Abstract
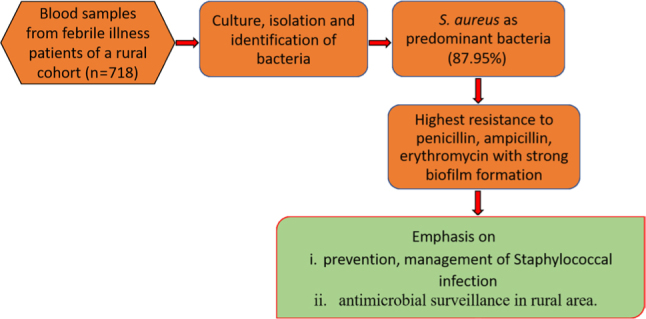
This study investigated the major pathogens in fever patients’ blood in a rural cohort and characterized its virulence. A total of 718 blood samples received from IPD/OPD (inpatient department/outpatient department) patients with H/O (history of) fever were cultured, and 73 out of 83 culture-positive samples were identified as Staphylococcus aureus. The isolates showed higher resistance to penicillin, most being multidrug resistant. They formed biofilm in vitro, and 27.4% of the isolates were strong biofilm producers. They were sensitive towards linezolid, gentamicin, and tetracycline. The findings emphasize the necessity of preventing and managing staphylococcal infection and regular antimicrobial surveillance in rural areas.
Keywords: S. aureus, biofilm, febrile cases, resistance, rural area
Staphylococcus aureus is a common bacterial pathogen responsible for community and hospital-acquired infections. The pathogen often becomes difficult to treat because of its property to develop resistance against antimicrobials. Infections by multidrug-resistant S. aureus result in a more extended hospital stay, increased treatment cost, further spreading of resistant bacteria, and higher mortality (Tarai et al. 2013; Chatterjee et al. 2018; Mehta et al. 2020). Factors such as overprescription of antimicrobials, over-the-counter purchase of drugs, medicines recommended by informal health care providers, polypharmacy practices, etc., influence and accentuate this problem further (Ansari et al. 2014). Such irrational antimicrobial use is even more common in remote and rural areas because of limited access to and availability of standard health care (Yau et al. 2021).
Many multidrug-resistant bacteria form biofilms to survive in unfavorable conditions and use them as a defense mechanism. Biofilms are sessile microbial colonies surrounded by extracellular matrix and attached to non-living or living subjects. They are associated with antibiotic tolerance and the production of virulence factors (Young et al. 2002). Methicillin-resistant S. aureus (MRSA) is often resistant to many other antibiotics. The isolates forming biofilm can be more resistant than those biofilm-no-forming isolates. In a study by Neopane et al. (2018), biofilm-producing S. aureus isolated from pus samples showed higher antibiotic resistance than non-biofilm-producing isolates. Notably, 86.7% of biofilm-producing S. aureus isolates were multidrug resistant, whereas all the biofilm-no-producing S. aureus isolates were non-MDR (Neopane et al. 2018). However, studies on characterizing these virulent properties of S. aureus in rural areas are scanty.
The present study was part of a surveillance project on “Enteric Fever” carried out among people of a rural cohort established under the model rural health research unit (MRHRU) in Odisha, India. In the current paper, we present our findings on the antimicrobial resistance patterns and biofilm-forming ability of S. aureus isolated from febrile illness patients of a rural cohort.
Under the study, 718 consented eligible participants (febrile patients suspected of enteric fever) visiting four public healthcare facilities in the study area were enrolled from December 2020 to December 2021. Patients over six months presenting with fever were included, and those who did not give written consent and were severely sick without fever were excluded. While the sample size was calculated to determine the prevalence of enteric fever, the primary objective of the present article focused on understanding the antimicrobial resistance pattern and the biofilm-forming characteristics of the isolates among the S. aureus-positive samples.
