-2-uninfected (Uninfected) controls are shown. CIs are shown as mean ± SD. ****P ≤ 0.0001.
Increased Durability of Vectored Immunoprophylaxis by a Decoy-Expressing Lentiviral Vector.
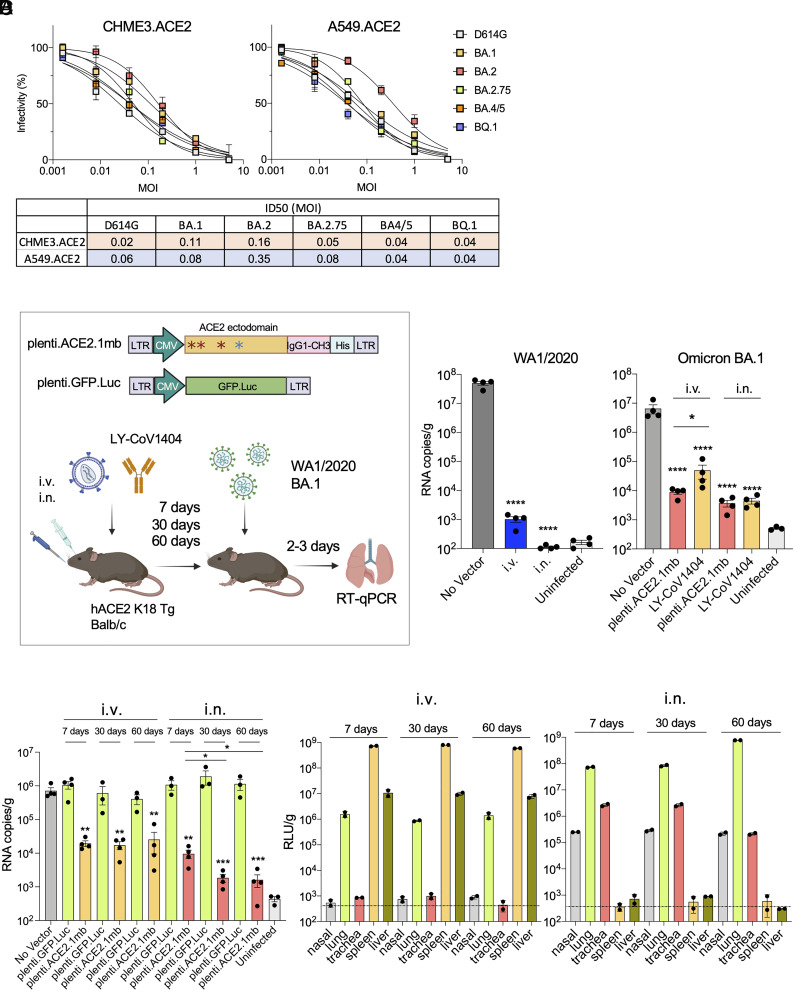
A concern in the clinical use of AAV vectors is that of preexisting neutralizing antibody to serotypes prevalent in the human population (46). It was therefore of interest to investigate an alternative vector that is not generally subject to pre-existing immunity. Lentiviral vectors are not generally subject to preexisting immunity in humans and because they integrate into the host cell genome, they form a stably integrated provirus in the transduced cell that is retained in any daughter cells, potentially providing long-term expression and increasing the durability of protection. In addition, pseudotyping by vesicular stomatitis virus (VSV-G) results in a broad target cell tropism of the vectors. To test the feasibility of lentiviral vectored immunoprophylaxis, we constructed a decoy-expressing lentiviral vector and compared its effectiveness to the AAV vectors. Pull-down of the lentiviral vector-produced decoy protein from transduced A549.ACE2 and CHME3.ACE2 cells showed that the decoy was secreted to levels similar to that of the AAV vectors (SI Appendix, Fig. S3). The potency of decoy protection was determined by transducing the cell lines with a serial dilution of the decoy-expressing lentiviral vector and then measuring resistance to infection by D614G and Omicron spike protein-pseudotyped reporter viruses. The results showed that vector was protective against all of the variants (Fig. 4A). Overall, the potency of neutralization, as calculated by the MOI required to decrease infection by 50%, was similar to that of the AAV vectors. As in the analysis of the AAV vectors, BA.2 was the most resistant to neutralization (8-fold in CHME3.ACE2 and 5.8-fold in A549.ACE2) (Fig. 4A, below). The ability of the vectors to neutralize the viruses at low MOIs confirmed that only a small fraction of the cells needed to be transduced to protect the entire population.
Fig. 4.
Long-term vectored immunoprophylaxis by decoy-expressing lentiviral vector. (A) A549.ACE2 and CHME3.ACE2 cells were transduced with a fivefold serial dilution of decoy-expressing lentiviral vectors and then challenged with D614G, Omicron BA.1, BA.2, 2.75, BA.4/5 and BQ.1 spike protein-pseudotyped lentiviral vectors. Luciferase activity was measured 2-dpi (above). The curves indicate infectivity based on luciferase activity normalized to mock vector-transduced cells. Measurements are the average of duplicates. ID50s calculated from the curves are shown in the table (below). (B) Structure of lentiviral vector and experimental scheme are shown. hACE2 K18 Tg mice or Balb/c were injected with lentiviral vector (5 × 106 IU) or LY-CoV1404 (100 μg) i.p., i.v. or i.n. injection. One week later, the mice were challenged with 2 × 104 PFU of SARS-CoV-2 WA1/2020 (hACE2 K18 Tg) or Omicron (Balb/c). Viral RNA in the lungs was quantified 3-dpi. (C) Decoy-expressing lentiviral vectors (5 × 106 IU) were administered i.v. or i.n. and after 7, 30 and 60 d challenged with SARS-CoV-2 (2 × 104 PFU). Viral RNA was quantified 3-dpi. CIs are shown as mean ± SD. **P ≤ 0.01, ****P ≤ 0.0001. (D) Mice were administered luciferase-expressing lentiviral vector i.v. or i.n. (n = 2). Tissues (nasal, lung, trachea, spleen, and liver) were harvested and luciferase activity was measured over 60 d at the indicated time-points.
