Abstract
Introduction
Recent studies have demonstrated that exosomes play roles in pathogenesis and in the treatment of various diseases. We explored the influence of exosomes released from Talaromyces marneffei (T. marneffei)‐infected macrophages on human macrophages to determine whether they play a role in the pathogenesis of T. marneffei infection.
Methods
Exosomes derived from macrophages infected with T. marneffei were extracted and characterized by transmission electron microscopy and western blot. Moreover, we examined exosomes that modulated IL‐10 and TNF‐α secretion and activation of p42 and p44 extracellular signal‐regulated kinase 1 and 2 (ERK1/2) and activation of autophagy.
Results
We found that exosomes promoted activation of ERK1/2 and autophagy, IL‐10 and TNF‐α secretion in human macrophages. Further, exosomes decreased the multiplication of T. marneffei in T. marneffei‐infected human macrophages. Interestingly, exosomes isolated from T. marneffei‐infected but not from uninfected macrophages can stimulate innate immune responses in resting macrophages.
Conclusion
Our studies are the first to demonstrate that exosomes isolated from T. marneffei‐infected macrophages can modulate the immune system to control inflammation, and we hypothesize that exosomes play significant roles in activation of ERK1/2 and autophagy, the replication of T. marneffei and cytokine production during T. marneffei infection.
Keywords: ERK1/2, exosomes, macrophages, Talaromyces marneffei
Our studies are the first to demonstrate that exosomes isolated from T. marneffei‐infected macrophages can induce a pro‐inflammatory response, and we hypothesize that exosomes play significant roles in activation of ERK1/2 and autophagy, the replication of T. marneffei and cytokine release during T. marneffei infection.

1. INTRODUCTION
Talaromyces marneffei, formerly named Penicillium marneffei is a significant emerging dimorphic fungus that can cause a severe systemic mycosis in humans across a narrow band of tropical South and Southeast Asia. 1 , 2 , 3 , 4 , 5
Recent studies have demonstrated that macrophages are the initial immune cells necessary for preventing and controlling infection with T. marneffei. 6 , 7 , 8 , 9 , 10 , 11 Further, previous study found that tumor necrosis factor‐a (TNF‐a) decreased the replication of T. marneffei in macrophages. 8 In addition, IL‐10 as a significant regulator of myeloid cells inhibits macrophages to produce cytokines. 12
ERK1/2 as a member of mitogen‐activated protein kinases (MAPKs) transduces the signals of various stimuli including mitogens, growth factors, and cytokines from the cell surface to the nucleus and regulate cytokine production. 13 ERK1/2 activation may limit the bacterial and fungal replication and plays important roles in many pathogens such as T. marneffei, Mycobacterium avium, Yersinia enterocolitica, and Candida albicans. 11 , 13 , 14 , 15 Besides, autophagy as a self‐degradative process was essential for the clearance of intracellular pathogens such as Shigella flexneri, Streptococcus pyogenes, Mycobacterium tuberculosis (M. tuberculosis) and T. marneffei. 16 , 17 , 18 , 19
Exosomes are small vesicles derived from many eukaryotic cells. Exosomes can be obtained from B cells, macrophages, dendritic cells, and natural killer cells, which are enriched in proteins of the tetraspanin family including CD63 and CD81. 20 Exosomes released from macrophages infected with M. tuberculosis promote both innate and acquired immune responses in vitro. 21 , 22
Although the modulatory effects of exosomes released from M. tuberculosis‐infected macrophages has been reviewed 23 and indicate that they can stimulate production of inflammatory mediators, little is known for the role of exosomes in ERK1/2 activation and autophagy, cytokine expression, and replication of T. marneffei in T. marneffei‐infected macrophages. In this research, the exosomes derived from T. marneffei‐infected macrophages were characterized and the effects of the exosomes on human macrophages in vitro were also explored.
2. MATERIALS AND METHODS
2.1. Reagents
Phospho‐ERK1/2, β‐actin, and ERK1/2 antibodies came from Cell Signaling Technology. Goat anti‐rabbit IgG and goat anti‐mouse IgG were obtained from Santa Cruz Biotechnology. LC‐3 antibody was purchased from Sigma Chemical Co.
