Abstract
Chemoenzymatic synthesis methods use organic and enzyme chemistry to synthesize a desired small molecule. Complementing organic synthesis with enzyme-catalyzed selective transformations under mild conditions enables more sustainable and synthetically efficient chemical manufacturing. Here, we present a multistep retrosynthesis search algorithm to facilitate chemoenzymatic synthesis of pharmaceutical compounds, specialty chemicals, commodity chemicals, and monomers. First, we employ the synthesis planner ASKCOS to plan multistep syntheses starting from commercially available materials. Then, we identify transformations that can be catalyzed by enzymes using a small database of biocatalytic reaction rules previously curated for RetroBioCat, a computer-aided synthesis planning tool for biocatalytic cascades. Enzymatic suggestions captured by the approach include ones capable of reducing the number of synthetic steps. We successfully plan chemoenzymatic routes for active pharmaceutical ingredients or their intermediates (e.g., Sitagliptin, Rivastigmine, and Ephedrine), commodity chemicals (e.g., acrylamide and glycolic acid), and specialty chemicals (e.g., S-Metalochlor and Vanillin), in a retrospective fashion. In addition to recovering published routes, the algorithm proposes many sensible alternative pathways. Our approach provides a chemoenzymatic synthesis planning strategy by identifying synthetic transformations that could be candidates for enzyme catalysis.
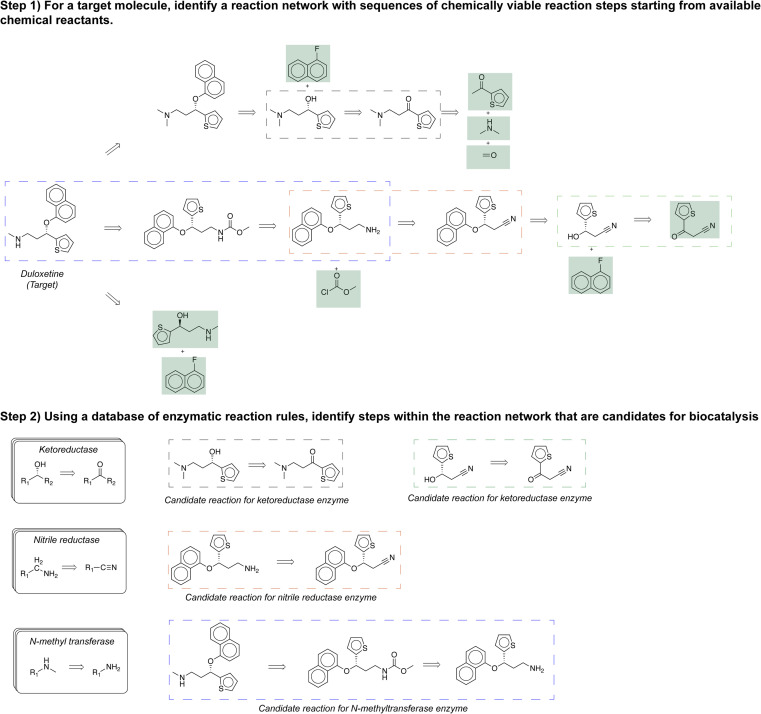
Given a target, our approach identifies a reaction network with sequences of chemically viable reaction steps starting from commercial compounds (in green). Then, it identifies steps within the reaction network that are candidates for biocatalysis.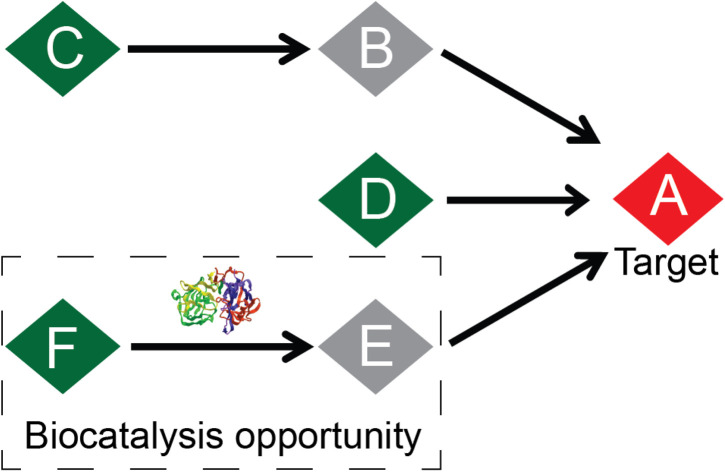
1. Introduction
Enzymes are essential tools employed in synthetic and process chemistry. They catalyze stereo-, regio-, and enantio-specific reactions; as a result, biocatalysis enables more efficient synthetic routes with less need for protection and deprotection reactions.1 Directed evolution has successfully enabled drastic changes to the substrate scope of enzymes to accept unnatural substrates to meet process needs.2 Further, enzymatic reactions typically occur in water or benign organic solvents under mild conditions.3–5 Moreover, multistep enzymatic reactions can be carried out in a single pot to avoid purifying intermediates and overcome equilibrium constraints.1,3 Enzymes are also more amenable to economic modeling than precious metal catalysts, whose prices fluctuate.6
Organic synthesis is the workhorse of the modern chemical manufacturing industry. It has been refined over centuries to allow efficient routes to creating human-made compounds. A subset of the organic reactions used can also be catalyzed by enzymes, for example, to reduce cost or employ milder reaction conditions. However, organic chemists' educational experience has only a small overlap with that of enzymologists, making it challenging for some synthetic chemists to tap into benefits at the interface between organic- and enzyme chemistry. Chemoenzymatic synthesis planning tools can efficiently help identify biocatalysis opportunities in the manufacture of small molecules.