Before the data and sample collection, the purpose of the study was explained to all participants, and their written informed consent was obtained. Socio-demographic data and clinical history were recorded using a standardized questionnaire developed by the National Surveillance System for Enteric Fever in India, with the help of the Android-based open data kit tool. Blood samples from the study participants were collected aseptically (3 ml from children and 5 ml from adults) by a trained phlebotomist, inoculated into BACTEC culture bottles, and immediately transported to the MRHRU laboratory for further processing. Blood cultures of S. aureus were included in the present study for further characterization. This study was approved by the Institutional Human Ethics Committee at ICMR-RMRC, Bhubaneswar, vide letter no. ICMR-RMRCB/IHEC-2019.
All the samples were incubated in BD BACTEC™ FX 40 Automated Blood Culture System (BD, USA). After incubation at 37°C, when the culture was positive, the colonies were streaked onto MacConkey agar and blood agar plates (Hi Media Laboratories, India). The inoculated plates were incubated for 48 h at 37°C, followed by Gram staining. The pure cultures were examined for colony morphology and biochemical characteristics (catalase and coagulase tests) to identify the bacteria.
Antibiotic susceptibility profiles of the isolates were determined by the Kirby-Bauer disk diffusion technique using Hi-media antibiotic disks following the criteria set by CLSI (CLSI 2018). Antibiotics, including penicillin (10 μg), erythromycin (15 μg/ml), ampicillin (10 μg), azithromycin (15 μg), cefoxitin (30 μg), co-trimoxazole (25 μg), clindamycin (30 μg), teicoplanin (30 μg), ciprofloxacin (5 μg), vancomycin (30 μg), chloramphenicol (30 μg), levofloxacin (5 μg), tetracycline (30 μg), gentamicin (10 μg), and linezolid (30 μg) were used to determine the antibiogram.
MRSA identification test was performed in accordance with the criteria of Clinical Laboratory Standard Institute (CLSI) 2020 using cefoxitin (30 μg) disk diffusion assay (Weinstein and Lewis 2020). The isolates showing the zone of inhibition of ≥ 22 mm, 21–22 mm, and ≤ 21 mm, respectively, were considered sensitive, intermediate, and resistant.
Biofilm formation assay for all isolates was carried out using the microplate method described by Aslantaş and Demir (2016). Individual isolates were grown in tryptic soy broth (TSB) overnight at 37°C. When the culture’s optical density (OD) reached 0.5, the suspension was diluted with 1:10 in TSB with 1% dextrose, and 200 μl was put in each 96-well polystyrene microtiter plate in triplicate. Wells with TSB served as a negative control. The plate was incubated at 37°C for 48 h without shaking. After incubation, the planktonic cells were carefully removed by micropipette aspiration and gently washed two times with sterile phosphate-buffered saline (PBS) to remove nonadherent cells. Methanol in the volume of 200 μl was added to each well for 10 min, and then, the biofilms were stained with 0.1% (w/v) crystal violet for 10 min. The plates were washed with water thrice and air-dried for 2 h. The stained biomass was dissolved with 200 μl 30% (v/v) acetic acid for 10 mins, and then, 150 μl from each well was transferred to new flat-bottomed microtiter plate to measure the OD at 570 nm using a microplate reader (Erba Mannheim, Germany). The result was analyzed as described by Stepanović et al. (2007). The biofilm formation ability was determined by comparing the OD of the isolates to that of the control and the cut-off value (ODc). The isolates were categorized into four groups: no biofilm producers (OD ≤ ODc), weak biofilm producers (ODc<OD≤2×ODc), moderate biofilm producers (2×ODc<OD≤4×ODc), and strong biofilm producers (OD>4×ODc) (Stepanović et al. 2007).
The univariate descriptive statistical analysis was carried out to determine the socio-demographic and clinical parameters’ distribution of the patients included in this study and those found positive for S. aureus.