To determine whether the decoy-expressing lentiviral vector could establish vectored immunoprophylaxis, the vector was administered i.v. or i.n. and after 7 d, the mice were challenged with WA1/2020 or Omicron BA.1. Virus loads in the lung were quantified 3-dpi (Fig. 4 B, Left). In mice challenged with WA1/2020, i.v. injection resulted in a nearly 5-log decrease in virus load while administration i.n. further decreased the virus load to undetectable levels (Fig. 4 B, Middle). A dose–response analysis, in which mice were administered decreasing amounts of the vector i.v. or i.n., confirmed the greater efficacy of i.n. administration. At a 1 X 106 IU dose of vector, i.n. administration decreased virus loads 400-fold more than i.v. administration (SI Appendix, Fig. S4). Administration of the vector i.v. or i.n. also suppressed Omicron BA.1 replication although not as effectively as WA1/2020, allowing a low level of virus replication in the treated mice (Fig. 4 B, Right). To compare the protective effect of the vector with that of a therapeutic monoclonal antibody, mice were administered the potent neutralizing monoclonal antibody LY-CoV1404 i.v. or i.n. and then challenged with Omicron BA.1. The results showed that by i.v. administration, the lentiviral vector was significantly more effective than LY-CoV1404; by i.n. administration, the two treatments were similarly effective (Fig. 4 B, Right). To test the durability of lentiviral vectored immunoprophylaxis, mice were treated i.v. or i.n. with decoy-expressing or control GFP-expressing lentiviral vector and challenged 7, 30, and 60 d later with SARS-CoV-2 WA1/2020. The was found to persist over the 60-d time-course by both routes of vector administration (Fig. 4C). Administration of the lentiviral vector i.n. more effectively suppressed virus loads and, interestingly, the effect became even more pronounced over the time-course. Analysis of proinflammatory and antiinflammatory cytokine levels (IFNγ, IL-10, TNFa, IL12-p70, IL-6 and MCP-1) in the lungs of the vector-treated, uninfected mice showed no alteration in cytokine levels, suggesting that the treatment itself was not inflammatory (SI Appendix, Fig. S1).
To understand the basis of the long-lasting protection provided by lentiviral vectored immunoprophylaxis, we administered a GFP/luciferase-expressing lentiviral vector i.v. or i.n. to mice and then determined the level of expression over a 60-d time-course by measuring both luciferase activity in cell lysates prepared from different tissues and by flow cytometry. I.v. administration resulted in high-level expression in the spleen at day-7, moderate levels in the lungs and liver (400-fold less in lung than in spleen and 50-fold less in liver) and undetectable levels in nasal tissue and trachea. Expression levels remained constant over the time-course in all of the tissues analyzed (Fig. 4D). I.n. administration of the vector resulted in high-level expression in the lung and moderate levels in the trachea (about 30-fold less on day 7) and nasal tissue and no detectable expression in spleen and liver. Expression levels remained constant over 30 d and increased about eightfold by 60 d, a finding that could explain the increasingly stronger virus load suppression at this time-point in mice that had been administered lentiviral the vector i.n. (Fig. 4C).
Comparison of Lung Cell Types Transduced by the AAV and Lentiviral Vectors.
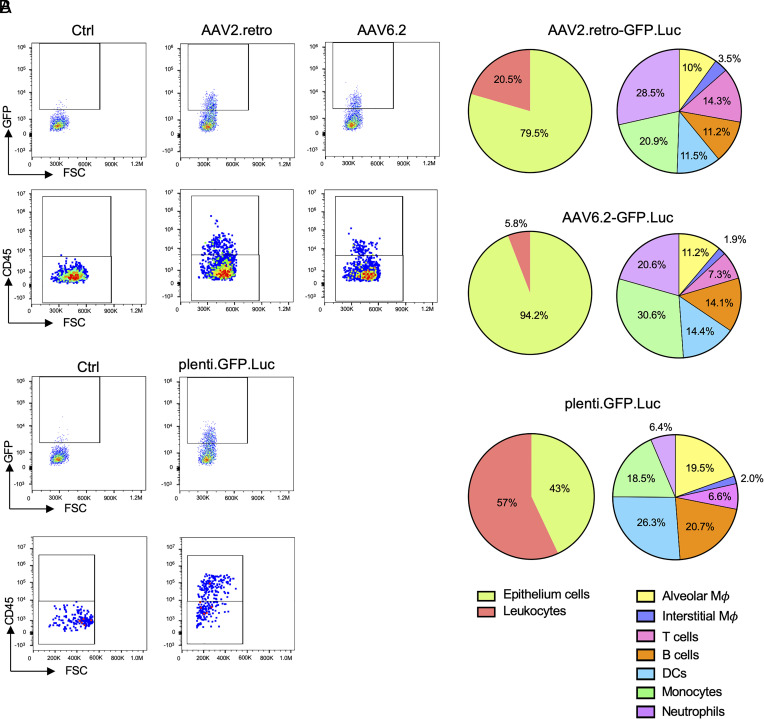
The effectiveness and longevity of the vectored immunoprophylaxis depends both upon the cell types and half-lives of the cells transduced by the vectors. To understand the basis of durable protection, we characterized the cell types transduced in the lung by the AAV and lentiviral vectors. Mice were administered GFP-expressing AAV and lentiviral vectors i.n. and killed after 3 d. The lungs were harvested and a single-cell suspension was prepared for analysis of GFP+ cells by flow cytometry using antibodies that distinguished pulmonary cell types. The results showed that the majority of cells transduced by the AAV2.retro and AAV6.2 vectors were epithelial (79.5% and 94.2%, respectively) (Fig. 5A). Of the cells transduced by AAV2.retro, 20.5% were leukocytes while the cells transduced by AAV6.2 included fewer leukocytes (5.8%). Analysis of the transduced leukocytes showed that the majority were interstitial macrophages and neutrophils with smaller proportions of T cells, B cells, DCs, monocytes, and alveolar macrophages. The distribution of leukocytes transduced by AAV6.2 was roughly similar. The lentiviral vector targeted a larger proportion of leukocytes (57%). Of the transduced leukocytes, the greatest proportion were DCs (26.3%) with substantial contributions from B cells (20.7%) and monocytes (18.5%) (Fig. 5B).