Retrosynthesis techniques in computer-aided synthesis planning (CASP) propose feasible multistep synthetic routes to a target from available starting materials by starting with the target and choosing appropriate disconnections recursively. From the early CASP tools in organic chemistry presented over 50 years ago,7,8 the methods have improved to predict realistic organic synthesis routes to a desired target using rule-based methods and machine learning to generalize known reactions.9–11 Recent developments in enzymatic retrosynthesis show a tremendous potential for developing similar CASP tools for enzymes.12–15 Finnigan et al. recently curated a small set of expertly encoded reaction rules to describe the enzyme toolbox for biocatalysis.13 These reaction rules implicitly reflect the established substrate promiscuity of the different enzyme classes. Enzymes that these rules represent have been shown to be amenable to enzyme engineering in many cases, for the acceptance of novel substrates.13 Furthermore, they were successfully employed to plan biocatalytic cascades to target molecules. Although RetroBioCat successfully plans multistep enzymatic routes, it cannot propose chemoenzymatic routes toward a desired target that synergistically involves both organic and enzymatic approaches.
Recently, Levin et al. created a chemoenzymatic synthesis planning tool that identifies enzymatic opportunities while performing retrosynthesis.14 This method employs the ‘BKMS-react’ database of enzymatic reactions mainly used for natural product biosynthesis to identify enzymatic opportunities. Limitations in this dataset result in two imperfections. First, it reduces the algorithm's capability to identify chemoenzymatic routes toward human-made molecules that are not similar to natural products. Specifically, the algorithm only identified a limited number of synthesis routes and did not capture the precedent chemoenzymatic routes for several model human-made compounds tested (Fig. S1†). However, this molecule class plays a significant role in pharmaceutical, commodity chemical, and agrochemical industries. Second, BKMS database is biased towards metabolic enzymes; this might lead to retrosynthetic suggestions with reduced chances of engineerability of the enzymes. Therefore, notwithstanding this impressive advance in chemoenzymatic synthesis planning, there remains a strong need for planning algorithms to identify enzymatic opportunities within synthetic routes toward human-made molecules.
Herein, we demonstrate a method facilitating chemoenzymatic synthesis by identifying enzymatic transformations within pathways proposed by the open-source CASP tool, ASKCOS.4 The procedure is illustrated in Fig. 1 using Duloxetine16 as an example target. First, a user specifies the target compound, duloxetine, by its SMILES string. An existing CASP algorithm proposes many possible synthetic routes from commercial starting materials to the target. We use ASKCOS for this step, but in principle, other CASP approaches would work. Second, a small database of biocatalytic reaction rules previously curated by Finnigan et al. enables the identification of synthetic reactions that are candidates for enzyme catalysis, including the possibility of shortened reaction pathways.
Fig. 1. Computer-assisted multistep chemoenzymatic retrosynthesis using a synthesis planner. Commercially available starting materials are highlighted in a green box.
By identifying biocatalysis opportunities in planned synthesis routes, we demonstrate the capability of our approach to propose chemoenzymatic steps. Enzymatic suggestions captured by our approach include ones capable of reducing the number of synthetic steps. We successfully plan chemoenzymatic routes for active pharmaceutical ingredients or their intermediates (e.g., Sitagliptin, Rivastigmine, and Ephedrine), commodity chemicals (e.g., acrylamide and glycolic acid), and specialty chemicals (e.g., S-Metalochlor and Vanillin), in a retrospective fashion. In addition to recovering published routes, the algorithm proposes sensible alternatives. A primary disadvantage of employing enzymes is the long lead time necessary for directed evolution campaigns.6 Our approach often proposes suitable chemo-catalytic alternatives to the enzymes to serve as placeholders during evolution campaigns, alleviating a practical bottleneck for applying enzymes in process chemistry.
2. Methods
2.1. Single-step biocatalysis opportunity identifier
Our single-step biocatalysis opportunity identifier asks the question: can an enzyme be utilized to catalyze the single-step transformation proposed by the synthesis planner? First, for a specific target molecule, the algorithm generates a diverse set of chemically viable single-step synthetic transformations that could produce the target. Second, the algorithm identifies candidate reactions that can be catalyzed by enzymes. This analysis treats biocatalysis opportunity identification as a catalyst prediction problem after performing retrosynthesis. The following paragraphs explain the workflow in detail.
First, we employ the single-step retrosynthesis model in ASKCOS to generate a set of reactants that could produce the target.9 Using information from 12.5 million single-step synthetic reactions tabulated in the Reaxys database, ASKCOS has approximately 160 000 generalized reaction rules summarizing retrosynthetic transformations commonly used in synthetic chemistry. A feedforward neural network predicts and ranks which of the 160 000 reaction rules most apply to a target based on its molecular structure. The top-1000 templates or top-N templates with a maximum cumulative predicted score of 0.999 (whichever set contains a lower number of templates) were considered for a typical analysis that could produce the target.