Detailed information on patients is provided in Table SI. The age of 718 study participants ranged between 11 months to 95 years, with a median age of 35. Among the participants, 397 (55.29%) were male. The median duration of fever was three days (Table SII A). Out of 718 blood samples, 73 (10.2%) showed the presence of S. aureus. The median age of those 73 patients was 42, and 39 (53.42%) were male (Table SII B).
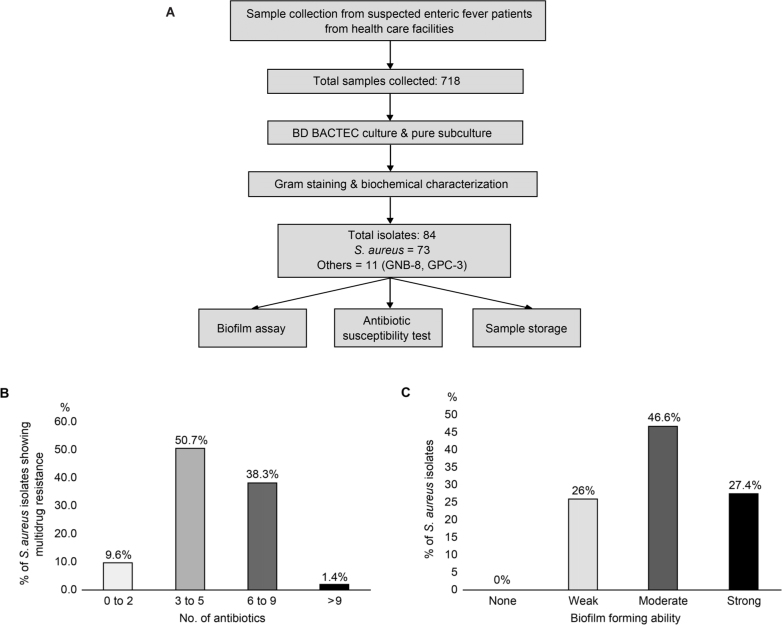
The workflow for sample collection and their investigation is presented in Fig. 1A. Out of 718 blood samples initially incubated in the BD BACTEC™ FX 40 Automated Blood Culture System, 84 (11.7%) were found positive for bacterial growth. Based on the colony characteristics, Gram stain, and biochemical properties S. aureus was identified in 73 samples.
Fig. 1.
A) Schematic presentation of methodology adopted for the study; B) figure represents the percentage of Staphylococcus aureus isolates showing resistance to multiple antibiotics. They were divided into four groups: resistant to two antibiotics, resistant to three to five antibiotics, resistant to six to nine antibiotics, and resistant to more than nine antibiotics; C) figure represents the percentage of S. aureus isolates showing biofilm forming ability as weak, moderate and strong biofilm producers.
All S. aureus isolates were found to be sensitive to linezolid (100%), whereas the sensitivity toward other antibiotics varied among isolates. The percentage of isolates sensitive, intermediate, or resistant to different antibiotics is detailed in Table I. The majority of S. aureus isolates were resistant to penicillin (65 isolates, 89%), followed by ampicillin (64 isolates, 88%), erythromycin (57 isolates, 78%), and azithromycin (52 isolates, 71%). Upon further analysis of S. aureus isolates resistant to multiple antibiotics, seven (9.6%) were found to be resistant to at least two antibiotics, 37 (50.7%) were resistant to three to five antibiotics, 28 (38.3%) were resistant to six to nine antibiotics, and one (1.4%) was resistant to nine antibiotics. The resistance of isolates to a number of antibiotics is outlined in Fig. 1B. Among all S. aureus isolates, 40 (54.8%) were found to be MRSA.
Table I.