Fig. 5.
Comparison of lung cell subpopulations transduced by AAV and lentiviral vectors. (A) GFP-expressing AAV vectors were administrated i.n. After 3 d, the lungs were harvested and the tissue were enzymatically disaggregated. The cells were analyzed by multi-color flow cytometry with cell-type-specific marker antibodies to distinguish subpopulations defined as follows: Leukocytes (CD45+), epithelial (CD45−), alveolar macrophages (CD45+, F4/80+, SiglecF+), interstitial macrophages (CD45+, F4/80+, SiglecF−), DCs (CD45+, F4/80−, CD11c+), T cells (CD45+, CD3+), B cells (CD45+, CD19+), monocytes (CD45+, CD11b+, CD14+), neutrophils (CD45+, CD62L+, Ly6C/Ly6G+). Representative flow cytometry plots of the GFP+ cells and GFP+/CD45+ populations are shown on the Left and the subpopulations within the GFP+/CD45+ leukocytes are shown in the pie charts on the Right. (B) Mice were administered GFP-expressing lentiviral vector i.n. GFP+ cells in the lung were analyzed by flow cytometry as in (A). Representative flow cytometry plots of the GFP+ cells and GFP+/CD45+ populations are shown on the Left and the subpopulations within the GFP+/CD45+ leukocytes are shown in the pie charts on the Right.
Discussion
We report here that viral vectored expression of an ACE2 microbody protein consisting of the ectodomain of ACE2, mutated to increase affinity for the viral spike protein and inactivate catalytic activity, fused to the CH3 domain of an IgG heavy chain Fc (17), established durable protection from SARS-CoV-2 infection in hACE2 K18 Tg and Balb/c mice. The vectors were also effective therapeutically when administered postinfection, suppressing virus replication to an extent comparable to that of a potent therapeutic monoclonal antibody. Decoy-expressing AAV vectors based on AAV2.retro and AAV6.2 serotypes and a decoy-expressing lentiviral vector established a high degree of protection to high-dose challenge with WA1/2020 isolate of SARS-CoV-2 and Omicron variants. Upon challenge with the WA1/2020 isolate, viral RNA in the lungs 3-dpi was undetectable, corresponding to a >10,000-fold decrease in virus load. The lungs of the treated mice were free of infiltrating leukocytes with no sign of pulmonary inflammation, and the mice did not experience weight loss. Delivery of the decoy by a lentiviral vector was as effective and appeared to be even more durable. The vectors were well tolerated; they did not disturb myeloid or lymphoid cell populations, did not induce proinflammatory cytokines, or cause T cell activation. The vectors established protection against a broad range of SARS-CoV-2 variants including the Omicron subvariants BA.1, BA.2, and BA.5. Protection was strongest against virus with the parental D614G spike protein and somewhat less effective against the BA.2 variant, an effect probably related to the decreased affinity of its spike protein for ACE2 (12).
Administration of the decoy-expressing AAV2.retro and AAV6.2 vectors by i.n. instillation suppressed virus replication in the lungs for at least 60 d. The AAV2.retro vector was somewhat more effective in suppressing virus replication than the AAV6.2 vector, which was surprising, as its capsid was selected for CNS tropism and retrograde transport in neurons (33, 34); its tropism for the respiratory tract has not, to our knowledge, been previously described. The tropism of the AAV2.retro vector for both lung and neuronal cells could be advantageous as a means to suppress SARS-CoV-2 replication both in the respiratory tract and in olfactory tissues.
In a previous report, Sims et al., used an AAV-expressed high-affinity ACE2 decoy to protect mice from SARS-CoV-2 infection (47). In that study, i.n. administration of a decoy-expressing AAVhu.68 vector caused at 30-fold decrease in Wuhan-Hu-1 SARS-CoV-2 virus load 7-dpi but at 4-dpi, close to the time at which virus loads peak, had no significant effect on virus loads. In contrast, we found that i.n. or i.m. administration of decoy-expressing AAV2.retro or AAV6.2 vectors decreased virus loads 1,000- to 10,000-fold at the time of peak virus load. The increased effectiveness of the therapy in our study does not appear to have resulted from differences in increased neutralizing activity of the decoys which appeared to be similar in both studies (IC50s (37 ng/mL vs. 20 ng/mL) (25) or differences in vector dosage, which also appeared to be similar. A potential explanation is that of more efficient transduction of respiratory tract cells by the AAV2.retro and AAV6.2 vectors.
