Abstract
The control of CD4 expression is linked to the signaling events that mediate T-cell development and is directly dependent on the CD4 promoter. The CD4 promoter does not contain functionally redundant sites: all four factor-binding sites must be intact to achieve wild-type activity. Here we demonstrate that the precise position of three factor-binding sites relative to each other is essential for promoter activity, indicating that they function together as an inseparable cassette for assembly of the transcription initiation complex. Small changes in either phasing or distance between any two sites in this cassette leads to complete abrogation of promoter function. In addition, we demonstrate that one of the factors that bind the promoter cassette is not present in CD8 SP TC cells. Thus, this factor is a candidate for mediating the relative subclass specificity of CD4 promoter function in activated CD4 SP TH cells.
INTRODUCTION
The control of expression of the CD4 gene is linked to T-cell development and thus can be used as a model system for studying the molecular mechanisms that drive thymopoiesis (1–4). Four transcriptional control elements have been identified in the CD4 locus: a mature enhancer that is induced to function after thymic selection (5,6), a silencer that represses transcription in CD4– T cells (7–9), an enhancer that induces expression in immature thymocytes (10) and the CD4 promoter (11–13). Genetic studies of the promoter both in transient transfection and transgenic assays have determined that the promoter functions at high levels only in activated CD4 SP TH cells in the absence of the locus enhancers, indicating that this element can function efficiently alone in a subclass-specific manner (12,14,15). In the presence of the enhancers and the silencer, however, specificity of transcription is determined primarily by these other elements acting directly upon the promoter (9). The CD4 promoter is therefore an important central regulatory element in the control of CD4 gene expression on which the other CD4 locus transcriptional control elements act. To understand how subclass-specific expression of CD4 is controlled, it is important to determine how the factors that bind to the different CD4 transcriptional control elements interact. Thus, characterizing CD4 promoter function is a critical first step in the study of the CD4 transcriptional control during development.
The correct positioning of factor-binding sites has been shown to be important for specificity and function of many promoters and enhancers (16–20). For example, alterations in the spacing of the transcription binding sites in the promoter of the cystatin B gene results in reduced gene expression leading to progressive myoclonus epilepsy of Unverricht–Lundborg type (17). Interestingly, the positioning of factor-binding sites in a transcriptional control element can also be important for the tissue specificity of element function, such as in the spacing in the Bz promoter of maize (16). The dependence of element function on the position of its factor-binding sites is believed to be the result of the requirement of transcription factors to maintain a particular position relative to each other on the DNA helix (21,22). Increasing the distance between two sites may make it difficult for the binding factors to interact physically with one another. Another consequence of changing the distance between two sites is that their phasing may be affected. Only binding sites spaced multiples of 10 bp apart are on the same face of the DNA helix, whereas 5 bp would place them on opposite faces of the DNA helix. Most interacting transcription factors require specific phasing of their binding sites to juxtapose their respective protein–protein interaction domains and affect function. Perturbing the phasing between two factor-binding sites will change the nature of the interaction and will affect the activity of the regulatory element dependent on that interaction (22,23).
These structural considerations are also important for understanding the structure of the tertiary transcription initiation complex. This view is especially important in the case when the transcription factors directly involved are always available, but their ability to form the correct tertiary structure can be influenced by modifications and by other positive or negative regulators, such as enhancers and silencers. In this study, we investigate the spatial relationship between the CD4 promoter factor-binding sites. We demonstrate that the distance and phasing between CD4 promoter binding sites P2, P3 and P4 are crucial for promoter activity in activated CD4 SP cells in that they form an inseparable cassette, necessary for the assembly of the transcription initiation complex. Furthermore, using biochemical analysis, we determine that the P3 binding factor is absent in CD8+ SP cells, indicating that this factor may contribute to CD4 promoter specificity. Based on these data and our previous work we propose a model: in the CD4 promoter, binding factors contribute both to the functional specificity and to the formation of the correct tertiary transcription initiation complex on the CD4 promoter.
MATERIALS AND METHODS
Cell transfection and maintenance
The CD4+CD8– TH2 clone D10, and the CD4–CD8+ TC clones L3 and B18 were maintained in EHAA medium supplemented with 10% fetal calf serum, 0.1 mg/ml penicillin/streptomycin, 2 mM l-glutamine, 50 µM 2-mercaptoethanol and 10–30 U/ml IL-2. AKR1G1, a double-positive lymphoma, and S49, a double-negative lymphoma, were maintained in the same medium without IL-2. The cells clones were stimulated every 2 weeks with antigen as described (12). Nuclear extracts were prepared using a modified Dignam protocol (24) or by the Schreiber protocol (25). Transient transfections were performed between days 4 and 6 after stimulation as described (12). Briefly, two plasmids were introduced into cells by the DEAE-dextran method: one experimental, containing the test CD4 promoter cloned upstream of the luciferase gene in pGL2; the second, the transfection control plasmid pRL-TK (Promega) containing the Renilla luciferase gene under the control of the tk promoter. Cells were harvested after 48 h and extracts were prepared for the Dual Luciferase assay as recommended by the manufacturer (Promega). All data points are corrected for transfection efficiency and are presented as a percent of the wild-type CD4 promoter activity. At least five independent transfections were performed for each construct and the percentages were averaged as described (11). The activity of the mutant promoters is described in the text as follows: 0–15%, complete abrogation of promoter activity; 15–40%, significant effect on promoter activity; 40–60%, partial effect on promoter activity; 60–80%, minor effect on promoter activity; 80–100%, no effect on promoter activity.