Antibiotic susceptibility pattern of Staphylococcus aureus (n = 73).
| Name of antibiotics | S | I | R |
|---|---|---|---|
| Penicillin | 8 (11%) | 0 (0%) | 65 (89%) |
| Ampicillin | 9 (12%) | 0 (0%) | 64 (88%) |
| Erythromycin | 9 (12%) | 7 (10%) | 57 (78%) |
| Azithromycin | 18 (25%) | 3 (4%) | 52 (71%) |
| Cefoxitin | 33 (45%) | 0 (0%) | 40 (55%) |
| Co-trimoxazole | 35 (48%) | 2 (3%) | 36 (49%) |
| Clindamycin | 52 (71%) | 11 (15%) | 10 (14%) |
| Teicoplanin | 54 (74%) | 8 (11%) | 11 (15%) |
| Ciprofloxacin | 59 (81%) | 2 (3%) | 12 (16%) |
| Vancomycin | 58 (81%) | 10 (12%) | 5 (7%) |
| Chloramphenicol | 62 (85%) | 5 (7%) | 6 (8%) |
| Levofloxacin | 62 (85%) | 5 (7%) | 6 (8%) |
| Tetracycline | 67 (92%) | 0 (0%) | 6 (8%) |
| Gentamicin | 71 (97%) | 0 (0%) | 2 (3%) |
| Linezolid | 73 (100%) | 0 (0%) | 0 (0%) |
S – susceptible, I – intermediate, R – resistant
All S. aureus isolates were found to form biofilm in the current study. The percentage of weak, moderate, and strong biofilm-forming isolates were 26% (19 isolates), 46.6% (34 isolates), and 27.4% (20 isolates) (Fig. 1C). Further analysis of the antibiotic sensitivity status in strong biofilm producers (20 isolates), the majority of them were found to be sensitive to linezolid (20 isolates, 100%), gentamicin (19 isolates, 95%), and tetracycline (19 isolates, 95%) (Table II).
Table II.
Details of antibiotic susceptibility pattern of strong biofilm-forming Staphylococcus aureus.
| Sample ID | Antibiotic |
|---|---|
| 38 | COT, VA, C, TEI, LZ, CD, TE |
| 116 | GEN, COT, CIP, C, LZ, TE, LE |
| 143 | GEN, COT, CIP, VA, C, LZ, TE, LE |
| 153 | GEN, COT, CIP, VA, C, CX, LZ, CD, TE, LE |
| 156 | GEN, COT, CIP, VA, C, TEI, CX, LZ, CD, TE, LE |
| 161 | GEN, COT, AMP, CIP, AZM, VA, C, TEI, P, CX, E, LZ, CD, TE, LE |
| 221 | GEN, CIP, C, TEI, LZ, CD, TE, LE |
| 229 | GEN, CIP, VA, C, TEI, LZ, CD, TE, LE |
| 231 | GEN, CIP, VA, C, TEI, CX, LZ, CD, TE, LE |
| 236 | GEN, CIP, VA, C, TEI, LZ, CD, TE, LE |
| 241 | GEN, COT, AMP, CIP, AZM, VA, C, TEI, LZ, TE, LE |
| 242 | GEN, CIP, VA, C, TEI, CX, LZ, CD, TE, LE |
| 271 | GEN, CIP, AZM, VA, C, TEI, LZ, CD, LE |
| 277 | GEN, CIP, C, LZ, CD, TE, LE |
| 350 | GEN, COT, CIP, VA, LZ, TE, LE |
| 352 | GEN, VA, TEI, LZ, TE |
| 354 | GEN, COT, CIP, AZM, VA, TEI, E, LZ, CD, TE, LE |
| 382 | GEN, CIP, VA, C, TEI, CX, LZ, CD, TE, LE |
| 498 | GEN, COT, CIP, VA, C, TEI, CX, LZ, CD, TE, LE |
| 515 | GEN, AMP, CIP, VA, C, TEI, P, CX, LZ, CD, TE, LE |
Most of the strong biofilm-producing S. aureus were sensitive to linezolid, gentamicin, and tetracycline.
