Summary:
Inflammatory bowel disease (IBD) is driven by host genetics and environmental factors, including commensal microorganisms. Speckled Protein 140 (SP140) is an immune-restricted chromatin ‘reader’ that is associated with Crohn’s disease (CD), multiple sclerosis (MS) and chronic lymphocytic leukemia (CLL). Yet, the disease-causing mechanisms of SP140 remain undefined. Here we identify an immune-intrinsic role for SP140 in regulating phagocytic defense responses to prevent the expansion of inflammatory bacteria. Mice harboring altered microbiota due to hematopoietic Sp140 deficiency exhibited severe colitis that was transmissible upon co-housing and ameliorated with antibiotics. Loss of SP140 results in blooms of Proteobacteria, including Helicobacter in Sp140−/− mice and Enterobacteriaceae in humans bearing the CD-associated SP140 loss-of-function variant. Phagocytes from patients with the SP140 loss-of-function variant and Sp140−/− mice exhibited altered antimicrobial defense programs required for control of pathobionts. Thus, mutations within this epigenetic reader may constitute a predisposing event in human diseases provoked by microbiota.
Graphical Abstract
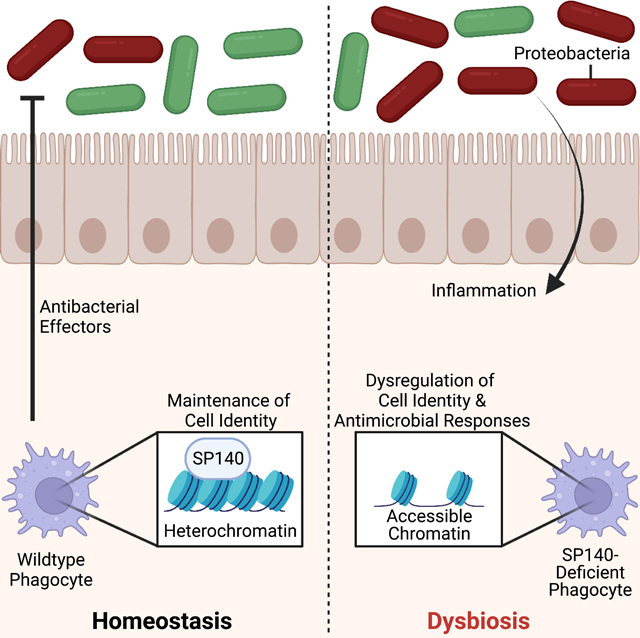
Introduction
Complex immune diseases, including inflammatory bowel disease (IBD) and multiple sclerosis (MS) are multifactorial diseases that develop as a result of gene-environment interactions (Graham and Xavier, 2020). Genome-wide association studies have identified over 240 genetic risk alleles that are associated with the two types of IBD: Ulcerative Colitis (UC) and Crohn’s disease (CD) (Jostins et al., 2012, Rivas et al., 2011, Liu et al., 2015). Many of these risk alleles overlap with loci that associate with MS (International Multiple Sclerosis Genetics et al., 2013) suggesting common disease initiating mechanisms. One shared CD and MS risk allele is SP140. SP140 is an immune-restricted member of the Speckled Protein (SP) epigenetic ‘reader’ family, consisting of SP100, SP110, SP140 and SP140L, which have high sequence homology with Autoimmune Regulator (AIRE) (Fraschilla and Jeffrey, 2020). Epigenetic readers are diverse proteins with specialized docking domains that ‘read’ covalent modifications primarily on histones to regulate transcription (Arrowsmith et al., 2012). To achieve this function, SPs all contain 3 ‘reader’ domains: 1) a SAND domain (named after the few SAND domain-containing proteins: SP100, AIRE, NucP41/P75, and DEAF) that interacts with DNA directly or through protein-protein interactions, 2) a plant homeodomain (PHD) that docks to histone methylation, and 3) a bromodomain that binds acetylated histones (Filippakopoulos et al., 2012, Bottomley et al., 2001, Waterfield et al., 2014, Bienz, 2006). Although, the PHD of SP140 may be atypical by facilitating SUMOylation of the bromodomain and associating with SETDB1, a histone methyltransferase of H3K9 that promotes gene silencing (Ivanov et al., 2007, Zucchelli et al., 2019, Peng and Wysocka, 2008, Garcia-Dominguez et al., 2008, Zhang et al., 2016). In addition, SP140 contains a caspase activation and recruitment domain (CARD) involved in multimerization and co-localization to ‘speckled’ promyelocytic leukemia (PML) nuclear bodies, macromolecular multiprotein complexes with diverse functionality including transcriptional repression (Hoischen et al., 2018, Corpet et al., 2020, Huoh et al., 2020).
Consistent with the predicted role for SP140 in gene silencing, in human macrophages, SP140 was found to predominantly occupy and maintain inaccessibility of promoters of developmentally silenced loci, such as the HOXA cluster (Mehta et al., 2017). HOXA9 is a known promoter of stem-like state in hematopoietic stem cells (HSCs) and an inhibitor of macrophage differentiation and glycolysis (Zhou et al., 2018, Thorsteinsdottir et al., 2002, Huang et al., 2012, Argiropoulos and Humphries, 2007). HOXA9 is therefore normally silenced in mature macrophages (De Santa et al., 2007), but not in SP140-deficient mouse or human macrophages, that ultimately display defective transcriptional responses to bacteria or viral ligands (Mehta et al., 2017). A global proteomics analysis subsequently found that SP140 directly represses topoisomerases to maintain heterochromatin, gene silencing, and macrophage responsiveness (Amatullah et al., 2022). Furthermore, an array of studies has now implicated SP140 as an essential factor in antibacterial, antiviral, and antiparasitic responses (Ji et al., 2021, Madani et al., 2002, Regad and Chelbi-Alix, 2001, Matsushita et al., 2021). However, despite some progress in understanding the function of this enigmatic epigenetic reader in macrophages, the role of SP140 in the immune response to intestinal microbiota, and how disruption of this function due to human genetic variation in SP140 may drive development of Crohn’s disease or MS remains unclear.
Development of complex immune disorders, such as IBD and MS, is dependent on both host genetics as well as cues derived from intestinal microbes and their metabolites (Wu et al., 2020, Graham and Xavier, 2020, Amatullah and Jeffrey, 2020, Schulthess et al., 2019, Blander et al., 2017, Vinolo et al., 2011, Kaiko et al., 2016, Iliev and Cadwell, 2021). Innate immune pathways are crucial for integrating bacterial, viral and fungal signals from the intestinal microenvironment to regulate gene expression, microbial balance and intestinal homeostasis (Iliev and Cadwell, 2021, Schulthess et al., 2019, Adiliaghdam et al., 2022). Moreover, many IBD-associated mutations are within genes essential for innate immune defense pathways, emphasizing their requirement for maintaining homeostatic host-microorganism relationships (Graham and Xavier, 2020, Jostins et al., 2012, Kugelberg, 2014). CD-associated mutations in SP140 result in defective mRNA splicing and a reduction in SP140 protein (Mehta et al., 2017, Matesanz et al., 2015) rendering innate immune cells hypo-responsive. Further, mouse models support that SP140 is normally required for intestinal homeostasis via an innate immune function, as hematopoietic knockdown (Mehta et al., 2017) or whole mouse deletion of Sp140 exacerbates dextran sulfate sodium (DSS)-induced colitis in a manner dependent on macrophages (Amatullah et al., 2022). In this study, we investigated the possible link between intestinal microbial communities and SP140 control of intestinal homeostasis. We found that disruptions in SP140 result in a defective microbicidal transcriptome in phagocytes but enhanced activation of CD8+ T cells in colonic lamina propria and exacerbated colitis. Remarkably, these intestinal malfunctions were dependent on the expansion of Proteobacteria, including Helicobacter, that was transmissible upon co-housing with wild-type mice and treated with antibiotics. This was further corroborated in CD patients bearing SP140 loss-of-function mutations that displayed significant elevation in Proteobacteria Enterobacteriaceae. Taken together, we have identified a key innate immune-intrinsic role for epigenetic reader SP140 in preventing intestinal inflammation by restricting the pathological expansion of a common member of the intestinal microbiota.
Results
SP140 is essential for antibacterial pathways in mice and human
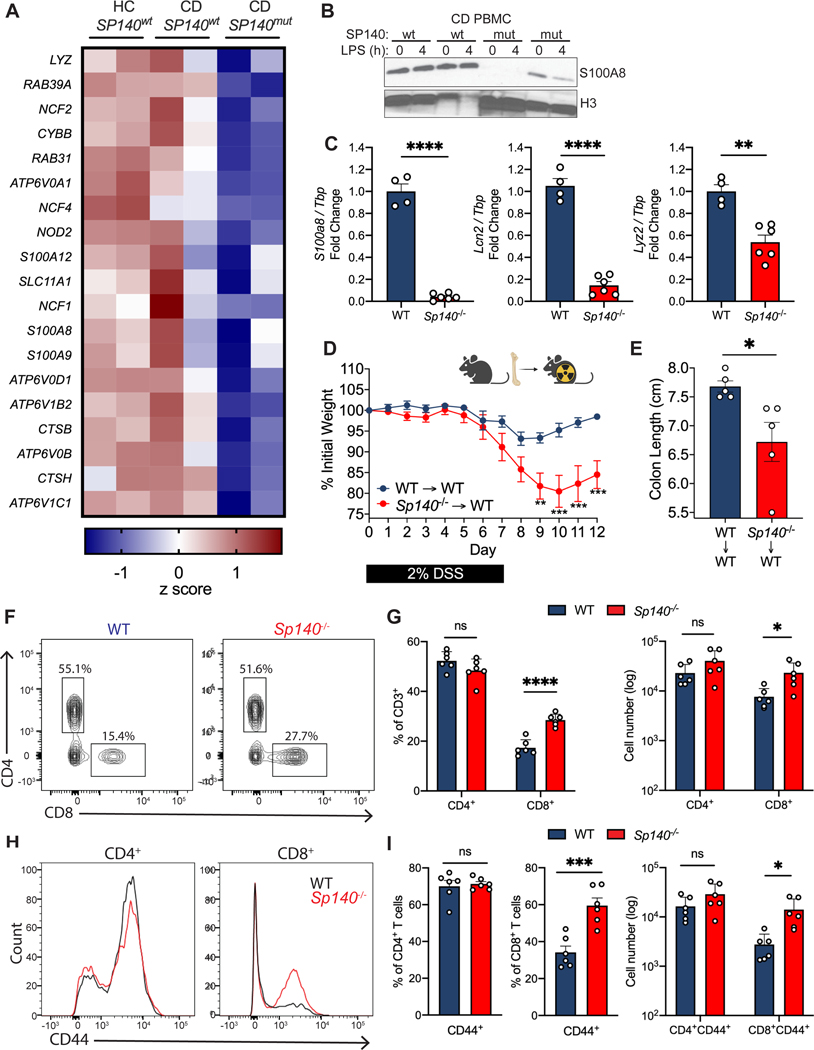
SP140 is an essential host regulator of antibacterial defense to Mycobacterium tuberculosis (Mtb) (Ji et al., 2021, Pan et al., 2005, Pichugin et al., 2009). We previously determined that SP140 deficient mouse and human macrophages have a defect in clearing gram negative bacteria (Salmonella enterica serovar Typhimurium, Escherichia coli, and Citrobacter rodentium) (Amatullah et al., 2022), but a loss of SP140 does not lead to defects in phagocytosis (S Fig. 1A,B). Therefore, we sought to understand the mechanism by which SP140 contributes to antibacterial host defense and how SP140 deficiency may impact intestinal microbial dysbiosis and inflammation in CD patients bearing loss-of-function mutations. We examined transcriptional programs of human peripheral blood mononuclear cells (PBMCs) obtained from healthy controls expressing wild-type SP140 (HC SP140wt), Crohn’s disease (CD) patients expressing wildtype SP140 (CD SP140wt), or CD patients that were homozygous for CD-associated SP140 mutations (CD SP140mut), obtained through the Prospective Registry in IBD Study at MGH (PRISM) cohort. A prominent gene set downregulated in CD SP140mut cells compared to both healthy controls and CD SP140wt PBMCs included genes associated with bacterial defense (Fig. 1A). CD SP140mut cells had reduced expression of transcripts encoding for subunits of NADPH oxidase (CYBB, NCF1, NCF2, NCF4) and H+-ATPase (ATP6V0A1, ATP6V0D1), which are known complexes that act at the phagosome to produce antibacterial reactive oxygen species (ROS) and low pH, respectively. Moreover, at steady state, genes essential for the degradation or killing of bacteria, including calprotectin subunits (S100A8, S100A9), lysozyme (LYZ), and cathepsins (CTSB, CTSH), were also reduced in CD SP140mut cells. Since calprotectin is a well-studied antimicrobial protein capable of enhancing bacterial killing, we confirmed reduced protein levels of calprotectin component S100A8 in CD SP140mut PBMCs compared to SP140wt CD patient donors (Fig. 1B).
Figure 1. SP140 is essential for antimicrobial gene programs in mouse and human.