Generation of promoter mutations
Site-directed mutagenesis was used to construct most of the promoter mutations. Oligonucleotides containing the mutations were obtained from GeneLink and used for site-directed mutagenesis in a pKS vector as described (26). Mutations were identified by DNA sequencing analysis or restriction digests. Mutant promoters were cloned into the pGL2 vector (Promega), which contains the luciferase gene. The promoter sequence and orientation were confirmed by sequencing. In the course of this work, we determined that the mutant 091 clone originally reported by Duncan et al. (11) was artifactual. The data reported in this manuscript are accurate. Mutant 023 was generated by restriction endonuclease cutting with PflMI, blunting by deleting the overhangs and reclosing. The sequences for the insertions in the insertion mutants are as follows: 020, GCTT; 014, ATCC; 070, CGACGCGTACG; 019, TCGAC; 053, GACTAGTCTGA; 080, GATCT; 089, GACTAGTCC. The precise point of insertion is indicated in the corresponding figures.
Electrophoretic mobility shift assays (EMSAs)
EMSAs were conducted as described previously (6,11,12,27). The P4-containing sequence from –20 to +4 of the murine CD4 promoter was subcloned and used as a radioactive probe (2–5 × 104 c.p.m. per reaction) for EMSA with equal protein amounts of nuclear extracts from D10, L3, S49 and AKR1G1 cell lines. The reaction also included 1 µg of dI–dC or 250 ng of herring sperm as a non-specific competitor, 1 mM spermidine and reaction buffer (10 mM HEPES pH 7.9, 50 mM NaCl, 5 mM Tris–HCl pH 7.5, 25 mM EDTA, 1 mM DTT and 10% glycerol). For competitions 50- or 250-fold molar excess of non-radioactive oligonucleotides was used. Competitors included: P3, the mutant P3 site D06, CRE consensus sequence 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ (28), NF-1 consensus sequence 5′-AATTGGCTTGAAGCCAACTAGATC-3′ (29) and P2 site C mutant probe as a non-specific competitor oligonucleotide. After 15 min of incubation at room temperature, the reactions were resolved on a non-denaturing 5% polyacrylamide gel and run at 150 V for 3 h at room temperature in glycine buffer (190 mM glycine, 25 mM Tris–HCl pH 8.5, 1 mM EDTA). Gels were dried and exposed on X-ray film overnight at –70°C with an intensifying screen.
RESULTS
Structural analysis of the CD4 promoter factor-binding sites
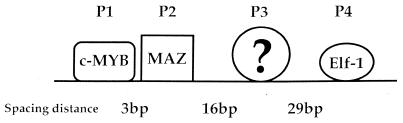
There are four factor-binding sites in the murine CD4 promoter (11; Fig. 1). There are several lines of evidence indicating that assembly of the correct tertiary structure at the CD4 promoter is an important level of promoter regulation. First, mutagenizing each one of the four binding sites produces a severe decrease in promoter activity, indicating that each of them is required for promoter function (11). Second, all of the identified CD4 promoter-binding factors, c-myb, Maz and Elf-1, are weak activators of transcription and function primarily by associating with other cofactors (30–32). Finally, the Maz-binding factor has been shown to participate in other TATA-less promoters (30) and to have DNA bending abilities (33), implying that tertiary structure could be important for promoter function.
Figure 1.
A schematic diagram of the CD4 promoter and its binding factors. P1, P2, P3 and P4 are the functionally important binding sites. The distance between the four binding sites is shown in base pairs. The known binding factors for each site are indicated.