The effectiveness of i.m. injection with the AAV2.retro vector was encouraging because clinically this route of administration may be more practical than i.n. instillation (48). By i.m. injection, the AAV2.retro vector was 3-logs more effective than AAV6.2. The reason for this difference is unclear; it is possible that retrograde transport of the AAV2.retro capsid in myocytes allows more efficient vector expression in these cells or that the vector has increased tropism for the cells of the respiratory tract. The effectiveness of i.m. injection suggests that decoy protein synthesized in transduced myocytes at the site of injection diffuses systemically, establishing a concentration in the respiratory tract sufficient to inhibit virus replication. Similarly, in the rhesus macaque model, i.m. administration of AAV vectors expressing broadly neutralizing antibodies suppressed the replication of SIV, a virus that replicates in secondary lymphoid organs (28, 38). The long-lasting suppression of SIV replication by the vector suggests that transduced terminally differentiated myocytes can produce AAV vector-encoded proteins for a period of several years. Similarly, i.m. administration of a decoy-expressing AAV vector could provide long-lived protection in humans that might be more durable than that of the extended half-life monoclonal antibodies that had been in clinical use (49).
Decoy-expressing AAVs were also effective therapeutically, a finding that was unexpected given the rapid time-course of disease progression in the mouse model. I.n. instillation of the vectors as late as 24 h post-SARS-CoV-2 infection suppressed virus replication, a time course similar to what is found in the treatment of mice with potent neutralizing monoclonal antibody (4). The effectiveness of the decoy therapeutically demonstrates the rapid kinetics with which the vectors transduce cells in the lung and program biosynthesis of the encoded protein. In clinical practice, monoclonal antibody therapy is effective when given several days postinfection (50). The similarity in the timing with which the AAV vectors and monoclonal antibodies are effective in mice suggests that in humans, a decoy-expressing AAV might similarly be effective up to several days postinfection. In clinical use, recombinant ACE2 decoy is expected to act more rapidly, as it does not require viral transduction or protein biosynthesis, decoy vectored immunoprophylaxis is expected to be more durable and obviates the need to administer large quantities of recombinant protein. AAV vectors are currently in use in a large number of clinical trials for a broad range of diseases and have a favorable safety profile.
Lentiviral vector delivery of the decoy was also effective in establishing vectored immunoprophylaxis. As with the AAV vectors, i.n. instillation was the most effective means of administration. Unlike the AAV vectors, the lentiviral vector was also effective by i.v. injection, a route that results mainly in the transduction of splenocytes, many of which are DCs (51). Lentiviral vectored immunoprophylaxis appeared to be more durable than AAV vector, remaining intact through the 60-d time course. Interestingly, the level of virus load suppression intensified with time, an effect that appeared to be the result of increased decoy expression levels in the lung over time as demonstrated with the luciferase-expressing lentiviral vector. The lentiviral vector transduced a high proportion of DCs in the lung, but these were not likely to be the source of long-lasting decoy expression as lung DCs are relatively short-lived with half-lives of only a few days (52). It is more likely that the durable expression by the vector was the result of the transduction of long-lived endothelial cells and the ability of the vector to integrate into the host cell genome. While the durability of i.n. administered AAV vectors was limited, it will be of interest to test additional AAV serotypes that may transduce cells with extended half-lives. Whether the durability of protection in the mouse translates directly to that in humans is unclear.
A potential clinical application of vectored immunoprophylaxis for SARS-CoV-2 is to provide a means of protection to immunocompromised individuals for whom vaccination is less effective. Evusheld, a mixture of two monoclonal antibodies formulated for slow release by i.m. injection, (49) had been highly effective for the long-term protection of immunocompromised individuals; however, both monoclonal antibodies have decreased neutralizing titers against Omicrons BA.1 and BA.2 (1–10) and may be inactive against the increasingly prevalent BQ.1 and BA.2.75 subvariants (53). The decreased neutralizing activity of the monoclonal antibodies contrasts with the decoy which maintains its effectiveness against BQ.1 and BA.2.75. A concern in the long-term expression of therapeutic proteins is that of an antidrug antibody response (54); however, the decoy is derived from human components (with the exception of the four introduced point mutations), its immunogenicity is expected to be low. A potential concern is that of selection for a SARS-CoV-2 with a variant spike with decreased affinity for the modified spike interaction site of the decoy; however, whether such a variant is possible is not known.
These analyses presented in this study are focused on SARS-CoV-2 but decoy vectored immunoprophylaxis could also be valuable in the case of a novel zoonotic coronavirus. Species such as bats and pangolins harbor large numbers of coronaviruses with the ability to use hACE2 (55, 56). If the novel coronavirus used ACE2 as its entry receptor, the decoy-expressing vectors reported here would be an “off-the-shelf” agent available prior to the production of a vaccine. The protection established by the vectors is more rapid than that of vaccination, which requires the induction of an adaptive immune response. In the event of the emergence of a novel SARS-CoV-2 variant that used an alternative receptor, the decoy receptor approach could be adapted to the new receptor. While a switch in receptor usage is possible, it has not happened to date.
Material and Methods
Study Design.
