Abstract
Nature has evolved eight different pathways for the capture and conversion of CO2, including the Calvin-Benson-Bassham cycle of photosynthesis. Yet, these pathways underlie constrains and only represent a fraction of the thousands of theoretically possible solutions. To overcome the limitations of natural evolution, we introduce the HydrOxyPropionyl-CoA/Acrylyl-CoA (HOPAC) cycle, a new-to-nature CO2-fixation pathway that was designed through metabolic retrosynthesis around the reductive carboxylation of acrylyl-CoA, a highly efficient principle of CO2 fixation. We realized the HOPAC cycle in a step-wise fashion and used rational engineering approaches and machine learning–guided workflows to further optimize its output by more than one order of magnitude. Version 4.0 of the HOPAC cycle encompasses 11 enzymes from six different organisms, converting ~3.0 mM CO2 into glycolate within 2 hours. Our work moves the hypothetical HOPAC cycle from a theoretical design into an established in vitro system that forms the basis for different potential applications.
Natural CO2-fixation pathways are expanded by a human-made alternative.
INTRODUCTION
Globally, more than 350 Gt of carbon dioxide (CO2) is fixed by autotrophic organisms each year (1). Of this, more than 95% is funneled through a single enzyme, ribulose-1,5-bisphosphate carboxylase-oxygenase (Rubisco), the carboxylating enzyme in the Calvin-Benson-Basham (CBB) cycle (2). In addition to Rubisco and the CBB cycle, seven other autotrophic cycles have been found, which revolve around eight alternative carbon-fixing enzymes (3).
Yet, these eight pathways represent only a small fraction of the possible thousands of autotrophic CO2-fixation pathways that have been available for nature to explore during evolution (4). Notably, there are several CO2-fixing enzymes, which are not known to operate in autotrophic CO2 fixation, but instead in other metabolic processes, such as the assimilation of organic compounds, anaplerosis, or biosynthesis (5). One prime example are enoyl–coenzyme A (CoA) carboxylase/reductases (Ecr, EC 1.3.1.85). These enzymes are the most efficient CO2-fixing enzymes described to date, outcompeting Rubisco by more than one order of magnitude (1, 6) and, unlike Rubisco, do not show any side reaction with oxygen. However, despite their highly favorable kinetic and biochemical properties, Ecrs have thus far only been described in the context of heterotrophic acetate assimilation via the ethylmalonyl-CoA pathway and polyketide biosynthesis but were apparently not recruited for autotrophic CO2 fixation.
The ostensible inability of evolution to systematically assemble the most promising enzymes into new pathways and cycles has inspired several approaches to overcome this roadblock by creating alternative, “synthetic” CO2 fixation pathways from the bottom-up. This effort has mainly been a theoretical exercise so far (1, 6–10) but has demonstrated that multiple pathways can indeed be designed that outcompete natural CO2 fixation in respect to kinetic and thermodynamic considerations.
In efforts to leverage the potential of Ecrs for synthetic CO2 fixation, we (and others) have drafted several new-to-nature CO2-fixation pathways that are based on this newly found CO2-fixation principle (1, 11). Notably, these designs are all superior to the natural CBB cycle. We recently realized one of these designs, the CETCH cycle (for Crotonyl-CoA/EThylmalonyl-CoA/Hydroxybutyryl-CoA cycle), in vitro and optimized it in several steps (1, 12). The CETCH version 5.4 cycle included 17 enzymes from nine different species including three re-engineered enzymes, and two reductive carboxylation reactions working in tandem to convert CO2 into glyoxylate/glycolate in vitro at fixation rates that are comparable to natural CO2 fixation pathways. Further optimization of CETCH using a machine learning–guided approach increased yield by almost an order of magnitude, achieving a system that can fix 5.9 mM CO2 within ~3 hours (from 100 μM starting substrate) in vitro.
Yet, the CETCH cycle is only one of the potential designs that we had originally envisioned (1). In ongoing efforts in our laboratory to realize other solutions, we present here the successful reconstruction of the HydrOxyPropionyl-CoA/Acrylyl-CoA (HOPAC) cycle (Fig. 1A). The overall topology of the HOPAC cycle is similar to the naturally existing 3-hydroxypropionyl-CoA (3HP) cycle (13, 14). However, the HOPAC cycle was designed to be energetically more efficient than the 3HP cycle. To realize the HOPAC cycle, we dissected the cycle into two partial pathways, a reductive and an oxidative part, which allowed for the convenient screening and prototyping of alternative variants of different enzyme chemistries (Fig. 1, B and C, and Table 1 for different combinations). Successful versions of the two parts were combined into a complete HOPAC cycle, which was further optimized through rational and machine learning–guided efforts to reach activities that compare to other in vitro CO2-fixation pathways (1, 11). Notably, the final, optimized version of the HOPAC cycle differs in several key reactions from the 3HP cycle, which makes it a truly new-to-nature CO2 fixation pathway with potential for different in vitro and in vivo applications.
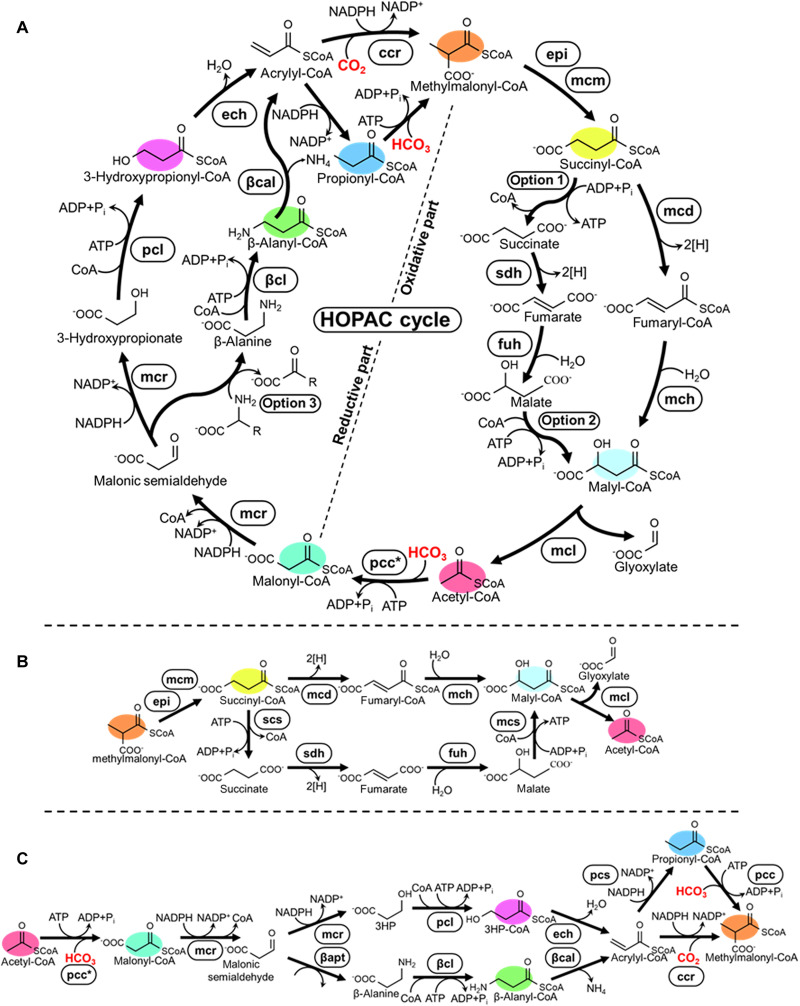
Fig. 1. The HOPAC cycle and its alternative designs explored in this study.
(A) Overall scheme of the HOPAC cycle. The cycle converts two molecules of inorganic carbon (HCO3− and/or CO2) into one molecule of glyoxylate. The HOPAC cycle can be divided into a reductive and an oxidative part, for each of which two different variants were developed in this study. Color coding indicates key metabolites that were analyzed in this study. Enzyme abbreviations are expanded in table S6. Option 1: Scs, Smt, or Sch; option 2: Smt or Mcs; option 3: GabT or βapt; option 4: βcl or βct. (B) Variants of the reductive part of the HOPAC cycle tested in this study with the respective cofactors (C) Variants of the oxidative part of the HOPAC cycle variants tested in this study with the respective cofactors. ATP and NADPH balances for all possible combinations of the reductive and oxidative parts of the HOPAC cycle are given in Table 1.
Table 1. Cofactor requirements for all possible combinations of the different reductive and oxidative part variants.
| HOPAC cycle combination | Cofactor | |||
|---|---|---|---|---|
| Reductive part variant | + | Oxidative part variant | ATP | NADPH |
| β-Alanine route | + | CoA route | 2 | 2* |
| β-Alanine route | + | Free acid route (Smt or Scs/Mcs variant) | 2 | 2* |
| 3HP route (Ccr variant) | + | CoA route | 2 | 3 |
| 3HP route (Ccr variant) | + | Free acid route (Smt or Scs/Mcs variant) | 2 | 3 |
| β-Alanine route | + | Free acid route (Sch variant) | 3 | 2* |
| 3HP route (Ccr variant) | + | Free acid route (Sch variant) | 3 | 3 |
| 3HP route (Pcs variant) | + | CoA route | 3† | 3 |
| 3HP route (Pcs variant) | + | Free acid route (Smt or Scs/Mcs variant)‡ | 3† | 3 |
| 3HP route (Pcs variant) | + | Free acid route (Sch variant) | 4† | 3 |
*These values relate to the number of NADPH consumed by the core chemistry of the cycle, an additional NADPH is required if the amino acid is regenerated.
†These values relate to the number of ATP consumed by the core chemistry of the cycle, an additional ATP is required if the AMP is regenerated to ATP.
‡This version of the HOPAC cycle, using the Pcs variant of the 3HP route and the Smt variant of the free acid route, is the option used in the first cycle of the natural 3-hydroxypropionate bicycle.