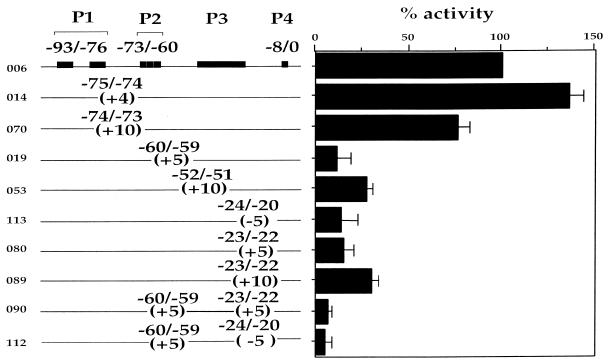
To determine whether distance and phasing between factor-binding sites are important for CD4 promoter function, we systematically introduced half or full helical turns on either side of each functional site. The mutations were made in regions of the CD4 promoter where linker-scanning mutagenesis indicated that no factor-binding sites were present (11). Half or full helical turns were inserted between P1 and the other sites (014 and 070; Fig. 2). Neither of these two mutations has an effect on promoter function, indicating that neither distance nor phasing between P1 and the rest of the promoter affects function significantly. Introducing similar mutations between P2 and P3 on the CD4 promoter produces a very different effect. As can be seen in Figure 2, mutations 019 and 053 introduce a half or full helical turn between P2 and P3. Both have deleterious effects on promoter function, leading to 71–95% decrease of promoter activity. Since site P3 has not been well characterized, it is still possible that mutation 053 is disrupting a functionally important site for the promoter. We believe that to be unlikely, because previous mutagenesis in the region (11) retains 75% of promoter activity, unlike our insertion mutation. In addition, oligonucleotides containing mutations in this region compete for P3 complex formation in EMSA as efficiently as the wild-type probe, indicating that sequences in this region are not important for P3 complex formation (data not shown). These data indicate that the distance and phasing between P2 and P3/4 are critical for promoter function. Similarly, mutations that introduce half or full helical turns between sites P3 and P4 also abrogate promoter activity completely (080 and 089; Fig. 2). Changing the phasing between P3 and P4 by a 5 bp deletion also abrogates promoter activity (113; Fig. 2). Taken together, these data indicate that the position of sites P2 and P4 relative to the rest of the promoter is very important for promoter function.
Figure 2.
Insertional and deletional mutagenesis of the CD4 promoter. The number of each mutant promoter in the pGL2 base vector is shown on the left panel. The solid lines represent the CD4 promoter from –101 to +10. The boxes indicate the functional binding sites and the numbers show their precise position relative to the transcription start site. For each mutation the position of the alteration and the number of base pairs inserted or deleted are given. The exact sequence of the insertions is shown in the Materials and Methods section. Functional activities of the mutant CD4 promoters in the CD4+CD8– cell clone D10 are shown in a bar graph form as percentage of the wild-type promoter activity, which is taken to be 100%. Transfection efficiency was controlled as described in Materials and Methods. The data are representative of five to seven independent transfections.
There are several possible interpretations of these data. It is possible that the distance and phasing between P2 and P4 is the important parameter for correct assembly of the tertiary complex on the CD4 promoter. An alternative explanation would be that the distance and phasing between P2 and P3 as well as P3 and P4 are structurally important and disturbing either one of the two leads to failure to assemble the correct transcription initiation complex. To distinguish between these possibilities we designed two additional mutations. The first contains two 5 bp half-helical insertions, one between P2 and P3 and another between P3 and P4 (090; Fig. 2). In this mutation, P2 and P4 are back in their correct phasing on the DNA, but a full helical turn apart, while P3 is phased out. The second mutation contains a 5 bp half-helical insertion between P2 and P3, but also contains a 5 bp half-helical deletion between P3 and P4 (112; Fig. 2). Thus, P2 and P4 are at the correct distance and phasing with respect to each other, whereas P3 is out of phase and at an incorrect distance with respect to both P2 and P4. If P2, P3 and P4 function as an inseparable functional cassette, then any mutation of the insertion/deletion type would produce a deleterious effect. In that case, our mutants 090 and 112 will not be able to restore promoter function. Alternatively, if the distance/phasing between P2 and P4 are the only critical parameters and the position of P3 is not important, then at least one of our composite mutants would be able to restore proper CD4 promoter function as it will orient P2 and P4 correctly. As can be seen in Figure 2, both mutations 090 and 112 have a severe effect on promoter function, indicating that the position of the P3 site relative to P2 and P4 is crucial for proper promoter activity. Thus, we can conclude that sites P2, P3 and P4 function together as a cassette and any disruption of the correct distance and phasing among the three sites results in a severe disruption of CD4 promoter activity. This implies that the factors binding to sites P2, P3 and P4 cooperate in the formation of the transcription initiation complex in a precise manner. To understand the individual contribution of each factor to the assembly of the initiation complex on the P2/3/4 functional cassette, a more detailed investigation of each factor-binding site is necessary.
