Abstract
We have investigated the effect of single amino acid substitutions of conserved arginines on the catalytic activities of the human Ogg1 protein (α-hOgg1-Ser326) (wild-type α-hOgg1). Mutant forms of hOgg1 with mutations Arg46→Gln (α-hOgg1-Gln46) and Arg154→His (α-hOgg1-His154) have previously been identified in human tumors. The mutant proteins α-hOgg1-Gln46 and α-hOgg1-His154 were expressed in Escherichia coli and purified to homogeneity. The substrate specificities of these proteins and wild-type α-hOgg1 were investigated using γ-irradiated DNA and the technique of gas chromatography/isotope-dilution mass spectrometry. All three enzymes excised 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) and 8-hydroxyguanine (8-OH-Gua) from γ-irradiated DNA containing a multiplicity of base lesions. Michaelis–Menten kinetics of excision were measured. Significant differences between excision kinetics of these three enzymes were observed. Excision of FapyGua and 8-OH-Gua by wild-type α-hOgg1 was greater than that by α-hOgg1-Gln46 and α-hOgg1-His154. The latter mutant protein was less active than the former. The diminished activity of the mutant proteins was more pronounced for 8-OH-Gua than for FapyGua. Cleavage assays were also performed using 32P-labeled 34mer oligonucleotide duplexes containing a single 8-OH-Gua paired to each of the four DNA bases. The results obtained with the oligonucleotide containing the 8-OH-Gua/Cyt pair were in good agreement with those observed with γ-irradiated DNA. Wild-type α-hOgg1 and its mutants repaired the three mismatches less efficiently than the 8-OH-Gua/Cyt pair. The substitution of Arg154, in addition to diminishing the activity on 8-OH-Gua, relaxes the selectivity found in the wild-type α-hOgg1 for the base opposite 8-OH-Gua. Taken together the results show that the mutant forms α-hOgg1-Gln46 and α-hOgg1-His154 found in human tumors are defective in their catalytic capacities.
INTRODUCTION
DNA damage generated by oxygen-derived free radicals, has been implicated as playing a role in mutagenesis, carcinogenesis and aging (1). Free radicals produce a variety of types of damage in DNA including base and sugar damage, DNA–protein crosslinks and strand breaks (2–4). Damaged bases in DNA are thought to be repaired in cells mainly by base-excision repair (5). In the first step of this type of repair, damaged bases are removed from DNA by DNA glycosylases, which catalyze the cleavage of the glycosidic bond between the damaged base and the sugar moiety, leaving an abasic site in DNA. Subsequently, the repair of DNA is completed by successive actions of other enzymes (6). Escherichia coli possesses several DNA glycosylases that remove oxidatively modified bases from DNA (5). These are pyrimidine-specific Nth and Nei proteins (endonucleases III and VIII, respectively) and purine-specific Fpg protein. In eukaryotes, functional homologs of these E.coli DNA enzymes were identified. In Saccharomyces cerevisiae, a DNA glycosylase, which is encoded by the OGG1 gene and named Ogg1 protein, catalyzes the excision of 8-hydroxyguanine (8-OH-Gua) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) from DNA (7–9). The biological function of E.coli Fpg and yeast Ogg1 proteins is to prevent mutations in the genome that may be induced by reactions of free radicals with DNA bases (10).
Two human cDNAs encoding Ogg1 proteins with 345 and 424 amino acids were cloned (10). Further studies showed that these cDNAs were the products of an alternative splicing of a same primary transcript (11,12). The two proteins named α-hOgg1 and β-hOgg1 show sequence similarities with yeast Ogg1 protein and possess 316 identical amino acids at the N-terminus, but different sequence at the C-terminus (10). They are targeted to the nucleus and mitochondrion, respectively (11,13). The nuclear α-hOgg1 protein possesses DNA glycosylase/AP lyase activities and removes 8-OH-Gua, FapyGua and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine (Me-FapyGua) from DNA (10). This form seems to be the most abundant (14). In analogy to E.coli Fpg and yeast Ogg1 proteins, hOgg1 may possess an antimutator function in human cells. Consistent with this, mice in which both OGG1 alleles are disrupted show an accumulation of premutagenic 8-OH-Gua lesions and display an elevated spontaneous mutation rate in non-proliferative tissues (15,16). Indeed, the analysis of human tumors for expression and mutation of the hOGG1 gene showed that somatic and polymorphic mutations of the hOGG1 gene exist in lung and kidney tumors (17,18). Furthermore, a genetic polymorphism at codon 326 (Ser326Cys) was found in the Japanese population, in both healthy individuals and lung cancer patients (17). European patients with head and neck or kidney cancer were shown to possess a similar polymorphism (19). The mutant α-hOgg1-Cys326 was reported to have a reduced activity when compared with the wild-type α-hOgg1-Ser326 (17). Recently, we showed that these two enzymes fused to the GST protein are functional and excise 8-OH-Gua, FapyGua and Me-FapyGua from damaged DNA (20). However, their specificity factors (kcat/KM) differ significantly by 2-fold for excision of 8-OH-Gua or FapyGua from damaged DNA, with the wild-type α-hOgg1-Ser326 being more active than the mutant α-hOgg1-Cys326 (20).
