Abstract
Background:
Pathologic response at the time of surgery after neoadjuvant therapy for HER2 positive early breast cancer impacts both prognosis and subsequent adjuvant therapy. Comprehensive descriptions of the tumor microenvironment (TME) in patients with HER2 positive early breast cancer is not well described. We utilized standard stromal pathologist-assessed TIL quantification, quantitative multiplex immunofluorescence, and RNA-based gene pathway signatures to assess pretreatment TME characteristics associated pathologic complete response in patients with hormone receptor positive, HER2 positive early breast cancer treated in the neoadjuvant setting.
Methods:
We utilized standard stromal pathologist-assessed TIL quantification, quantitative multiplex immunofluorescence, and RNA-based gene pathway signatures to assess pretreatment TME characteristics associated pathologic complete response in 28 patients with hormone receptor positive, HER2 positive early breast cancer treated in the neoadjuvant setting.
Results:
Pathologist-assessed stromal TILs were significantly associated with pathologic complete response (pCR). By quantitative multiplex immunofluorescence, univariate analysis revealed significant increases in CD3+, CD3+CD8-FOXP3-, CD8+ and FOXP3+ T-cell densities as well as increased immune cell aggregates in pCR patients. In subsets of paired pre/post-treatment samples, we observed significant changes in gene expression signatures in non-pCR patients and significant decreases in CD8+ densities after treatment in pCR patients. No RNA based pathway signature was associated with pCR.
Conclusion:
TME characterization HER2 positive breast cancer patients revealed several stromal T-cell densities and immune cell aggregates associated with pCR. These results demonstrate the feasibility of these novel methods in TME evaluation and contribute to ongoing investigations of the TME in HER2+ early breast cancer to identify robust biomarkers to best identify patients eligible for systemic de-escalation strategies.
Keywords: Breast cancer, HER2 positive, tumor microenvironment, quantitative multiplex immunofluorescence, tumor infiltrating lymphocytes, neoadjuvant therapy, gene expression
Micro abstract
We studied the tumor microenvironment in patients with HER2 positive early breast cancer treated in the neoadjuvant setting. We evaluated associations of pathologist-assessed stromal tumor infiltrating lymphocyte counting, quantitative multiplex immunofluorescence, and RNA-based gene pathway signatures with pathologic complete response. Our results show significant associations between immune cell densities and their spatial organization with pathologic response.
Introduction
Patients with HER2+ early breast cancer (HER2+ EBC) are often treated with neoadjuvant chemotherapy and trastuzumab +/− pertuzumab (NACT), and these combinations result in pathologic complete response rates (pCR) of 40–70% [1, 2]. Achieving a pCR to NACT is predictive of outcomes and is clinically significant given future adjuvant therapy can be tailored to the biology of the tumor [3].
Higher densities of pre-treatment tumor infiltrating lymphocytes (TILs) have been shown to correlate with higher pCR rates in some [4, 5], but not all studies [6], evaluating TIL levels and responses to NACT. In addition, outside of standard TIL analysis, a more comprehensive description of the tumor microenvironment (TME) and its impact on pCR is not well described in HER2+ EBC. We hypothesized that a more granular assessment of the TME using novel methods may identify pretreatment TME characteristics associated with pCR. Identification of reliable characteristics associated with pCR may better select patients for systemic de-escalation strategies in HER2+ EBC.
We utilized standard stromal pathologist-assessed TILs [7], quantitative multiplex immunofluorescence (qmIF), and RNA-based gene pathway signatures to assess pretreatment TME characteristics associated with pCR to NACT in HR+/HER2+ EBC (npCR=12, nnon-pCR=16).
Methods
Patient Cohort
We performed a single institution retrospective review of women diagnosed with invasive breast cancer between January 2004 and February 2016 who were treated in the neoadjuvant setting with an anthracycline, taxane or both. All patients received oncologic care at Columbia University Medical Center. Clinicopathologic data were abstracted from the medical records by four independent researchers, which was then double verified; any discrepancies were resolved by EC and KK.
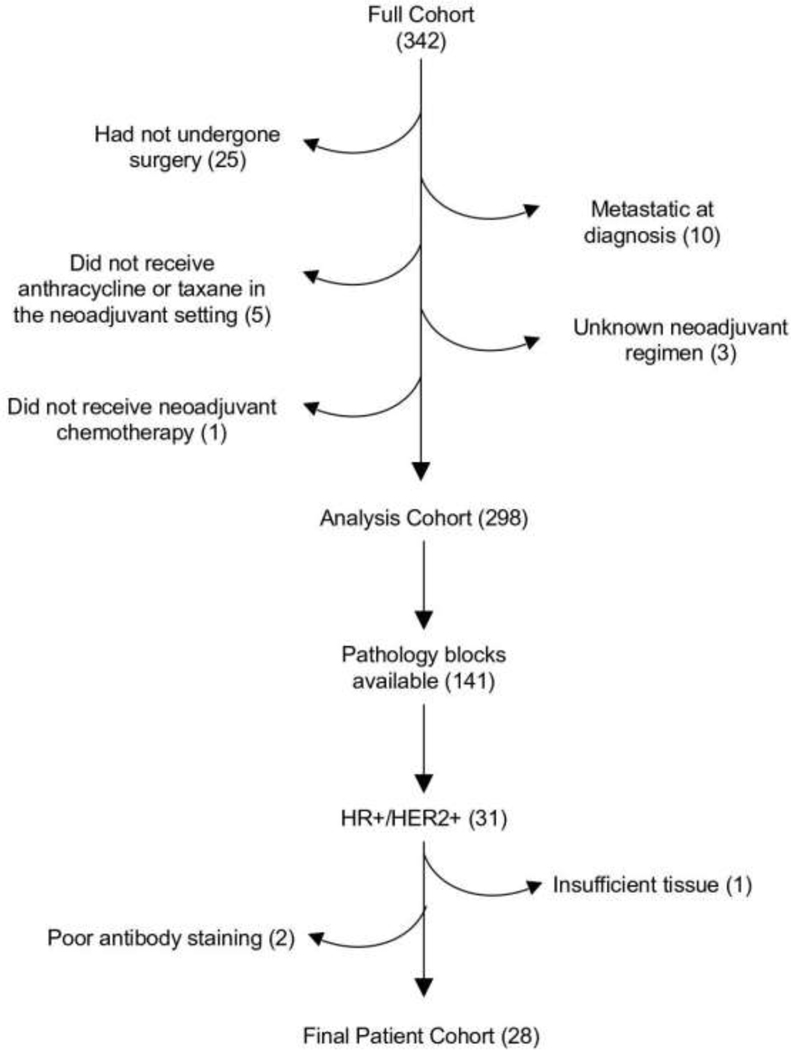