MAZ binding of the P2 site
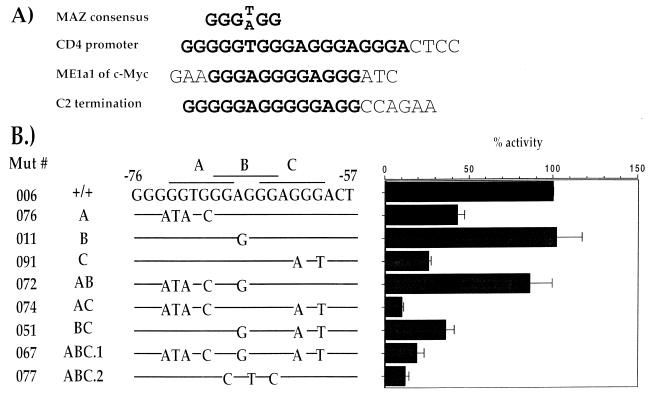
One of the predictions that comes out of the apparent strict distance/phasing requirement of the P2/3/4 functional cassette is that only one correct binding site exists at site P2. This is unexpected, since site P2 contains three overlapping consensus binding sites for the Zn finger transcription factor Maz (11). The consensus sequence for Maz binding is GGG(T/A)GG (Fig. 3A) (33), but the requirements for occupancy of the site remain unknown. Thus, it is possible that only one of the Maz-binding sites is preferentially utilized. To determine which of the sites is preferred for promoter function, we have generated both single and combination promoter mutations in the three sites and tested them in our functional assay (Fig. 3B).
Figure 3.
Functional analysis of the P2 site. (A) Alignment of the P2 sequence with the MAZ consensus sequence and other known MAZ-binding sites (33,40). (B) The sequence of the P2 probe, representing CD4 promoter sequence between –76 and –57 is shown. The three overlapping MAZ consensus sites A, B and C are indicated by a line above the sequence. Mutations in these sites are shown and aligned with the wild-type sequence. The mutations were generated in the context of the entire CD4 promoter –101 to +171. Functional activities of the mutant CD4 promoters are presented as in Figure 2.
The first set of mutations—076, 011 and 091—were designed to alter only one of the three consensus binding sites A, B or C, respectively (Fig. 3B). The mutation in site A produces only a partial decrease in promoter function, whereas a mutation in site B has no effect on promoter function. Interestingly, the site C mutation 091 produces a very significant effect on promoter function, resulting in an ∼75% decrease of promoter activity, indicating that site C is likely to be the critical MAZ binding site. The importance of site C is also supported by the effect of the double mutants 072, 074 and 051 on promoter function (Fig. 3B). These mutations combine in pairs the single mutations in P2. Mutation of the A and B sites shows no effect on promoter function, while mutations in the A and C or B and C sites abrogate promoter function (072, 074 and 051; Fig. 3B). Thus, promoter function is most severely affected if site C is mutated. Finally, mutation of all three sites significantly decreases promoter activity (076 and 077; Fig. 3B). These data taken together indicate the C site in P2 is the functional Maz-binding site.
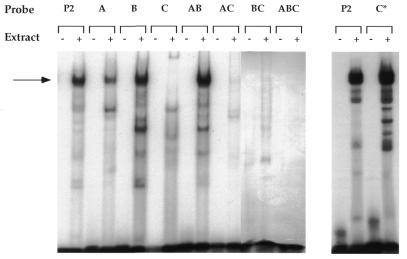
To address the question biochemically, we also analyzed the P2 mutants to determine their ability to bind MAZ by EMSA (Fig. 4). We have previously shown that MAZ binds to the P2 probe to form one major complex (11). Probes with mutations in site A or site B are capable of forming the same complex as the wild-type probe P2, while a probe with a mutation in site C results in complete inability to bind MAZ, indicating that MAZ binds to site C (Fig. 4). We cannot explain the decreased ability of mutant A (076) to bind MAZ, especially since the double mutant AB (072) binds Maz with equal strength to the wild-type P2 probe. It is possible that in the particular context of the CD4 promoter, site A can cooperate with site C for Maz binding and we are seeing the effect of the loss of cooperativity. Still, the mutant probes AC, BC and the triple mutant ABC cannot form the Maz complex, indicating that site C is the necessary and sufficient site for Maz binding (Fig. 4). These data are consistent with our hypothesis that MAZ binds to site C in the P2 site of the CD4 promoter. Taken together, these data indicate that consensus site C in P2 is the functional binding site for MAZ.
Figure 4.
EMSA of the P2 site. EMSA, using the CD4+CD8– D10 cell extract and radioactively labeled probes of P2 and its mutants. The probes are indicated above each pair of lanes and correspond to the mutations in Figure 3B; C* probe sequence corresponds to the mutant 019 sequence from Figure 2, including the five inserted base pairs. For each pair, the presence or absence of protein extract in the binding reaction is indicated by + or – respectively. The MAZ/P2 complex is indicated by an arrow.
In light of the insertional mutagenesis performed in close proximity to site C, it is important to determine if such mutation might affect the binding of Maz to P2. To determine if our insertion mutant 019 has any effect on binding, we introduced the same mutation in the probe, named C* in Figure 4. As can be seen, the binding of Maz to this probe is as good as to the wild-type P2 probe. Thus, we can conclude that not only the correct position of P2, but also the correct positioning of MAZ on the functional cassette of the CD4 promoter is important for its function.