(A) Heat map of genes down-regulated (log2FC>2) in human peripheral blood mononuclear cells (PBMC) from healthy controls (HC), Crohn’s disease (CD) patients bearing wildtype SP140 (SP140wt) or CD-risk SP140 genetic variant (SP140mut) (n=2, RNA-seq) from GSE89876. (B) Immunoblot of S100A8 in PBMC from SP140wt or SP140mut CD patients after 0 or 4 hours LPS stimulation (100 ng/mL). (C) Expression of mouse S100a8, Lcn2, and Lyz2 mRNA relative to Tbp, as determined by qPCR, in mouse bone marrow-derived macrophages (BMDMs) from wildtype (WT) and Sp140−/− mice. (D) Daily body weight of WT and Sp140−/− mice and (E) quantification of day 12 colon lengths after 2% dextran sulfate sodium (DSS) administration (n=5). (F-I) Flow cytometry analysis of WT and Sp140−/− mouse colon lamina propria (n=6). (F) Representative flow cytometry plots of live CD45+CD3+ cells gated on CD4+ and CD8+ T cells. (G) Quantification of frequency and total count of CD4+ and CD8+ T cells. (H) Representative histogram overlay of CD44 expression in CD4+ and CD8+ T cells. (I) Quantification of frequency and total count of CD44+CD4+ and CD8+ T cells. Data are mean of 2–6 biological replicates. Error bars represent means ± SEM. n.s., not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; unpaired t tests for (C), (E), (F), (H), two-way ANOVA for (D).
Human and mouse SP140 are only 54% homologous at the amino acid level, owing to inclusion of an intrinsically disorder region (IDR) in human SP140 (Fraschilla and Jeffrey, 2020). However, we found that antimicrobial transcripts, such as S100a8, Lyz2 and Lcn2, were also downregulated in Sp140−/− bone-marrow derived macrophages (BMDMs) (Fig. 1C). The conserved role of SP140 in controlling antimicrobial gene programs suggests that this chromatin reader plays an essential role in host defense. Since previous analysis of genome-wide occupancy of SP140 in human macrophages found that SP140 did not directly occupy antimicrobial gene loci and promoted gene repression at silenced chromatin regions (Mehta et al., 2017), SP140 likely promotes an antibacterial gene program via maintenance of heterochromatin and macrophage identity.
SP140 expression has been shown to be immune-restricted and particularly high in macrophages (Mehta et al., 2017). Intestinal macrophages, which differentiate from circulating monocytes (Bain et al., 2014), and other phagocytes in the intestine are responsible for bactericidal responses, namely production of ROS and antimicrobial proteins (Schulthess et al., 2019, Smythies et al., 2005). Furthermore, other IBD-associated loci have similarly been shown to regulate microbial responses in the intestine via epithelial cells (Graham and Xavier, 2020, Ramanan et al., 2014, Conway et al., 2013), which are resistant to irradiation. To determine whether SP140 expression in immune cells is sufficient to protect against intestinal inflammation, we irradiated wildtype (WT) mice and reconstituted them with WT or Sp140−/− bone marrow. Mice with specific Sp140 deficiency in the hematopoietic compartment displayed significantly exacerbated DSS-colitis compared to WT controls, as determined by weight loss (Fig. 1D) and colon length (Fig. 1E). Thus, Sp140 within the immune compartment is essential for intestinal homeostasis.
To extend our ex vivo characterization of antibacterial programs in mouse and human primary cells, we next investigated whether Sp140 deficiency alters the development of intestinal phagocyte populations that are necessary for bacterial clearance. We performed immunophenotyping of colonic lamina propria CD45+ cells by flow cytometry analysis in Sp140−/− and WT mice. Sp140 deficiency did not affect the proportions of mononuclear phagocyte populations (Fig. S2C) or B cells (Fig. S2A, B) suggesting that recruitment and development of these cell types is intact in Sp140−/− mice. However, we observed a significant and specific expansion of total CD8+ T cells (Fig. 1F, G) as well as a significant increase in the frequency and number of activated CD8+CD44+ T cells (Fig. 1H, I) in the colonic lamina propria of Sp140−/− mice. Intriguingly, this defect was specific to CD8+ T cells as CD4+ T cells and CD4+CD44+ T cell frequencies and numbers were unaltered with Sp140 deficiency (Fig. 1F-I). Taken together, these results show that Sp140 is required for macrophage antibacterial responses and a loss of Sp140 leads to CD8+ T cell activation in the gut lamina propria prior to the manifestation of disease.
Cohousing wild-type mice with Sp140−/− mice increased severity of induced colitis
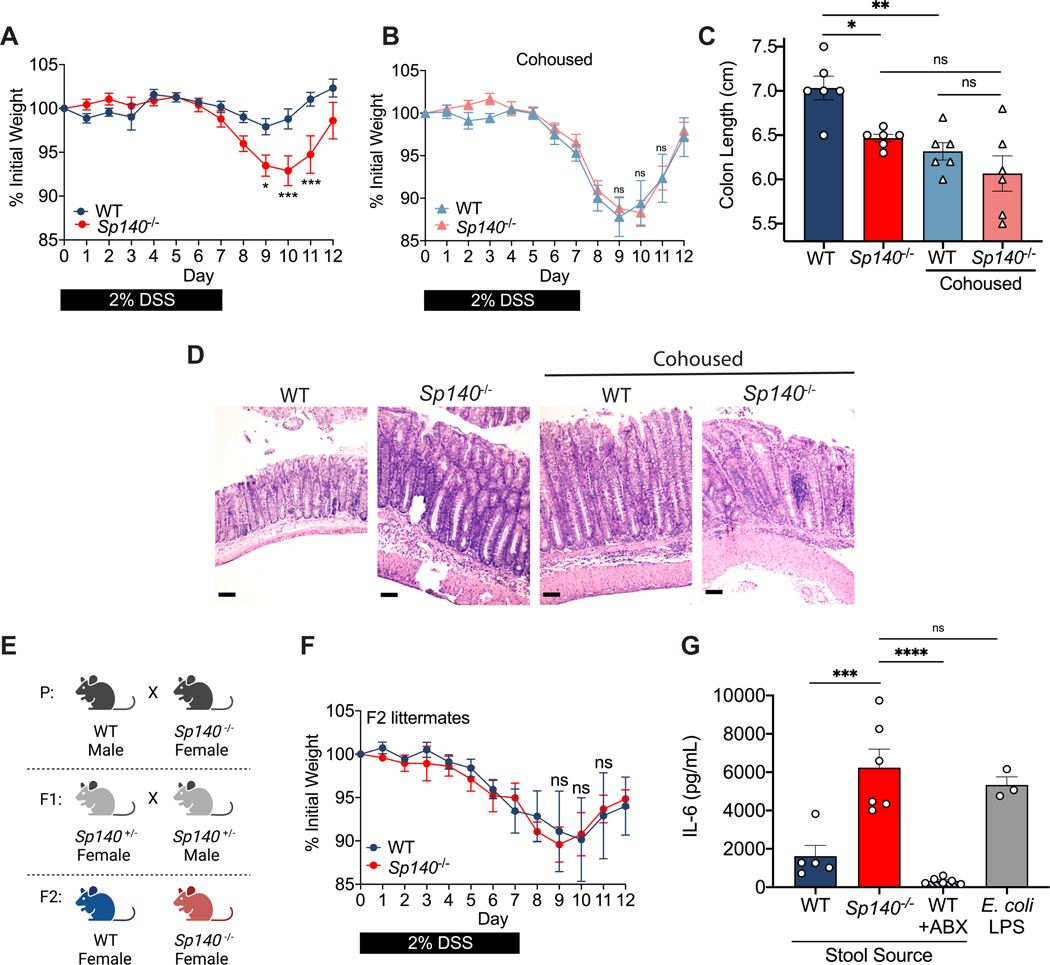
Mice with an shRNA-mediated hematopoietic knockdown of Sp140 and Sp140−/− mice generated by CRISPR/Cas9 targeting are more susceptible to DSS-mediated colitis, in a manner dependent on Sp140 control of cytokine production and bacterial killing in macrophages (Mehta et al., 2017, Amatullah et al., 2022). Further, an abundance of studies has demonstrated that mice deficient in innate immune system components, or cytokines, have altered microbiota with increased inflammatory capacity (Ramanan et al., 2014, Elinav et al., 2011, Zenewicz et al., 2013, Frederic et al., 2012, Caruso et al., 2019). Moreover, presence of activated CD8+ T cells in the colon frequently associates with microbiota dysbiosis (Yu et al., 2020, Tanoue et al., 2019, Ramanan et al., 2014). We therefore questioned whether the microbiota of Sp140−/− mice could contribute to the exacerbated colitis phenotype. We designed an experiment in which WT mice were cohoused with Sp140−/− mice or were housed separately. After 4 weeks of cohousing, a period previously established to be sufficient for the transmission of microbiota between mice via coprophagia (Zenewicz et al., 2013, Stappenbeck and Virgin, 2016a), we administered DSS and examined the resulting weight loss and inflammation associated with colitis. Whereas Sp140−/− mice exhibit exacerbated colitis compared to WT mice when housed separately, cohousing WT mice with Sp140−/− mice led to enhanced weight loss of WT mice, similar to separated or cohoused Sp140−/− mice (Fig. 2A, B). To examine inflammation, we measured colon lengths at day 12 post-DSS administration and found significantly shorter colons in WT mice that were cohoused with the Sp140−/− mice than WT mice that were housed separately (Fig. 2C). Histological examination of colons confirmed an increase in inflammation in WT mice specifically upon co-housing as shown by crypt elongation, thickening of the mucosa, and a leukocyte infiltrate to the level of the lamina submucosa (Fig. 2D). The transmission of the altered gut microbiota from Sp140−/− mice to cohoused WT mice, along with increased susceptibility to DSS-induced colitis, indicates that the altered gut microbiota works as a contributing factor for, rather than a consequence of, the disease. Beyond extended cohousing, we also induced DSS-colitis in WT and Sp140−/− F2 littermates from a heterozygous cross (Fig. 2E), another method used to standardize the microbiota between mice (Robertson et al., 2019, Stappenbeck and Virgin, 2016b). Like cohoused mice, unseparated WT and Sp140−/− littermates displayed similar weight loss as a result of intestinal injury (Fig. 2F). Finally, to understand whether the microbiome of Sp140−/− mice is colitogenic due to increased inflammatory capacity, we collected and filtered stool homogenates from untreated Sp140−/− mice and controls and stimulated BMDMs in vitro. Stool homogenates obtained from Sp140−/− mice proved to be hyperinflammatory compared to WT stool, as shown by a significant increase in the production of the pro-inflammatory cytokine interleukin (IL)-6 (Fig. 2G).
Figure 2. The pro-inflammatory intestinal microbiome of Sp140−/− mice is transferable and exacerbates colitis.
(A) Daily body weight of separately housed wildtype (WT) and Sp140−/− mice after 2% dextran sulfate sodium (DSS) administration (n=6). (B) Daily body weight of cohoused WT and Sp140−/− mice after 2% DSS administration (n=6). (C) Day 12 colon lengths of separated or cohoused WT and Sp140−/− mice after 2% DSS administration (n=6). (D) Representative hematoxylin and eosin-stained sections of day 12 distal colon tissue from separated or cohoused WT and Sp140−/− mice after 2% DSS administration. (E) Breeding scheme for F2 littermates used to normalize microbiota between WT and Sp140−/− mice. (F) Daily body weight of WT and Sp140−/− F2 littermates after 2% DSS administration (n=5). (G) IL-6 production, as determined by ELISA, of bone marrow-derived macrophages (BMDMs) supernatants after 16 hours stimulation with LPS (1mg/mL) or stool homogenates (1mg/mL) from WT (n=5), Sp140−/− (n=6), or broad-spectrum antibiotic treated WT mice (n=3); representative of 3 experiments. Error bars represent means ± SEM. n.s., not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; two-way ANOVA for (A) and (B), one-way ANOVA for (C) and (E).
