Significance
The gaseous hormone ethylene regulates many aspects of plant growth and development; however, the molecular basis by which ethylene receptors bind and respond to ethylene concentrations at less than one part per billion has remained an unresolved question. Here, evolutionary and computational modeling approaches were used to develop a new molecular model for the ethylene-binding site of the receptor ETR1 and key features of this model validated experimentally. Results shed light on the basis for high-affinity ethylene binding and how ethylene binding is transduced to mediate changes in signal output by the receptor. The new model is also relevant to our understanding of bacterial chemotaxis, convergent receptor evolution, and the development of ethylene nanosensors for agricultural and industrial applications.
Keywords: ethylene, ethylene receptor, ligand binding, copper cofactor, structural model
Abstract
The gaseous hormone ethylene is perceived in plants by membrane-bound receptors, the best studied of these being ETR1 from Arabidopsis. Ethylene receptors can mediate a response to ethylene concentrations at less than one part per billion; however, the mechanistic basis for such high-affinity ligand binding has remained elusive. Here we identify an Asp residue within the ETR1 transmembrane domain that plays a critical role in ethylene binding. Site-directed mutation of the Asp to Asn results in a functional receptor that has a reduced affinity for ethylene, but still mediates ethylene responses in planta. The Asp residue is highly conserved among ethylene receptor-like proteins in plants and bacteria, but Asn variants exist, pointing to the physiological relevance of modulating ethylene-binding kinetics. Our results also support a bifunctional role for the Asp residue in forming a polar bridge to a conserved Lys residue in the receptor to mediate changes in signaling output. We propose a new structural model for the mechanism of ethylene binding and signal transduction, one with similarities to that found in a mammalian olfactory receptor.
The gaseous hormone ethylene regulates multiple aspects of plant growth and development, ripening being the best known of these, as well as responses to biotic and abiotic factors (1–3). Ethylene is perceived in plants by membrane-bound receptors, the first identified and best studied of these being ETR1 from Arabidopsis (2, 4–6). Most plants contain families of ethylene receptors, the five-member ethylene-receptor family of Arabidopsis consisting of ETR1, ETR2, EIN4, ERS1, and ERS2 (2, 6). The plant ethylene receptors have similar overall structures with transmembrane (TM) domains near their N-termini and signaling motifs in their C-terminal regions (2, 6). The N-terminal TM domains contain the ethylene-binding site (5, 7, 8), and also serve in membrane localization of the receptor, the majority of the receptors being found associated with the endoplasmic reticulum (9–12). Following the TM domain is a GAF domain (named after the proteins cGMP-specific phosphodiesterase, adenylyl cyclase, and FhlA in which it was initially identified) implicated in receptor interactions (12, 13). The C-terminal portions of each receptor contain domains with similarity to histidine kinases and in some cases the receiver domains of response regulators (14, 15). Histidine kinases and receiver domains are signaling elements originally identified as components in bacterial phosphorelays and are now known to be present in plants, fungi, and slime molds (16). The plant ethylene receptors are negative regulators of ethylene signal transduction, such that the receptors are “on” in the absence of ethylene and actively repress the ethylene response, and “off ” when bound to ethylene, allowing for derepression of the ethylene response (2, 6, 17–19). As a result, higher-order loss-of-function mutants such as the etr1 etr2 ein4 triple mutant and the etr1 ers1 double mutant result in constitutive ethylene-response phenotypes, the etr1 ers1 mutant resulting in infertility (17–19). Since the initial identification of ETR1 in plants, similar proteins with the conserved features of the ethylene-binding domain (EBD) have also been identified in prokaryotes, notably in cyanobacteria (7, 20, 21).
Due in part to the difficulty in obtaining high-resolution structural information from TM domains, much of what is known about the requirements for ethylene binding by the receptors comes from a coupling of biochemical and genetic analyses (5, 7, 20, 22, 23). Through these analyses, the receptors have been determined to function as homodimers, with ethylene binding mediated through an associated Cu(I) co-factor (7, 22–25). A set of highly conserved Cys and His residues in the TM domain is implicated in chelating the copper cofactor (5, 7, 25). Initial analysis indicated the existence of one copper cofactor per receptor dimer, suggesting a model in which the copper is chelated by two Cys and two His residues, thereby resulting in a single ethylene-binding site per receptor dimer (7). However, recent analysis is consistent with the existence of one copper cofactor per receptor monomer, which supports a model with two copper cofactors and potentially two ethylene-binding sites per receptor dimer (24). The well-characterized missense mutation etr1-1 arises due to a mutation in the liganding Cys residue (Cys65Tyr), resulting in a receptor that no longer binds the copper cofactor and as a result also no longer binds ethylene (4, 5, 7, 26). The etr1-1 mutation confers dominant ethylene insensitivity on plants because of this inability to perceive the ethylene signal. Additional missense mutations in the receptor have further refined our understanding of ethylene binding and signal transduction (20), as has computational modeling and tryptophan scanning mutagenesis (24, 27).
A major and still unresolved question is how the ethylene receptors bind ethylene with such high affinity. Ethylene binds to ETR1 with a calculated dissociation constant (Kd) of 2.4 × 10−9 M, and with a half-life for dissociation of over 12 h (5), consistent with plants responding to ethylene concentrations as low as 0.2 nL L−1 (28). Here we identify a highly conserved aspartate within the ETR1 TM domain (Asp25) as playing a critical role in copper and ethylene binding. Of particular interest, we determine that a natural variant of Asp25 (Asp25Asn) is still functional but has a reduced affinity for ethylene, pointing to the key role Asp25 plays in modulating high-affinity ethylene binding by the receptors. Additionally, we identify a highly conserved lysine residue (Lys91) that we propose forms a polar bridge to Asp25 to internally transduce the ethylene signal within the receptor to mediate changes in signaling output. Taking advantage of evolutionary and computational modeling approaches, combined with experimental verification, we propose a new structural model for the mechanism of ethylene binding and signal transduction, one with similarities to that found in a mammalian olfactory receptor.
Results
A Highly Conserved Asp Residue in the EBD Modulates Copper and Ethylene Binding by ETR1.
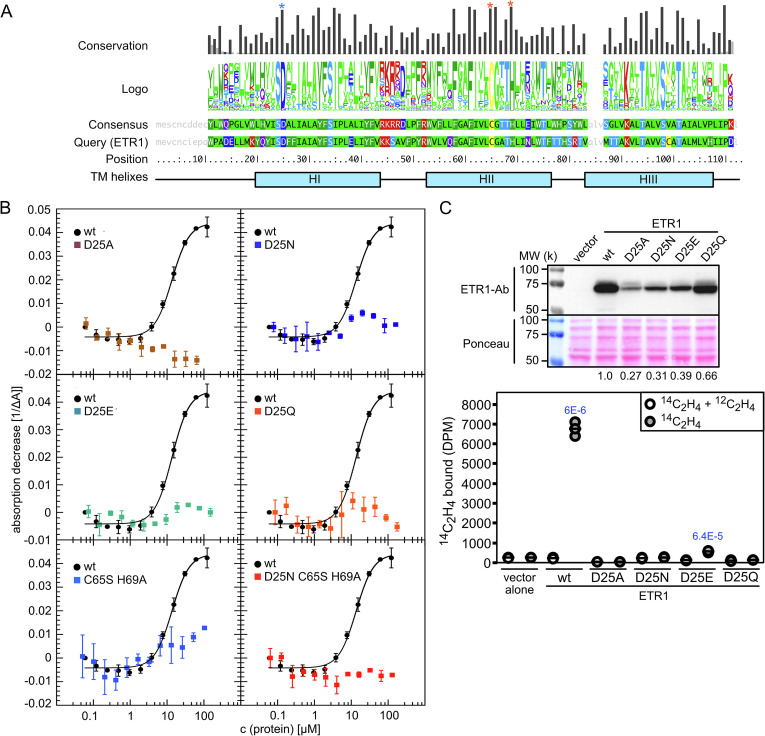
The EBD of the receptor ETR1 of Arabidopsis is contained within the N-terminal TM domain of the protein (7, 22). This TM domain contains three predicted TM helixes, with the Cys65 and a His69 residues of TM helix II directly implicated in coordinating the copper cofactor required for ethylene binding (7, 22). Similar EBDs have been identified in a wide variety of organisms, including prokaryotes, the ethylene-binding capability of receptors from Arabidopsis, tomato, and the cyanobacterium Synechocystis sp PCC 6803 having all been confirmed (20, 22, 29, 30). Fig. 1A indicates the degree of amino acid conservation found in a comparison of EBDs from 1,221 eukaryotic and prokaryotic sequences related to ETR1. As predicted, the Cys and His residues of TMII implicated in coordinating the copper cofactor of ETR1 are highly conserved in EBDs.
Fig. 1.
Copper and ethylene binding of wildtype and Asp25 mutant versions of ETR1. (A) Amino acid conservation of the EBD in ETR1-like proteins. The three conserved TM (TM) helixes (H1, HII, and HIII) are indicated. Red asterisks indicate Cys65 and His69 of ETR1 HII implicated in coordinating the copper cofactor; the blue asterisk indicates the highly conserved Asp25 of HI. (B) Copper binding of wildtype and Asp25 mutant versions of ETR1 TM domain (ETR1-TMD; n = 3). Purified ETR1-TMD was titrated to the BCA2-Cu(I) complex, and copper binding monitored spectrophotometrically based on the change in absorbance at 562 nm. For comparison copper binding of a Cys65Ser His69Ala mutation was examined alone and in combination with the Asp25Asn mutation. For site-directed mutations, the single letter abbreviations for the amino acids Asp (D), Ala (A), Asn (N), Cys (C), Glu (E), Gln (Q), His (H), and Ser (S) are used (SI Appendix, Fig. S1). (C) Ethylene binding to yeast transgenically expressing wildtype and Asp25 mutant versions of ETR1. ETR1 protein levels were determined by immunoblot analysis with an anti-ETR1 antibody (quantification is relative to ETR1-wt), and the proteins on the blot staining with Ponceau-S as a loading control. To analyze ethylene-binding activity, transgenic yeast samples (n = 3; horizontal line = mean) were incubated with 0.21 µL L−1 [14C]ethylene, in the presence or absence of excess [12C]ethylene, the difference between the two values representing the saturable binding; P values for significant saturable binding, as determined by t test are given for P < 0.05.