Second, for every retrosynthetic suggestion, we exhaustively apply the biocatalytic templates from RetroBioCat to determine whether enzymes could potentially catalyze the reaction. RetroBioCat's 135 biocatalytic templates summarize the transformations catalyzed by different classes of enzymes and implicitly describe their established substrate scopes.13 Many templates are promiscuous with respect to stereochemical preferences of enzymes; this is intended to capture the potential to engineer enzymes for meeting process specific stereochemical needs.17–20 First, the product SMILES string and a list of separated reactant SMILES strings are obtained for every ASKCOS proposed retrosynthetic suggestion. Then, biocatalytic templates with reaction rules that apply to the target product are applied to the product SMILES using RDChiral, an open-source tool for retrosynthetic template application.21 For every applied template, the algorithm subsequently checks whether the resulting enzymatic reactants are present within the list of reactants associated with the original ASKCOS proposed retrosynthetic suggestion using an exact SMILES string match. If true, the enzyme class associated with the biocatalytic template is recorded as applicable to the ASKCOS proposed retrosynthetic suggestion. By ensuring the biocatalytic template applies to the proposed retrosynthetic suggestion, this approach intends to propose enzymes whose active site can catalyze the proposed synthetic transformation and whose binding pockets can accommodate the substrate through protein engineering.
2.2. Multistep chemoenzymatic retrosynthesis
Complex target molecules often require a series of organic and enzymatic transformations, which sequentially build molecular complexity starting from simpler building blocks. Herein, we developed two complementary methods for performing multistep chemoenzymatic retrosynthesis. First, we recursively apply the combined single-step retrosynthesis model and biocatalysis opportunity identifier to plan multistep chemoenzymatic synthesis routes. This first workflow simply extends the single-step biocatalysis opportunity identifier described in Section 2.1. Second, we developed an automated computer-aided synthesis planning tool that plans multistep chemoenzymatic reaction pathways in two sequential steps. Given a target compound as input, ASKCOS proposes multistep synthetic pathways starting from commercial starting materials. Then, we identify candidate reactions within ASKCOS proposed routes well suited for biocatalysis. The following paragraphs detail the second workflow.
First, we employ the MCTS-based multistep chemical synthesis planning tool in ASKCOS to generate a reaction network comprising different pathways to synthesize the target starting from commercially available materials.9 Retrosynthetic expansion occurs recursively for a predetermined amount of time (typically about 2 minutes) or up to a specified depth (typically about 5 steps) before all resulting pathways are returned. ASKCOS uses the Monte Carlo tree search algorithm to balance exploitation of branches thought to be promising and exploration of less frequently visited branches. Our database of buyables contains commercially available compounds from eMolecules, LabNetwork, or Sigma Aldrich. This buyables database can be customized for individual user needs.22 In this study, all compounds with an average price per gram listed at $100 or lower were included in the database. This analysis yielded a reaction network with different routes to produce the target staring from commercially available materials.
Second, we developed a method to process the resulting reaction network for identifying opportunities to employ biocatalysts. Because of the chemo-, regio-, and enantio-selectivity of enzyme-catalyzed reactions, enzymes offer opportunities to reduce the overall number of steps in the synthesis. This reduction was encouraged by allowing the algorithm to identify multistep synthetic transformations that a single enzymatic reaction can replace. Since a typical reaction network comprises O(10)–O(100) possible pathways to produce a target from commercial materials, the algorithm first parses the reaction network to identify the individual routes before each one is subsequently further analyzed. For every molecule in the pathway, the algorithm identifies single-step and multistep synthetic transformations to produce the molecule using precursors in the route. Then, the algorithm uses the single-step biocatalysis opportunity identifier described previously to determine whether the molecule can be created using the initial reactants of the identified single-step or multistep synthetic transformations. Importantly, this technique is a ‘post-processing method’ that uses pathways proposed by a synthesis planning tool; therefore, it can be integrated into other open-source or proprietary retrosynthetic workflows efficiently to identify biocatalysis opportunities.
3. Results
3.1. Single-step biocatalysis opportunity identifier
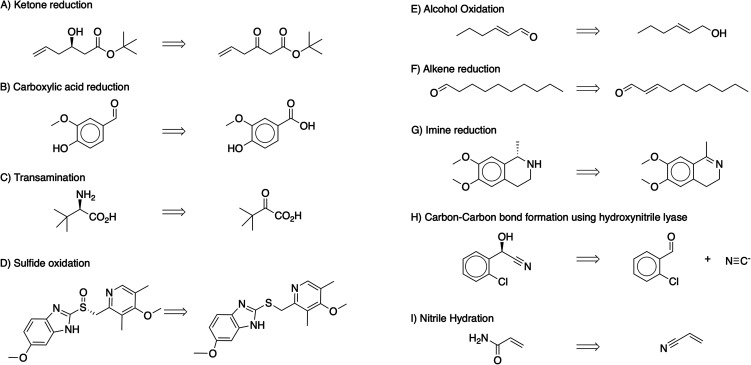
We tested our algorithm on diverse model compounds or intermediates produced using enzymes in different chemical manufacturing industries, and it successfully recovered their recorded precursors. Tert-butyl (R)-3-hydroxyl-5-hexenoate,23 Esomeprazole,24 (S)-salsolidine,25l-(S)-tert-leucine,26 and Clopidogrel intermediate27 are pharmaceutical intermediates. Vanillin,28trans-2-hexen-1-al,29 and decanal30 are flavor/food compounds. Acrylonitrile is a commodity chemical.31 These compounds employ a range of biocatalysts for their production, including ketoreductase (Fig. 2A),23 carboxylic acid reductase (Fig. 2B),28 transaminase (Fig. 2C),26 prazole sulfide monooxygenase (Fig. 2D),24 alcohol oxidase (Fig. 2E),29 ene reductase (Fig. 2F),30 imine reductase (Fig. 2G),25 hydroxynitrile lyase (Fig. 2H),27 and nitrile hydratase (Fig. 2I).31 The approach correctly proposes the recorded reactants and identifies suitable enzymes in all cases. The diversity of these transformations highlights the power of using the collective knowledge contained in a synthetic retrosynthesis prediction tool (i.e., ASKCOS single-step predictor) and a biocatalysis template database to identify strategic retrosynthetic steps and enzyme catalysts that might otherwise be overlooked, particularly by someone less familiar with enzymatic synthesis.