Sp140−/− mice have altered commensal microbiota that is transmissible
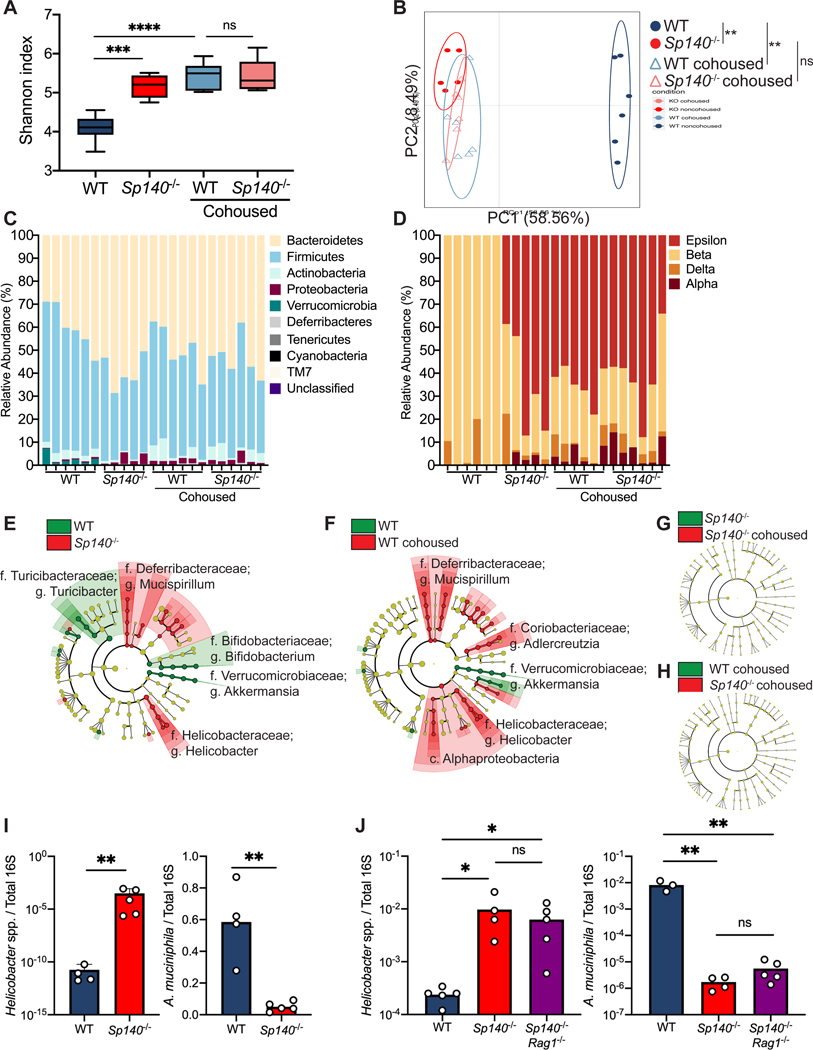
Our data support the hypothesis that Sp140−/− mice have an altered microbiota that is transmissible to wild-type mice. Thus, we undertook bacterial 16S rRNA gene pyrosequencing to examine the microbiome in four groups of mice: WT mice, Sp140−/− mice, and WT mice or Sp140−/− mice cohoused with each other. Prior to DSS treatment we collected fecal samples from each mouse, prepared fecal DNA, and used barcoded pyrosequencing of the 16S rRNA gene V4 region to explore the change in the microbiome. A significant increase in community richness in Sp140−/− mice was observed. Furthermore, this increase in richness was transferred to cohoused WT mice compared to non-cohoused WT controls (Fig. 3A). UniFrac-based principal component analysis (PCA) determined that WT and Sp140−/− microbiome communities were significantly dissimilar from each other (Fig. 3B). Moreover, this difference was lost upon cohousing whereby WT microbiomes now overlapped with Sp140−/− communities, demonstrating a dominance of Sp140−/− microbiota (Fig. 3B). Proportional taxonomy analysis revealed that Sp140−/− mouse stool contained a higher percentage of Proteobacteria phyla than WT mice, and Proteobacteria then bloomed in WT cohoused mice (Fig. 3C) thus identifying a putative, transmissible bacteria of the Sp140−/− mouse microbiome that may confer sensitivity to colitis. Proteobacteria expansion is associated with worsened colitis in mouse models (Garrett et al., 2010, Ni et al., 2017). Furthermore, we specifically determined that Sp140−/− intestines were colonized by a greater percentage of classes Epsilonproteobacteria and Alphaproteobacteria (Fig. 3D). Conversely, Sp140−/− exhibited reduced Verrucomicrobia phyla that are present in wild-type controls (Fig. 3C). Unbiased Linear discriminant analysis Effect Size (LEfSe) analysis determined a significant elevation of colitis-driving Helicobacter genera of the Epsilonproteobacteria class and Mucispirillum genera of the Deferribacteria phylum in Sp140−/− mice compared to WT controls (Fig. 3E). Notably, a significant reduction in the protective Akkermansia muciniphila was also observed in Sp140−/− mice (Fig. 3E). Furthermore, the Sp140 deficient microbiome was efficiently transferred to cohoused WT mice (Fig. 3F) to the point where no significant differences were detected between Sp140−/− mice and WT cohoused mice by LEfSe analysis (Fig. 3G-H). Thus, Sp140 deficiency triggers the development of a colitogenic microbiome, that is efficiently transferred upon cohousing.
Figure 3. Sp140 deficiency in mice permits a Helicobacter bloom and a reduction in Akkermansia.
(A) Diversity of fecal bacterial communities of separately housed or cohoused wildtype (WT) and Sp140−/− mice as determined by Shannon Index. (B) Principal Coordinate Analysis (PCA), P<0.05, PERMANOVA on unweighted UniFrac distance. (C) Distribution of bacterial phyla operational taxonomic units presented as relative abundance in each sample. (D) Distribution of bacterial class operational taxonomic units presented as relative abundance of Proteobacteria in each sample. (E) The compositional differences between separately housed WT and Sp140−/− mice, (F) separately housed WT and WT cohoused with Sp140−/− mice, (G) separately house Sp140−/− and cohoused Sp140−/− mice, and (H) cohoused WT and Sp140−/− mice were determined by a linear discriminant analysis using LEfSe (https://huttenhower.sph.harvard.edu/galaxy/). (I) Expression of Helicobacter species (spp.) and Akkermansia muciniphila 16S rRNA relative to total 16S rRNA, as determined by qPCR, in stool from WT and Sp140−/− mice (n=4–5). (J) Expression of Helicobacter species (spp.) and Akkermansia muciniphila 16S rRNA relative to total 16S rRNA in stool from WT, Sp140−/−, and Sp140−/−Rag1−/− mice (n=3–5). Error bars represent means ± SEM. n.s., not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; (I) unpaired t tests. (A, J) one-way ANOVA.
The Helicobacter genus can drive colitis and inflammation in mice (Yang et al., 2013, Cahill et al., 1997, Kullberg et al., 1998, Chai et al., 2017) while A. muciniphila promotes mucosal wound healing and ameliorates colitis in mouse models (Alam et al., 2016, Ansaldo et al., 2019). We therefore confirmed the significant elevation in Helicobacter and reduction in A. muciniphila in Sp140−/− feces by qPCR (Fig. 3I). To assess the contribution of lymphocytes in regulating Helicobacter and A. muciniphila proportions, we crossed Sp140−/− mice with lymphocyte-deficient Rag1−/− mice and found that feces from double mutant Sp140−/−Rag1−/− mice still displayed significantly elevated Helicobacter and reduced A. muciniphila (Fig. 3J). This data suggests that the expansion of activated CD8+ T cells in the colonic lamina propria of Sp140−/− mice (Fig. 1F-I) is a consequence of microbiota dysbiosis and not a cause.
Crohn’s disease patients with SP140 mutations exhibit increase in intestinal Enterobacteriaceae
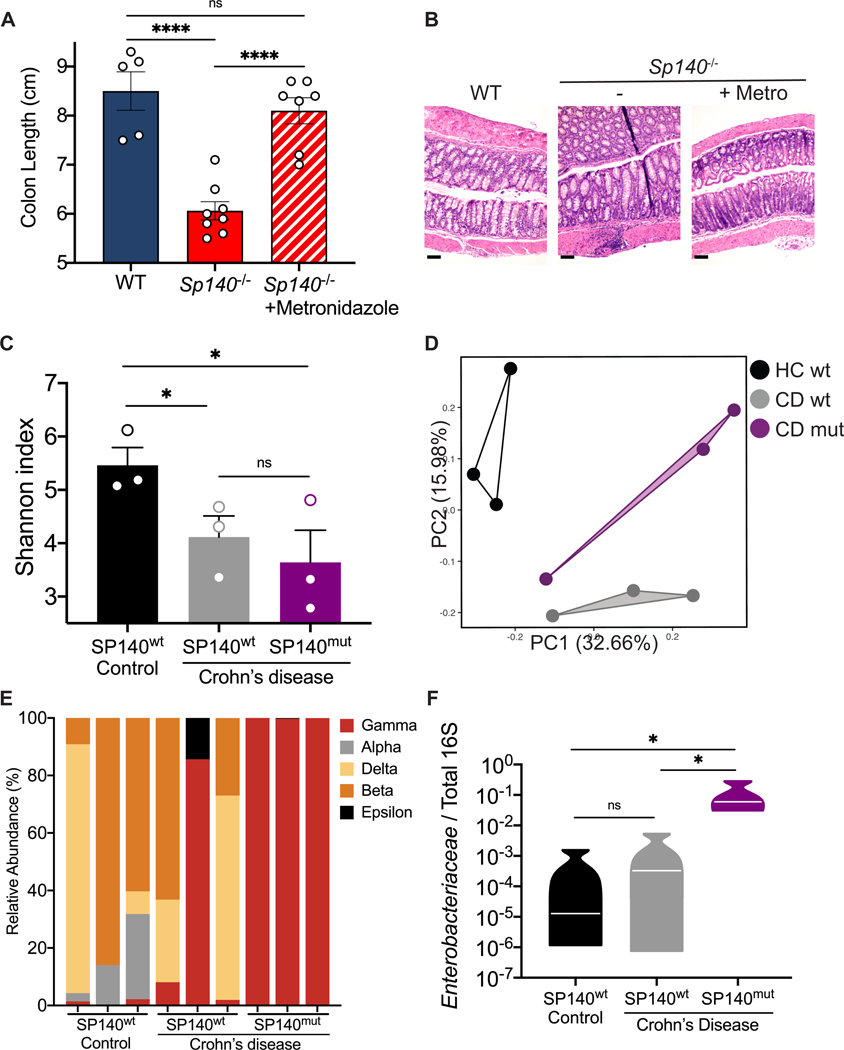
Our analysis of intestinal bacterial populations in Sp140−/− mice suggest that a bloom of pathogenic Proteobacteria results in exacerbated colitis. To demonstrate that expanded Proteobacteria was a driving factor of exacerbated colitis in Sp140−/− mice, we administered mice the antibiotic metronidazole via drinking water for 2 weeks before DSS-colitis induction. Indeed, metronidazole treatment protected Sp140−/− mice from inflammation associated with colitis as determined by increased colon length (Fig. 4A) and reduced crypt hyperplasia (Fig. 4B) compared to Sp140−/− mice that did not receive antibiotics. Metronidazole targets gram-negative anaerobes, including species within the Proteobacteria phylum, and is used in the clinic to treat CD. In order to determine whether manipulation of the microbiota might be a beneficial route of treatment in CD patients bearing SP140 loss-of-function mutations, we sought to determine whether the architecture of microbial communities in Sp140−/− mice recapitulated species dominance in humans with SP140 loss-of-function. We performed 16S sequencing analysis of stool obtained from healthy controls, CD SP140wt, and CD SP140mut individuals (Fig. S3A). Alpha-diversity analysis determined that stool from CD patients exhibited reduced Shannon diversity compared to healthy controls (Fig. 4C) as previously reported (Franzosa et al., 2019). However, each group segregated from each other in principal component analysis (Fig. 4D) demonstrating that while both CD SP140wt and CD SP140mut individuals lose bacterial diversity, there are compositional differences between these two groups of CD patients. Proportional taxonomy analysis demonstrated that Proteobacteria diversity was lost in CD SP140mut individuals whereby Gammaproteobacteria dominates (Fig. 4E). Specifically, we observed a significant increase in Enterobacteriaceae, a family of Gammaproteobacteria, in CD patients bearing SP140 loss-of-function mutations (Fig. 4F). The Enterobacteriaceae family of the Proteobacteria phylum encompasses gram negative facultative anaerobes including Escherichia coli, Citrobacter rodentium, and Salmonella enterica. Blooming of facultative anaerobes, particularly Enterobacteriaceae, with a concomitant depletion of obligate anaerobes associated with short chain fatty acid production is a common feature of IBD gut microbiomes (Lloyd-Price et al., 2019, Morgan et al., 2012, Knights et al., 2014). Furthermore, Enterobacteriaceae blooms are associated with chronic inflammation and treatment failure in IBD (Olbjørn et al., 2019, Walujkar et al., 2014). We predict that the inability to initiate production of toxic reactive oxygen species and antibacterial proteins in SP140-deficient CD patients permits expansion of Enterobacteriaceae as has been suggested for individuals with loss-of-function mutations in NAPDH oxidase subunits (Plichta et al., 2019, Denson et al., 2018, Muise et al., 2012).
Figure 4. CD-associated SP140 loss-of-function variant associates with increased intestinal Enterobacteriaceae in humans.
(A) Day 12 colon lengths of WT, Sp140−/−, and Sp140−/− mice treated with or without metronidazole and administered 2% dextran sulfate sodium (DSS). (B) Representative hematoxylin and eosin-stained sections of day 12 distal colon tissue after 2% DSS administration to WT, Sp140−/−, and Sp140−/− mice treated with metronidazole (metro). (C) Diversity of fecal bacterial communities of healthy controls, Crohn’s disease (CD) patients expressing wildtype SP140 (SP140wt), and CD patients bearing the common SP140 genetic variant (SP140mut). (D) Principal Coordinate Analysis (PCA), P<0.05, PERMANOVA on unweighted UniFrac distance. (E) Distribution of bacterial class operational taxonomic units presented as relative abundance of Proteobacteria in each sample. (F) Expression of Enterobacteriaceae 16S rRNA relative to total 16S rRNA in human stool. Error bars represent means ± SEM. n.s., not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; (A-B) n=5–8; (C-F) n=3, one-way ANOVA.
