Abstract
Tumor derived exosomes (TEXs) have emerged as promising biomarkers for cancer liquid biopsy. Conventional methods (such as ELISA and qRT-PCR) and emerging biosensing technologies mainly detect a single type of exosomal biomarker due to the distinct properties of different biomolecules. Sensitive detection of two different types of TEX biomarkers, i.e., protein and microRNA combined biomarkers, may greatly improve cancer diagnostic accuracy. We developed an exosome protein microRNA one-stop (Exo-PROS) biosensor that not only selectively captured TEXs but also enabled in situ, simultaneous detection of TEX protein–microRNA pairs via a surface plasmon resonance mechanism. Exo-PROS assay is a fast, reliable, low sample consumption, and user-friendly test. With a total of 175 cancer patients and normal controls, we demonstrated that TEX protein–microRNA pairs measured by Exo-PROS assay detected lung cancer and breast cancer with 99% and 96% accuracy, respectively. Exo-PROS assay also showed superior diagnostic performance to conventional ELISA and qRT-PCR methods. Our results demonstrated that Exo-PROS assay is a potent liquid biopsy assay for cancer diagnosis.
Keywords: exosomes, proteins, microRNAs, cancer liquid biopsy, surface plasmon resonance, biosensor
Graphical Abstract
Exosomes are one type of cell-secreted extracellular vesicle with a size between approximately 40 and 160 nm.1 They mediate intercellular communications by transporting various cargos including proteins, RNAs, DNAs, and lipids between cells. Exosomes originated from the endosomal pathway, therefore their cargos closely mimic the parent cells.1 Exosomes play important roles in cancer development, immune response regulation, metastasis, and drug resistance.2,3 They present abundantly in various body fluids including blood, urine, and saliva. Because of these favorable features, exosomes have emerged as promising biomarkers for cancer liquid biopsy.1–4
Exosomal proteins and microRNAs are the most widely investigated cancer biomarkers. Exosomal proteins are usually measured by Western blot, enzyme-linked immunosorbent assay (ELISA), flow cytometry, and liquid chromatography–mass spectrometry (LC-MS). Exosomal microRNAs are typically detected by quantitative reverse transcription polymerase chain reaction (qRT-PCR), next-generation sequencing, and microarray. However, the clinical utility of these conventional methods is limited by low sensitivity, tedious and time-consuming processes, large sample volumes, and high cost. Most importantly, a major challenge faced by these methods is that they cannot distinguish tumor derived exosomes (TEXs) from their nontumor counterparts. Since TEXs are a small portion of total exosomes, the interferences from non-TEXs may result in the tests having poor sensitivity and specificity in cancer diagnosis.
To overcome these limitations, many biosensors have been developed for exosomal protein and microRNA detection.5–8 However, due to the distinct properties of biomolecules, these methods are only able to detect a single type of exosomal biomarker, i.e., either exosomal proteins or exosomal micro-RNAs. It is widely accepted that multiple biomarkers are required to achieve high sensitivity and specificity in cancer diagnosis, but most studies only employed multiple biomarkers of the same type. The use of two different types of biomarkers, i.e., protein and microRNA combined biomarkers, to further improve cancer diagnostic accuracy has just begun.9–11 However, traditional methods with poor sensing performances and tedious processes were applied in these studies to characterize the combined biomarkers, which greatly limit the clinical translational potential of these biomarkers.9–11 Considering the large population of cancer patients and patients at high risk of cancer, new and sensitive biosensors for simultaneous detection of TEX protein and microRNA biomarkers in a simple, fast, and user-friendly way are urgently needed to facilitate the translation of these combined biomarkers into clinical settings for cancer screening and diagnosis.
Herein, we report an exosome protein microRNA one-stop (Exo-PROS) biosensor that selectively captures TEXs from all exosomes, and, in situ, simultaneously detects TEX protein–microRNA pairs on a single platform. In an Exo-PROS assay, antibodies against tumor-overexpressed proteins are first used to capture TEXs. A surface plasmon resonance (SPR) sensing mechanism is then used to quantitatively measure the expression of TEX proteins. Next, molecular beacons (MBs), the sensing probes for microRNAs, are added to detect microRNAs of captured TEXs. After the hybridization of MBs with target microRNAs, the restored fluorescence signals from MBs are detected via surface plasmon enhanced fluorescence spectroscopy (SPEFS) on the same platform. The expression of TEX microRNAs is proportionally related with the fluorescence intensities of MBs.
The analytical sensing performances of the Exo-PROS assay were first evaluated. Our results showed that Exo-PROS assay is a highly sensitive, fast (assay time: 4 h), reliable (coefficient of variation <10%), low sample consumption (as low as 1 μL serum), user-friendly (only a pipet is required), and cost-effective test. The clinical utility of the Exo-PROS assay was then demonstrated using lung cancer and breast cancer as the disease models. In this work, Exo-PROS assay was used to measure the levels of TEX epidermal growth factor receptor (EGFR)-miR-21 pair and galectin-3-binding protein (LG3BP)-miR-210 pair in serum samples from 60 lung cancer patients (stage I–IV) and 35 normal controls for lung cancer diagnosis. Exo-PROS assay was also used to detect the TEX Annexin A8 (ANXA8)-miR-342 pair in serum samples from 60 breast cancer patients (stage 0–IV) and 20 normal controls for breast cancer diagnosis. In both cancer models, TEX protein–microRNA pairs provided high sensitivity and specificity and >96% accuracy in distinguishing cancer groups from normal controls. TEX protein and microRNA combined biomarkers also showed higher diagnostic accuracy than single type biomarkers. Our results demonstrated that the Exo-PROS assay together with the use of two types of exosomal biomarkers are a potent and effective strategy to improve cancer diagnosis.
RESULTS AND DISCUSSION
Selection of TEX Protein–MicroRNA Pairs for Cancer Diagnosis.
The potential of Exo-PROS assay in cancer diagnosis was demonstrated in both lung cancer and breast cancer. For lung cancer diagnosis, we selected EGFR and LG3BP as the model protein biomarkers because they facilitate tumor growth and metastasis,12,13 and they are promising diagnostic biomarkers validated in >350 lung cancer patients and lung cancer cell-derived exosomes.14–22 We selected miR-21 and miR-210 as the model microRNA biomarkers because both microRNAs regulate important signaling pathways of lung cancer.23,24 Recent meta-analyses involving >7000 lung cancer patients and >7000 healthy controls strongly demonstrated the diagnostic values of these microRNAs in lung cancer.25–28
For breast cancer diagnosis, we selected ANXA8 as the protein biomarker because tissue microarray analyses of >1700 breast cancer cases showed ANXA8 is upregulated in breast cancer and closely correlated with tumor stage, grade, and positive lymph nodes.29,30 We selected miR-342 as the microRNA biomarker because miR-342 is downregulated in breast cancer. It regulates breast cancer progression, metastasis, and drug resistance, and the loss of miR-342 is associated with worse overall survival in breast cancer patients.31–34 Besides, miR-342 directly targets ANXA8 mRNA and down-regulates ANXA8 protein expression.35 Because of the inverse relationship between the expression of ANXA8 and miR-342, the ratio between ANXA8 and miR-342 levels can be used to further enhance the differences between cancer and normal controls, and thus increase diagnostic sensitivity and specificity.
Detection Mechanism of Exo-PROS Biosensor.
The Exo-PROS biosensor had a compact design and used SPR and SPEFS as the sensing mechanism (Figures 1a and 1b). To detect TEX protein–microRNA pairs, a biochip was first prepared by coating a glass slide with 2 nm Ti and 49 nm Au films. Then the surface of the biochip was modified with a mixture of PEG200-SH and biotin-PEG1000-SH. Finally, neutravidin and biotinylated antibodies against tumor overexpressed proteins including EGFR, LG3BP, and ANXA8 were sequentially applied to attach the antibodies on the surface of biochip through biotin–avidin interaction.
Figure 1.
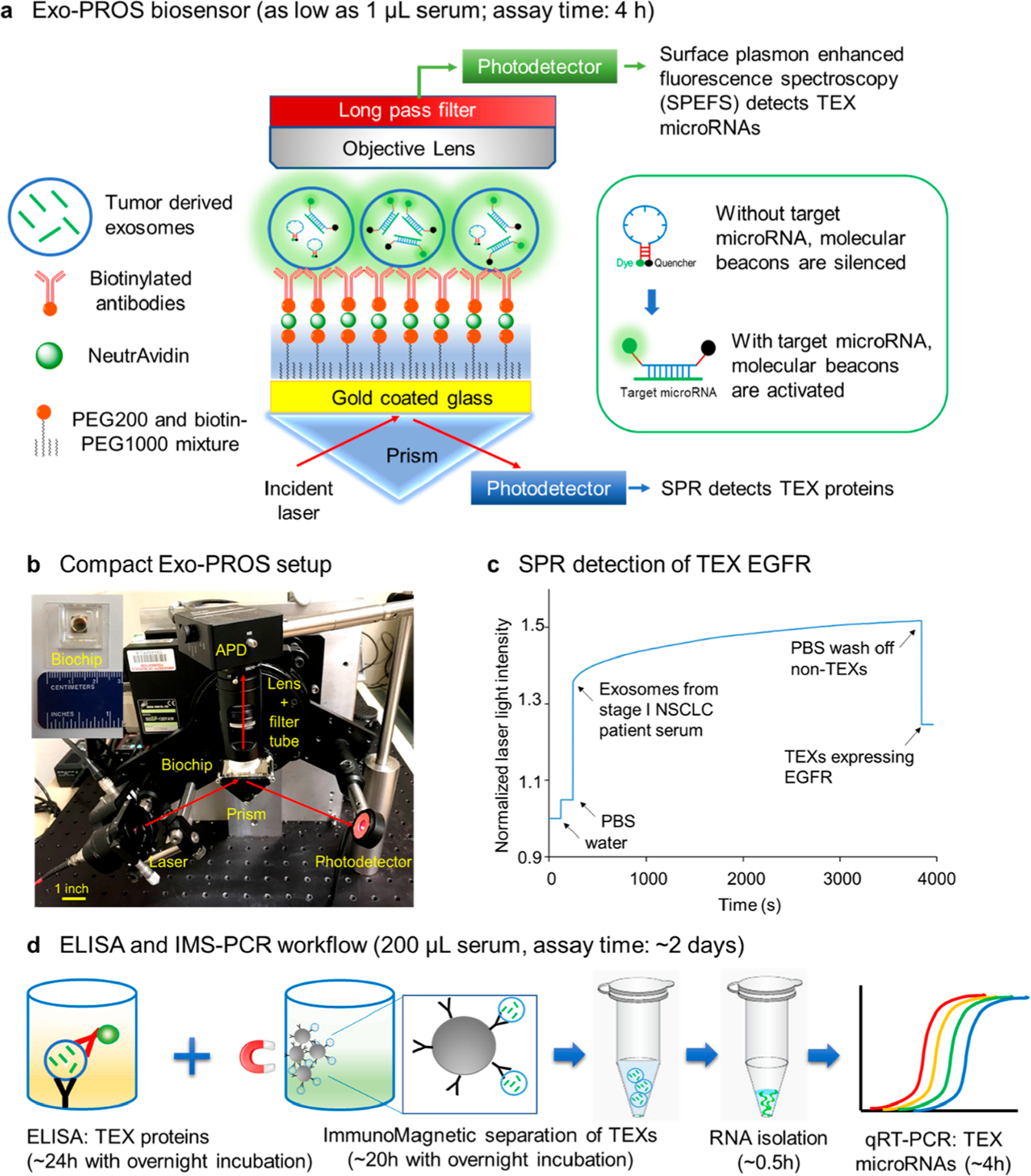
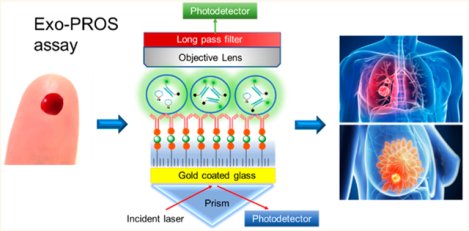
Overview of the Exo-PROS assay. (a) Sensing mechanism of Exo-PROS assay. A gold coated glass slide is coated with PEG and antibodies against tumor-overexpressed proteins. The capture of TEXs by the antibodies changes the local refractive index and allows the detection of TEX proteins via SPR mechanism. Next, molecular beacons are added to diffuse into TEXs and bind to target microRNAs. The restored fluorescence signals from molecular beacons allow the detection of TEX microRNAs. Insert: Sensing mechanism of molecular beacons. (b) A photo of the Exo-PROS biosensor. (c) A representative SPR curve for measuring TEX EGFR expression in the serum from a Stage I NSCLC patient. (d) Conventional ELISA and TEX ImmunoMagnetic Separation-RNA Isolation-qRT-PCR (IMS-PCR for short) workflow to detect TEX protein–microRNA pairs.
To measure the expression of TEX proteins, water and PBS were applied on the biochip to collect background signals (Iwater and IPBS). Then exosomes were added to allow the capture of TEXs by the antibodies. The capture of TEXs changed the local refractive index, affected the optical properties of the surface plasmon modes, and permitted optical detection of TEX proteins. After unbounded exosomes were washed off using PBS, the intensity of reflected laser light (ITEX) was recorded. A representative SPR curve for the measurement of TEX EGFR expression in the serum of a nonsmall cell lung cancer (NSCLC) patient is shown in Figure 1c. The expression of TEX proteins was calculated using the following equation:
| (1) |
where ITEX, IPBS, and Iwater were the reflected laser intensities of TEXs, PBS, and water. The difference between IPBS and Iwater was used as the normalization factor to remove the biochip-to-biochip variation.
