Abstract
DNA damage repair (DDR) is a double-edged sword with different roles in cancer susceptibility and drug resistance. Recent studies suggest that DDR inhibitors affect immune surveillance. However, this phenomenon is poorly understood. We report that methyltransferase SMYD2 plays an essential role in nonhomologous end joining repair (NHEJ), driving tumor cells adaptive to radiotherapy. Mechanically, in response to DNA damage, SMYD2 is mobilized onto chromatin and methylates Ku70 at lysine-74, lysine-516, and lysine-539, leading to increased recruitment of Ku70/Ku80/DNA-PKcs complex. Knockdown of SMYD2 or its inhibitor AZ505 results in persistent DNA damage and improper repair, which sequentially leads to accumulation of cytosolic DNA, and activation of cGAS-STING pathway and triggers antitumor immunity via infiltration and activation of cytotoxic CD8+ T cells. Our study reveals an unidentified role of SMYD2 in regulating NHEJ pathway and innate immune responses, suggesting that SMYD2 is a promising therapeutic target for cancer treatment.
SMYD2 may be a promising therapeutic target of cancer by regulating NHEJ pathway and immune responses.
INTRODUCTION
Both intrinsic and extrinsic stimulations such as oxidative damage, irradiation, and replication stress can ultimately lead to DNA damage. Improper DNA damage repair (DDR) can result in genome instability and diseases, including cancer (1, 2). In addition to its role in tumorigenesis, loss of DNA repair capacity has important implications in therapeutic response. Therefore, various agents targeting the DDR pathway have been used as anticancer therapies (3).
DNA double-strand breaks are one of the most serious and complex damages and are repaired by two main pathways, nonhomologous end joining (NHEJ) and homologous recombination (HR). As one of the predominant pathways, NHEJ is a compensatory mechanism for a separate DNA repair defect and is frequently up-regulated in several cancers, making it an effective target for synthetic lethality (4). The Ku70/Ku80/DNA-DNA- dependent protein kinase catalytic subunit (PKcs) complex plays an essential role in NHEJ, with Ku70 and Ku80 forming a Ku- heterodimer, is rapidly recruited to sites of damaged DNA, and activates DNA-PKcs to initiate the NHEJ DNA damage response (5). NHEJ deficiency appears to be a risk factor for the development of malignancy (6, 7). However, the posttranslational regulations of this core component are still unclear. From a translational perspective, directly targeting the DDR pathway has long been a strategy for cancer therapy, but many classical chemotherapeutic agents resulted in numerous side effects, requiring drugs that can regulate the DDR process with improved targeting specificity (8).
Posttranslational modifications (PTMs) are diverse and include phosphorylation, methylation, acetylation, glycosylation and ubiquitination. As one of the most important protein modifications, methylation is dynamically mediated by methyltransferase or demethylase (9) and functional in various biological processes. Among the various methyltransferases, there are five members of the SMYD [Su (Var) 3-9, enhancer of zeste, and trithorax (SET) and myeloid, Nervy, and DEAF-1 (MYND) domain-containing] protein family, SMYD1-5. They are a very specific class of protein lysine methyltransferases that methylate both histone and nonhistone targets. SMYD2 was originally described as H3K36- and H3K4-specific methyltransferase (10, 11). With more in-depth research, nonhistone lysine methylation substrates of SMYD2 have begun to be explored. For example, SMYD2 was reported to methylate p53 or Phosphatase and tensin homolog deleted on chromosome ten (PTEN) and inhibit their tumor suppressive activities (12, 13). SMYD2 also carried out its function of cyst growth in autosomal dominant polycystic kidney disease via methylation and activation of signal transducer and activator of transcription 3 and the p65 subunit of nuclear factor κB (14). These diverse substrates broaden the function of SMYD2 in a variety of diseases. Recent evidence has also indicated that SMYD2 is a bona fide oncogene because of its essential role in promoting tumorigenesis (15–18). However, whether the oncogenic potential of SMYD2 has some relationship with genome stability and has potential as a therapeutic target remains unknown.
Recently, the interaction between DDR and immune response has drawn increased attention. Deficiency in the DNA repair of double-stranded breaks (DSBs) leads to the accumulation of DNA damage, accompanied by leakage of damaged DNA into the cytoplasm, which is an initial factor to trigger the cyclic guanosine monophosphate (GMP)–adenosine monophosphate (AMP) synthase (cGAS)–stimulator of interferon (IFN) genes (STING) pathway activation; cGAS interacts with cytosolic DNA and catalyzes cyclic GMP-AMP and 3′,3′-cGAMP (cGAMP) synthesis, followed by the sequential phosphorylation of STING, TBK1 (TANK-binding kinase 1), and IRF3 (IFN regulatory factor 3), and then enhanced the production of type I IFNs and the expression of IFN-stimulated genes (19). Accumulated DNA damage or incorrect DNA repair results in the alteration of the tumor genome and unbalances the immune system within the tumor microenvironment (TME) (3). For example, targeting KDM4A (lysine-specific demethylase 4A), a H3K9 trimethylation demethylase, induces replication stress and DNA damage, followed by activated cGAS-STING signaling and antitumor immunity in squamous cell carcinoma (20). Loss of KEAP1 (Kelch-like ECH-associated protein l) mediated the stabilization of EMSY, a part of chromatin remodeling complexes, elicits genome instability through HR repair defects, and results in increased tumor mutation burden, which prompts an innate immune response, fostering antitumor immunity in lung cancer (21). However, the mechanisms of how chromatin regulators modulate DNA repair and its potential link to immunity are still poorly understood.
In our current study, we report that lysine methyltransferase SMYD2 is recruited to chromatin in response to DSBs and promotes NHEJ repair. Mechanistically, SMYD2 interacts with Ku70 and mediates Ku70 methylation directly and then promotes the formation of Ku70/Ku80/DNA-PKcs complex assembly at DSBs. In addition, we find that blockage of DNA repair by SMYD2 inhibition leads to an accumulation of cytosolic DNA and STING/TBK1/IRF3 pathway activation, boosting the production of chemokines such as chemokine (C-C motif) ligand 5 (CCL5) and CXCL10. Genetic or pharmacological inhibition of SMYD2 alters the immunosuppressive TME through the infiltration of activated cytotoxic CD8+ T cells. Moreover, the SMYD2 inhibitor enhanced radiation therapy efficiency in vivo, demonstrating its translational significance.
RESULTS
SMYD2 is required for DDR
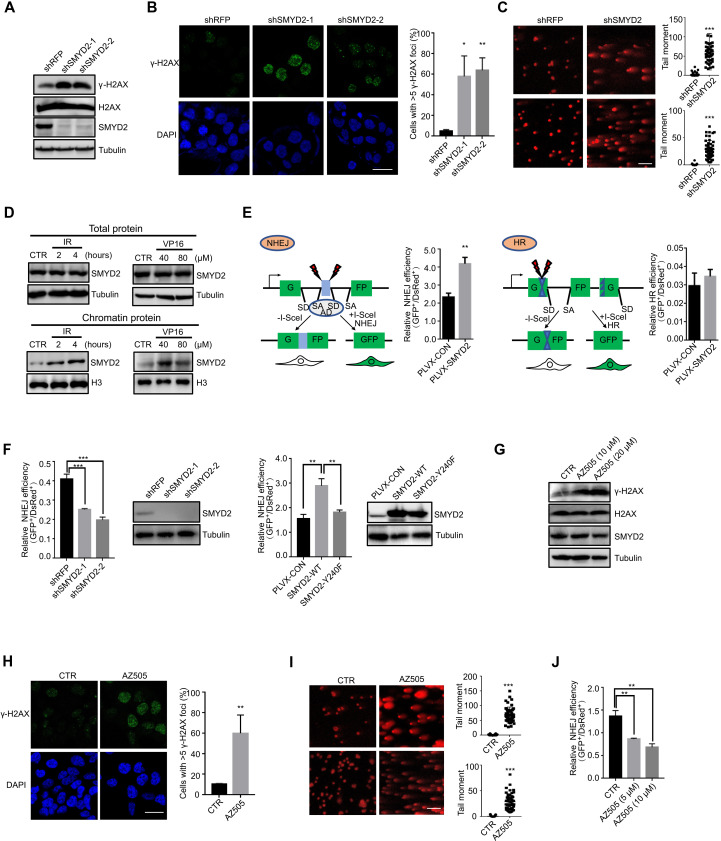
To determine whether the SMYD family is involved in the cellular response to DNA damage, we used short hairpin RNA (shRNA) mini-screen to target SMYD1-SMYD5 family members, with γ-H2AX staining and comet assay measuring DNA damage and repair efficiency (fig. S1A). As shown in fig. S1 (B and C), knockdown of SMYD1, SMYD3, SMYD4, SMYD5 does not efficiently increase γ-H2AX foci formation and comet tails. However, we observed the critical role of SMYD2 in regulating the DDR pathway. We first used γ-H2AX, to quantify DNA damage in cells. As shown in Fig. 1A and fig. S1D, the level of γ-H2AX was substantially increased in SMYD2 knockdown cells compared to control, with two shRNAs in two cell lines. We also observed robust foci formation of γ-H2AX following SMYD2 deletion in different cells (Fig. 1B and fig. S1E). In addition, the comet assay also showed that the repair of DNA breaks was delayed in SMYD2 knockdown cells (Fig. 1C). These results suggest that SMYD2 participates in DNA damage response caused by genotoxic stress. Consistent with the data above, we also found that the expression level of SMYD2 was not affected by x-ray irradiation (IR)/etoposide (VP16)–induced DNA damage in human colon carcinoma cells (HCT116) and mouse colon cancer (CT26) cells. However, DNA damage strongly induced the enrichment of SMYD2 onto chromatin. This phenomenon was confirmed by chromatin fractionation assays in a dose- and time-dependent manner (Fig. 1D and fig. S1F). Moreover, we used two model cell lines to monitor the role of SMYD2 in the DSB repair pathway, HR or NHEJ (22). Unexpectedly, overexpression of SMYD2 led to increased efficiency of NHEJ-mediated DSB repair but not HR (Fig. 1E). We next investigated the role of SMYD2 in the NHEJ repair pathway. We found decreased NHEJ repair efficiency in two stable SMYD2 knockdown cell lines (Fig. 1F). To further confirm whether this process is dependent on SMYD2 enzymatic activity, we introduced SMYD2–wild type (WT) or SMYD2-Y240F (enzymatic dead mutant) plasmids into I9a cells. As shown in Fig. 1F, WT SMYD2 markedly increased the efficacy of NHEJ repair, but enzymatic-dead Y240F mutant failed to do so. In addition to the genetic approach, we used AZ505, a classical effective and specific inhibitor of SMYD2, to further validate our results. As expected, AZ505 led to accumulated DNA damage and significantly inhibited NHEJ repair in a dose-dependent manner (Fig. 1, G to J, and fig. S1, G and H). Together, our results demonstrate a role for SMYD2 in regulating the DNA damage response/NHEJ repair pathway, in which inhibition of SMYD2 causes the persistent generation of DSBs and impairs repair.