RESULTS
Design of the HOPAC cycle
In its initial design, the HOPAC cycle combines two carboxylation reactions to build up one molecule of glyoxlate from CO2. Starting from the C2 acceptor molecule acetyl-CoA, the first carboxylation reaction yields malonyl-CoA, which is converted into 3HP through subsequent reduction steps and further into acrylyl-CoA. In the second CO2-fixation step, acrylyl-CoA is reductively carboxylated into the C4 compound (2S)-methylmalonyl-CoA, which is subsequently converted into succinyl-CoA and further into malyl-CoA. Malyl-CoA is cleaved into glyoxylate, the primary output molecule of the cycle, as well as acetyl-CoA the initial acceptor molecule, thus closing the cycle.
The overall reaction sequence of the HOPAC cycle is similar to the 3HP bicycle of Chloroflexus aurantiacus. However, the use of a reductive carboxylation reaction that directly converts acrylyl-CoA into (2S)-methylmalonyl-CoA renders the cycle more adenosine triphosphate (ATP) efficient compared to the 3HP bicycle, where the conversion of acrylyl-CoA into (2S)-methylmalonyl-CoA is separated into two steps: first reduction into propionyl-CoA, followed by an ATP-dependent carboxylation of the latter into (2S)- methylmalonyl-CoA. Thus, the overall reaction of the HOPAC cycle requires one fewer ATP compared to the 3HP and can be summarized as 2 CO2 + 3 NADPH +2 ATP + FAD ➔ glyoxylate +3 NADP+ + H+ + 2 ADP + 2 Pi + FADH2. To realize the HOPAC cycle in vitro, we decided to separate the overall reaction sequence into two parts: a reductive part, converting acetyl-CoA into (2S)-methylmalonyl-CoA, and an oxidative part that regenerates (2S)-methylmalonyl-CoA into acetyl-CoA (and glyoxylate).
The reductive part: Overall design
To realize the reductive part of the HOPAC (Fig. 1B), we mainly relied on reactions from the 3HP bicycle of C. aurantiacus. However, because many of the enzymes are thermophilic, we replaced these enzymes with mesophilic alternatives, whenever necessary and/or possible.
For the conversion of malonyl-CoA to malonic semialdehyde, we selected malonyl-CoA reductase (Mcr) from C. aurantiacus (WP_012258473) (15). Mcr is a bifunctional enzyme that has a dual activity for the reduction of both malonyl-CoA as well as malonic semialdehyde and sufficiently active at mesophilic temperatures (24 U/mg; app. Km-malonyl-CoA, 1.3 μM; app. Km-NADPH, 10 μM). For the conversion of malonyl-CoA into malonic semialdehyde, we produced a C-terminal version of Mcr (McrC), which has been shown to catalyze the reduction of malonyl-CoA but not malonic semialdehyde (Fig. 2B) (16).
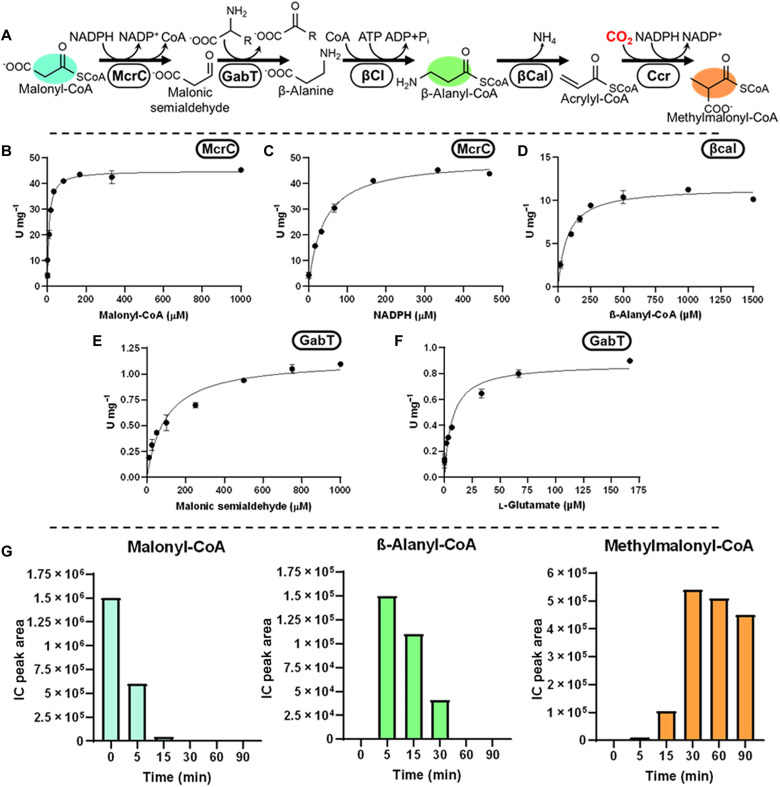
Fig. 2. Construction of the β-alanine route.
(A) Reaction sequence includes reduction of malonyl-CoA into malonic semialdehyde by the C-terminal domain of Mcr (McrC), transamination of malonic semialdehyde to β-alanine by GabT (or βApt), CoA ligation of β-alanine by βCl (or βCt), and lastly deamination of β-alanyl-CoA by βCal. (B and C) Michaelis-Menten kinetics of Chloroflexus aurantiacus McrC for Malonyl-CoA (B) and NADPH (C), respectively (D) Michaelis-Menten kinetics of S. aurantiaca βCal on β-alanyl-CoA. (E and F) Michaelis-Menten kinetics of Escherichia coli GabT for malonic semialdehyde (E) and l-glutamate (F), respectively. (G) Validation of the β-alanine route using GabT and Gdh (assay with βApt and Adh can be found in fig. S18). Values refer to relative concentrations of intermediates as determined by LCMS. Colors correspond to compounds in (A).
For the subsequent multistep conversion of malonic semialdehyde to acrylyl-CoA, we explored two alternative routes. In the first route (“β-alanine route”), malonic semialdehyde is transaminated to β-alanine, which is ligated to CoA, deaminated to acrylyl-CoA, and further carboxylated into (2S)-methylmalonyl-CoA. In the second route (“3HP route”), malonic semialdehyde is directly reduced to 3HP, which is ligated to CoA, subsequently dehydrated to acrylyl-CoA, and further converted into (2S)-methylmalonyl-CoA.
The β-alanine route of the reductive part
For the β-alanine route, we established β-alanine–pyruvate transaminase (βApt, PON06840) from Rhizobium hidalgonense and 4-aminobutyrate-2-oxoglutarate transaminase (GabT, AKK18527) from Escherichia coli (17, 18). We confirmed activity of βApt with l-alanine and GabT with l-glutamate (1.1 U/mg, app. Km-malonic-semialdehyde, 98 μM, app. Km-L-glutamate, 7 μM), giving rise to two different options to produce β-alanine from malonic semialdehyde (Fig. 2 and fig. S18). To maintain the amino acid pool, we tested l-alanine dehydrogenase from Cereibacter (formerly Rhodobacter) (19) sphaeroides (ABA79899) (l-alanine variant) and l-glutamate dehydrogenase from Paracoccus denitrificans (MBB4626159, 47 U/mg; app. Km-2-oxoglutarate, 36 μM; app. Km-NADPH, 11 μM; app. Km-NH4, 86 mM) (l-glutamate variant) that regenerate the respective amino acid from pyruvate and α-ketoglutarate (fig. S10).
To ligate β-alanine to CoA, we screened 10 synthetases with primary activities ranging from acetate to malonate but failed to establish this activity (fig. S13). Instead, we used a previously identified β-alanine CoA-transferase from Anaerotignum propionicum (CAG29276) (fig. S14) (20). Unfortunately, the enzyme showed promiscuous activity with l-alanine in addition to β-alanine, which ruled out the l-alanine dehydrogenase variant. Hence, we focused solely on the l-glutamate variant. For the conversion of β-alanyl-CoA into acrylyl-CoA, we established β-alanyl-CoA:ammonia lyase from Stigmatella aurantiaca (ADO72524; 12 U/mg, Km-β-alanyl-CoA, 70 μM; Fig. 2D) and for the final carboxylation step, Ccr from Methylorubrum (formerly Methylobacterium) (21) extorquens AM1 (ACS38140, 274.4 U/mg; app. Km-acrylyl-CoA, 0.78 mM; app. Km-NADPH, 0.21 mM), which showed 40% carboxylating activity with acrylyl-CoA compared with crotonyl-CoA (22).
The 3HP route of the reductive part
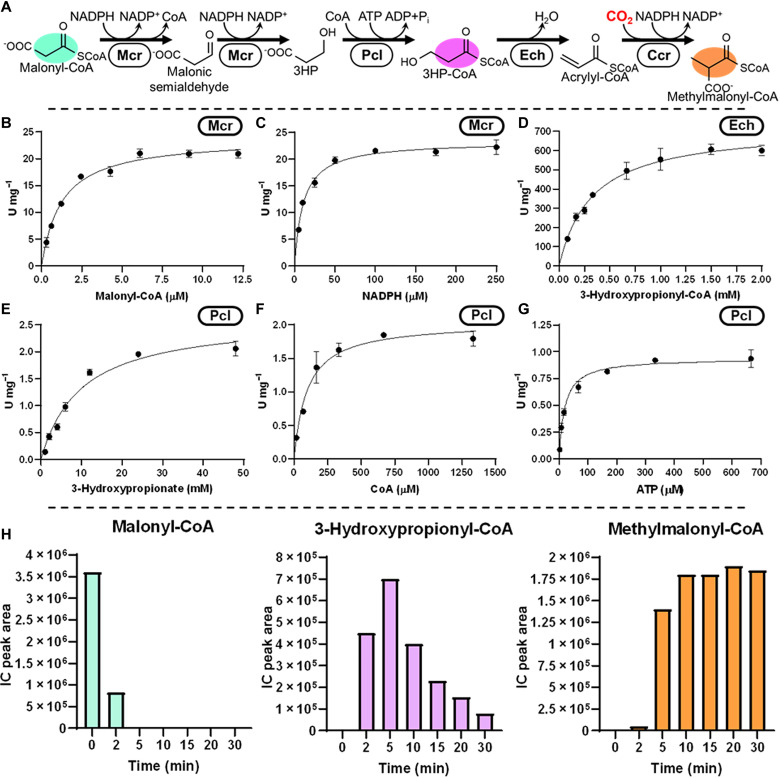
For the 3HP route, we used the full-length construct of Mcr from C. aurantiacus that reduces malonyl-CoA in two subsequent steps into 3HP (Fig. 3, B and C) (15). For the activation of 3HP, we screened 10 synthetases and three transferases and decided for propionyl-CoA ligase from Cupriavidus necator (QCC01349) (Fig. 3, E to G). For the dehydration of 3HP, we chose Ech (CDI91698, 720 U/mg; Km-3HP, 330 μM) from Pseudomonas aeruginosa (Fig. 3D) which was able to produce acrylyl-CoA at favorable kinetics for the subsequent carboxylation by Ccr (23).