Fig. 2. Example biocatalysis opportunities identified by combining ASCKOS single-step retrosynthesis model and the newly developed biocatalysis opportunity identifier.
3.2. Interactive multistep chemoenzymatic pathway planning
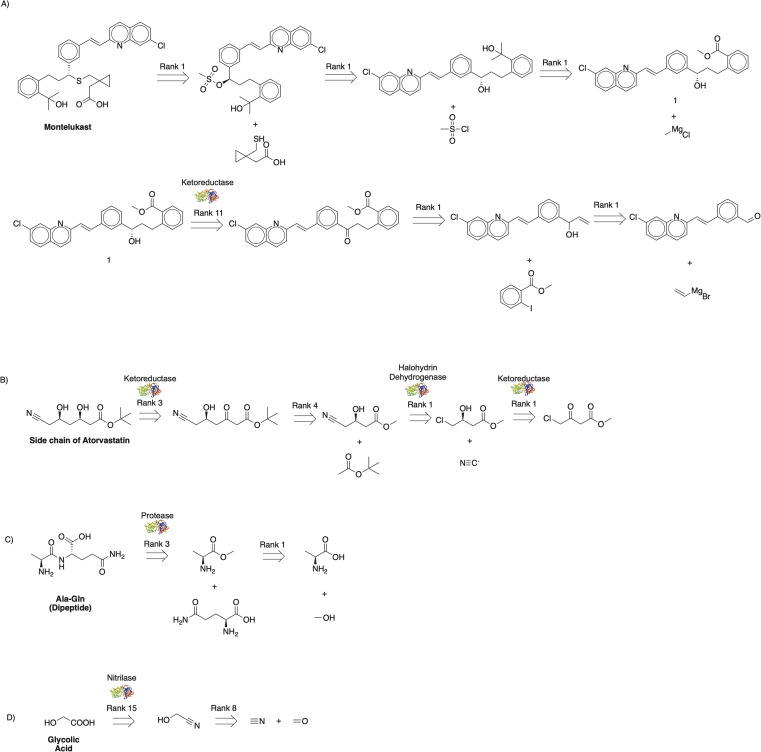
We then sought to recursively apply the combined single-step retrosynthesis model and biocatalysis opportunity identifier to plan multistep chemoenzymatic synthesis routes for industrially relevant small molecule targets starting from commercial materials. The complex pharmaceutical target Montelukast (Fig. 3A),32 chiral pharmacophore of Atorvastatin (Fig. 3B),33 dipeptide Ala–Gln (Fig. 3C),34 and commodity chemical glycolic acid (Fig. 3D)35 served as model targets. This interactive pathway planning approach first employs the newly developed single-step chemoenzymatic retrosynthesis approach for brainstorming synthesis strategies for every step and subsequently relies on human expertise to guide the overall multistep chemoenzymatic synthesis plan towards commercial starting materials. The suggested disconnections are consistent with published paths, except for one slight difference. The published biocatalytic route to the side chain of Atorvastatin by Codexis starts with ethyl 4-chloroacetoacetate rather than methyl 4-chloroacetoacetate;33 however, the proposed route (Fig. 3B) is sensible and captures the overall synthesis strategy.
Fig. 3. Multistep chemoenzymatic synthesis planning with human intervention. Routes are constructed by recursively applying the single-step chemoenzymatic synthesis methodology to (a) Montelukast, (b) the key side chain of Atorvastatin, (c) dipeptide Ala–Gln, and (d) commodity chemical glycolic acid. The suggested disconnections are consistent with published pathways, with slight differences discussed in the text.
3.3. Computer-aided chemoenzymatic synthesis planning using MCTS
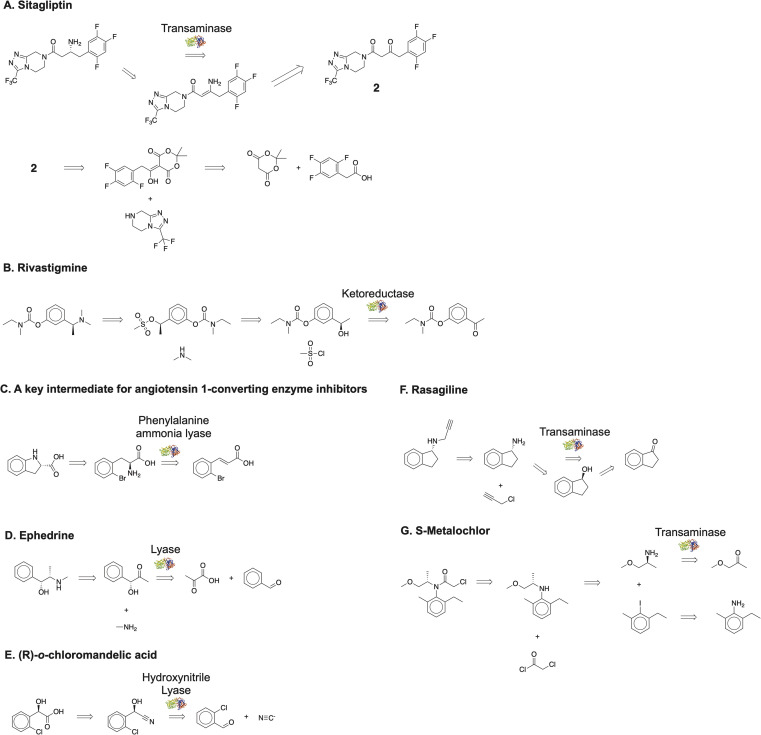
Chemically diverse compounds produced in multiple steps using chemocatalysts and enzymes served as model compounds to test the algorithm. Sitagliptin (Fig. 4A),2,36 Rivastigmine (Fig. 4B),37 a chiral intermediate for angiotensin 1 converting enzyme inhibitor (Fig. 4C),31 Ephedrine (Fig. 4D),38 (R)-o-chloromandelic acid (Fig. 4E),31 Rasagiline (Fig. 4F),39 (S)-Metalochlor (Fig. 4G)40 served as model compounds in the pharmaceutical and agrochemical industry. The algorithm capitalized on the ability of enzymes to catalyze stereoselective transformations to introduce chirality into the molecules. In all cases, we recovered the literature pathways successfully. We removed all intermediates in the literature pathways from our buyable database to prevent premature termination at expensive starting materials. In practice, users can recursively run the algorithm if proposed routes start from costly but commercially available starting materials.