We identified 342 patients that met these criteria. 317 patients had undergone surgery at the time of this analysis. 19 patients were subsequently excluded: 10 patients with metastatic disease at diagnosis, five patients who received neither an anthracycline nor a taxane in the neoadjuvant setting, three whom the neoadjuvant regimen was unknown and one patient who was pregnant and did not receive neoadjuvant chemotherapy, which resulted in a final cohort of 298 patients. From this cohort, 141 patients had pathology blocks available for analysis, 31 of whom were HR+/HER2+. One additional patient was excluded given the available tissue was only lymphoid tissue and two patients were excluded due to poor antibody staining. A flowchart outlining the patients used for the study is shown in Figure 1. The resulting patient characteristics are shown in Table 1.
Figure 1:
Patient selection
Table 1:
Patient characteristics;
| Characteristic | n (%) - Total Population | n (%) - pCR | n (%) - non-pCR |
|---|---|---|---|
| Age - median (range) | 51 (29–77) | 51 (29–64) | 59 (41–77) |
| Race | |||
| Hispanic | 12 (43) | 6 (50) | 6 (38) |
| African American | 7 (25) | 3 (25) | 4 (25) |
| Causcausian | 6 (21) | 1 (8) | 5 (31) |
| Other | 3 (11) | 2 (17) | 1 (6) |
| Hormone receptor status | |||
| ER positive | 23 (82) | 8 (67) | 15 (94) |
| PR positive | 28 (100) | 12 (100) | 16 (100) |
| Stage | |||
| I | 1 (4) | 1 (8) | 0 (0) |
| II | 18 (64) | 8 (67) | 10 (63) |
| III | 9 (32) | 3 (25) | 6 (38) |
| HER2 targered therapy | |||
| Trastuzumab | 25 (89) | 11 (92) | 14 (88) |
| Pertuzumab | 13 (46) | 8 (67) | 5 (31) |
| Chemothreapy | |||
| Taxane | 26 (93) | 12 (100) | 14 (88) |
| Anthracycline | 14 (50) | 5 (42) | 9 (56) |
| pCR | |||
| yes | 12 (43) | 12 (100) | 0 (0) |
| no | 16 (57) | 0 (0) | 16 (100) |
| PAM50 | |||
| Luminal A | 3 (11) | 1 (8) | 2 (13) |
| Luminal B | 3 (11) | 1 (8) | 2 (13) |
| HER2-enriched | 19 (68) | 9 (75) | 10 (63) |
| Basal | 2 (7) | 0 (0) | 2 (13) |
| Not Avaliable | 1 (4) | 1 (8) | 0 (0) |
ER = estrogen receptor; PR = progesterone receptor; PAM50 = pretreatment PAM50 as determined by nCounter Breast Cancer 360 panel
Tumor infiltrating lymphocytes assessment
Stromal TILs (sTILs) were evaluated on whole slide hematoxylin and eosin (H&E) sections. The area of the tumor stroma occupied by mononuclear inflammatory cells was divided by the total tumor stromal area according to the International TILs Working Group guidelines [7, 8]. TIL quantification post-NACT was done according to the International Immuno-Oncology Biomarker Working Group on Breast Cancer [9]. sTILs were assessed within the boundaries of the residual tumor bed as defined by the RCB (Residual Cancer Burden)-index [9, 10]. Pathologists (HG and HH) scanned the tumor bed (50–100× magnification) and estimated average TILs across microscopic fields (200–400× magnification). Necrotic areas were excluded. Ambiguous and discrepant cases were resolved by discussing and reviewing at multi-headed microscopes by at least two pathologists.
Multiplex staining and analysis
Multiplex staining was performed with diagnostic tissue samples and surgical specimens (pre and post therapy). After harvesting, tumors were immediately fixed overnight in 10% neutral-buffered formalin. Fixed tumors were dehydrated using an ethanol series, embedded in paraffin, and sections were cut at a thickness of 5 μm. Full-section slides of tumor tissues were stained using Opal multiplex 6-plex kits, according to the manufacturer’s protocol (Akoya). Staining was performed using an automated staining system (BOND-Rx; Leica Microsystems, Vista, CA) with Opal kits (Perkin Elmer) for CD3 (Leica, clone:LN10), CD8 (Leica, 4B11), FOXP3 (Abcam, 236A/E7), CD68 (Biogenex, KP1), HLA-DR (Abcam, LN3), pancytokeratin (Biolegend, AE1/AE3) and DAPI (Akoya Biosciences). Single color controls and an unstained slide were also included for proper spectral unmixing. Multispectral image capture was done at 20X magnification using Vectra (PerkinElmer, Hopkinton, MA). Pathologists (HG and HH) divided the whole slide image into fields of view optimized for the tumor and stroma content as well as representativeness of the sample. Five random fields of view per sample were then chosen for high-resolution scanning. Images were exported using inForm software version 2.4.6 (PerkinElmer). Subsequent analysis on the resulting images was performed with HALO (Indica labs) including tissue segmentation, cellular segmentation, and phenotyping. We did not include results from HLA-DR due to inadequate staining quality.