Fig. 1. SMYD2 is required for DDR.
(A) Immunoblot of the expression levels of γ-H2AX in SMYD2-depleted HCT116 cells. (B) Representative fluorescence images and quantification of γ-H2AX immunostaining in cells with and without SMYD2. Scale bars, 25 μm. Green, γ-H2AX; blue, 4′,6-diamidino-2-phenylindole (DAPI). (C) Representative fluorescence images and quantification of tail moments in HCT116 and CT26 cells with and without SMYD2 as determined by a comet assay. Scale bars, 100 μm. (D) Immunoblot of endogenous SMYD2 in total proteins treated with IR at 10 grays (Gy) and released for 2 or 4 hours and etoposide (VP16) at 40 or 80 μM for 4 hours of HCT116 cells (top). Immunoblot of endogenous SMYD2 in chromatin proteins treated with IR at 10 Gy and released for 2 or 4 hours and etoposide (VP16) at 40 or 80 μM for 4 hours of HCT116 cells (bottom). (E) Diagram of the NHEJ and HR reporter assay. Effects of SMYD2 overexpression on the efficiency of NHEJ and HR in I9a and H15c cells. (F) Effects of SMYD2 stably knockdown on the efficiency of NHEJ in I9a cells and immunoblot showing the knockdown efficiency of SMYD2 (left graph). Effects of SMYD2-WT/Y240F on NHEJ efficiency in I9a cells and immunoblot showing the expressed protein (right graph). (G) Immunoblot of the expression levels of γ-H2AX in AZ505-treated HCT116 cells. (H) Representative fluorescence images and quantification of γ-H2AX immunostaining in cells treated with and without SMYD2 inhibitor AZ505. Scale bars, 25 μm. Green, γ-H2AX; blue, DAPI. (I) Representative fluorescence images and quantification of tail moments in cells treated with and without SMYD2 inhibitor AZ505 at 20 μM as determined by a comet assay. Scale bars, 100 μm. (J) NHEJ efficiency of SMYD2 inhibition with its inhibitor AZ505 at indicated concentrations.
SMYD2 interacts with Ku70 and is responsible for the NHEJ complex assembly
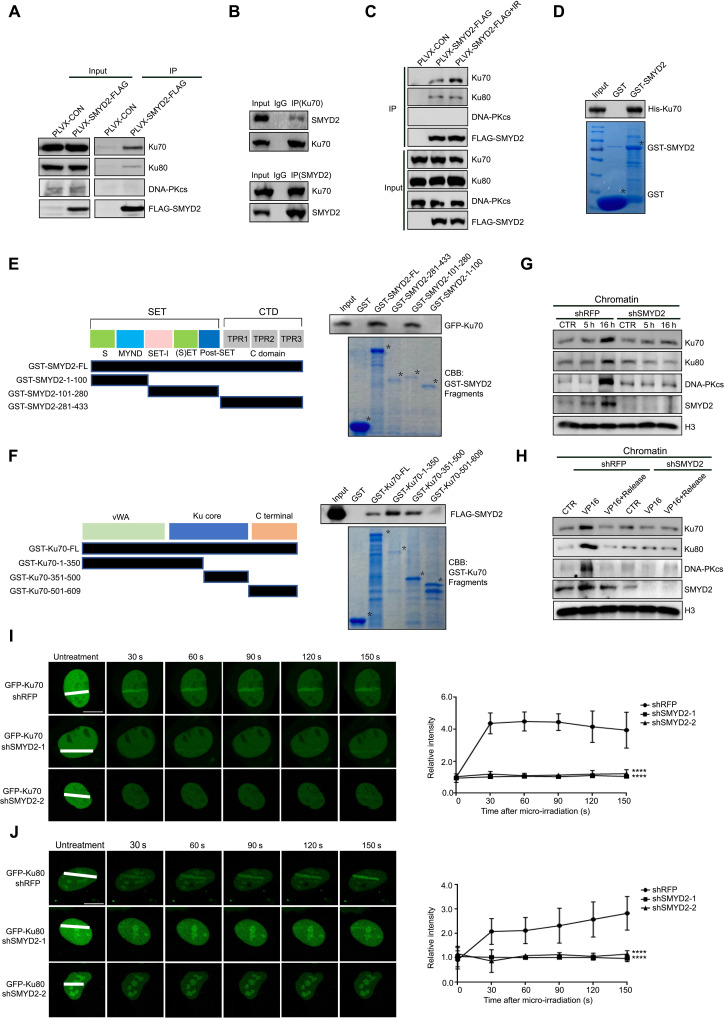
To gain further insights into how SMYD2 regulates NHEJ repair, we investigated the expression level of the key components (Ku70, Ku80, DNA-PKcs, LIG4, XRCC4, and XLF) in the NHEJ repair pathway. As expected, no obvious changes were observed upon SMYD2 knockdown or inhibition with an AZ505 inhibitor (fig. S2, A and B), while coimmunoprecipitation (Co-IP) assays using exogenous FLAG-SMYD2 detected a clear interaction between SMYD2 and Ku70 or Ku80, but not DNA-PKcs (Fig. 2A). To further confirm this interaction, plasmids expressing FLAG-tagged SMYD2 and a green fluorescent protein (GFP)–tagged Ku70 were cotransfected into cells. Lysates of transfected cells were subjected to a Co-IP assay with anti-FLAG or anti-GFP (fig. S2C). Similarly, endogenous Ku70 also showed a strong interaction with endogenous SMYD2 in HCT116 cells (Fig. 2B). The interaction between SMYD2 and Ku70 or Ku80 was not affected by the treatment of lysates with Benzonase nuclease to block the nonspecific protein-DNA interactions, indicating that the interaction is not mediated through DNA (fig. S2D). Moreover, only the interaction between SMYD2 and Ku70 was obviously increased upon IR/VP16–induced DNA damage, indicating that Ku70 is the major substrate of SMYD2 in the NHEJ repair pathway (Fig. 2C and fig. S2E). To investigate whether SMYD2 directly interacts with Ku70, a His-tagged Ku70 or Ku80 protein was expressed and purified from bacteria and incubated with glutathione S-transferase (GST) or GST-SMYD2. As shown in Fig. 2D, Ku70 directly interacts with GST-SMYD2, but not with GST. However, SMYD2 did not directly interact with Ku80 as determined by GST pull-down in vitro (fig. S2F). To further map the domains of SMYD2 responsible for its interaction with Ku70, we fragmented SMYD2 according to its functional domains. It was clearly shown that both full-length (FL) and the SET domain of SMYD2 interacted with Ku70 (Fig. 2E). Similarly, we also fragmented Ku70 and confirmed that the Ku core domain of Ku70 interacted with SMYD2 (Fig. 2F). These results raise the possibility that a direct interaction between SMYD2 and Ku70 may be essential for NHEJ repair.
Fig. 2. SMYD2 interacts with Ku70 and is responsible for the NHEJ complex assembly.
(A) Immunoblot of indicated proteins in the lysates or anti-FLAG immunoprecipitates of HEK293FT cells transfected with FLAG-SMYD2. (B) Immunoblot of SMYD2 or Ku70 protein that was immunoprecipitated with anti-Ku70 or anti-SMYD2 antibodies for endogenous interaction. IgG, immunoglobulin G. (C) Immunoblot of indicated proteins in the lysates or anti-FLAG immunoprecipitates of HEK293FT cells transfected with FLAG-SMYD2 after irradiation at 10 Gy. (D) In vitro GST pull-down assay of his-Ku70 combined with GST or GST-SMYD2. (E) Left graph: Fragments of SMYD2. Right graph: GST alone, GST-SMYD2-FL, GST-SMYD2-1-100, GST-SMYD2-101-280, or GST-SMYD2-281-433 was incubated with GFP-Ku70–transfected HEK293FT cell lysates for the GST pull-down assay. CBB, Coomassie brilliant blue. Asterisk (*) represents specific protein bands. (F) Left graph: Fragments of Ku70. Right graph: GST alone, GST-Ku70-FL, GST-Ku70-1-350, GST-Ku70-351-500, or GST-Ku70-501-609 was incubated with FLAG-SMYD2–transfected HEK293FT cell lysates for the GST pull-down assay. (G) Immunoblot analysis of indicated chromatin proteins in cells with or without SMYD2 and treated with irradiation at 10 Gy, release for 5 or 16 hours. (H) Immunoblot analysis of indicated chromatin proteins in cells with or without SMYD2 and treated with VP16 at 40 μM for 1 hour or release for 24 hours. (I) Left graph: Accumulation of exogenous GFP-Ku70 in 5-bromo-2′-deoxyuridine (BrdU)–sensitized U2OS cells stably expressing shSMYD2 or shRFP with GFP-Ku70. Right graph: Relative intensity of GFP-Ku70 at micro-irradiated sites in the experiments described in the left graph. Scale bars, 10 μm. (J) Left graph: Accumulation of exogenous GFP-Ku80 in BrdU-sensitized U2OS cells stably expressing shSMYD2 or shRFP with GFP-Ku80. Right graph: Relative intensity of GFP-Ku80 at micro-irradiated sites in the experiments described in the left graph. Scale bars, 10 μm.
Consistently, depletion of SMYD2 by shRNAs led to a marked decreased accumulation of Ku70, Ku80, and DNA-PKcs onto chromatin upon IR/VP16 treatment (Fig. 2, G and H), indicating that the recruitment of the NHEJ repair complex is SMYD2 dependent. Consistent with the chromatin recruitment results, the deficiency of SMYD2 also caused the impaired loading of Ku70 and Ku80 to DNA damage sites by using a micro-irradiation assay (Fig. 2, I and J). Moreover, for the later time points, we also observed low levels of Ku70 or Ku80 recruitment to laser-generated DSBs (fig. S2, G and H). Collectively, the data support that SMYD2 is a bone fide partner of Ku70, which loads the NHEJ repair complex onto chromatin in response to DNA damage. However, the interaction between SMYD2 and Ku80 does not change upon DNA damage, and there is no physical interaction between SMYD2 and DNA-PKcs. Thus, how SMYD2 functions in NHEJ repair complex assembly will be deeply investigated.