2.2. T. marneffei fungal strain
T. marneffei strain, named B33w was purchased from Chinese Medicine Mycology Database. It grew on potato dextrose agar (PDA) at 25°C for 14 days. T. marneffei conidia were collected and washed with phosphate‐buffered saline before each experiment.
2.3. Isolation and culture of human macrophages
Peripheral blood mononuclear cells were isolated according to standard procedures described in our previous study by Ficoll‐Paque. The cells were incubated for 1 h at 37°C, and nonadherent cells were removed. Macrophages were obtained by culturing the monocytes for 7–8 days.
2.4. In vitro infection
Macrophages were infected with T. marneffei conidia at a multiplicity of infection of 5:1. Further, macrophages were treated with or without exosomes.
2.5. Exosomes isolation
Macrophages were stimulated with T. marneffei conidia for 48 h and then supernatants were centrifuged at 3000g for 15 min to remove cells and cell debris. The ExoQuick Exosome Precipitation Solution (System Biosciences [SBI]) was added to supernatants according to the procedure. Exosomes were quantified using the Micro BCA Protein Assay and stored at −80°C for following experiments.
2.6. Transmission electron microscopy
We detected exosomes by transmission electron microscopy. The prepared exosome eluate was stained with 3% phosphotungstic acid solution for 3–5 min and viewed on a JEOL‐Jem‐1200EX (JEOL) transmission electron microscope.
2.7. Western blot analysis
Macrophages were treated with 25 μg/mL exosomes within 120 min. For western blot analysis, proteins from cell lysates, as determined by the Micro BCA Protein Assay, were loaded on 12% SDS‐PAGE gels, electrophoresed, and transferred onto PVDF membrane. The membranes were probed for the primary antibody (anti‐CD63, anti‐CD9, anti‐CD81 anti‐ERK1/2, anti‐phospho‐ERK1/2, anti‐LC3, or anti‐β‐actin). Immunodetected bands were developed using chemiluminescent substrate.
2.8. Cytokine assays
Macrophages were treated with or without 12.5, 25, and 50 μg/mL exosomes for 24 h. Supernatants were harvested and kept at −80°C until analysis and were measured for TNF‐α and IL‐10 levels by enzyme‐linked immunosorbent assay (ELISA) using a commercial kit (R&D Systems).
2.9. Determination of colony forming units (CFUs)
To study the impact of exosomes on the multiplication of T. marneffei, macrophages were infected with T. marneffei containing with or without 25 and 50 μg/mL exosomes for 24 h. Lysates were serially diluted and plated on Sabouraud dextrose agar (SDA) at 37°C in triplicate. CFUs were counted after 3 days.
2.10. Statistical analysis
Data are shown as mean ± standard deviation by means of a Student's t test or one‐way analysis of variance using GraphPad Prism 5.0 (GraphPad Software, Inc.). Values of p < .05 were defined as statistically significant. All experiments were repeated at least three times.
3. RESULTS
3.1. Identification of exosomes derived from macrophages
TEM images showed that exosomes exhibited vesicle morphology and ranged in size from 30 to 100 nm (Figure 1A). The results of western blot analysis confirmed the expression of CD9, CD63, and CD81 in exosomes (Figure 1B).
Figure 1.
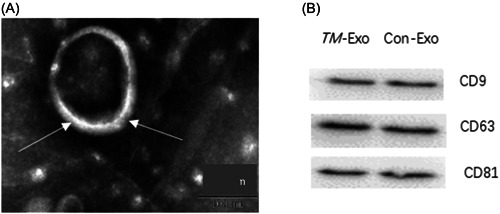
Identification of exosomes derived from Talaromyces marneffei‐infected macrophages. (A) The morphology of exosomes, as shown by transmission electron microscopy. (B) Expression levels of CD9, CD63, and CD81 in macrophage‐derived exosomes stimulated by T. marneffei (TM‐Exo), as shown by western blot analysis. Con‐Exo, exosomes derived from naïve macrophages.
3.2. Exosomes induced TNF‐α and IL‐10 production in human macrophages
To examine whether exosomes could promote macrophages activation, the production of TNF‐α (Figure 2A) and IL‐10 (Figure 2B) from human macrophages treated with exosomes was detected. The results indicated that TNF‐α and IL‐10 secretion was increased when compared with controls.