The result was a manifest of cells with their corresponding phenotypic assignments and locations, which was used to derive density estimates and perform spatial analysis using the Spatial Profiling Toolbox [11]. Estimates were limited to the stromal compartment given cellular segmentation was not reliable intratumorally because of the aggregation of cell populations in this compartment. Densities were computed by summing the cell areas in the stromal compartment for each phenotype and dividing by the sum of all cell areas in the same compartment. Spatial analysis was performed for every possible pair of phenotypes by computing the number of cells of a phenotype within 30 microns of each cell of another phenotype and computing the average.
Gene expression
A section of the formalin-fixed paraffin-embedded (FFPE) breast tissue was examined by a breast pathologist to confirm the presence of invasive tumor cells and determine the minimum tumor surface area. At least four 5-μm FFPE slides were used to purify total RNA using the Qiagen miRNeasy FFPE kit (Cat# 217504). The concentration of the extracted RNA was estimated by ultraviolet-visible spectrophotometry to ensure sample purity. Eluted RNA was then tested with NanoDrop 8000 (Thermo Scientific) and Bioanalyzer 2100 (Agilent) for quality. Degree of RNA integrity was assessed using the smear analysis function in the Agilent 2100 Expert Software to measure the percentage of RNA molecules > 300 bp. One sample needed to be excluded due to the poor RNA quality and another sample needed to be excluded due to lack of tumor. Final RNA concentration was normalized across all samples before input. From each sample, 100 ng of total RNA(>300bp) was used to measure the expression of 776 breast cancer-related genes by Breast 360™ Codeset using the nCounter platform (Nanostring Technologies). Data was normalized using a group of housekeeping genes and log2 transformed. Intrinsic molecular subtypes were identified using the research-based PAM50 predictor from the Nanostring database.
Statistical analysis
Multiplex derived densities are compared between patients who achieved pCR with those who did not using t-tests. Pre and post therapy samples are compared using paired t-tests.
We used logistic regression models to explore how a variety of measures may be associated with the odds of pCR. Measures of interest included multiplex derived metrics (immune densities and spatial metrics) and nCounter Breast Cancer 360 panel derived metrics (gene signatures and individual gene expressions). Other variables included: pathologist-assessed sTIL and iTIL estimates, clinical stage, PAM50, whether or not Perjeta was used, HR expression, and demographic factors.