SMYD2 mediates Ku70 methylation at K74, K516, and K539 and influences its function in NHEJ repair
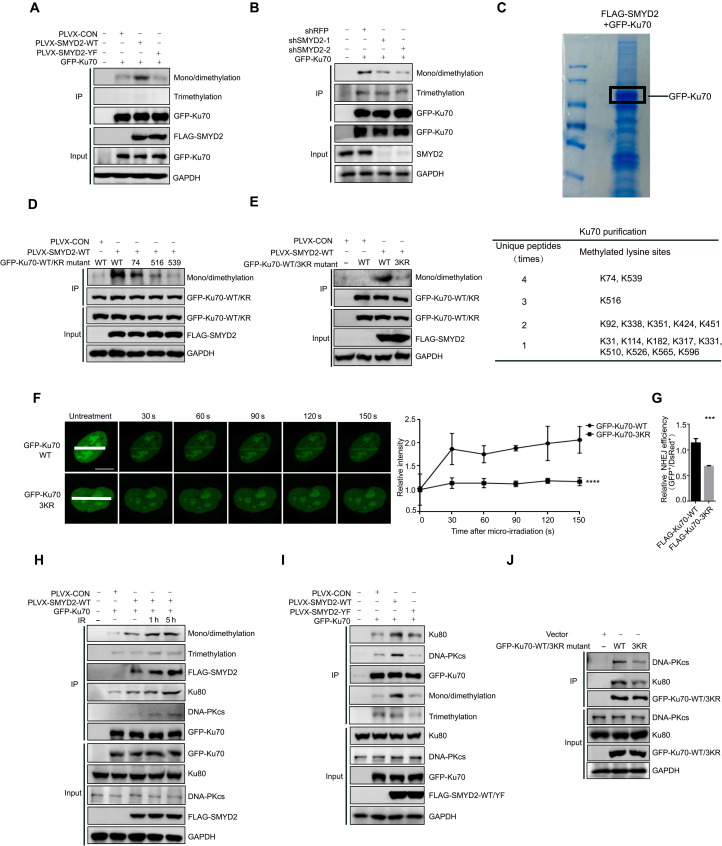
As a methyltransferase, we first asked whether SMYD2 could methylate Ku70, thereby affecting the NHEJ complex assembly. To this end, we performed IP assays and found that the methylation of Ku70 could be enhanced after SMYD2 overexpression by probing with an anti-pan-mono/di methylation antibody. However, the enzymatic-dead Y240F mutant of SMYD2 exhibited no activity in catalyzing Ku70 (Fig. 3A). As expected, depletion of SMYD2 by two independent shRNAs led to the mono/di methylation of Ku70 abolishment (Fig. 3B). To further map the sites of Ku70 that are methylated by SMYD2, we performed a mass spectroscopy analysis of immunoprecipitated GFP-Ku70, and 17 sites were identified to be the potential methylation sites (Fig. 3C). To determine the precise methylation sites of Ku70, we mutated those residues from lysine (K) to arginine (R). Only the mutation of K74, K516, and K539 obviously abolished SMYD2 mediated Ku70 mono/di methylation, indicating that those three sites may be predominantly involved in the methylation of Ku70 (Fig. 3D and fig. S3, A to E). In addition, we generated a Ku70-3KR mutant (K74R, K516R, and K539R) and tested its methylation possibility. Consistently, the methylation of Ku70 is largely abolished by the 3KR mutant (Fig. 3E). Compared to WT Ku70, 3KR mutant Ku70 showed a decreased recruitment to DNA damage sites, as shown by micro-irradiation experiments (Fig. 3F and fig. S3F). To further confirm that SMYD2 regulates the NHEJ repair through Ku70 methylation, we overexpressed WT or 3KR Ku70 in a NHEJ reporter cell line. As shown in Fig. 3G, overexpression of 3KR mutant Ku70 markedly inhibited NHEJ repair efficiency compared to WT Ku70, suggesting that SMYD2-mediated Ku70 methylation is essential for correctly loading Ku70 to chromatin.
Fig. 3. SMYD2 mediates Ku70 methylation at K74, K516, and K539 and influences its function in NHEJ repair.
(A) Immunoblot analysis of pan-methylation (mono/dimethylation and trimethylation) of Ku70 in the anti-GFP immunoprecipitates HEK293FT cells cotransfected with GFP-Ku70 and FLAG-SMYD2-WT/Y240F. (B) Immunoblot analysis of pan-methylation of Ku70 in the anti-GFP immunoprecipitates SMYD2 knockdown HEK293FT cells, which transfected with GFP-Ku70. (C) Mass spectrometry analysis of methylation sites of Ku70 following transfection with SMYD2 and GFP-Ku70. The table in the lower graph shows the times of unique peptides and the detailed lysine methylation sites. (D) Immunoblot analysis of mono/dimethylation of Ku70-WT/mutants in anti-GFP immunoprecipitates. HEK293FT cells cotransfected with GFP-Ku70-WT or K74R/K516R/K539R and FLAG-SMYD2-WT. (E) Immunoblot analysis of mono/dimethylation of Ku70-WT/3KR mutants in anti-GFP immunoprecipitates HEK293FT cells cotransfected with GFP-Ku70-WT or 3KR and FLAG-SMYD2-WT. (F) Left graph: Accumulation of exogenous GFP-Ku70-WT/3KR mutant in BrdU-sensitized U2OS cells. Scale bars, 10 μm. Right graph: Relative intensity of GFP-Ku70-WT/3KR at micro-irradiated sites in the experiments described in the left graph. (G) Effects of overexpression of Ku70-WT or 3KR mutant on the efficiency of NHEJ in I9a cells. The I9a cells were transfected with FLAG-Ku70 WT/3KR lentivirus with puromycin selection. (H) Immunoblot analysis of mono/dimethylation and trimethylation of Ku70 and the interaction between Ku70 and Ku80 or DNA-PKcs in anti-GFP immunoprecipitates. Plasmids encoding GFP-Ku70 and FLAG-SMYD2 were cotransfected in HEK293FT for 72 hours, treated with irradiation at 10 Gy, and released for the indicated time. (I) Immunoblot analysis of the interaction between Ku70 and Ku80 or DNA-PKcs in anti-GFP immunoprecipitates. Plasmids encoding GFP-Ku70 and FLAG-SMYD2-WT or Y240F were cotransfected in HEK293FT for 72 hours. (J) Immunoblot analysis of the interaction between WT/3KR-Ku70 and Ku80 or DNA-PKcs in anti-GFP immunoprecipitates. Plasmids encoding GFP-Ku70-WT/3KR were transfected in HEK293FT for 72 hours.
To further explain how SMYD2 regulates the loading of other NHEJ repair members such as Ku80 and DNA-PKcs onto chromatin, we did Co-IP to determine the binding affinity among these proteins. In response to IR/VP16 treatment, the mono/di methylation of Ku70 substantially increased as did the interaction between Ku70 and Ku80 or DNA-PKcs (Fig. 3H and fig. S3G). Unlike the enzymatic-dead Y240F mutant of SMYD2, WT SMYD2 mediated methylation of Ku70 and enhanced the interaction between Ku70 and Ku80 or DNA-PKcs, enhancing the DNA repair complex assembly (Fig. 3I). Eventually, 3KR mutant of Ku70 decreased the binding affinity between Ku70 and Ku80 or DNA-PKcs (Fig. 3J), suggesting that methylation of Ku70 is beneficial for binding Ku80. With a DNA pull-down assay, we found that these site mutations of Ku70-3KR did not affect the Ku70 binding to DNA by using double-stranded DNA (dsDNA) with 3′ overhangs, a common substrate found at DNA breaks. However, we incubated Ku70-WT/3KR and Ku80 with DNA and found that the binding activity of Ku70-Ku80 heterodimer to DNA was decreased in Ku70-3KR group (fig. S3H). To further confirm the SMYD2-Ku70 axis in response to DNA damage reagents, we first depleted SMYD2, or treated with its inhibitor, AZ505, in HCT116 cells and found that its inhibition sensitized cells to radiation or etoposide treatment (fig. S3, I and J). Consistently, we also observed that Ku70-3KR mutant exhibited sensitivities to radiation or etoposide when comparing with Ku70-WT (fig. S3K). Together, this evidence demonstrates that SMYD2 methylates multiple sites of Ku70 and the novel modification of Ku70 plays an important role in recruiting the NHEJ repair complex onto DNA damage sites, promoting the high efficiency of NHEJ repair.