Discussion
Previous findings from our group determined that chromatin reader SP140 suppresses the expression of lineage inappropriate transcription factors in macrophages, and a loss of SP140 ultimately results in reduced pro-inflammatory cytokine production in response to microbial ligands (Mehta et al., 2017). Furthermore, an array of studies has now implicated SP140 as an essential factor in antibacterial, antiviral, and antiparasitic responses (Ji et al., 2021, Madani et al., 2002, Regad and Chelbi-Alix, 2001, Matsushita et al., 2021). Based on the data presented herein, we extend the role of SP140 to regulation of intestinal microbiota. We demonstrate that SP140 deficient mouse and human phagocytes are incapable of mounting antibacterial effector programs at steady state. Both human and mouse SP140-deficient phagocytes have defects in the production of antibacterial proteins, such as calprotectin. In addition to production by neutrophils, calprotectin is also secreted by human macrophages at baseline (Schulthess et al., 2019) and chelates Mn2+ and Zn2+ metals that are a nutrient source for bacteria (Corbin et al., 2008). Furthermore, human phagocytes bearing SP140 mutations display a transcriptional defect at NADPH oxidase subunit genes. Mutations in CYBB, CYBA, NCF1, NCF2, or NCF4 result in chronic granulomatous disease (CGD) in which recurrent bacterial and fungal infections occur due to an inability to produce ROS for microbial killing (Holland, 2010). Production of antibacterial effectors and NADPH subunits at baseline may also be necessary for homeostatic barrier defenses and the regulation of the microbiota ecosystem. Furthermore, SP140 deficient phagocytes may demonstrate defects in the clearance of bacterial intruders during infection or upon injury, such as during DSS-induced colitis models in mice. We propose that antibacterial gene expression is essential to control pathobiont expansion, such as expansion of Helicobacter in mice and Enterobacteriaceae in humans.
In addition to phagocyte functional defects, we observed an expansion of activated CD8+ T cells in the lamina propria of Sp140−/− mice at steady state. Single cell RNA-sequencing analysis of lamina propria immune populations in human biopsies have demonstrated expansions of CD8+ T cells in CD or ulcerative colitis (UC) patients compared to healthy controls (Martin et al., 2019, Corridoni et al., 2020, Jaeger et al., 2021). However, it is unknown whether this expansion is a consequence of disease or driver of inflammation. Further work is necessary to determine whether this expansion also occurs in individuals with SP140 mutations or dysbiosis. In a cell intrinsic manner, SP140 deficiency within CD8+ T cells may also result in altered proliferation or activation phenotypes. These questions may begin to be answered by examining CD8+ T cell populations in Helicobacter containing WT mice that were cohoused with Sp140−/− mice. As we observe elevated levels of Helicobacter in Sp140−/−Rag−/− mice, we hypothesize that CD8+ T cell expansion is a result of dysbiosis.
IBD symptoms and severity are a spectrum, suggesting that subtypes of disease are dependent on an axis of genetics, environmental cues, and epigenetics. Understanding how risk alleles contribute to pathogenicity and how epigenetic dysregulation is a contributing factor to IBD is essential for identifying disease subtypes as well as developing precision medicine treatment options. Here we have identified a key regulatory role for an epigenetic reader protein, SP140, that is also a risk allele for IBD, in the maintenance of macrophage function and prevention of intestinal microbe expansion, specifically Proteobacteria members. These findings have important implications for not only CD patients with loss-of-function SP140 mutations but also for MS that is driven by an altered microbiota. Genetic loss of SP140 has drastic consequences on the composition of the microbial communities leading to a shift towards a pro-inflammatory configuration that could predispose, and contribute to phenotypes of, both CD and MS (Berer et al., 2011, Britton et al., 2019, Britton et al., 2020, Berer et al., 2017, Cekanaviciute et al., 2017), as well as other diseases influenced by genetics and microbe-derived factors. These results shed light on the etiology of CD and provide insight into the relationship between genetic susceptibility, epigenetics, microbial dysbiosis and the immune response in the gut. Patients with SP140 mutations may benefit from targeting pro-inflammatory commensals through targeted antibiotics or fecal transplants.
Limitations of Study
Homozygosity for the loss of function rs28445040 SP140 variant that associates with CD occurs in ∼2% of patients with CD, including our PRISM cohort. Therefore, future investigations will benefit from using larger patient cohorts to ascertain the role of SP140 in antibacterial host defense mechanisms and microbiota shifts. We determined that WT mice cohoused with Sp140−/− mice exhibit exacerbated colitis and increased colonization by Helicobacter, but this study could be extended further by examining the fecal microbiota of WT irradiated mice that are reconstituted with Sp140−/− hematopoietic cells and conversely whether reconstitution of Sp140−/− irradiated mice with WT hematopoietic cells or macrophages is able to rescue from dysbiosis. Furthermore, examining whether CD8+ T cells are expanded in the colonic lamina propria of WT mice cohoused with Sp140−/− mice will be beneficial in determining whether this expansion is caused my microbiota changes. Finally, future studies will benefit from the creation of an Sp140 conditional knockout mouse to determine the effects of Sp140 depletion within innate versus adaptive immune cells.
STAR Methods.
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kate L. Jeffrey (Kate.Jeffrey@modernatx.com)
Materials Availability
All reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
16S rRNA sequencing of mice and de-identified patient stool have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA818684 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-CF594 anti-NK-1.1 (Myeloid Panel) | BD | Cat#: 562864; RRID: AB_2737850 |
| PE-CF594 anti-γδ TCR (Myeloid Panel) | BD | Cat#: 563532; RRID: AB_2661844 |
| PE-CF594 anti-TCRβ (Myeloid Panel) | BD | Cat#: 562841; RRID: AB_2737831 |
| PE-CF594 anti-CD45R/B220 (Myeloid Panel) | BD | Cat#: 562313; RRID: AB_11151901 |
| FITC anti-Ly6C (Myeloid Panel) | BD | Cat#: 553104; RRID: AB_394628 |
| AF700 anti-I-A/I-E (Myeloid Panel) | BioLegend | Cat#: 107621; RRID: AB_493726 |
| APC/Cy7 anti-CD11b (Myeloid Panel) | BioLegend | Cat#: 101226; RRID: AB_830642 |
| PE anti-CCR2 (Myeloid Panel) | R&D | Cat#: FAB5538P; RRID: AB_10718414 |
| BV510 anti-CD24 (Myeloid Panel) | BioLegend | Cat#: 101831; RRID: AB_2563894 |
| PerCP/Cy5.5 anti-CD64 (Myeloid Panel) | BioLegend | Cat#: 139307; RRID: AB_2561963 |
| APC anti-CD11c (Myeloid Panel) | BioLegend | Cat#: 117309; RRID: AB_313778 |
| BV785 anti-CX3CR1 (Myeloid Panel) | BioLegend | Cat#: 149029; RRID: AB_2565938 |
| BV650 anti-Ly6G (Myeloid Panel) | BioLegend | Cat#: 127641; RRID: AB_2565881 |
| BV605 anti-CD103 (Myeloid Panel) | BioLegend | Cat#: 121433; RRID: AB_2629724 |
| Pacific Blue anti-CD45 (Myeloid/Adaptive Panels) | BioLegend | Cat#: 103126; RRID: AB_493535 |
| FITC anti-CD62L (Adaptive Panel) | BioLegend | Cat#: 104405; RRID: AB_313092 |
| APC/Cy7 anti-NK-1.1 (Adaptive Panel) | BioLegend | Cat#: 108723; RRID: AB_830870 |
| PE/Cy7 anti-CD44 (Adaptive Panel) | BioLegend | Cat#: 103029; RRID: AB_830786 |
| APC anti-CD45R/B220 (Adaptive Panel) | BioLegend | Cat#: 100206; RRID: AB_312663 |
| BV605 anti-CD4 (Adaptive Panel) | BioLegend | Cat#: 100451; RRID: AB_2564591 |
| BV711 anti-CD8α (Adaptive Panel) | BioLegend | Cat#: 100747; RRID: AB_11219594 |
| TruStain FcX | BioLegend | Cat#: 101319; RRID: AB_1574973 |
| Zombie UV Fixable Viability Kit | BioLegend | Cat#: 423107 |
| Fixation Buffer | eBioscience | Cat#: 00-5523-00 |
| Anti-Histone H3 | abcam | Cat#: ab1791; RRID: AB_302613 |
| Anti-S100A8 | abcam | Cat#: ab92331; RRID: AB_2050283 |
| Biological samples | ||
| Crohn’s disease peripheral blood mononuclear cells | PRISM Cohort, MGH | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI media | Life technologies | Cat#: 11875-093 |
| DMEM media | Life technologies | Cat#: 11995-073 |
| Dextran Sulfate Sodium (DSS) | MP Biomedicals | Cat#: 160110; CAS: 9011-18-1 |
| Hyclone fetal bovine serum | GE Healthcare | S#30396.03 |
| Penicillin-Streptomycin | Gibco | P4333 |
| Ficoll-Paque Plus | GE Healthcare | 17-1440-02 |
| RNAiMAX | Life Technologies | 13778075 |
| HiPerfect Transfection Reagent | Qiagen | 1029975 |
| LPS 0111:B4 | Sigma | L4391 |
| Metronidazole | RPI | Cat#: M81000; CAS: 443-48-1 |
| Ampicilin | RPI | Cat#: 69-52-3; CAS: 69-52-3 |
| Neomycin | Sigma Aldrich | Cat#: N6386; CAS: 1405-10-3 |
| Vancomycin | Chem Impex | Cat#: 00315; CAS: 1404-93-9 |
| Dispase II | Sigma | Cat#: D4693; CAS: 42613-33-2 |
| Collagenase, Type IV | Sigma | Cat#: 17104019 |
| Recombinant murine M-CSF | Peprotech | 315-02 |
| Recombinant murine IL-3 | Peprotech | 213-13 |
| Critical commercial assays | ||
| PureLink Microbiome DNA Purification Kit | Invitrogen | A29790 |
| RNeasy mini kit | Qiagen | 74104 |
| Mouse IL-6 ELISA kit | R&D | DY406 |
| Deposited data | ||
| SP140 16S rRNA sequencing data | This paper | SRA: PRJNA818684 |
| SP140 PBMC RNA sequencing data | Mehta et al., 2017 | GEO: GSE89876 |
| Experimental models: Cell lines | ||
| Human: THP1 | Laboratory of Hans-Christian Reinecker | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL6/J | The Jackson Laboratory | JAX: 000664 |
| Mouse: SP140 Knockout (on C57BL6/J background) | Laboratory of Russell Vance | Ji et al. 2021 |
| Oligonucleotides | ||
| ON-TARGETplus Non-targeting Control Pool UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA | Dharmacon/Horizon s Discovery | D-001810-10-05 |
| ON-TARGETplus Human SP140 siRNA – SMARTpool CCGAGCAGAUGUAUGAACA, CAAGAGUGAUGUAUUGUGU, GAACGUAGAGGGUCAGAAC, GGAUUAACCUGAUGGCCUA | Dharmacon/Horizon s Discovery | L-016508-00-0005 |
| Primer for Stool qPCR Total 16S F-GTGCCAGCMGCCGCGGTAA R-GACTACCAGGGTATCTAAT | IDT, this paper | N/A |
| Primer for Stool qPCR A. muciniphila F-CAGCACGTGAAGGTGGGGAC R-CCTTGCGGTTGGCTTCAGAT | IDT, this paper | N/A |
| Primer for Stool qPCR Helicobacter spp. F-GCTATGACGGGTATCC R-GATTTTACCCCTACACCA | IDT, this paper | N/A |
| Primer for Stool qPCR Enterobacteriaceae F-CATTGACGTTACCCGCAGAAGAAGC R-CTCTACGAGACTCAAGCTTGC | IDT, this paper | N/A |
| Primer for Mouse qPCR Lcn2 F-TCCTCAGGTACAGAGCTACAA R-GCTCCTTGGTTCTTCCATACA | IDT, this paper | N/A |
| Primer for Mouse qPCR Lyz2 F-ATGGAATGGCTGGCTACTATG | IDT, this paper | N/A |
| R-GGTCTCCACGGTTGTAGTTT | ||
| Primer for Mouse qPCR S100a8 F-CTTTGTCAGCTCCGTCTTCA R-TGTAGAGGGCATGGTGATTTC | IDT, this paper | N/A |
| Primer for Mouse qPCR Tbp F-TGATCAAACCCAGAATTGTTCT R-TGGTCTTCCTGAATCCCTTTA | IDT, this paper | N/A |
| Software and algorithms | ||
| QIIME 2 | Bolyen et al., 2019 | https://qiime2.org/ |
| DADA2 | Callahan et al., 2016 | N/A |
| MAFFT | Katoh et al., 2002 | N/A |
| UniFrac | Lozupone et al., 2005 | N/A |
| q2-feature-classifier | Bokulich et al., 2018 | N/A |
| LEfSe | Segata et al., 2011 | http://huttenhower.sph.harvard.edu/galaxy |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| FlowJo software | BD | http://flowjo.com |
| Graphpad Prism v9.1.2 | Graphpad software | https://www.graphpad.com/ |
| Other | ||
| iTaq Universal SYBR Green supermix | Bio Rad | 1725124 |
| iScript cDNA Synthesis Kit | Bio Rad | 1708841 |
| Beadbug prefilled homogenizer tubes | Sigma-Aldrich | Z763799 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Subjects
Healthy human Peripheral Blood Mononuclear Cells (PBMCs) were isolated from 20–30 mL of blood buffy coats from human volunteers (Blood Components Lab, Massachusetts General Hospital). All patient blood samples were collected under Institutional Review Board (IRB)-approved protocol by Massachusetts General Hospital (MGH) from patients that were enrolled in the “Prospective Registry in IBD Study at Massachusetts General Hospital (PRISM, IRB# FWA00003136) and genotyped by CD-risk SP140 SNPs rs28445040 and rs6716753 Study research coordinators obtained consent and medical history and data was confirmed by review of the electronic medical record. The average age of patients from which samples were used in this study was 39 (age range: 22–66) with a male/female distribution of 1:4.5. Patient metadata is provided in Supplementary Figure 2. Briefly, mononuclear cells were isolated by density gradient centrifugation of PBS-diluted buffy coat/blood (1:2) over Ficoll-Paque Plus (GE Healthcare). The PBMC layer was carefully removed and washed 3 times with PBS. In order to differentiate into macrophages, PBMCs were re-suspended in X-VIVO medium (Lonza) containing 1% penicillin/streptomycin (Gibco) and incubated at 37°C, 5% CO2 for 1h to adhere to the tissue culture dish. After 1h, adherent cells were washed 3 times with PBS and differentiated in complete X-VIVO medium containing 100 ng/mL human M-CSF (Peprotech) for 7 days at 37°C, 5% CO2. On day 4, cultures were supplemented with one volume of complete X-VIVO medium containing 100 ng/mL human M-CSF. PBMC were unstimulated or treated with 0.1mg/mL LPS treatment for 4 hours.