C57BL/6 mice were from Taconic. Balb/c, IFNAR KO mice [B6(Cg)-Ifnar1tm1.2Ees/J(028288)], hACE2-knock-in (KI) [B6.129S2(Cg)-Ace2tm1(ACE2)Dwnt/J] and hACE2 K18 Tg [B6.Cg-Tg(K18-ACE2)2Prlmn/J] were from The Jackson Laboratory.SAMHD1 KO mice were provided by Axel Roers, Technische Universität Dresden (Dresden, Germany). To produce hACE2/IFNAR KO mice, hACE2-KI mice were crossed with IFNAR KO mice. Subsequently, heterozygous mice were crossed to produce homozygous (hACE2/IFNAR KO). All animal experiments were done under protocols approved by the NYU Langone Institutional Animal Care and Use Committee (#170304) in accord with the standards set by the Animal Welfare Act. The study was approved by the NYU School of Medicine Division of Comparative Medicine Standard Operating Protocol (40-008-17).
Cells.
First, 293T cells were cultured in DMEM/10% FBS. Clonal cell lines 293T.ACE2, CHME3.ACE2 and A549.ACE2 were established by lipofection of 293T, CHME3, and A549 cells with plenti.ACE2 (17) using lipofectamine 2000 (Invitrogen). The cells were selected in 1 μg/mL puromycin and single-cell clones were evaluated by flow cytometry for high ACE2 expression. hSABCi-NS1.1 cells were differentiated in air–liquid interface cultures in transwell dishes at 1.5 × 105 cells/well. The cells were plated onto inserts coated with human type IV collagen (Sigma) in PneumaCult Ex Plus medium (Stemcell Technologies). The medium in the upper and lower chambers was changed 1 d after plating, and the medium in the lower chamber was replaced every 2 d. The medium in the upper chamber was removed the apical surface was washed with PBS weekly for 2 wk.
Plasmids.
The expression vectors used for the production of AAV vectors were AAV2.retro Rep/Cap2 (Addgene 81070), Rep/Cap6 (Addgene 110770), pAdDeltaF6 (Addgene 112867) and pAAV-CAG-tdTomato. Rep/Cap6.2 expression plasmid was generated by overlap extension PCR using Rep/Cap6 template to introduce the F129L mutation (35). The amplicon was cloned into the EcoR-I and Nru-I sites of Rep/Cap6. To construct GFP/nanoluciferase-expressing AAV vectors pAAV-GFP.nLuc, pAAV-CAG-ACE2.1mb.nLuc and pAAV-CAG-ACE2.1mb, DNA fragments encoding GFP.nLuc, ACE2.1mb.nLuc and ACE2.1mb were amplified by PCR and joined by overlap extension PCR using primers containing Kpn-I and EcoR-I sites. The insert was removed from pAAV-CAG-tdTomato by cleavage with Kpn-I and EcoR-I and replaced with similarly cleaved amplicon. Decoy expression vector pcACE2.1mb has been previously described (17). Expression plasmids used to produce lentiviral pseudotypes were pMDL, pcVSV.G, pRSV.Rev, the lentiviral transfer vector plenti.GFP.nLuc (17). Expression vectors for the SARS-CoV-2 D614G, Omicron BA.1, Omicron BA.2 spike proteins have been previously described (5, 8, 17). Expression vectors for the novel Omicron spike proteins were constructed by overlap extension PCR mutagenesis using the D614G spike protein plasmid (17) as template and cloned into pcDNA6.
AAV Vector Stocks.
AAV vector stocks were produced by cotransfection of 293T cells with pAAV-CAG-ACE2.1mb, pAdDeltaF6, and AAV.retro RepCap2 or Rep/Cap6.2 at a ratio of 25:25:30 by the calcium phosphate method. Virus-containing supernatant was harvested 2 d posttransfection and filtered through a 0.45-μm filter. The virus was concentrated by ultracentrifugation on 40% sucrose cushion at 4 °C for 16 h at 30,000 × g, resuspended in PBS and concentrated 500-fold on an Amicon Ultra Centrifugal Filter Unit (Millipore). Virus titers were measured by RT-qPCR with a primer pair and probe that hybridized to the AAV2 ITR sequences (57).
SARS-CoV-2 Virus Stocks.
SARS-CoV-2 WA1/2020 (BEI Resources, NR-52281), Omicron BA.1 (BEI Resources, NR-56461), BA.2 (BEI Resources, NR-56781), and BA.5 virus (BEI Resources, NR-58616) stocks were prepared by infection of ACE2.TMPRSS2.Vero E6 cells at an MOI = 0.05 (BEI Resources, NR-56781). Two hours postinfection, the input virus was removed and a day later, the virus-containing supernatant was filtered through a 0.45-μm filter, concentrated on an Amicon Ultra centrifugal filter unit (Millipore) and frozen at −80 °C in aliquots.
Decoy Pull-Down.
CHME3.ACE2 or A549.ACE2 cells (1 × 106) were infected with AAV2.retro, AAV6.2-ACE2.1mb or plenti.ACE2.1mb at MOI = 0.5. The virus was removed the following day, and the supernatant was harvested 3 d later. The decoy protein was pulled-down by a 1-h incubation with 30-μL nickel-nitrilotriacetic acid-agarose beads (QIAGEN) and eluted in Laemmle loading buffer. The protein was then analyzed on an immunoblot probed with anti-His antibody and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG secondary antibody (Sigma-Aldrich). The signals were developed with Immobilon Crescendo Western HRP substrate (Millipore) and visualized on an iBright imaging system (Invitrogen).
Pseudotype Neutralization Assay.