Inhibition of SMYD2 activates the cGAS-STING pathway
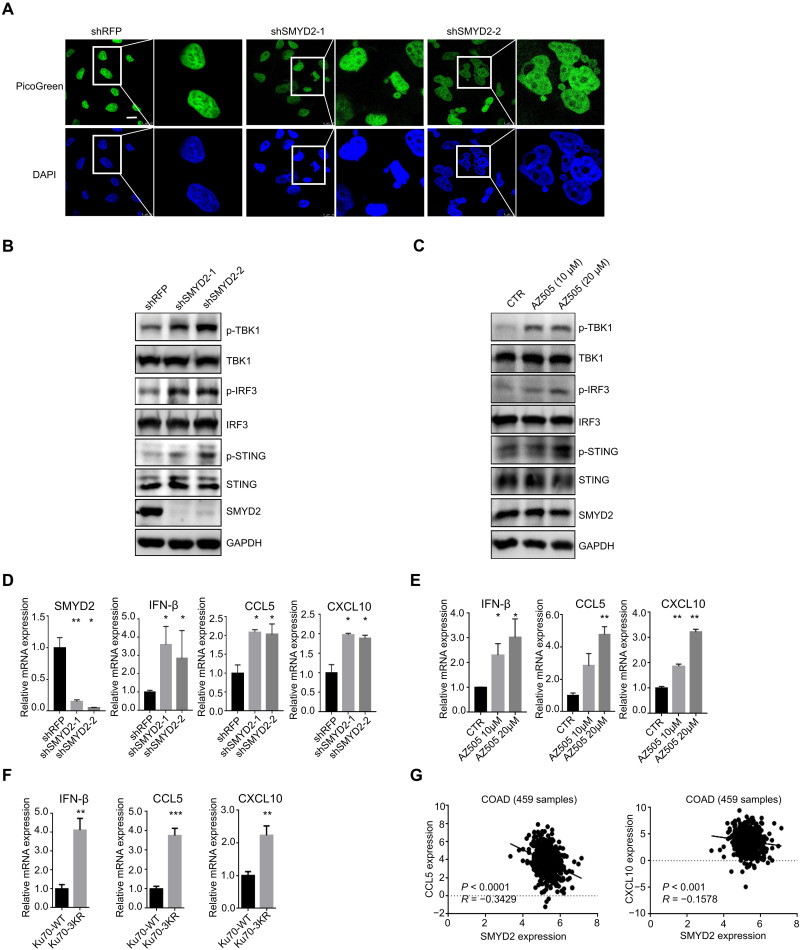
Previous studies have shown that deficiency in the DNA repair of DSBs leads to persistent DNA damage, accompanied by leakage of damaged nuclear DNA into the cytoplasm (23, 24). Under exogenous and endogenous conditions, cytosolic DNA can be functional as a powerful stimulus to activate the innate immune response. When we examined the accumulation of unrepaired endogenous DNA damage, we were surprised to find that the percentage of cells with accumulated cytosolic DNA was higher in the SMYD2 knockdown group compared with the control group. The data were confirmed by imaging with PicoGreen, a widely used fluorescent stain that selectively binds to dsDNA (Fig. 4A). The cytosolic DNA is recognized by the cGAS-STING sensing pathway, which is a key axis for innate and adaptive immunity. The transduced signaling then increases the phosphorylation level of STING, TBK1, and IRF3, followed by the induction of IFN-stimulated genes (ISGs) (25, 26). To determine whether the observed cytosolic DNA leads to cGAS-STING pathway activation, two independent shRNAs were used to knockdown the expression of SMYD2. As expected, loss of SMYD2 increased the phosphorylation level of TBK1, IRF3, and STING in HCT116 and CT26 cells (Fig. 4B and fig. S4A). Consistent with the genetic approach, the SMYD2 inhibitor AZ505 also up-regulated the phosphorylation level of TBK1, IRF3, and STING (Fig. 4C and fig. S4B). The activated STING/TBK1/IRF3 innate immune pathway can lead to the production of proinflammatory cytokines, including type I IFNs, CCL5, and CXCL10. As expected, we found that the deficiency of SMYD2 triggered type I IFN IFNβ production and the levels of chemokine CCL5 and CXCL10 (Fig. 4D and fig. S4C), as well as the inhibition by its specific inhibitor AZ505 (Fig. 4E and fig. S4D). Moreover, compared with Ku70-WT, Ku70-3KR mutant led to the increased production of the type I IFNs IFNβ, CCL5, and CXCL10 (Fig. 4F), which is consistent with the deficient repair capacity of Ku70-3KR mutant (Fig. 3G). Through analyzing the The Cancer Genome Atlas (TCGA) database, we found that the expression levels of CCL5 and CXCL10 are both negatively correlated with SMYD2 expression level in colon cancer [TISIDB website (27) and Fig. 4G]. These results reveal that inhibition of SMYD2 results in impaired DDR, causing the release of cytosolic DNA, which further triggers cGAS-STING pathway activation.
Fig. 4. Inhibition of SMYD2 activates the cGAS-STING pathway.
(A) Representative fluorescence images and quantification of cytosolic DNA in cells with and without SMYD2. Scale bars, 10 μm. Green, PicoGreen; blue, DAPI. DNA was detected using the PicoGreen fluorescence dye selectively binding dsDNA. (B) Immunoblot of markers in the STING pathway including total and phospho-TBK1 (p-TBK1) (S172), total and p-IRF3 (S396), and total and p-STING (S366) in lysates collected from cells with and without SMYD2 in HCT116 cells. (C) Immunoblot of markers in the STING pathway including total and p-TBK1 (S172), total and p-IRF3 (S396), and total and p-STING (S366) in lysates collected from HCT116 cells with and without SMYD2 inhibitor AZ505 at 10 and 20 μM for 7 days. (D) The expression of SMYD2, IFN-β, CCL5, and CXCL10 were measured via quantitative polymerase chain reaction (qPCR) in cells with and without SMYD2 in HCT116 cells. (E) The expression of IFN-β, CCL5, and CXCL10 were measured via qPCR in cells with and without SMYD2 inhibitor AZ505 at 10 and 20 μM in HCT116 cells. (F) The expression of IFN-β, CCL5, and CXCL10 were measured via qPCR in cells with Ku70-WT or 3KR mutant in HCT116 cells. The cells were transfected with FLAG-Ku70 WT/3KR lentivirus with puromycin selection. (G) The correlation of SMYD2 with CCL5 or CXCL10 expression in TCGA colon adenocarcinoma (COAD) database. The primary data were downloaded from the TISIDB website.
Inhibition of SMYD2 elicits an antitumor adaptive immune response
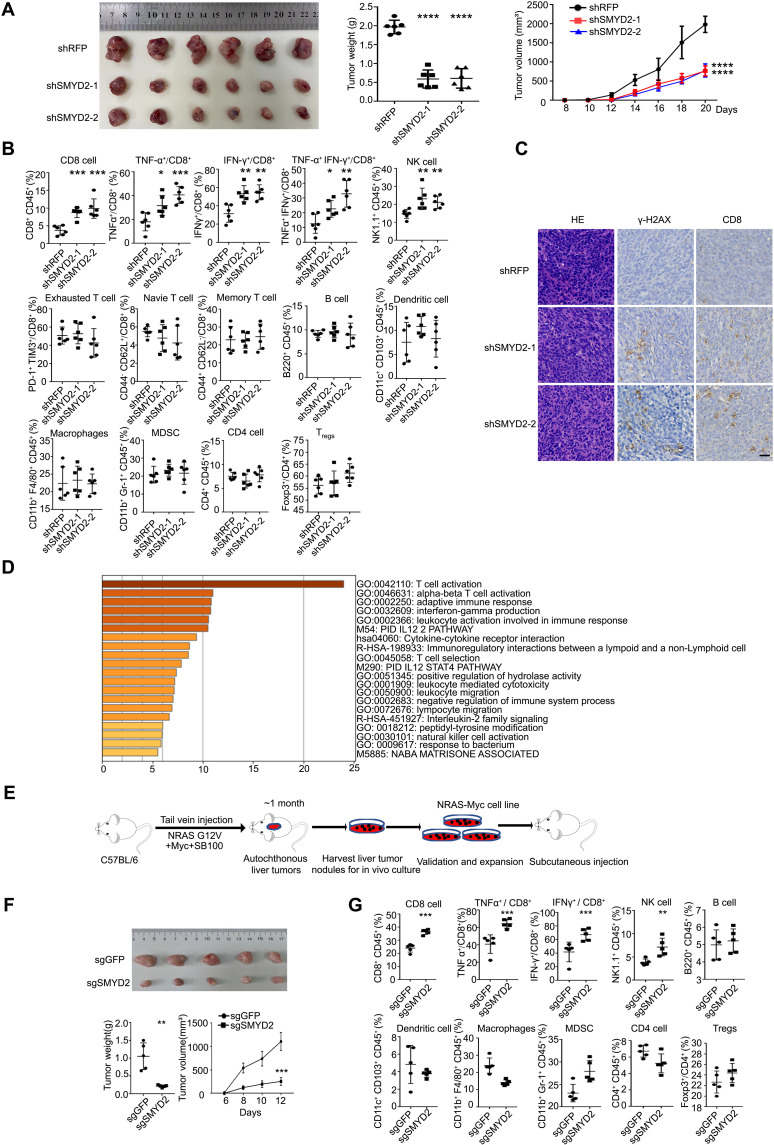
The cGAS-STING pathway has been established as a critical activator of the antitumor immune response. We first compared the in vivo tumor growth of CT26 cells with or without SMYD2 and found that tumor weight and volume were significantly decreased upon SMYD2 depletion (Fig. 5A). Next, to assess the effects of SMYD2 deficiency on the tumor microenvironment, we examined multiple immune cell lineage markers from CT26 shRFP or shSMYD2 tumors by flow cytometry analysis. We observed the robustly enhanced infiltration of CD8+ T cells and natural killer (NK) cells in SMYD2-deficient tumors compared to control. In addition to the increased overall percentage of CD8+ T cells, IFN-γ–expressing CD8+ T cells and tumor necrosis factor α (TNF-α)–expressing CD8+ T cells were also increased upon SMYD2 knockdown (Fig. 5B), indicating the elevated level of T cell activity. Through immunohistochemistry (IHC), we also found the increased DNA damage as stained by γ-H2AX and infiltrated CD8+ T cells (Fig. 5C). In addition, through pathway analysis (http://metascape.org/) with the SMYD2-correlated down-regulated genes in patient samples from the TCGA liver cancer database, we found that SMYD2 is also closely associated with T cell activation (Fig. 5D). To confirm this correlation, we applied a transposon system with the hydrodynamic injection of NRAS-G12V and Myc in combination with CMV-SB100 into 6-week-old C57BL/6 mice that developed autochthonous liver tumors in approximately 1 month (28, 29), as shown in Fig. 5E. We then derived mouse liver cancer cells from the bulk tumor for a secondary subcutaneous injection. Knockout of SMYD2 in the NRAS-Myc–based cell line markedly decreased tumor weight and tumor volume in C57BL/6 immunocompetent mice (Fig. 5F) without any change in immune-deficient nude mice (fig. S5A). These data indicated that immune cells play a critical role in SMYD2-dependent tumor growth. In addition, the enhanced infiltration of CD8+ T cells, NK cells, and IFN-γ– and TNF-α–expressing CD8+ T cells was also detected in the group of SMYD2 deficient liver tumors compared to control tumors (Fig. 5G). Consistent with the role of SMYD2 in tumor growth, we also performed the in vivo experiments using Ku70-WT or 3KR mutant stable cells. As shown in fig. S5B, the tumor weight and volume were remarkably decreased in Ku70-3KR cell–injected mice as compared with that of Ku70-WT. In addition, by the IHC assay, a higher percentage of DNA damage and infiltrated CD8+ T cells were accumulated in Ku70-3KR–bearing tissues (fig. S5C), suggesting that SMYD2 affects tumor growth through mediating Ku70 methylation and NHEJ repair.
Fig. 5. Inhibition of SMYD2 elicits an antitumor adaptive immune response.
(A) Tumor weight and tumor volume of CT26 tumors with or without SMYD2 in BALB/c mice. (B) Flow cytometry analysis of immune cells in CT26 tumors at day 13 with or without SMYD2, shown by percent of parent gates. (C) Representative images of hematoxylin and eosin (H&E) and IHC staining of γ-H2AX and CD8 in tissues from CT26 tumors at day 13 with or without SMYD2. Scale bars, 30 μm. (D) The pathway analysis based on the SMYD2 expression correlated down-regulated genes from cBioPortal of liver hepatocellular carcinoma (LIHC) in TCGA database. (E) Scheme for the establishment of mouse liver cancer cell line NRAS-Myc. (F) Tumor weight and tumor volume of NRAS-Myc tumors with or without SMYD2 in C57BL/6 mice. (G) Flow cytometry analysis of immune cells in NRAS-Myc tumors with or without SMYD2, shown by percent of parent gates.