Fig. 4. Multistep chemoenzymatic synthesis planning with minimal human intervention. Using our algorithm, we plan chemoenzymatic synthetic routes for (A) Sitagliptin (B) Rivastigmine (C) a key intermediate for angiotensin 1-converting enzyme inhibitors (D) Ephedrine (E) (R)-o-chloromandelic acid, (F) Rasagiline, and (G) S-Metalochlor. The suggested disconnections are consistent with published pathways.
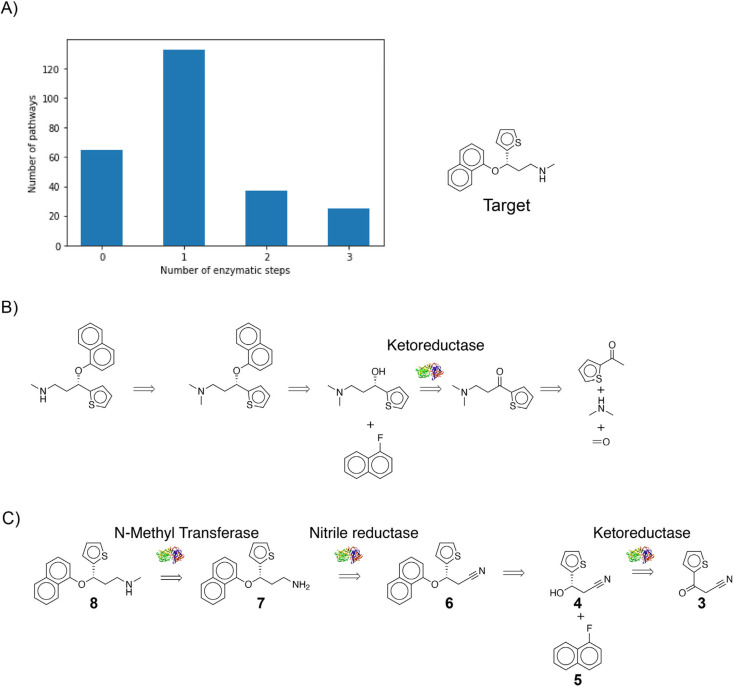
In addition to recovering the known literature precedent, the algorithm suggests other sensible chemoenzymatic routes to produce the target. (S)-duloxetine is a versatile pharmaceutical in treating major depressive disorders, stress urinary incontinence, and obsessive-compulsive disorder.16 In a few minutes, the algorithm identified 260 different pathways starting from commercially available materials that could produce this compound. 195 out of 260 pathways were identified as candidates for using enzymes as catalysts (Fig. 5A). To begin with, the known literature precedent to produce this compound was successfully identified by the algorithm (Fig. 5B).16 Second, the algorithm identifies many alternative routes for producing this molecule, and one such pathway is presented (Fig. 5C). This pathway starts with commercially available starting material 2-theonylacetonitrile (3). In the first synthetic step, a ketoreductase converts the ketone (3) to the chiral alcohol (4). Then, a nucleophilic aromatic substitution reaction transforms 1-fluoronaphthalene (4) and (5) into (6). Third, a nitrile reductase converts the nitrile (6) into the amine (7). Finally, an N-methyltransferase methylates the amine (7) to produce the target (8). A commercial process could potentially develop a biocatalytic cascade around the last two transformations; these reactions could be run in a single pot to avoid purifying the intermediate and reduce the overall number of steps in the process. Compared to the published route (Fig. 5B), this alternative pathway (Fig. 5C) is predominantly catalyzed by enzymes and reduces the overall number of steps. Similarly, the algorithm proposes additional chemoenzymatic alternatives for producing duloxetine. Ultimately, the final choice of production process comes down to many factors, including costs, impurity profiles, kinetics, and available equipment resources.
Fig. 5. (A) Chemoenzymatic synthesis planning for the exemplary pharmaceutical compound duloxetine yields 260 total routes that start from commercial materials, including 195 routes that employ at least one enzyme. (B) The published pathway to duloxetine is recovered by the approach. (C) A chemically sensible alternative route to duloxetine proposed by the approach. In this alternative route, three out of four proposed reactions are enzyme-catalyzed; therefore, enzymes play a dominant role in producing the target.