Figure 2.
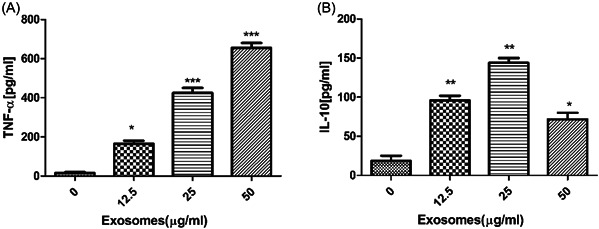
Exosomes induced TNF‐α and IL‐10 production in human macrophages. Macrophages were treated with different doses of exosomes. The levels of TNF‐α (A) and IL‐10 (B) were measured after the times indicated by ELISA. Data are expressed as the mean ± standard deviation by a one‐way analysis of variance. All experiments were repeated at least three times. TNF‐α, tumor necrosis factor‐a. *p < .05, **p < .01, ***p < .001.
3.3. Exosomes induced phosphorylation of ERK1/2 in human macrophages
To study the influence of exosomes on ERK1/2, the phosphorylation of ERK1/2 in human macrophages treated with exosomes was therefore evaluated (Figure 3A,B). Exosomes induced strong phosphorylation of ERK1/2 within 120 min. The peak of phosphorylation of ERK1/2 occurred within 60–120 min.
Figure 3.
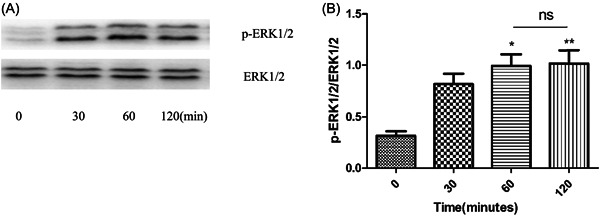
Exosomes induced phosphorylation of ERK1/2 in human macrophages. Human macrophages were stimulated with or without 25 μg/mL exosomes for the time interval indicated. The phosphorylation of ERK1/2 in macrophages were measured by Western blot (A) and quantification of ERK1/2 (B) was performed by densitometric analysis. Membranes were stripped and reprobed for total ERK 1/2. Data are expressed as the mean ± standard deviation by a one‐way analysis of variance. All experiments were repeated at least three times. ns, not significant. *p < .05, **p < .01.
3.4. Autophagy induction increased in human macrophages upon the exosomes
To investigate increased autophagy stimulated by exosomes, macrophages were incubated with or without exosomes for the time interval indicated. Along with the higher conversion of LC3B‐I to LC3B‐II in exosomes‐stimulated macrophages also increased, suggesting that exosomes promote autophagic flux in macrophages. Exosomes induced activation of autophagy within 120 min. The peak of autophagic flux occurred within 30–60 min (Figure 4A,B).
Figure 4.
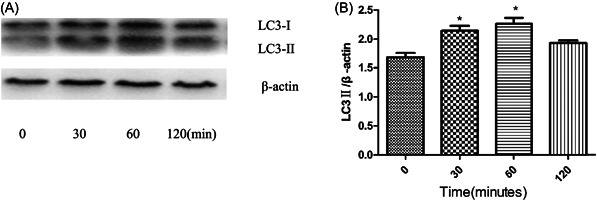
Autophagy induction increased in human macrophages upon the exosomes. Human macrophages were incubated with or without 25 μg/mL exosomes for the time interval indicated. (A) The levels of LC3B‐I, LC3B‐II protein in human macrophages were examined by Western blot and quantification of LC3B‐II (B) was performed by densitometric analysis. Data are expressed as the mean ± standard deviation by a one‐way analysis of variance. All experiments were repeated at least three times. *p < .05.
3.5. Effect of exosomes on intracellular replication of T. marneffei in human macrophages
As shown above, T. marneffei induced the release of exosomes within macrophages. To examine the effect of exosomes on the replication of T. marneffei, macrophages were pretreated with different concentrations of exosomes for 1 h, and then cultured with T. marneffei for 24 h. Exosomes decreased the replication of T. marneffei in human macrophages (Figure 5).