Mice
C57BL/6J mice were originally purchased from Jackson Laboratory and bred in-house. All mice were bred and housed under specific pathogen-free conditions according to the National Institutes of Health (NIH). All animal experiments were conducted under protocols approved by the MGH Institutional Animal Care and Use Committee (IACUC), and in compliance and appropriate ethical regulations. For all experiments, 6–10 week old females were used. Sp140−/− mice were made on C57BL/6J background as previously characterized (Ji et al., 2021). Rag1−/− mice were purchased from Jackson Laboratory and crossed to Sp140−/− mice. For cohousing experiments, WT and Sp140−/− mice were cohoused from weaning for 4 weeks at a 2:2 ratio per cage.
Cells and treatments.
THP-1 human monocytes were maintained in RPMI media (Life Technologies) with 1% penicillin-streptomycin (Gibco) and 10% heat-inactivated Hyclone fetal bovine serum (FBS, GE Healthcare). Mouse bone marrow-derived macrophages (BMDMs) were produced from WT or Sp140−/− mice. Bone marrow was flushed from tibia and femur and allowed to adhere to a non-treated tissue culture plate for 1 day. Non-adherent cells were then differentiated to macrophages in DMEM containing 10% FBS, 1% L-Glutamine, 1% penicillin/streptomycin, 0.1% β-mercaptoethanol, 5ng/mL of interleukin 3 (IL-3, Peprotech) and macrophage colony stimulating factor (M-CSF, Peprotech) for 7 days.
METHOD DETAILS
Quantitative PCR.
RNA was extracted from cells using the RNeasy Mini Kit (Qiagen) with on-column DNase digest (Qiagen) according to manufacturer’s instructions. cDNA was synthesized from isolated RNA by reverse transcription using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. In the case of quantitative PCR performed on stool DNA, QIAamp DNA Stool Mini Kit was used to extract DNA from feces. Quantitative PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. To analyze relative mRNA levels, derived values were normalized to indicated housekeeping genes. qPCR primers were purchased from Sigma-Aldrich. A complete list of primer sequences is provided in STAR Methods chart.
Western blotting
For Immunoblotting of PBMCs or human macrophages, 2.5 million cells were used and cell lysates were prepared by incubation and sonication of cells in RIPA buffer (10 mM Tris-Cl (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl, 1% protease/phosphatase inhibitor cocktai). 20 μg protein was electrophoresed per lane on NuPAGE Novex Bis-Tris 4–12% Gels (Invitrogen) and transferred to PVDF membranes using iBlot dry blotting system (Invitrogen). Insoluble debris from this lysate was removed by centrifugation at 21000g for 20 minutes at 4°C and remaining supernatant was taken as whole cell lysates. Antibodies are provided in STAR Methods chart.
Flow cytometry of lamina propria cells
For flow cytometric analysis, colons were dissected, and caeca were removed. The tissue was placed in RPMI containing 5% FBS and fat was removed by careful dissection followed by gentle rolling along a moist paper towel. The tissue was cut open longitudinally and rinsed to remove loose fat and fecal matter. Colon was cut into 1–2 cm pieces and transferred to RPMI containing 5% FBS, 5mM EDTA, and 1mM DTT and incubated at 37C for 20 minutes with shaking (400rpm). To remove epithelial cells, colons were next vigorously shaken in complete RPMI with 2mM EDTA. Colon was minced into fine pieces and digested in complete RPMI containing 0.5 mg/mL Dispase II (Gibco) and 1.5 mg/mL type IV collagenase (Gibco) for 45 minutes at 37C shaking (400rpm). Single-cell suspensions were prepared using a 40um nylon cell strainer. Cells were stained with fixable live/dead stain, Fc Block, counting beads, and fluorochrome-conjugated antibodies for 20 min on ice then fixed on ice for 30 min using Fixation/Permeabilization Solution Kit. Data were acquired using a BD FACSAria running FACSDiva software and analyzed with FlowJo software. Mononuclear phagocytes were analyzed using a previously described gating strategy (Adiliaghdam et al., 2022). Antibodies are provided in STAR Methods chart. Flow cytometry samples were acquired using FACSDiva on an LSRII (BD) and analyzed using FlowJo version 10.
Mouse bone marrow reconstitution
Bone marrow progenitor cells obtained from C57BL/6 donor mice were placed in 24-well tissue culture dishes (1 × 106 cells per well) in fresh media containing 15% fetal bovine serum, IL-3 (20 ng/ml), IL-6 (50 ng/ml), and stem cell factor (SCF) (50 ng/ml). On day 4, the cells were harvested and washed twice in PBS. For bone marrow transplantation, C57BL/6 recipient mice were irradiated with 131Cs at 10 gray on day 0 and injected retro-orbitally with 2.5 × 106 bone marrow cells in 200 μl of Dulbecco’s modified Eagle’s medium, after which the mice were monitored for 6 weeks to allow for immune reconstitution.
Dextran sulfate sodium (DSS)-colitis
Mice were administered 2% DSS salt (DSS, MW = 36,000–50,000 Da; MP Biomedicals) in drinking water ad libitum for 7 days (freshly prepared every other day), followed by regular drinking water for 5 days. Mice were sacrificed on day 12. Colon length was measured. For histology, tissue was collected from the distal part of the colon, and hematoxylin and eosin staining were performed. Images were obtained on an EVOS XL Core Imaging System microscope (ThermoFisher). Metronizadole treated mice were administered 1g/L metronidazole drinking water with 8mg/mL Sweet’N Low for 2 weeks prior to the start of DSS administration and continued through day 12.
16S rRNA gene sequencing
PureLink Microbiome DNA Purification Kit (Invitrogen) was used to extract DNA from mouse and human feces. The hypervariable region (V4) of the 16S rRNA gene sequences were amplified by using PCR with adaptor primer set containing 12bp multiplex identifier sequences (Integrated DNA Technologies, Coralville, IA). Each reaction contained template DNA, each primer (0.2 μM), and 2× Platinum™ Hot Start PCR Master Mix (Invitrogen, Carlsbad, CA). PCR conditions were as follows: 94°C for 3 min, followed by 22 cycles at 94°C for 45 sec, 52°C for 1 min, and 72°C for 1.5 min and a final extension step at 72°C for 10 min. Triplicate reactions were prepared, pooled, and purified using SpeedBead™ carboxylate magnetic bead solution (GE Healthcare, Marlborough, MA). The amplicons were quantified using KAPA Library Quantitation kit (KAPA, Cape Town, South Africa) and an equimolar amount of each sample was sequenced on the MiSeq System (Illumina, San Diego, CA) with 2×150 paired-end run parameters. Microbiome bioinformatics analysis was performed with QIIME 2 release 2018.11. In brief, cutadapt was used to trim 515F/806R primers in the paired end sequences. Subsequently, the sequences were stripped of noise by using the DADA2 denoiser program. The denoised, trimmed and higher quality amplicon sequence variants (ASVs) were aligned with MAFFT and used to construct a phylogeny with fasttree2. All ASVs were assigned taxonomy using the q2-feature-classifier. In brief, a naïve Bayes classifier was trained on the Greengenes 13_8 99% OTUs reference sequences to assign taxonomy to each ASV. The compositional differences among the groups were determined by a linear discriminant analysis using LEfSe (Segata et al., 2011) with a threshold of 2.0 on the logarithmic score and alpha value of 0.01 for the factorial Kruskal-Wallis test among classes using Galaxy (https://huttenhower.sph.harvard.edu/galaxy/). Mouse and human 16S rRNA sequencing data are deposited at Sequencing Read Archive (SRA) under accession number PRJNA818684.
Macrophage stimulations with stool homogenates
Stool pellets were weighed and resuspended in 40mg/mL complete DMEM then homogenized using a BeadBug 3 microtube homogenizer (Benchmark Scientific) for 9 minutes at highest speed in BeadBug tubes containing 1.5mm Zirconium beads. Homogenized stool was centrifuged at 400g for 1 min. Supernatant was collected and passed through 70μm strainer. Stool homogenates were then added to 0.5×106 bone marrow-derived macrophages (BMDMs) in a 96 well plate at a final concentration of 1mg/mL. Antibiotic treated mice were administered a broad-spectrum antibiotic cocktail (1g/L metronidazole, 0.5g/L vancomycin, 1g/L ampicillin, 1g/L neomycin) containing 8mg/mL Sweet’N Low via drinking water. Stool was collected after 4 weeks of antibiotic treatment. Mouse IL-6 was measured in cell supernatants using a DuoSet ELISA kit per manufacturer instructions (R&D Systems).
Phagocytosis Assay.
THP-1 human monocytes or mature primary human peripheral blood derived macrophages were transfected with control (D-001810-10-05, Dharmacon) or SP140 (L-016508-00-0005, Dharmacon) SMARTpool siRNA (100nM) using RNAiMAX (Invitrogen) for 48 hours followed by 1 hour treatment with pHrodo Green E. coli BioParticles (Invitrogen) for phagocytosis assay. Day 7 mouse BMDMs were treated 1 hour with pHrodo Green E. coli BioParticles (Invitrogen). In all experiments, 0.05 mg of E. coli BioParticles were used to treat 0.5 × 106 cells. Phagocytosis of E. coli BioParticles was assessed by flow cytometry. Flow cytometry samples were acquired using FACSDiva on an LSRII (BD) and analyzed using FlowJo version 10.
QUANTIFICATION AND STATISTICAL ANALYSIS.
Results are shown as mean ± s.e.m. Visual examination of the data distribution as well as normality testing demonstrated that all variables appeared to be normally distributed. Comparisons and statistical tests were performed as indicated in each figure legend. For comparisons of two groups, two-tailed unpaired t tests were used, except where indicated. For comparison of multiple groups, one-way ANOVA was used. Statistical analyses were performed in the GraphPad Prism 8 software. The P values denoted throughout the manuscript highlight biologically relevant comparisons. A P value of less than 0.05 was considered significant, denoted as *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 for all analyses.
Supplementary Material
Epigenetic reader SP140 facilitates antibacterial programs in macrophages
Exacerbated colitis in Sp140 deficient mice is dependent on microbiota dysbiosis
Pro-inflammatory Proteobacteria expand in SP140 deficient mice and humans
A loss-of-function mutation in the chromatin reader SP140 is associated with Crohn’s disease. Fraschilla et al. demonstrate that SP140-deficient phagocytes lack the ability to mount homeostatic antibacterial responses, and expansion of pathobionts occurs in the intestine of mice and humans that lack SP140.
Acknowledgements
We sincerely thank the clinical coordinators and patients enrolled in the Prospective Registry in IBD study at Massachusetts General Hospital (PRISM), Russell Vance (University of California, Berkeley) for Sp140−/− mice, Amy Avery (Massachusetts General Hospital) for technical assistance with 16S sequencing, the entire Jeffrey lab for valuable scientific discussions as well as Robert Anthony, James Moon, Charles Evavold, and Jonathan Kagan for critical reading of the manuscript. This study was supported by F31DK127518 (I.F.), the Kenneth Rainin Foundation (Innovator and Synergy Awards to K.L.J), NIH R01DK119996 (K.L.J), K.L.J is a John Lawrence MGH Research Scholar 2020–2025. Figures were created using BioRender.
Diversity and Inclusion.