4. Discussion
In this study, we have developed a tool for identifying opportunities to employ biocatalysis in organic syntheses toward synthetic human-made molecules. Chemists tasked with producing a new molecule can utilize this platform as a brainstorming tool. In a few minutes, this planning tool identifies O(10)–O(100) chemoenzymatic synthesis pathways to the target starting from commercial materials. Enzymes are successfully proposed to build the necessary stereochemical and molecular complexity associated with the target. Further, this synthesis planning tool identifies multistep synthetic reactions that could potentially be replaced by a single enzymatic step because of the selectivity of enzyme-catalyzed transformations.
Two complementary multistep chemoenzymatic retrosynthesis tools were developed in this study. The interactive multistep chemoenzymatic pathway planning tool leverages human expertise to guide the synthesis planner. It allows human experts to control the biocatalytic transformations employed in the synthesis. As a result, these human experts can choose to prioritize biocatalytic transformations that their organizations have significantly invested in previously. On the other hand, computer-aided chemoenzymatic synthesis planning using MCTS automatically considers the use of 135 commonly employed biocatalytic transformations. This second tool can algorithmically generate many different chemoenzymatic routes to the target with minimal human intervention. Further, this task can easily be parallelized to compute chemoenzymatic synthesis routes to O(100)–O(1000) different targets. Therefore, this CASP platform can be employed for long-term investment planning to identify critical biocatalytic transformations needed to produce a portfolio of small molecules. As a result, strategic biocatalysis investments can be made to promote the use of enzymes in the manufacture of human-made small molecules.
The presented chemoenzymatic algorithms are open-source and widely available to practitioners. Identifying biocatalysis opportunities can promote collaborations among organic chemists, chemical and protein engineers, and enzymologists to facilitate the sustainable manufacture of small molecules. Furthermore, it could complement many textbooks and review articles on this topic,31,39,41–43 with an emphasis on solving real-world synthesis challenges important to individual practitioners.
To our knowledge, there is currently only one other approach in the literature that synergistically employs both organic and enzymatic transformations for planning multistep synthesis of complex target compounds. Levin et al. has introduced enzymatic transformations during the retrosynthetic analysis using reaction templates associated with natural product biosynthesis.14 In our method, we focus on identifying synthetic transformations that can potentially be catalyzed by enzymes, which complements the accomplishment of Levin et al. by capturing suggestions towards synthetic, human-made chemicals. We performed a comparison between the two methods for the three model compounds Sitagliptin, (S)-Duloxetine, and (S)-Metalochlor (Fig. S2†). In this comparison, the primary difference is the technique employed to identify biocatalytic transformations in organic syntheses. In all three case studies, our method exceeds that of Levin et al. by successfully recovering the known literature precedents. Further, our approach also proposes a greater number of synthetic routes to the target. We hypothesize that by limiting the biocatalysis search to a small expert-curated biocatalytic template set after completion of the synthetic tree search, we avoid exploring a large number of algorithmically extracted enzymatic reaction templates commonly used in the biosynthesis of natural products during the tree search. Such templates are less likely to be productive when the target is dissimilar to natural products. In the future, computer-aided synthesis planners integrating both approaches could exploit the advantages of the individual methods.
This algorithm can identify candidate transformations for enzyme catalysis but does not propose specific enzyme sequences for directed evolution efforts. Large biocatalysis reaction databases are not widely available like metabolic reaction datasets44 or organic reaction datasets.45 Therefore, chemical similarity-based methods for selecting enzyme sequences for directed evolution12 are less applicable to biocatalysis opportunities associated with human-made compounds. The expert curated reaction rules from RetroBioCat are associated with enzymes that generally have a track record for showing promiscuous substrate specificity.13 Therefore, identifying these biocatalysis opportunities can be valuable on their own even if specific enzyme sequences are not proposed by the algorithm; enzyme screening panels can be employed to find the right enzyme for a specific reaction.
This tool relies on the synthesis planner ASKCOS to propose retrosynthetic suggestions.9 ASKCOS predictions are driven by an underlying database of synthetic transformations from Reaxys, and most of these transformations are organic. At every step in a retrosynthetic analysis, beneficial enzymatic reaction templates that do not overlap with ASKCOS predicted templates are not considered. Biocatalytic and chemoenzymatic routes that do not align with the same order of chemical transformations as classical synthetic approaches might not be favored by our tool. On the other hand, suggestions resulting from our tool might have alternative classical synthetic approaches for the enzyme catalyzed steps. Because of the possibility of accomplishing some of the enzyme catalyzed steps through alternative approaches, the inherent risk of pursuing risk-prone and time-intensive biocatalysis opportunities is greatly mitigated. This could encourage risk-averse practitioners in highly regulated industries (e.g. pharmaceutical manufacturing) to explore biocatalysis opportunities.
5. Conclusion
We have developed a computer-assisted chemoenzymatic synthesis planner for synthetic human-made molecules. Our approach can capture selective enzymatic transformations that reduce the number of synthetic steps. Our route modification strategy identifies synthetic transformations that could be candidates for enzyme catalysis to plan chemoenzymatic syntheses. This post-processing step could, in principle, be integrated with other open source or proprietary organic synthesis planners to identify opportunities for biocatalysis. Efforts to incorporate beneficial enzymatic reaction templates into the retrosynthetic analysis that don't overlap with synthetic templates already in Reaxys will further improve the synthetic efficiency of the proposed routes. Notwithstanding this limitation, the stage is set for computational chemoenzymatic synthesis planning for human-made compounds.