In the present study, we investigated the substrate specificity of the free form (not fused to the GST protein) of the wild-type α-hOgg1-Ser326 (wt-hOgg1) and its two mutants with Arg46→Gln (α-hOgg1-Gln46) and Arg154→His (α-hOgg1-His154) found in human kidney tumor (M.Audebert, S.Chevillard, C. Levalois, G.Gyapay, A.Vieillfond, J.Klijanienko, P.Vieth, A.El Naggar, S.Oudard, S.Boiteux et al., submitted for publication) and a gastric cancer cell line (21), respectively. DNA exposed to γ-irradiation was used as a substrate. The technique of gas chromatography/isotope-dilution mass spectrometry (GC/IDMS) was used to determine the excision of modified bases from DNA and to measure their excision kinetics. The results of the activities of the different α-hOgg1 alleles on oligonucleotides carrying a single 8-OH-Gua residue opposite each of the four DNA bases are also reported.
MATERIALS AND METHODS
Materials
Modified DNA bases, their stable isotope-labeled analogs and other materials for GC/IDMS were obtained as described previously (22,23). For preparation of DNA samples, calf thymus DNA (Sigma) was dissolved in phosphate buffer (pH 7.4) at a concentration of 0.3 mg/ml. An aliquot of this solution was bubbled with N2O and irradiated with γ-rays in a 60Co γ-source at a dose of 80 Gy (dose rate 35.5 Gy/min). Subsequently, unirradiated and irradiated DNA solutions were dialyzed against 10 mM phosphate buffer (pH 7.4) for 18 h. Phosphate buffer outside the dialysis tubes was changed three times during the course of dialysis.
Enzyme purification
Plasmid pPR71, coding for glutathione S-transferase (GST) fused to α-hOgg1, was mutagenized using the QuickChange site-directed mutagenesis kit (Stratagene), as directed by the manufacturer, to generate plasmids pPR217 and pPR220. The oligonucleotides used were: 5′-CTGGACAATCTTTCCAGTGGAGGGAGCAAAG and its complementary oligonucleotide for pPR217; 5′-CAACAACATCGCCCACATCACTGGCATGGTG and its complementary oligonucleotide for pPR220. Plasmids pPR217 and pPR220 code then for GST-hOgg1-Glu46 and GST-hOgg1-His154, respectively.
Escherichia coli PR195 (fpg mutY) harboring a pPR71, pPR217 or pPR220 plasmid was grown at 37°C in LB-broth medium (2 l) containing 150 µg/ml ampicillin, until the absorbance at 600 nm reached 0.3, and was induced for 16 h at 20°C in the presence of IPTG (1 mM). Cells were collected and stored at –80°C. Cell pellets were resuspended in 10 ml/g lysis buffer (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 250 mM NaCl, 0.8 µg/ml antipain, 0.8 µg/ml leupeptine, 0.8 µg/ml aprotinin) and sonicated. After centrifugation of the cell lysate, the supernatant fraction (fraction 1) was dialyzed against PBS buffer and applied to glutathione–Sepharose 4B (Pharmacia Biotech) equilibrated with PBS buffer. The Sepharose was washed with PBS and proteins eluted with a buffer containing 50 mM Tris–HCl pH 8.0 and 30 mM reduced glutathione. Fractions containing the enzyme activity were pooled and dialyzed overnight against a buffer containing 50 mM Tris–HCl pH 8.0 and 50 mM NaCl and, subsequently, for 4 h against 50 mM Tris–HCl pH 8.0 with 150 mM NaCl and 2.5 mM CaCl2. Thrombin (ICN) was added (10 U/mg of GST–hOgg1 fusion protein) and the mixture incubated for 2 h at 25°C. Reactions were stopped by adding Na2EDTA to a final concentration of 5 mM. Proteins were dialyzed against a buffer containing 20 mM Tris–HCl pH 8.0, 2 mM Na2EDTA, 50 mM NaCl and 2% glycerol and applied to a MonoS column (FPLC system, Pharmacia Biotech). Proteins were eluted with a linear salt gradient (50–800 mM NaCl). The active fractions were pooled and their protein concentration determined by the method of Bradford (24).