Each model had a dichotomized outcome variable, pCR. Due to the small sample size, we limited models to include 2 covariates. Continuous variables were scaled by their mean to prevent extreme estimates caused by incomparable variable ranges. We computed odds ratios, confidence intervals, and p-values for each estimated coefficient in all of the models.
We first considered each variable in the univariate case, and then developed multivariate models with combinations of selected variables. Statistical significance is defined using an alpha cut-off of p<0.05.
All analyses were performed using R version 4.0.5.
Results
Histological immune assessments are associated with pathologic response
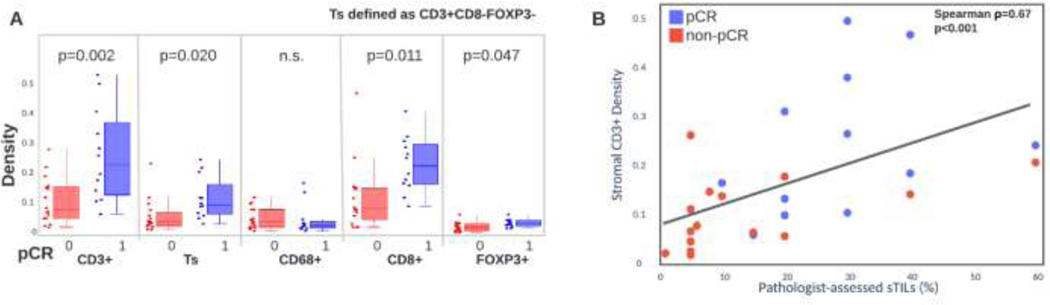
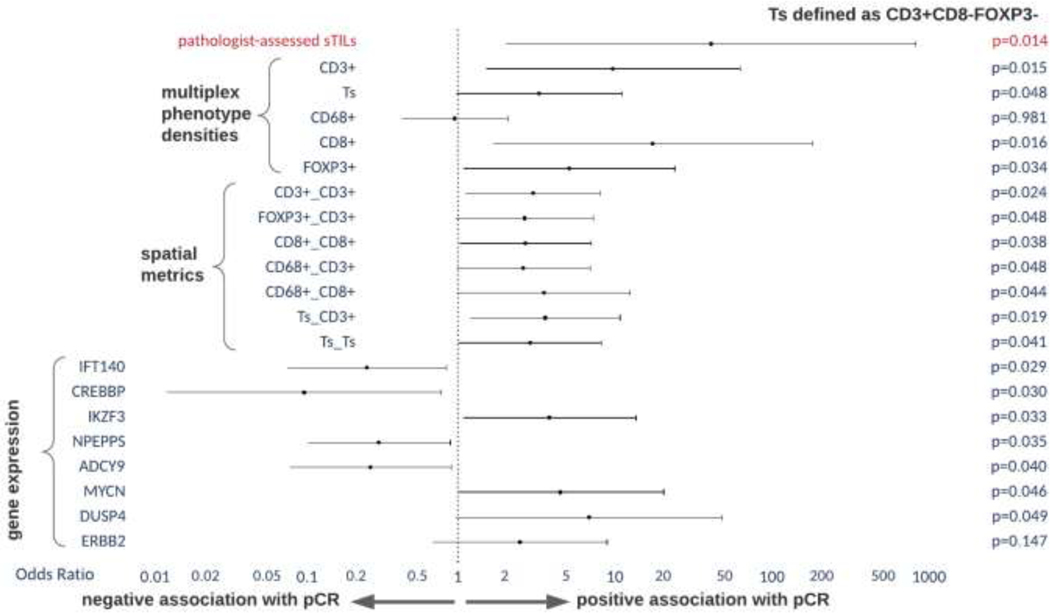
We found standard H&E-based assessment of sTILs to be significantly associated with pCR (OR 45.1, 95% CI 2.1–951.3, p=0.014). We then used multiplex immunofluorescence to evaluate the association of stromal cellular densities to pathologic response including T-cells (CD3+, CD3+CD8-FOXP3-, CD8+, FOXP3+) and macrophages (CD68+) (Figure 2). We found that CD3+ (OR 10.5, 95% CI 1.6–69.6, p=0.015), CD3+CD8-FOXP3- (OR 3.5, 95% CI 1.0–11.9, p=0.048), CD8+ (OR 18.9, 95% CI 1.7–204.1, p=0.016) and FOXP3+ (OR 5.5, 95% CI 1.1–26.1, p=0.034) T-cells were significantly associated to pCR (Figure 3A). We did not find stromal macrophage density to be significantly associated with response (OR 1.0, 95% CI 0.4–2.2, p=0.981). All populations significantly associated with response were higher in those that achieved pCR. We also found that sTIL counts were strongly correlated to multiplex derived CD3+ T-cell densities (Spearman ⍴=0.67, p<0.001) (Figure 3B).
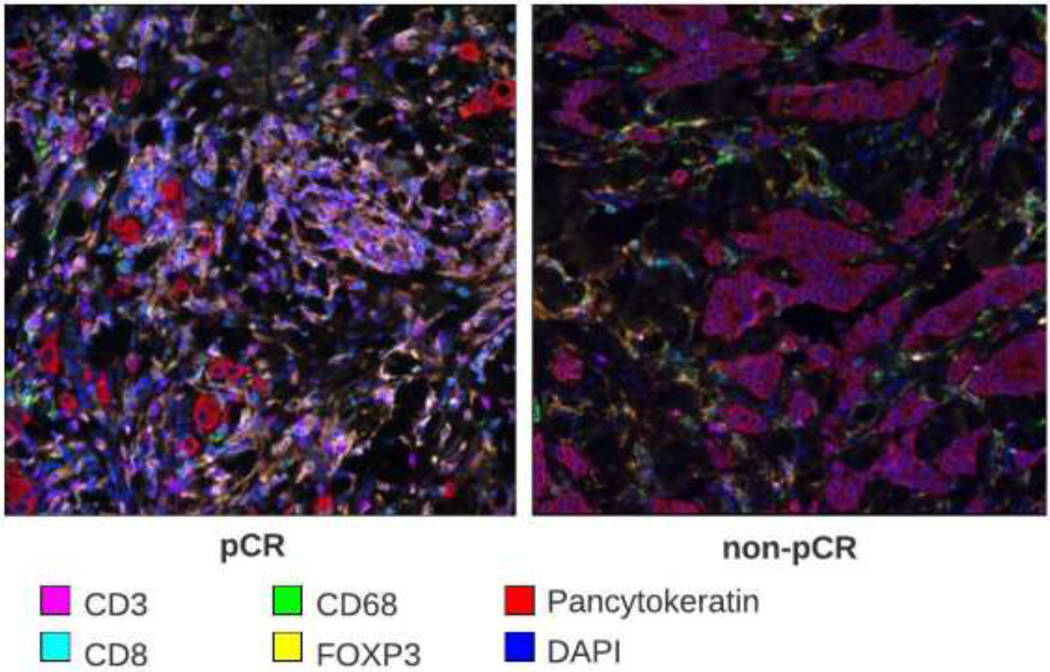
Figure 2:
Example pre-therapy biopsy tissue stained with multiplex immunofluorescence of a patient who achieved pCR and another patient who did not achieve pCR.
Figure 3:
A. Stromal immune densities for patients who did/did not achieve pCR. Significance of immune differences are computed with a t-test. B. Correlation of stromal CD3+ T-cell density with H&E-based pathologist assessed stromal tumor infiltrating lymphocytes. Correlation is computed with Spearman correlation.
Spatial immune aggregates are associated with pathologic response
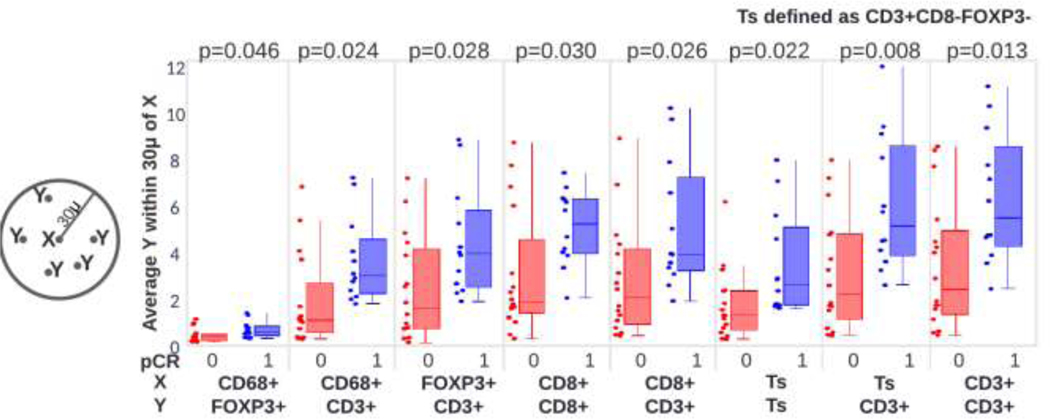
Analysis of the spatial distribution of immune populations also showed associations to pCR using univariate analysis (Figure 4). Aggregation of CD3+ T-cells (OR 3.2, 95% CI 1.2–8.7, p=0.024) and the average number of CD3+ T-cells within 30 microns of CD3+CD8-FOXP3-(OR 3.8, 95% CI 1.2–11.7, p=0.019) were both found to be higher in cases which achieved pCR. A complete list of univariate results including significantly associated spatial aggregates is shown in Figure 5.