Of particular interest are the additional conserved polar and charged residues found in the TM helixes, such as Asp25 (D25) of helix I in ETR1 (Fig. 1A), because such residues are likely to play significant roles in ethylene binding and/or signal transduction. Mutation of Asp25 to Ala (D25A) abolishes ethylene binding by the receptor when analyzed in a heterologous yeast expression system and also confers dominant ethylene insensitivity when expressed in Arabidopsis (20). Computational modeling places Asp25 of helix I in proximity to Cys65 and His69 of helix II (24, 27), suggesting that it could play a role in coordinating the copper cofactor. Interestingly, although Asp is found in 93.55% of the sequences examined, in some cases (3.67%) it is substituted by an Asn residue, most commonly in cyanobacteria but also in several plants, Pyrus communis (Pear) and Cajanus cajan (Pigeon pea).
To characterize the role of Asp25 in copper and ethylene binding, we generated four site-directed mutant versions of ETR1. ETR1D25A was previously found to eliminate ethylene binding (20). ETR1D25N represents a relatively conserved change of the Asp R-group from a carboxylic acid to a carboxamide, one that will preserve the general size of the side group, but which eliminates the negative charge. As noted above, although Asp25 is highly conserved in EBDs, an Asn residue is found at that position in a few EBDs (Fig. 1A). ETR1D25E preserves the carboxylic acid and its negative charge found in the Asp R-group, but Glu has a longer sidechain than does Asp. The ETR1D25Q mutation is analogous to that for ETR1D25N, exchanging a carboxyamide for a carboxylic acid on the R-group of Glu, and so eliminating the negative charge of the Glu.
We tested the effects of Asp25 mutants on copper binding to the ETR1 TM domain following expression and purification from E. coli (24). Purified receptors lack the copper cofactor, allowing for their reconstitution with copper using this in vitro assay. Cu(I) was stabilized by the copper chelator bicinchoninic acid (BCA), and then titrated with increasing ETR1 protein concentration. As shown in Fig. 1B, titration with ETR1wt results in copper binding and a concomitant decrease in absorbance at 562 nm of the purple BCA2-Cu(I) complex. In contrast, no copper binding was observed for ETR1D25A and only minimal residual binding observed for ETR1D25N, ETR1D25E, and ETR1D25Q (Fig. 1B). These data thus support a model in which Asp25 contributes to copper binding. Furthermore, we found that the Asp25 mutants had an additive effect on copper binding when combined with an ETR1C65S;H69A mutant, because the ETR1C65S;H69A mutant exhibited residual binding that was eliminated when combined with the Asp25 mutations (ETR1D25X;C65S;H69A) (Fig. 1B and SI Appendix, Fig. S1).
Saturable ethylene binding of the ETR1 Asp25 mutants was examined by heterologous expression in yeast, with binding to [14C]ethylene determined in the presence or absence of excess [12C]ethylene (Fig. 1C) (5, 31). Expression in yeast results in the production of functional ethylene receptors as membrane-associated disulfide-linked homodimers containing the copper cofactor, facilitating an in vivo analysis of ethylene binding due to the cell permeability of ethylene (5, 22). Saturable binding of [14C]ethylene was observed for ETR1wt, as the positive control, and not with the pYCDE2 vector negative control (Fig. 1C). Binding of ethylene by ETR1D25E was still observed but was substantially reduced compared to that observed with ETR1wt, likely arising due to steric problems from the larger size of the Glu R-group as well as differences in protein expression levels (Fig. 1C). No saturable [14C]ethylene binding was detected for the other D25 mutants. This included the ETR1D25A mutant, consistent with previous observations (20), as well as for the ETR1D25N and ETR1D25Q mutants (Fig. 1C). Results from the ethylene-binding assay are therefore consistent with the negative charge at D25 playing a significant role in mediating high-affinity ethylene binding.
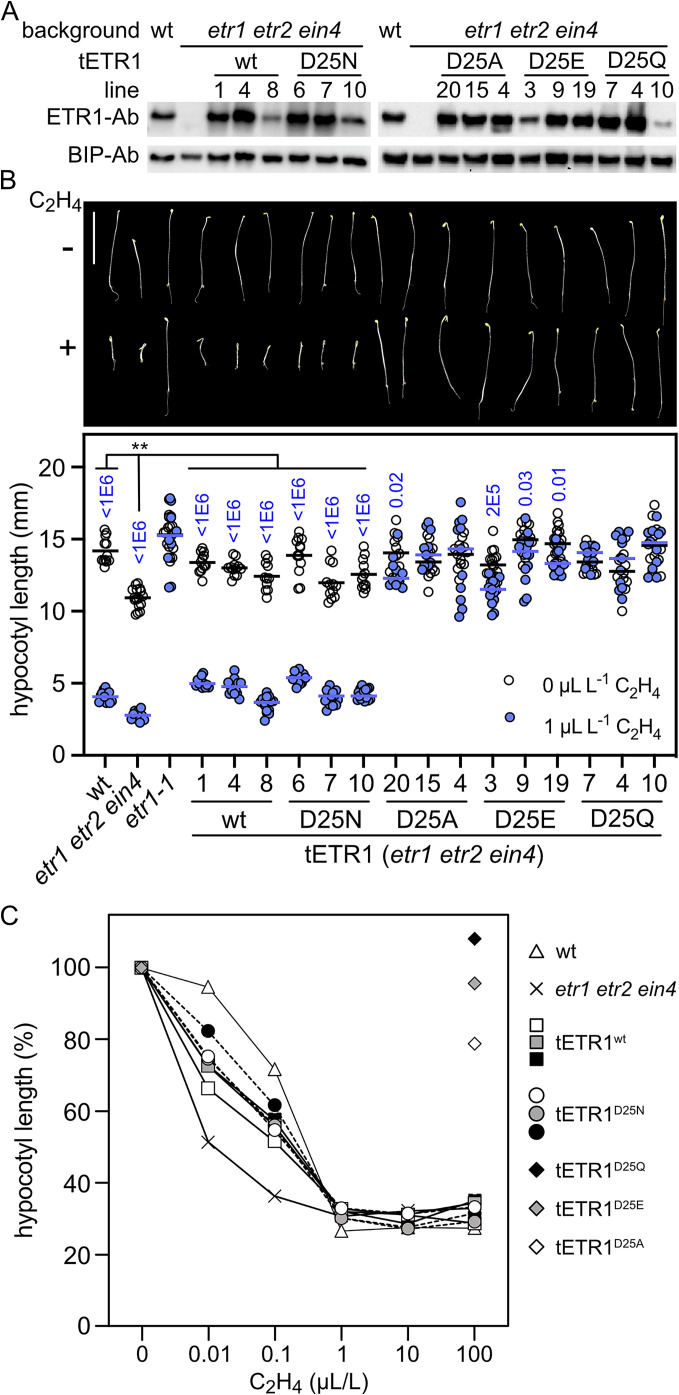
Functional Analysis of Asp25 Mutations on the ETR1 Responses In Planta.
Functionality of the ETR1 Asp25 mutants was tested by transgenic expression in the etr1 etr2 ein4 Arabidopsis background. The rationale for this approach is that the etr1 etr2 ein4 triple mutant exhibits a partial constitutive ethylene-response phenotype, resulting in reduced shoot growth as well as a shorter hypocotyl in the air than is found in the wildtype (Fig. 2 A and B and SI Appendix, Fig. S2) (18). We can thus exploit the triple mutant to assess the ability of the ETR1 transgenes to rescue growth in the absence of ethylene as well as their ability to mediate a response to ethylene (32). Such an analysis indicates whether the encoded receptors can assume the “on” conformation that represses the ethylene response in air, as well as the “off” conformation that occurs in response to ethylene binding. All transgenes were expressed based on immunological detection of the tETR1 protein, and all rescued growth of the etr1 etr2 ein4 triple mutant in air based on hypocotyl and adult shoot growth analysis, indicating that all the tETR1 proteins (tETR1wt, tETR1D25N, tETR1D25Q, tETR1D25E, tETR1D25A) can assume the “on” conformation (Fig. 2 A and B and SI Appendix, Fig. S2).
Fig. 2.
Ethylene sensitivity of Arabidopsis seedlings expressing Asp25 mutants of ETR1. Wildtype (wt) and mutant versions of ETR1 were transgenically (tETR1) expressed in the etr1 etr2 ein4 background. (A) ETR1 protein levels as determined by immunoblot analysis with an anti-ETR1 antibody in dark-grown seedlings. BIP serves a loading control. (B) Triple-response seedling growth assay to ethylene. Dark-grown seedlings were treated with 0 or 1 µL L−1 ethylene. Images of seedlings are shown along with quantification of the hypocotyl growth response (n ≥ 10; horizontal line = mean). The mutant etr1-1 serves as an ethylene-insensitive control. P values (blue) for significant differences in the ethylene responsiveness of seedlings, as determined by t test, are given for P < 0.05. For data comparison of etr1 etr2 ein4 to wt, tETR1(wt), and tETR1(D25N) lines grown in the absence of ethylene, ANOVA was performed with post hoc Holm multiple comparison calculation (**P < 0.01). (C) Ethylene dose response curves of hypocotyl growth in dark grown seedlings for the three lines of ETR1wt and ETR1D25N compared to wt and etr1 etr2 ein4. Representative ethylene-insensitive tETR1D25A-#20, tETR1D25E-#19, and tETR1D25Q-#4 lines are also included and examined at 0 and 100 µL L−1 ethylene. The ethylene response is normalized for each line relative to its hypocotyl length at 0 µL L−1 ethylene (n ≥ 17; SE < 3% of hypocotyl length, not shown for clarity) (SI Appendix, Figs. S3 and S4).