Enzymatic assays
Aliquots of DNA substrates (100 µg) were dried in a SpeedVac under vacuum and were then dissolved in 100 µl of the incubation buffer consisting of phosphate buffer (final concentration 50 mM, pH 7.4), 100 mM KCl, 1 mM EDTA and 0.1 mM dithiothreitol. For the determination of the dependence of excision on the enzyme amount, 0.5, 1 or 2 µg of an hOgg1 were added to the mixture and three replicates of each mixture were incubated at 37°C for 30 min. Time dependence of excision was determined by incubation of the samples with 1 µg of an hOgg1 for 10, 20, 30 and 45 min. As controls, DNA samples were incubated with the heat-inactivated enzyme or without the enzyme. Inactivation of the enzyme was achieved by heating at 140°C for 30 min. After incubation, 260 µl of cold ethanol (–20°C) were added to stop the reaction and precipitate DNA. The samples were kept at –20°C for 2 h. Aliquots of stable isotope-labeled analogs of modified DNA bases and an additional 180 µl of cold ethanol (–20°C) were added. The samples were centrifuged at 15 000 g for 30 min at 4°C. DNA pellets and supernatant fractions were separated. Supernatant fractions were freed from ethanol in a SpeedVac under vacuum, frozen in liquid nitrogen and then lyophilized for 18 h.
For the measurement of excision kinetics, 15, 25, 35, 50 and 75 µg of irradiated DNA were supplemented with 85, 75, 65, 50 and 25 µg of unirradiated DNA, respectively. Additional samples containing 100 µg of irradiated or unirradiated DNA were also used. Two sets of these samples with three replicates of each mixture were prepared. One set of samples was used to determine the amounts of modified DNA bases in each sample. For this purpose, stable isotope-labeled analogs of modified bases as internal standards were added to the samples. Subsequently, they were dried in a SpeedVac under vacuum and hydrolyzed with 0.5 ml of 60% formic acid in evacuated and sealed tubes for 30 min at 140°C. The hydrolysates were frozen in liquid nitrogen and lyophilized for 18 h. The second set of samples was used for the measurement of the amounts of modified bases released by hOgg1 proteins. Three replicates of these samples were dried in a SpeedVac under vacuum and then dissolved in 100 µl of the incubation buffer. The samples were incubated with or without 1 µg of an hOgg1 protein at 37°C for 30 min. The amount of each hOgg1 corresponded to an enzyme concentration of 245 nM. After incubation, cold ethanol was added and then the samples were treated as described above for determination of enzyme amount and time dependence of excision.
Analysis by GC/IDMS
An aliquot (0.1 ml) of a mixture of nitrogen-bubbled bis(trimethylsilyl)trifluoroacetic acid [containing trimethylchlorosilane (1% v/v)] and pyridine (1:1 v/v) was added to vials containing lyophilized supernatant fractions of enzyme-digested samples or lyophilized formic acid hydrolysates of DNA samples. The samples were vortexed and purged individually with ultra-high-purity nitrogen, and the vials were tightly sealed under nitrogen with Teflon-coated septa. The supernatant fractions were derivatized by heating the vials at 120°C for 30 min. Formic acid-hydrolysates of DNA samples were derivatized by vigorously shaking the vials for 2 h at room temperature (25). After cooling, the samples treated with hOgg1 proteins were centrifuged at 5000 g for 30 min to precipitate the salt. The clear supernatant fractions were removed and placed in vials used for injection of samples onto the GC-column. Derivatized formic acid-hydrolysates were transferred to injection vials without further treatment. Vials were purged with nitrogen and tightly sealed with septa. Aliquots (4 µl) of derivatized samples were analyzed by GC/IDMS with selected-ion monitoring under the experimental conditions described previously (25). The oven temperature of the gas chromatograph was programmed from 130 to 280°C at a rate of 8°C/min after 2 min at 130°C.
Cleavage assays
Cleavage assays were performed on 32P-labeled 34mer oligonucleotide duplexes containing a single 8-OH-Gua at position 16 paired with each of the four normal bases on the complementary strand (26). In a standard reaction (14 µl final volume), 25 fmol of 32P-labeled 8-OH-Gua containing oligonucleotide duplex were incubated in reaction buffer (25 mM Tris–HCl pH 7.6, 2 mM Na2EDTA, 0.4 mg/ml BSA) with the amounts of hOgg1 protein indicated. Reactions were carried out at 37°C and stopped by adding 4 µl of formamide dye and subjected to 7 M urea–20% PAGE as described (26). Gels were scanned and quantified using a Storm PhosphorImager (Molecular Dynamics).
RESULTS
In order to compare the enzymatic activities of the wild-type and mutant forms of the human Ogg1 protein, the OGG1 cDNA cloned into a bacterial expression vector was mutagenized to result in ORFs coding for GST-hOgg1-Gln46 and GST-hOgg1-His154. The proteins expressed as GST fusions were purified from bacterial lysates by their affinity to glutathione. Treatment with thrombine allowed the cleavage of the native protein from the GST. The site of cleavage was confirmed by N-terminal sequencing of the protein with an apparent molecular mass of 38 kDa (data not shown). The Ogg1 proteins were further purified to homogeneity by FPLC (data not shown).