Figure 4:
Spatial aggregation of immune populations for patients who did/did not achieve pCR. Significance of aggregation differences are computed with a t-test.
Figure 5:
Univariate associations H&E-based assessment of stromal tumor infiltrating lymphocytes, multiplex immunofluorescence derived immune densities and spatial statistics and RNA-based gene expressions to pCR.
Gene expression signatures are not associated with pathologic response
We searched for associations between pCR and gene pathway signatures as well as individual gene expressions characterized by the Nanostring IO360 Breast panel. While none of the gene pathway signatures were significant, we found associations between pCR and increased expression of several genes (Figure 5), including: IKZF3 (OR 4.1, 95% CI 1.1–14.8, p=0.03), MYCN (OR 4.8, 95% CI 1.0–22.3, p=0.05) and DUSP4 (OR 7.3, 95% CI 1.0–53.7, p=0.05). Decreased expression of IFT140 (OR 0.3, 95% CI 0.1–09, p=0.03), CREBBP (OR 0.1, 95% CI 0.0–0.8, p=0.03), NPEPPS (OR 0.3, 95% CI 0.1–0.9, p=0.03) and ADCY9 (OR 0.3, 95% CI 0.1–0.9, p=0.04) were also associated with pCR.
Multivariate analysis reveals only H&E-based sTILs significantly associated with pathologic response
Using multivariate analysis including pathologist-assessed sTILs, qmIF populations, RNA-based pathway signatures and individual gene expressions, stage and PAM50 subtype, only pathologist-assessed sTILs remain a significant predictor of reponse.
Analysis of pre/post paired samples shows significant decrease in immune populations for responding patients
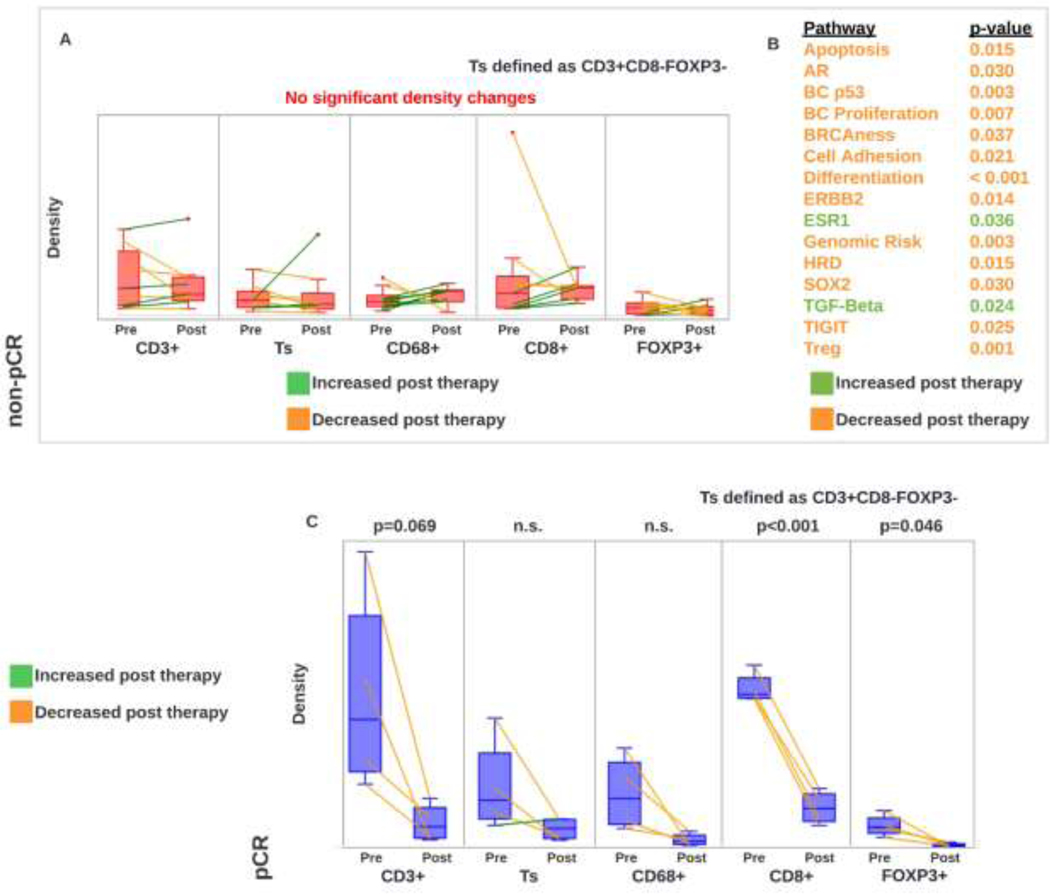
Analysis of 13 paired pretreatment and posttreatment samples (npCR=4, nnon-pCR=9) revealed no significant changes in immune subset populations in non-pCR patients (Figure 6A). Pre/post paired RNA-based gene expression data was available for 11 patients. Excluding 2 pCR patients, several RNA-based gene expression pathway signatures change after therapy in the remaining 9 non-pCR patients (Figure 6B). In pCR patients, CD8+ and FOXP3+ populations were significantly decreased in posttreatment samples (Figure 6C). These results are consistent with prior publications [16].