D614G and Omicron spike protein-pseudotyped lentiviruses were generated by cotransfection of 293T cells with pMDL, pRSV.Rev, pLenti.GFP.nLuc and spike protein expression vector and normalized for reverse transcriptase activity as previously described (17, 25). CHME3.ACE2 and A549.ACE2 cells were transduced with serially diluted decoy-expressing AAV or lentiviral vector. The medium was removed the following day and the cells were challenged with pseudotyped viruses (MOI = 0.2). ACE2.1mb protein was purified as previously described (17, 25). Serially diluted ACE2.1mb protein was incubated with D614G spike protein-pseudotyped lentiviruses for 30 min and then added on 293T.ACE2 cells. Luciferase activity in duplicate samples was measured 2-dpi in an Envision 2103 microplate luminometer (PerkinElmer).
Flow Cytometry and Antibodies.
GFP-expressing AAV or lentiviral vectors were administrated i.n. into hACE2 K18 Tg (AAV) or SAMHD1 KO mice (lentivirus). At 3-dpi, the lungs were homogenized in ACK buffer and the cells were disaggregated by a 30-min treatment with 1.5 mg/mL collagenase and 0.1 mg/mL DNase followed by passage through a 100-μm mesh. The cells were blocked with anti-CD16/CD32 (RRID:AB_1574975) and stained with Alexa 700-anti-CD45 (RRID:AB_493715), PerCP-Cy5.5-anti-F4/80 (RRID:AB_893496), APC-Cy7-SiglecF (RRID:AB_2904295), PE-Cy7-anti-CD11c (RRID:AB_493569), PE-Cy7-anti-CD19 (RRID:AB_313655), APC-anti-CD3 (RRID:AB_2561456), Pacblue-anti-CD11b (RRID:AB_755985), PE-Cy5.5-anti-CD62L (RRID:AB_313097), APC-anti-CD14 (RRID:AB_940574) and PE-Ly6C/Ly6G (Gr1) (RRID:AB_313372) (BioLegend) and analyzed on a Beckman CytoFLEX flow cytometer using with FlowJo software. Cell types were classified as epithelial (CD45−), alveolar macrophages (CD45+, F4/80+, SiglecF+), interstitial macrophages (CD45+, F4/80+, SiglecF−), DCs (CD45+, F4/80−, CD11c+), T cells (CD45+, CD3+), B cells (CD45+, CD19+), monocytes (CD45+, CD11b+, CD14+) and neutrophils (CD45+, CD62L+, Ly6C/Ly6G+). LY-CoV1404 was obtained from discarded vials of Bebtelovimab.
Antiinflammatory Cytokine Assay.
hACE2 K18 Tg were administered 1 × 1012 vg decoy-expressing AAV or 5 × 106 IU decoy-expressing lentiviral vector. Mice treated with AAV or lentiviral vector were challenged 3 or 7 d later, respectively, with 2 × 104 plaque-forming unit (PFU) SARS-CoV-2 WA1/2020. Sera were harvested 3-dpi and IFNγ, MCP-1, TNFα, IL-10, IL-12, and IL-6 were measured by cytokine bead array using the BD Cytometric Bead Array Mouse Inflammation Kit (BD Biosciences).
In Vivo and in Vitro Luciferase Assays.
Balb/c or SAMHD1 KO mice were administered AAV2.retro or AAV6.2-ACE2.1mb.nLuc (1 × 1012 vg) or pLenti.GFP.nLuc (5 × 106 IU) by i.n. instillation. The mice were imaged over 60 d by the injection of 100 μL 1:40 diluted Nano-Glo substrate (Nanolight) and visualization on an IVIS Lumina III XR (PerkinElmer). To measure luciferase activity in the tissues, organs were harvested and homogenized in lysing matrix D tubes with a FastPrep-24 5G homogenizer (MP Biomedicals). The Nano-Glo Luciferase Assay Reagent (Nanolight) was added and luminescence was measured on an Envision 2103 plate reader (PerkinElmer).
Live Virus Infection of Cell Lines.
CHME3.ACE2, A549.ACE2, and hSABCi-NS1.1 cells (2 × 105) were infected with AAV2.retro or AAV6.2-ACE2.1mb at MOI = 0.5. The medium was replaced 1-dpi and the following day, the cells were infected with SARS-CoV-2 at MOI = 0.01. The cultures were lysed 2-dpi after which RNA was prepared and cell-associated viral RNA copies were quantified by reverse transcriptase RT-qPCR. Absolute RNA copy numbers were calculated using a standard curve generated by the analysis of a serially diluted in vitro transcribed synthetic subgenomic viral RNA using the 2−ΔΔCT method.
Prophylactic and Therapeutic Administration of Decoy-Expressing Vectors.
For prophylaxis experiment, 6 to 8-wk-old hACE2 K18 Tg or Balb/c mice were anesthetized with isoflurane or ketamine–xylazine cocktail and injected with 80 μL (i.n. or i.v. or i.m.) (1 × 1012 vg) of AAV2.retro or AAV6.2-decoy or 5 × 106 IU of plenti.ACE2.1mb or LY-CoV1404 (100 μg). After 1 to 60 d of infection, the mice were infected i.n. with 2 × 104 PFU of SARS-CoV-2 WA1/2020 (hACE2 K18 Tg) or Omicron BA.1 or BA.2 or BA.5 (Balb/c). At 2-dpi (Omicron) or 3-dpi (SARS-CoV-2 WA1/2020), the mice were killed and RNA was prepared from 200 µL lung lysate using the Quick-RNA MiniPrep kit (Zymo Research). For therapeutic testing, hACE2 K18 Tg were infected i.n. with 2 × 104 PFU SARS-CoV-2 WA1/2020. The mice were infected 0 to 48 h postinfection i.n. with 80 μL (1 × 1012 vg) AAV2.retro or AAV6.2-decoy. 3-dpi (SARS-CoV-2 WA1/2020), the mice were killed and RNA was prepared from 200 µL of the lung lysate using a Quick-RNA MiniPrep kit.