Fig. 3. Construction of the 3HP route.
(A) Reaction sequence of the 3HP route includes reduction of malonyl-CoA to malonic semialdehyde and 3-hydroxypropionate by Mcr, CoA ligation of 3-hydroxypropionate by Pcl (or Pct), and dehydration of 3HP by Ech. (B and C) Michaelis-Menten kinetics of Chloroflexus aurantiacus Mcr for Malonyl-CoA (B) and NADPH (C), respectively. (D) Michaelis–Menten kinetics of P. aeruginosa Ech on 3HP. Michaelis-Menten kinetics of Cupriavidus necator Pcl (E) 3-hydroxypropionate, (F) CoA, and (G) ATP. (H) Validation of the 3HP route using Ccr (assay with Pcs/Pcc can be found in fig. S19). Values refer to relative concentrations of intermediates as determined by LCMS. Colors correspond to compounds in (A).
As an alternative to the above multienzyme variant, which we termed the “Ccr variant,” we also considered the use of the multifunctional propionyl-CoA synthetase (Pcs) from Erythrobacter NAP1 (EAQ29651; 140 mU/mg; app. Km-3-hydroxypropionate, 85 μM; app. Km-ATP, 10 μM; app. Km-CoA, 0.34 mM, and app. Km-NADPH, 0.14 mM) (fig. S7). Pcs forms a multicatalytic nanocompartment that activates 3HP to 3HP and dehydrates the latter to acrylyl-CoA, which it further reduces to propionyl-CoA (24). Notably, Pcs was recently engineered to catalyze the reductive carboxylation of acrylyl-CoA (see Discussion) (25). However, for initial tests, we used wild-type Pcs in combination with ATP-dependent propionyl-CoA carboxylase (Pcc, ACS40950/ AMB43430) to generate (2S)-methylmalonyl-CoA, albeit this came at the cost of one additional ATP per carboxylation of propionyl-CoA. For this reaction, we selected the Pcc of Methylobacterium extorquens AM1 (26) (22 U/mg; app. Km-propionyl-CoA, 55 μM; app. Km-acetyl-CoA, 0.27 mM; and app. Km-ATP, 0.33 mM) after which we named this variant the “Pcs/Pcc variant.”
Having established the individual enzyme activities for the β-alanine and 3HP routes, as well as their variants, we tested the different versions of the reductive part of the HOPAC cycle (Figs. 2G and 3H and fig. S19). While all versions were able to convert malonyl-CoA into (2S)-methylmalonyl-CoA, the Ccr variant and the Pcs/Pcc variant of the 3HP route showed rapid conversion without obvious bottlenecks, which made these two variants viable options for the construction of the full HOPAC cycle.
The oxidative part: Overall design
The first step in the oxidative part is the conversion of methylmalonyl-CoA into succinyl-CoA. For this reaction, we relied on methylmalonyl-CoA mutase (Mcm, ABA80144; 450 U/mg, Km-methylmalonyl-CoA, 19 μM) and methylmalonyl-CoA epimerase (Epi, ABA79990; 440 U/mg; Km-methylmalonyl-CoA, 80 μm) (27) from C. sphaeroides that had been successfully used already in the CETCH cycle (1, 27). We also tested MeaB (WP_012640779), a native Mcm chaperone (28). MeaB protects Mcm from inactivation and stimulates the guanosine triphosphate–dependent ejection of aquacobalamin (OH2Cbl), the oxidation product of adenosyl- cobalamin (AdoCbl), which is generated during suicide inactivation. In standalone assays, MeaB markedly extended the life span of Mcm in the presence of its substrates (fig. S16). However, when tested in the context of the full HOPAC cycle, addition of MeaB resulted only in a minor improvement on overall productivity (see below, HOPAC 3.0).
For the subsequent multistep conversion of succinyl-CoA to (S)-malyl-CoA, we investigated two potential routes. In the first variant, analogous to the tricarboxylic acid (TCA) cycle (“free acid route”), succinyl-CoA is cleaved into CoA and succinate, which is oxidized to fumarate and hydrated to malate (Fig. 4G), before being religated to CoA. In the second variant (“CoA route”), we sought to oxidize succinyl-CoA directly into fumaryl-CoA and further into (S)- malyl-CoA without release of the CoA ester.
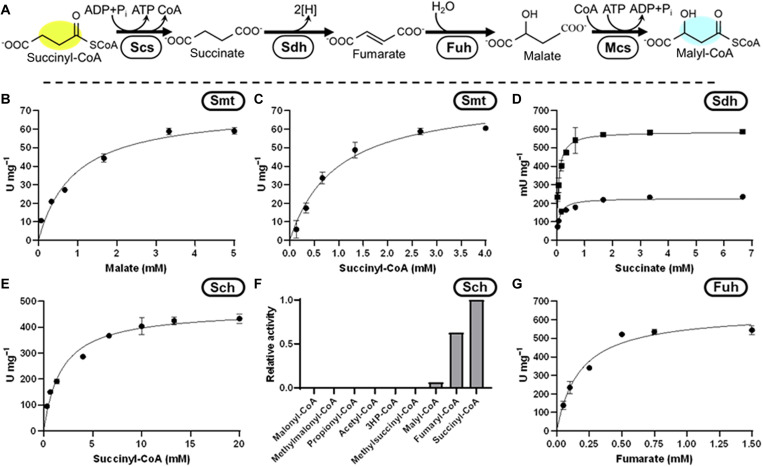
Fig. 4. Construction of the free acid route.
(A) Reaction sequence, cleavage of succinyl-CoA to succinate by #1 (Scs, Smt, or Sch), oxidation of succinate to fumarate by Sdh, hydration of fumarate to malate by Fuh, and ligation of malate to malyl-CoA by #2 (Smt or Mcs). Michaelis-Menten kinetics of Chloroflexus aurantiacus Smt (B) malate and (C) succinyl-CoA. (D) Michaelis-Menten kinetics of Escherichia coli Sdh, circles are SdhA, squares are SdhAB on succinate. (E) Michaelis-Menten kinetics of Mus musculus Sch on succinyl-CoA. (F) Promiscuity assay of Sch. (G) Michaelis-Menten kinetics of E. coli Fuh on fumarate.
The free acid route of the oxidative part
To realize the free acid route, we considered three options to cleave succinyl-CoA (Fig. 4). First, analogous to the TCA cycle, we used a succinyl-CoA synthetase (Scs, P0AGE9/ AVS48775, 37.7 U/mg; app. Km-succinate, 0.25 mM; app. Km-CoA, 4.0 μM; app. Km-ATP, 70 μM) (29) from E. coli. Second, as in the 3HP-bicycle, we used a succinyl-CoA:L-malate CoA transferase (Smt, Q1KLK1/ Q1KLK0, 78 U/mg; app. Km-succinyl-CoA, 0.96 mM; app. Km-malate, 0.91 mM; Fig. 4, B and C) from C. aurantiacus. Last, we also considered succinyl-CoA hydrolase (Sch, NP_599008, 468 U/mg; Km-succinyl-CoA, 1.8 mM; Fig. 4E) from Mus musculus. When tested, all three enzymes were active, however, their integration into the HOPAC cycle posed different challenges. Scs requires a delicate balancing of nucleoside triphosphate (NTP) to nucleoside diphosphate (NDP) pools. This, however, interferes with other reactions in the HOPAC cycle that require a constantly high NTP:NDP ratio, in particular the carboxylation by Pcc and the ligations by malyl-CoA synthetase (Mcs, P53595/ P53594) (30), Hps, and Pcs. Smt, on the other hand, preserves the energy of the CoA thioester bond in an NTP- and NDP-independent fashion but requires a constant malate concentration, which would interfere with the HOPAC cycle when it is not in its steady state. Last, the use of Sch would increase the net NTP requirements of the HOPAC cycle, due to its hydrolysis of the CoA-thioester bond which is exacerbated by promiscuity (Fig. 4F).
For succinate oxidation, we tested membrane-bound succinate dehydrogenase (Sdh) and/or soluble fumarate reductase (Frd, AAN40014). Soluble Frd from Trypanosoma brucei was incapable of performing this reaction because of the highly endergonic nature of this reaction using NAD+ as an electron acceptor (31). We also purified soluble versions of the Sdh complex from E. coli (32), in particular, Sdh subunit A alone (STL43511), which contains the active site and a flavin adenine dinucleotide (FAD) cofactor, and a construct of Sdh subunits A and B (P07014), which contains additional iron-sulfur clusters. In both cases, the enzyme was active with ferrocenium hexafluorophosphate as electron acceptor (Fig. 4D) at specific activities of 230 mU/mg (SdhA) and 590 mU/mg (SdhAB), respectively.
Although we could demonstrate all individual reactions for the succinyl-CoA cleaving enzymes and Sdh, none of the combinations allowed for production of acetyl-CoA from methylmalonyl-CoA, which led us to focus on the CoA route (Fig. 5) in the following section.