Previous studies have found that the site-directed mutation of residues that result in a loss of ethylene-binding activity typically confer dominant ethylene insensitivity on the seedlings, due to an inability of the etr1 mutant protein to switch from its “on” to its “off” conformation (7, 20, 22). Based on this we anticipated that those Asp25 mutants that resulted in a loss of high-affinity ethylene binding (tETR1D25N, tETR1D25Q, and tETR1D25A) would confer dominant ethylene insensitivity. Since the tETR1D25E protein still exhibited reduced ethylene binding, it was possible that it would respond similarly to wildtype, but it was also possible that tETR1D25E would confer dominant insensitivity due to the reduced ability to bind and/or the perturbation of the ethylene binding site. As shown in Fig. 2 A and B, the ETR1D25A, ETR1D25E, and ETR1D25Q transgenes all conferred ethylene insensitivity based on a hypocotyl growth response analysis, consistent with predictions (i.e., less than a 20% decrease in hypocotyl growth in response to ethylene). Surprisingly, the ETR1D25N transgene rescued the hypocotyl growth response to ethylene to the same degree as ETR1wt even though, based on our previous analyses, ETR1D25N was compromised in its copper and ethylene-binding ability. Hypocotyl ethylene dose-response analyses confirmed a similar ethylene responsiveness for both the ETR1wt and ETR1D25N lines, their responsiveness being similar to that found with an etr2 ein4 double mutant (Fig. 2C and SI Appendix, Figs. S3 and S4).
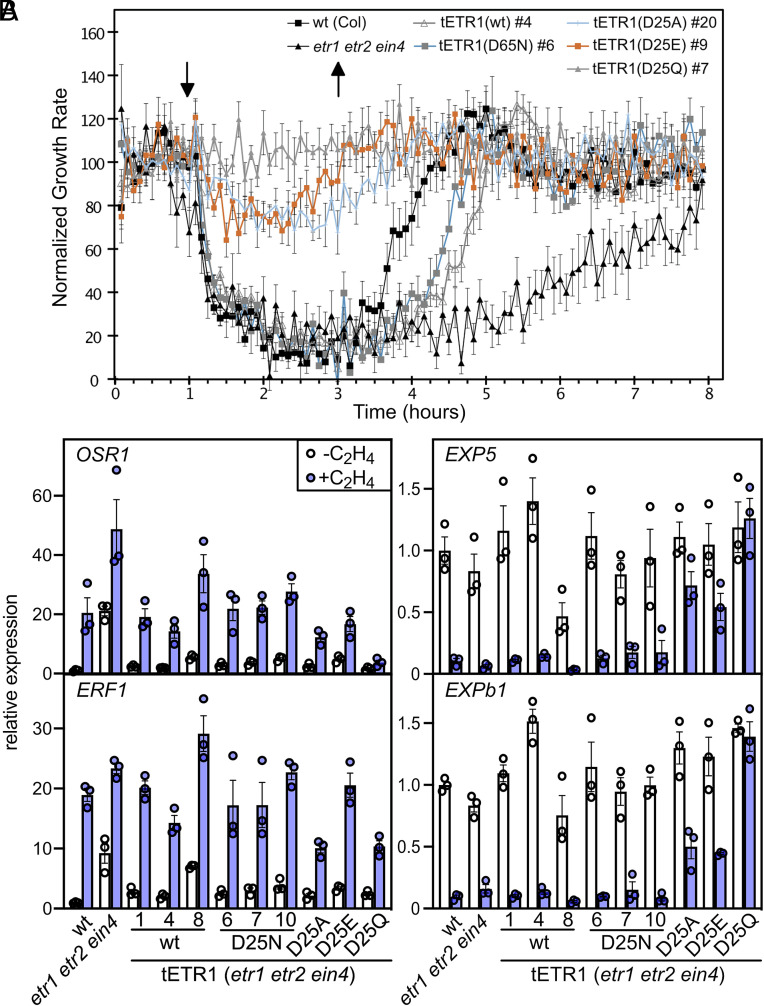
Based on the differences, we uncovered for the long-term ethylene growth responses for the Asp25 mutants, we also analyzed their short-term hypocotyl growth and molecular responses to ethylene. To this end, we performed a short-term kinetic analysis (28, 33), analyzing the initial hypocotyl growth inhibition in response to 10 µL/L ethylene and the growth recovery following the removal of ethylene after 2 h of treatment (Fig. 3A). As shown in Fig. 3A, both wildtype and the triple mutant etr1 etr2 ein4 exhibit a rapid inhibition of hypocotyl growth in response to ethylene, but the growth recovery for the etr1 etr2 ein4 mutant following removal of ethylene is substantially slower than that observed for the wildtype (33). The ethylene-insensitive mutants ETR1D25A, ETR1D25E, and ETR1D25Q all exhibit ethylene insensitivity based on the short-term kinetic analysis (Fig. 3A). In contrast, both the ETR1wt and ETR1D25N lines exhibit a rapid growth response to ethylene and, upon ethylene removal, a similar growth recovery intermediate between that exhibited by the wildtype and the etr1 etr2 ein4 seedlings (Fig. 3A and SI Appendix, Fig. S5). To examine the short-term molecular response, we characterized ethylene-dependent gene expression for the ETR1 Asp25 mutants (Fig. 3B and SI Appendix, Fig. S6). The expression of OSR1 and ERF1 is induced, whereas the expression of EXP5 and EXPb1 is repressed, in response to ethylene (Fig. 3B). The molecular response to ethylene is similar in the ETR1wt and ETR1D25N transgenic lines for all four genes, and consistent with what is observed in the wildtype control. In contrast, the ethylene-insensitive mutants ETR1D25A, ETR1D25E, and ETR1D25Q all exhibit various levels of hyposensitivity for ethylene-dependent gene expression, this being most apparent in the analysis of EXP5 and EXPb1 expression. Similar effects were found on the expression of additional ethylene-regulated genes, including induction of ARGOS, ERS1, and ERS2 and repression of CAPE2 (SI Appendix, Fig. S6).
Fig. 3.
Short-term ethylene responses of Arabidopsis seedlings expressing Asp25 mutants of ETR1. (A) Kinetics of growth response to ethylene of ETR1wt and ETR1D25 mutant lines. Ethylene dose-response kinetics were analyzed in hypocotyls of 2-d–old etiolated seedlings for wildtype, the etr1 etr2 ein4 triple mutant, and for the triple mutant complemented with ETR1wt or various ETR1D25 mutants as indicated. Measurements were made in air for 1 h, followed by a 2-h exposure to 10 µL L−1 ethylene, and then a 5-h recovery in air. Growth rates for each line are normalized to the growth rate during the first hour in the air. Arrows indicate the time points for the addition and removal of ethylene. Error bars represent SE (n ≥ 7) (SI Appendix, Fig. S5). (B) Effect of ETR1 Asp25 mutants on ethylene-dependent gene expression. Dark-grown seedlings were treated with 0 or 1 µL L−1 ethylene for 2 h, and gene expression examined by qRT-PCR (n = 3). Expression was normalized to a tubulin control and is presented as relative to the untreated wildtype control. Three tETR1-wt and three tETR1D25N lines were examined. The ethylene-insensitive tETR1D25A#20, tETR1D25E#19, and tETR1D25Q#4 lines were also included. See SI Appendix, Fig. S6 for expression of additional genes.
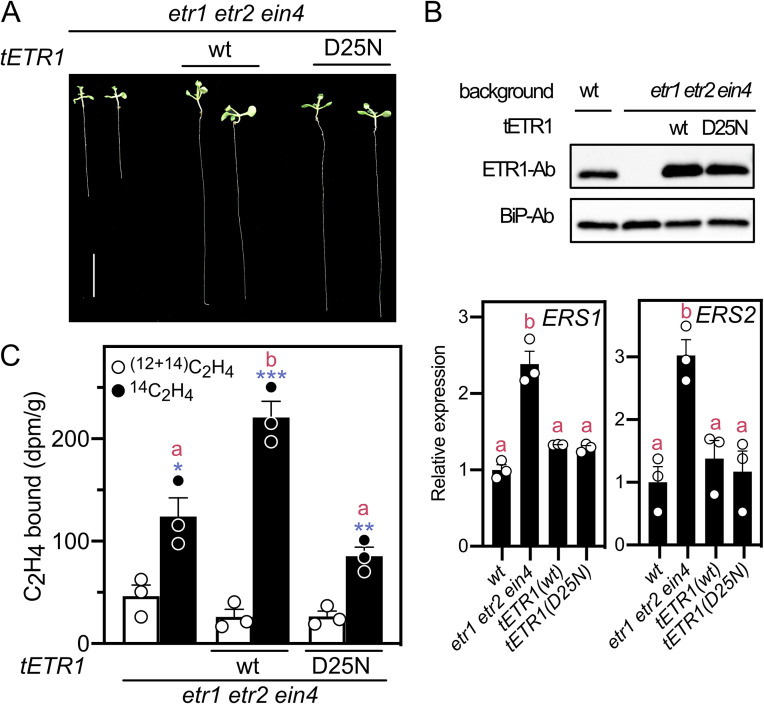
To confirm that the results obtained in the heterologous yeast expression system for ETR1D25N reflected the condition in planta, we compared ethylene binding of ETR1wt and ETRD25N in the etr1 etr2 ein4 Arabidopsis background, making use of lines that exhibited a similar seedling phenotype, ETR1 protein levels, and expression levels of the remaining receptors ERS1 and ERS2 (Fig. 4 A and B). [14C]ethylene binding was examined in 2-week–old green seedlings grown on media containing 5 µM aminoethoxyvinylglycine (AVG) to inhibit ethylene biosynthesis. As shown in Fig. 4C, the triple mutant etr1 etr2 ein4 exhibits a basal level of ethylene binding due to the presence of the receptors ERS1 and ERS2; however, transgenic expression of ETR1wt results in a significant increase in ethylene binding. In contrast, no increase in ethylene binding was observed following transgenic expression of ETRD25N.