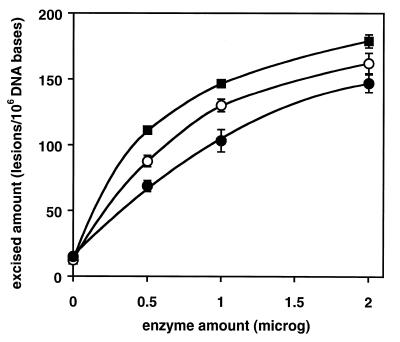
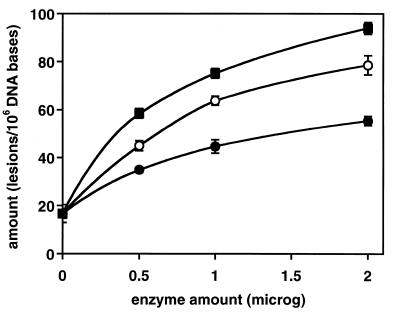
We investigated the ability of wt-hOgg1 and the two mutants found in human tumors to excise modified bases from irradiated DNA. Using GC/IDMS, 17 modified bases were identified and quantified in DNA γ-irradiated under N2O (27). Of these modified bases, wt-hOgg1 and its mutants efficiently excised FapyGua and 8-OH-Gua. The excision of FapyGua and 8-OH-Gua by wt-hOgg1 is in agreement with previous work (20). No other modified base was excised significantly under the conditions used in this work. Figures 1 and 2 illustrate excision as a function of the enzyme amount. Greatest excision was observed with wt-hOgg1 followed by α-hOgg1-Gln46 and α-hOgg1-His154. In the case of 8-OH-Gua, the diminished activity of the mutant proteins was more dramatic than in the case of FapyGua.
Figure 1.
Excision of FapyGua by wt-hOgg1, hOgg1-Gln46 and hOgg1-His154 as a function of the enzyme amount. DNA γ-irradiated under N2O (100 µg) was used as a substrate. The incubation time was 30 min at 37°C. The amounts given on the y-axis represent those found in the supernatant fractions. Closed squares, wt-hOgg1; open circles, hOgg1-Gln46; closed circles, hOgg1-His154.
Figure 2.
Excision of 8-OH-Gua by wt-hOgg1, hOgg1-Gln46 and hOgg1-His154 as a function of the enzyme amount. Other details are as in Figure 1.
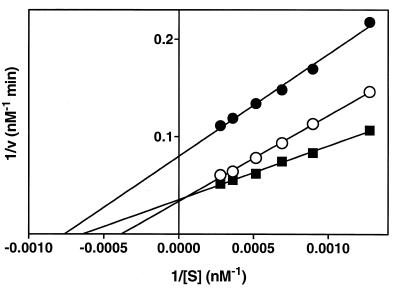
Kinetic parameters were determined by measurement of excision at six different concentrations of FapyGua and 8-OH-Gua with the total amount of DNA remaining constant in each sample. Concentration ranges of FapyGua and 8-OH-Gua were 0.8–3.8 and 0.38–2.3 µM, respectively. The excised amounts of these products in supernatant fractions were used for the determination of the initial velocity. Excision followed Michaelis–Menten kinetics (28). Kinetic constants and standard deviations (n = 6) were calculated using Lineweaver–Burk plots (28), and a linear least-squares analysis of the data. Initial velocities were estimated by using the plots of excision as a function of incubation time. As an example, Figure 3 illustrates the Lineweaver–Burk plot of the excision of FapyGua. Kinetic parameters calculated from Lineweaver–Burk plots are given in Table 1.
Figure 3.
Lineweaver–Burk plots for excision of FapyGua by wt-hOgg1, hOgg1-Gln46 and hOgg1-His154 from DNA γ-irradiated under N2O. The incubation time was 30 min at 37°C. The enzyme amount was 1 µg/100 µg of DNA. The amounts of products found in supernatant fractions were used for initial velocity. S, concentration of FapyGua; v, initial velocity of FapyGua excision. Closed squares, wt-hOgg1; open circles, hOgg1-Gln46; closed circles, hOgg1-His154.
Table 1. Kinetic constants for excision of FapyGua and 8-OH-Gua by wt-hOgg1, hOgg1-Gln46 and hOgg1-His154 from DNA γ-irradiated under N2O.
| Protein | Vmax (nM min–1)a | KM (nM)a | kcat/KM × 105 (min–1 nM–1)a | |||
|---|---|---|---|---|---|---|
| FapyGua | 8-OH-Gua | FapyGua | 8-OH-Gua | FapyGua | 8-OH-Gua | |
| wt-hOgg1 |
28.3 ± 1.1b,c |
10.3 ± 0.2b |
1558 ± 76c,d |
513 ± 15 |
7.4 ± 0.15b,d |
8.2 ± 0.08b,d |
| hOgg1-Gln46 |
29.6 ± 1.3b,c |
8.7 ± 0.3b |
2601 ± 124b,c |
600 ± 27 |
4.7 ± 0.11 |
5.9 ± 0.10b |
| hOgg1-His154 | 12.5 ± 0.5c | 4.5 ± 0.1 | 1311 ± 81c | 426 ± 14 | 3.9 ± 0.09 | 4.3 ± 0.04 |
aValues represent the means ± SD (n = 6). kcat = Vmax/[enzyme]; [enzyme] = 245 nM. The concentration ranges of the FapyGua and 8-OH-Gua were 0.8–3.8 and 0.38–2.3 µM, respectively.
bStatistically different from the value in line 3 (P < 0.05).
cStatistically different from the value in column 2 (P < 0.05).
dStatistically different from the value in line 2 (P < 0.05).