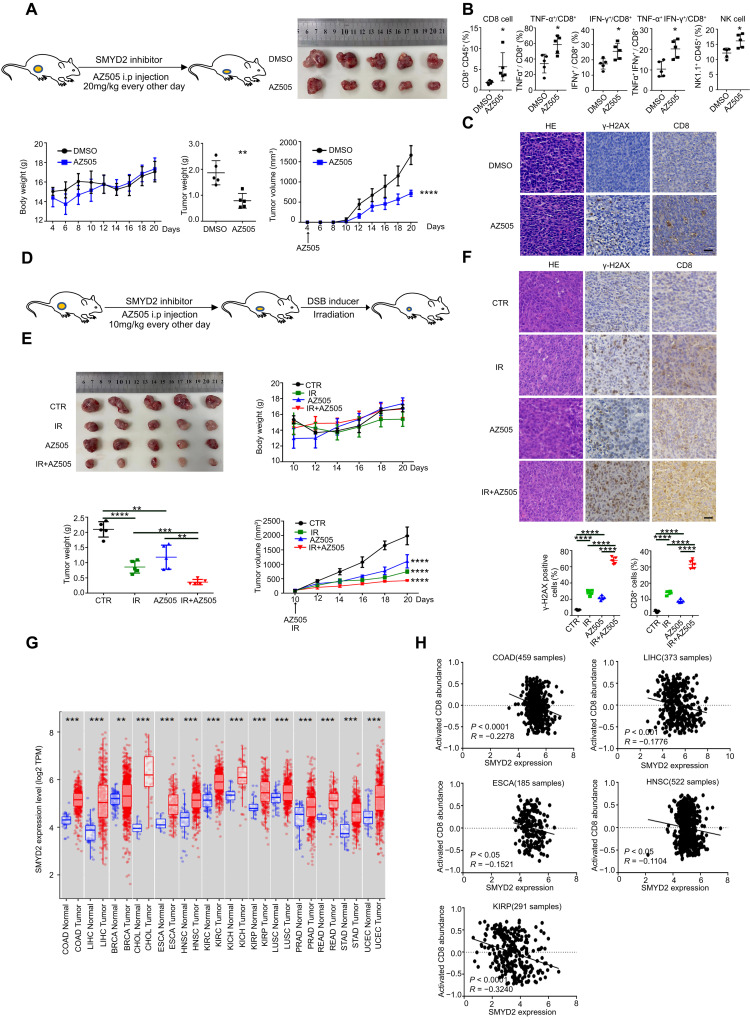
We next sought to investigate whether pharmacological inhibition of SMYD2 has translational significance in vivo. To this end, we used the SMYD2 inhibitor, AZ505, in tumor-bearing mice. As shown in Fig. 6A, AZ505 treatment resulted in decreased tumor weight and tumor volume of CT26 cells in BALB/c mice with no decrease in body weight. This demonstrated that the SMYD2 inhibitor has no overt toxicity but high efficiency. In vivo flow data showed the increased tumor infiltration of CD8+ T cells, NK cells, and IFN-γ– and TNF-α–expressing CD8+ T cells (Fig. 6B) in AZ505-treated mice, with no change in the spleen (fig. S6A). Consistently, the IHC assay showed more DNA damage accumulation and infiltration of CD8+ T cells in AZ505-treated tumors compared with the control group (Fig. 6C). The above data demonstrate that inhibition of SMYD2 results in damage accumulation and decreased efficiency of repair, which eventually led to the antitumor immune response. We next tried to examine the effect of combining SMYD2 inhibition with irradiation (Fig. 6D); AZ505 and irradiation were initiated 10 days after tumor implantation, with no effect on body weight (Fig. 6E). After tumors were harvested, we found that tumor weight and volume were significantly decreased in the combination group as compared with AZ505 alone or irradiation alone. Moreover, consistent with the above results, the IHC and quantified data also showed a higher percentage of DNA damage accumulation and infiltration of CD8+ T cells in tumors treated with AZ505 and irradiation combination (Fig. 6F). Last, we assessed whether this therapeutic strategy might be applicable in a broad range of cancers. By analyzing SMYD2 expression profiles of multiple cancers in the TCGA database, as shown in Fig. 6G, elevated SMYD2 was observed in tumor tissues compared with adjacent normal tissues in most cancers, such as colon adenocarcinoma (COAD), liver hepatocellular carcinoma (LIHC), breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC). The specificity of SMYD2 expression in tumors would have fewer off-target effects. We also found a negative correlation between SMYD2 expression and activated CD8 abundance in COAD, LIHC, ESCA, HNSC, and KIRP cancers based on the TISIDB database (Fig. 6H). The clinical relevance suggested that SMYD2 expression is correlated with tumor development and promoting immune surveillance. In conclusion, SMYD2 inhibition provokes a robust antitumor immune program in vivo, which is further enhanced by combination with radiotherapy. Targeting SMYD2 could be applied to developing combined antitumor therapies for several major cancer types.
Fig. 6. SMYD2 inhibition provokes a robust antitumor immune program and further enhanced by combination with radiotherapy.
(A) Scheme for the BALB/c mice bearing CT26 cells treated with SMYD2 inhibitor AZ505 or vehicle. IP injection every other day at a dose of 20 mg/kg from day 4. Body weight, tumor weight and tumor volume of CT26 tumors treated with or without AZ505. (B) Flow cytometry analysis of immune cells in CT26 tumors collected at day 13 with or without AZ505 from day 5, shown by percent of parent gates. (C) Representative images of H&E and IHC staining of γ-H2AX and CD8 in CT26 tumor tissues collected at day 13 with or without AZ505 from day 5. Scale bars, 30 μm. (D) Scheme for the BALB/c mice bearing CT26 cells treated with AZ505 and irradiation or AZ505 alone or irradiation alone or vehicle. IP injection of AZ505 every other day at a dose of 10 mg/kg from day 10 and local irradiation at a dose of 6 Gy once at day 10. (E) Body weight, tumor weight, and tumor volume of CT26 tumors from four groups above. (F) Representative images of H&E and IHC staining of γ-H2AX and CD8 in tissues from four groups above. IP injection every other day at a dose of 10 mg/kg and local irradiation at a dose of 6 Gy once at day 7. The tumor tissues were collected at day 13. Lower graphs of statistics for the percentage of positive cells within the tumors. Scale bars, 30 μm. (G) The expression of SMYD2 in tumors and adjacent normal tissues for indicated cancers from TCGA database of TIMER website. (H) Correlation of SMYD2 expression and the activated CD8 abundance for indicated cancers. The immune-related activated CD8 signatures were from the study of Charoentong et al. (55) and TISIDB website.
DISCUSSION
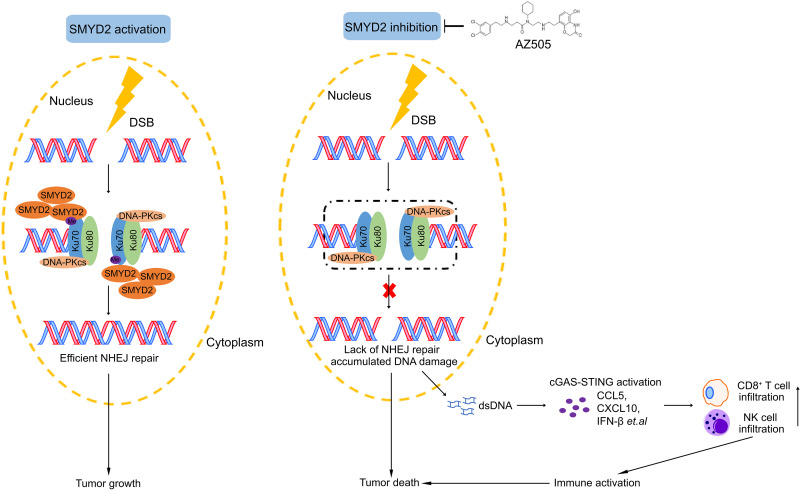
In this study, we reveal an important role of SMYD2 in mediating the methylation of Ku70, which is required for NHEJ repair and immune response. Our reports support that SMYD2 inhibition leads to hypomethylation of Ku70 and therefore influences the inefficient recruitment of the Ku70/Ku80/DNA-PKcs complex onto DSB sites, resulting in the deficiency of NHEJ repair. The compromised DNA repair caused cytoplasmic DNA accumulation and sequentially induced cGAS-STING–dependent innate immune response (Fig. 7). Moreover, our study highlights the possibility that the SMYD2 inhibitor AZ505 sensitizes tumors to irradiation and can serve as another combination strategy for cancer.
Fig. 7. Schematic model of SMYD2 inhibition–mediated hypomethylation of Ku70 contributes to impaired NHEJ repair and antitumor immunity.
In response to DNA damage, SMYD2 is recruited to DNA damage sites and sequentially mediates Ku70 methylation at lysine-74, lysine-516, and lysine-539, which promotes the efficient recruitment of Ku70/Ku80/DNA-PKcs complex and efficient NHEJ repair. Genetic or pharmacological inhibition of SMYD2 lead to the lack of NHEJ repair and accumulation of DNA damage, which leads to the increased cytoplasmic dsDNA, and activates the cGAS-STING pathway, boosting the production of chemokines and then triggers the antitumor immunity via infiltration and activation of cytotoxic CD8+ T cells and NK cells. SMYD2 inhibitor AZ505-enhanced radiation therapy efficiency indicates its translational significance.