Fig. 4.
Ethylene binding of Arabidopsis seedlings expressing ETR1wt and ETR1D25N. (A) Representative 2-wk–old green seedlings of the ETR1wt-#4 and ETR1D25N-#6 lines expressed in etr1 etr2 ein4 background, as well as the etr1 etr2 ein4 triple mutant itself (Scale bar, 1 cm). (B) Expression levels of the tETR1 receptor versions as determined by immunoblot, and of ERS1 and ERS2 as determined by qRT-PCR. Significant differences in gene expression between lines were determined by ANOVA with post-hoc Holm multiple comparison calculation; different red letters indicate a significant difference at P < 0.05. (C) Saturable ethylene binding (n = 3; error bar = SE) of the seedlings. Seedlings were incubated with 0.31 µL L−1 [14C]ethylene, in the presence or absence of excess [12C]ethylene, the difference between the two values representing the saturable binding. Significant saturable binding as determined by t test for each line is indicated (blue; *P < 0.05, **P < 0.01, ***P < 0.001). Significant differences in expression between lines are ANOVA-based analyses (red; P < 0.01).
Mechanism by Which ETR1D25N Mediates Ethylene Signaling In Planta.
We considered two hypotheses, not mutually exclusive, as to how ETR1D25N could mediate ethylene signaling in planta: residual ethylene binding of the ETR1D25N mutant and/or cooperative interactions with other wildtype ethylene receptors in Arabidopsis. The first hypothesis is based on the affinity of the receptors for ethylene. ETR1 has a half-life for ethylene dissociation of over 12 h (5), allowing for the ready detection of [14C]ethylene binding in yeast or in planta; however, if the ETR1D25N mutant still retained the copper cofactor and bound ethylene with lower affinity (less tightly), then it would be substantially more difficult to detect with the [14C]ethylene binding assays. The second hypothesis is based on the existence of ethylene receptor families in Arabidopsis. Even in the etr1 etr2 ein4 background, used for expression of the ETR1D25N mutant, the ERS1 and ERS2 wildtype receptors are still present. These could bind ethylene and, as part of a receptor complex, potentially pass on a conformational information to ETR1D25N, causing it to adopt a signaling conformation even when it has not bound ethylene. This type of signaling interaction has been found for bacterial chemoreceptors, and has also been proposed to occur for the ethylene receptors which, like chemoreceptors, form higher-order receptor complexes (12, 13, 28, 34).
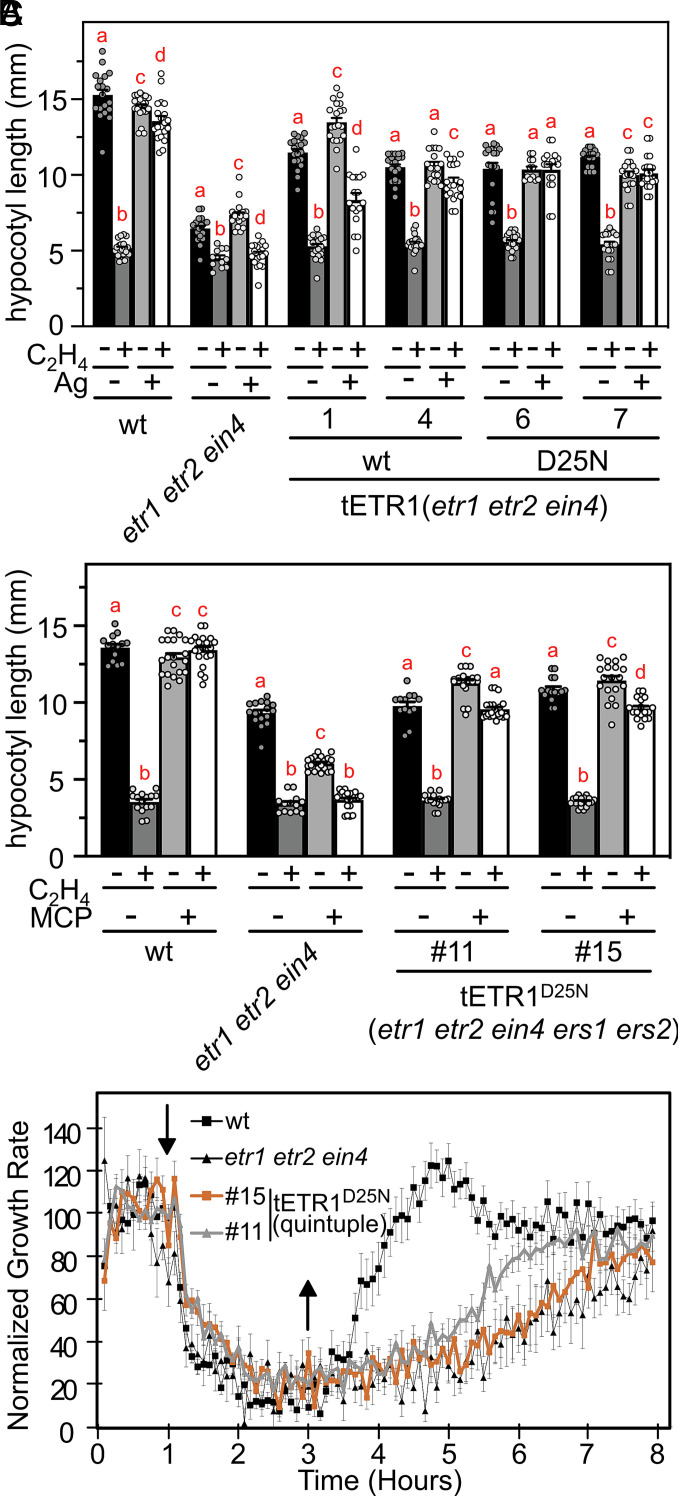
As an initial test for the feasibility of the first hypothesis, we asked if ETR1D25N could still bind a metal cofactor, and therefore potentially ethylene, in planta. For this purpose, we examined the effects of silver (Ag) on ethylene sensitivity. Silver is thought to substitute for the copper cofactor and, although receptors containing silver can still bind ethylene, they no longer transmit the signal, resulting in ethylene insensitivity (7, 23, 35, 36). As shown in Fig. 5A, the dark-grown seedling response of wildtype to 10 µL/L ethylene is blocked when seedlings are grown on silver, but the etr1 etr2 ein4 triple mutant is still responsive to ethylene in the presence of silver. We find that expression of either tETR1wt or tETR1D25N in the etr1 etr2 ein4 background restores the ability of silver to block the ethylene response, consistent with ETR1D25N retaining an ability to bind its metal cofactor in vivo, even though it showed reduced affinity in the in vitro assay (Fig. 1B).
Fig. 5.
Responsiveness of the ETR1D25N mutant to ethylene, silver, and 1-MCP. (A) The ethylene responsiveness of ETR1wt and ETR1D25N, transgenically expressed in the etr1 etr2 ein4 background, is blocked in the presence of 100 µM silver (Ag). The hypocotyl growth of dark-grown seedlings was analyzed in the absence or presence of 10 µM ethylene (n ≥ 11; error bars = SE). (B) ETR1D25N responds to 1 µL L−1 ethylene in a background lacking all five native ethylene receptors (etr1 etr2 ein4 ers1 ers2), and has this response is blocked by 1 µL L−1 1-MCP (n ≥ 13; error bars = SE). Loss-of-function mutations in the ERS1 and ERS2 genes were generated by a CRISPR-Cas9 approach in the ETR1D25N (etr1 etr2 ein4) line #6. Significant differences in hypocotyl length following treatment were determined for each line by ANOVA with post hoc Holm multiple comparison calculation; different red letters indicate a significant difference for each line at P < 0.05. (C) Kinetics of growth response to ethylene of wildtype, etr1 etr2 ein4, and two lines of ETR1D25N in the etr1 etr2 ein4 ers1 ers2 (quintuple) background. Error bars represent SE (n ≥ 8).
As an alternative approach to test the two hypotheses, we used CRISPR-cas9 methodology to knock out ERS1 and ERS2 in the ETR1D25N (etr1 etr2 ein4) line to generate an ETR1D25N (etr1 etr2 ein4 ers1 ers2) line (SI Appendix, Fig. S7), the prediction being that signaling by ETR1D25N will be lost if dependent on the presence of other wildtype receptors. Two independent ETR1D25N (etr1 etr2 ein4 ers1 ers2) lines were generated (#11 and #15). However, contrary to the second hypothesis, the ETR1D25N lines still responded to ethylene (Fig. 5B), consistent with an ability for ETR1D25N to bind ethylene, in agreement with residual copper binding (Fig. 1B) and the silver sensitivity assay (Fig. 5A). Furthermore, the ethylene response of the ETR1D25N (etr1 etr2 ein4 ers1 ers2) lines was blocked by the competitive inhibitor 1-methylcyclopropene (1-MCP) (Fig. 5B) (36), also consistent with ETR1D25N retaining its c opper cofactor and ability to bind its gaseous ligand. Kinetic analysis supports the ability of the ETR1D25N (etr1 etr2 ein4 ers1 ers2) lines to rapidly mediate the ethylene response (Fig. 5C). These data thus support a key role for Asp25 of ETR1 in regulating the kinetics of ethylene binding by the receptor.
Effect of Lys91 Mutations of ETR1 on Seedling Growth and Ethylene Sensitivity.