All three enzymes excised both FapyGua and 8-OH-Gua. However, there were significant differences between kinetic parameters. Maximum velocities of excision of FapyGua or 8-OH-Gua by wt-hOgg1 and hOgg1-Gln46 were similar, but they were significantly greater than the maximum velocity of excision by hOgg1-His154 (Table 1). The maximum velocity and Michaelis constant (KM) of FapyGua excision by each enzyme were greater than those of 8-OH-Gua excision. The KM value of FapyGua excision by hOgg1-Gln46 was greater than those of FapyGua excision by wt-hOgg1 and hOgg1-His154, whereas no significant differences between the KM values of 8-OH-Gua excision by three enzymes were noted. The specificity constants (kcat/KM) of the excision of FapyGua and 8-OH-Gua by wt-hOgg1 were significantly greater than those by the mutant enzymes. This indicates the reduced specificity of the mutant enzymes for excision of FapyGua and 8-OH-Gua from DNA when compared with wt-hOgg1. A significant difference was also noted between the kcat/KM values of the excision of 8-OH-Gua by hOgg1-Gln46 and hOgg1-His154. In the case of the same enzyme, there were no significant differences between the specificity factors of the excision of FapyGua and 8-OH-Gua, indicating the same specificity of each enzyme for these two modified bases.
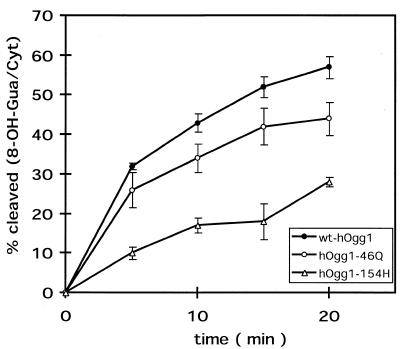
The catalytic properties of the wild-type and mutant proteins were also compared using 34mer oligonucleotide duplexes harboring a single 8-OH-Gua residue as substrates. Figure 4 shows that both mutant forms of hOgg1 were less active than the wild-type protein in this cleavage assay using an oligonucleotide with a 8-OH-Gua/Cyt pair. The kinetics shown are in good agreement with the GC/IDMS results on irradiated DNA (Fig. 2). Indeed, hOgg1-Gln46 exhibited an intermediate activity between the wild-type protein and hOgg1-His154, which is 4- to 5-fold less active than the wild-type form in this test for 8-OH-Gua DNA glycosylase/AP lyase activity.
Figure 4.
Cleavage of DNA duplex oligonucleotides harboring a single 8-OH-Gua/Cyt pair by wt-hOgg1, hOgg1-Gln46 and hOgg1-His154 as a function of time. For each enzyme, 10 ng of protein were incubated at 37°C with substrate for the times indicated. The results correspond to the averages of three independent experiments. Q, Gln; H, His.
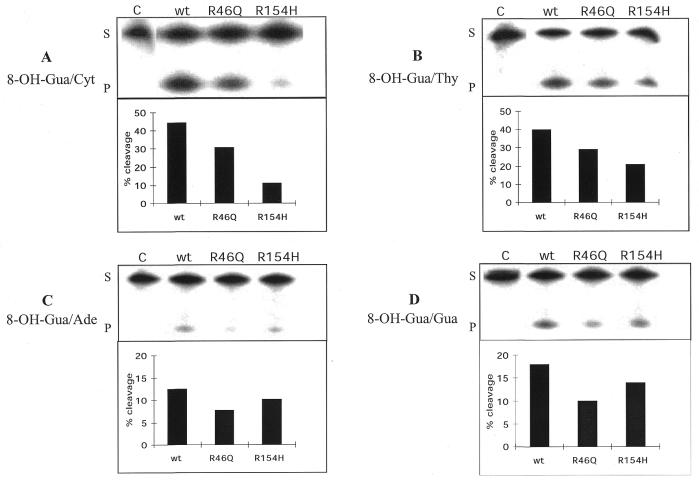
Oligonucleotide substrates were also used to analyze the opposite base dependence of the hOgg1 activity on 8-OH-Gua. For this purpose, the 32P-labeled 34mer harboring this lesion was hybridized to complementary oligonucleotides carrying each of the four normal DNA bases opposite 8-OH-Gua. The cleavage activity on the strand harboring the lesion was analyzed for each of the four DNA duplexes. All three mismatches were repaired by wt-hOgg1 and the two mutant forms less efficiently than the 8-OH-Gua/Cyt pair. For all three proteins, the substrate specificity hierarchy was: 8-OH-Gua/Cyt > 8-OH-Gua/Thy >> 8-OH-Gua/Gua > 8-OH-Gua/Ade. The specific activities for each of the 8-OH-Gua pairs are shown in Table 2. These values were calculated from at least four independent experiments. Figure 5 illustrates a series of gels and the percentages of cleavage obtained for a representative experiment. These results show that hOgg1-Gln46 consistently exhibited a diminished activity on all the substrates when compared with wt-hOgg1. However, there was a more significant loss of the specificity for hOgg1-His154. Indeed, whereas there is a 4-fold reduction of the activity of the mutant form with respect to the wild-type when the substrate has an 8-OH-Gua/Cyt pair (Figs 4 and 5A), the activity of the mutant protein on an 8-OH-Gua/Thy substrate was only 2-fold lower (Fig. 5B). More strikingly, when the base opposite the lesion was a purine, there was no difference in the efficiency of repair between wt-hOgg1 and hOgg1-His154 (Fig. 5C and D). These results are in agreement with the role proposed for amino acid 154 in the recognition of the base opposite the 8-OH-Gua (29).