Data availability
ASKCOS is publicly available at https://askcos.mit.edu/. The tool's algorithms are available at https://github.com/karthiksankar93/ChemoEnzymaticSynthesis. Additional information on the supporting tables and figures are provided in the ESI.†
Author contributions
KS: conceptualization, methodology, software, validation, formal analysis, investigation, data curation, writing – original draft, review, and editing. KFJ: conceptualization, writing – review and editing. supervision and funding acquisition.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work is supported by the DARPA PANACEA program grant HR0011-19-2-0022. This work is also supported by the consortium for Machine Learning in Pharmaceutical Discovery and Synthesis (MLPDS). We thank Deeptak Verma, Colin Lam, Willow Ross Carretero Chavez, Andrew Zahrt, Jason Mustakis, and Ania Fryszkowska for helpful discussions.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc01355c
References
- Huffman M. A. Fryszkowska A. Alvizo O. Borra- Garske M. Campos K. R. Canada K. A. Devine P. N. Duan D. Forstater J. H. Grosser S. T. Halsey H. M. Hughes G. J. Jo J. Joyce L. A. Kolev J. N. Liang J. Maloney K. M. Mann B. F. Marshall N. M. McLaughlin M. Moore J. C. Murphy G. S. Nawrat C. C. Nazor J. Novick S. Patel N. R. Rodriguez-Granillo A. Robaire S. A. Sherer E. C. Truppo M. D. Whittaker A. M. Verma D. Xiao L. Xu Y. Yang H. Science. 2019;366:1255–1259. doi: 10.1126/science.aay8484. [DOI] [PubMed] [Google Scholar]
- Savile C. K. Janey J. M. Mundorff E. C. Moore J. C. Tam S. Jarvis W. R. Colbeck J. C. Krebber A. Fleitz F. J. Brands J. Devine P. N. Huisman G. W. Hughes G. J. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan K. Antaris X. X. Palanski B. A. El Gamal A. Kao C. M. Fitch W. L. Fischer C. R. Khosla C. J. Am. Chem. Soc. 2019;141:9474–9478. doi: 10.1021/jacs.9b03066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry B. Robbins T. Weng C.-H. O'Brien R. V. Cane D. E. Khosla C. J. Am. Chem. Soc. 2013;135:16809–16812. doi: 10.1021/ja409048k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. Liu T. Zhu F. Khosla C. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18643–18648. doi: 10.1073/pnas.1110852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truppo M. D. ACS Med. Chem. Lett. 2017;8:476–480. doi: 10.1021/acsmedchemlett.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey E. J. Wipke W. T. Science. 1969;166:178–192. doi: 10.1126/science.166.3902.178. [DOI] [PubMed] [Google Scholar]
- Vléduts G. É. Finn V. K. Inf. Storage Retr. 1963;1:101–116. doi: 10.1016/0020-0271(63)90012-3. [DOI] [Google Scholar]
- Coley C. W. Thomas D. A. Lummiss J. A. M. Jaworski J. N. Breen C. P. Schultz V. Hart T. Fishman J. S. Rogers L. Gao H. Hicklin R. W. Plehiers P. P. Byington J. Piotti J. S. Green W. H. Hart A. J. Jamison T. F. Jensen K. F. Science. 2019;365:eaax1566. doi: 10.1126/science.aax1566. [DOI] [PubMed] [Google Scholar]
- Segler M. H. S. Preuss M. Waller M. P. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- Genheden S. Thakkar A. Chadimová V. Reymond J.-L. Engkvist O. Bjerrum E. J. Cheminf. 2020;12:70. doi: 10.1186/s13321-020-00472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan K. Heid E. Coley C. W. Verma D. Green W. H. Jensen K. F. Chem. Sci. 2022;13:6039–6053. doi: 10.1039/D2SC01588A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnigan W. Hepworth L. J. Flitsch S. L. Turner N. J. Nat. Catal. 2021;4:98–104. doi: 10.1038/s41929-020-00556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin I. Liu M. Voigt C. A. Coley C. W. Nat. Commun. 2022;13:7747. doi: 10.1038/s41467-022-35422-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst D. Manica M. Nana Teukam Y. G. Castrogiovanni A. Paratore F. Laino T. Nat. Commun. 2022;13:964. doi: 10.1038/s41467-022-28536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Liu Z.-Q. Lin C.-P. Zheng Y.-G. Bioorg. Chem. 2016;65:82–89. doi: 10.1016/j.bioorg.2016.02.002. [DOI] [PubMed] [Google Scholar]
- May O. Nguyen P. T. Arnold F. H. Nat. Biotechnol. 2000;18:317–320. doi: 10.1038/73773. [DOI] [PubMed] [Google Scholar]
- Jaeger K.-E. Eggert T. Curr. Opin. Biotechnol. 2004;15:305–313. doi: 10.1016/j.copbio.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Reetz M. T. J. Org. Chem. 2009;74:5767–5778. doi: 10.1021/jo901046k. [DOI] [PubMed] [Google Scholar]