As a SET domain–containing methyltransferase, SMYD2 controls a wide range of cellular functions through methylation of its substrates (10, 12, 30–32). For example, SMYD2 is firstly identified as a methyltransferase that can dimethylate H3K36 as a transcriptional repressor (10). SMYD2 also methylates p53 at lysine-370, providing regulatory cross-talk between PTM and tumor-suppressive function (12). In addition, SMYD2 potentiates the maintenance and function of skeletal muscle by controlling the cytoplasmic lysine methylation of heat shock protein 90 (Hsp90) (30). Here, we show that Ku70 is a bona fide partner of SMYD2. Although previous study has reported that the phosphor-Ku70 upon DNA damage interacts with RNA polymerase II and promotes the formation of phosphor-53BP1 foci for classical NHEJ (33). Our study identifies a previously unidentified PTM of Ku70 and expands our understanding of the regulatory network of classical NHEJ. Specifically, SMYD2 mediates Ku70 methylation at lysine-74, lysine-516, and lysine-539 and then influences the recruitment of Ku70/Ku80/DNA-PKcs complex, participating in NHEJ repair. Previous work in Chinese hamster ovary (CHO) and human cells has shown that knockout of the canonical NHEJ factors (Ku, DNA-PKcs, LIG4, etc.) results in complete loss (>95%) of NHEJ in GFP reporter assays (34). In our study, during knockdown of SMYD2, we observed a 50% decrease of NHEJ efficiency. We consider that Ku70/80 was still weakly recruited to the DSBs at even a later stage in SMYD2 knockdown cells. In addition, knockdown of SMYD2 is not completely 100%, and thus, part of Ku70 is methylated and recruited to DSBs. We also observed a 50% decrease of NHEJ efficiency that may be due to the existence of endogenous Ku70 when overexpression of Ku70-WT/3KR, which may also lead to an attenuated effect of Ku70-3KR on decreasing NHEJ repair efficiency. Our data provide the first evidence that the SMYD2-dependent NHEJ pathway regulates genome instability. We also deeply investigated the function of SMYD2 in tumor development and radiation therapy. However, further explorations into the interplay between phosphorylation and methylation on Ku70 are needed.
Recently, the interaction between DNA damage and the immune system has drawn increased attention. For example, targeting DNA damage response through checkpoint kinase 1 (CHK1) and poly(ADP-ribose) polymerase (PARP) inhibition results in the activation of tumor-infiltrating T lymphocytes via STING/TBK1/IRF3 innate immune response pathway in small cell lung cancer or BRCA-associated breast cancer (24, 35, 36). Ataxia telangiectasia mutated (ATM) inhibition can induce an IFN-mediated innate immune response, which leads to more efficient anti–programmed cell death-ligand 1 (PD-L1) therapy (37). In our study, we demonstrate that SMYD2 is an important pharmacological target for the modulation of innate immune response. The deficiency of SMYD2 leads to the activation of the STING/TBK1/IRF3 pathway, which relied on increased cytosolic DNA. On one hand, the activation of the STING pathway can play a critical role in promoting inflammation-induced tumorigenesis (38); on the other hand, its activation can create a favorable tumor microenvironment accompanied with activated CD4+ or CD8+ cells (35, 39, 40). Our findings are consistent with the model that deficiency of a DNA repair pathway can stimulate STING-mediated innate immune response, IFN signaling, and antitumor activities. As reported before, cytoplasmic-translocated Ku70 senses intracellular DNA and promotes IFN-λ induction (41). Our observations here reveal the chromatin binding ability of Ku70, but whether the cytosolic translocation of Ku70 serves as another noncanonical mechanism for the activation of the cGAS-STING pathway needs further investigations.
Targeting tumor DNA repair deficiency has been proposed as a reasonable strategy in cancer for more than a century, as demonstrated by DNA damage–related chemotherapeutic agents and irradiation. The accumulated damage or improper repair can lead to changes both in the tumor genome and the tumor microenvironment (3). However, this synthetic lethality-based strategy is narrowly applied in subtypes of cancer that are driven by unique genetic alterations such as BRAC1/2, ATM/ATR, etc. There is currently an urgent need for other anti-DDR compounds that can be widely applied. Preclinical data from tumor-associated epigenetic enzymes have already shown promising results, especially inhibitors of histone methylases (42). There are currently several SMYD2 inhibitors such as AZ505, LLY-507, and BAY-598 (43–46), which were discovered and characterized for use as therapeutics. On the basis of our study, we provide evidence of the efficacy of SMYD2 inhibitors in combination with radiation as a strategy for the treatment of liver and colon cancer. Notably, our results showed that SMYD2 inhibition together with radiation substantially increased CD8+ T cell presence in tumor tissues, suggesting a potential implication for improving tumor immunity. However, more experiments based on immune-deficient mice and CD8+ T cell deletion are warranted in the future to clarify the involvement of T cell–mediated control of tumor growth in the combination therapy. By analyzing the correlation between SMYD2 and activated CD8 abundance, we also broaden the same rationale of targeting SMYD2 to other types of cancer. With the connection between targeting DNA damage–associated proteins with the immune response, we also provide the evidence that there could be a therapeutic advantage in combining chemoradiotherapy and immunotherapy. DNA damage, STING activation, and production of type I IFN have also been linked with PD-L1 expression, as has adaptive resistance to IFN-γ produced by infiltrating T cells (47, 48). Previous studies have demonstrated that targeting DNA damage response proteins, such as PARP1 and CHK1, substantially increase PD-L1 expression and infiltration of cytotoxic T cells and potentiate the antitumor effect of PD-L1 blockade through the STING/TBK1/IRF3 pathway (35). In addition, the study that tumors defective in Ku70/80 complex exhibit up-regulation of PD-L1 after DSBs (49) implies that SMYD2 deficiency, which impairs Ku70-mediated repair, may also result in increased PD-L1 expression. We speculate that type I IFN induced by STING activation with these treatments may contribute to PD-L1 up-regulation. These data suggest that targeting SMYD2 may be a potential strategy to improve the efficacy of immune checkpoint blockade therapy.
In summary, we have shown that SMYD2 promotes DDR and functions through directly methylating Ku70 and affects the NHEJ repair pathway. Because of its effects on DNA repair and immune response, SMYD2 is a promising target for combined therapy. Inhibition of SMYD2 caused cytosolic DNA accumulation and activation of the cGAS-STING pathway, recruiting activated cytotoxic CD8+ T cells to block tumor development. Given that SMYD2 is overexpressed in different types of cancer, SMYD2 represents as a promising target for cancer therapy.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney (HEK) 293FT, U2OS, HCT116, and CT26 cells were obtained from the American Type Culture Collection (ATCC; USA). These cell lines were cultured in Dulbecco’s modified Eagle’s medium (Shanghai BasalMedia Technologies, China) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Israel) and 1% penicillin-streptomycin (Shanghai BasalMedia Technologies, China), 1% nonessential amino acid (HyClone, catalog no. SH3023801) according to ATCC guidelines and maintained in a 37°C incubator with a humidified, 5% CO2 atmosphere. Inhibitor AZ505 was purchased from MCE (HY-15226, MedChemExpress, USA).
Plasmids and shRNA
All plasmids were transfected with polyethylenimine (Polysciences, USA) according to the manufacturer’s instructions. The packaging plasmids pMD2.G (Addgene plasmid, 12259), psPAX2 (Addgene plasmid, 12260), and HEK293FT were used for producing lentivirus. The lentivirus was generated by transfecting HEK293FT cells with packaging plasmids pMD2.G and psPAX2 and targeting shRNAs or plasmids with polyethylenimine. The medium containing lentivirus was harvested with a 0.45-μm filter at 72 hours after transfection. SMYD2, Ku70, and Ku80 cDNAs were amplified, at full length, and various fragments were cloned into pLVX-Puro, pGEX4T1, pET-28b, or pEGFP-C2 vectors (Addgene, USA). Site-specific mutations were generated using a site-directed mutagenesis kit (Vazyme, China). The shRNA sequences targeted human SMYD2 (sequence 1, CGGCAAAGATCATCCATATAT; sequence 2, ACTTAGTTCAGAAACCTTAAA). The shRNA sequences targeted mouse SMYD2 (sequence 1, CCATTTGGGATCGGCGATATT; sequence 2, CCGGCTAAGAGACTCCTATTT).
SDS–polyacrylamide gel electrophoresis and Western blotting
Western blotting was used to evaluate the levels of protein as previously described with minor modifications. Briefly, equal amounts of protein were sized-fractionated on 6 to 15% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels. The antibodies used were SMYD2 (A6474, ABclonal), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 10494-1-AP, Proteintech), tubulin (11224-1-AP, Proteintech), H3 (17168-1-AP, Proteintech), γ-H2AX (#2577S, Cell Signaling Technology), H2AX (#7631, Cell Signaling Technology), DNA-PKcs (28534-1-AP, Proteintech), Ku70 (A0883, ABclonal), Ku80 (A5862, ABclonal), LIG4 (A1743, ABclonal), XLF (A4985, ABclonal), XRCC4 (A1677, ABclonal), mono/dimethylation (ab23366, Abcam), trimethylation (PTM-601, PTM BIO), FLAG (AE063, AE005, ABclonal), GFP (66002-1-Ig, Proteintech), p-TBK1-Ser172 (#5483S, Cell Signaling Technology), TBK1 (28397-1-AP, Proteintech), p-IRF3-Ser396 (#29047, Cell Signaling Technology), IRF3 (11312-1-AP, Proteintech), p-STING-Ser365 (#72971S, Cell Signaling Technology), p-STING-Ser366 (50907, Cell Signaling Technology), and STING (19851-1-AP, Proteintech). The fluorochrome-conjugated antibodies were used for fluorescence-activated cell sorting as follows: CD45-PerCP-Cy5.5 (45-0451-82, eBioscience), Gr-1-APC (17-5931-81, eBioscience), CD11b-FITC (11-0112-81, eBioscience), CD11c (12-0114-81, eBioscience), NK1.1-APC (17-5941-63, eBioscience), CD8-FITC (11-0081-81, eBioscience), PD-1–PE (12-9985-81, eBioscience), CD44-PE (12-0441-81, eBioscience), CD62L-APC (17-0621-81, eBioscience), Foxp3-PE (12-5773-80, eBioscience), IFN-γ–PE (12-7331-81, eBioscience), CD16/CD32 (14-0161-82, eBioscience), F4/80-PE (123109, BioLegend), B220-PE-Cy7 (103221, BioLegend), CD103-FITC (156911, BioLegend), Tim-3–APC (134006, BioLegend), TNF-α–APC (506307, BioLegend), and CD4-PE-Cy7 (100421, BioLegend).
Chromatin fractionation
Cells were harvested and washed by phosphate-buffered saline (PBS) at 10,000 rpm at 4°C for 30 s. The cell pellet was resuspended in 500 μl of buffer I [50 mM Hepes (pH 7.5), 1 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, and 1% protease inhibitor cocktail] on ice for 3 min and then centrifuged at 13,000 rpm for 3 min, and the insoluble pellet was washed twice in buffer I without 0.1% Triton X-100 at 4°C for 3 min. The remaining pellet was resuspended in 1× SDS loading buffer and boiled 10 min for Western blotting.