For studies involving human subjects, recruited in the Prospective Registry in IBD study at Massachusetts General Hospital (PRISM), we worked to ensure gender balance in the recruitment of human subjects, ensured ethnic or other types of diversity in the recruitment of human subjects and ensured that the study questionnaires were prepared in an inclusive way. For studies involving non-human subjects or material, we ensured diversity in experimental samples through the selection of the cell lines. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Footnotes
Declaration of Interests. K.L.J. is a shareholder and employee of Moderna Inc., 200 Technology Square, Cambridge MA 02138, since November 2021. K.L.J is a member of the scientific advisory board for Ancilia Biosciences. None of these relationships influenced the work in this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ADILIAGHDAM F, AMATULLAH H, DIGUMARTHI S, SAUNDERS TL, RAHMAN RU, WONG LP, SADREYEV R, DROIT L, PAQUETTE J, GOYETTE P, RIOUX JD, HODIN R, MIHINDUKULASURIYA KA, HANDLEY SA & JEFFREY KL. 2022. Human enteric viruses autonomously shape inflammatory bowel disease phenotype through divergent innate immunomodulation. Sci Immunol, 7, eabn6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALAM A, LEONI G, QUIROS M, WU H, DESAI C, NISHIO H, JONES RM, NUSRAT A. & NEISH AS. 2016. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nature Microbiology, 1, 15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMATULLAH H, FRASCHILLA I, DIGUMARTHI S, HUANG J, ADILIAGHDAM F, BONILLA G, WONG LP, RIVARD ME, BEAUCHAMP C, MERCIER V, GOYETTE P, SADREYEV RI, ANTHONY RM, RIOUX JD & JEFFREY KL. 2022. Epigenetic reader SP140 loss of function drives Crohn’s disease due to uncontrolled macrophage topoisomerases. Cell. [DOI] [PMC free article] [PubMed]
- AMATULLAH H. & JEFFREY KL. 2020. Epigenome-metabolome-microbiome axis in health and IBD. Curr Opin Microbiol, 56, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANSALDO E, SLAYDEN LC, CHING KL, KOCH MA, WOLF NK, PLICHTA DR, BROWN EM, GRAHAM DB, XAVIER RJ, MOON JJ & BARTON GM. 2019. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science, 364, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARGIROPOULOS B. & HUMPHRIES RK. 2007. Hox genes in hematopoiesis and leukemogenesis. Oncogene, 26, 6766–6776. [DOI] [PubMed] [Google Scholar]
- ARROWSMITH CH, BOUNTRA C, FISH PV, LEE K. & SCHAPIRA M. 2012. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov, 11, 384–400. [DOI] [PubMed] [Google Scholar]
- BAIN CC., BRAVO-BLAS A., SCOTT CL., GOMEZ PERDIGUERO E., GEISSMANN F., HENRI S., MALISSEN B., OSBORNE LC., ARTIS D. & MOWAT AM. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nature Immunology, 15, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERER K, GERDES LA, CEKANAVICIUTE E, JIA X, XIAO L, XIA Z, LIU C, KLOTZ L, STAUFFER U, BARANZINI SE, KÜMPFEL T, HOHLFELD R, KRISHNAMOORTHY G. & WEKERLE H. 2017. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proceedings of the National Academy of Sciences, 114, 10719–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERER K, MUES M, KOUTROLOS M, RASBI ZA, BOZIKI M, JOHNER C, WEKERLE H. & KRISHNAMOORTHY G. 2011. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature, 479, 538–41. [DOI] [PubMed] [Google Scholar]
- BIENZ M. 2006. The PHD finger, a nuclear protein-interaction domain. Trends in Biochemical Sciences, 31, 35–40. [DOI] [PubMed] [Google Scholar]
- BLANDER JM, LONGMAN RS, ILIEV ID, SONNENBERG GF & ARTIS D. 2017. Regulation of inflammation by microbiota interactions with the host. Nature Immunology, 18, 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOTTOMLEY MJ, COLLARD MW, HUGGENVIK JI, LIU Z, GIBSON TJ & SATTLER M. 2001. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nature Structural Biology, 8, 626–633. [DOI] [PubMed] [Google Scholar]
- BRITTON GJ, CONTIJOCH EJ, MOGNO I, VENNARO OH, LLEWELLYN SR, NG R, LI Z, MORTHA A, MERAD M, DAS A, GEVERS D, MCGOVERN DPB, SINGH N, BRAUN J, JACOBS JP, CLEMENTE JC, GRINSPAN A, SANDS BE, COLOMBEL J-F, DUBINSKY MC & FAITH JJ. 2019. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORγt+ Regulatory T Cells and Exacerbate Colitis in Mice. Immunity, 50, 212–224.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRITTON GJ, CONTIJOCH EJ, SPINDLER MP, AGGARWALA V, DOGAN B, BONGERS G, SAN MATEO L, BALTUS A, DAS A, GEVERS D, BORODY TJ, KAAKOUSH NO, KAMM MA, MITCHELL H, PARAMSOTHY S, CLEMENTE JC, COLOMBEL JF, SIMPSON KW, DUBINSKY MC, GRINSPAN A. & FAITH JJ. 2020. Defined microbiota transplant restores Th17/RORgammat(+) regulatory T cell balance in mice colonized with inflammatory bowel disease microbiotas. Proc Natl Acad Sci U S A, 117, 21536–21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAHILL RJ, FOLTZ CJ, FOX JG, DANGLER CA, POWRIE F. & SCHAUER DB. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun, 65, 3126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARUSO R, MATHES T, MARTENS EC, KAMADA N, NUSRAT A, INOHARA N. & NUNEZ G. 2019. A specific gene-microbe interaction drives the development of Crohn’s disease-like colitis in mice. Sci Immunol. [DOI] [PMC free article] [PubMed]
- CEKANAVICIUTE E, YOO BB, RUNIA TF, DEBELIUS JW, SINGH S, NELSON CA, KANNER R, BENCOSME Y, LEE YK, HAUSER SL, CRABTREE-HARTMAN E, SAND IK, GACIAS M, ZHU Y, CASACCIA P, CREE BAC, KNIGHT R, MAZMANIAN SK & BARANZINI SE. 2017. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proceedings of the National Academy of Sciences, 114, 10713–10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAI JN., PENG Y., RENGARAJAN S., SOLOMON BD., AI TL., SHEN Z., PERRY JSA., KNOOP KA., TANOUE T., NARUSHIMA S., HONDA K., ELSON CO., NEWBERRY RD., STAPPENBECK TS., KAU AL., PETERSON DA., FOX JG. & HSIEH CS. 2017. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci Immunol. [DOI] [PMC free article] [PubMed]
- CONWAY KL, KUBALLA P, SONG JH, PATEL KK, CASTORENO AB, YILMAZ OH, JIJON HB, ZHANG M, ALDRICH LN, VILLABLANCA EJ, PELOQUIN JM, GOEL G, LEE IA, MIZOGUCHI E, SHI HN, BHAN AK, SHAW SY, SCHREIBER SL, VIRGIN HW, SHAMJI AF, STAPPENBECK TS, REINECKER HC & XAVIER RJ. 2013. Atg16l1 is Required for Autophagy in Intestinal Epithelial Cells and Protection of Mice From Salmonella Infection. Gastroenterology, 145, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORBIN BD, SEELEY EH, RAAB A, FELDMANN J, MILLER MR, TORRES VJ, ANDERSON KL, DATTILO BM, DUNMAN PM, GERADS R, CAPRIOLI RM, NACKEN W, CHAZIN WJ & SKAAR EP. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science, 319, 962–5. [DOI] [PubMed] [Google Scholar]
- CORPET A, KLEIJWEGT C, ROUBILLE S, JUILLARD F, JACQUET K, TEXIER P. & LOMONTE P. 2020. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Research, 48, 11890–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORRIDONI D, ANTANAVICIUTE A, GUPTA T, FAWKNER-CORBETT D, AULICINO A, JAGIELOWICZ M, PARIKH K, REPAPI E, TAYLOR S, ISHIKAWA D, HATANO R, YAMADA T, XIN W, SLAWINSKI H, BOWDEN R, NAPOLITANI G, BRAIN O, MORIMOTO C, KOOHY H. & SIMMONS A. 2020. Single-cell atlas of colonic CD8+ T cells in ulcerative colitis. Nature Medicine, 26, 1480–1490. [DOI] [PubMed] [Google Scholar]
- DE SANTA F, TOTARO MG, PROSPERINI E, NOTARBARTOLO S, TESTA G. & NATOLI G. 2007. The Histone H3 Lysine-27 Demethylase Jmjd3 Links Inflammation to Inhibition of Polycomb-Mediated Gene Silencing. Cell, 130, 1083–1094. [DOI] [PubMed] [Google Scholar]
- DENSON LA, JURICKOVA I, KARNS R, SHAW KA, CUTLER DJ, OKOU DT, DODD A, QUINN K, MONDAL K, ARONOW BJ, HABERMAN Y, LINN A, PRICE A, BEZOLD R, LAKE K, JACKSON K, WALTERS TD, GRIFFITHS A, BALDASSANO RN, NOE JD, HYAMS JS, CRANDALL WV, KIRSCHNER BS, HEYMAN MB, SNAPPER S, GUTHERY SL, DUBINSKY MC, LELEIKO NS, OTLEY AR, XAVIER RJ, STEVENS C, DALY MJ, ZWICK ME & KUGATHASAN S. 2018. Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients With Crohn’s Disease. Gastroenterology, 154, 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELINAV E, STROWIG T, ANDREW HENAO-MEJIAJ, CARMEN CHRISTOPH, DAVID BERTINJ, JEFFREY STEPHANIE & RICHARD 2011. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell, 145, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FILIPPAKOPOULOS P, PICAUD S, MANGOS M, KEATES T, LAMBERT JP, BARSYTE-LOVEJOY D, FELLETAR I, VOLKMER R, MULLER S, PAWSON T, GINGRAS AC, ARROWSMITH CH & KNAPP S. 2012. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell, 149, 214–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANZOSA EA, SIROTA-MADI A, AVILA-PACHECO J, FORNELOS N, HAISER HJ, REINKER S, VATANEN T, HALL AB, MALLICK H, MCIVER LJ, SAUK JS, WILSON RG, STEVENS BW, SCOTT JM, PIERCE K, DEIK AA, BULLOCK K, IMHANN F, PORTER JA, ZHERNAKOVA A, FU J, WEERSMA RK, WIJMENGA C, CLISH CB, VLAMAKIS H, HUTTENHOWER C. & XAVIER RJ. 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nature Microbiology, 4, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRASCHILLA I. & JEFFREY KL. 2020. The Speckled Protein (SP) Family: Immunity’s Chromatin Readers. Trends in Immunology, 41, 572–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREDERIC KOREN, JULIA MALINO, NALBANTOGLU I, JESSE SUY, CHASSAING B, WILLIAM GONZÁLEZ, JOSE A, TYLER BARNICHN, DARFEUILLE-MICHAUD A, VIJAY-KUMAR M, KNIGHT R, RUTH & ANDREW 2012. Transient Inability to Manage Proteobacteria Promotes Chronic Gut Inflammation in TLR5-Deficient Mice. Cell Host & Microbe, 12, 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARCIA-DOMINGUEZ M, MARCH-DIAZ R. & REYES JC. 2008. The PHD Domain of Plant PIAS Proteins Mediates Sumoylation of Bromodomain GTE Proteins. Journal of Biological Chemistry, 283, 21469–21477. [DOI] [PubMed] [Google Scholar]
- GARRETT WS, GALLINI CA, YATSUNENKO T, MICHAUD M, DUBOIS A, DELANEY ML, PUNIT S, KARLSSON M, BRY L, GLICKMAN JN, GORDON JI, ONDERDONK AB & GLIMCHER LH. 2010. Enterobacteriaceae Act in Concert with the Gut Microbiota to Induce Spontaneous and Maternally Transmitted Colitis. Cell Host & Microbe, 8, 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRAHAM DB & XAVIER RJ. 2020. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature, 578, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOISCHEN C, MONAJEMBASHI S, WEISSHART K. & HEMMERICH P. 2018. Multimodal Light Microscopy Approaches to Reveal Structural and Functional Properties of Promyelocytic Leukemia Nuclear Bodies. Front Oncol, 8, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLAND SM. 2010. Chronic Granulomatous Disease. Clinical Reviews in Allergy & Immunology, 38, 3–10. [DOI] [PubMed] [Google Scholar]