A subclass-specific P3 binding factor
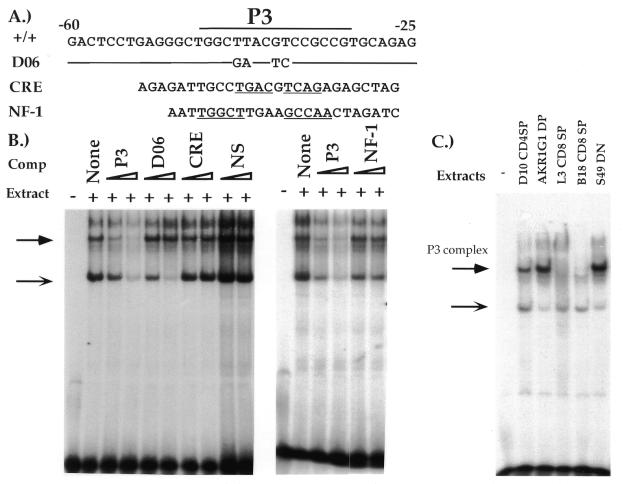
The P3 factor remains unknown; however, our functional data indicate that it plays an important role in CD4 promoter function. To characterize this factor further, we conducted biochemical experiments using the P3 site as a probe. As shown in Figure 5A, there are two consensus sequence motifs for known transcription factors in P3. At position –44, there is a CRE site; its cognate transcription factor, CREB-1, has been reported to bind to and transactivate the human CD4 promoter. In addition, there is an NF-1 recognition site at position –49, overlapping the CRE site. Thus, it is possible that both or either of these factors are binding to the murine CD4 promoter at P3 and mediating its function. Previously, we determined that the D06 mutation leads to a significant 84% decrease in promoter activity; as this mutation alters the consensus CRE and NF-1 sites, these data are also consistent with the hypothesis that these factors bind to P3 and mediate promoter function. To determine if this is the case, we conducted competition EMSA experiments with competitor oligonucleotides containing the D06 mutation, and consensus CRE and NF-1 sites (Fig. 5B). Using the P3 radioactive probe, we can detect two complexes that compete with non-radioactive probe, but not with non-specific DNA sequence, indicating that they bind specifically. The oligonucleotide containing the D06 mutation competes for the lower complex efficiently but not the upper, indicating that the factor forming the upper complex recognizes the functionally relevant region containing the consensus CRE and NF-1 sites. Interestingly, neither the consensus CRE nor the NF-1 oligonucleotides can compete for the formation of any of the P3-binding complexes, suggesting that CREB and NF-1 in fact do not bind to the P3 region. These conclusions are supported by the fact that we cannot supershift any of the complexes with anti-CREB-1 and anti-NF-1 antibodies (data not shown).
Figure 5.
Biochemical analysis of the P3 site. (A) The P3 sequence from the CD4 promoter is aligned with the consensus sequences for CREB-1 and NF-1. The two half sites of each consensus are underlined. Mutant D06, which causes a significant decrease of promoter activity (84%), is shown, aligned to the wild-type sequence. (B) EMSA of P3 with CD4+CD8– D10 cell extract. Two complexes are indicated with a filled arrow and with a thin arrow. Non-radioactively labeled competitors are indicated above the lanes and are used in 50–200-fold molar excess. The sequence of the competitors is the same as in (A). (C) EMSA with extracts from five different cell lines and the P3 probe. The cell lines and their developmental stage are indicated above each lane. Complexes are labeled with arrows as in (B).
To determine the cell-type specificity of the P3-binding factor, we conducted EMSA analyses with nuclear extracts purified from T cells of different phenotypes. Extracts from the DP cell line AKR1G1, the DN cell line S49, and the CD4 SP T cell clone D10 are all able to form the upper complex in EMSA experiments (Fig. 5C). However nuclear extracts from two different CD8 SP T cell clones form only the lower but not the upper complex. Taken together these data imply that there is a sequence-specific transcription factor important for CD4 promoter function that displays subclass-specific differences in its ability to bind to the P3 site.