Next, to quantify the expression of TEX microRNAs, molecular beacons (MBs), the oligonucleotide hybridization probes for miR-21, miR-210, or miR-342 were added. MBs are oligonucleotides with a stem-loop structure. The loop has a complementary sequence to the target microRNA. The stems have a fluorophore and a quencher attached at each end. MBs can diffuse into captured TEXs and interact with micro-RNAs.10,36,37 The binding between MBs and target micro-RNAs separates the fluorophores from the quenchers and restores fluorescence signals from MBs (Figure 1a insert). In Exo-PROS sensor, the fluorescence signals are enhanced ~100 times by the SPR system, and therefore can be easily measured by a low-cost avalanche photodetector (APD) above the biochip (Figure 1b). The fluorescence intensities before and after the addition of MBs were used to calculate the expression of TEX microRNAs via the following equation:
| (2) |
where Ibefore MB and Iafter MB were fluorescence intensities collected before and after the addition of MBs.
Conventional techniques, such as ELISA and qRT-PCR can be used to measure the levels of TEX protein–microRNA pairs but in a much more tedious and high sample consumption way (Figure 1d). ELISA is typically used to measure the levels of exosomal proteins, but it requires at least 100 μL serum samples for one measurement. To measure the expression of microRNAs in TEXs, immunomagnetic separation needs to be performed first to separate TEXs from other exosomes, then RNA is extracted from isolated TEXs, and finally qRT-PCR is performed to measure the expression of microRNAs. This TEX immunomagnetic separation-RNA isolation-qRT-PCR (IMS-PCR for short) workflow requires at least 100 μL serum samples and approximately 2 days to complete. Compared with ELISA and IMS-PCR workflow, the Exo-PROS assay offers one-stop, in situ characterization of TEX protein–microRNA pairs, and is a highly sensitive, simple, fast (~4 h vs 2 days), low sample consumption (as low as 1 μL vs 200 μL serum), and cost-effective test.
Characterization of Exosomes.
The exosomes were isolated from cell culture-conditioned medium and human serum samples. The size, size distribution and number concentration of exosomes were characterized by nanoparticle tracking analysis (NTA). Figure 2a and Supplementary Figure S1a showed the representative size distributions of exosomes isolated from the serum of a late-stage NSCLC patient and the cell culture medium of A549 NSCLC cells. The size of exosomes ranged from 64 to 165 nm, and the averaged concentration of exosomes isolated from serum samples was 1.61 × 1013 exosomes/mL (Supplementary Table S1). The morphology of exosomes was observed by cryotransmission electron microscopy (Cryo-TEM) (Figure 2b). Exo-Check exosome antibody arrays and Western blotting were used to measure exosome positive markers (such as CD63 and CD81) and exosome negative markers (such as GM130 and GRP94). Results confirmed the presence and high purity of exosomes (Supplementary Figure S1b and S1c).
Figure 2.
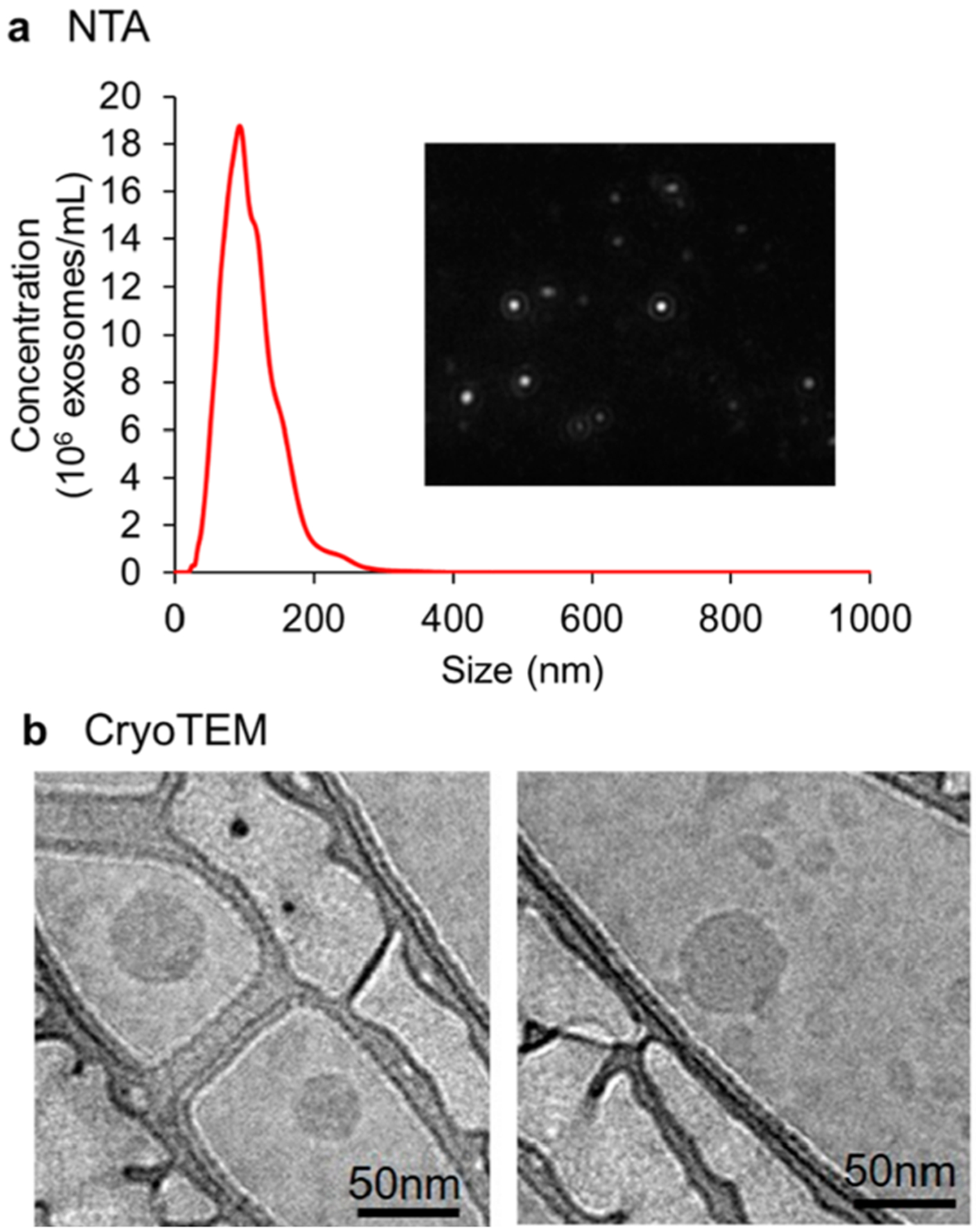
Characterization of exosomes. Exosomes were isolated from 50 μL of serum from a late stage NSCLC patient. (a) Size and size distribution of exosomes (10,000× dilution) were characterized by nanoparticle tracking analysis (NTA). Insert: Screenshot from the recorded video of exosomes during NTA. (b) Morphology of exosomes was characterized by CryoTEM.
Sensing Performances of Exo-PROS Assay.
The analytical sensitivity, specificity, dynamic range, and repeatability of the Exo-PROS assay were characterized using exosomes derived from A549 NSCLC cells and MDA-MB-231 breast cancer cells. Cell-derived exosomes were applied on the biochip at various concentrations ranging from 0 (blank control) to 1.25 × 1012 exosomes/mL. The blank control was the nanovesicles isolated from the plain cell culture medium. The levels of TEX EGFR-miR-21, LG3BP-miR-210, and ANXA8-miR-342 pairs were measured. Representative SPR curves that measured the levels of EGFR in A549 cell-derived exosomes at concentrations of 5 × 1010 and 2.5 × 1011 exosomes/mL were shown in Supplementary Figure S2a. Scanning electron microscopy (SEM) was used to confirm the capture of exosomes expressing EGFR (i.e., EGFR+ exosomes) on the biochip surface (Supplementary Figure S2b). Approximately 5-fold more exosomes were captured on the biochip when exosomes were applied at 5-fold higher concentration (i.e., 2.5 × 1011 exosomes/mL vs 5 × 1010 exosomes/mL). The limit of detection (LOD) was calculated as the mean of the blank control plus 3-fold of standard deviation.38 As shown in Figures 3a, 3b and Supplementary Figure S2, the LOD of Exo-PROS assay for exosomal EGFR sensing was 3.5 × 109 exosomes/mL and for EGFR+ exosomal miR-21 detection was 5 × 109 exosomes/mL. The dynamic range of Exo-PROS assay was from 5 × 109 to 1.25 × 1012 exosomes/mL, offering approximately 3 orders of magnitude. The LODs for TEX LG3BP-miR-210 pair were 2 × 1010 and 5 × 1010 exosomes/mL, respectively (Supplementary Figure S3). The LODs for TEX ANXA8-miR-342 pair were 5 × 109 and 2 × 1010 exosomes/mL respectively (Supplementary Figure S4). The typical exosome concentration in serum is 1012–1013 exosomes/mL (Supplementary Table S1);39–41 therefore, the Exo-PROS biosensor may require as low as 1 μL serum sample for one assay, demonstrating its high sensitivity.
Figure 3.
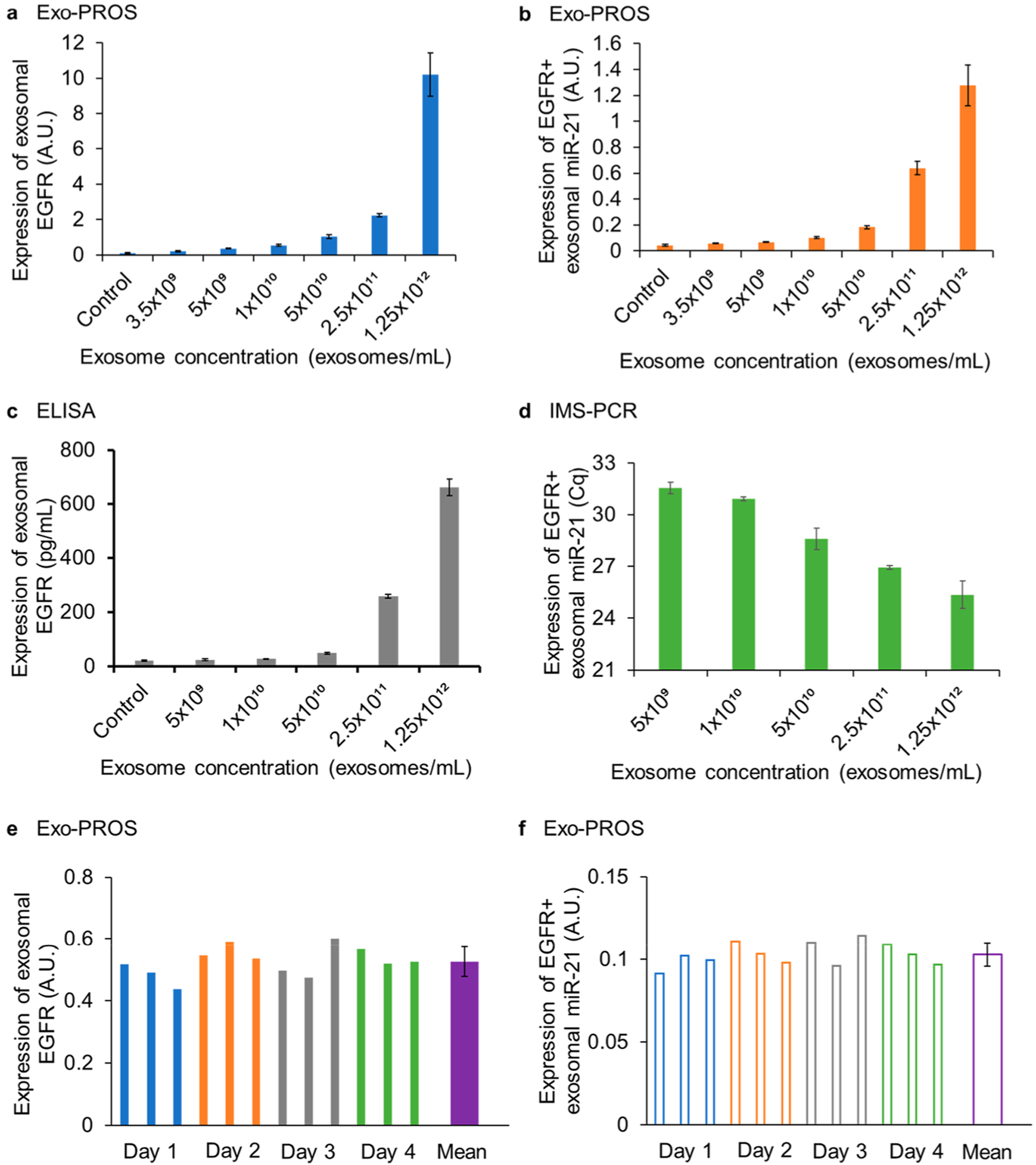
Sensing performance of Exo-PROS assay. To determine the limit of detection and linear range of Exo-PROS assay, the levels of EGFR and miR-21 in A549 cell-derived exosomes at concentrations of 0 (blank control) to 1.25 × 1012 exosomes/mL were measured by (a, b) Exo-PROS assay, (c) ELISA, and (d) IMS-PCR workflow. For exosomal EGFR, the LOD of Exo-PROS assay was 14-fold lower than ELISA (3.5 × 109 exosomes/mL vs 5 × 1010 exosomes/mL). For EGFR+ exosomal miR-21, the LOD of Exo-PROS assay was same as IMS-PCR (5 × 109 exosomes/mL). Exo-PROS assay showed 3-log linear range. (e, f) To determine the repeatability of Exo-PROS assay, the levels of EGFR and miR-21 of A549 cell-derived exosomes (1 × 1010 exosomes/mL) were measured on 4 different days, 3 replicates per day. For exosomal EGFR, the interday CV and intraday CV were 9.01% and 7.84%, respectively. For EGFR+ exosomal miR-21, the interday and intraday CVs were 6.73% and 6.66%, respectively.