Fig. 5. Construction of the fumaryl-CoA route.
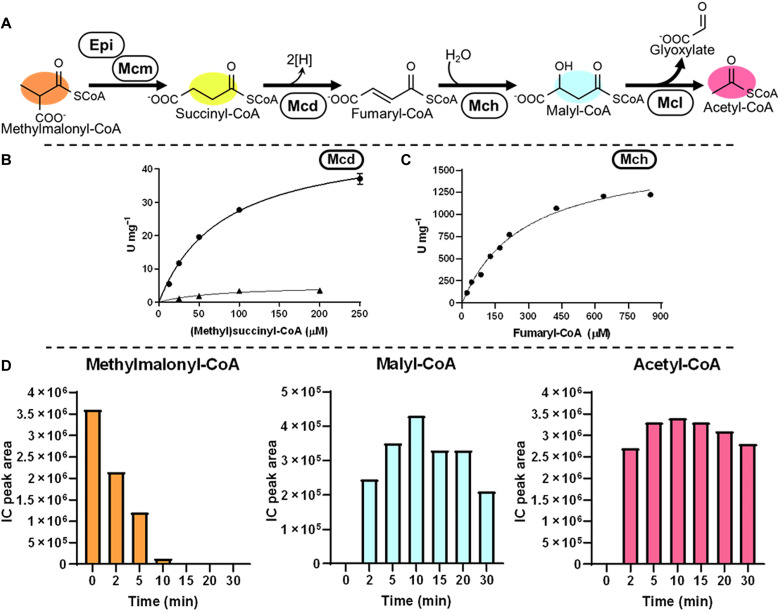
(A) Reaction sequence, oxidation of succinyl-CoA to fumaryl-CoA by Mcd and hydration of fumaryl-CoA to malyl-CoA by Mch. (B) Michaelis-Menten kinetics of P. migulae Mcd on succinyl-CoA (triangles) and methylsuccinyl-CoA (circles). (C) Michaelis-Menten kinetics of C. sphaeroides Mch on fumaryl-CoA. (D) Validation of the fumaryl-CoA route using Etf and EtfQO. Values refer to relative concentrations of intermediates as determined by LCMS. Colors correspond to compounds in (A).
The CoA route of the oxidative part
To establish this route, we needed to identify two new-to-nature enzyme activities. One enzyme that could oxidize succinyl-CoA to fumaryl-CoA and another to hydrate fumaryl-CoA to (S)-malyl-CoA. To establish the latter, we tested Ech from P. aeruginosa, which had previously shown high activity on 3HP, as well as mesaconyl-CoA hydratase (Mch, Q3IZ78, 1.7 × 103 U/mg; Km-fumaryl-CoA, 280 μM) from C. sphaeroides (Fig. 5C (33). As only the latter showed activity with fumaryl-CoA, we used this enzyme moving forward.
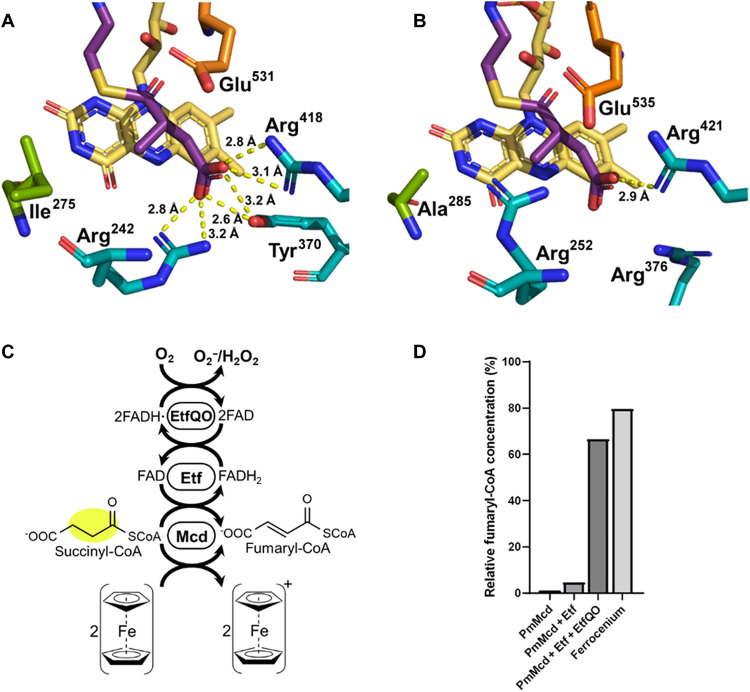
For the oxidation of succinyl-CoA to fumaryl-CoA, we screened different acyl-CoA dehydrogenases, including nondecarboxylating glutaryl-CoA dehydrogenase (C3UVB0) (34), as well as different (2S)-methylsuccinyl-CoA dehydrogenase (Mcd) homologs and active site variants with ferrocenium hexafluorophosphate as electron acceptor (figs. S11 and S12). We identified one homolog from Pseudomonas migulae (PmMcd, D3JV03) that showed very favorable side activity with succinyl-CoA, outcompeting all other enzymes by more than one order of magnitude (5.2 U/mg; Km-succinyl-CoA, 78 μM; Fig. 5B). To understand the factors influencing substrate specificity in PmMcd, we determined the structure of the enzyme in complex with FAD and methylsuccinyl-CoA at 1.9-Å resolution [Protein Data Bank (PDB): 8CIW, Fig. 6 and fig. S25). Residue I275 caught our attention, as mutation of the analogous residue A282 in C. sphaeroides Mcd (ADC44452) to valine had increased oxidase activity with succinyl-CoA. We explored several mutations of this residue in PmMcd including I275H/K/L/Q/S/T. However, all variants showed a marked loss in catalytic activity (fig. S26) necessitating the use of the wild-type enzyme. PmMcd in combination with its native electron transfer protein (EtfAB, SEE84602/ MCP1497298) had a modest activity with O2 as an electron acceptor. Notably, addition of EtfAB together with its cognate quinone reductase EtfQO (MCP1497299) (35) (solubilized with detergents) enabled the rapid recycling of reduced FAD cofactors in the presence of O2 (Fig. 6C). This allowed us to use the enzyme without natural or artificial electron acceptors in solution, which we exploited especially during optimization of the HOPAC cycle (see below, Fig. 7B). With all required enzyme activities in hand, we reconstructed the oxidative part of the HOPAC cycle and confirmed formation of acetyl-CoA from (2S)-methylmalonyl-CoA (Fig. 5D).
Fig. 6. Active site of methylsuccinyl-CoA dehydrogenase (Mcd).
(A) P. migulae Mcd in complex with FAD and methylsuccinyl-CoA, as determined in this study (PDB: 8CIW). (B) Paracoccus denitrificans Mcd in complex with FAD (PDB: 6ES9) (methylsuccinyl-CoA modeled). Residues in cyan are responsible for substrate binding, orange is the catalytic glutamate, and green was targeted for mutagenesis to improve succinyl-CoA dehydrogenase activity. (C) Reaction scheme for electron transport. FADH2 can be oxidized by ferrocenium, or as part of a chain through Etf terminating in molecular O2. (D) Fumaryl-CoA production by Mcd alone, with Etf, with Etf and EtfQO, or with ferrocenium.
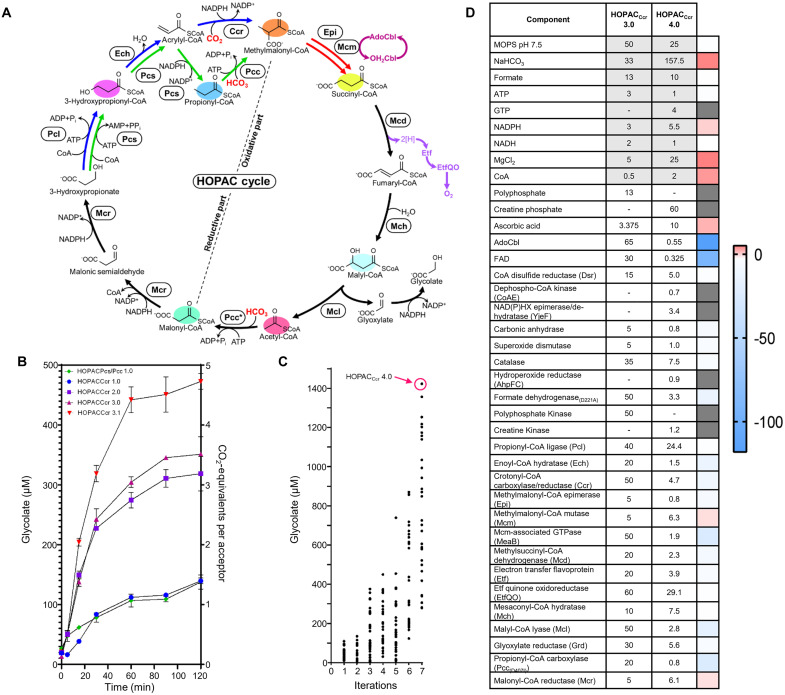
Fig. 7. Uniting and optimizing the HOPAC cycle.