Figure 5.
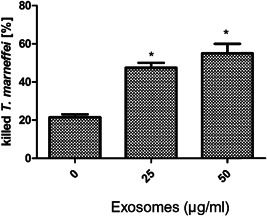
Effect of exosomes on intracellular replication of Talaromyces marneffei in human macrophages. Macrophages were pretreated with or without exosomes for 1 h, and then infected with T. marneffei for 24 h. The cells were lysed and plated onto SDA in serial dilutions for CFU assay. Data are expressed as the mean ± standard deviation by a one‐way analysis of variance. All experiments were repeated at least three times. *p < .05.
4. DISCUSSION
Exosomes are derived from cell endosomal membrane system and play a role in regulating immune responses. Previous studies have shown that exosomes released from M. tuberculosis‐infected cells can induce a pro‐inflammatory response when exposed to macrophages. 24 Further, exosomes were verified containing many marker proteins, such as heat shock proteins, CD63, CD9, CD81, and CD82. There were several studies on virulence factors of T. marneffei such as heat shock proteins, antioxidant enzymes, MP1p, and nutritional metabolism‐related enzymes. 25 , 26 , 27 , 28 However, the pathogenesis of T. marneffei has not been fully elucidated. Also, a recent study found that some proven or putative virulence factors, including heat shock proteins, MP1p, and peroxidase, were also identified in the proteome of T. marneffei‐derived extracellular vesicles. 29 In the study, we found that exosomes were derived from T. marneffei‐infected macrophages and expressed CD63, CD9, and CD81. Importantly, exosomes promoted activation of autophagy and ERK1/2, IL‐10, and TNF‐α secretion in human macrophages. Moreover, exosomes reduced the replication of T. marneffei in T. marneffei‐infected human macrophages.
In our previous study, we have found that T. marneffei causes a significant increase in the secretion of both IL‐10 and TNF‐a by macrophages. 11 Interestingly, we found similar results, particularly regarding TNF‐a and IL‐10, when macrophages were treated with exosomes from T. marneffei‐infected macrophages. This is similar to the results that exosomes released from infected cells contain M. avium glycopeptidolipids and are pro‐inflammatory. 30
ERK1/2 is activated by a variety of infections and takes part in the induction of innate immunity. Previous studies showed that both C. albicans and Saccharomyces cerevisiae blastoconidia in J774 cells activated the ERK1/2 pathway. 31 Also, T. marneffei induced ERK1/2 phosphorylation in human macrophages. 11 Further, the ERK signaling pathway has been reported to be the major positive regulator of autophagy. 15 In addition, inhibition of ERK1/2 pathway increases the replication of T. marneffei. Numerous studies reported that exosomes isolated from cells infected with various intracellular pathogens contain microbial components and could promote antigen presentation and macrophage activation. 32 , 33 To determine the role of exosomes in activation to macrophages, we examined the impact of exosomes on macrophages. In this study, we found that exosomes induced phosphorylation of ERK1/2. Besides, exosomes could inhibit the replication of T. marneffei. These results showed that exosomes, causing of ERK1/2 activation and translocation to nucleus, which might be a significant regulator of host‐pathogen interactions.
As we know, autophagy is a nonapoptotic form of programmed cell death and it has played an important role in cell immune responses. In this study, we explored the impact of exosomes on the formation of autophagosomes in macrophages. LC3 is the only known mammalian protein that associates with the autophagosome membrane. Upon activation of autophagy, LC3 transformed from LC3I into LC3II. 34 , 35 In this research, exosomes not only induced the LC3II protein, but also importantly increased the LC3II/β‐actin ratio. Because the amount of LC3II is connected with the number of autophagosomes, it may be as a good indicator of autophagosome formation. 36
In general, our study indicated that exosomes released from T. marneffei‐infected macrophages can promote macrophage activation, release of cytokines, and killing T. marneffei (Figure 6). Our results suggest that exosomes play vital roles in immune response. The study could provide a novel mediator for host‐pathogen interactions and the development of novel approaches to defend this pathogen. Further studies are needed to test whether virulence factors were from T. marneffei‐derived exosomes.