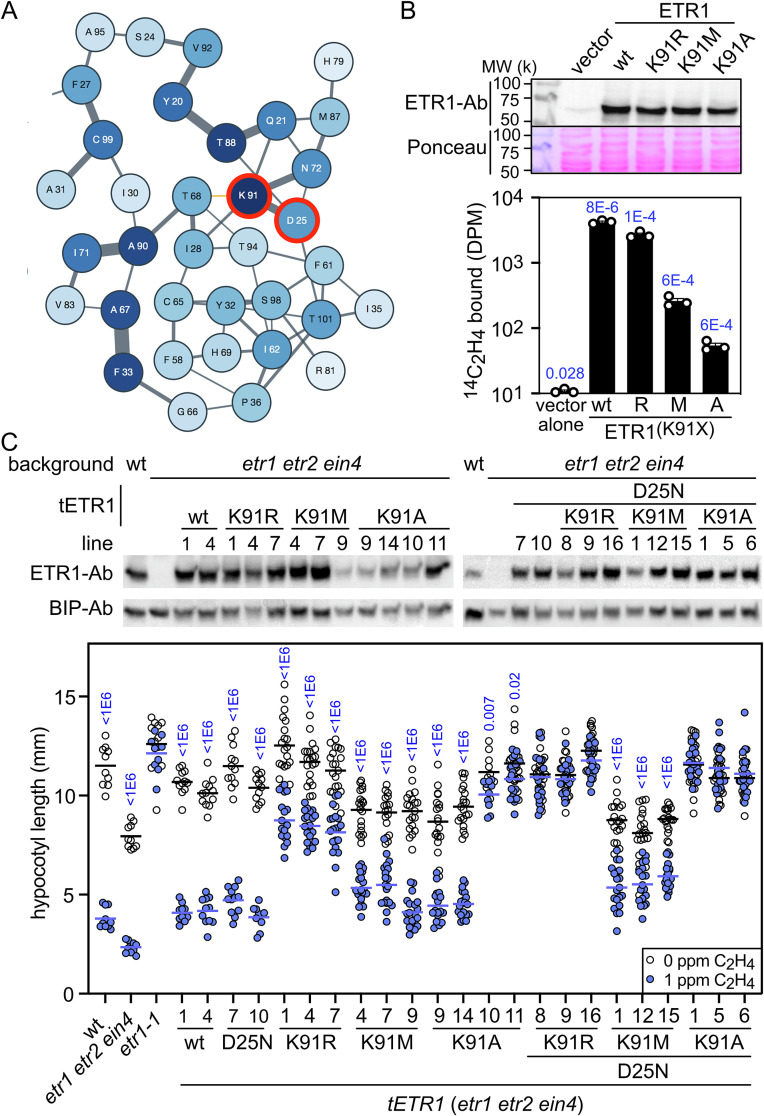
Coevolutionary analysis can be used to predict interactions/contacting residues in proteins (37, 38). For example, the EVCOUPLINGs algorithm derives residue–residue evolutionary couplings from deep multiple sequence alignment by a pseudolikelihood maximization method. Therefore, to gain further information about the structure of the EBD, we used the 1 to 112 amino-acid sequence of ETR1 as an input for both the EVCOUPLINGs and GREMLIN servers (37, 38). Sequence coevolution analysis predicts a coupling between Asp25 of TM helix I with Lys91 of TM helix III (GREMLIN probability of 0.960; EVcouplings probability of 0.997; Fig. 6A and SI Appendix, Table S1) (37, 38). This predicted Asp25-Lys91 coupling is of interest because, in contrast to the first and second TM helixes of ETR1 that play substantial roles in copper and ethylene binding, the third TM helix is implicated in signal transduction (20). Like Asp25, Lys91 is highly conserved (Fig. 1A) but Lys is not among the amino acids favored to coordinate copper ions (39, 40). A role for Asp25 in both copper binding and in forming a polar bond to TM helix III would provide a potential mechanism by which to couple ethylene binding to receptor signal output.
Fig. 6.
Characterization of Lys91 mutants of ETR1. (A) Evolutionary couplings of ETR1 amino acid residues from the EBD, based on the analysis of 1,221 ETR1-related sequences using the EVcouplings framework (37). The thickness of the lines represents the strength of individual couplings. The intensity of the blue shading represents the overall strength of couplings for that specific residue. Lys91 and Asp25 are highlighted in red. (B) Ethylene binding to yeast transgenically expressing wildtype and Lys91 mutant versions of ETR1. Immunoblot analysis was used to determine the ETR1 protein levels with an anti-ETR1 antibody; the proteins on the blot were stained with Ponceau-S as a loading control. To determine saturable ethylene-binding activity of the ETR1 Lys91 mutants, transgenic yeast samples (n = 3) were incubated with 0.21 µL L−1 [14C]ethylene, in the presence or absence of excess [12C]ethylene, the difference between the two values representing the saturable binding; P values for significant saturable binding, as determined by t test are given for P < 0.05. (C) Characterization of Arabidopsis seedlings expressing Lys91 mutants of ETR1. Lys91 mutant versions of ETR1 alone or with Asp25Asn (D25N) were transgenically (tETR1) expressed in the etr1 etr2 ein4 background. Immunoblot analysis was used to determine ETR1 protein levels with an anti-ETR1 antibody in dark-grown seedlings; BIP serves a loading control. For the hypocotyl growth response of dark-grown seedlings to ethylene, seedlings were treated with 0 or 1 µL L−1 ethylene (n ≥ 10; horizontal line = mean). P values (blue) for significant differences in the ethylene responsiveness of seedlings, as determined by t test, are given for P < 0.05.
To examine the role of Lys91 in signaling by ETR1, we made the site-directed mutations ETR1K91R, ETR1K91M, and ETR1K91A, and examined their copper-binding ability using the in vitro assay (SI Appendix, Fig. S8), their ethylene-binding ability using the yeast expression system (Fig. 6B) and their functionality by transgenic expression in the etr1 etr2 ein4 Arabidopsis background (Fig. 6C). All three Lys91 mutants of ETR1 bound copper similarly to the wildtype control (SI Appendix, Fig. S8), consistent with Lys91 not playing a direct role in copper binding. In contrast, as described below, differing effects of the Lys91 ETR1 mutants were found on ethylene binding and functionality in planta.
In ETR1K91R, the basic Lys91 residue is replaced with another basic residue (Arg) which should preserve the ability to form a polar bond to Asp25. The ETR1K91R mutant exhibited an ethylene binding ability similar to that of ETR1wt (63% of wildtype binding; Fig. 6B) but like some other previously characterized site-directed mutations in TM segment three (20), conferred ethylene hyposensitivity (reduced ethylene sensitivity rather than insensitivity) in Arabidopsis (Fig. 6C), potentially due to the larger Arg sidechain perturbing the receptor structure so that it does not effectively turn “off ” upon ethylene binding.
In ETR1K91M, the basic sidechain of Lys91 is replaced with a nonpolar sidechain of similar size; this mutation would no longer be able to participate in a polar bridge to mediate an ethylene-dependent change in conformation. ETR1K91M exhibited substantially reduced binding of approximately 6% of that found in ETR1wt (Fig. 6B). Interestingly, the ETR1K91M mutant exhibited a seedling growth response not previously noted for ETR1 mutants (20). The ETR1K91M mutant failed to rescue seedling growth efficiently but also exhibited partial ethylene hyposensitivity (Fig. 6C), suggesting that the receptor is less capable of maintaining the “on” and “off” conformations typically found in the absence and presence of ethylene, respectively, consistent with a lack of “communication” between the ethylene binding site involving TM helixes I and II, and the output domain of TM helix III.
In ETR1K91A, as with ETR1K91M, the basic sidechain of Lys is replaced with a nonpolar sidechain but one that is smaller than that found with Lys; the ETR1K91A mutation was previously found to confer ethylene insensitivity on Arabidopsis seedlings but still retained a low level of ethylene binding ability (20). Like ETR1K91M, we found that ETR1K91A retained a minimal ability to bind ethylene (only 1% of that found in ETR1wt; Fig. 6B). Thus, preservation of the basic nature of the Lys91 sidechain is important to ethylene binding, although the finding that substitution with a nonpolar sidechain does not eliminate ethylene binding is consistent with Lys91 not playing a direct role in chelating the copper cofactor. ETR1K91A conferred two different ethylene response phenotypes in the seedling lines we examined (Fig. 6C): a phenotype similar to that of the ETR1K91M mutant (lines 9 and 14) or ethylene insensitivity (lines 10 and 11) such as previously reported for the mutation (20). The ETR1K91A lines that exhibited ethylene insensitivity generally also had higher receptor protein levels (Fig. 6C), suggesting that insensitivity could arise due to increases in the number of misfolded receptors, these receptors potentially unable to bind ethylene or unable to change conformation upon ethylene binding.
To gain further information on the interaction of Asp25 and Lys91, we combined the Asp25Asn mutation with the ETR1K91X mutations to generate ETR1D25N, K91R, ETR1D25N, K91M, and ETR1D25N, K91A, and examined their functionality in the etr1 etr2 ein4 Arabidopsis background (Fig. 6C). The ETR1D25N, K91X combinations tended to accentuate the mutant growth phenotypes noted for the ETR1K91X single mutants. The ETR1D25N, K91R mutant is ethylene insensitive rather than hyposensitive as found in the ETR1K91R mutant; the ETR1D25N, K91M mutant rescued seedling growth even less efficiently than the ETR1K91M mutant; and the ETR1D25N, K91A mutant lines are all ethylene insensitive rather than just a subset as found with the ETR1K91A mutant lines. The heightening of the ETR1K91X phenotypes when combined with Asp25Asn indicates that the Asp25Asn mutation does affect signaling by ETR1, even though the individual ETR1D25N mutant could not be distinguished from the ETR1wt by standard growth response assays (Figs. 2 and 3).