Figure 6:
A. Stromal immune density changes in pre/post paired samples in non-pCR patients. B. RNA-based gene expression changes in pre/post paired samples in non-pCR patients. C. Stromal immune density changes in pre/post paired samples in pCR patients.
Discussion
Assessment of the TME is not well characterized in HER2+ EBC. To our knowledge this is the first analysis of its kind in a histologically homogenous cohort of HR+/HER2+ patients, an important distinction given the impact that HR expression has on disease biology and responses to neoadjuvant therapy. We present an analysis of a cohort of 28 HR+/HER2+ patients and observed significant associations between several immune subpopulations and pCR. Due to the exploratory nature of our analysis, we do not correct p-values for multiple hypotheses.
Increased densities of CD3+, CD8+, CD3+CD8-FOXP3- and FOXP3+ T-cells by qmIF were all significantly associated with pCR on univariate analysis, indicating that baseline immune infiltrates contribute to responses to NACT in HER2+ EBC. While the association of FOXP3+ T-cells and pCR may seem counterintuitive given their classically immunosuppressive properties, prior studies have demonstrated that the location of FOXP3+ T-cells relative to tumor are more important than the presence of this subpopulation alone [17]; however, intratumoral FOXP3+ T-cells were not investigated in this cohort. In contrast with previous studies [18], macrophage density was not significantly associated with pCR. However, spatial analysis shows higher aggregation of CD3+ T-cells in the proximity of macrophages in patients with pCR. Prior studies have demonstrated that these immune subsets are impacted by neoadjuvant anti-HER2 based therapy. In an analysis of the TME of 49 patients treated on the PAMELA trial, two weeks of neoadjuvant dual HER2 targeted therapy was associated with increases in sTILs and associated increases in CD3+, CD3+CD8-FOXP3-, CD8+ and FOXP3+ T-cell subpopulations [19]. However, this change was only significant in patients with HR- disease, highlighting distinct TME differences dependent on concomitant HR status. Similarly, in an analysis of on treatment biopsies 29 patients treated on TRIO-US B07 who received neoadjuvant trastuzumab and/or lapatinib, T-cell markers using digital spatial profiling were increased relative to pretreatment levels to a more significant degree in patients achieving a pCR [20].
Pathologist-assessed sTIL were significantly associated with pCR rates in our cohort in a univariate analysis and all multivariate analysis configurations regardless of which covariates were included. Importantly, qmIF-derived CD3+ T-cell estimate significantly correlates with the pathologist-assessed sTILs. Since the qmIF-based estimates of TILs involve significantly lower tissue area than the pathologist assessment, it is possible that sTIL estimates could be made from smaller representative areas. In a meta-analysis of 5 trials evaluating the association of TILs and pCR to neoadjuvant therapy, high baseline TILs (defined as >=60% TILs) were associated with higher pCR rates, but only when data from the NeoALTTO trial was excluded, which did not use anthracycline and defined high baseline TILs as >=30% [21]. In patients treated with dual anti-HER2 therapy (trastuzumab and lapatinib) high TIL levels were not associated with increases in pCR. Notably, none of the trials included in this meta-analysis included trails in which pertuzumab was used in the neoadjuvant setting. A separate analysis revealed that in patients with HER2+ breast cancer treated with NACT, increased TIL concentrations were associated with longer disease free survival but not overall survival [5]. The International Immuno-Oncology Biomarker Working Group notes an ongoing challenge in interpreting the data regarding TILs and their impact on breast cancer outcomes remains the threshold to define “high” TILs, which have been highly variable between studies [22].
In our cohort, univariate analysis showed that pCR was significantly associated with CD3+, CD8+ and CD3+CD8-FOXP3- T-cell aggregates as well as macrophages, FOXP3+, CD8+, and CD3+CD8-FOXP3- cells aggregating around CD3+ T-cells. These observations further contribute to emerging data that spatial organization of immune infiltrates affects responses to NACT in breast cancer [23]. However, none of these findings were significant on multivariate analysis. In our cohort nearly 50% of patients were treated with both trastuzumab and pertuzumab. It is possible that in the presence of effective dual anti-HER2 inhibition, the impact of immune infiltration and organization in responses to chemotherapy is lessened. These biomarkers may have more importance in subtypes that are not primarily driven by a single molecular pathway alteration.
No RNA-based gene expression pathway signature was associated with pCR in our cohort, including ERBB2 and various immunogenicity and immune cell pathways. While we did find several genes significantly associated with pCR, they have not been extensively evaluated in breast cancer. Increased DUSP4 expression by NanoString gene expression profiling has been shown to correlate with increased sensitivty to chemotherapy in basal-like breast cancers [13]. Depletion of DUSP4 was hypothesized to results in increased RAS-ERK pathway activation. In a cohort of patients with triple negative breast cancer (TNBC), MYCN expression was associated with a decreased likelihood of acheiveing a pCR to NACT; MYCN expression is classically considered to be the driver of neuroendocrine prostate cancer [14]. CREBBP plays a role in epigenetic modifications and is commonly mutated in small cell lung cancer but there is a lack of data regarding its role in breast cancer [15]. Identification of biomarkers in HER2+ EBC via gene expression has shown mixed results. In an analysis of 117 patients treated with neoadjuvant therapy on the ADAPT trial using the same Nanostring panel, ERBB2 and estrogen receptor signaling were significantly associated with pCR, while PTEN was an unfavorable factor for pCR [24]. In a large analysis of the WSG-ADAPT HER2+/HR+ trial in EBC, high expression of HER2 and GRB7 and low expression of ESR1 and PgR by RNA based expression was associated with pCR independent of PAM50 subtype [25]. In contrast, RNA based gene expression patterns of 59 patients treated on TRIO-US B07 using CIBERSORT deconvolution did not find gene expression patterns that predicted pCR despite numerical reductions in ERBB2 signaling and increased immune infiltration [26].