Measurement of Decoy Concentration in the Sera.
C57/B6 Tg mice (n = 2) were administered decoy-expressing AAV2.retro vector i.m. (1 × 1012 vg). The mice were bled 1, 2, and 4 wk postinjection, and sera were prepared. The sera were serially diluted and incubated with pseudotyped lentivirus. Neutralizing activity in the sera was determined using D614G pseudotyped lentivirus on ACE2.293T cells. Pseudotyped virus infectivity is displayed as the percent infection normalized to untreated pseudotype. Neutralizing titers were converted to decoy concentration using a standard curved generated with recombinant decoy protein serially diluted into normal mouse serum.
Virus Loads.
SARS-CoV-2 E gene subgenomic RNA levels were measured by reverse transcriptase RT-qPCR with a TaqMan probe. Lung RNA was mixed with TaqMan Fast Virus 1-step Master Mix (Applied Biosystems), 10 mM forward and reverse primers, and 2 mM probe. PCR cycles were 5 min at 50 °C, 20 s at 95 °C, 40 cycles of 3 s at 95 °C, 3 s at 60 °C). E gene subgenomic RNA copies were measured using forward primer subgenomic F (CGATCTCTTGTAGATCTGTTCTC), reverse primer E Sarbeco R and probe E Sarbeco P1) (58, 59). Tissue analyses were normalized to GAPDH mRNA copies measured using the mouse GAPDH.forward (CAATGTGTCCGTCGTGGATCT) and mouse GAPDH.reverse (GTCCTCAGTGTAGCCCAAGATG) with mouse GAPDH probe (FAM-CGTGCCGCCTGGAGAAACCTGCC-BHQ) or human GAPDH.forward (GTCTCCTCTGACTTCAACAGCG) and human GAPDH.reverse (ACCACCCTGTTGCTGTAGCCAA) with human GAPDH probe (FAM-TAGGAAGGACAGGCAAC-IBFQ). Absolute RNA copy numbers were calculated using a standard curve generated by the analysis of a serially diluted in vitro transcribed synthetic subgenomic viral RNA containing the E gene sequence (2019-nCoV_E Positive Control, IDT: 10006896) using the 2−ΔΔCT method.
Histology.
The lungs of SARS-CoV-2-infected mice were harvested 3-dpi. The tissue was fixed in 10% neutral buffered formalin for 72 h at room temperature and then processed through graded ethanol, xylene, and into paraffin in a Leica Peloris automated processor. Then, 5-μm paraffin-embedded sections were deparaffinized and stained with hematoxylin (Leica, 3801575) and eosin (Leica, 3801619) on a Leica ST5020 automated histochemical strainer. Slides were scanned at 40× magnification on a Leica AT2 whole slide scanner, and the images were transferred to the NYULH Omero web-accessible image database.
Statistical Analysis.
Statistical significance was determined by the Kruskal–Wallis test with post hoc Dunn’s test. Significance was calculated based on two-sided testing and is shown in the Figures as mean ±SD with CIs listed as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Meike Dittmann for ACE2.TMPRSS2.Vero E6 cells, Belinda Dcosta for technical assistance, and the NYU Langone Laura and the Isaac Perlmutter Cancer Center Experimental Pathology Research Laboratory for histology. We thank David J. Simon (Weill Cornell Medicine) for AAV plasmids. Funding was provided by the NIH (DA046100, AI122390, and AI120898). The Experimental Pathology Research Laboratory is partially supported by the Cancer Center Support Grant P30CA016087.
Author contributions
T.T. and N.R.L. designed research; T.T. and J.M. performed research; T.T. contributed new reagents/analytic tools; T.T. analyzed data; and T.T. and N.R.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and SI Appendix.
Supporting Information
References
- 1.VanBlargan L. A., et al. , An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 28, 490–495 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L., et al. , Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Planas D., et al. , Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Dragoni F., et al. , Impact of SARS-CoV-2 omicron BA.1 and delta AY.4.2 variants on the neutralization by sera of patients treated with different authorized monoclonal antibodies. Clin. Microbiol. Infect. 28, 1037–1039 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H., Dcosta B. M., Landau N. R., Tada T., Resistance of SARS-CoV-2 Omicron BA.1 and BA.2 variants to vaccine-elicited sera and therapeutic monoclonal antibodies. Viruses 14, 1334 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iketani S., et al. , Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604, 553–556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen R. E., et al. , In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature 596, 103–108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tada T., et al. , Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. EBioMedicine 78, 103944 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond M., et al. , The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res. Sq., 10.21203/rs.3.rs-1211792/v1 (2021). [DOI] [Google Scholar]
- 10.Bruel T., et al. , Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat. Med. 28, 1297–1302 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Touret F., et al. , In vitro activity of therapeutic antibodies against SARS-CoV-2 Omicron BA.1, BA.2 and BA.5. Sci. Rep. 12, 12609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y., et al. , BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608, 593–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai M., et al. , Efficacy of antiviral agents against Omicron subvariants BQ.1.1 and XBB. N. Engl. J. Med. 388, 89–91 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czajkowsky D. M., Hu J., Shao Z., Pleass R. J., Fc-fusion proteins: New developments and future perspectives. EMBO Mol. Med. 4, 1015–1028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges T. L., et al. , Phase 1 study of recombinant human CD4-immunoglobulin G therapy of patients with AIDS and AIDS-related complex. Antimicrob. Agents Chemother. 35, 2580–2586 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gershon D., Genentech sheds gp120 vaccine. Nat. Med. 2, 370–371 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Tada T., et al. , An ACE2 microbody containing a single immunoglobulin Fc domain is a potent inhibitor of SARS-CoV-2. Cell Rep. 33, 108528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan K. K., et al. , Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369, 1261–1265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jing W., Procko E., ACE2-based decoy receptors for SARS coronavirus 2. Proteins 89, 1065–1078 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., et al. , An ACE2 decoy can be administered by inhalation and potently targets omicron variants of SARS-CoV-2. EMBO Mol. Med. 14, e16109 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linsky T. W., et al. , De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science 370, 1208–1214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higuchi Y., et al. , Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat. Commun. 