Figure 6.
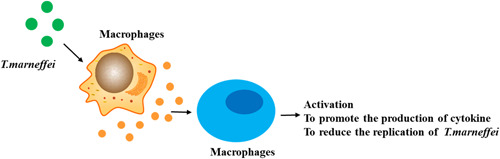
A model that shows the effects of exosomes from Talaromyces marneffei‐infected macrophages on human macrophages. Exosomes released from T. marneffei‐infected macrophages can promote macrophage activation, release of cytokines, and killing T. marneffei.
AUTHOR CONTRIBUTIONS
Shan Feng: Conceptualization; methodology. Hong Ren: Conceptualization; resources. Wenhao Cheng: Methodology; software. Guangquan Ji: Investigation; methodology; writing—original draft. Renqiong Chen: writing—original draft; software.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This study was supported by grants (81602769) from the National Natural Science Foundation of China and Science Foundation and Lianyungang Science and Technology Development Project (SF2236) and Science Foundation and Research and Development Fund project, Kangda College of Nanjing Medical University (KD2021KYJJZD003 and KD2021KYJJZD013).
Ji G, Feng S, Ren H, Chen W, Chen R. Exosomes released from macrophages infected with Talaromyces marneffei activate the innate immune responses and decrease the replication. Immun Inflamm Dis. 2023;11:e881. 10.1002/iid3.881
Contributor Information
Shan Feng, Email: fengshan@163.com.
Hong Ren, Email: renhong417@163.com.
Renqiong Chen, Email: chenrenqiong718@126.com.
REFERENCES
- 1. Vanittanakom N, Cooper Jr., CR , Fisher MC, Sirisanthana T. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev. 2006;19:95‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ustianowski AP, Sieu TP, Day JN. Penicillium marneffei infection in HIV. Curr Opin Infect Dis. 2008;21:31‐36. [DOI] [PubMed] [Google Scholar]
- 3. Houbraken J, de Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199‐249. [DOI] [PubMed] [Google Scholar]
- 4. Hu Y, Zhang J, Li X, et al. Penicillium marneffei infection: an emerging disease in mainland China. Mycopathologia. 2013;175:57‐67. [DOI] [PubMed] [Google Scholar]
- 5. Cooper CR, Vanittanakom N. Insights into the pathogenicity of Penicillium marneffei . Future Microbiol. 2008;3:43‐55. [DOI] [PubMed] [Google Scholar]
- 6. Chen R, Ji G, Ma T, Huang X, Ren H, Xi L. Role of intracellular free calcium in killing Penicillium marneffei within human macrophages. Microb Pathog. 2015;83‐84:29‐34. [DOI] [PubMed] [Google Scholar]
- 7. Cogliati M, Roverselli A, Boelaert JR, Taramelli D, Lombardi L, Viviani MA. Development of an in vitro macrophage system to assess Penicillium marneffei growth and susceptibility to nitric oxide. Infect Immun. 1997;65:279‐284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rongrungruang Y, Levitz SM. Interactions of Penicillium marneffei with human leukocytes in vitro. Infect Immun. 1999;67:4732‐4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen R, Ji G, Wang L, Ren H, Xi L. Activation of ERK1/2 and TNF‐α production are regulated by calcium/calmodulin signaling pathway during Penicillium marneffei infection within human macrophages. Microb Pathog. 2016;93:95‐99. [DOI] [PubMed] [Google Scholar]
- 10. Kudeken N, Kawakami K, Saito A. Role of superoxide anion in the fungicidal activity of murine peritoneal exudate macrophages against Penicillium marneffei . Microbiol Immunol. 1999;43:323‐330. [DOI] [PubMed] [Google Scholar]
- 11. Chen R, Li X, Lu S, et al. Role of extracellular signal‐regulated kinases 1 and 2 and p38 mitogen‐activated protein kinase pathways in regulating replication of Penicillium marneffei in human macrophages. Microb Infect. 2014;16:401‐408. [DOI] [PubMed] [Google Scholar]
- 12. Mosmann TR. Properties and functions of interleukin‐10. Adv Immunol. 1994;56:1‐26. [PubMed] [Google Scholar]
- 13. Shiratsuchi H, Ellner JJ, Basson MD. Extracellular‐regulated kinase activation regulates replication of Mycobacterium avium intracellularly in primary human monocytes. Cell Tissue Res. 2008;332:237‐244. [DOI] [PubMed] [Google Scholar]