Discussion
A key but unresolved structural question for the ethylene receptors is how the copper cofactor(s) required for ethylene binding are coordinated within the TM domain. The Cu(I) oxidation state is known to exist in a variety of coordination geometries, with coordination numbers anywhere from two to six (41, 42). Based on initial evidence for a single Cu(I) in the ethylene-binding site, a tetrahedral geometry for the copper binding site was proposed involving Cys65 (as the thiolate form) and His69 of ETR1: (Cys65)2(His69)2, hereafter referred to as a CCHH coordination model (7, 22). The tetrahedral geometry of the proposed CCHH copper-binding site was considered consistent with the homodimeric nature of ethylene receptors and the fact that Cys65 and His69 are on the same face of the second TM helix, one helical turn apart.
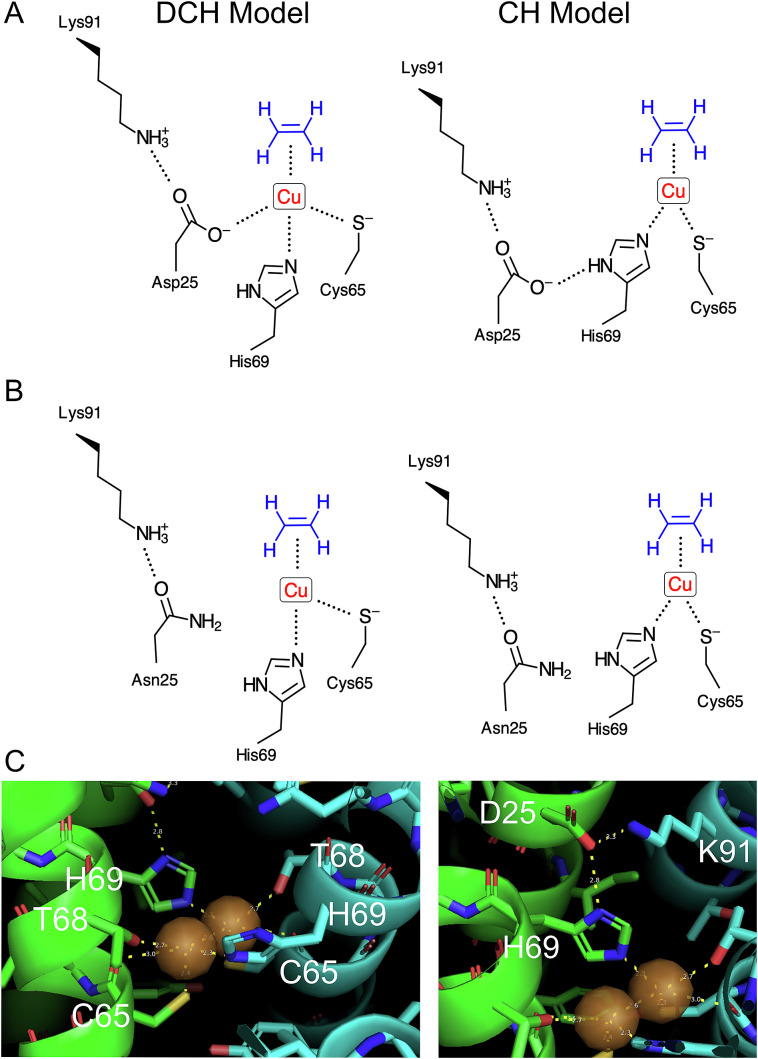
Recent data point to the existence of two coppers per receptor dimer (i.e. one copper per receptor monomer) (24), a possibility not inconsistent with data from the earlier study in which it was unclear if all the purified receptors were competent for copper binding (7), a finding that necessitates the consideration of new coordination structures for the copper cofactors. Here we implicate Asp25 of ETR1 as playing a critical role in copper binding based on the same principles that implicate Cys65 and His69. Based on our current understanding that there is one copper per monomer, we consider two potential models by which Asp25 could contribute to ethylene binding by ETR1 (Fig. 7 A and B). First, Asp25 could directly participate in copper binding along with Cys65 and His69: a tridentate Asp-Cys-His (DCH) model for copper coordination. Alternatively, Asp25 could form a hydrogen bond to His69 to orient and polarize it for copper binding: a bidentate Cys-His (CH) model for copper coordination.
Fig. 7.
Models for ETR1 interactions with copper(I) and ethylene. (A) Asp-Cys-His (DCH) and Cys-His (CH) models for how Asp25 contributes to copper(I) and ethylene binding and signal transmission via bonding with Lys91 in DCH and CH models. (B) Predicted effect of the Asn25 variant on molecular interactions at the ethylene-binding site. (C) AlphaFold-Multimer-based model of the ETR1 dimer (interacting coppers), with views highlighting copper-binding geometry (Left; H69-Cu = 2.3 Å, C65-Cu = 2.4 Å, T68-Cu = 2.7 Å, C65 backbone carbonyl-Cu = 3.0 Å) and interactions between His69, Asp25, and Lys91 (Right; D25-H69 = 2.8 Å, D25-K91 = 3.3 Å). (SI Appendix, Fig. S9 and Movies S1 and S2).
In addition to results from the recent in vitro study in which one copper per ETR1 monomer was detected (24), the DCH and CH models are favored over the earlier CCHH model for the following reasons. First, chemical analysis has demonstrated that both tridentate and bidentate ancillary ligands can effectively bind Cu(I) and form a Cu(I)–ethylene complex, the primary consideration being that anionic, electron-donating ancillary ligands foster the strongest backbonding of the filled Cu(I) 3d orbital to the unfilled ethylene π* orbital (43, 44). Second, lower coordination numbers favor Cu(I) binding over that of Cu(II) and other divalent metal ions (45), supporting the existence of two or three copper-coordinating ligands, rather than four, with the ethylene receptors. Third, modeling of Cu(I) interactions with 1-methycyclopropene (1-MCP), a potent competitive inhibitor for ethylene binding, support Cu(I) being coordinated by no more than three ligands in addition to 1-MCP (46). We note that, even should subsequent studies provide support for the CCHH model with one Cu(I) per receptor dimer, the role(s) for Asp25 identified here in copper binding are still relevant.
Structural modeling supports the CH model over the DCH model, with Asp25 playing an indirect rather than a direct role in chelation of Cu(I). Support for the CH model is found with the previous ab initio structural model of the EBD (24) as well as our modeling the ETR1 homodimer with AlphaFold-Multimer (Fig. 7C and SI Appendix, Fig. S9, and Movies S1 and S2) (47, 48). AlphaFold-Multimer builds on the neural network-based AlphaFold to generate structural models of protein complexes, taking advantage of related amino-acid sequences and experimentally determined protein structures (47, 48). Using AlphaFold-Multimer, we generated structural models for the ETR1 homodimer for full-length ETR1 as well as for the EBD (Fig. 7C and SI Appendix, Fig. S9). The copper cofactors were modeled under two potential coordinations involving Cys65 and His69 of the ETR1 homodimer, one in which the two coppers are bound independently and do not share an interaction with each other, and another where they are closely bonded. Both the ab initio and AlphaFold models place Asp25 of helix I in proximity to Cys65 and His69 of helix II but not optimally positioned to interact with Cu(I). However, as shown in Fig. 7C for the AlphaFold model, the Asp25 carboxylate is well positioned to interact with the protonated nitrogen of the His sidechain. Thr68 and the backbone carbonyl of Cys65 are also close enough to potentially contribute to copper chelation, but an ETR1T68A mutant still bound ethylene when expressed in yeast (20), indicating that Thr68 does not play a major role in copper chelation.
The CH model, in which the interaction of the Asp carboxylate with His contributes to Cu(I) binding may help explain the high binding affinity of the receptors for ethylene (5, 28). Such carboxylate–His–metal interactions are fairly common in proteins, being a form of indirect carboxylate–metal coordination, with the carboxylate thought to modulate the histidine to make it a more effective Lewis base and strengthen the metal complexation (49). This carboxylate-His structure is similar to that found in the well-characterized “catalytic triads” of serine proteases as well as in a host of other enzymes that also make use of an Asp–His interactions to facilitate hydrolytic and other enzymatic activities (50). The increase in the effectiveness of such enzymes may not just be due to increased nucleophilicity of the carboxylate-His, but also due to maintenance of the correct tautomer of His (e.g., N-1H vs N-3H) for the reaction (51, 52), another consideration that may apply to the ability of His69 of ETR1 to interact with the Cu(I) cofactor. Our enhanced understanding for mechanisms underlying high-affinity ethylene binding should facilitate the development of ethylene nanosensors for agricultural and industrial use (53, 54).
An unexpected but physiologically relevant finding from our studies was that the substitution of Asn for Asp25 of ETR1, unlike the other site-directed mutations examined, still allowed for ethylene binding but affected the binding kinetics. Experimental analyses of [14C]ethylene binding in transgenic yeast and in planta are dependent on the extended half-life for dissociation of ethylene from the receptors, this being of over 12 h for ETR1wt (5). Our inability to detect [14C]ethylene binding by ETR1D25N when assayed in transgenic yeast suggests a half-life for dissociation on the order of minutes or less (koff increasing at least 100-fold), the consistency of the in planta results with those from yeast indicating that this difference in ethylene binding between ETR1wt and ETR1D25N is not an artifact of the transgenic yeast system. Interestingly, although Asp in found in 93.55% of the ETR1-like sequences examined (Fig. 1A), in some cases (3.67% of the sequences examined) it is substituted by an Asn residue, most commonly in cyanobacteria but also reportedly in the plants P. communis (Pear) and C. cajan (Pigeon pea). We consider it likely that the Asn-containing ethylene receptor-like proteins of cyanobacteria facilitate chemotaxis (55), the ability to rapidly detect and respond to changes in ligand concentration not being compatible with the extended binding kinetics associated with the typical plant receptors. Plants, with their slower release kinetics for ethylene, rely upon proteasome-dependent degradation of ethylene-bound receptors and transcriptional induction of new receptors to facilitate resensitization once environmental ethylene levels decrease (9, 33, 34, 56, 57).