Co-IP assay
Cells were harvested and washed by PBS at 10,000 rpm at 4°C for 30 s. The cell pellet was resuspended in 1 ml of lysis buffer [137 mM NaCl, 20 mM tris-HCl (pH 8.0), 10% glycerol, 1% NP-40, 2 mM EDTA, and 1% protease inhibitor cocktail] on ice for 30 min and sonicated using a microtip at 35% amplitude for 1 s 10 times. The sonicated samples were then centrifuged at 12,000 rpm at 4°C for 15 min. Antibodies (1 to 2 μg) and protein A/G agarose beads were added to the supernatant lysate and incubated at 4°C overnight with a rotary shaker. For Benzonase treatment, 1-U Benzonase (M046-01A, Novoprotein) was added in the lysates. The immunoprecipitants were washed with wash buffer [20 mM tris-HCl (pH 8.0), 0.1% NP-40, 100 mM NaCl, and 1 mM EDTA] three times and eluted by boiling for 5 min with loading buffer. After centrifugation, the supernatant was subjected to Western blotting.
GST pull-down
GST and GST-fusion/His fusion proteins were constructed and induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside overnight at 16°C in Escherichia coli and purified using glutathione–Sepharose 4B beads (GE Healthcare, NY, USA). Equal proteins were incubated with GST fusion proteins in TEN buffer [10 mM tris-HCl (pH 8.0), 1 mM EDTA, and 100 mM NaCl] at 4°C for 4 hours. After washing with TEN buffer three times and then precipitation, components were analyzed by Western blotting.
Immunofluorescence
Forty to 50% confluence of cells was seeded on glass-bottom dishes and incubated overnight in medium in humidified incubator at 37°C. The cells were transfected with the indicated plasmids or treated with chemicals according to the manufacturer’s protocol. The cells were then fixed with 4% paraformaldehyde at room temperature and permeabilized with methanol for 10 min at −20°C. The cells were then incubated with blocking buffer (0.8 mg of bovine serum albumin in 100 ml of PBS) and incubated with indicated primary antibodies (1:100 dilution) overnight at 4°C. After being washed three times with blocking buffer, the cells were incubated with secondary antibodies (1:100 dilution) conjugated to tetramethyl rhodamine isothiocyanate/FITC for 1 hour at room temperature in the dark and washed three times with blocking buffer and then embedded with 4′,6-diamidino-2-phenylindole. The immunofluorescence signal was observed under a fluorescence microscope (Leica, Germany).
Laser micro-irradiation–coupled live-cell imaging
Laser micro-irradiation was performed as previously described (50). Briefly, cells were grown on a glass-bottom dish and transfected with GFP-tagged plasmids. The cells were then treated with 10 μM 5-bromo-2′-deoxyuridine for 24 hours and irradiated with a pulsed nitrogen laser (365 nm, 16-Hz pulse, and 65% laser output) generated from a MicroPoint system (MicroPoint Ablation Laser System, Andor). The system was directly coupled to the Leica confocal image system, and time-lapse images were captured for the indicated time. The signal intensity of the irradiated path was calculated using ImageJ software.
Comet assay
Comet assay was performed as previously described (51). Briefly, cells were treated as indicated and mixed gently with premelted low temperature–melting agarose at a volume ratio of 1:1 (v/v) and spread on glass slides. The slides were then submerged in precooled lysis buffer at 4°C for 2 hours and electrophoresed in running buffer [1 mM Na2EDTA and 300 mM NaOH (pH 13.0)] at 1.0 V/cm for 20 min and then stained with propidium iodide (5 μg/ml). The immunofluorescence signal was observed under a fluorescence microscope.
Flow cytometry
For HR or NHEJ analysis, cells were harvested at 72 hours after transfection using an electroporator with DsRed and I-SceI and then resuspended in 200 μl of PBS for flow cytometry analysis. GFP-based repair efficiency was calculated as the GFP-positive cells over the DsRed-positive cells.
For tumor-infiltrated immune cell analysis, tumors were cut into small pieces, minced with scissors, and digested at 37°C in a rotor for 30 min in RPMI 1640 supplemented with 10% FBS, collagenase type I (0.05 mg/ml; C5894, Sigma-Aldrich), collagenase type IV (0.05 mg/ml; C1889, Sigma-Aldrich), deoxyribonuclease I (0.01 mg/ml; 11284932001, Roche), and hyaluronidase (0.025 mg/ml; H4272, Sigma-Aldrich). The digested samples were passed through a 70-μm mesh and then lysed in RBC lysis buffer (RT122-02, TIANGEN) for 3 min. For cytokine staining, cells were incubated with ionomycin (S1672, Beyotime Biotechnology), phorbol 12-myristate 13-acetate (5 ng/ml; P1585, Sigma-Aldrich), and brefeldin A (00-4506-51, eBioscience) at 37°C for 4 hours in a CO2 incubator. Cells were then stained with fixable viability dye eFluor450 (65-0863-14, eBioscience) for 30 min and blocked with CD16/CD32 antibody. Cell labeling was analyzed with fluorescently conjugated antibodies. For intracellular staining, commercial Fixation/Permeabilization regents (00-5123-43, 00-8333-56, and 00-5223-56, eBioscience) was used.
Colony formation assay
Equal numbers of cells were seeded in the medium containing 10% FBS and 1% penicillin-streptomycin in dishes with indicated treatments and continued to culture at 37°C and 5% CO2 for 2 weeks and stained with crystal violet.
Quantitative real-time polymerase chain reaction
Total RNA was extracted using TRIzol reagent, isolated RNA using chloroform, and precipitated the upper liquid using isopropanol at 12,000 rpm at 4°C and washed with 75% ethanol. The extracted RNA was transcribed to cDNA using an RT kit (Yeasen, Shanghai, China). The relative expression of target genes was measured by ABI 7500 (Life Technologies, NY, USA) with SYBR Green dye. The primers used for reverse transcription polymerase chain reaction were as follows: mouse Ccl5-forward-5′, ATATGGCTCGGACACCACTC; mouse Ccl5-reverse-3′, ACTTGGCGGTTCCTTCGAG; mouse Cxcl10-forward-5′, TCAAGCCATGGTCCTGAGAC; mouse Cxcl10-reverse-3′, CGCACCTCCACATAGCTTACA; mouse Ifnβ-forward-5′, GGTGGAATGAGACTATTGTTG; mouse Ifnβ-reverse-3′, AGGACATCTCCCACGTC; mouse Smyd2-forward-5′, GGTGGAAGTCCGAAAGCTCA; mouse Smyd2-reverse-3′, GCAGCTCACTAGGGGATTTGT; mouse Gapdh-forward-5′, AGGTCGGTGTGAACGGATTTG; mouse Gapdh-reverse-3′, TGTAGACCATGTAGTTGAGGTCA; human CCL5-forward-5′, ATGACTCCCGGCTGAACAAG; human CCL5-reverse-3′, CAGGTTCAAGGACTCTCCATCC; human CXCL10-forward-5′, AAGTGGCATTCAAGGAGTACCT; human CXCL10-reverse-3′, ACGTGGACAAAATTGGCTTGC; human IFNβ-forward-5′, GCCATCAGTCACTTAAACAGC; human IFNβ-reverse-3′, GAAACTGAAGATCTCCTAGCCT; human SMYD2-forward-5′, GCCGGGAGAGGAGGTTTTTA; human SMYD2-reverse-3′, GGTACACTCCTGGCACTCAC; human GAPDH-forward-5′, TCCTGTTCGACAGTCAGCCGCA; and human GAPDH-reverse-3′, ACCAGGCGCCCAATACGACCA.
Chemoradiation treatment
Cells or mice were irradiated with a biological x-ray irradiator RS2000pro (Rad Source Technologies, USA) with a radiation output of 160 kV and 25 mA at a dose rate of 4.125 grays/min.
Immunohistochemistry
Paraffin-embedded tumor tissue was first deparaffinized with xylene, followed by rehydration using gradient ethanol and immersion in 3% H2O2 at room temperature for 10 min. Antigens were retrieved in 0.01 M citric buffer (pH 6.0) at 97°C for 30 min. Slides were cooled down and blocked by blocking buffer (P0103, Beyotime Biotechnology) for 1 hour. Staining with diluted primary antibodies (1:100 dilution) was conducted at 4°C overnight, followed by secondary antibodies at room temperature for 1 hour. After Diaminobenzidine staining, the slides were counterstained with hematoxylin, dehydrated with ethanol, and mounted with coverslips.
Mass spectrometry analysis
Cell precipitates were subjected to IP assays, separated by SDS-PAGE, and then stained with Coomassie blue. The gel of the 100-kDa (the position of GFP-Ku70) band was excised, digested with trypsin, and analyzed with mass spectrometry analysis. Mass spectrometry referred to the previously described procedure (52, 53).
DNA pull-down assay
Primers (primer1: 5′Biotin-GCGATCATGACGTAGACGATAGCGTTCTTTTTTTTTCTTTTTCT-3′; primer2: 5′-CGCTATCGTCTACGTCATGATCGC-3′) for 3′ overhangs were synthetized for an annealing experiment. DNA binding reactions were done in reaction buffer [150 mM KCl, 5 mM MgCl2, 10 mM tris-HCl (pH7.5), 5% glycerol, 0.25% Tween-20, 3.5 mM dithiothreitol, and 0.1%BSA] with 10-pmol 3′ overhangs biotinylated oligo and purified Ku70-WT or Ku70-3KR and Ku80 for 30 min at 4°C. After 30 min, 10 μl of Dynabeads M-280 streptavidin (11206D, Thermo Fisher Scientific) in reaction buffer was added and incubated for 30 min at 4°C. After this incubation, the beads were washed with reaction buffer three times and then subjected to Western blotting. DNA pull-down assay was performed as previous described (54).
Animal experiments
Four- to six-week-old female BALB/c mice, C57BL/6 mice, and nude mice were purchased from Shanghai Jiesijie Laboratory Animal (Shanghai, China). CT26 or NRAS-Myc cells (3 × 105 or 5 × 105 cells in 100 μl of PBS per mouse) were injected into mice through subcutaneous injection. Tumor volume was calculated as follows: volume (mm3) = width2 × length/2.
Statistics
Statistical comparisons were carried out using the two-tailed Student’s t test. One-way or two-way analysis of variance (ANOVA) was used to analyze statistical differences between multiple-group comparisons. P < 0.05 was considered statistically significant (not significant, P > 0.05; **P < 0.01, ***P < 0.001, and ****P < 0.0001). At least three independent experiments were performed in all cases. GraphPad was used to analyze all data.
Study approval
The study was approved by the ethics committee of Shanghai First Maternity and Infant Hospital of Tongji University. Each animal study was conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by Tongji University School of Medicine Animal Care and Use Committee.