(A) Scheme of the different final HOPAC cycle variants and versions. The best-performing reductive parts (HOPACCcr and HOPACPcs/Pcc) were combined with the CoA route of the oxidative part to create HOPACCcr version 1.0 and HOPACPcs/Pcc version 1.0. Both variants follow the same topology except that HOPACPcs/Pcc uses two ATP-dependent carboxylation reactions (green reactions), while HOPACCcr uses one ATP-dependent carboxylation and one reductive carboxylation reaction (blue reactions). In HOPACCcr version 2.0, ETFQ was added to improve ETF oxidation (dark purple reactions). In HOPACCcr version 3.0, AdoCBl recycling (light purple reactions) was introduced to maintain high Mcm activities. HOPACCcr version 3.1, Mcm was added in regular intervals (red reactions) to maintain high Mcm activities. (B) Carbon fixation efficiency of the HOPAC cycle. Right axis: Number of CO2/HCO3− fixed per molecule of acetyl-CoA in the assay. Left axis: Absolute concentration of glycolate produced. Concentrations were determined with triplicate assays measured by LCMS. Colors correspond to variants and versions of the HOPAC cycles as described in (A). (C) Further optimization of HOPACCcr (starting from version 3.0) through the active learning–based workflow “METIS” over eight rounds featuring 240 different combinations of individual enzyme and cofactor concentrations. Right axis: Absolute glycolate concentrations of triplicate assays after 2 hours measured by LCMS. Each dot represents a HOPACCcr assay with different setup. Per round, 30 different combinations of individual enzyme and cofactor concentrations were tested. The best combination (HOPACCcr version 4.0), producing almost 1500 μM glycolate in 2 hours is highlighted in hot pink. (D) Compositions and changes between HOPACCcr version 3.0 and 4.0. All enzymes, factors, and their respective concentrations are provided for version 3.0 and 4.0, and the respective fold changes are indicated in color. Concentrations are given in mM (gray shading) and μM (no shading).
Engineering acetyl-CoA carboxylase activity
Last, we sought to identify a suitable acetyl-CoA carboxylase (Acc) for the HOPAC cycle. Acc are commonplace in nature; however, they are typically multisubunit enzymes that are prone to dissociation, which complicates their purification and in vitro handling (36). Pcc, on the other hand, are often fusion proteins and are more robust. Pcc from M. extorquens (see above) showed some basal activity with acetyl-CoA (1.1 U/mg; app. Km, 270 μM), which we sought to improve further through active site mutation. Acc from Streptomyces coelicolor that is structurally similar to Pcc from M. extorquens (26) was recently engineered for improved activity with propionyl-CoA by a D422I mutation (37). Installing the analogous mutation in Pcc from M. extorquens increased the enzyme’s activity with acetyl-CoA substantially (15.5 U/mg; app. Km-acetyl-CoA, 560 µM; app. Km-ATP, 50 μM) yielding variant Pcc(D407I) (fig. S6), which we subsequently used to construct the full HOPAC cycle.
Uniting and optimizing the HOPAC cycle
Having successfully established the CoA route for the oxidative part of the HOPAC and two different routes for the reductive part (Ccr route and Pcs/Pcc route), we next assembled the Ccr variant of HOPAC (HOPACCcr), as well as the Pcs/Pcc variant (HOPACPcs/Pcc) and assessed their performance.
To test HOPACCcr, we combined all 11 enzymes of the basic variant (i.e., without EtfQO and MeaB) in 150 μl of MOPS (pH 7.5) with 3 mM ATP, 3 mM NADPH (reduced form of nicotinamide adenine dinucleotide phosphate), and 0.5 mM CoA. We added ATP (38) and NADPH-regeneration systems, (39) as well as enzymes for the repair of hydrated NADPH, (40) reduction of oxidized CoA dimers (41), and phosphorylation of dephosphorylated CoA (42), carbonic anhydrase to maintain the CO2/HCO3− equilibrium, and superoxide dismutase and catalase to decompose reactive oxygen species. In addition, we provided a glyoxylate reductase to reduce the output molecule of the cycle, glyoxylate, to glycolate, additionally driving the cycle forward.
The Ccr-based variant of HOPAC was functional in its initial version but further improved with the implementation of Etf and EtfQO as well as MeaB and Mcm supplementation (Fig. 7). The first version of HOPACCcr (HOPACCcr 1.0) produced ~100 μM glycolate from 200 μM acetyl-CoA, which could be increased to ~300 μM with the addition of EtfQO (HOPACCcr 2.0). Addition of MeaB increased glycolate production to ~350 μM (HOPACCcr 3.0). Glycolate production could be further increased to ~500 μM with Mcm supplementation (every 15 min, HOPACCcr 3.1), suggesting that MeaB alone was not sufficient to prevent Mcm inactivation in the complex setup of the HOPAC cycle.
Although the Pcs/Pcc variant of the HOPAC cycle was also functional (HOPACPcs/Pcc), it failed to outperform HOPACCcr 1.0 even with EtfQO and supplemented Mcm, so we focused exclusively on the Ccr variant of the HOPAC. To further improve HOPACCcr, we took advantage of an active learning–guided workflow for optimization of complex in vitro systems (“METIS”) that we had recently established in our laboratory (12). This workflow combines laboratory automation with active learning to explore the combinatorial space of a given system in iterative cycles for (local) optima.
To set up the METIS workflow, we miniaturized/reduced the assay volume to 10 μl, constrained each of the 36 compounds in the HOPACcr 3.0 cycle to a maximum of five different concentrations and tested 30 different combinations in triplicate in each round. After eight rounds of optimization and a total of 240 different tested HOPACCcr conditions, we achieved a maximum production of ~1500 μM glycolate (Fig. 7C) from 200 μM acetyl-CoA as substrate (HOPACCcr 4.0), which represents a ~3-fold improvement compared to the starting point (HOPACCcr 3.0) and ~15-fold compared to HOPACCcr 1.0. Notably, with 2.4 nmol CO2 min−1 mg−1 protein, CO2-fixation productivity of the HOPAC is comparable to the CETCH (1), the reductive glyoxylate and pyruvate synthesis-malyl-CoA-glycerate (rGPS-MCG) cycle (11), and the THETA cycle (table S8) which lifts the HOPAC cycle from a theoretical design to a functional in vitro system.
DISCUSSION
In this work, we prototyped and optimized a new-to-nature CO2-fixation pathway, the HOPAC cycle. The core cycle consists of 11 enzymes from six different organisms and is similar to the naturally existing 3HP bicycle. However, for the conversion of acrylyl-CoA into (2S)-methylmalonyl-CoA, the HOPAC cycle uses a reductive carboxylation instead of a combination of acrylyl-CoA reduction and ATP-dependent carboxylation. This makes the HOPACCcr 33% more ATP-efficient than the 3HP, which underlines the potential of synthetic biology to realize more efficient metabolic pathways than natural evolution.
Beyond this difference in acrylyl-CoA carboxylation, the HOPACCcr cycle also differs in respect to other enzyme chemistries from the 3HP. The central oxidation of succinyl-CoA to malyl-CoA proceeds through a new-to-nature metabolite, fumaryl-CoA, and involves two new-to-nature reactions: one reaction producing fumaryl-CoA (through oxidation of succinyl-CoA) and another reaction that consumes fumaryl-CoA (through hydration into malyl-CoA). We succeeded in establishing these reactions through screening multiple enzymes candidates for promiscuous activities with these metabolites that we could potentially exploit for construction of the cycle. Thus, the final implementation of our design differs not only from 3HP but also from other natural reaction sequences and pathways, qualifying the HOPACCcr as a bona fide “new-to-nature” pathway.
During our design, we explored other variants to the HOPAC, such as the free-acid variant in the oxidative part or the β-alanine variant in the reductive part. Although we did not pursue these options further in the framework of this study, these pathway variants are still viable and could be used in the context of other in vitro setups or efforts to establish the HOPAC in vivo. One of the most promising alternatives is the Pcs/Pcc variant of the HOPAC cycle, in which we used Pcs in combination with Pcc for the conversion of 3HP into (2S)-methylmalonyl-CoA—at the cost of one extra ATP. The acrylyl-CoA reductase domain of Pcs has been recently engineered toward a reductive carboxylase, reaching 97% carboxylation activity at saturating CO2 concentrations, albeit at lowered overall catalytic activity of the enzyme. Using a “carboxylating” Pcs (with improved overall rates) could omit the need for Pcc in the future. This would lower the extra ATP need for carboxylation and make the Pcs/Pcc-based variant energetically equivalent to HOPACCcr.
Having established a first version of the HOPAC cycle, we could further optimize the system through iterative rounds of rational and machine learning–guided efforts. Addition of auxiliary enzymes, such as MeaB, and adjusting the concentration of individual components was crucial to improve the system by 10-fold compared to version 1.0, which strongly emphasizes the potential and the need of design-build-test-cycles in complex in vitro biology. In its current form, HOPACCcr fixes CO2 at a rate of 2.4 nmol min−1 mg−1, which matches well the efficiencies of other in vitro CO2-fixation pathways and provides the basis for the further development of the HOPAC cycle toward different in vitro and in vivo applications.
MATERIALS AND METHODS
Protein expression and purification
General expression protocol
Whenever possible, enzymes were produced in E. coli BL21 DE3 ArcticExpress (DE3)RIL (unless stated otherwise below). Transformations were plated on LB agar supplemented with chloramphenicol (34 μg/ml) for the selection of the ArcticExpress cells and the antibiotic corresponding to each plasmid [ampicillin (100 μg/ml), kanamycin (50 μg/ml), and streptomycin (100 μg/ml)] and incubated at 37°C overnight. One-liter cultures were grown in Terrific Broth (TB) medium containing the same antibiotics as above at 37°C with moderate agitation until an OD600 (optical density at 600 nm) of ~1.5 was reached. Cultures were chilled to 20°C, induced with 500 μM isopropyl-β-d-thiogalactopyranoside, and incubated for an additional 16 hours before harvest. After expression, cultures were harvested by centrifugation at 8000 g for 10 min. The resulting pellets were resuspended in resuspension buffer containing 20 mM MOPS (pH 7.5), 500 mM NaCl, 5 mM MgCl2, and 20% (v/v) glycerol. Suspensions were sonicated to homogeneity and centrifuged at 100,000g for 1 hour. Last, lysates were decanted and passed through 0.45-μm cutoff filters. His-tagged protein was purified with 5-ml HisTrap FF columns (GE Healthcare, Freiburg, Germany). Bound columns were washed with resuspension buffer and eluted in a stepwise manner with resuspension buffer supplemented with 500 μM imidazole.The resulting protein was concentrated to ~1 ml before being passed through two HiTrap 5 ml Desalting Columns (GE Healthcare, Freiburg, Germany) for buffer exchange into 20 mM MOPS (pH 7.5), 200 mM NaCl, 5 mM MgCl2, and 20% glycerol (v/v). Proteins were concentrated again, and the concentration was determined by measuring the absorbance at 280 nm with extinction coefficients determined with ExPASy ProtParam (43).