Table 2. Specific activities of the hOgg1 proteins for the cleavage of oligonucleotides harboring each of the four bases opposite the 8-OH-Gua residue.
| 8-OH-Gua/Cyt | 8-OH-Gua/Thy | 8-OH-Gua/Gua | 8-OH-Gua/Ade | |
|---|---|---|---|---|
| wt-hOgg1 | 1300 ± 66 | 458 ± 36 | 240 ± 38 | 150 ± 15 |
| hOgg1-Gln46 | 1050 ± 117 | 402 ± 33 | 157 ± 27 | 88 ± 8 |
| hOgg1-His154 | 450 ± 113 | 256 ± 29 | 176 ± 1 | 115 ± 20 |
The specific activities and standard deviations were calculated from at least four independent experiments of the kind shown in Figure 5 and are expressed as pmol of substrate cleaved/mg of protein in 15 min at 37°C.
Figure 5.
Effect of the base opposite 8-OH-Gua on the cleavage activity of the Ogg1 proteins. Duplex 34mers carrying each of four DNA bases opposite 8-OH-Gua were incubated for 15 min at 37°C in the presence of 10 ng (A) or 20 ng (B–D) of each of the proteins. Products of the reaction were resolved on denaturing 20% PAGE (upper panels). S, substrate; P, product. The gels were quantified and the results were plotted (bottom panels). Note the different scales for (A) and (B) and (C) and (D). R, Arg; Q, Gln; H, His.
DISCUSSION
The results show that single amino acid substitutions in the human Ogg1 protein involving Arg46→Gln or Arg154→His significantly affect the specificity of the Ogg1 protein for the repair of FapyGua and 8-OH-Gua in free-radical damaged DNA. However, hOgg1-His154 was less effective than hOgg1-Gln46 for both FapyGua and 8-OH-Gua. This indicates that these two types of mutations have different effects on excision. We have shown previously that the hOgg1-Cys326 protein was also found to exhibit a significantly reduced specificity for both FapyGua and 8-OH-Gua when compared with the wild-type protein (20). Interestingly, the GST-tagged wt-hOgg1 had a similar kcat/KM value for FapyGua excision to that for FapyGua excision by the free wt-hOgg1 used in this work. On the other hand, the latter enzyme exhibited a 2-fold greater specificity for the excision of 8-OH-Gua than the GST-tagged wt-hOgg1. This indicates the importance of the investigation of the free enzyme in addition to the GST-tagged enzyme as was done in the present study.
The results revealed that the mutant forms of hOgg1 found in tumors, both hOgg1-Gln46 and hOgg1-His154, are defective in their catalytic capacities. A statistically significant difference of ∼1.5–2-fold between the kcat/KM value of the wt-hOgg1 and those of its mutants was observed. This difference may not be profound. However, the mutant forms were detected in tumors, where the loss of heterozygosity is generally found on the chromosome 3p25, which contains the OGG1 gene. These mutant proteins may therefore be the only forms of Ogg1 left. This means that the removal of 8-OH-Gua may then depend on the mutant enzymes only. In such cases, ‘small’ differences between the activities of the mutant enzymes and their wild-type enzyme can be important. In agreement with this notion, it was shown that cells having only one functional allele of the hOGG1 gene have significantly greater levels of 8-OH-Gua in their DNA (30).