Functional analysis of the P1 site
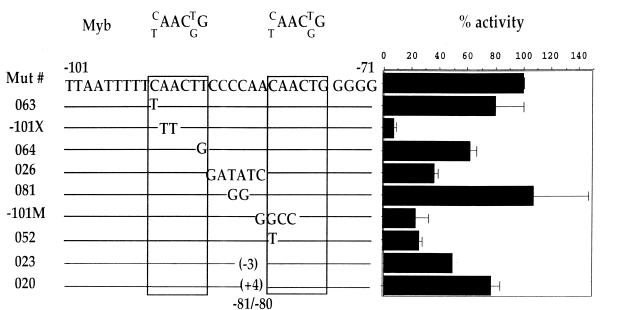
The P1 site contains two c-Myb consensus binding sites. Previously, it was shown that mutation of either one of the two sites causes a severe drop (80–90%) in CD4 promoter activity (–101X and –101M; Fig. 6) (12). To examine the relationship of the two Myb sites with promoter function in more detail, we generated a series of mutations around and within the P1 site. First, we generated mutations within the consensus Myb sites that do not change the sequence from consensus Myb sites and thus in principle do not affect the binding of c-Myb; as can be seen in Figure 6, most of these mutations do not alter CD4 promoter function appreciably. Interestingly, one of the mutations that alters a single base pair in the 3′ c-Myb site leads to a significant decrease in CD4 promoter activity, even though this mutation is normally tolerated by c-Myb (052; Fig. 6). It is possible that c-Myb binds only the 5′ recognition site, and a different factor binds to this 3′ region. Alternatively, it is possible that the 3′ site is a true c-Myb recognition sequence and that the binding of c-Myb to the 3′ site is preferred within the context of the CD4 promoter. These data demonstrate that the sequence context surrounding this site is also important for promoter function, suggesting that other functionally relevant factors may also be binding to this region. To test this hypothesis directly, we generated additional mutations that alter the adjacent nucleotides, but do not change the c-Myb site itself. As can be seen in Figure 6, mutation of all six bases between the two c-Myb sites (mutant 026) leads to significantly reduced promoter activity, whereas a smaller mutation of the central two bases does not, indicating that sequences directly adjacent to the c-Myb sites are important for promoter function, consistent with our hypothesis.
Figure 6.
Functional analysis of the P1 site. The sequence of the P1 probe, representing CD4 promoter sequence between –101 and –72 is shown. The c-Myb consensus sequence is given and aligned with the P1 sequence. The two c-Myb consensus sites are boxed. Mutations in these sites, as well as in the spacer sequence are shown and aligned with the wild-type sequence. In the mutants where insertions or deletions were generated, the number and position of the affected base pairs is given. The mutations were generated in the context of the entire CD4 promoter –101 to +171. Functional activities of the mutant CD4 promoters are presented as in Figure 2.
To investigate whether the spacing between the two Myb sites is important, mutations changing the distance/phasing between the two Myb sites were generated (020 and 023; Fig. 6). Increasing the distance between the two c-Myb sites by 4 bp, while preserving the context around the sites (020; Fig. 6), has a minor effect on promoter activity, indicating that neither the distance nor the phasing between the two c-Myb sites are important. The second mutation decreases the distance between the two Myb sites in P1 by 3 bp (023; Fig. 6). This has a partial effect on promoter function, suggesting either that there are some steric requirements for binding, or that this mutation deletes an additional factor-binding site between the c-Myb sites. Thus, the P1 sequences required for full promoter function contain more than just the Myb recognition sites, indicating that additional factors may be binding to this region.
DISCUSSION
The P2/3/4 functional cassette
Our mutagenesis experiments revealed that the CD4 promoter sites P2, P3 and P4 are dependent on their position relative to each other. Previous work in other systems has described how the precise spacing between two factor-binding sites can be critical for transcriptional control element function (16,17,19,22,23). In these cases, the dependence of the spacing distance is attributed to the specific interaction between the two DNA-binding factors. The CD4 promoter is different in that multiple factor-binding sites are required to be positioned precisely in relation to each other, in effect forming an inseparable functional cassette. The precise requirements of assembly enforced by the P2/3/4 cassette make it difficult to assemble a functional transcription initiation complex unless all of the factors are present and binding to the promoter in their correct modification state. The deletion of any one of the factors will cripple promoter function and prohibit the transcription of the CD4 gene. In contrast to the promoter, a high level of redundancy exists in the other transcriptional control elements of the CD4 gene, notably in the silencer (9). Three functionally redundant factor-binding sites have been described in the CD4 silencer (9,34,35). Deleting any one of the three sites, which in some cases produces alterations in distance/phasing, does not affect silencer function. Thus, the silencer has very relaxed requirements for function and a large number of configurations of a putative silencer complex can be productively assembled without sacrificing function. This property is important for the silencer since it must be able to act in many different situations at several different stages of T-cell development. In contrast, the promoter functions on its own only in activated CD4 SP T cells, and thus does not require flexibility in inducing its function (12,14). This flexibility may in fact be detrimental to the cell. For example, should CD4 promoter function during activation be dependent on the induction of only one of the promoter-binding factors, the induction of CD4 could occur inappropriately in response to the coincidental induction of a single factor for other purposes. The requirement that all promoter-binding factors be present and binding in the appropriate configuration insures that the CD4 promoter is induced to function only by the appropriate stimulatory signals.