Access to high-quality medical services is a great challenge in low-income and middle-income countries and often leads to lower rate of disease screening, diagnosis, and treatment (Wilson et al. 2018). To the best of our knowledge, this is the first study carried out in a rural cohort of eastern India to understand the antibiotic resistance and biofilm-forming characteristics of S. aureus isolated from febrile cases. Among total of 718 febrile cases, 84 (11.7%) showed blood culture positive for bacterial infection showing a higher prevalence of bacterial infection among febrile cases. S. aureus isolates were found in 73 (86.9%) samples, implying the predominance of this species among febrile patients. In contrast to our finding, a study of hospital-visiting patients in Nepal reported that out of 666 samples showing bacterial growth, 133 (20%) were S. aureus (Sapkota et al. 2019). Another study reported the presence of 36.26% S. aureus among culture-positive clinical samples collected from patients of a tertiary healthcare hospital in India (Pappu et al. 2020). The higher prevalence in our study could be specific to our rural study setting.
The antimicrobial stewardship program has been restricted owing to the challenges concerning data collection, access, awareness, and lack of accurate diagnosis tools, thereby limiting information on Anti-Microbial Resistance (AMR) burden. A higher resistance of S. aureus to the penicillin group of antibiotics found in our study was similar to the previous studies (Yılmaz and Aslantaş 2017; Islam et al. 2021). The most probable reason for this phenomenon might be the overprescribing such drugs in our study areas. The resistance of S. aureus isolates to ampicillin and erythromycin found in the present study was higher than in other studies. The difference could be attributed to the situation of our settings (Duran et al. 2012; Yılmaz and Aslantaş 2017). Another important finding of our study is the high prevalence of MRSA, which certainly requires further investigation to explore the reasons and take action to prevent it. Another study reported that 47.4% of S. aureus phenotype was MRSA, and 59.2% were MDR among isolates from patients visiting the hospital. Importantly, the rate of multi-drug and methicillin resistance was high among biofilm-producing S. aureus compared to biofilm-non-producing S. aureus (Belbase et al. 2017).
Biofilm formation in clinical isolates of S. aureus has been studied widely because of the association between biofilm, disease chronicity, and antimicrobial resistance. A recent study reported that 67.07% of MRSA obtained from different clinical samples in India produce biofilms (Chaudhary et al. 2021). Gupta et al. (2014) carried out a similar study on fever patients in India and found 84.28% of Staphylococci isolates as biofilm producers. Among those, 21.4%, 62.8%, and 15.8% were strong, moderate, and non-biofilm producers, respectively (Gupta et al. 2014). It is interesting to note that, in our study, all the isolates formed biofilm, and 27.4% were strong biofilm producers. Most of the strong biofilm producers were sensitive to linezolid, gentamicin, and tetracycline, and this finding resonates with another study (Mottola et al. 2016). Linezolid has been found to be an efficient agent against biofilms formed by MRSA isolates in vitro (Martínez et al. 2016). With insufficient knowledge of the anti-biofilm mechanism of action of linezolid, more studies are required to understand it better. In another study, strong biofilm-producing S. aureus showed the highest sensitivity towards cefoxitin, followed by clindamycin whereas non-strong biofilm producers mainly were susceptible to cefoxitin and ciprofloxacin (Derakhshan et al. 2021). Gaire et al. (2021) reported that the highest percentage of weak biofilm-producing S. aureus were resistant to penicillin, followed by erythromycin. Likely, all the moderate biofilm producers showed resistance to penicillin, followed by cefoxitin and erythromycin. However, the strong biofilm producer was found to be resistant to most of the antibiotics used in the study (Gaire et al. 2021).
We found S. aureus as the primary cause of bacteremia in rural areas. Its high antimicrobial resistance with biofilm-producing ability is highly alarming. Prescription of antibiotics such as penicillin, ampicillin, erythromycin, and azithromycin must be done judiciously in the locality. Regular investigation of S. aureus infections and antibiotic sensitivity would help the physicians prescribe the correct antibiotic if required.
Supplementary Material
Supplementary Material Details
Acknowledgments
The authors are grateful to the medical officers, doctors, and nurses of the hospitals included in the study for their support in collecting blood samples. The authors also thank Mr. Soumya Ranjan Nayak for his help with the statistical analysis carried out in the study.