Consistent with the importance of Arg46 and Arg154 in the normal activity of hOgg1, these two residues are conserved among all the eukaryotic Ogg1 proteins described so far. The recent elucidation of the structure of hOgg1 allows the positioning of the two amino acids with respect to the active site (29). In fact, amino acid 46 is only three residues away from Gly42, which is in direct contact with the 8-OH-Gua in the hOgg1–DNA complex crystal. The effect on the activity suggests that the substitution at position 46 may affect the structure of the active site. As for Arg154, Bruner et al. (29) have shown that this amino acid is directly involved in the recognition of the estranged cytosine located opposite 8-OH-Gua. In this work, we confirm that the change of this Arg154 to His relaxes the requirement for a pyrimidine opposite the lesion. We also show that this amino acid change affects the hOgg1 activity on 8-OH-Gua/Cyt substrates. Therefore, the presence of this mutant form in a cell would have a double mutator effect by reducing the repair rate of 8-OH-Gua in the context of an 8-OH-Gua/Cyt pair without affecting the repair rate of 8-OH-Gua/Ade, which would then lead to G/C→T/A transversion.
Past in vitro and in vivo studies demonstrated that FapyGua is produced in DNA by free radical-generating systems as abundantly as 8-OH-Gua depending on experimental conditions (reviewed in 2,3). FapyGua is the other major product of hydroxyl radical attack on guanine and is formed by one-electron reduction of the thus formed hydroxyl-adduct radical of guanine, whereas 8-OH-Gua is formed by one-electron oxidation of the same adduct radical (reviewed in 2,3). Moreover, repair enzymes such as Fpg and Ogg1 proteins of different origin remove FapyGua from damaged DNA as efficiently as 8-OH-Gua (for example, see 9,20; this study). In fact, all DNA glycosylases studied thus far, which remove 8-OH-Gua from DNA, also remove FapyGua with similar excision kinetics. In the past, much attention has been directed to 8-OH-Gua because of its premutagenic properties and its easy detection by HPLC (reviewed in 10). In contrast, FapyGua has not been investigated for its biological properties and it is not known whether this lesion is premutagenic or lethal, or both (5). The fact that all the known repair enzymes, which are specific for 8-OH-Gua, are also specific for FapyGua indicates the importance of the latter lesion in biological effects of free radical-induced DNA damage. Future work should also be directed toward the investigation of the biological properties of this important lesion.
DNA damage generated by free radicals produced as by-products of cellular metabolism has been proposed as a key factor in mutagenesis and cancer as well as in the process of aging (1). It is widely accepted that mutation events are at the origin of the cancer development process. Therefore, it is likely that a mutator phenotype might be involved at some point in the multistage process of carcinogenesis. This model has been actually confirmed by the finding that the hereditary non-polyposis colorectal cancer is associated with defects in genes coding for homologs to the bacterial mismatch repair proteins (31). Indeed, cells from these tumors have a hypermutator phenotype and the biochemical defect in the mismatch repair process was established (32,33). Similarly, impairments of the mechanisms of repair of oxidative DNA damage can lead to mutations in oncogenes or inactivation of tumor suppressor genes, altering the cell growth control. The enzymatic activities displayed by hOgg1 and its antimutator effect suggest that cells in which there is no functional allele of the hOGG1 gene could have a hypermutator phenotype and therefore accelerate the carcinogenic process. The generation of OGG1–/– mice (15,16) has demonstrated that the lack of a functional Ogg1 protein indeed generates a mutator phenotype in mammalian cells. Moreover, tumors having lost one allele of the hOGG1 gene may have twice as much 8-OH-Gua in their DNA (30). The results presented in this paper, where the impairment of mutant alleles of the hOGG1 gene found in tumors was demonstrated, point to an important role of hOgg1 in maintaining the stability of the genetic information and therefore in the prevention of cancer.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Claudine Dhérin for her excellent technical assistance, and Serge Boiteux for fruitful discussions and critical reading of the manuscript. This work was supported by the Commissariat à l’Energie Atomique (CEA), Conseil National pour la Recherche Scientifique (CNRS) and Association pour la Recherche sur le Cancer (ARC). Certain commercial equipment or materials are identified in this paper in order to specify adequately the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
REFERENCES
- 1.Halliwell B. and Gutteridge,J.M.C. (1999) Free Radicals in Biology and Medicine. Oxford University Press, New York.