To compare the sensing performance of Exo-PROS assay with conventional methods, the expression of EGFR and miR-21 in A549 cell-derived exosomes at concentrations from 0 (blank control) to 1.25 × 1012 exosomes/mL were measured by ELISA (Figure 3c) and IMS-PCR workflow (Figure 3d and Supplementary Figure S5). The LOD of ELISA for exosomal EGFR was 5 × 1010 exosomes/mL, which was 14-fold higher than Exo-PROS assay. The expression of EGFR+ exosomal miR-21 was successfully detected by IMS-PCR at the concentration of 5 × 109 exosomes/mL, which was comparable to Exo-PROS assay. However, no amplification process is required in Exo-PROS assay.
To evaluate the repeatability of the Exo-PROS assay, A549 cell-derived exosomes were applied on the biochip at the concentration of 1010 exosomes/mL. The expression of TEX EGFR-miR-21 pair was measured 12 times independently on 4 different days, 3 replicates per day. No significant difference was observed between measurements (Figure 3e, 3f). For exosomal EGFR, the interday coefficient of variation (CV) was 9.01% and the intraday CV was 7.84%. For EGFR+ exosomal miR-21, the interday and intraday CVs were 6.73% and 6.66%, respectively. These results demonstrated that the Exo-PROS assay had great repeatability.
To evaluate the specificity of Exo-PROS assay in exosomal protein detection, the biochip was modified with anti-IgG control antibodies. A549 cell-derived exosomes were applied at the concentration of 1.25 × 1012 exosomes/mL. MDA-MB-231 cell-derived exosomes were applied at the concentration of 1011 exosomes/mL. The expression of exosomal EGFR, LG3BP, and ANXA8 was measured. Little signals were observed from the anti-IgG control antibodies (Supplementary Figure S6). To evaluate the specificity of Exo-PROS assay in exosomal microRNA detection, the glass slides were modified with anti-EGFR, anti-LG3BP, or anti-ANXA8 antibodies to capture exosomes expressing EGFR, LG3BP or ANXA8. Then, molecular beacons with sequence complementary to cel-miR-39 (MB-cel-miR-39) were added. Compared with MB-miR-21, MB-miR-210, and MB-miR-342, negligible signals were detected with the MB-cel-miR-39 (Supplementary Figure S6). In addition, in our previous studies, we designed MBs that had mismatched sequence to miR-21 and demonstrated the sensing specificity in distinguishing target microRNAs from other microRNAs in the exosomes.40,42 These results showed that Exo-PROS assay had excellent sensing specificity in detecting TEX protein–microRNA pairs.
Evaluation of Diagnostic Performance of Exo-PROS Assay Using Cell-Derived Exosomes.
The diagnostic performance of Exo-PROS assay in detecting lung cancer and breast cancer was evaluated using cell-derived exosomes. For lung cancer diagnosis, exosomes isolated from the culture media of A549 NSCLC cells and BEAS-2B human normal bronchial epithelial cells were applied on the biochips at the concentration of 1.25 × 1012 exosomes/mL. The expression of TEX EGFR-miR-21 and LG3BP-miR-210 pairs was measured (Figure 4a, 4b). Significant differences were observed for both TEX protein–microRNA pairs between A549 NSCLC cells and BEAS-2B normal cells. Exosomes derived from A549 cells had 4.12-fold and 3.56-fold higher levels of EGFR and miR-21 than those from BEAS-2B cells. A549 cell-derived exosomes had 2.49-fold and 2.82-fold higher levels of LG3BP and miR-210 than BEAS-2B cell-derived exosomes. For breast cancer diagnosis, exosomes isolated from the culture media of MDA-MB-231 breast cancer cells and MCF-10A normal cells were applied on the biochips at the concentration of 1011 exosomes/mL. The expression of TEX ANXA8-miR-342 pair was measured. The expression of ANXA8 was 2.07-fold higher in exosomes from MDA-MB-231 cells than those from MCF-10A cells (Figure 4c). The expression of ANXA8+ exosomal miR-342 was 2.40-fold lower in MDA-MB-231 cell-derived exosomes than MCF-10A cell-derived exosomes (Figure 4d). These results demonstrated that Exo-PROS assay successfully distinguished cancer groups from normal controls by detecting significantly different levels of exosomal protein–microRNA pairs in cancer cell-derived exosomes than normal cell-derived exosomes.
Figure 4.
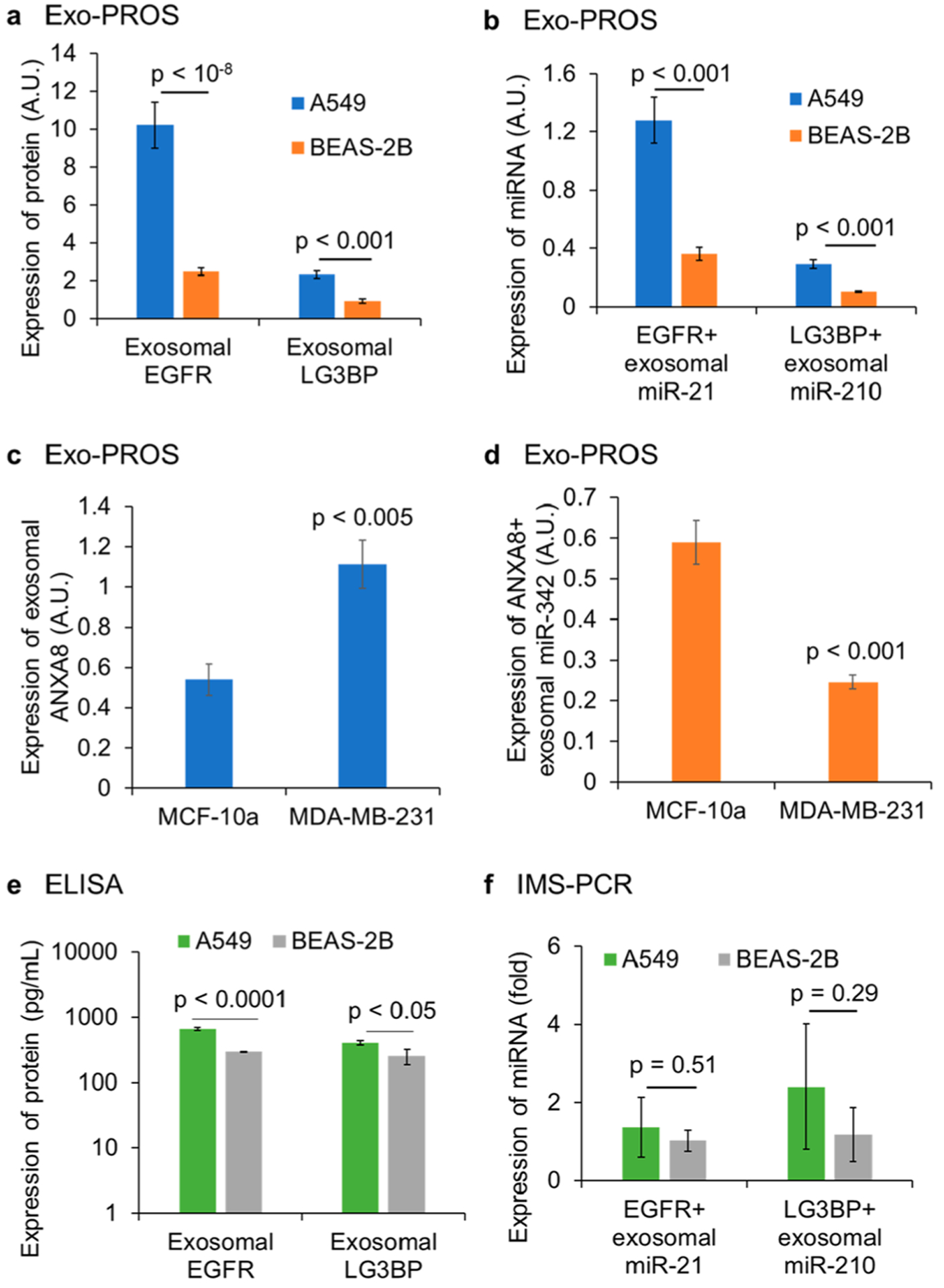
Evaluation of diagnostic performance of Exo-PROS assay using cell-derived exosomes. Exosomes from A549 NSCLC cells and BEAS-2B normal cells were applied at the concentration of 1.25 × 1012 exosomes/mL (n = 3). Exosomes from MCF-10A normal cells and MDA-MB-231 breast cancer cells were applied at the concentration of 1011 exosomes/mL (n = 3). (a, b) Exo-PROS assay detected significantly higher levels of EGFR-miR-21 pair and LG3BP-miR-210 pair in A549 cell-derived exosomes than BEAS-2B cell-derived exosomes. (c, d) Exo-PROS assay detected higher expression of ANXA8 and lower expression of miR-342 in MDA-MB-232 cell-derived exosomes than those from MCF-10A cells. (e, f) To demonstrate that the Exo-PROS assay is superior to conventional methods, ELISA and IMS-PCR were used to measure the expression of EGFR-miR-21 and LG3BP-miR-210 pairs in the same exosome samples from A549 and BEAS-2B cells (1.25 × 1012 exosomes/mL, n = 3). ELISA detected significantly higher levels of EGFR and LG3BP levels in A549 cell-derived exosomes than BEAS-2B cell-derived exosomes; however, IMS-PCR workflow detected no significant difference in the expression of EGFR+ exosomal miR-21 and LG3BP+ exosomal miR-21 between A549 cell-derived exosomes and BEAS-2B cell-derived exosomes. Results showed that the Exo-PROS assay distinguished lung cancer and breast cancer groups from normal controls and showed better diagnostic performance than ELISA and IMS-PCR workflows.
To demonstrate Exo-PROS assay is superior to conventional methods, ELISA and IMS-PCR workflow were used to quantify the levels of EGFR-miR-21 pair and LG3BP-miR-210 pair in the same exosome samples from A549 and BEAS-2B cells (1.25 × 1012 exosomes/mL). In A549 cell-derived exosomes, the levels of EGFR and LG3BP were 2.24-fold and 1.61-fold higher than those of BEAS-2B cell-derived exosomes (Figure 4e). The levels of EGFR+ exosomal miR-21 and LG3BP+ exosomal miR-210 of A549 cell-derived exosomes were 1.33-fold and 2.04-fold higher than those of BEAS-2B cell-derived exosomes, however, the differences were not significant (Figure 4f). Compared with ELISA and IMS-PCR workflow, Exo-PROS assay detected larger differences in the expression of exosomal protein and microRNA pairs between A549 NSCLC cells and BEAS-2B normal cells, indicating that Exo-PROS assay had higher sensitivity than conventional methods.
Evaluation of Diagnostic Performance of Exo-PROS Assay in Lung Cancer Diagnosis.
The clinical utility of Exo-PROS assay was demonstrated in lung cancer diagnosis. A total of 35 normal controls and 60 treatment naïve lung cancer patients (stages I–IV) were enrolled in this study. The patient characteristics were provided in Supplementary Table S1. Normal controls were 15 patients at low risk of lung cancer and 20 patients at high risk of lung cancer. All patients at high risk of lung cancer had >30 pack years of smoking, and a majority of them had pulmonary diseases including pneumonia, asthma, chronic bronchitis, and emphysema/COPD. For lung cancer group, both NSCLC patients (n = 52) and small cell lung cancer (SCLC) patients (n = 8) were included. For NSCLC, both adenocarcinoma and squamous cell carcinoma were included. Patients in the normal and cancer groups were matched in gender, age, and race. Exosomes were isolated from 50 μL serum samples, resuspended in 50 μL PBS and characterized by NTA. As shown in Supplementary Figure S7, no significant difference was observed in the size and number concentration of serum-derived exosomes between normal controls at low risk of lung cancer (n = 15) and lung cancer patients (n = 60). Exosomes from normal controls at high risk of lung cancer (n = 20) had larger size than those from lung cancer patients (n = 60) (134 nm vs 108 nm; p < 0.001) but had similar number concentration as those from lung cancer patients. The levels of TEX EGFR-miR-21 and LG3BP-miR-210 pairs were measured using the Exo-PROS assay (Figures 5a, 5b). The expression of all four biomarkers was significantly higher in exosomes from lung cancer patients than normal controls. The expression of EGFR and miR-21 was 1.76-fold and 2.64-fold higher in exosomes from lung cancer patients than normal controls. The expression of LG3BP and miR-210 was 2.74-fold and 2.64-fold higher in exosomes from lung cancer patients than normal controls. These results demonstrated that TEX EGFR-miR-21 and LG3BP-miR-210 pairs measured by Exo-PROS assay were able to distinguish lung cancer patients from normal controls. Power analysis indicated that we had enough samples to achieve 80% power to detect a fold change of 1.5 when the fold change under the null hypothesis was 1 and the significance level was 0.05.
Figure 5.