Our analysis has several limitations. First, despite the population homogeneity it only includes 28 patients which may limit detection of significance. Secondly, given the changes in standard of care for HER2+ disease over the period of which our cohort was treated, there was heterogeneity in the NACT patients received, which likely impacted pCR rates. However, nearly all patients in our cohort received trastuzumab. The use of CD4 antibody clone in addition to CD3 and CD8 would have allowed for a more precise identification of T-cell subpopulations; however, our marker selection was limited by the panel depth of the Vectra platform and availability of antibody clones. Lastly, qmIF is associated with disadvantages, and despite our experience with this approach, it is possible that issues relating to assay optimization and antibody staining or a limited set of multiplex markers impacted the results in this cohort.
Despite these limitations, our results demonstrate the feasibility of utilizing these novel methods to evaluate the TME, particularly with qmIF which was found to be more sensitive than RNA based gene expression profiling In a larger cohort of patients, these methods can be applied to detect biomarkers that are associated with pCR to identify subsets of patients with HER2+ EBC that can be safely de-escalated while maintaining oncologic outcomes.
Conclusion
Our study shows that pathologist-assessed sTILs are a significant predictor of pCR in HER2 positive EBC. We also find associations between pCR and several stromal immune populations including CD3+, CD8+, CD3+CD8-FOXP3- and FOXP3+ T-cells. Moreover, spatial analysis revealed spatial immune cell aggregates were significantly associated with pCR. We did not find a significant association between pCR and gene pathway signatures.
Clinical Practice Points.
Pathologist-assessed stromal tumor infiltrating lymphocytes are a robust predictor of pathologic response in hormone receptor/HER2+ early stage breast cancer.
We found no association between gene pathway signatures and response.
Multiplex immunofluorescence can be used to characterize immune densities and their spatial relationships to develop predictive biomarkers for pCR.
Our study motivates larger studies to describe biomarkers associated with pCR in HER2+ EBC to better select patients eligible for systemic de-escalation strategies.
Acknowledgements
This work was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number KL2 TR000081.
Footnotes
Conflicts of interest
KMK reports grants/contracts to his institution from Incyte, Novartis, Genentch, Eli-Lilly, Pfizer, Calithera, Immunomedics, Acetylon, Seattle Genetics, Amgen, Zentalis, and CytomX Therapeutics; consulting fees/consultant honorarium from Daiichi Sankyo, Eli-Lilly, Pfizer, Novartis, Eisai, AstraZeneca, Immunomedics, Merck, Seattle Genetics, Cyclocel, Oncosec, and 4D Pharma; speakers bureau: Eli-Lilly; support for attending meetings and/or travel from Eli-Lilly, AstraZeneca, Pfizer; participation on advisory board (steering committee): Immunomedics, AstraZeneca, Ambryx, Genentech; and stock options/employment: Grail (spouse), Array BioPharma (spouse), and Pfizer (spouse). TJH receives grant funding from Bristol Myers Squibb and Calico labs. EPC receives grant funding from Merck and Eisai; travel from Zeiss. The remaining authors declare no competing interests.
Ethics
Tissue samples were collected after approval by the Columbia University Irving Medical Center Institutional Review Board (IRB). The IRB determined that written consent was not necessary due to the retrospective nature of the study.
CRediT authorship contribution statement.
Rami Vanguri: Formal analysis, Investigation, Methodology
Kathleen M. Fenn: Formal analysis, Investigation, Methodology
Matthew R. Kearney: : Formal analysis, Investigation, Methodology, Writing - original draft
Qi Wang: Data curation, Investigation
Hua Guo: Formal analysis, Investigation
Douglas K. Marks: Formal analysis, Investigation
Christine Chin: Formal analysis, Investigation
Claire F. Alcus: Formal analysis, Investigation
Julia B. Thompson: Formal analysis, Methodology
Cheng-Shiun Leu: Methodology
Hanina Hibshoosh: Formal analysis, Investigation
Kevin M. Kalinsky: Formal analysis, Investigation
James C. Mathews: Methodology, Software
Saad Nadeem: Methodology
Travis J. Hollmann: Methodology
Eileen P. Connolly: Conceptualization, Supervision
All authors: Writing - Review & Editing
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
The data used for this study, including quantitative multiplex immunofluorescence cell manifests and gene expression values, is available upon reasonable request from the corresponding author.