- HUANG Y, SITWALA K, BRONSTEIN J, SANDERS D, DANDEKAR M, COLLINS C, ROBERTSON G, MACDONALD J, CEZARD T, BILENKY M, THIESSEN N, ZHAO Y, ZENG T, HIRST M, HERO A, JONES S. & HESS JL. 2012. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood, 119, 388–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUOH Y-S, WU B, PARK S, YANG D, BANSAL K, GREENWALD E, WONG WP, MATHIS D. & HUR S. 2020. Dual functions of Aire CARD multimerization in the transcriptional regulation of T cell tolerance. Nature Communications. [DOI] [PMC free article] [PubMed]
- ILIEV ID & CADWELL K. 2021. Effects of Intestinal Fungi and Viruses on Immune Responses and Inflammatory Bowel Diseases. Gastroenterology, 160, 1050–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INTERNATIONAL MULTIPLE SCLEROSIS GENETICS, C., BEECHAM AH, PATSOPOULOS NA, XIFARA DK, DAVIS MF, KEMPPINEN A, COTSAPAS C, SHAH, SPENCER C, BOOTH D, GORIS A, OTURAI A, SAARELA J, FONTAINE B, HEMMER B, MARTIN C, ZIPP F, D’ALFONSO S, MARTINELLI-BONESCHI F, TAYLOR B, HARBO HF, KOCKUM I, HILLERT J, OLSSON T, BAN M, OKSENBERG JR, HINTZEN R, BARCELLOS LF, WELLCOME TRUST CASE CONTROL C, INTERNATIONAL IBDGC, AGLIARDI C, ALFREDSSON L, ALIZADEH M, ANDERSON C, ANDREWS R, SONDERGAARD HB, BAKER A, BAND G, BARANZINI SE, BARIZZONE N, BARRETT J, BELLENGUEZ C, BERGAMASCHI L, BERNARDINELLI L, BERTHELE A, BIBERACHER V, BINDER TM, BLACKBURN H, BOMFIM IL, BRAMBILLA P, BROADLEY S, BROCHET B, BRUNDIN L, BUCK D, BUTZKUEVEN H, CAILLIER SJ, CAMU W, CARPENTIER W, CAVALLA P, CELIUS EG, COMAN I, COMI G, CORRADO L, COSEMANS L, COURNU-REBEIX I, CREE BA, CUSI D, DAMOTTE V, DEFER G, DELGADO SR, DELOUKAS P, DI SAPIO A, DILTHEY AT, DONNELLY P, DUBOIS B, DUDDY M, EDKINS S, ELOVAARA I, ESPOSITO F, EVANGELOU N, FIDDES B, FIELD J, FRANKE A, FREEMAN C, FROHLICH IY, GALIMBERTI D, GIEGER C, GOURRAUD PA, GRAETZ C, GRAHAM A, GRUMMEL V, GUASCHINO C, HADJIXENOFONTOS A, HAKONARSON H, HALFPENNY C, HALL G, HALL P, HAMSTEN A, HARLEY J, HARROWER T, et al. 2013. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet, 45, 1353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IVANOV AV, PENG H, YURCHENKO V, YAP KL, NEGOREV DG, SCHULTZ DC, PSULKOWSKI E, FREDERICKS WJ, WHITE DE, MAUL GG, SADOFSKY MJ, ZHOU MM & RAUSCHER FJ 3RD 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell, 28, 823–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAEGER N, GAMINI R, CELLA M, SCHETTINI JL, BUGATTI M, ZHAO S, ROSADINI CV, ESAULOVA E, DI LUCCIA B, KINNETT B, VERMI W, ARTYOMOV MN, WYNN TA, XAVIER RJ, JELINSKY SA & COLONNA M. 2021. Single-cell analyses of Crohn’s disease tissues reveal intestinal intraepithelial T cells heterogeneity and altered subset distributions. Nature Communications, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JI DX, WITT KC, KOTOV DI, MARGOLIS SR, LOUIE A, CHEVÉE V, CHEN KJ, GAIDT M, DHALIWAL HS, LEE AY, NISHIMURA SL, ZAMBONI DS, KRAMNIK IA, PORTNOY D, DARWIN KH & VANCE RE. 2021. Role of the transcriptional regulator SP140 in resistance to bacterial infections via repression of type I interferons. eLife: eLife Sciences Publications, Ltd. [DOI] [PMC free article] [PubMed]
- JOSTINS L., RIPKE S., WEERSMA RK., DUERR RH., MCGOVERN DP., HUI KY., LEE JC., SCHUMM LP., SHARMA Y., ANDERSON CA., ESSERS J., MITROVIC M., NING K., CLEYNEN I., THEATRE E., SPAIN SL., RAYCHAUDHURI S., GOYETTE P., WEI Z., ABRAHAM C., ACHKAR JP., AHMAD T., AMININEJAD L., ANANTHAKRISHNAN AN., ANDERSEN V., ANDREWS JM., BAIDOO L., BALSCHUN T., BAMPTON PA., BITTON A., BOUCHER G., BRAND S., BUNING C, COHAIN A, CICHON S, D’AMATO M, DE JONG D, DEVANEY KL, DUBINSKY M, EDWARDS C, ELLINGHAUS D, FERGUSON LR, FRANCHIMONT D, FRANSEN K, GEARRY R, GEORGES M, GIEGER C, GLAS J, HARITUNIANS T, HART A, HAWKEY C, HEDL M, HU X, KARLSEN TH, KUPCINSKAS L, KUGATHASAN S, LATIANO A, LAUKENS D, LAWRANCE IC, LEES CW, LOUIS E, MAHY G, MANSFIELD J, MORGAN AR, MOWAT C, NEWMAN W, PALMIERI O, PONSIOEN CY, POTOCNIK U, PRESCOTT NJ, REGUEIRO M, ROTTER JI, RUSSELL RK, SANDERSON JD, SANS M, SATSANGI J, SCHREIBER S, SIMMS LA, SVENTORAITYTE J, TARGAN SR, TAYLOR KD, TREMELLING M, VERSPAGET HW, DE VOS M, WIJMENGA C, WILSON DC, WINKELMANN J, XAVIER RJ, E S, ZHANG B, ZHANG CK, ZHAO H, INTERNATIONAL IBDGC, SILVERBERG MS, ANNESE V, HAKONARSON H, BRANT SR, RADFORD-SMITH G, MATHEW CG, RIOUX JD, et al. 2012. Hostmicrobe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAIKO GE, RYU SH, KOUES OI, COLLINS PL, SOLNICA-KREZEL L, PEARCE EJ, PEARCE EL, OLTZ EM & STAPPENBECK TS. 2016. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell, 165, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KNIGHTS D, SILVERBERG MS, WEERSMA RK, GEVERS D, DIJKSTRA G, HUANG H, TYLER AD, VAN SOMMEREN S, IMHANN F, STEMPAK JM, HUANG H, VANGAY P, AL-GHALITH GA, RUSSELL C, SAUK J, KNIGHT J, DALY MJ, HUTTENHOWER C. & XAVIER RJ. 2014. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med, 6, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUGELBERG E. 2014. Curbing gut inflammation. Nature Reviews Immunology, 14, 583–583. [DOI] [PubMed] [Google Scholar]
- KULLBERG MC, WARD JM, GORELICK PL, CASPAR P, HIENY S, CHEEVER A, JANKOVIC D. & SHER A. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun, 66, 5157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU JZ, VAN SOMMEREN S, HUANG H, NG SC, ALBERTS R, TAKAHASHI A, RIPKE S, LEE JC, JOSTINS L, SHAH T, ABEDIAN S, CHEON JH, CHO J, DARYANI NE, FRANKE L, FUYUNO Y, HART A, JUYAL RC, JUYAL G, KIM WH, MORRIS AP, POUSTCHI H, NEWMAN WG, MIDHA V, ORCHARD TR, VAHEDI H, SOOD A, SUNG JJY, MALEKZADEH R, WESTRA H-J, YAMAZAKI K, YANG S-K, BARRETT JC, FRANKE A, ALIZADEH BZ, PARKES M, B K T, DALY MJ, KUBO M, ANDERSON CA & WEERSMA RK. 2015. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics, 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLOYD-PRICE J., ARZE C., ANANTHAKRISHNAN AN., SCHIRMER M., AVILA-PACHECO J., POON TW., ANDREWS E., AJAMI NJ., BONHAM KS., BRISLAWN CJ., CASERO D., COURTNEY H., GONZALEZ A., GRAEBER TG., HALL AB., LAKE K., LANDERS CJ., MALLICK H., PLICHTA DR., PRASAD M., RAHNAVARD G., SAUK J., SHUNGIN D., VÁZQUEZ-BAEZA Y., WHITE RA., BRAUN J., DENSON LA., JANSSON JK., KNIGHT R., KUGATHASAN S., MCGOVERN DPB., PETROSINO JF., STAPPENBECK TS., WINTER HS., CLISH CB., FRANZOSA EA., VLAMAKIS H., XAVIER RJ. & HUTTENHOWER C. 2019. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature, 569, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MADANI N, MILLETTE R, PLATT EJ, MARIN M, KOZAK SL, BLOCH DB & KABAT D. 2002. Implication of the Lymphocyte-Specific Nuclear Body Protein Sp140 in an Innate Response to Human Immunodeficiency Virus Type 1. Journal of Virology, 76, 11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN JC, CHANG C, BOSCHETTI G, UNGARO R, GIRI M, GROUT JA, GETTLER K, CHUANG L-S, NAYAR S, GREENSTEIN AJ, DUBINSKY M, WALKER L, LEADER A, FINE JS, WHITEHURST CE, MBOW ML, KUGATHASAN S, DENSON LA, HYAMS JS, FRIEDMAN JR, DESAI PT, KO HM, LAFACE I, AKTURK G, SCHADT EE, SALMON H, GNJATIC S, RAHMAN AH, MERAD M, CHO JH & KENIGSBERG E. 2019. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell, 178, 1493–1508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATESANZ F, POTENCIANO V, FEDETZ M, RAMOS-MOZO P, ABAD-GRAU MDEL M, KARAKY M, BARRIONUEVO C, IZQUIERDO G, RUIZ-PENA JL, GARCIA-SANCHEZ MI, LUCAS M, FERNANDEZ O, LEYVA L, OTAEGUI D, MUNOZ-CULLA M, OLASCOAGA J, VANDENBROECK K, ALLOZA I, ASTOBIZA I, ANTIGUEDAD A, VILLAR LM, ALVAREZ-CERMENO JC, MALHOTRA S, COMABELLA M, MONTALBAN X, SAIZ A, BLANCO Y, ARROYO R, VARADE J, URCELAY E. & ALCINA A. 2015. A functional variant that affects exon-skipping and protein expression of SP140 as genetic mechanism predisposing to multiple sclerosis. Hum Mol Genet, 24, 5619–27. [DOI] [PubMed] [Google Scholar]
- MATSUSHITA K, LI X, NAKAMURA Y, DONG D, MUKAI K, TSAI M, MONTGOMERY SB & GALLI SJ. 2021. The role of Sp140 revealed in IgE and mast cell responses in Collaborative Cross mice. JCI Insight, 6, 2379–3708. [DOI] [PMC free article] [PubMed]
- MEHTA S, CRONKITE DA, BASAVAPPA M, SAUNDERS TL, ADILIAGHDAM F, AMATULLAH H, MORRISON SA, PAGAN JD, ANTHONY RM, TONNERRE P, LAUER GM, LEE JC, DIGUMARTHI S, PANTANO L, HO SUI SJ, JI F, SADREYEV R, ZHOU C, MULLEN AC, KUMAR V, LI Y, WIJMENGA C, XAVIER RJ, MEANS TK & JEFFREY KL. 2017. Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. 20170303 ed. Sci Immunol. [DOI] [PMC free article] [PubMed]
- MORGAN XC, TICKLE TL, SOKOL H, GEVERS D, DEVANEY KL, WARD DV, REYES JA, SHAH SA, LELEIKO N, SNAPPER SB, BOUSVAROS A, KORZENIK J, SANDS BE, XAVIER RJ & HUTTENHOWER C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol, 13, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUISE AM, XU W, GUO C-H, WALTERS TD, WOLTERS VM, FATTOUH R, LAM GY, HU P, MURCHIE R, SHERLOCK M, GANA JC, RUSSELL RK, GLOGAUER M, DUERR RH, CHO JH, LEES CW, SATSANGI J, WILSON DC, PATERSON AD, GRIFFITHS AM, SILVERBERG MS & BRUMELL JH. 2012. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut, 61, 1028–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N J., SHEN TD., CHEN EZ., BITTINGER K., BAILEY A., ROGGIANI M., SIROTA-MADI A., FRIEDMAN ES., CHAU L., LIN A., NISSIM I., SCOTT J., LAUDER A., HOFFMANN C., RIVAS G., ALBENBERG L., BALDASSANO RN., BRAUN J., XAVIER RJ., CLISH CB., YUDKOFF M., LI H., GOULIAN M., BUSHMAN FD., LEWIS JD. & WU GD. 2017. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med. [DOI] [PMC free article] [PubMed]
- OLBJØRN C, CVANCAROVA SMÅSTUEN M, THIIS-EVENSEN E, NAKSTAD B, VATN MH, JAHNSEN J, RICANEK P, VATN S, MOEN AE, TANNÆS TM, LINDSTRØM JC, SÖDERHOLM JD, HALFVARSON J, GOMOLLÓN F, CASÉN C, KARLSSON MK, KALLA R, ADAMS AT, SATSANGI J. & PERMINOW G. 2019. Fecal microbiota profiles in treatment-naive pediatric inflammatory bowel disease - associations with disease phenotype, treatment, and outcome. Clinical and Experimental Gastroenterology, Volume 12, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAN H, YAN B-S, ROJAS M, SHEBZUKHOV YV, ZHOU H, KOBZIK L, HIGGINS DE, DALY MJ, BLOOM BR & KRAMNIK I. 2005. Ipr1 gene mediates innate immunity to tuberculosis. Nature, 434, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENG J. & WYSOCKA J. 2008. It takes a PHD to SUMO. Trends Biochem Sci, 33, 191–4. [DOI] [PubMed] [Google Scholar]
- PICHUGIN AV, YAN BS, SLOUTSKY A, KOBZIK L. & KRAMNIK I. 2009. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am J Pathol, 174, 2190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PLICHTA DR, GRAHAM DB, SUBRAMANIAN S. & XAVIER RJ. 2019. Therapeutic Opportunities in Inflammatory Bowel Disease: Mechanistic Dissection of Host-Microbiome Relationships. Cell, 178, 1041–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMANAN D, TANG MS, BOWCUTT R, LOKE P. & CADWELL K. 2014. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity, 41, 311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REGAD T. & CHELBI-ALIX MK. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene, 20, 7274–7286. [DOI] [PubMed] [Google Scholar]