- 14. Ruckdeschel K, Machold J, Roggenkamp A, et al. Yersinia enterocolitica promotes deactivation of macrophage mitogen‐activated protein kinases extracellular signal‐regulated Kinase‐1/2, p38, and c‐Jun NH2‐terminal kinase. J Biol Chem. 1997;272:15920‐15927. [DOI] [PubMed] [Google Scholar]
- 15. Ibata‐Ombetta S, Jouault T, Trinel PA, Poulain D. Role of extracellular signal‐regulated protein kinase cascade in macrophage killing of Candida albicans . J Leukoc Biol. 2001;70:149‐154. [PubMed] [Google Scholar]
- 16. Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727‐731. [DOI] [PubMed] [Google Scholar]
- 17. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753‐766. [DOI] [PubMed] [Google Scholar]
- 18. Nakagawa I, Amano A, Mizushima N, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037‐1040. [DOI] [PubMed] [Google Scholar]
- 19. Chen R, Ji G, Xi L, et al. Role of autophagy in regulating the immune response of dendritic cells to Talaromyces marneffei infection. Microb Pathog. 2018;123:120‐125. [DOI] [PubMed] [Google Scholar]
- 20. Kalra H, Simpson RJ, Ji H, et al. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10:e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh PP, LeMaire C, Tan JC, Zeng E, Schorey JS. Exosomes released from. M. tuberculosis infected cells can suppress IFN‐γ mediated activation of naive macrophages. PLoS ONE. 2011;6:e18564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giri PK, Kruh NA, Dobos KM, Schorey JS. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis‐infected and culture filtrate protein treated macrophages. Proteomics. 2010;10:3190‐3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh PP, Li L, Schorey JS. Exosomal RNA from Mycobacterium tuberculosis‐infected cells is functional in recipient macrophages. Traffic. 2015;16:555‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234‐3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kummasook A, Kummasook A, Pongpom P, Kummasook A, Pongpom P, Vanittanakom N. Cloning, characterization and differential expression of an hsp70 gene from the pathogenic dimorphic fungus, Penicillium marneffei . DNA Seq. 2007;18:385‐394. [DOI] [PubMed] [Google Scholar]
- 26. Vanittanakom N, Pongpom M, Praparattanapan J, Cooper CR, Sirisanthana T. Isolation and expression of heat shock protein 30 gene from Penicillium marneffei . Med Mycol. 2009;47:521‐526. [DOI] [PubMed] [Google Scholar]
- 27. Woo PCY, Tam EWT, Chong KTK, et al. High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei . FEBS J. 2010;277:3750‐3758. [DOI] [PubMed] [Google Scholar]
- 28. Sze KH, Lam WH, Zhang H, et al. Talaromyces marneffei Mp1p is a virulence factor that binds and sequesters a key proinflammatory lipid to dampen host innate immune response. Cell Chem Biol. 2017;24:182‐194. [DOI] [PubMed] [Google Scholar]
- 29. Yang B, Wang J, Jiang H, et al. Extracellular vesicles derived from Talaromyces marneffei yeasts mediate inflammatory response in macrophage cells by bioactive protein components. Front Microbiol. 2021;11:603183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain mycobacterium avium glycopeptidolipids and are proinflammatory. J Biol Chem. 2007;282:25779‐25789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu P, Lai D, Lu P, Gao J, He H. ERK and Akt signaling pathways are involved in advanced glycation end product‐induced autophagy in rat vascular smooth muscle cells. Int J Mol Med. 2012;29:613‐618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blaser MJ, Berg DE. Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest. 2001;107:767‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181‐206. [DOI] [PubMed] [Google Scholar]
- 35. Sato K, Tsuchihara K, Fujii S, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677‐9684. [DOI] [PubMed] [Google Scholar]
- 36. Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720‐5728. [DOI] [PMC free article] [PubMed] [Google Scholar]