Amplifying the basal function of the CD4 promoter: c-Myb and Maz
c-Myb is important for T-cell development (36) and plays an important role in CD4 promoter and silencer function (12, R.Allen, S.D.Sarafova and G.Siu, submitted for publication). Our studies indicate that the P1 site is likely to consist of additional factor-binding sites in addition to the two c-Myb recognition sites. These observations in turn suggest that the P1-binding complex contains additional as-yet uncharacterized factors that are critical for promoter function. As c-Myb binds to both positive and negative regulatory elements in the CD4 locus (7, R.Allen, S.D.Sarafova and G.Siu, submitted for publication), the requirement for functional cofactors indicates that whether or not c-Myb activates or represses transcription depends on the context in which it binds to the element. In binding to the promoter, c-Myb may bind with closely associated cofactors to induce transcription; in binding to the silencer, c-Myb may bind with a different set of cofactors, or on its own. The nature of this complex is currently being studied; although c-Myb is known to associate with several different cofactors, we have been unable to demonstrate that these interactions are playing a role in CD4 promoter function (data not shown).
Although there are three directly abutting consensus MAZ sites in the P2 region, only one is important for promoter function. In addition to the MAZ consensus sequence itself, the position of the site relative to the other factor-binding sites in the promoter is critical for function. The requirement of precise position and phasing of the MAZ site with respect to the other sites in a transcriptional control element are indicative of a structural role for MAZ. Consistent with this, work by Bossone and others has demonstrated that MAZ is capable of inducing a bend in the DNA helix upon binding to its site (30,33). Thus, in our model, the induction of a bend in the DNA by MAZ places the c-Myb-containing P1-binding complex adjacent to the other factors, thus assisting in the construction of the initiation complex assembly. MAZ thus plays primarily a structural role in CD4 promoter function, insuring that all of the promoter-binding factors are capable of interacting to form the initiation complex. A functional role for MAZ is not ruled out and is compatible with our model.
A model for CD4 promoter function
Our studies on the CD4 promoter have enabled us to propose an overall model for its role in mediating CD4 expression. As discussed above, the CD4 promoter functions either alone or with the other CD4 transcriptional control elements at different stages of T-cell development and activation (37,38). In activated CD4 SP TH cells, the CD4 promoter is induced to function at high levels in a subclass-specific manner independent of the CD4 mature enhancer, implying the existence of subclass-specific promoter-binding factors (14). We have reported previously that Elf-1 is capable of transactivating the CD4 promoter in CD4 but not CD8 SP T cells (27). Here we report that the P3-binding factor is not present in CD8 SP TC cells; thus, in our model a combination of these factors is inducing the subclass-specific function of the CD4 promoter. In resting CD4 SP TH cells, the mature enhancer induces promoter function, and thus the function of Elf-1 and the P3-binding factor are not limiting. In activated CD8 SP TC cells, the absence of the P3-binding factor and the non-functional state of Elf-1 (26) prevents the induction of CD4 promoter function, since a productive initiation complex is difficult to assemble. In resting CD8 SP TC cells, the promoter is also non-functional, and mature enhancer function is silenced; thus, CD4 transcription is turned off. Therefore, the specificity of control of CD4 gene expression differs in resting as opposed to activated mature T cells: in resting T cells, specificity is mediated by the enhancer and the silencer, whereas in activated T cells, specificity is mediated by the Elf-1 and P3 promoter-binding factors. This promoter-induced activation of CD4 transcription is consistent with the physiological expression of CD4; Ridgway et al. (39) have demonstrated that CD4 expression is upregulated upon antigenic stimulation in peripheral TH cells during the development of memory T cells. These data suggest that in specific situations, the CD4 promoter is capable of overriding the function of the other CD4 transcriptional control elements and inducing CD4 expression. The study of the factors that mediate this induced expression will enable us to characterize better the mechanisms in which activated TH cells mediate their response to antigen.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Kathryn Calame, Chris Schindler and Konstantina Alexandropoulos for helpful discussions and critical reading of the manuscript. We also thank Wendy Rokach for technical assistance. This work was supported by a grant from the National Institutes of Health (RO1 AI34925). Core facilities were provided by the Columbia-Rockefeller Center for AIDS Research (P30 AI42848 from the NIH).