Although deamidation of Asn and conversion to Asp has been reported to sometimes occur spontaneously in vivo (58, 59), such post-translational processing is not supported for the ETR1D25N mutant based on structural and experimental considerations. First, ETR1D25N lacks the Asn-Gly motif associated with deamidation but has a stabilizing Phe residue at position 26 (58, 59). Second, deamidation of Asn is suppressed in alpha-helices (60), such as are found in the TMD of ETR1. Third, we do not recover detectable ethylene binding for the ETR1D25N mutant when assayed in yeast or in planta, such as would be expected if deamidation had occurred. Fourth, a small additive effect of the Asp25Asn mutation was observed when combined with the ETR1K91X mutations, consistent with the presence of the Asp25Asn mutation and that it has functional consequences outside of its effect on ethylene binding.
Based on our findings, Asp25 of ETR1 plays a dual role in signaling, functioning in copper/ethylene binding as well as in internally transmitting information on ethylene binding to TM helix III through an association with Lys91 (Fig. 7). Coevolution analysis predicts a strong association between these two residues, both of which are highly conserved, the one residue acidic and the other basic, suggestive not only of physical proximity but the ability to make a strong polar bond. Mutations of Lys91did not affect copper binding of ETR1 but loss of its basic amino acid character resulted in decreased ethylene binding, potentially due to the receptor being unable to maintain the optimal conformation for ethylene binding and/or due to altered kinetics for ethylene binding as found for the Asp25Asn mutation. Of particular interest is the reduced functionality in the absence and presence of ethylene observed with the ETR1K91M, ETR1D25N, K91M, and some of the ETR1K91A mutant lines, a phenotype consistent with a necessity to transmit conformational information from the ethylene binding site to TM helix III. These mutants may be at an equilibrium between the conformations typically found in the absence (“on”) and presence (“off ”) of ethylene, with ethylene no longer able to stabilize one conformation over the other. Alternatively, if the receptors can take on intermediate conformations, the receptors may be stuck in such an intermediate conformation. Critically, the results of these mutations indicate the importance of the basic character of Lys91 in maintaining functionality of ETR1, and how loss of this character uncouples ETR1 from the conformations needed to mediate responses in the absence and presence of ethylene.
The dual role for Asp25 supported by our study of ETR1 is remarkably like the role proposed for Asp180 in a model for the mouse olfactory receptor MOR244-3 (61). This olfactory receptor is responsive to organosulfur odorants and has a binding site with a required Cu(I) cofactor coordinated by Cys, His, and possibly an Asn residue (61, 62). Like the ethylene receptors, the copper cofactor is required for the MOR244-3 receptor to assume its active conformation, and the ligands (ethylene or methylthio-methanethiol) exert inverse agonist effects on receptor activity (18, 20, 61–64). In the model of the MOR244-3 binding site, Asp180 forms hydrogen bonds to the Cu-coordinating His105 and also to Lys269 (61). These two evolutionarily distinct receptors, both employing a Cu(I) cofactor and exhibiting high affinity for their ligands, may have converged on a similar mechanism to enhance the performance of the Cu(I)-chelating His and relaying intramolecular changes in response to ligand binding.
Materials and Methods
Detailed information is provided in SI Appendix, Materials and Methods. All Arabidopsis lines were of the Columbia (Col-0) accession. The etr1-1 and etr1-6 etr2-3 ein4-4 mutant lines have been described (4, 18, 26). Analyses of the triple response and short-term kinetic response of dark-grown Arabidopsis seedlings to ethylene were performed as described (28, 33, 65). Primers used for mutagenesis of ETR1 are listed in SI Appendix, Table S2. For plant transformation, constructs were introduced into Agrobacterium tumefaciens strain GV3101 and transformed into the etr1-6 etr2-3 ein4-4 background (18) by the floral-dip method (66). Yeast constructs were transformed into the yeast Saccharomyces cerevisiae strain FY834 (MATα his3Δ200 ura3-52 leu2Δ1 lys2Δ202 trp1Δ63 GAL2+) (67).
For generation of CRISPR-Cas9 mutant lines targeting ERS1 and ERS2, guide RNAs were designed using CRISPR-P 2.0 (68) and the gRNA cassette cloned into the pCAMBIA2300-Cas9 vector (69). Heat stress treatment of transgenic lines was used to increase the efficiency of CRISPR-Cas9 mutagenesis (70). Genomic DNA was isolated (71), and the region surrounding the CRISPR target sequence sequenced using primers given in SI Appendix, Table S2. Characteristics of the indel mutations are given in SI Appendix, Fig. S7.
Copper binding of ETR1 in vitro was monitored spectrophotometrically by measuring absorbance of the purple BCA2-Cu(I) complex as described (24). Saturable ethylene binding to Arabidopsis seedlings and transgenic yeast expressing ETR1 was determined by analyzing binding to [14C]ethylene in the presence or absence of excess [12C]ethylene (5, 31). For ethylene-binding assays in yeast, ETR1 was expressed in the yeast S. cerevisiae (strain FY834) (5, 67, 72). For ethylene binding assays with Arabidopsis seedlings, 2-wk–old green seedlings were used.
Immunodetection of ETR1 in plants and transgenic yeast was performed as described (56), using an anti-ETR1 antibody generated against amino acids 401-738 of ETR1 (10). RNA isolation and RT-qPCR was performed as described (73) with three biological replicates. Relevant primers are listed in SI Appendix, Table S2.
For the coevolutionary analysis, we used EVCOUPLINGS and GREMLIN web servers to predict interactions/contacting residues in the EBD of ETR1 (37, 38). Results from EVCOUPLINGS are reported for the recommended result, which is based on the analysis of 1,221 sequences and an overall quality score of 9 (SI Appendix, Table S1). For GREMLIN, the multiple sequence alignment was performed by HHBLITS and the alignment then filtered to remove regions where the gap was greater than 75. An ab initio structural model of the EBD has been previously described (24). New structural models for the ETR1 homodimer were generated with AlphaFold-Multimer (47, 48). Coppers were modeled under two potential coordinations involving Cys65 and His69 of the ETR1 homodimer, one in which the two coppers are bound independently and do not share an interaction with each other, and another where they are closely bonded. Coordinates of the full-length ETR1 structural models with copper are available as Protein Data Bank (PDB) and PyMOL (74) files at https://digitalcommons.dartmouth.edu/facoa/4313 (75).
Statistical analyses were performed in Prism (GraphPad Software, Inc.) or using an online calculator (astatsa.com/OneWay_Anova_with_TukeyHSD/).
Supplementary Material
Appendix 01 (PDF)
AlphaFold-Multimer-based model of the ETR1 homodimer (coppers interacting).
AlphaFold-Multimer-based model of the ETR1 homodimer (coppers not interacting).
Acknowledgments
This work was supported by grants from the NSF (MCB-1517032 to G.E.S., G. Grigoryan, and B.M.B.; IOS-1856513 to G.E.S.; MCB-1817304 to B.M.B.), by the Deutsche Forschungsgemeinschaft (German Research Foundation) project no. 267205415/CRC 1208 grant to G. Groth (TP B06), and by the International Research Support Initiative Program of Higher Education Commission of Pakistan to B.J.A. and S. Abbas.
Author contributions
G. Grigoryan and G.E.S. designed research; B.J.A., S. Abbas, S. Aman, M.V.Y., W.C., L.M., B.U., D.A.J., J.D., S.N.S., B.M.B., G. Grigoryan, and G.E.S. performed research; B.J.A., S. Abbas, S. Aman, M.V.Y., W.C., L.M., B.U., D.A.J., S.N.S., G. Groth, B.M.B., G. Grigoryan, and G.E.S. analyzed data; G. Groth, G. Grigoryan, and G.E.S. supervised research; and B.J.A., G. Grigoryan, and G.E.S. wrote the paper.
Competing interests
A patent application related to this work has been filed (#PCT/US2023/017522. Ethylene receptors and binding domains with modified binding kinetics for ethylene).
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Gevorg Grigoryan, Email: gevorg.grigoryan@gmail.com.
G. Eric Schaller, Email: george.e.schaller@dartmouth.edu.
Data, Materials, and Software Availability
All other data are included in the manuscript and/or supporting information. Coordinates for the ETR1 structural models are available at the Dartmouth Digital Commons (https://digitalcommons.dartmouth.edu/facoa/4313) (75).
Supporting Information
References
- 1.Abeles F. B., Morgan P. W., Saltveit M. E. Jr., Ethylene in Plant Biology (Academic Press, San Diego, ed. 2, 1992). [Google Scholar]
- 2.Binder B. M., Ethylene signaling in plants. J. Biol. Chem. 295, 7710–7725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang K. N., et al. , Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2, e00675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M., Arabidopsis ethylene response gene ETR1: Similarity of product to two-component regulators. Science 262, 539–544 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Schaller G. E., Bleecker A. B., Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Binder B. M., Chang C., Schaller G. E., “Perception of ethylene by plants: Ethylene receptors” in Annual Plant Reviews: The Plant Hormone Ethylene, McManus M. T., Ed. (Wiley-Blackwell, 2012), vol. 44, pp. 117–145. [Google Scholar]
- 7.Rodriguez F. I., et al. , A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283, 996–998 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Hall A. E., Chen Q. G., Findell J. L., Schaller G. E., Bleecker A. B., The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 121, 291–299 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y.-F., et al. , Ligand-induced degradation of the ethylene receptor ETR2 through a proteasome-dependent pathway in Arabidopsis. J. Biol. Chem. 282, 24752–24758 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Chen Y. F., Randlett M. D., Findell J. L., Schaller G. E., Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 277, 19861–19866 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Dong C.-H., Rivarola M., Resnick J. S., Maggin B. D., Chang C., Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 53, 275–286 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grefen C., et al. , Subcellular localization and in vivo interaction of the Arabidopsis thaliana ethylene receptor family members. Mol. Plant 1, 308–320 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Gao Z., et al. , Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J. Biol. Chem. 283, 23081–23810 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaller G. E., Kieber J. J., "Ethylene", The Arabidopsis Book, C. Somerville, E. Meyerowitz, Eds., 1–18 (2002).