Acetyl-/propionyl-CoA carboxylase
Pcc and Pcc(D407I) are biotin-dependent enzymes and as such, require biotinylation of their active sites for activity. Biotinylation by native E. coli enzymes is possible; however, incomplete reaction typically yields a pool of inactive enzyme. Instead, we coexpressed the cognate biotin ligase (BirA), and activity is increased several fold (44). To maximize biotinylation, 5 μmol biotin was added to the growth medium during induction.
Mcds, Etf, and CoA-disulfide reductase
FAD-containing proteins were purified as described above, except that an approximately equimolar volume of FAD was added before desalting. Because FAD absorbs significantly, it results in inaccurate concentrations if determined from absorbance at 280 nm (A280) alone. Instead, absorbance was measured at 280 and 450 nm. The A450/A280 ratio for FAD alone is 0.57 between A450 0.04 and 0.3. Using this ratio to determine the absorbance at 280 nm caused by FAD, we used the remaining absorbance to determine the protein concentration as above.
Etf-QO
Because ETF-QO in a membrane-bound protein, it first needs to be extracted from the membrane. It was extracted as described previously (35). Briefly, we maintained our lysis protocol except the pellet was recovered rather than the supernatant after centrifugation. The pellet was homogenized in resuspension buffer supplemented with 5% (w/v) Dodecyl-β-d-maltoside (CAS 69227-93-6) by sonication. The resulting suspension was incubated on ice for 1 hour and centrifuged at 100,000g for 1 hour. The supernatant from this step was treated as the lysate in the general expression protocol.
Synthesis and purification of acyl-CoA
Acetyl-, propionyl-, crotonyl-, and succinyl-CoA
Acetic, propionic, crotonic, or succinic anhydride were used to synthesize their respective acyl-CoA using the symmetric anhydride method (45). First, 200 mg of Coenzyme A was dissolved in 5 ml of 500 mM NaHCO3 on ice. An approximately 1.5× molar excess of anhydride (acetic, 42 mg; propionic, 53 mg; crotonic, 63 mg; or succinic, 41 mg) was added, and the solution incubated on ice with constant agitation for 1 hour. Completion was confirmed with Ellman’s reagent (46).
3HP and methylsuccinyl-CoA
3HP and methylsuccinyl-CoA were synthesized from CoA and their respective acids using the carbonyldiimidazole method (45). CoA (200 mg) was dissolved in 2 ml of 500 mM NaHCO3 on ice. An approximately 5× molar excess of each acid (methylsuccinate160 mg, 3-hydroxypropionate 350 μl, 30%) was dissolved in 8 ml of tetrahydrofuran (THF) with 160 mg of carbonyldiimidazole (CDI) and incubated for 45 min with constant agitation. The solutions were combined and incubated for a final hour after which completion was confirmed with Ellman’s reagent (46). Reactions were lyophilized before purification.
Fumaryl-CoA
Fumaryl-CoA was synthesized from fumarate and CoA using the ethylchloroformate method (45). CoA (190 mg) was dissolved in 10 ml of 500 mM NaHCO3 on ice. An approximately 10× molar excess of fumarate (290 mg) was dissolved 10 ml of THF on ice. Triethylamine (180 μl) and 140 mg of ethylchloroformate were added and incubated for 45 min with constant agitation. The solutions were combined and incubated for a final hour after which completion was confirmed with Ellman’s reagent (46). Reactions were lyophilized before purification.
Acrylyl-, malonyl-, and methylmalonyl-CoA
Acrylate, malonate, and methylmalonate were used to synthesize their respective acyl-CoA using acyl-CoA synthetases. CoA (20 mg), 15 mg of ATP, and 50 μmol acid (3.6 mg of acrylate, 5 mg of malonate, 6 mg of methylmalonate) were dissolved in 10 ml of 50 mM MOPS (pH 7.5) with 15 mM MgCl2. The reaction was initiated with the addition of 10 μM Pcl from C. necator (acrylate) or MatB from Rhizobium leguminosarum (45) (malonate/methylmalonate) and was incubated at 30°C. Completeness of reaction was monitored using Ellman’s reagent. Reactions were stopped by the addition of 4% formic acid, centrifuged, and the supernatant was lyophilized before purification.
(S)-malyl- and (2R,3S)-β-methylmalyl-CoA
(S)-Malyl- and (2R,3S)-β-methylmalyl-CoA were synthesized from glyoxylate and acetyl-CoA or propionyl-CoA respectively using the Mcl method (47). Acetyl- or propionyl-CoA (4 mg) was dissolved in 10 ml of 50 mM MOPS (pH 7.5) with 10 mM glyoxylate. The reaction was initiated with the addition of 10 μM Mcl from C. sphaeroides and the was incubated at 30°C. Completeness of reaction was monitored using Ellman’s reagent. Reactions were stopped by the addition of 4% formic acid and centrifuged, and the supernatant was lyophilized before purification.
β-Alanyl-CoA
β-alanyl-CoA was synthesized from β-alanine and propionyl-CoA using the A. propionicum β-alanyl-CoA transferase. Propionyl-CoA (10 mg) was dissolved in 10 ml of 50 mM MOPS (pH 7.5) with 10 mM β-alanine. The reaction was initiated with the addition of 10 μM βct and the solution was incubated at 30°C. Completeness of reaction was monitored using Ellman’s reagent. Reactions were stopped by the addition of 4% formic acid and centrifuged, and the supernatant was lyophilized before purification.
Purification
Samples were purified with a Gemini 10 μm NX-C18 110 Å Column (Phenomenex, Aschaffenburg, Germany) in a 1260 Infinity HPLC (Agilent Technologies GmbH) using a methanol/sodium formate (pH 4.2) gradient. Purified fractions were flash-frozen in liquid nitrogen, lyophilized, and stored with desiccant at 20°C for future use. CoA-thioester concentration was determined by spectrophotometric absorbance at 260 nm with ε = 22.4 mM−1 cm−1 for unsaturated (fumaryl-CoA) and ε = 16.4 mM−1 cm−1 for saturated CoA-thioesters (all others).
Kinetics
ATP-dependent reactions
The activity of ATP-dependent reactions was determined using the pyruvate kinase, lactate dehydrogenase–coupled reaction (Sigma-Aldrich, P0294). Triethanolamine (1 mM), 1.6 mM phosphoenolpyruvate, 4.2 mM magnesium sulfate, 6.8 mM potassium chloride, 300 μM NADH, 40 U of pyruvate kinase, and 100 U of lactic dehydrogenase were used. The oxidation of NADH was monitored at 340 nm ɛNADH = 6.22 M−1 cm−1. This was used for Pcc, Pcc(D407I), Pcs, Pcl, and Mcs. For activity screens, reactions were quenched with formic acid and samples were analyzed by liquid chromatography–mass spectrometry (LCMS).
NADPH-dependent reactions
The activity of NADPH-dependent reactions was determined by directly monitoring the oxidation of NADPH at 360-nm ɛNADPH = 3.4 mM−1 cm−1. This was used for Ccr, Gdh, Mcr, and Pcs. In the case of Mcr, the enzyme performs the reduction of malonyl-CoA and malonic semialdehyde, it was assumed that for every two molecules of NADPH consumed, one molecule of malonyl-CoA was reduced to 3-hydroxypropionate. In the case of Pcs, the rate of NADPH oxidation was used as a proxy for the complete reaction of ligation, dehydration, and reduction of 3-hydroxypropionate and CoA to propionyl-CoA.
Enoyl-CoA hydratase
The activity of Ech was determined in a coupled assay using Ccr. Ech was incubated with 3HP, NADPH, NaHCO3 and an excess of Ccr. As 3HP was dehydrated to acrylyl-CoA, it was reductively carboxylated by Ccr and the NADPH oxidation was monitored as above.
β-Alanyl-CoA lyase
The activity of βCal was determined in a coupled assay using Ccr. βCal was incubated with β-alanyl-CoA, NADPH, NaHCO3, and an excess of Ccr. As β-alanyl-CoA was deamiated to acrylyl-CoA, it was reductively carboxylated by Ccr and the NADPH oxidation was monitored as above.
Mcm and MeaB
The activity of Mcm was determined in a coupled assay using a succinyl-CoA reductase from Clostridium kluyveri. Mcm was incubated with methylmalonyl-CoA, NADPH, and an excess of Scr. As methylmalonyl-CoA was converted to succinyl-CoA, it was reduced by Scr and the simultaneous oxidation of NADPH was monitored as above. The effect of MeaB was determined by incubating methylmalonyl-CoA alone, with Mcm, or with Mcm and MeaB. Time points were incubated with Scr and NADPH to consume any succinyl-CoA produced before being quenched with formic acid and analyzed by LCMS.
(S)-malyl-CoA lyase
The activity of Mcl was determined with a coupled assay using Grd. Mcl was incubated with either (S)-malyl- or (2R,3S)-β-methylmalyl-CoA, NADPH, and an excess of Grd. As (methyl)malyl-CoA were cleaved, the glyoxylate was reduced by Grd and the NADPH oxidation was monitored as above.
Succinyl-CoA:L-malate CoA transferase
The activity of Smt was determined in a coupled assay using Mcl and Grd. Smt was incubated with succinyl-CoA, malate, NADPH, and an excess of Mcl and Grd. As malyl-CoA was generated, it was cleaved by Mcl and monitored as above.
Succinyl-CoA hydrolase
The activity of Sch was determined using the Ellman’s Assay (46). Sch was incubated with 2.5 mM sodium acetate, 100 μM 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), and succinyl-CoA. 2-Nitro-5-thiobenzoic acid (TNB) formation was monitored at 412 nm ɛTNB = 13.6 mM−1 cm−1. To screen for promiscuity, Sch was incubated with various acyl-CoAs. Time points were quenched with formic acid and analyzed by LCMS.