REFERENCES
- 1.Sarafova S.D. and Siu,G. (1999) Braz. J. Med. Biol. Res., 32, 785–803. [DOI] [PubMed] [Google Scholar]
- 2.Littman D.R. (1987) Annu. Rev. Immunol., 5, 561–584. [DOI] [PubMed] [Google Scholar]
- 3.Perlmutter R.M., Marth,J.D., Ziegler,S.F., Garvin,A.M., Pawar,S., Cooke,M.P. and Abraham,K.M. (1988) Biochim. Biophys. Acta, 948, 245–262. [DOI] [PubMed] [Google Scholar]
- 4.Parnes J.R. (1989) Adv. Immunol., 44, 265–311. [DOI] [PubMed] [Google Scholar]
- 5.Sawada S. and Littman,D.R. (1991) Mol. Cell. Biol., 11, 5506–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawada S. and Littman,D.R. (1993) Mol. Cell. Biol., 13, 5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siu G., Wurster,A.L., Duncan,D.D., Soliman,T.M. and Hedrick,S.M. (1994) EMBO J., 13, 3570–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawada S., Scarborough,J.D., Killeen,N. and Littman,D.R. (1994) Cell, 77, 917–929. [DOI] [PubMed] [Google Scholar]
- 9.Duncan D.D., Adlam,M. and Siu,G. (1996) Immunity, 4, 301–311. [DOI] [PubMed] [Google Scholar]
- 10.Adlam M., Duncan,D.D., Ng,D.K. and Siu,G. (1997) Int. Immunol., 9, 877–887. [DOI] [PubMed] [Google Scholar]
- 11.Duncan D.D., Stupakoff,A., Hedrick,S.M., Marcu,K.B. and Siu,G. (1995) Mol. Cell. Biol., 15, 3179–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siu G., Wurster,A.L., Lipsick,J.S. and Hedrick,S.M. (1992) Mol. Cell. Biol., 12, 1592–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salmon P., Giovane,A., Wasylyk,B. and Klatzmann,D. (1993) Proc. Natl Acad. Sci. USA, 90, 7739–7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna Z., Simard,C., Laperriere,A. and Jolicoeur,P. (1994) Mol. Cell. Biol., 14, 1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao-Emonet J.C., Boyer,O., Cohen,J.L. and Klatzmann,D. (1998) Biochim. Biophys. Acta, 1442, 109–119. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan T.D., Schiefelbein,J.W.,Jr and Nelson,O.E.,Jr (1989) Dev. Genet., 10, 412–424. [DOI] [PubMed] [Google Scholar]
- 17.Lalioti M.D., Scott,H.S. and Antonarakis,S.E. (1999) Hum. Mol. Genet., 8, 1791–1798. [DOI] [PubMed] [Google Scholar]
- 18.Arcot S.S. and Deininger,P.L. (1992) Gene, 111, 249–254. [DOI] [PubMed] [Google Scholar]
- 19.Nikolajczyk B.S., Nelsen,B. and Sen,R. (1996) Mol. Cell. Biol., 16, 4544–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolajczyk B.S., Cortes,M., Feinman,R. and Sen,R. (1997) Mol. Cell. Biol., 17, 3527–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellomy G.R., Mossing,M.C. and Record,M.T.,Jr (1988) Biochemistry, 27, 3900–3906. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z. and Little,J.W. (1998) J. Mol. Biol., 278, 331–338. [DOI] [PubMed] [Google Scholar]
- 23.Merkel T.J., Dahl,J.L., Ebright,R.H. and Kadner,R.J. (1995) J. Bacteriol., 177, 1712–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterman M.L. and Jones,K.A. (1990) New Biol., 2, 621–636. [PubMed] [Google Scholar]
- 25.Schreiber E., Matthias,P., Muller,M.M. and Schaffner,W. (1989) Nucleic Acids Res., 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- 27.Sarafova S. and Siu,G. (1999) J. Biol. Chem., 274, 16126–16134. [DOI] [PubMed] [Google Scholar]
- 28.Flamand L., Romerio,F., Reitz,M.S. and Gallo,R.C. (1998) J. Virol., 72, 8797–8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goyal N., Knox,J. and Gronostajski,R.M. (1990) Mol. Cell. Biol., 10, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks C.L. and Shenk,T. (1996) J. Biol. Chem., 271, 4417–4430. [DOI] [PubMed] [Google Scholar]
- 31.Crepieux P., Coll,J. and Stehelin,D. (1994) Crit. Rev. Oncog., 5, 615–638. [PubMed] [Google Scholar]
- 32.Oh I.H. and Reddy,E.P. (1999) Oncogene, 18, 3017–3033. [DOI] [PubMed] [Google Scholar]
- 33.Bossone S.A., Asselin,C., Patel,A.J. and Marcu,K.B. (1992) Proc. Natl Acad. Sci. USA, 89, 7452–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H.K. and Siu,G. (1998) Mol. Cell. Biol., 18, 7166–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim W.W. and Siu,G. (1999) J. Exp. Med., 190, 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen R.D.III, Bender,T.P. and Siu,G. (1999) Genes Dev., 13, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robey E. and Fowlkes,B.J. (1994) Annu. Rev. Immunol., 12, 675–705. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki H., Punt,J.A., Granger,L.G. and Singer,A. (1995) Immunity, 2, 413–425. [DOI] [PubMed] [Google Scholar]
- 39.Ridgway W., Fasso,M. and Fathman,C.G. (1998) J. Immunol., 161, 714–720. [PubMed] [Google Scholar]
- 40.Ashfield R., Patel,A.J., Bossone,S.A., Brown,H., Campbell,R.D., Marcu,K.B. and Proudfoot,N.J. (1994) EMBO J., 13, 5656–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]