12, 3802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Case J. B., et al. , Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485.e5. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, et al. , Engineered ACE2 decoy mitigates lung injury and death induced by SARS-CoV-2 variants. Nat. Chem. Biol. 18, 342–351 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tada T., Dcosta B. M., Zhou H., Landau N. R., Prophylaxis and treatment of SARS-CoV-2 infection by an ACE2 receptor decoy in a preclinical animal model. iScience 26, 106092 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson P. R., et al. , Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15, 901–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner M. R., Promise and progress of an HIV-1 cure by adeno-associated virus vector delivery of anti-HIV-1 biologics. Front. Cell Infect. Microbiol. 10, 176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner M. R., et al. , AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519, 87–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guy J. L., Jackson R. M., Jensen H. A., Hooper N. M., Turner A. J., Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis. FEBS J. 272, 3512–3520 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer-Berg H., et al. , Identification of AAV serotypes for lung gene therapy in human embryonic stem cell-derived lung organoids. Stem Cell Res. Ther. 11, 448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Lieshout L. P., Domm J. M., Wootton S. K., AAV-mediated gene delivery to the lung. Methods Mol. Biol. 1950, 361–372 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Halbert C. L., Allen J. M., Miller A. D., Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75, 6615–6624 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tervo D. G., et al. , A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., et al. , Functional analysis of mutations endowing rAAV2-retro with retrograde tracing capacity. Neurosci. Lett. 784, 136746 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Limberis M. P., Vandenberghe L. H., Zhang L., Pickles R. J., Wilson J. M., Transduction efficiencies of novel AAV vectors in mouse airway epithelium in vivo and human ciliated airway epithelium in vitro. Mol. Ther. 17, 294–301 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halfmann P. J., et al. , SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 603, 687–692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y. N., et al. , Different pathogenesis of SARS-CoV-2 Omicron variant in wild-type laboratory mice and hamsters. Signal Transduct. Target. Ther. 7, 62 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spitsin S., et al. , Protection against SIV in Rhesus macaques using albumin and CD4-based vector-mediated gene transfer. Mol. Therapy. Methods Clin. Dev. 17, 1088–1096 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goertsen D., Goeden N., Flytzanis N. C., Gradinaru V., Targeting the lung epithelium after intravenous delivery by directed evolution of underexplored sites on the AAV capsid. Mol. Ther. Methods Clin. Dev. 26, 331–342 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korbelin J., et al. , Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol. Ther. 24, 1050–1061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond M. S., Kanneganti T.-D., Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 23, 165–176 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diamond M. S., Lambris J. D., Ting J. P., Tsang J. S., Considering innate immune responses in SARS-CoV-2 infection and COVID-19. Nat. Rev. Immunol. 22, 465–470 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiber G., The role of type I interferons in the pathogenesis and treatment of COVID-19. Front. Immunol. 11, 595739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boudewijns R., et al. , STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 11, 5838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang D., Tai P. W. L., Gao G., Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discovery 18, 358–378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mingozzi F., High K. A., Immune responses to AAV vectors: Overcoming barriers to successful gene therapy. Blood 122, 23–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sims J. J., et al. , Intranasal gene therapy to prevent infection by SARS-CoV-2 variants. PLoS Pathog. 17, e1009544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tosolini A. P., Sleigh J. N., Intramuscular delivery of gene therapy for targeting the nervous system. Front. Mol. Neurosci. 13, 129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levin M. J., et al. , Intramuscular AZD7442 (Tixagevimab-Cilgavimab) for prevention of Covid-19. N. Engl. J. Med. 386, 2188–2200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miguez-Rey E., Choi D., Kim S., Yoon S., Sandulescu O., Monoclonal antibody therapies in the management of SARS-CoV-2 infection. Expert Opin. Invest. Drugs 31, 41–58 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tada T., Norton T. D., Leibowitz R., Landau N. R., Directly injected lentiviral vector-based T cell vaccine protects mice against acute and chronic viral infection. JCI Insight 7, e161598 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamath A. T., Henri S., Battye F., Tough D. F., Shortman K., Developmental kinetics and lifespan of dendritic cells in mouse lymphoid organs. Blood 100, 1734–1741 (2002). [PubMed] [Google Scholar]
- 53.Akerman A., et al. , Emergence and antibody evasion of BQ, BA.2.75 and SARS-CoV-2 recombinant sub-lineages in the face of maturing antibody breadth at the population level. EBioMedicine 90, 104545 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martinez-Navio J. M., et al. , Long-term delivery of an anti-SIV monoclonal antibody with AAV. Front. Immunol. 11, 449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Demogines A., Farzan M., Sawyer S. L., Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 86, 6350–6353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wacharapluesadee S., et al. , Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12, 972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aurnhammer C., et al. , Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods 23, 18–28 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Winkler E. S., et al. , SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Corman V. M., et al. , Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro. Surveill. 25, 2000045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and SI Appendix.