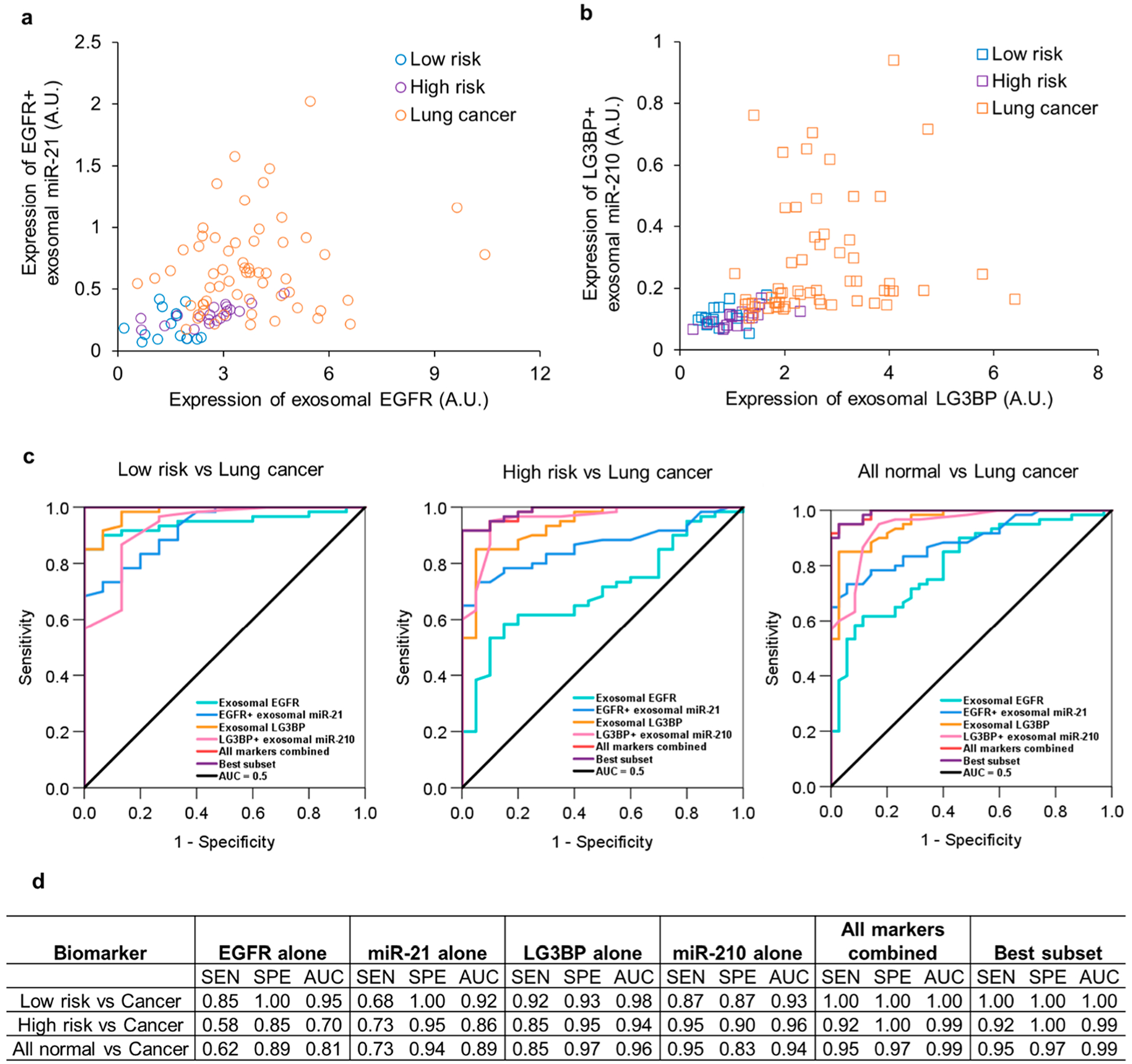
Evaluation of the diagnostic performance of Exo-PROS assay in lung cancer. Exosomes were isolated from 50 μL sera from normal controls (n = 35, 15 low risk and 20 high risk of lung cancer) and lung cancer patients (n = 60, 52 NSCLC and 8 SCLC, stage I–IV). The levels of TEX EGFR-miR-21 and LG3BP-miR-210 pairs were measured by Exo-PROS assay (a, b). The Exo-PROS assay detected significantly higher levels of TEX EGFR-miR-21 and LG3BP-miR-210 pairs in sera from lung cancer patients than normal controls (p < 10−5). (c, d) ROC analysis was performed to determine the sensitivity (SEN), specificity (SPE), and AUC of each biomarker, best subset of the biomarkers, and all markers combined in distinguishing lung cancer patients from normal controls at a low risk of lung cancer, normal controls at a high risk of lung cancer, and all normal controls. The Exo-PROS assay showed high diagnostic accuracy in distinguishing lung cancer patients from normal controls.
Receiver operating characteristic (ROC) analysis was performed to determine the sensitivity, specificity, and area under curve (AUC) of each biomarker and combined biomarkers measured by Exo-PROS assay in distinguishing normal controls from lung cancer patients (Figure 5c and 5d). When comparing normal controls at low risk of lung cancer with lung cancer patients, each individual biomarker showed AUC ≥ 0.92. When all four biomarkers were combined, the Exo-PROS assay successfully discriminated lung cancer patients from normal controls at low risk of lung cancer with sensitivity of 1.00, specificity of 1.00 and AUC of 1.00. When comparing normal controls at high risk of lung cancer with lung cancer patients, LG3BP-miR-210 pair showed higher diagnostic accuracy than EGFR-miR-21 pair and achieved AUC ≥ 0.94. With all four biomarkers, the Exo-PROS assay differentiated lung cancer patients from normal controls at high risk of lung cancer with sensitivity of 0.92, specificity of 1.00 and AUC of 0.99. When both low risk and high risk normal controls were considered, each individual biomarker measured by Exo-PROS assay detected lung cancer with AUC ≥ 0.81. When all four biomarkers were combined, Exo-PROS assay detected lung cancer from normal controls (both low risk and high risk) with sensitivity of 0.95, specificity of 0.97 and AUC of 0.99. In addition, best subset selection analysis using the bestglm R package43 was performed to determine the best combination of these biomarkers for lung cancer diagnosis. TEX EGFR-miR-21 pair and LG3BP+ exosomal miR-210 were the best biomarker combination that provided sensitivity of 0.95, specificity of 0.97, and AUC of 0.99 in differentiating lung cancer patients from normal controls (both low risk and high risk patients).
To demonstrate that Exo-PROS assay outperformed conventional methods, the levels of TEX EGFR-miR-21 and LG3BP-miR-210 pairs in selected normal controls (n = 20) and NSCLC patients (n = 40) were measured using both Exo-PROS assay and the ELISA and IMS-PCR workflow (Supplementary Figure S8a–d). For ELISA and IMS-PCR workflow, exosomes were isolated from 200 μL serum and split evenly between ELISA and IMS-PCR. Among all biomarkers, only exosomal LG3BP showed significantly higher expression (1.87-fold increase) in NSCLC patients than normal controls. The expression of exosomal EGFR, miR-21, and miR-210 was not significantly different between NSCLC patients and normal controls. ROC analysis was performed to compare the diagnostic performances of each biomarker and combined biomarkers measured by Exo-PROS assay and conventional ELISA and IMS-PCR workflow (Supplementary Figure S8e–g). Not only each individual biomarker but also the combined biomarkers measured by Exo-PROS assay showed superior sensitivity, specificity and AUC to ELISA and IMS-PCR workflow in distinguishing normal controls from lung cancer patients. These results demonstrated that Exo-PROS assay is more sensitive, specific, and accurate than conventional ELISA and IMS-PCR methods in lung cancer diagnosis.
Evaluation of Diagnostic Performance of Exo-PROS Assay in Breast Cancer Diagnosis.
Breast cancer was used as the second cancer model to evaluate the clinical utility of Exo-PROS assay in cancer diagnosis. Total 20 normal controls and 60 treatment naïve patients with stage 0–IV breast cancer were recruited in this study. The patient characteristics were provided in Supplementary Table S2. Patients with HR +/HER2−, HR−/HR2+, and triple negative breast cancer were included. All patients and controls were female because breast cancer in male is rare. Patients in both groups were matched in age and race. To demonstrate that Exo-PROS assay can directly capture exosomes from serum with no need for exosome isolation, 10 μL serum were diluted in 40 μL PBS and immediately applied on the biochip without exosome isolation step. The expression of TEX ANXA8-miR-342 pair was measured by the Exo-PROS assay. Figure 6a shows the representative SPR curve for the detection of exosomal ANXA8 in the serum from a stage I breast cancer patient. SEM was used to confirm the capture of exosomes expressing ANXA8 (i.e., ANXA8+ exosomes) on the biochip surface (Figure 6b). Significantly higher levels of ANXA8 and lower levels of miR-342 were detected in sera from breast cancer patients than those from normal controls (Figure 6c). The expression of exosomal ANXA8 in sera from breast cancer patients was 1.69-fold higher than that of normal controls. After normalization by the expression of ANXA8 to further increase the difference between cancer patients and normal controls, the levels of ANXA8+ exosomal miR-342 in sera from breast cancer patients was 5.00-fold lower than that of normal controls. Power analysis indicated that we had enough samples to achieve 80% power to detect a fold change of 2 when the fold change under the null hypothesis was 1 and the significance level was 0.05. Results from ROC analysis showed that exosomal ANXA8 had sensitivity of 0.67, specificity of 0.85 and AUC of 0.83 in distinguishing breast cancer from normal controls. ANXA8+ exosomal miR-342 differentiated breast cancer from normal controls with sensitivity of 0.95, specificity of 0.90 and AUC of 0.96. When these two biomarkers were combined, the sensitivity, specificity and AUC were 0.98, 0.90, and 0.96 respectively (Figure 6d and 6e). All results collectively demonstrated that Exo-PROS assay can directly capture exosomes from serum samples without requiring exosome isolation and is an effective assay for breast cancer diagnosis.
Figure 6.
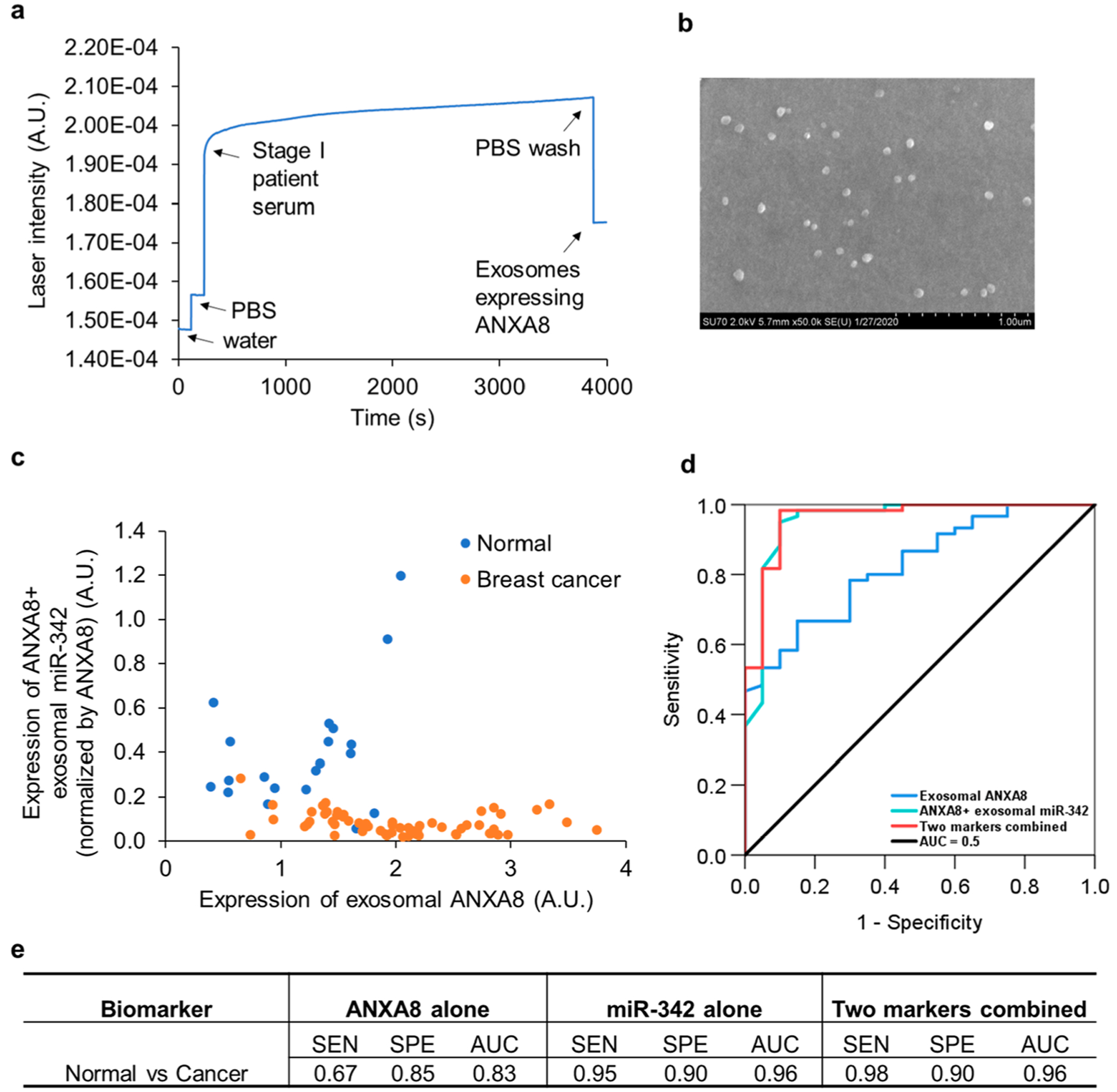
Evaluation of the diagnostic performance of Exo-PROS assay in breast cancer. 10 μL sera from normal controls (n = 20) and breast cancer patients (n = 60, stage 0–IV) were diluted in 40 μL PBS and directly applied on the biochip. The levels of TEX ANXA8-miR-342 pair were measured by Exo-PROS assay. (a) A representative SPR curve for the detection of exosomal ANXA8 in the serum of a stage I breast cancer patient. (b) SEM confirmed the capture of ANXA8+ exosomes on the biochip. (c) The Exo-PROS assay detected significantly higher levels of exosomal ANXA8 and lower levels of ANXA8+ exosomal miR-342 in sera from breast cancer patients than normal controls (p < 10−5). (e) ROC analysis was performed to determine the sensitivity (SEN), specificity (SPE), and AUC of each biomarker and two markers combined. (e) Exo-PROS assay successfully distinguished breast cancer patients from normal controls. When two biomarkers were combined, the Exo-PROS assay detected breast cancer with a sensitivity of 0.98, specificity of 0.90, and AUC of 0.96.