Hydratases
The activity of Mesaconyl-C1-CoA hydratase was determined by directly monitoring the alkenyl formation/depletion at 290 nm ɛMesaconyl-CoA = 3.1 mM−1 cm−1 (48). As (methyl)fumaryl-CoA is hydrated to (methyl)malyl-CoA, the absorbance at 290 nm decreases. The activity of fumarate hydratase was determined by directly monitoring the alkenyl formation/depletion at 240 nm ɛFumarate = 2.4 mM−1 cm−1 (49). As fumarate is hydrated to malate, the absobance at 240 nm decreases.
Aminotransferase
Malonyl-CoA was preincubated with McrC to generate malonic semialdehyde. l-alanine or l-glutamate was added with NH4Cl and Adh or Gdh, respectively. To maintain consistent NADPH concentration, sufficient volumes were added to reduce all malonyl-CoA and to have reported concentrations remaining.
Dehydrogenases
The activity of Mcd and Sdh were determined using ferrocenium hexafluorophosphate as an electron acceptor and monitoring ferrocenium oxidation at 300 nm ɛFerrocenium = 2.75 mM−1 cm−1. Because (methyl)succinyl-CoA are prone to intramolecular general base catalysis, they were synthesized regularly and monitored for integrity using Ellman’s reagent (46).
Etf and EtfQO
The effect of Etf and EtfQO were determined by incubating methylsuccinyl-CoA with Mcd, Mcd and Etf, or Mcd, Eft, and EtfQO. Timepoints were quenched with formic acid and analyzed by LCMS.
CoA kinase
Acetyl-CoA, malonyl-CoA, 3HP, propionyl-CoA, methylmalonyl-CoA, fumaryl-CoA, malyl-CoA, or β-alanyl-CoA were incubated with shrimp alkaline phosphatase from Sigma-Aldrich (GEE70092Y) to generate dephosphorylated analogs. To remove phosphatase, samples were then passed through a 5-kDa cutoff filter. These samples were then incubated with CoAE and ATP. Samples were quenched with formic acid and analyzed by LCMS.
Partial cycle assays
To determine the compatibility of various enzyme combinations, the cycle was split into two pathways allowing for a simplified workflow to test different options. Reductive paths were started with malonyl-CoA, time points were quenched with formic acid, and malonyl-CoA, 3HP, β-alanyl-CoA, propionyl-CoA, and methylmalonyl-CoA were detected using LCMS. Oxidative paths were started with methylmalonyl-CoA, and methylmalonyl-CoA, succinyl-CoA, malyl-CoA, and acetyl-CoA were detected with LCMS. Concentration of enzymes and cofactors in reductive pathway assays are listed in table S1. Concentration of enzymes and cofactors in oxidative pathway assays are provided in table S2.
Full cycle assays
Full cycle variants were tested similarly to the partial cycles except ferrocenium could not be used as an electron acceptor due to cross-reactivity with NADPH, assays were started with acetyl-CoA, and in addition to acyl-CoA intermediates, glycolate was quantified. 13C labeled HCO3− and formate were used in assays for acyl-CoA measurements, which allowed for the tracking of compounds across multiple cycles because one net 13C was incorporated per cycle. 12C HCO3− and formate were used in assays for glycolate quantification which allowed for the use of an internal 13C glycolate standard. In addition, a glycolate standard curve ranging from 10 μM to 2 mM were used for quantification. Concentration of enzymes and cofactors in full HOPAC cycle assays are listed in table S3.
Optimization of HOPAC
Optimization of HOPAC 3.0 was performed with METIS (12). Concentrations of enzymes and cofactors from previous assays were taken as a baseline and two and four additional concentrations were fed to METIS for recombination. Thirty conditions were selected by METIS in each iteration, tested in triplicate, and analyzed by LCMS after 120-min reactions. Concentrations for METIS optimization are provided in table S4.
Ultraperformance liquid chromatography–high resolution MS
Acyl-CoA
Detection of acyl-CoAs was performed using isotope dilution mass spectrometry. The chromatographic separation was performed on an Agilent Infinity II 1290 HPLC system using a Kinetex EVO C18 column (150 mm by 1.7 mm, 3-μm particle size, 100-Å pore size, Phenomenex) connected to a guard column of similar specificity (20 mm by 2.1 mm, 5-μm particle size, Phenomoenex) a constant flow rate of 0.25 ml/min with mobile phase A being 50 mM Ammonium Acetate in water at a pH of 8.1 and phase B being 100% methanol (Honeywell, Morristown, New Jersey, USA). The injection volume was 5 μl. The mobile phase profile consisted of the following steps and linear gradients: 0 to 1 min constant at 2.5% B; 1 to 6 min from 2.5 to 95% B; 6 to 8 min constant at 95% B; 8 to 8.1 min from 95 to 2.5% B; 8.1 to 10 min constant at 2.5% B. An Agilent 6550 ion funnel QTOF mass spectrometer was used in positive mode with an electrospray ionization source and the following conditions: ESI spray voltage 3500 V, nozzle voltage 500 V, sheath gas 400°C at 12 liters/min, nebulizer pressure 20 psig and drying gas 225°C at 13 liters/min. Compounds were identified on the basis of their accurate mass (mass error < 5 ppm) and retention time compared to standards. Chromatograms were integrated using MassHunter software (Agilent, Santa Clara, CA, USA).
Glycolate
Quantitative determination of Glycolate was performed using isotope dilution mass spectrometry. The chromatographic separation was performed on an Agilent Infinity II 1290 HPLC system using a Kinetex EVO C18 column (150 mm by 1.7 mm, 3-μm particle size, 100-Å pore size, Phenomenex) connected to a guard column of similar specificity (20 mm by 2.1 mm, 5-μm particle size, Phenomoenex) a constant flow rate of 0.1 ml/min with mobile phase A being 0.1% formic acid in water and phase B being 0.1% formic acid methanol (Honeywell, Morristown, New Jersey, USA) at 25°C. The injection volume was 0.5 μl. The mobile phase profile consisted of the following steps and linear gradients: 0 to 4 min constant at 0% B; 4 to 6 min from 0 to 100% B; 6 to 7 min constant at 100% B; 7 to 7.1 min from 100 to 0% B; 7.1 to 12 min constant at 0% B. An Agilent 6495B ion funnel mass spectrometer was used in negative mode with an electrospray ionization source and the following conditions: ESI spray voltage 2000 V, nozzle voltage 500 V, sheath gas 400°C at 11 liters/min, nebulizer pressure 50 psig and drying gas 80°C at 16 liters/min. Compounds were identified on the basis of their mass transition and retention time compared to standards. Chromatograms were integrated using MassHunter software (Agilent, Santa Clara, CA, USA). Absolute concentrations were calculated on the basis of an external calibration curve prepared in sample matrix, using uniformly 13C-labeled Glycolate as internal standard. Mass transitions, collision energies, Cell accelerator voltages and Dwell times have been optimized using chemically pure standards. Parameter settings of all targets are given in table S8.
Acknowledgments
We thank I. Bernhardsgrütter for providing the PCs expression construct and C. Seng for experimental assistance.
Funding: This work was supported by the Max Planck Society.
Author contributions: R.M., T.J.E., and T.S. conceptualized the study. T.J.E. drafted the initial version of the HOPAC cycle, which was further developed into different variants by R.M. R.M. and T.S. prototyped and characterized the proteins used for construction of the HOPAC cycle. J.Z. solved crystal structures. R.M. constructed the different variants of the HOPAC cycle. C.D. collaborated with R.M. for machine learning–guided optimization of the HOPAC cycle. N.S.C. and N.P. performed and supervised mass spectrometric analysis. R.M. analyzed all data and discussed it with the authors. The initial manuscript draft was written by R.M. and T.J.E. All authors edited the manuscript, contributed to the interpretation of results, and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data are provided in the manuscript, the Supplementary Materials and Supplementary Data, as well as relevant repositories (crystal structure 8CIW in the RCSB PDB).