- Reetz M. T. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5716–5722. doi: 10.1073/pnas.0306866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coley C. W. Green W. H. Jensen K. F. J. Chem. Inf. Model. 2019;59:2529–2537. doi: 10.1021/acs.jcim.9b00286. [DOI] [PubMed] [Google Scholar]
- Buyable Compounds, https://askcos.mit.edu/buyables/, accessed 29 April 2023
- Hu C. Liu M. Yue X. Huang Z. Chen F. Org. Process Res. Dev. 2020;24:1700–1706. doi: 10.1021/acs.oprd.0c00320. [DOI] [Google Scholar]
- Xu N. Zhu J. Wu Y.-Q. Zhang Y. Xia J.-Y. Zhao Q. Lin G.-Q. Yu H.-L. Xu J.-H. Org. Process Res. Dev. 2020;24:1124–1130. doi: 10.1021/acs.oprd.0c00115. [DOI] [Google Scholar]
- Leipold F. Hussain S. Ghislieri D. Turner N. J. ChemCatChem. 2013;5:3505–3508. doi: 10.1002/cctc.201300539. [DOI] [Google Scholar]
- Seo Y.-M. Yun H. J. Microbiol. Biotechnol. 2011;21:1049–1052. doi: 10.4014/jmb.1105.05049. [DOI] [PubMed] [Google Scholar]
- Glieder A. Weis R. Skranc W. Poechlauer P. Dreveny I. Majer S. Wubbolts M. Schwab H. Gruber K. Angew. Chem., Int. Ed. 2003;42:4815–4818. doi: 10.1002/anie.200352141. [DOI] [PubMed] [Google Scholar]
- Park J. Lee H.-S. Oh J. Joo J. C. Yeon Y. J. Biochem. Eng. J. 2020;161:107683. doi: 10.1016/j.bej.2020.107683. [DOI] [Google Scholar]
- de Almeida T. P. van Schie M. M. C. H. Ma A. Tieves F. Younes S. H. H. Fernández-Fueyo E. Arends I. W. C. E. Riul Jr A. Hollmann F. Adv. Synth. Catal. 2019;361:2668–2672. doi: 10.1002/adsc.201801312. [DOI] [Google Scholar]
- Papadopoulou A. Peters C. Borchert S. Steiner K. Buller R. Org. Process Res. Dev. 2022;26:2102–2110. doi: 10.1021/acs.oprd.2c00096. [DOI] [Google Scholar]
- Wu S. Snajdrova R. Moore J. C. Baldenius K. Bornscheuer U. T. Angew. Chem., Int. Ed. 2021;60:88–119. doi: 10.1002/anie.202006648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollikonda S. Mohanarangam S. Jinna R. R. Kandirelli V. K. K. Makthala L. Sen S. Chaplin D. A. Lloyd R. C. Mahoney T. Dahanukar V. H. Oruganti S. Fox M. E. J. Org. Chem. 2015;80:3891–3901. doi: 10.1021/acs.joc.5b00197. [DOI] [PubMed] [Google Scholar]
- Turner N. J. O'Reilly E. Nat. Chem. Biol. 2013;9:285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- Yagasaki M. Hashimoto S. Appl. Microbiol. Biotechnol. 2008;81:13–22. doi: 10.1007/s00253-008-1590-3. [DOI] [PubMed] [Google Scholar]
- Panova A. Mersinger L. J. Liu Q. Foo T. Roe D. C. Spillan W. L. Sigmund A. E. Ben-Bassat A. Wagner L. W. O'Keefe D. P. Wu S. Petrillo K. L. Payne M. S. Breske S. T. Gallagher F. G. DiCosimo R. Adv. Synth. Catal. 2007;349:1462–1474. doi: 10.1002/adsc.200700061. [DOI] [Google Scholar]
- Ye F. Zhang Z. Zhao W. Ding J. Wang Y. Dang X. RSC Adv. 2021;11:4805–4809. doi: 10.1039/D0RA10273C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P.-C. Zhu G.-L. Xie J.-H. Zhang X.-D. Zhou Q.-L. Li Y.-Q. Shen W.-H. Che D.-Q. Org. Process Res. Dev. 2013;17:307–312. doi: 10.1021/op3003147. [DOI] [Google Scholar]
- Winkler C. K. Schrittwieser J. H. Kroutil W. ACS Cent. Sci. 2021;7:55–71. doi: 10.1021/acscentsci.0c01496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N. J. and Humphreys L., Biocatalysis in Organic Synthesis: The Retrosynthesis Approach, Royal Society of Chemistry, 2018 [Google Scholar]
- Jackson D. A., in Biocatalysis for Green Chemistry and Chemical Process Development, John Wiley & Sons, Ltd, 2011, pp. 255–276 [Google Scholar]
- Turner N. J. O'Reilly E. Nat. Chem. Biol. 2013;9:285–288. doi: 10.1038/nchembio.1235. [DOI] [PubMed] [Google Scholar]
- Patel R. N. Bioorg. Med. Chem. 2018;26:1252–1274. doi: 10.1016/j.bmc.2017.05.023. [DOI] [PubMed] [Google Scholar]
- Reetz M. T. J. Am. Chem. Soc. 2013;135:12480–12496. doi: 10.1021/ja405051f. [DOI] [PubMed] [Google Scholar]
- Bansal P. Morgat A. Axelsen K. B. Muthukrishnan V. Coudert E. Aimo L. Hyka-Nouspikel N. Gasteiger E. Kerhornou A. Neto T. B. Pozzato M. Blatter M.-C. Ignatchenko A. Redaschi N. Bridge A. Nucleic Acids Res. 2022;50:D693–D700. doi: 10.1093/nar/gkab1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe D. M., Doctor of Philosophy (PhD thesis), University of Cambridge, 2012 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ASKCOS is publicly available at https://askcos.mit.edu/. The tool's algorithms are available at https://github.com/karthiksankar93/ChemoEnzymaticSynthesis. Additional information on the supporting tables and figures are provided in the ESI.†