Current cancer screening and diagnostic tests, such as low dose computed tomography (CT) for lung cancer and mammography for breast cancer, are challenged by high false positive/negative rates, limited tumor information, uncomfortable procedures, and risk of radiation exposure.44–46 Tissue biopsy is the gold standard for cancer diagnosis; however, it is an invasive procedure and repeated sampling is quite challenging. Liquid biopsy detects circulating cancer biomarkers, complements medical imaging and risk factor data, and allows sequential monitoring of cancer development, and thus it has shown great promise as in vitro diagnostics for cancer.47,48
TEXs play important roles in every step of cancer development, immune regulation, metastasis and drug resistance, therefore, TEXs are promising biomarkers for cancer liquid biopsy.1–4 Among various cargos, exosomal proteins and microRNAs have shown high sensitivity and specificity in diagnosing many cancers. Conventional technologies such as ELISA and qRT-PCR are widely used to characterize exosomal proteins and RNAs. However, these methods are limited by low sensitivity, tedious processes, and high sample consumption. To overcome these limitations, many biosensors have been developed for exosomal protein analysis, such as SPR sensors,39,49 nPLEX sensor,50 Exo-Screen,51 EV array,52,53 iMEX sensor,54 uNMR sensor,55 hMFEX sensor,56 SERS sensors,57,58 FET sensors,59,60 and nanopatterned microfluidic chips.61,62 For exosomal microRNA analysis, biosensors such as tethered cationic nanoparticles (tCLN)-based sensors,41,42,63 SPR sensors,64,65 electrochemical biosensors,66–68 SERS sensors,69,70 and micro-fluidic devices71–73 have been developed. Even though these emerging techniques have greatly improved detection performances over conventional methods, they were only able to detect one type of exosomal biomarkers, i.e., either exosomal proteins or exosomal microRNAs. None of them established in situ and integrated sensing of two types of TEX biomarkers. Recently, Zhou et al. further developed the tCLN based assay reported by our group42 to detect exosomal microRNAs, mRNAs, and proteins simultaneously.74 However, this advanced tCLN assay lacked the capability of capturing TEXs, instead, it only measured the levels of RNAs and proteins from all exosomes. To date, three studies demonstrated that the use of protein and microRNA combined biomarkers greatly improved diagnostic accuracy in cervical cancer,9 prostate cancer,10 and pancreatic cancer.11 Among them, two studies used conventional methods, such as ELISA, beads-based immunoassay and qRT-PCR to characterize proteins and microRNAs.9,11 Cho et al. developed a magnetic beads-based assay, in which magnetic beads were first used to capture TEXs from prostate cancer cell-derived exosomes, and fluorescent dye-conjugated antibodies and molecular beacons were then used to detect TEX protein and microRNA combined biomarkers.10 However, Cho et al. only tested this assay using cell-derived exosomes and did not validate its diagnostic performances using blood samples from cancer patients. In blood samples, interferences from exosomes released by normal cells represent a major challenge in TEX marker detection and impair the diagnostic accuracy. We showed in this study that ELISA and magnetic beads-based assay are less sensitive in cancer diagnosis, especially when detecting TEX protein and microRNA biomarkers in serum samples from patients (Figure 4 and Supplementary Figure S8). Therefore, the clinical translational potential of the magnetic beads-based assay developed by Cho et al. is uncertain.
We developed the Exo-PROS biosensor to enable sensitive detection of TEX protein–microRNA combined biomarkers and thus further improve cancer diagnosis accuracy. Exo-PROS biosensor effectively separates TEXs from other exosomes using cancer overexpressed markers. It then utilizes SPR sensing mechanism to allow one-stop, in situ characterization of TEX protein and microRNA biomarkers in a sensitive manner (Figure 1). The removal of interferences from nontumor-derived exosomes and the use of TEX protein and microRNA combined biomarkers offer superior sensitivity and specificity in cancer diagnosis. Besides, the Exo-PROS assay has high analytical sensitivity with the LOD as low as 3.5 × 109 exosomes/mL, suggesting that this assay may require as low as 1 μL serum (Figure 3). We note that although A549 cell-derived exosomes were used in LOD determination, cell-derived exosomes are also heterogeneous and carry different biomarkers, which resemble the exosomes in blood.75 We recognize that the LOD of Exo-PROS assay is not the lowest among all emerging biosensors, however, the major advantage of Exo-PROS assay is detecting two different types of TEX biomarkers simultaneously to significantly improve diagnostic accuracy, and the use of 10 μL serum per test is considered quite feasible in clinical settings. Exo-PROS assay also showed great analytical specificity and repeatability with intra- and interday CV less than 10% (Figure 3 and Supplementary Figure S6). Compared with ELISA and IMS-PCR workflow, Exo-PROS assay is ~14-fold more sensitive (3.5 × 109 vs 5 × 1010 exosomes/mL), ~10-fold faster (4 h vs 2 days), and consumes ~20-fold less sample volume (10 vs 200 μL). The Exo-PROS assay has a user-friendly design. Only a micro-pipette is needed to perform this assay without requiring intensive training. The Exo-PROS biosensor is a compact system and has small footprint. These features allow the quick adaptation of Exo-PROS assay to clinical settings.
The diagnostic performance of Exo-PROS biosensor was demonstrated using both lung cancer and breast cancer as disease models. For lung cancer diagnosis, TEX EGFR-miR-21 and LG3BP-miR-210 pairs were selected as the biomarkers because they are actively involved in lung cancer development and have been widely reported as diagnostic and prognostic markers in lung cancer.12–28 Exo-PROS assay detected significantly higher levels of EGFR-miR-21 and LG3BP-miR-210 in A549 cell-derived exosomes than normal BEAS-2B cell-derived exosomes (Figure 4a and 4b). Similarly, the levels of these four biomarkers were significantly higher in exosomes isolated from sera of lung cancer patients (n = 60, stage I–IV) compared with normal controls (n = 35 including patients at both low risk and high risk of lung cancer) (Figure 5). ROC analysis showed that each individual biomarker detected lung cancer with high sensitivity and specificity. When TEX EGFR-miR-21 pair and LG3BP+ exosomal miR-210 were used as combined biomarkers, the Exo-PROS assay achieved diagnostic sensitivity of 0.95, specificity of 0.97, and AUC of 0.99 in distinguishing lung cancer patients from normal controls. Among 60 lung cancer patients, two major lung cancer subtypes, i.e., NSCLC (n = 52) and SCLC (n = 8) were included, and 30 out of 60 patients were patients with early stage (stage I/II) tumors. Among 35 normal controls, 20 patients were smokers at high risk of lung cancer with pulmonary diseases. The Exo-PROS assay was able to distinguish both lung cancer subtypes from normal controls especially smokers at high risk of lung cancer, and detect small, early stage tumors, demonstrating that it is a potent liquid biopsy test for lung cancer screening and early detection.
For breast cancer diagnosis, TEX ANXA8-miR-342 pair was used as the biomarkers. ANXA8 is up-regulated in breast cancer and plays a key role in inducing ductal carcinoma in situ (DCIS), suggesting that it is a promising biomarker for early detection of breast cancer.29,30 However, miR-342 is down-regulated in breast cancer and directly regulates the expression of ANXA8.31–35 This inverse relationship between ANXA8 and miR-342 further increases the difference between breast cancer patients and normal controls, and thus may help improve diagnostic accuracy. As expected, Exo-PROS assay detected higher levels of ANXA8 but lower levels of miR-342 in exosomes derived from MDA-MB-231 than those from MCF-10A cells (Figure 4c and 4d). Exo-PROS assay can directly capture exosomes from serum without requiring exosome isolation. With 10 μL serum, Exo-PROS assay detected significantly higher expression of ANXA8 and lower expression of ANXA8+ exosomal miR-342 in sera from breast cancer patients (n = 60, stage 0–IV) than those from normal controls (n = 20) (Figure 6). The ANXA8 and miR-342 combined biomarker differentiated breast cancer patients from normal controls with sensitivity of 0.98, specificity of 0.90 and AUC of 0.96. For the 60 breast cancer patients, all molecular subtypes (i.e., HR+/HER2−, HR−/HR2+, and triple negative) were included. Among them, 15 were stage 0 patients and 15 were early stage (stage I/II) patients. The Exo-PROS assay successfully detected breast cancer with various molecular subtypes, especially those at early stage, suggesting that it is an effective assay for breast cancer diagnosis.
We note that based on literature review, EGFR-miR-21 and LG3BP-miR-210 pairs were selected as the biomarkers for lung cancer diagnosis and ANXA8-miR-342 pair was used for breast cancer diagnosis. However, these biomarkers are not cancer specific biomarkers. For example, EGFR, miR-21, LG3BP, and miR-210 were also reported to be dysregulated in breast cancer and may serve as biomarkers for breast cancer diagnosis.76–82 We measured the levels of TEX EGFR-miR-21 and LG3BPmiR-210 pairs in sera from breast cancer patients. As shown in Supplementary Figure S9, significantly higher levels of TEX EGFR-miR-21 and LG3BP-miR-210 pairs were observed in sera from breast cancer patients (n = 10, stage 0–IV) than female normal controls (n = 16) (p < 0.05). When all four biomarkers were used as the combined biomarkers, the Exo-PROS assay distinguished breast cancer patients from female normal controls with sensitivity of 1.00, specificity of 1.00 and AUC of 1.00. We recognize that the sample size of breast cancer patients was small, therefore, future studies with large cohort of breast cancer patients and normal controls are needed to further validate these biomarkers in breast cancer diagnosis. Because many biomarkers are cancer associated biomarkers instead of cancer specific biomarkers, in real world cancer screening and diagnosis, Exo-PROS assay can be used together with medical imaging, such as low dose CT for lung cancer and mammography for breast cancer, to further improve diagnostic accuracy.
The diagnostic performance of Exo-PROS assay was also compared with conventional methods. In lung cancer diagnosis, ELISA detected significantly higher levels of EGFR and LG3BP in exosomes derived from A549 NSCLC cells than BEAS-2B normal cells (Figure 4e). However, ELISA detected less difference than Exo-PROS assay in the expression of EGFR and LG3BP between A549 cell-derived exosomes and BEAS-2B cell-derived exosomes. No significant difference was observed in the expression of EGFR+ exosomal miR-21 and LG3BP+ exosomal miR-210 measured by IMS-PCR between A549 cell-derived exosomes and BEAS-2B cell-derived exosomes (Figure 4f). With serum samples from NSCLC patients (n = 40) and normal controls (n = 20), TEX EGFR-miR-21 pair and LG3BP+ exosomal miR-210 measured by ELISA and IMS-PCR workflow showed poor performances in detecting lung cancer with AUCs ranging from 0.55 to 0.61 (Supplementary Figure S8). Only TEX LG3BP showed relatively good diagnostic accuracy with AUC of 0.83, which is still lower than that of Exo-PROS assay (AUC = 0.98). These results demonstrated that Exo-PROS assay has better diagnostic performance than ELISA and IMS-PCR workflow. The reason may be because the sensing target (i.e., exosomes) has a diameter of ~100 nm, which is relatively large and easier to be detected by SPR mechanism than the colorimetric detection mechanism in ELISA. For IMS-PCR workflow, multiple steps are involved so material loss is a big challenge. We used 100 μL serum in IMS-PCR workflow, which is 10-fold higher than that of Exo-PROS assay. However, the Cq values were typically high (~35) and close to the detection limit of qRT-PCR, which may make IMS-PCR workflow less sensitive than Exo-PROS assay.
CONCLUSION
In summary, we developed an Exo-PROS biosensor to selectively capture TEXs from all other exosomes and utilize a SPR sensing mechanism to perform in situ, one-stop quantification of TEX protein and microRNA combined biomarkers for cancer diagnosis. The Exo-PROS biosensor is a sensitive (low LOD and 3-log linear range), fast (~4 h), low sample consumption (as low as 1 μL), reliable (<10% CV), and user-friendly (only requires a pipet) test. With a total of 175 patients and controls, we demonstrated that the Exo-PROS assay detects both lung cancer and breast cancer with AUC ≥ 0.96. The Exo-PROS assay also outperformed conventional methods, such as ELISA and IMS-PCR workflows. In the future, we aim to further improve the sensing performance of Exo-PROS assay to realize multiplex and high throughput sensing. Different combinations of TEX protein–microRNA pairs, such as the LG3BP-miR-21 pair, will be evaluated to investigate which combinations may offer further improved diagnostic accuracy. The clinical utility of the Exo-PROS assay will be validated using a larger cohort of patients. Exo-PROS assay is a universal sensing platform, antibodies and molecular beacons can be easily changed to detect various TEX proteins and microRNAs, therefore, we will also explore the applications of Exo-PROS assay in the diagnosis of other cancers and in other aspects of cancer care, such as treatment response monitoring and prognosis.
MATERIALS AND METHODS
Materials.