Supplementary Materials
This PDF file includes:
Tables S1 to S8
Figs. S1 to S26
Legend for data S1
Amino Acid Sequences
References
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Schwander T., Schada von Borzyskowski L., Burgener S., Cortina N. S., Erb T. J., A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erb T. J., Zarzycki J., A short history of RubisCO: The rise and fall (?) of Nature’s predominant CO2 fixing enzyme. Curr. Opin. Biotechnol. 49, 100–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos Correa S., Schultz J., Lauersen K. J., Soares Rosado A., Natural carbon fixation and advances in synthetic engineering for redesigning and creating new fixation pathways. J. Adv. Res. 47, 75–92 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurtzel E. T., Vickers C., Hanson A. D., Millar A. H., Cooper M., Voss-Fels K. P., Nikel P. I., Erb T. J., Revolutionizing agriculture with synthetic biology. Nat. Plants 5, 1207–1210 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Erb T. J., Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 77, 8466–8477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bar-Even A., Noor E., Lewis N. E., Milo R., Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U.S.A. 107, 8889–8894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotton C. A., Edlich-Muth C., Bar-Even A., Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation. Curr. Opin. Biotechnol. 49, 49–56 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Yu H., Li X., Duchoud F., Chuang D. S., Liao J. C., Augmenting the Calvin-Benson-Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway. Nat. Commun. 9, 2008 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X., Yang Y., Wang S., Wang Q., Wang X., Yan Z., Cheng J., Liu C., Yang X., Luo H., Yang S., Gou J., Ye L., Lu L., Zhang Z., Guo Y., Nie Y., Lin J., Li S., Tian C., Cai T., Zhuo B., Ma H., Wang W., Ma Y., Liu Y., Li Y., Jiang H., Constructing a synthetic pathway for acetyl-coenzyme A from one-carbon through enzyme design. Nat. Commun. 10, 1378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel J. B., Smith A. B., Poust S., Wargacki A. J., Bar-Even A., Louw C., Shen B. W., Eiben C. B., Tran H. M., Noor E., Gallaher J. L., Bale J., Yoshikuni Y., Gelb M. H., Keasling J. D., Stoddard B. L., Lidstrom M. E., Baker D., Computational protein design enables a novel one-carbon assimilation pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 3704–3709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo S., Lin P. P., Nieh L.-Y., Liao G.-B., Tang P.-W., Chen C., Liao J. C., A cell-free self-replenishing CO2-fixing system. Nat. Catal. 5, 154–162 (2022). [Google Scholar]
- 12.Pandi A., Diehl C., Kharrazi A. Y., Scholz S. A., Bobkova E., Faure L., Nattermann M., Adam D., Chapin N., Foroughijabbari Y., Moritz C., Paczia N., Cortina N. S., Faulon J.-L., Erb T. J., A versatile active learning workflow for optimization of genetic and metabolic networks. Nat. Commun. 13, 3876 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarzycki J., Fuchs G., Coassimilation of organic substrates via the autotrophic 3-hydroxypropionate bi-cycle in Chloroflexus aurantiacus. Appl. Environ. Microbiol. 77, 6181–6188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarzycki J., Brecht V., Müller M., Fuchs G., Identifying the missing steps of the autotrophic 3-hydroxypropionate CO2 fixation cycle in Chloroflexus aurantiacus. Proc. Natl. Acad. Sci. U.S.A. 106, 21317–21322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hügler M., Menendez C., Schägger H., Fuchs G., Malonyl-coenzyme A reductase from Chloroflexus aurantiacus, a key enzyme of the 3-hydroxypropionate cycle for autotrophic CO2 fixation. J. Bacteriol. 184, 2404–2410 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C., Wang Q., Xian M., Ding Y., Zhao G., Dissection of malonyl-coenzyme A reductase of Chloroflexus aurantiacus results in enzyme activity improvement. PLOS ONE 8, e75554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakano Y., Tokunaga H., Kitaoka S., Two omega-amino acid transaminases from Bacillus cereus. J. Biochem. 81, 1375–1381 (1977). [PubMed] [Google Scholar]
- 18.Hayaishi O., Nishizuka Y., Tatibana M., Takeshita M., Kuno S., Enzymatic studies on the metabolism of β-Alanine. J. Biol. Chem. 236, 781–790 (1961). [PubMed] [Google Scholar]
- 19.Hördt A., López M. G., Meier-Kolthoff J. P., Schleuning M., Weinhold L.-M., Tindall B. J., Gronow S., Kyrpides N. C., Woyke T., Göker M., Analysis of 1,000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 11, 468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann G., Selmer T., Jessen H. J., Gokarn R. R., Selifonova O., Gort S. J., Buckel W., Two beta-alanyl-CoA:ammonia lyases in Clostridium propionicum. FEBS J. 272, 813–821 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Green P. N., Ardley J. K., Review of the genus Methylobacterium and closely related organisms: A proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int. J. Syst. Evol. Microbiol. 68, 2727–2748 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Erb T. J., Brecht V., Fuchs G., Müller M., Alber B. E., Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proc. Natl. Acad. Sci. U.S.A. 106, 8871–8876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuge T., Taguchi K., Seiichi T., Doi Y., Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: Metabolic tools for synthesis of polyhydroxyalkanoates via fatty acid beta-oxidation. Int. J. Biol. Macromol. 31, 195–205 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Bernhardsgrütter I., Vögeli B., Wagner T., Peter D. M., Cortina N. S., Kahnt J., Bange G., Engilberge S., Girard E., Riobé F., Maury O., Shima S., Zarzycki J., Erb T. J., The multicatalytic compartment of propionyl-CoA synthase sequesters a toxic metabolite. Nat. Chem. Biol. 14, 1127–1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernhardsgrütter I., Schell K., Peter D. M., Borjian F., Saez D. A., Vöhringer-Martinez E., Erb T. J., Awakening the sleeping carboxylase function of enzymes: Engineering the natural CO2-binding potential of reductases. J. Am. Chem. Soc. 141, 9778–9782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chistoserdova L. V., Lidstrom M. E., Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquens AM1. Microbiol. Read. Engl. 142, 1459–1468 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Erb T. J., Rétey J., Fuchs G., Alber B. E., Ethylmalonyl-CoA mutase from Rhodobacter sphaeroides defines a new subclade of coenzyme B12-dependent acyl-CoA mutases. J. Biol. Chem. 283, 32283–32293 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Padovani D., Banerjee R., Assembly and protection of the radical enzyme, methylmalonyl-CoA mutase, by its chaperone. Biochemistry 45, 9300–9306 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Hidber E., Brownie E. R., Hayakawa K., Fraser M. E., Participation of Cys123α of Escherichia coli succinyl-CoA synthetase in catalysis. Acta Crystallogr. D Biol. Crystallogr. 63, 876–884 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Chistoserdova L. V., Lidstrom M. E., Genetics of the serine cycle in Methylobacterium extorquens AM1: Identification, sequence, and mutation of three new genes involved in C1 assimilation, orf4, mtkA, and mtkB. J. Bacteriol. 176, 7398–7404 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besteiro S., Biran M., Biteau N., Coustou V., Baltz T., Canioni P., Bringaud F., Succinate secreted by Trypanosoma brucei is produced by a novel and unique glycosomal enzyme, NADH-dependent fumarate reductase. J. Biol. Chem. 277, 38001–38012 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Cecchini G., Schröder I., Gunsalus R. P., Maklashina E., Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta 1553, 140–157 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Zarzycki J., Schlichting A., Strychalsky N., Müller M., Alber B. E., Fuchs G., Mesaconyl-coenzyme A hydratase, a new enzyme of two central carbon metabolic pathways in bacteria. J. Bacteriol. 190, 1366–1374 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wischgoll S., Demmer U., Warkentin E., Günther R., Boll M., Ermler U., Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases. Biochemistry 49, 5350–5357 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Usselman R. J., Fielding A. J., Frerman F. E., Watmough N. J., Eaton G. R., Eaton S. S., Impact of mutations on the midpoint potential of the [4Fe-4S]+1,+2 cluster and on catalytic activity in electron transfer flavoprotein-ubiquinone oxidoreductase (ETF-QO). Biochemistry 47, 92–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong L., Acetyl-coenzyme A carboxylase: Crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. CMLS 62, 1784–1803 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diacovich L., Mitchell D. L., Pham H., Gago G., Melgar M. M., Khosla C., Gramajo H., Tsai S. C., Crystal structure of the beta-subunit of acyl-CoA carboxylase: Structure-based engineering of substrate specificity. Biochemistry 43, 14027–14036 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Mordhorst S., Singh J., Mohr M. K. F., Hinkelmann R., Keppler M., Jessen H. J., Andexer J. N., Several polyphosphate kinase 2 enzymes catalyse the production of adenosine 5′‐polyphosphates. Chembiochem. 20, 1019–1022 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Hoelsch K., Sührer I., Heusel M., Weuster-Botz D., Engineering of formate dehydrogenase: Synergistic effect of mutations affecting cofactor specificity and chemical stability. Appl. Microbiol. Biotechnol. 97, 2473–2481 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Marbaix A. Y., Noël G., Detroux A. M., Vertommen D., Van Schaftingen E., Linster C. L., Extremely conserved ATP- or ADP-dependent enzymatic system for nicotinamide nucleotide repair. J. Biol. Chem. 286, 41246–41252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.delCardayre S. B., Stock K. P., Newton G. L., Fahey R. C., Davies J. E., Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J. Biol. Chem. 273, 5744–5751 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Wadler C., Cronan J. E., Dephospho-CoA kinase provides a rapid and sensitive radiochemical assay for coenzyme A and its thioesters. Anal. Biochem. 368, 17–23 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.E. Gasteiger, C. Hoogland, A. Gattiker, A. Duvaud, M. R. Wilkins, R. D. Appel, A. Bairoch, Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handbook, J. M.Walker, Ed. (Humana Press, 2005), pp. 571–607. [Google Scholar]
- 44.Rodrı Guez E., Gramajo H., Genetic and biochemical characterization of the α and β components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3(2) The GenBank accession numbers for the accA1, aacA2 and pccB sequences determined in this work are AF113603, AF113604 and AF113605, respectively. Microbiol. Read. Engl. 145, 3109–3119 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Peter D. M., Vögeli B., Cortina N. S., Erb T. J., A chemo-enzymatic road map to the synthesis of CoA Esters. Mol. Basel Switz. 21, 517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellman G. L., Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 (1959). [DOI] [PubMed] [Google Scholar]
- 47.Herter S., Busch A., Fuchs G., L-malyl-coenzyme A Lyase/β-methylmalyl-coenzyme A lyase from chloroflexus aurantiacus, a bifunctional enzyme involved in autotrophic CO2 fixation. J. Bacteriol. 184, 5999–6006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Erb T. J., Frerichs-Revermann L., Fuchs G., Alber B. E., The apparent malate synthase activity of Rhodobacter sphaeroides is due to two paralogous enzymes, (3S)-Malyl-coenzyme A (CoA)/β-methylmalyl-CoA lyase and (3S)- Malyl-CoA thioesterase. J. Bacteriol. 192, 1249–1258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kronen M., Berg I. A., Mesaconase/Fumarase FumD in Escherichia coli O157:H7 and promiscuity of Escherichia coli class I Fumarases FumA and FumB. PLOS ONE 10, e0145098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao L., Liu G., Gong F., Zhu H., Zhang Y., Cai Z., Li Y., A minimized synthetic carbon fixation cycle. ACS Catal. 12, 799–808 (2022). [Google Scholar]
- 51.Gale N. L., Beck J. V., Evidence for the Calvin cycle and hexose monophosphate pathway in thiobacillus ferrooxidans. J. Bacteriol. 94, 1052–1059 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S8
Figs. S1 to S26
Legend for data S1
Amino Acid Sequences
References
Data S1