- 2.Dizdaroglu M. (1992) Mutat. Res., 275, 331–342. [DOI] [PubMed] [Google Scholar]
- 3.Breen A.P. and Murphy,J.A. (1995) Free Rad. Biol. Med., 18, 1033–1077. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J., Berger,M., Douki,T. and Ravanat,J.L. (1997) Rev. Physiol. Biochem. Pharmacol., 131, 1–87. [DOI] [PubMed] [Google Scholar]
- 5.Wallace S.S. (1998) Radiat. Res., 150, S60–S79. [PubMed] [Google Scholar]
- 6.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 7.Auffret van der Kemp P., Thomas,D., Barbey,R., de Oliveira,R. and Boiteux,S. (1996) Proc. Natl Acad. Sci. USA, 93, 5197–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nash H.M., Lu,R., Lane,W.S. and Verdine,G.L. (1997) Chem. Biol., 4, 693–702. [DOI] [PubMed] [Google Scholar]
- 9.Karahalil B., Girard,P.M., Boiteux,S. and Dizdaroglu,M. (1998) Nucleic Acids Res., 26, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radicella P. and Boiteux,S. (1999) Biochimie, 81, 59–67. [DOI] [PubMed] [Google Scholar]
- 11.Nishioka K., Ohtsubo,T., Oda,H., Fujiwara,T., Kang,D.C., Sugimachi,K. and Nakabeppu,Y. (1999) Mol. Biol. Cell, 10, 1637–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida T., Takashima,R., Fukayama,M., Hamada,C., Hippo,Y., Fujii,T., Moriyama,S., Matsuba,C., Nakahori,Y., Morita,H., Yazaki,Y., Kodama,T., Nishimura,S. and Aburatami,H. (1999) Int. J. Cancer, 80, 18–21. [DOI] [PubMed] [Google Scholar]
- 13.Takao M., Aburatani,H., Kobayashi,K. and Yasui,A. (1998) Nucleic Acids Res., 26, 2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monden Y., Arai,T., Asano,M., Ohtsuka,E., Aburatani,H. and Nishimura,S. (1999) Biochem. Biophys. Res. Commun., 258, 605–610. [DOI] [PubMed] [Google Scholar]
- 15.Klungland A., Rossewell,I., Hollenbach,S., Larsen,E., Daly,G., Epe,B., Seeberg,E., Lindahl,T. and Barnes,D.E (1999) Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minowa O., Tsuyochi,A., Hirano,M., Monden,Y., Nakai,S., Fukuda,M., Itoh,M., Takano,H., Hippou,Y., Aburatani,H., Masumura,K., Nohmi,T., Nishimura,S. and Noda,T. (2000) Proc. Natl Acad. Sci. USA, 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohno T., Shinmura,K., Tosaka,M., Tani,M., Kim,S.R., Sugumura,H., Nohmi,T., Kasai,H. and Yokota,J. (1998) Oncogene, 16, 3219–3225. [DOI] [PubMed] [Google Scholar]
- 18.Chevillard S., Radicella,J.P., Levalois,C., Lebeau,J., Poupon,M.F., Oudard,S., Dutrillaux,B. and Boiteux,S. (1998) Oncogene, 16, 3083–3086. [DOI] [PubMed] [Google Scholar]
- 19.Blons H., Radicella,J.P., Laccoureye,O., Brasnu,D., Beaune,P., Boiteux,S. and Laurent-Puig,P. (1999) Mol. Carcinog., 26, 254–260. [PubMed] [Google Scholar]
- 20.Dherin C., Radicella,J.P., Dizdaroglu,M. and Boiteux,S. (1999) Nucleic Acids Res., 27, 4001–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinmura K., Kohno,T., Kasai,H., Koda,K., Sugimura,H. and Yokota,J. (1998) Jpn. J. Cancer Res., 89, 825–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dizdaroglu M. (1994) Methods Enzymol., 234, 3–16. [DOI] [PubMed] [Google Scholar]
- 23.Nelson V.C. (1996) J. Label. Comp. Radiopharm., 38, 713–723. [Google Scholar]
- 24.Bradford M.M.(1976) Anal. Biochem. 72248–254. [DOI] [PubMed] [Google Scholar]
- 25.Sentürker S. and Dizdaroglu,M. (1999) Free Radic. Biol. Med., 27, 370–380. [DOI] [PubMed] [Google Scholar]
- 26.Girard P.M., Guibourt,N. and Boiteux,S. (1997) Nucleic Acids Res., 23, 3404–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dizdaroglu M., Bauche,C., Rodriguez,H. and Laval,J. (2000) Biochemistry, 39, 5586–5592. [DOI] [PubMed] [Google Scholar]
- 28.Gutfreund H. (1972) Enzymes: Physical Principals. Wiley-Interscience, London, UK, pp. 116–175.
- 29.Bruner S.D., Norman,D.P.G. and Verdine,G.L. (2000) Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 30.Hardie L.J., Briggs,J.A., Davidson,L.A., Allan,J.M., King,R.F.G.J., Williams,G.I. and Wild,C.P. (2000) Carcinogenesis, 21, 167–172. [DOI] [PubMed] [Google Scholar]
- 31.Jiricny J. (1994) Trends Gen., 10, 164–168. [DOI] [PubMed] [Google Scholar]
- 32.Fishel R., Lescoe,R.K., Rao,M.R.S., Copeland,N.G., Jenkins,N.A., Garber,J., Kane,M. and Kolodner,R. (1993) Cell, 75, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 33.Parsons R., Li,G.M., Longley,M.J., Papadopoulos,N., Jen,J., de la Chapelle,A., Kinzler,K.W., Vogelstein,B. and Modrich,P. (1993) Cell, 75, 1227–1236. [DOI] [PubMed] [Google Scholar]