SYLGARD 184 silicone elastomer kit (PDMS, 2065622) was purchased from Dow Corning Corp (Midland, MI, United States). Methyl-(polyethylene glycol)4-thiol (PEG200, MW 224 Da, 26132) and NeutrAvidin protein (31000) were purchased from Thermo Fisher Scientific (Waltham, MA, United States). Thiol-PEG1000-biotin (biotin-PEG1000, MW 1000 Da, PG2-BNTH-1k) was purchased from Nanocs (Boston, MA, United States). Biotinylated anti-EGFR antibodies (ab24293), biotinylated mouse IgG2b (kappa monoclonal Isotype Control) antibodies (ab18418) and human EGFR ELISA kit (ab100505) were purchased from Abcam (Cambridge, MA, United States). Biotinylated anti-LG3BP antibodies (LS-C686310–1), human LG3BP ELISA kit (LS-F39884–1), and anti-ANXA8 antibodies (LS-C403822–200) were purchased from LSBio (Seattle, WA, United States). The MBs were synthesized by Alpha DNA (Quebec, Canada). The sequence of miR-21-sensing MB is 5′-Cy5-CGCGATCTCAACATCAGTCTGATAAGCTAG-ATCGCG-BHQ3–3′. The sequence of miR-210-sensing MB is 5′-Cy5-CGCGATCTCAGCCGCTGTCACACGCACAGGATCGCG-BHQ3–3′. The sequence of miR-342-sening MB is 5′-Cy5-CGCGAT-CACGGGTGCGATTTCTGTGTGAGAGATCGCG-BHQ3–3′. The sequence of cel-miR-39-sensing MB is 5′-Cy5-CGCGATCCAAGC-TGATTTACACCCGGTGAGATCGCG-BHQ3–3′. RPMI 1640 medium (11875093), DMEM medium (11995073), DMEM/F-12 medium (11330032), fetal bovine serum (FBS, 26140–079), penicillin–streptomycin (PS, 15140–122), total exosome isolation (from cell culture medium) kit (4478359), total exosome isolation (from serum) kit (4478360), exosome–streptavidin isolation/detection reagent (Dynabeads magnetic beads, 10608D), and EZ-Link Micro Sulfo-NHS-LC-Biotinylation kit (21935) were purchased from Thermo Fisher Scientific. Exo-Check Exosome Antibody Arrays (EXORAY200A-4) was purchased from System Biosciences (Palo Alto, CA, United States). WesternBright Sirius HRP substrate (K-12043-C20) was purchased from Advansta (San Jose, CA, United States). Total RNA Purification Kit (37500) was purchased from Norgen Biotek (Thorold, Canada). MiRCURY LNA RT Kit (339340), miRCURY LNA SYBR Green PCR Kit (339346), miR-21–5p miRCURY LNA miRNA PCR Assay (YP00204230), miR-210–3p miRCURY LNA miRNA PCR Assay (YP00204333), and miR-191–5p miRCURY LNA miRNA PCR Assay (YP00204306) were purchased from Qiagen (Germantown, MD, United States).
Setup of Exo-PROS Biosensor.
As shown in Figure 1, the Exo-PROS biosensor was consisted with a 647 nm polarized laser, a motorized rotor, a prism, a biochip, a photodetector, and a home-built fluorescence photodetector consisting of a lens/filter tube and an APD. The prism was held in the center of the motorized rotor, on top of which sit the biochip. The laser was shed through the prism, and the reflected light intensity was recorded by the photodetector on the other side to characterize the expression of exosomal proteins. The home-built fluorescence photodetector was placed on top of the biochip to detect the restored fluorescence signals from MBs to quantify the expression of exosomal microRNAs.
Fabrication of Exo-PROS Biochip.
Sapphire glass slides (25.4 mm (L) × 25.4 mm (W) × 0.5 mm (H); Suzhou Crystal Silicon Electronic & Technology, China) were cleaned with acetone, methanol and deionized water (DI water) with 10 min sonication for each washing step. Then a 2 nm Ti adhesion layer and a 49 nm Au film was deposited on the sensing area of the sapphire glass slides via electron-beam evaporation (Kurt J. Lesker Company, USA). The deposition rate was controlled at 1 Å/s. Finally, A PDMS layer with a 5 mm hole was bound on the glass slide to serve as the sample reservoir.
Surface Modification of Exo-PROS Biochip.
The surface of the biochip was first coated with PEG by incubating with 100 μL PEG mixture (PEG200:biotin-PEG1000 = 3:1 molar ratio, 10 mM) for 1 h at RT. After washing off excess PEG with PBS, 100 μL 50 μg/mL NeutrAvidin was added and incubated for 1 h at RT. After washing off excess NeutrAvidin with PBS, 50 μL 50 μg/mL biotinylated anti-EGFR antibodies, anti-LG3BP antibodies, anti-ANXA8 antibodies or anti-IgG control antibodies were added and incubated at 4 °C overnight to conjugate antibodies on the surface of the biochip. After washing off excess antibodies with PBS, the biochip was ready for use.
Cell Culture.
A549 human NSCLC cells, BEAS-2B human normal bronchial epithelial cells, MDA-MB-231 breast cancer cells and MCF-10A normal breast epithelial cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, United States). A549 and BEAS-2B cells were cultured in RPMI 1640 medium containing 10% v/v FBS and 1× PS. A549 and BEAS-2B cells were seeded at 1.0 × 106 and 1.5 × 106 cells/P100 dish, respectively, and subcultured every 2–3 days. MDA-MB-231 cells were cultured in DMEM medium containing 10% v/v FBS and 1× PS. MDA-MB231 cells were seeded at 1.5 × 106 cells/P100 dish and subcultured every 2–3 days. MCF-10A cells were cultured in DMEM/F-12 medium containing 10% v/v FBS and 1× PS. MCF-10A cells were seeded at 2 × 106 cells/P100 dish and subcultured every 2–3 days.
Isolation of Exosomes from Cell Culture-Conditioned Media.
To obtain cell-derived exosomes, A549, BEAS-2B, MDA-MB-231, or MCF-10A cells were cultured in 10 mL cell culture medium for 2 days to allow the cells to reach ~80% confluence. Then the cell culture medium was removed, and the cells were washed with PBS twice to fully remove the cell culture medium. The cells were then cultured in 11 mL starving cell culture medium containing no FBS for 2 days. The cell-culture conditioned medium was collected, centrifuged at 4000g for 30 min at 4 °C to remove cells, and centrifuged at 10,000g for 1 h at 4 °C to remove cell debris. The supernatant was collected to isolate exosomes using total exosome isolation (from cell culture medium) kit. Following the manufacturer’s protocol, the total exosome isolation kit was thoroughly mixed with cell culture-conditioned medium at a volume ratio of 1:2 and incubated at 4 °C overnight. The mixture was then centrifuged at 10,000g for 1 h at 4 °C. The precipitated exosomes were resuspended in PBS for further analysis. To obtain the blank control, the above procedure was repeated with no cells seeded in the Petri dishes.
Human Serum Samples.
Deidentified serum samples and clinical data from normal controls, lung cancer patients, and breast cancer patients were obtained from the Data Bank and BioRepository Shared Resource (DBBR) at Roswell Park Comprehensive Cancer Center. The use of human serum samples in this study has been approved by Institutional Review Boards (IRBs) of both Roswell Park Comprehensive Cancer Center and University at Buffalo.
Isolation of Exosomes from Human Sera.
Exosomes were isolated from human serum samples by the total exosome isolation kit (from serum). Following the manufacturer’s protocol, the total exosome isolation kit was thoroughly mixed with human serum at volume ratio of 1:5, and incubated at 4 °C for 30 min. After incubation, the mixture was centrifuged at 10,000g for 10 min at RT to precipitate exosomes. The exosome pellet was resuspended in the same volume of PBS as the volume of the input serum.
Characterization of Exosomes.
The nanoparticle tracking analysis (NTA) system (NanoSight, LM10, Malvern Instruments Ltd.) was used to determine the size, size distribution and concentrations of exosomes. Exosomes isolated from cell cultureconditioned media and human sera were first diluted in PBS until 50–100 nanoparticles could be tracked in the field of view of NTA system. The equipment parameters of NTA system were identical for all measurements, where the detection threshold, the screen gain and the camera level were set at 6, 8, and 14, respectively.
The morphology of exosomes was characterized by Cryo-Transmission Electron Microcopy (Cryo-TEM). Briefly, 5 μL exosomes were first added on glow discharged lacey carbon coated copper grids (400 mesh, Pacific Grid-Tech, San Francisco, CA, United States), and then snap-frozen in liquid ethane using the automated vitrification device (FEI Vitrobot Mark IV, FEI, Hillsboro, OR, United States). The vitrified exosomes were moved to a Gatan Cryo holder (Model 626.DH) and characterized by an FEI Tecnai G2 F20 ST TEM (FEI, Hillsboro, OR, United States).
The quality of exosomes was examined using Exo-Check Exosome Antibody Arrays following the manufacturer’s protocol and Western blotting. Exosome markers including CD63, CD81, ALIX, FLOT1, ICAM1, EpCam, ANXA5, and TSG101 were measured to confirm the presence of exosomes. The GM130, GRP94, and β-tubulin were used to confirm no cellular contamination in exosome samples.
The exosomes captured on the biochip were characterized by the field emission Scanning Electron Microscope (SEM, SU-70, Hitachi High Technologies) with an Energy-Dispersive X-ray Spectrometer (EDS, Oxford Instruments, UK). Briefly, after the capture of exosomes via antibodies, the biochip was washed with PBS 3 times to remove unbound exosomes. Then the PBS was thoroughly aspirated from the biochip. The biochip was air-dried at 4 °C and carbon-coated. The surface-bound exosomes were observed by SEM.
Characterization of TEX Protein–MicroRNA Pair by Exo-PROS Assay.
To measure TEX proteins by the Exo-PROS biosensor, 50 μL DI water was applied on the biochip. The SPR mode was then initiated, and the reflected light intensity of water (Iwater) was measured. Then the DI water was replaced with 50 μL PBS, and the reflected light intensity of PBS (IPBS) was measured. After removing the PBS, 50 μL exosomes or serum samples (10 μL serum and 40 μL PBS mixture) were applied and incubated for 1 h at RT to allow the capture of TEXs via antibodies. After unbound exosomes were removed by 3 times of PBS washing, the reflected light intensity of TEXs was collected (ITEX). The expression of TEX proteins was calculated using eq 1.
Following the quantification of TEX proteins, MBs were added to detect TEX microRNAs. Briefly, the baseline fluorescence intensity before MBs addition was first measured (Ibefore MB). Then 100 μL 3 μM MBs were added on the biochip. After the biochip was incubated at RT for 1.5 h, excess MBs were removed by 3 times of PBS washing. 50 μL PBS was added and the fluorescence intensity was measured (Iafter MB). The expression of TEX microRNAs was calculated using eq 2.
Characterization of TEX Proteins by ELISA.
The expression of TEX EGFR and LG3BP was measured by ELISA following the manufacturers’ protocols. Briefly, exosomes isolated from cell culture-conditioned media or serum samples were diluted and applied on the ELISA plates. For EGFR ELISA assay, the plate was incubated at 4 °C overnight. For LG3BP ELISA assay, the plate was incubated at 37 °C for 90 min. After thoroughly washing off unbound exosomes, biotinylated detection antibodies were added. For EGFR, the plate was incubated at RT for 1 h. For LG3BP, the plates were incubated at 37 °C for 1 h. After thoroughly washing off excessive detection antibodies, horseradish peroxidase (HRP)–streptavidin (i.e., Avidin–Biotin HRP complex) was added. For EGFR, the plate was incubated at RT for 45 min. For LG3BP, the plate was incubated at 37 °C for 30 min. After thoroughly washing off excessive HRP–streptavidin, 3,3′,5,5′-tetramethylbenzidine (TMB) substrates were added and incubated for 30 min in dark. Stop solution was finally added and the absorbance at 450 nm of each well was immediately measured by a microplate reader (Sunrise, Tecan, Morrisville NC).
Characterization of Exosomal MicroRNAs by IMS-PCR Workflow.
Exosomes were first isolated from cell culture-conditioned media or 100 μL serum samples as described above. Exosomes expressing EGFR and LG3BP (i.e., EGFR+ exosomes and LG3BP+ exosomes) were then isolated from total exosomes by exosome–streptavidin isolation/detection reagent (Dynabeads magnetic beads) following the manufacturer’s protocol. Briefly, biotinylated anti-EGFR antibodies or biotinylated anti-LG3BP antibodies were conjugated on the Dynabeads magnetic beads by incubating 4 μg of biotinylated antibodies with 1 mL bead solution containing 107 Dynabeads magnetic beads for 1 h at RT. Then 2 × 106 antibody-conjugated magnetic beads were mixed with 100 μL exosomes at 4 °C overnight on the shaker with gentle mixing. After incubation, the bead-bound EGFR+ exosomes or LG3BP+ exosomes were separated from the unbound exosomes on the magnetic rack and resuspended in 100 μL PBS. Total RNA from EGFR+ exosomes or LG3BP+ exosomes was extracted using total RNA purification kit following the manufacturer’s protocol. The extracted RNA was eluted in 45 μL water. Then 6.3 μL extracted RNAs were reverse transcribed into cDNAs by miRCURY LNA RT Kit. The cDNA is diluted 20 folds with water and 6 μL diluted cDNA is used per 15 μL PCR reaction using miRCURY LNA SYBR Green PCR kit. The expression of miR-21 and miR-210 was measured using miR-21–5p miRCURY LNA miRNA PCR Assay and miR-210–3p miRCURY LNA miRNA PCR Assay. miR-191–5p was used as the endogenous control83–85 and its expression was measured using miR-191–5p miRCURY LNA miRNA PCR Assay.
Statistical Analysis.
PASS software was used to perform power analysis. Binary logistic regression and ROC analysis were performed on the comparisons between cancer groups and normal controls using biomarkers individually or jointly. The pROC package in the R software was used.86 Cut-offs with the highest sum of sensitivity and specificity were first identified, and then AUC, sensitivity, and specificity corresponding to the selected cutoff were calculated. In addition, for data from lung cancer patients, we did the best subset selection analysis using the bestglm R package43 where the best subset of biomarkers that discriminates normal controls from lung cancer patients were identified using the best subset model selection approach and the Akaike information criterion.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge funding support from National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award numbers R21CA235305 (YW and QG), R33CA191245 (YW), and R01CA272827 (YW). Deidentified human serum samples and clinical data were provided by the Data Bank and BioRepository (DBBR), which is funded by NCI under award number P30CA16056 and is a Roswell Park Cancer Institute Cancer Center Support Grant shared resource. The authors thank the support from National Science Foundation under award number CBET-1337860, which funds the nanoparticle tracking analysis system (Nano-Sight, LM10, Malvern Instruments Ltd.). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH and NSF. The authors thank Dr. Min Gao and the TEM facility at the Advanced Materials and Liquid Crystal Institute at Kent State University for the cryo-TEM characterization of exosomes.