- 15.Gamble R. L., Coonfield M. L., Schaller G. E., Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7825–7829 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller G. E., Shiu S. H., Armitage J. P., Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 21, R320–R330 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Wang W., Hall A. E., O’Malley R., Bleecker A. B., Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc. Natl. Acad. Sci. U.S.A. 100, 352–357 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua J., Meyerowitz E. M., Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Qu X., Hall B. P., Gao Z., Schaller G. E., A strong constitutive ethylene response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 7, 3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., et al. , Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18, 3429–3442 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacey R. F., Binder B., Ethylene regulates the physiology of the Cyanobacterium Synechocystis sp. PCC 6803 via an ethylene receptor. Plant Physiol. 171, 2798–2809 (2016), 10.1104/pp.16.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaller G. E., Ladd A. N., Lanahan M. B., Spanbauer J. M., Bleecker A. B., The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J. Biol. Chem. 270, 12526–12530 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Binder B. M., Rodriguez F. I., Bleecker A. B., Patterson S. E., The effects of Group 11 transition metals, including gold, on ethylene binding to the ETR1 receptor and growth of Arabidopsis thaliana. FEBS Lett. 581, 5105–5109 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Schott-Verdugo S., Muller L., Classen E., Gohlke H., Groth G., Structural model of the ETR1 ethylene receptor transmembrane sensor domain. Sci. Rep. 9, 8869 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutsail G. 3rd, et al. , Spectroscopic and QM/MM studies of the Cu(I) binding site of the plant ethylene receptor ETR1. Biophys J. 121, 3862–3873 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bleecker A. B., Estelle M. A., Somerville C., Kende H., Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Kugele A., et al. , Mapping the helix arrangement of the reconstituted ETR1 ethylene receptor transmembrane domain by EPR spectroscopy. RSC Adv. 12, 7352–7356 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binder B. M., Mortimore L. A., Stepanova A. N., Ecker J. R., Bleecker A. B., Short-term growth responses to ethylene in Arabidopsis seedlings are EIN3/EIL1 independent. Plant Physiol. 136, 2921–2927 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Malley R. C., et al. , Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J. 41, 651–659 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Lacey R. F., Binder B. M., Ethylene regulates the physiology of the cyanobacterium synechocystis sp. PCC 6803 via an ethylene receptor. Plant Physiol. 171, 2798–2809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binder B. M., Schaller G. E., Analysis of ethylene receptors: Ethylene-binding assays. Methods Mol. Biol. 1573, 75–86 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Qu X., Schaller G. E., Requirement of the histidine kinase domain for signal transduction by the ethylene receptor ETR1. Plant Physiol. 136, 2961–2970 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binder B. M., et al. , Arabidopsis seedling growth response and recovery to ethylene. A kinetic analysis. Plant Physiol. 136, 2913–2920 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azhar B. J., Zulfiqar A., Shakeel S. N., Schaller G. E., Amplification and adaptation in the ethylene signaling pathway. Small Methods 4, 1900452 (2019), 10.1002/smtd.201900452. [DOI] [Google Scholar]
- 35.McDaniel B. K., Binder B. M., ETHYLENE RECEPTOR 1 (ETR1) Is Sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J. Biol. Chem. 287, 26094–26103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaller G. E., Binder B. M., Inhibitors of ethylene biosynthesis and signaling. Methods Mol. Biol 1573, 223–235 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Hopf T. A., et al. , The EVcouplings Python framework for coevolutionary sequence analysis. Bioinformatics 35, 1582–1584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ovchinnikov S., Kamisetty H., Baker D., Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife 3, e02030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch K. A., Pena M. M., Thiele D. J., Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem. Biol. 4, 549–560 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Sharma A., Sharma D., Verma S. K., In silico study of iron, zinc and copper binding proteins of Pseudomonas syringae pv. lapsa: Emphasis on secreted metalloproteins. Front Microbiol. 9, 1838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm R. H., Kennepohl P., Solomon E. I., Structural and functional aspects of metal sites in biology. Chem. Rev. 96, 2239–2314 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Balamurugan R., Palaniandavar M., Gopalan R. S., Trigonal planar copper(I) complex: Synthesis, structure, and spectra of a redox pair of novel copper(II/I) complexes of tridentate bis(benzimidazol-2’-yl) ligand framework as models for electron-transfer copper proteins. Inorg Chem. 40, 2246–2255 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Light K. M., Wisniewski J. A., Vinyard W. A., Kieber-Emmons M. T., Perception of the plant hormone ethylene: Known-knowns and known-unknowns. J. Biol. Inorg Chem. 21, 715–728 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Geri J. B., Pernicone N. C., York J. T., Comparing the impact of different supporting ligands on copper(I)-ethylene interactions. Polyhedron 52, 207–215 (2013). [Google Scholar]
- 45.Davis A. V., O’Halloran T. V., A place for thioether chemistry in cellular copper ion recognition and trafficking. Nat. Chem. Biol. 4, 148–151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joy S., Periyasamy G., Binding mode of 1-methylcyclopropene, an ethylene antagonist in various copper models. Comput. Theor. Chem. 1171, 112662 (2020). [Google Scholar]
- 47.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2021). 10.1101/2021.10.04.463034 (Accessed 10 March 2022). [DOI]
- 48.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christianson D. W., Alexander R. S., Carboxylate-histidine-zinc interactions in protein structure and function. J. Am. Chem. Soc. 111, 6412–6419 (1989). [Google Scholar]
- 50.Rauwerdink A., Kazlauskas R. J., How the same core catalytic machinery catalyzes 17 different reactions: The serine-histidine-aspartate catalytic triad of alpha/beta-hydrolase fold enzymes. ACS Catal. 5, 6153–6176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cramer K. D., Zimmerman S. C., Kinetic effect of a syn-oriented carboxylate on a proximate imidazle in catalysis: A model for the histidine-aspartate couple in enzymes. J. Am. Chem. Soc. 112, 3680–3682 (1990). [Google Scholar]
- 52.Sprang S., et al. , The three-dimensional structure of Asn102 mutant of trypsin: Role of Asp102 in serine protease catalysis. Science 237, 905–909 (1987). [DOI] [PubMed] [Google Scholar]
- 53.Vong K., et al. , An artificial metalloenzyme biosensor can detect ethylene gas in fruits and Arabidopsis leaves. Nat. Commun. 10, 5746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esser B., Schnorr J. M., Swager T. M., Selective detection of ethylene gas using carbon nanotube-based devices: Utility in determination of fruit ripeness. Angew. Chem. Int. Ed. Engl. 51, 5752–5756 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Mutalipassi M., et al. , Symbioses of cyanobacteria in marine environments: Ecological insights and biotechnological perspectives. Mar. Drugs 19, 227 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shakeel S. N., et al. , Ethylene regulates levels of ethylene receptor/CTR1 signaling complexes in Arabidopsis thaliana. J. Biol. Chem. 290, 12415–12424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kevany B. M., Tieman D. M., Taylor M. G., Cin V. D., Klee H. J., Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 51, 458–467 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Robinson N. E., Robinson A. B., Prediction of protein deamidation rates from primary and three-dimensional structure. Proc. Natl. Acad. Sci. U.S.A. 98, 4367–4372 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson N. E., Robinson A. B., Molecular clocks. Proc. Natl. Acad. Sci. U.S.A. 98, 944–949 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosky A. A., Razzaq U. O., Treuheit M. J., Brems D. N., The effects of alpha-helix on the stability of Asn residues: Deamidation rates in peptides of varying helicity. Protein Sci. 8, 2519–2523 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sekharan S., et al. , QM/MM model of the mouse olfactory receptor MOR244-3 validated by site-directed mutagenesis experiments. Biophys. J. 107, L5–L8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan X., et al. , Crucial role of copper in detection of metal-coordinating odorants. Proc. Natl. Acad. Sci. U.S.A. 109, 3492–3497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirayama T., et al. , RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97, 383–393 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Woeste K. E., Kieber J. J., A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell 12, 443–455 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall B. P., et al. , Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol. 159, 682–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Winston F., Dollard C., Ricupero-Hovasse S. L., Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Liu H., et al. , CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Denbow C. J., et al. , Gateway-compatible CRISPR-Cas9 vectors and a rapid detection by high-resolution melting curve analysis. Front. Plant Sci. 8, 1171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.LeBlanc C., et al. , Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 93, 377–386 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Kasajima I., Ide Y., Ohkama-Ohtsu N., Yoneyama T., Fujiwara T., A protocol for rapid DNA extraction from Arabidopsis thaliana for PCR analysis. Plant Mol. Biol. Rep. 22, 49–52 (2004). [Google Scholar]
- 72.Hadfield C., Cashmore A. M., Meacock P. A., An efficient chloramphenicol-resistance marker for Saccharomyces cerevisiae and Escherichia coli. Gene 45, 149–158 (1986). [DOI] [PubMed] [Google Scholar]
- 73.Rai M. I., et al. , The ARGOS gene family functions in a negative feedback loop to desensitize plants to ethylene. BMC Plant Biol. 15, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Azhar B.J., et al. , Structural files for the ETR1 ethylene-receptor dimer based on computational modeling. Dartmouth Scholarship. 4313. (2023) https://digitalcommons.dartmouth.edu/facoa/4313. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
AlphaFold-Multimer-based model of the ETR1 homodimer (coppers interacting).
AlphaFold-Multimer-based model of the ETR1 homodimer (coppers not interacting).
Data Availability Statement
All other data are included in the manuscript and/or supporting information. Coordinates for the ETR1 structural models are available at the Dartmouth Digital Commons (https://digitalcommons.dartmouth.edu/facoa/4313) (75).