Funding Statement
Funding The study was supported by funding received from the Indian Council of Medical Research (No. VIR/28/2019/ECD-I).
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Contributor Information
Subrata Kumar Palo, Email: drpalsubrat@gmail.com.
Sanghamitra Pati, Email: drsanghamitra12@gmail.com.
Literature
- Ansari S, Nepal HP, Gautam R, Rayamajhi N, Shrestha S, Upadhyay G, Acharya A, Chapagain ML Threat of drug resistant Staphylococcus aureus to health in Nepal BMC Infect Dis. 2014 NaN14:157. doi: 10.1186/1471-2334-14-157. . . ; : [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslantaş Ö, Demir C Investigation of the antibiotic resistance and biofilm-forming ability of Staphylococcus aureus from subclinical bovine mastitis cases J Dairy Sci. 2016 NaN99(11):8607. doi: 10.3168/jds.2016-11310. . . ; ( ): –. [DOI] [PubMed] [Google Scholar]
- Belbase A, Pant ND, Nepal K, Neupane B, Baidhya R, Baidya R, Lekhak B Antibiotic resistance and biofilm production among the strains of Staphylococcus aureus isolated from pus/wound swab samples in a tertiary care hospital in Nepal Ann Clin Microbiol Antimicrob. 2017 NaN16(1):15. doi: 10.1186/s12941-017-0194-0. . . ; ( ): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Rai S, Guddattu V, Mukhopadhyay C, Saravu K Is methicillin-resistant Staphylococcus aureus infection associated with higher mortality and morbidity in hospitalized patients? A cohort study of 551 patients from South Western India Risk Manag Healthc Policy. 2018 NaN11:243. doi: 10.2147/RMHP.S176517. . . ; : –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary BL, Bisht D, Faujdar SS Biofilm formation and its association with antibiotic susceptibility pattern in methicillin-resistant Staphylococcus aureus isolates J Pure Appl Microbiol. 2021;15(4):2041. doi: 10.22207/JPAM.15.4.26. . . ; ( ): –. [DOI] [Google Scholar]
- CLSI . Performance standards for antimicrobial disk susceptibility tests, 13th ed. CLSI standard M02. Wayne (USA): Clinical and Laboratory Standards Institute; 2018. . . : ; . [Google Scholar]
- Derakhshan S, Navidinia M, Haghi F Antibiotic susceptibility of human-associated Staphylococcus aureus and its relation to agr typing, virulence genes, and biofilm formation BMC Infect Dis. 2021 NaN21(1):627. doi: 10.1186/s12879-021-06307-0. . . ; ( ): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran N, Ozer B, Duran GG, Onlen Y, Demir C Antibiotic resistance genes and susceptibility patterns in staphylococci Indian J Med Res. 2012 NaN135(3):389. . . ; ( ): –. . [PMC free article] [PubMed] [Google Scholar]
- Gaire U, Thapa Shrestha U, Adhikari S, Adhikari N, Bastola A, Rijal KR, Ghimire P, Banjara MR Antibiotic susceptibility, biofilm production, and detection of mecA gene among Staphylococcus aureus isolates from different clinical specimens Diseases. 2021 NaN9(4):80. doi: 10.3390/diseases9040080. . . ; ( ): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Gupta P, Mittal G, Agarwal RK, Goyal R Detection of biofilm production in blood culture isolates of Staphylococci Int J Med Res Health Sci. 2015;4(1):22. doi: 10.5958/2319-5886.2015.00004.1. . . ; ( ): –. [DOI] [Google Scholar]
- Islam MA, Uddin MS, Islam MJ, Ahmed MU, Alam MM Investigation of antibiotic resistance pattern of Staphylococcus aureus in clinical samples of animals and humans from selective areas of Bangladesh Bangl J Vet Med. 2021;19(1):9. doi: 10.33109/bjvmjj21vph1. . . ; ( ): –. [DOI] [Google Scholar]