Footnotes
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.2c10970
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.2c10970.
Characteristics of normal controls, lung cancer patients, and breast cancer patients; Characterization of exosomes isolated from cell culture conditioned media and human serum samples; Limit of detection and linear range of Exo-PROS assay in detecting exosomal EGFR-miR-21, LG3BP-miR-210, and ANXA8-miR-342 pairs; Characterization of EGFR+ exosomal miR-21 using IMS-PCR workflow; Specificity of Exo-PROS assay; Size and number concentration of exosomes isolated from serum samples; Diagnostic performance comparison between Exo-PROS assay and ELISA-IMS-PCR workflow; Exo-PROS assay detected TEX EGFR-miR-21 and LG3BP-miR-210 pairs in sera from both lung cancer patients and breast cancer patients. (PDF)
Contributor Information
Chang-Chieh Hsu, Department of Biomedical Engineering, University at Buffalo, The State University of New York, Buffalo, New York 14260, United States.
Yunchen Yang, Department of Biomedical Engineering, University at Buffalo, The State University of New York, Buffalo, New York 14260, United States.
Eric Kannisto, Department of Thoracic Surgery, Roswell Park Comprehensive Cancer Center, Buffalo, New York 14263, United States.
Xie Zeng, Department of Electrical Engineering, University at Buffalo, The State University of New York, Buffalo, New York 14260, United States.
Guan Yu, Department of Biostatistics, University at Buffalo, The State University of New York, Buffalo, New York 14263, United States.
Santosh K. Patnaik, Department of Thoracic Surgery, Roswell Park Comprehensive Cancer Center, Buffalo, New York 14263, United States
Grace K. Dy, Department of Medicine, Roswell Park Comprehensive Cancer Center, Buffalo, New York 14263, United States
Mary E. Reid, Department of Medicine, Roswell Park Comprehensive Cancer Center, Buffalo, New York 14263, United States
Qiaoqiang Gan, Department of Electrical Engineering, University at Buffalo, The State University of New York, Buffalo, New York 14260, United States; Materials Science Engineering, King Abdullah University of Science and Technology, Thuwal 23955-6900, Saudi Arabia.
Yun Wu, Department of Biomedical Engineering, University at Buffalo, The State University of New York, Buffalo, New York 14260, United States.
REFERENCES
- (1).Kalluri R; LeBleu VS The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, No. eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Möller A; Lobb RJ The Evolving Translational Potential of Small Extracellular Vesicles in Cancer. Nat. Rev. Cancer 2020, 20, 697–709. [DOI] [PubMed] [Google Scholar]
- (3).LeBleu VS; Kalluri R Exosomes As a Multicomponent Biomarker Platform in Cancer. Trends Cancer. 2020, 6, 767–774. [DOI] [PubMed] [Google Scholar]
- (4).Zhou B; Xu K; Zheng X; Chen T; Wang J; Song Y; Shao Y; Zheng S Application of Exosomes As Liquid Biopsy in Clinical Diagnosis. Signal Transduct Target Ther. 2020, 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Xiong H; Huang Z; Yang Z; Lin Q; Yang B; Fang X; Liu B; Chen H; Kong J Recent Progress in Detection and Profiling of Cancer Cell-Derived Exosomes. Small. 2021, 17, 2007971. [DOI] [PubMed] [Google Scholar]
- (6).Xu L; Shoaie N; Jahanpeyma F; Zhao J; Azimzadeh M; Al-Jamal KT Optical, Electrochemical and Electrical (Nano) Biosensors for Detection of Exosomes: A Comprehensive Overview. Biosens. Bioelectron 2020, 161, 112222. [DOI] [PubMed] [Google Scholar]
- (7).Cheng N; Du D; Wang X; Liu D; Xu W; Luo Y; Lin Y Recent Advances in Biosensors for Detecting Cancer-Derived Exosomes. Trends Biotechnol. 2019, 37, 1236–1254. [DOI] [PubMed] [Google Scholar]
- (8).Wu Y; Zhang Y; Zhang X; Luo S; Yan X; Qiu Y; Zheng L; Li L Research Advances for Exosomal Mirnas Detection in Biosensing: From the Massive Study to the Individual Study. Biosens Bioelectron. 2021, 177, 112962. [DOI] [PubMed] [Google Scholar]
- (9).Du S; Zhao Y; Lv C; Wei M; Gao Z; Meng X Applying Serum Proteins and Microrna As Novel Biomarkers for Early-Stage Cervical Cancer Detection. Sci. Rep 2020, 10, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cho S; Yang HC; Rhee WJ Simultaneous Multiplexed Detection of Exosomal Micrornas and Surface Proteins for Prostate Cancer Diagnosis. Biosens Bioelectron. 2019, 146, 111749. [DOI] [PubMed] [Google Scholar]
- (11).Yuan W; Tang W; Xie Y; Wang S; Chen Y; Qi J; Qiao Y; Ma J New Combined Microrna and Protein Plasmatic Biomarker Panel for Pancreatic Cancer. Oncotarget. 2016, 7, 80033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bethune G; Bethune D; Ridgway N; Xu Z Epidermal Growth Factor Receptor (EGFR) in Lung Cancer: An Overview and Update. J. Thorac. Dis 2010, 2, 48. [PMC free article] [PubMed] [Google Scholar]
- (13).Capone E; Iacobelli S; Sala G Role of Galectin 3 Binding Protein in Cancer Progression: A Potential Novel Therapeutic Target. J. Transl. Med 2021, 19, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yamashita T; Kamada H; Kanasaki S; Maeda Y; Nagano K; Abe Y; Inoue M; Yoshioka Y; Tsutsumi Y; Katayama S Epidermal Growth Factor Receptor Localized to Exosome Membranes As a Possible Biomarker for Lung Cancer Diagnosis. Pharmazie 2013, 68, 969–973. [PubMed] [Google Scholar]
- (15).Jakobsen KR; Paulsen BS; Bæk R; Varming K; Sorensen BS; Jørgensen MM Exosomal Proteins As Potential Diagnostic Markers in Advanced Non-Small Cell Lung Carcinoma. J. Extracell Vesicles 2015, 4, 26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sandfeld-Paulsen B; Aggerholm-Pedersen N; Baek R; Jakobsen K; Meldgaard P; Folkersen B; Rasmussen T; Varming K; Jørgensen M; Sorensen B Exosomal Proteins As Prognostic Biomarkers in Non-Small Cell Lung Cancer. Mol. Oncol 2016, 10, 1595–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Clark DJ; Fondrie WE; Yang A; Mao L Triple SILAC Quantitative Proteomic Analysis Reveals Differential Abundance of Cell Signaling Proteins Between Normal and Lung Cancer-Derived Exosomes. J. Proteomics 2016, 133, 161–169. [DOI] [PubMed] [Google Scholar]
- (18).Huang S.-h.; Li Y; Zhang J; Rong J; Ye S Epidermal Growth Factor Receptor-Containing Exosomes Induce Tumor-Specific Regulatory T Cells. Cancer Invest. 2013, 31, 330–335. [DOI] [PubMed] [Google Scholar]
- (19).Li X.-j.; Hayward C; Fong P-Y; Dominguez M; Hunsucker SW; Lee LW; McLean M; Law S; Butler H; Schirm M; et al. A Blood-Based Proteomic Classifier for the Molecular Characterization of Pulmonary Nodules. Sci. Transl. Med 2013, 5, 207ra142–207ra142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Vachani A; Pass HI; Rom WN; Midthun DE; Edell ES; Laviolette M; Li X-J; Fong P-Y; Hunsucker SW; Hayward C; et al. Validation of a Multiprotein Plasma Classifier to Identify Benign Lung Nodules. J. Thorac. Oncol 2015, 10, 629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Silvestri GA; Tanner NT; Kearney P; Vachani A; Massion PP; Porter A; Springmeyer SC; Fang KC; Midthun D; Mazzone PJ Assessment of Plasma Proteomics Biomarker’s Ability to Distinguish Benign from Malignant Lung Nodules: Results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) Trial. Chest 2018, 154, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Tanner NT; Springmeyer SC; Porter A; Jett JR; Mazzone P; Vachani A; Silvestri GA Assessment of Integrated Classifier’s Ability to Distinguish Benign from Malignant Lung Nodules: Extended Analyses and 2-Year Follow-Up Results of the PANOPTIC (Pulmonary Nodule Plasma Proteomic Classifier) Trial. Chest. 2021, 159, 1283–1287. [DOI] [PubMed] [Google Scholar]
- (23).Bica-Pop C; Cojocneanu-Petric R; Magdo L; Raduly L; Gulei D; Berindan-Neagoe I Overview Upon mir-21 in Lung Cancer: Focus on NSCLC. Cell. Mol. Life Sci 2018, 75, 3539–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).He R.-q.; Cen W.-l.; Cen J.-m.; Cen W.-n.; Li J.-y.; Li M.-w.; Gan T.-q.; Hu X.-h.; Chen G Clinical Significance of mir-210 and Its Prospective Signaling Pathways in Non-Small Cell Lung Cancer: Evidence From Gene Expression Omnibus and the Cancer Genome Atlas Data Mining With 2763 Samples and Validation Via Real-Time Quantitative PCR. Cell Physiol Biochem. 2018, 46, 925–952. [DOI] [PubMed] [Google Scholar]
- (25).Yang Y; Hu Z; Zhou Y; Zhao G; Lei Y; Li G; Chen S; Chen K; Shen Z; Chen X; et al. The Clinical Use of Circulating Micrornas as Non-Invasive Diagnostic Biomarkers for Lung Cancers. Oncotarget 2017, 8, 90197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pop-Bica C; Pintea S; Magdo L; Cojocneanu R; Gulei D; Ferracin M; Berindan-Neagoe I The Clinical Utility of miR-21 and Let-7 in Non-Small Cell Lung Cancer (NSCLC). A Systematic Review and Meta-Analysis. Front Oncol. 2020, 10, 516850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wu J; Shen Z Exosomal miRNAs as Biomarkers for Diagnostic and Prognostic in Lung Cancer. Cancer Med. 2020, 9, 6909–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Yang H; Wang H; Zhang C; Tong Z The Accuracy of microRNA-210 in Diagnosing Lung Cancer: a Systematic Review and Meta-Analysis. Oncotarget. 2016, 7, 63283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Rossetti S; Bshara W; Reiners JA; Corlazzoli F; Miller A; Sacchi N Harnessing 3D Models of Mammary Epithelial Morphogenesis: an Off the Beaten Path Approach to Identify Candidate Biomarkers of Early Stage Breast Cancer. Cancer letters. 2016, 380, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Stein T; Price KN; Morris JS; Heath VJ; Ferrier RK; Bell AK; Pringle M-A; Villadsen R; Petersen OW; Sauter G; et al. Annexin A8 Is Up-Regulated During Mouse Mammary Gland Involution and Predicts Poor Survival in Breast Cancer. Clin. Cancer Res 2005, 11, 6872–6879. [DOI] [PubMed] [Google Scholar]
- (31).Young J; Kawaguchi T; Yan L; Qi Q; Liu S; Takabe K Tamoxifen Sensitivity-Related microRNA-342 Is a Useful Biomarker for Breast Cancer Survival. Oncotarget. 2017, 8, 99978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Bitaraf A; Babashah S; Garshasbi M Aberrant Expression of a Five-microRNA Signature in Breast Carcinoma As a Promising Biomarker for Diagnosis. J. Clin. Lab. Anal 2020, 34, No. e23063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Romero-Cordoba SL; Rodriguez-Cuevas S; Bautista-Pina V; Maffuz-Aziz A; D’Ippolito E; Cosentino G; Baroni S; Iorio MV; Hidalgo-Miranda A Loss of Function of miR-342–3p Results in MCT1 Over-Expression and Contributes to Oncogenic Metabolic Reprogramming in Triple Negative Breast Cancer. Sci. Rep 2018, 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lindholm EM; Leivonen S-K; Undlien E; Nebdal D; Git A; Caldas C; Børresen-Dale A-L; Kleivi K miR-342–5p As a potential Regulator of HER2 Breast Cancer Cell Growth. MicroRNA 2019, 8, 155–165. [DOI] [PubMed] [Google Scholar]
- (35).Rossetti S; Sacchi N 3D Mammary Epithelial Cell Models: a Goldmine of DCIS Biomarkers and Morphogenetic Mechanisms. Cancers. 2019, 11, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Lee JH; Kim JA; Jeong S; Rhee WJ Simultaneous and Multiplexed Detection of Exosome MicroRNAs Using Molecular Beacons. Biosens Bioelectron. 2016, 86, 202–210. [DOI] [PubMed] [Google Scholar]
- (37).Lee JH; Kim JA; Kwon MH; Kang JY; Rhee WJ In Situ Single Step Detection of Exosome Microrna Using Molecular Beacon. Biomaterials. 2015, 54, 116–125. [DOI] [PubMed] [Google Scholar]
- (38).Shrivastava A; Gupta VB Methods for the Determination of Limit of Detection and Limit of Quantitation of the Analytical Methods. Chron. Young Sci 2011, 2, 21–25. [Google Scholar]