References
- 1.Gianni L, Pienkowski T, Im YH, Roman L, Tseng LM, Liu MC, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 2.Schneeweiss A, Chia S, Hickish T, Harvey V, Eniu A, Hegg R, et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA). Ann Oncol. 2013;24(9):2278–84. doi: 10.1093/annonc/mdt182. [DOI] [PubMed] [Google Scholar]
- 3.von Minckwitz G, Huang CS, Mano MS, Loibl S, Mamounas EP, Untch M, et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N Engl J Med. 2019;380(7):617–28. doi: 10.1056/NEJMoa1814017. [DOI] [PubMed] [Google Scholar]
- 4.Salgado R, Denkert C, Campbell C, Savas P, Nuciforo P, Aura C, et al. Tumor-Infiltrating Lymphocytes and Associations With Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated With Lapatinib and Trastuzumab: A Secondary Analysis of the NeoALTTO Trial. JAMA Oncol. 2015;1(4):448–54. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19(1):40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 6.Bianchini G, Pusztai L, Pienkowski T, Im YH, Bianchi GV, Tseng LM, et al. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann Oncol. 2015;26(12):2429–36. doi: 10.1093/annonc/mdv395. [DOI] [PubMed] [Google Scholar]
- 7.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–71. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv Anat Pathol. 2017;24(5):235–51. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dieci MV, Radosevic-Robin N, Fineberg S, van den Eynden G, Ternes N, Penault-Llorca F, et al. Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: A report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin Cancer Biol. 2018;52(Pt 2):16–25. doi: 10.1016/j.semcancer.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol. 2007;25(28):4414–22. doi: 10.1200/JCO.2007.10.6823. [DOI] [PubMed] [Google Scholar]
- 11.Mathews J, Nadeem, Saad, & Vanguri Rami. nadeemlab/SPT: SPT prerelease v0.7.1 (v0.7.1r). Zenodo. 2021. doi: 10.5281/zenodo.5576219. [DOI] [Google Scholar]
- 12.Morice WG. The immunophenotypic attributes of NK cells and NK-cell lineage lymphoproliferative disorders. Am J Clin Pathol. 2007;127(6):881–6. doi: 10.1309/Q49CRJ030L22MHLF. [DOI] [PubMed] [Google Scholar]
- 13.Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012;18(7):1052–9. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer JM, Lehmann BD, Gonzalez-Ericsson PI, Marshall CB, Beeler JS, Redman LN, et al. Targeting MYCN-expressing triple-negative breast cancer with BET and MEK inhibitors. Sci Transl Med. 2020;12(534). doi: 10.1126/scitranslmed.aaw8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, et al. Crebbp Loss Drives Small Cell Lung Cancer and Increases Sensitivity to HDAC Inhibition. Cancer Discov. 2018;8(11):1422–37. doi: 10.1158/2159-8290.CD-18-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Warren S, Pelekanou V, Wali V, Cesano A, Liu M, et al. Immune profiling of pre- and post-treatment breast cancer tissues from the SWOG S0800 neoadjuvant trial. J Immunother Cancer. 2019;7(1):88. doi: 10.1186/s40425-019-0563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Lang R, Zhao J, Zhang X, Pringle GA, Fan Y, et al. CD8(+) cytotoxic T cell and FOXP3(+) regulatory T cell infiltration in relation to breast cancer survival and molecular subtypes. Breast Cancer Res Treat. 2011;130(2):645–55. doi: 10.1007/s10549-011-1647-3. [DOI] [PubMed] [Google Scholar]
- 18.García-Martínez E, Gil GL, Benito AC, González-Billalabeitia E, Conesa MA, García García T, et al. Tumor-infiltrating immune cell profiles and their change after neoadjuvant chemotherapy predict response and prognosis of breast cancer. Breast cancer research : BCR. 2014;16(6):488. doi: 10.1186/s13058-014-0488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19..!!! INVALID CITATION !!! [17].
- 20.McNamara KL, Caswell-Jin JL, Joshi R, Ma Z, Kotler E, Bean GR, et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nature Cancer. 2021;2(4):400–13. doi: 10.1038/s43018-021-00190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solinas C, Ceppi M, Lambertini M, Scartozzi M, Buisseret L, Garaud S, et al. Tumor-infiltrating lymphocytes in patients with HER2-positive breast cancer treated with neoadjuvant chemotherapy plus trastuzumab, lapatinib or their combination: A meta-analysis of randomized controlled trials. Cancer Treat Rev. 2017;57:8–15. doi: 10.1016/j.ctrv.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 22.El Bairi K, Haynes HR, Blackley E, Fineberg S, Shear J, Turner S, et al. The tale of TILs in breast cancer: A report from The International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer. 2021;7(1):150. doi: 10.1038/s41523-021-00346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez K, Kim I, Chun B, Pucilowska J, Redmond WL, Urba WJ, et al. Multiplex immunofluorescence to measure dynamic changes in tumor-infiltrating lymphocytes and PD-L1 in early-stage breast cancer. Breast Cancer Research. 2021;23(1):2. doi: 10.1186/s13058-020-01378-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graeser M, Gluz O, Biehl C, Ulbrich-Gebauer D, Palatty J, Christgen M, et al. LBA2 Impact of RNA expression signatures and tumour infiltrating lymphocytes (TILs) for pathological complete response (pCR) and survival after 12 week de-escalated neoadjuvant pertuzumab + trastuzumab ± paclitaxel in the WSG-HER2+/HR- ADAPT trial. Annals of Oncology. 2021;32:S48. doi: 10.1016/j.annonc.2021.03.215. [DOI] [Google Scholar]
- 25.Harbeck N, von Schumann R, Kates RE, Braun M, Kuemmel S, Schumacher C, et al. Immune Markers and Tumor-Related Processes Predict Neoadjuvant Therapy Response in the WSG-ADAPT HER2-Positive/Hormone Receptor-Positive Trial in Early Breast Cancer. Cancers (Basel). 2021;13(19). doi: 10.3390/cancers13194884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurvitz SA, Caswell-Jin JL, McNamara KL, Zoeller JJ, Bean GR, Dichmann R, et al. Pathologic and molecular responses to neoadjuvant trastuzumab and/or lapatinib from a phase II randomized trial in HER2-positive breast cancer (TRIO-US B07). Nat Commun. 2020;11(1):5824. doi: 10.1038/s41467-020-19494-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used for this study, including quantitative multiplex immunofluorescence cell manifests and gene expression values, is available upon reasonable request from the corresponding author.