- RIVAS MA, BEAUDOIN M, GARDET A, STEVENS C, SHARMA Y, ZHANG CK, BOUCHER G, RIPKE S, ELLINGHAUS D, BURTT N, FENNELL T, KIRBY A, LATIANO A, GOYETTE P, GREEN T, HALFVARSON J, HARITUNIANS T, KORN JM, KURUVILLA F, LAGACÉ C, NEALE B, LO KS, SCHUMM P, TÖRKVIST L, DUBINSKY MC, BRANT SR, SILVERBERG MS, DUERR RH, ALTSHULER D, GABRIEL S, LETTRE G, FRANKE A, D’AMATO M, MCGOVERN DPB, CHO JH, RIOUX JD, XAVIER RJ & DALY MJ. 2011. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature Genetics, 43, 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTSON SJ, LEMIRE P, MAUGHAN H, GOETHEL A, TURPIN W, BEDRANI L, GUTTMAN DS, CROITORU K, GIRARDIN SE & PHILPOTT DJ. 2019. Comparison of Co-housing and Littermate Methods for Microbiota Standardization in Mouse Models. Cell Reports, 27, 1910–1919.e2. [DOI] [PubMed] [Google Scholar]
- SCHULTHESS J, PANDEY S, CAPITANI M, RUE-ALBRECHT KC, ARNOLD I, FRANCHINI F, CHOMKA A, ILOTT NE, JOHNSTON DGW, PIRES E, MCCULLAGH J, SANSOM SN, ARANCIBIA-CÁRCAMO CV, UHLIG HH & POWRIE F. 2019. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity, 50, 432–445.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEGATA N, IZARD J, WALDRON L, GEVERS D, MIROPOLSKY L, GARRETT WS & HUTTENHOWER C. 2011. Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMYTHIES LE, SELLERS M, CLEMENTS RH, MOSTELLER-BARNUM M, MENG G, BENJAMIN WH, ORENSTEIN JM & SMITH PD. 2005. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. 115, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STAPPENBECK TS & VIRGIN HW. 2016a. Accounting for reciprocal host-microbiome interactions in experimental science. Nature, 534, 191–9. [DOI] [PubMed] [Google Scholar]
- STAPPENBECK TS & VIRGIN HW. 2016b. Accounting for reciprocal host–microbiome interactions in experimental science. Nature, 534, 191–199. [DOI] [PubMed] [Google Scholar]
- TANOUE T., MORITA S., PLICHTA DR., SKELLY AN., SUDA W., SUGIURA Y., NARUSHIMA S., VLAMAKIS H., MOTOO I., SUGITA K., SHIOTA A., TAKESHITA K., YASUMA-MITOBE K., RIETHMACHER D., KAISHO T., NORMAN JM., MUCIDA D., SUEMATSU M., YAGUCHI T., BUCCI V., INOUE T., KAWAKAMI Y., OLLE B., ROBERTS B., HATTORI M., XAVIER RJ., ATARASHI K. & HONDA K. 2019. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature, 565, 600–605. [DOI] [PubMed] [Google Scholar]
- THORSTEINSDOTTIR U, MAMO A, KROON E, JEROME L, BIJL J, LAWRENCE HJ, HUMPHRIES K. & SAUVAGEAU G. 2002. Overexpression of the myeloid leukemia– associatedHoxa9 gene in bone marrow cells induces stem cell expansion. Blood, 99, 121–129. [DOI] [PubMed] [Google Scholar]
- VINOLO MAR, RODRIGUES HG, NACHBAR RT & CURI R. 2011. Regulation of Inflammation by Short Chain Fatty Acids. Nutrients, 3, 858–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALUJKAR SA, DHOTRE DP, MARATHE NP, LAWATE PS, BHARADWAJ RS & SHOUCHE YS. 2014. Characterization of bacterial community shift in human Ulcerative Colitis patients revealed by Illumina based 16S rRNA gene amplicon sequencing. Gut Pathogens, 6, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATERFIELD M, KHAN IS, CORTEZ JT, FAN U, METZGER T, GREER A, FASANO K, MARTINEZ-LLORDELLA M, POLLACK JL, ERLE DJ, SU M. & ANDERSON MS. 2014. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat Immunol, 15, 258–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU SE, HASHIMOTO-HILL S, WOO V, ESHLEMAN EM, WHITT J, ENGLEMAN L, KARNS R, DENSON LA, HASLAM DB & ALENGHAT T. 2020. Microbiota-derived metabolite promotes HDAC3 activity in the gut. Nature, 586, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG I, EIBACH D, KOPS F, BRENNEKE B, WOLTEMATE S, SCHULZE J, BLEICH A, GRUBER AD, MUTHUPALANI S, FOX JG, JOSENHANS C. & SUERBAUM S. 2013. Intestinal Microbiota Composition of Interleukin-10 Deficient C57BL/6J Mice and Susceptibility to Helicobacter hepaticus-Induced Colitis. PLoS ONE, 8, e70783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU AI, ZHAO L, EATON KA, HO S, CHEN J, POE S, BECKER J, GONZALEZ A, MCKINSTRY D, HASSO M, MENDOZA-CASTREJON J, WHITFIELD J, KOUMPOURAS C, SCHLOSS PD, MARTENS EC & CHEN GY. 2020. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Reports, 31, 107471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENEWICZ LA, YIN X, WANG G, ELINAV E, HAO L, ZHAO L. & FLAVELL RA. 2013. IL-22 Deficiency Alters Colonic Microbiota To Be Transmissible and Colitogenic. The Journal of Immunology, 190, 5306–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, ZHAO D, XIONG X, HE Z. & LI H. 2016. Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C. Journal of Biological Chemistry, 291, 12786–12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU L., WANG Y., ZHOU M., ZHANG Y., WANG P., LI X, YANG J, WANG H. & DING Z. 2018. HOXA9 inhibits HIF-1α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nature Communications: Springer Science and Business Media LLC. [DOI] [PMC free article] [PubMed]
- ZUCCHELLI C, TAMBURRI S, FILOSA G, GHITTI M, QUILICI G, BACHI A. & MUSCO G. 2019. Sp140 is a multi-SUMO-1 target and its PHD finger promotes SUMOylation of the adjacent Bromodomain. Biochim Biophys Acta Gen Subj, 1863, 456–465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA sequencing of mice and de-identified patient stool have been deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA818684 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-CF594 anti-NK-1.1 (Myeloid Panel) | BD | Cat#: 562864; RRID: AB_2737850 |
| PE-CF594 anti-γδ TCR (Myeloid Panel) | BD | Cat#: 563532; RRID: AB_2661844 |
| PE-CF594 anti-TCRβ (Myeloid Panel) | BD | Cat#: 562841; RRID: AB_2737831 |
| PE-CF594 anti-CD45R/B220 (Myeloid Panel) | BD | Cat#: 562313; RRID: AB_11151901 |
| FITC anti-Ly6C (Myeloid Panel) | BD | Cat#: 553104; RRID: AB_394628 |
| AF700 anti-I-A/I-E (Myeloid Panel) | BioLegend | Cat#: 107621; RRID: AB_493726 |
| APC/Cy7 anti-CD11b (Myeloid Panel) | BioLegend | Cat#: 101226; RRID: AB_830642 |
| PE anti-CCR2 (Myeloid Panel) | R&D | Cat#: FAB5538P; RRID: AB_10718414 |
| BV510 anti-CD24 (Myeloid Panel) | BioLegend | Cat#: 101831; RRID: AB_2563894 |
| PerCP/Cy5.5 anti-CD64 (Myeloid Panel) | BioLegend | Cat#: 139307; RRID: AB_2561963 |
| APC anti-CD11c (Myeloid Panel) | BioLegend | Cat#: 117309; RRID: AB_313778 |
| BV785 anti-CX3CR1 (Myeloid Panel) | BioLegend | Cat#: 149029; RRID: AB_2565938 |
| BV650 anti-Ly6G (Myeloid Panel) | BioLegend | Cat#: 127641; RRID: AB_2565881 |
| BV605 anti-CD103 (Myeloid Panel) | BioLegend | Cat#: 121433; RRID: AB_2629724 |
| Pacific Blue anti-CD45 (Myeloid/Adaptive Panels) | BioLegend | Cat#: 103126; RRID: AB_493535 |
| FITC anti-CD62L (Adaptive Panel) | BioLegend | Cat#: 104405; RRID: AB_313092 |
| APC/Cy7 anti-NK-1.1 (Adaptive Panel) | BioLegend | Cat#: 108723; RRID: AB_830870 |
| PE/Cy7 anti-CD44 (Adaptive Panel) | BioLegend | Cat#: 103029; RRID: AB_830786 |
| APC anti-CD45R/B220 (Adaptive Panel) | BioLegend | Cat#: 100206; RRID: AB_312663 |
| BV605 anti-CD4 (Adaptive Panel) | BioLegend | Cat#: 100451; RRID: AB_2564591 |
| BV711 anti-CD8α (Adaptive Panel) | BioLegend | Cat#: 100747; RRID: AB_11219594 |
| TruStain FcX | BioLegend | Cat#: 101319; RRID: AB_1574973 |
| Zombie UV Fixable Viability Kit | BioLegend | Cat#: 423107 |
| Fixation Buffer | eBioscience | Cat#: 00-5523-00 |
| Anti-Histone H3 | abcam | Cat#: ab1791; RRID: AB_302613 |
| Anti-S100A8 | abcam | Cat#: ab92331; RRID: AB_2050283 |
| Biological samples | ||
| Crohn’s disease peripheral blood mononuclear cells | PRISM Cohort, MGH | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI media | Life technologies | Cat#: 11875-093 |
| DMEM media | Life technologies | Cat#: 11995-073 |
| Dextran Sulfate Sodium (DSS) | MP Biomedicals | Cat#: 160110; CAS: 9011-18-1 |
| Hyclone fetal bovine serum | GE Healthcare | S#30396.03 |
| Penicillin-Streptomycin | Gibco | P4333 |
| Ficoll-Paque Plus | GE Healthcare | 17-1440-02 |
| RNAiMAX | Life Technologies | 13778075 |
| HiPerfect Transfection Reagent | Qiagen | 1029975 |
| LPS 0111:B4 | Sigma | L4391 |
| Metronidazole | RPI | Cat#: M81000; CAS: 443-48-1 |
| Ampicilin | RPI | Cat#: 69-52-3; CAS: 69-52-3 |
| Neomycin | Sigma Aldrich | Cat#: N6386; CAS: 1405-10-3 |
| Vancomycin | Chem Impex | Cat#: 00315; CAS: 1404-93-9 |
| Dispase II | Sigma | Cat#: D4693; CAS: 42613-33-2 |
| Collagenase, Type IV | Sigma | Cat#: 17104019 |
| Recombinant murine M-CSF | Peprotech | 315-02 |
| Recombinant murine IL-3 | Peprotech | 213-13 |
| Critical commercial assays | ||
| PureLink Microbiome DNA Purification Kit | Invitrogen | A29790 |
| RNeasy mini kit | Qiagen | 74104 |
| Mouse IL-6 ELISA kit | R&D | DY406 |
| Deposited data | ||
| SP140 16S rRNA sequencing data | This paper | SRA: PRJNA818684 |
| SP140 PBMC RNA sequencing data | Mehta et al., 2017 | GEO: GSE89876 |
| Experimental models: Cell lines | ||
| Human: THP1 | Laboratory of Hans-Christian Reinecker | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL6/J | The Jackson Laboratory | JAX: 000664 |
| Mouse: SP140 Knockout (on C57BL6/J background) | Laboratory of Russell Vance | Ji et al. 2021 |
| Oligonucleotides | ||
| ON-TARGETplus Non-targeting Control Pool UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA, UGGUUUACAUGUUUUCCUA | Dharmacon/Horizon s Discovery | D-001810-10-05 |
| ON-TARGETplus Human SP140 siRNA – SMARTpool CCGAGCAGAUGUAUGAACA, CAAGAGUGAUGUAUUGUGU, GAACGUAGAGGGUCAGAAC, GGAUUAACCUGAUGGCCUA | Dharmacon/Horizon s Discovery | L-016508-00-0005 |
| Primer for Stool qPCR Total 16S F-GTGCCAGCMGCCGCGGTAA R-GACTACCAGGGTATCTAAT | IDT, this paper | N/A |
| Primer for Stool qPCR A. muciniphila F-CAGCACGTGAAGGTGGGGAC R-CCTTGCGGTTGGCTTCAGAT | IDT, this paper | N/A |
| Primer for Stool qPCR Helicobacter spp. F-GCTATGACGGGTATCC R-GATTTTACCCCTACACCA | IDT, this paper | N/A |
| Primer for Stool qPCR Enterobacteriaceae F-CATTGACGTTACCCGCAGAAGAAGC R-CTCTACGAGACTCAAGCTTGC | IDT, this paper | N/A |
| Primer for Mouse qPCR Lcn2 F-TCCTCAGGTACAGAGCTACAA R-GCTCCTTGGTTCTTCCATACA | IDT, this paper | N/A |
| Primer for Mouse qPCR Lyz2 F-ATGGAATGGCTGGCTACTATG | IDT, this paper | N/A |
| R-GGTCTCCACGGTTGTAGTTT | ||
| Primer for Mouse qPCR S100a8 F-CTTTGTCAGCTCCGTCTTCA R-TGTAGAGGGCATGGTGATTTC | IDT, this paper | N/A |
| Primer for Mouse qPCR Tbp F-TGATCAAACCCAGAATTGTTCT R-TGGTCTTCCTGAATCCCTTTA | IDT, this paper | N/A |
| Software and algorithms | ||
| QIIME 2 | Bolyen et al., 2019 | https://qiime2.org/ |
| DADA2 | Callahan et al., 2016 | N/A |
| MAFFT | Katoh et al., 2002 | N/A |
| UniFrac | Lozupone et al., 2005 | N/A |
| q2-feature-classifier | Bokulich et al., 2018 | N/A |
| LEfSe | Segata et al., 2011 | http://huttenhower.sph.harvard.edu/galaxy |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| FlowJo software | BD | http://flowjo.com |
| Graphpad Prism v9.1.2 | Graphpad software | https://www.graphpad.com/ |
| Other | ||
| iTaq Universal SYBR Green supermix | Bio Rad | 1725124 |
| iScript cDNA Synthesis Kit | Bio Rad | 1708841 |
| Beadbug prefilled homogenizer tubes | Sigma-Aldrich | Z763799 |