- (39).Liu C; Zeng X; An Z; Yang Y; Eisenbaum M; Gu X; Jornet JM; Dy GK; Reid ME; Gan Q; Wu Y Sensitive Detection of Exosomal Proteins Via a Compact Surface Plasmon Resonance Biosensor for Cancer Diagnosis. ACS Sens. 2018, 3, 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yang Y; Kannisto E; Yu G; Reid ME; Patnaik SK; Wu Y An Immuno-Biochip Selectively Captures Tumor-Derived Exosomes and Detects Exosomal RNAs for Cancer Diagnosis. ACS Appl. Mater. Interfaces 2018, 10, 43375–43386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Liu C; Kannisto E; Yu G; Yang Y; Reid ME; Patnaik SK; Wu Y Non-Invasive Detection of Exosomal MicroRNAs Via Tethered Cationic Lipoplex Nanoparticles (tCLN) Biochip for Lung Cancer Early Detection. Front. Genet 2020, 11, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wu Y; Kwak KJ; Agarwal K; Marras A; Wang C; Mao Y; Huang X; Ma J; Yu B; Lee R; et al. Detection of Extracellular RNAs in Cancer and Viral Infection via Tethered Cationic Lipoplex Nanoparticles Containing Molecular Beacons. Anal. Chem 2013, 85, 11265–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).McLeod AI; Xu C; Lai Y Package BestGLM. cran.rproject.org. https://cran.r-project.org/web/packages/bestglm/bestglm.pdf (accessed on March 13, 2020). [Google Scholar]
- (44).Oudkerk M; Liu S; Heuvelmans MA; Walter JE; Field JK Lung Cancer LDCT Screening and Mortality Reduction—Evidence, Pitfalls and Future Perspectives. Nat. Rev. Clin Oncol 2021, 18, 135–151. [DOI] [PubMed] [Google Scholar]
- (45).Jonas DE; Reuland DS; Reddy SM; Nagle M; Clark SD; Weber RP; Enyioha C; Malo TL; Brenner AT; Armstrong C; et al. Screening for Lung Cancer with Low-Dose Computed Tomography: An Evidence Review for the U.S. Preventive Services Task Force; Agency for Healthcare Research and Quality (US): Rockville, MD, 2021. Report No.: 20–05266-EF-1. [PubMed] [Google Scholar]
- (46).Sprague BL; Arao RF; Miglioretti DL; Henderson LM; Buist DS; Onega T; Rauscher GH; Lee JM; Tosteson AN; Kerlikowske K; Lehman CD Breast Cancer Surveillance Consortium. National Performance Benchmarks for Modern Diagnostic Digital Mammography: Update from the Breast Cancer Surveillance Consortium. Radiology. 2017, 283, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ignatiadis M; Sledge GW; Jeffrey SS Liquid biopsy Enters the Clinic—Implementation Issues and Future Challenges. Nat. Rev. Clin Oncol 2021, 18, 297–312. [DOI] [PubMed] [Google Scholar]
- (48).Siravegna G; Marsoni S; Siena S; Bardelli A Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin Oncol 2017, 14, 531–548. [DOI] [PubMed] [Google Scholar]
- (49).Sina AAI; Vaidyanathan R; Wuethrich A; Carrascosa LG; Trau M Label-Free Detection of Exosomes Using a Surface Plasmon Resonance Biosensor. Anal Bioanal Chem. 2019, 411, 1311–1318. [DOI] [PubMed] [Google Scholar]
- (50).Im H; Shao H; Park YI; Peterson VM; Castro CM; Weissleder R; Lee H Label-Free Detection and Molecular Profiling of Exosomes with a Nano-Plasmonic Sensor. Nat. Biotechnol 2014, 32, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yoshioka Y; Kosaka N; Konishi Y; Ohta H; Okamoto H; Sonoda H; Nonaka R; Yamamoto H; Ishii H; Mori M; et al. Ultra-Sensitive Liquid Biopsy of Circulating Extracellular Vesicles Using Exoscreen. Nat. Commun 2014, 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jorgensen M; Baek R; Pedersen S; Sondergaard EKL; Kristensen SR; Varming K Extracellular Vesicle (EV) Array: Microarray Capturing of Exosomes and Other Extracellular Vesicles for Multiplexed Phenotyping. J. Extracell. Vesicles 2013, 2, 20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Jørgensen MM; Bæk R; Varming K Potentials and Capabilities of the Extracellular Vesicle (EV) Array. J. Extracell Vesicles 2015, 4, 26048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jeong S; Park J; Pathania D; Castro CM; Weissleder R; Lee H Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano 2016, 10, 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Shao H; Chung J; Balaj L; Charest A; Bigner DD; Carter BS; Hochberg FH; Breakefield XO; Weissleder R; Lee H Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med 2012, 18, 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Li B; Pan W; Liu C; Guo J; Shen J; Feng J; Luo T; Situ B; Zhang Y; An T; et al. Homogenous Magneto-Fluorescent Nanosensor for Tumor-Derived Exosome Isolation and Analysis. ACS Sens. 2020, 5, 2052–2060. [DOI] [PubMed] [Google Scholar]
- (57).Zong S; Wang L; Chen C; Lu J; Zhu D; Zhang Y; Wang Z; Cui Y Facile Detection of Tumor-Derived Exosomes Using Magnetic Nanobeads and SERS Nanoprobes. Anal Methods. 2016, 8, 5001–5008. [Google Scholar]
- (58).Fan C; Zhao N; Cui K; Chen G; Chen Y; Wu W; Li Q; Cui Y; Li R; Xiao Z Ultrasensitive Exosome Detection by Modularized SERS Labeling for Postoperative Recurrence Surveillance. ACS Sens. 2021, 6, 3234–3241. [DOI] [PubMed] [Google Scholar]
- (59).Yu Y; Li Y-T; Jin D; Yang F; Wu D; Xiao M-M; Zhang H; Zhang Z-Y; Zhang G-J Electrical and Label-Free Quantification of Exosomes with a Reduced Graphene Oxide Field Effect Transistor Biosensor. Anal. Chem 2019, 91, 10679–10686. [DOI] [PubMed] [Google Scholar]
- (60).Kwong Hong Tsang D; Lieberthal TJ; Watts C; Dunlop IE; Ramadan S; del Rio Hernandez AE; Klein N Chemically Functionalised Graphene FET Biosensor for the Label-Free Sensing of Exosomes. Sci. Rep 2019, 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Zhang P; Zhou X; He M; Shang Y; Tetlow AL; Godwin AK; Zeng Y Ultrasensitive Detection of Circulating Exosomes with a 3D-Nanopatterned Microfluidic Chip. Nat. Biomed Eng 2019, 3, 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhang P; Wu X; Gardashova G; Yang Y; Zhang Y; Xu L; Zeng Y Molecular and Functional Extracellular Vesicle Analysis Using Nanopatterned Microchips Monitors Tumor Progression and Metastasis. Sci. Transl. Med 2020, 12, No. eaaz2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Hu J; Sheng Y; Kwak KJ; Shi J; Yu B; Lee LJ A Signal-Amplifiable Biochip Quantifies Extracellular Vesicle-Associated RNAs for Early Cancer Detection. Nat. Commun 2017, 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Joshi GK; Deitz-McElyea S; Johnson M; Mali S; Korc M; Sardar R Highly Specific Plasmonic Biosensors for Ultrasensitive Microrna Detection in Plasma From Pancreatic Cancer Patients. Nano Lett. 2014, 14, 6955–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Wu W; Yu X; Wu J; Wu T; Fan Y; Chen W; Zhao M; Wu H; Li X; Ding S Surface Plasmon Resonance Imaging-Based Biosensor for Multiplex and Ultrasensitive Detection of NSCLC-Associated Exosomal miRNAs Using DNA Programmed Heterostructure of Au-on-Ag. Biosens Bioelectron. 2021, 175, 112835. [DOI] [PubMed] [Google Scholar]
- (66).Zhang J; Wang L-L; Hou M-F; Xia Y-K; He W-H; Yan A; Weng Y-P; Zeng L-P; Chen J-H A Ratiometric Electro-chemical Biosensor for the Exosomal MicroRNAs Detection Based on Bipedal DNA Walkers Propelled by Locked Nucleic Acid Modified Toehold Mediate Strand Displacement Reaction. Biosens Bioelectron. 2018, 102, 33–40. [DOI] [PubMed] [Google Scholar]
- (67).Boriachek K; Umer M; Islam MN; Gopalan V; Lam AK; Nguyen N-T; Shiddiky MJ An Amplification-Free Electro-chemical Detection of Exosomal miRNA-21 in Serum Samples. Analyst. 2018, 143, 1662–1669. [DOI] [PubMed] [Google Scholar]
- (68).Luo L; Wang L; Zeng L; Wang Y; Weng Y; Liao Y; Chen T; Xia Y; Zhang J; Chen J A Ratiometric Electrochemical DNA Biosensor for Detection of Exosomal MicroRNA. Talanta. 2020, 207, 120298. [DOI] [PubMed] [Google Scholar]
- (69).Lee JU; Kim WH; Lee HS; Park KH; Sim SJ Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-Flocked Gold Nanopillars. Small. 2019, 15, 1804968. [DOI] [PubMed] [Google Scholar]
- (70).Pang Y; Wang C; Lu L; Wang C; Sun Z; Xiao R Dual-SERS Biosensor for One-Step Detection of MicroRNAs in Exosome and Residual Plasma of Blood Samples for Diagnosing Pancreatic Cancer. Biosens Bioelectron. 2019, 130, 204–213. [DOI] [PubMed] [Google Scholar]
- (71).Ramshani Z; Zhang C; Richards K; Chen L; Xu G; Stiles BL; Hill R; Senapati S; Go DB; Chang H-C Extracellular Vesicle MicroRNA Quantification From Plasma Using an Integrated Microfluidic Device. Commun. Biol 2019, 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Cao H; Zhou X; Zeng Y Microfluidic Exponential Rolling Circle Amplification for Sensitive MicroRNA Detection Directly from Biological Samples. Sens Actuators B Chem. 2019, 279, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Yang Y; Kannisto E; Patnaik SK; Reid ME; Li L; Wu Y Ultrafast Detection of Exosomal RNAs Via Cationic Lipoplex Nanoparticles in a Micromixer Biochip for Cancer Diagnosis. ACS Appl. Nano Mater 2021, 4, 2806–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Zhou J; Wu Z; Hu J; Yang D; Chen X; Wang Q; Liu J; Dou M; Peng W; Wu Y; et al. High-Throughput Single-EV Liquid Biopsy: Rapid, Simultaneous, and Multiplexed Detection of Nucleic Acids, Proteins, and Their Combinations. Sci. Adv 2020, 6, No. eabc1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Lee K; Fraser K; Ghaddar B; Yang K; Kim E; Balaj L; Chiocca EA; Breakefield XO; Lee H; Weissleder R Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Gonzalez-Conchas GA; Rodriguez-Romo L; Hernandez-Barajas D; Gonzalez-Guerrero JF; Rodriguez-Fernandez IA; Verdines-Perez A; Templeton AJ; Ocana A; Seruga B; Tannock IF; Amir E; Vera-Badillo FE Epidermal growth factor receptor overexpression and outcomes in early breast cancer: A systematic review and a meta-analysis. Cancer Treat Rev. 2018, 62, 1–8. [DOI] [PubMed] [Google Scholar]
- (77).Ding Y; Wu W; Ma Z; Shao X; Zhang M; Wang Z Potential value of MicroRNA-21 as a biomarker for predicting the prognosis of patients with breast cancer: A protocol for meta-analysis and bioinformatics analysis. Medicine. 2021, 100, No. e25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Liu M; Mo F; Song X; He Y; Yuan Y; Yan J; Yang Y; Huang J; Zhang S Exosomal hsa-miR-21–5p is a biomarker for breast cancer diagnosis. PeerJ. 2021, 9, No. e12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Iacobelli S; Sismondi P; Giai M; D’Egidio M; Tinari N; Amatetti C; Di Stefano P; Natoli C Prognostic value of a novel circulating serum 90K antigen in breast cancer. Br J. Cancer 1994, 69, 172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Tinari N; Lattanzio R; Querzoli P; Natoli C; Grassadonia A; Alberti S; Hubalek M; Reimer D; Nenci I; Bruzzi P; Piantelli M; Iacobelli S Consorzio Interuniversitario Nazionale per la Bio-Oncologia (CINBO). High expression of 90K (Mac-2 BP) is associated with poor survival in node-negative breast cancer patients not receiving adjuvant systemic therapies. Int. J. Cancer 2009, 124, 333–338. [DOI] [PubMed] [Google Scholar]
- (81).Nashtahosseini Z; Aghamaali MR; Sadeghi F; Heydari N; Parsian H Circulating status of microRNAs 660–5p and 210–3p in breast cancer patients. J. Gene Med 2021, 23, No. e3320. [DOI] [PubMed] [Google Scholar]
- (82).Lopes BC; Braga CZ; Ventura FV; de Oliveira JG; Kato-Junior EM; Bordin-Junior NA; Zuccari DA MiR-210 and miR-152 as biomarkers by liquid biopsy in invasive ductal carcinoma. J. Pers Med 2021, 11, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Hu Z; Dong J; Wang L-E; Ma H; Liu J; Zhao Y; Tang J; Chen X; Dai J; Wei Q; et al. Serum MicroRNA Profiling and Breast Cancer Risk: the Use of miR-484/191 As Endogenous Controls. Carcinogenesis 2012, 33, 828–834. [DOI] [PubMed] [Google Scholar]
- (84).Li Y; Zhang L; Liu F; Xiang G; Jiang D; Pu X Identification of Endogenous Controls for Analyzing Serum Exosomal miRNA in Patients with Hepatitis B Or Hepatocellular Carcinoma. Dis. Markers 2015, 2015, 893594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Zheng G; Wang H; Zhang X; Yang Y; Wang L; Du L; Li W; Li J; Qu A; Liu Y; Wang C Identification and Validation of Reference Genes for qPCR Detection of Serum MicroRNAs in Colorectal Adenocarcinoma Patients. PLoS One 2013, 8, No. e83025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Robin X; Turck N; Hainard A; Tiberti N; Lisacek F; Sanchez J-C; Muüller M pROC: an Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinf. 2011, 12, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