- Martínez SR, Rocca DM, Aiassa V, Becerra MC Linezolid as an eradication agent against assembled methicillin-resistant Staphylococcus aureus biofilms RSC Adv. 2016;6(103):101023. doi: 10.1039/C6RA19670E. . . ; ( ): –. [DOI] [Google Scholar]
- Mehta Y, Hegde A, Pande R, Zirpe KG, Gupta V, Ahdal J, Qamra A, Motlekar S, Jain R Methicillin-resistant Staphylococcus aureus in intensive care unit setting of India: a review of clinical burden, patterns of prevalence, preventive measures, and future strategies Indian J Crit Care Med. 2020 NaN24(1):55. doi: 10.5005/jp-journals-10071-23337. . . ; ( ): –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottola C, Matias CS, Mendes JJ, Melo-Cristino J, Tavares L, Cavaco-Silva P, Oliveira M Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections BMC Microbiol. 2016 NaN16(1):119. doi: 10.1186/s12866-016-0737-0. . . ; ( ): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y In vitro biofilm formation by Staphyloc occus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance Int J Gen Med. 2018 NaN11:25. doi: 10.2147/IJGM.S153268. . . ; : –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu RK, Kumar R, Poddar CK, Singh MN Methicillin resistance Staphylococcus aureus isolated from surgical site infection in a tertiary care hospital, Koshi region (Northern Bihar), India J Evid Based Med Healthc. 2020 NaN7(19):927. doi: 10.18410/jebmh/2020/203. . . ; ( ): –. [DOI] [Google Scholar]
- Sapkota J, Sharma M, Jha B, Bhatt CP Prevalence of Staphylococcus aureus isolated from clinical samples in a tertiary care hospital: a descriptive cross-sectional study JNMA J Nepal Med Assoc. 2019 NaN57(220):398. doi: 10.31729/jnma.4673. . . ; ( ): –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Cirković I, Ruzicka F Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci APMIS. 2007 NaN115(8):891. doi: 10.1111/j.1600-0463.2007.apm_630.x. . . ; ( ): –. [DOI] [PubMed] [Google Scholar]
- Tarai B, Das P, Kumar D Recurrent challenges for clinicians: emergence of methicillin-resistant Staphylococcus aureus, vancomycin resistance, and current treatment options J Lab Physicians. 2013 NaN5(2):71. doi: 10.4103/0974-2727.119843. . . ; ( ): –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein MP, Lewis JS, 2nd The Clinical and Laboratory Standards Institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes J Clin Microbiol. 2020 NaN58(3):e01864. doi: 10.1128/JCM.01864-19. . . ; ( ): - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ML, Fleming KA, Kuti MA, Looi LM, Lago N, Ru K Access to pathology and laboratory medicine services: a crucial gap Access to pathology and laboratory medicine services: a crucial gap. 2018 NaN391(10133):1927. doi: 10.1016/S0140-6736(18)30458-6. . . ; ( ): –. [DOI] [PubMed] [Google Scholar]
- Yau JW, Thor SM, Tsai D, Speare T, Rissel C Antimicrobial stewardship in rural and remote primary health care: a narrative review Antimicrob Resist Infect Control. 2021 NaN10(1):105. doi: 10.1186/s13756-021-00964-1. . . ; ( ): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yılmaz EŞ, Aslantaş Ö Antimicrobial resistance and underlying mechanisms in Staphylococcus aureus isolates Asian Pac J Trop Med. 2017 NaN10(11):1059. doi: 10.1016/j.apjtm.2017.10.003. . . ; ( ): –. [DOI] [PubMed] [Google Scholar]
- Young D, Hussell T, Dougan G Chronic bacterial infections: living with unwanted guests Nat Immunol. 2002 NaN3(11):1026. doi: 10.1038/ni1102-1026. . . ; ( ): –. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material Details