Acknowledgments
We thank J. Zhang from Shenzhen University for discussions.
Funding: This study was supported by the National Natural Science Foundation of China (grant numbers 32170602, 32090033, 81971338, and 82273475) and Shanghai Science and Technology Development Funds-Shanghai Rising-Star Program (grant numbers 22QA1407400).
Author contributions: M.T., W.L., and W.-G.Z. conceived, designed, and performed the experiments and wrote the manuscript. M.T., G.C., and B.T. analyzed the data and performed material preparation. B.T., Z.H., Y.H., C.C.D., X.W., Z.M., Y.L., W.L., and W-G.Z. discussed the results and commented on the manuscript. M.T. supervised the project.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs S1 to S6
REFERENCES AND NOTES
- 1.Tubbs A., Nussenzweig A., Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccia A., Elledge S. J., The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mouw K. W., Goldberg M. S., Konstantinopoulos P. A., D'Andrea A. D., DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 7, 675–693 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weterings E., Gallegos A. C., Dominick L. N., Cooke L. S., Bartels T. N., Vagner J., Matsunaga T. O., Mahadevan D., A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair 43, 98–106 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Zhao B., Rothenberg E., Ramsden D. A., Lieber M. R., The molecular basis and disease relevance of nonhomologous DNA end joining. Nat. Rev. Mol. Cell Biol. 21, 765–781 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodbine L., Gennery A. R., Jeggo P. A., The clinical impact of deficiency in DNA nonhomologous end-joining. DNA Repair 16, 84–96 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Teoh N. C., Dan Y. Y., Swisshelm K., Lehman S., Wright J. H., Haque J., Gu Y., Fausto N., Defective DNA strand break repair causes chromosomal instability and accelerates liver carcinogenesis in mice. Hepatology 47, 2078–2088 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Kloeber J. A., Lou Z., Critical DNA damaging pathways in tumorigenesis. Semin. Cancer Biol. 85, 164–184 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamamoto R., Saloura V., Nakamura Y., Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 15, 110–124 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Brown M. A., Sims R. J. III, Gottlieb P. D., Tucker P. W., Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 5, 26 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abu-Farha M., Lambert J. P., al-Madhoun A. S., Elisma F., Skerjanc I. S., Figeys D., The tale of two domains. Mol. Cell Proteomics 7, 560–572 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Kubicek S., Opravil S., Jenuwein T., Berger S. L., Repression of p53 activity by Smyd2-mediated methylation. Nature 444, 629–632 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Nakakido M., Deng Z., Suzuki T., Dohmae N., Nakamura Y., Hamamoto R., Dysregulation of AKT pathway by SMYD2-mediated lysine methylation on PTEN. Neoplasia 17, 367–373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L. X., Fan L. X., Zhou J. X., Grantham J. J., Calvet J. P., Sage J., Li X., Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J. Clin. Invest. 127, 2751–2764 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynoird N., Mazur P. K., Stellfeld T., Flores N. M., Lofgren S. M., Carlson S. M., Brambilla E., Hainaut P., Kaznowska E. B., Arrowsmith C. H., Khatri P., Stresemann C., Gozani O., Sage J., Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreatic cancer. Genes Dev. 30, 772–785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L., Ding B., Liu H., Zhang Y., Zeng J., Hu J., Yao W., Yu G., An R., Chen Z., Ye Z., Xing J., Xiao K., Wu L., Xu H., Inhibition of SMYD2 suppresses tumor progression by down-regulating microRNA-125b and attenuates multi-drug resistance in renal cell carcinoma. Theranostics 9, 8377–8391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L. X., Zhou J. X., Calvet J. P., Godwin A. K., Jensen R. A., Li X., Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 9, 326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y., Qiu R., Yang Y., Gao T., Zheng Y., Huang W., Gao J., Zhang K., Liu R., Wang S., Hou Y., Yu W., Leng S., Feng D., Liu W., Zhang X., Wang Y., Regulation of EZH2 by SMYD2-mediated lysine methylation is implicated in tumorigenesis. Cell Rep 29, 1482–1498.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Hopfner K. P., Hornung V., Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Zhang W., Liu W., Jia L., Chen D., Chang I., Lake M., Bentolila L. A., Wang C. Y., Targeting KDM4A epigenetically activates tumor-cell-intrinsic immunity by inducing DNA replication stress. Mol. Cell 81, 2148–2165.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marzio A., Kurz E., Sahni J. M., di Feo G., Puccini J., Jiang S., Hirsch C. A., Arbini A. A., Wu W. L., Pass H. I., Bar-Sagi D., Papagiannakopoulos T., Pagano M., EMSY inhibits homologous recombination repair and the interferon response, promoting lung cancer immune evasion. Cell 185, 169–183.e19 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao Z., Bozzella M., Seluanov A., Gorbunova V., Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 7, 1765–1771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan J., Lu C., Jin Q., Lu H., Chen X., Tian L., Zhang Y., Ortega J., Zhang J., Siteni S., Chen M., Gu L., Shay J. W., Davis A. J., Chen Z. J., Fu Y. X., Li G. M., MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS-STING pathway. Cancer Cell 39, 109–121.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen J., Zhao W., Ju Z., Wang L., Peng Y., Labrie M., Yap T. A., Mills G. B., Peng G., PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res 79, 311–319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T., Chen Z. J., The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon J., Bakhoum S. F., The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 10, 26–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ru B., Wong C. N., Tong Y., Zhong J. Y., Zhong S. S. W., Wu W. C., Chu K. C., Wong C. Y., Lau C. Y., Chen I., Chan N. W., Zhang J., TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 35, 4200–4202 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Calvisi D. F., Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 184, 912–923 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen L., Xin B., Wu P., Lin C. H., Peng C., Wang G., Lee J., Lu L. F., Feng G. S., An efficient combination immunotherapy for primary liver cancer by harmonized activation of innate and adaptive immunity in mice. Hepatology 69, 2518–2532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donlin L. T., Andresen C., Just S., Rudensky E., Pappas C. T., Kruger M., Jacobs E. Y., Unger A., Zieseniss A., Dobenecker M. W., Voelkel T., Chait B. T., Gregorio C. C., Rottbauer W., Tarakhovsky A., Linke W. A., Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 26, 114–119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saddic L. A., West L. E., Aslanian A., Yates J. R. III, Rubin S. M., Gozani O., Sage J., Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 285, 37733–37740 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L. X., Zhou J. X., Wang X., Zhang H., Harris P. C., Calvet J. P., Li X., Cross-talk between CDK4/6 and SMYD2 regulates gene transcription, tubulin methylation, and ciliogenesis. Sci. Adv. 6, eabb3154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schellenbauer A., Guilly M. N., Grall R., le Bars R., Paget V., Kortulewski T., Sutcu H., Mathé C., Hullo M., Biard D., Leteurtre F., Barroca V., Corre Y., Irbah L., Rass E., Theze B., Bertrand P., Demmers J. A. A., Guirouilh-Barbat J., Lopez B. S., Chevillard S., Delic J., Phospho-Ku70 induced by DNA damage interacts with RNA Pol II and promotes the formation of phospho-53BP1 foci to ensure optimal cNHEJ. Nucleic Acids Res. 49, 11728–11745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhfittig-Kulle S., Feldmann E., Odersky A., Kuliczkowska A., Goedecke W., Eggert A., Pfeiffer P., The mutagenic potential of nonhomologous end joining in the absence of the NHEJ core factors Ku70/80, DNA-PKcs and XRCC4-LigIV. Mutagenesis 22, 217–233 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Sen T., Rodriguez B. L., Chen L., Corte C. M. D., Morikawa N., Fujimoto J., Cristea S., Nguyen T., Diao L., Li L., Fan Y., Yang Y., Wang J., Glisson B. S., Wistuba I. I., Sage J., Heymach J. V., Gibbons D. L., Byers L. A., Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 9, 646–661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantelidou C., Sonzogni O., de Oliveria Taveira M., Mehta A. K., Kothari A., Wang D., Visal T., Li M. K., Pinto J., Castrillon J. A., Cheney E. M., Bouwman P., Jonkers J., Rottenberg S., Guerriero J. L., Wulf G. M., Shapiro G. I., PARP inhibitor efficacy depends on CD8(+) T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q., Green M. D., Lang X., Lazarus J., Parsels J. D., Wei S., Parsels L. A., Shi J., Ramnath N., Wahl D. R., Pasca di Magliano M., Frankel T. L., Kryczek I., Lei Y. L., Lawrence T. S., Zou W., Morgan M. A., Inhibition of ATM increases interferon signaling and sensitizes pancreatic cancer to immune checkpoint blockade therapy. Cancer Res. 79, 3940–3951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn J., Xia T., Konno H., Konno K., Ruiz P., Barber G. N., Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkes E. E., Walker S. M., Taggart L. E., McCabe N., Knight L. A., Wilkinson R., McCloskey K. D., Buckley N. E., Savage K. I., Salto-Tellez M., McQuaid S., Harte M. T., Mullan P. B., Harkin D. P., Kennedy R. D., Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J. Natl. Cancer Inst. 109, djw199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei B., Xu L., Guo W., Wang Y., Wu J., Li X., Cai X., Hu J., Wang M., Xu Q., Liu W., Gu Y., SHP2-mediated inhibition of DNA repair contributes to cGAS-STING activation and chemotherapeutic sensitivity in colon cancer. Cancer research 81, 3215–3228 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Sui H., Chen Q., Imamichi T., Cytoplasmic-translocated Ku70 senses intracellular DNA and mediates interferon-lambda1 induction. Immunology 163, 323–337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner T., Jung M., New lysine methyltransferase drug targets in cancer. Nat. Biotechnol. 30, 622–623 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Ferguson A. D., Larsen N. A., Howard T., Pollard H., Green I., Grande C., Cheung T., Garcia-Arenas R., Cowen S., Wu J., Godin R., Chen H., Keen N., Structural basis of substrate methylation and inhibition of SMYD2. Structure 19, 1262–1273 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Nguyen H., Allali-Hassani A., Antonysamy S., Chang S., Chen L. H., Curtis C., Emtage S., Fan L., Gheyi T., Li F., Liu S., Martin J. R., Mendel D., Olsen J. B., Pelletier L., Shatseva T., Wu S., Zhang F. F., Arrowsmith C. H., Brown P. J., Campbell R. M., Garcia B. A., Barsyte-Lovejoy D., Mader M., Vedadi M., LLY-507, a cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J. Biol. Chem. 290, 13641–13653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eggert E., Hillig R. C., Koehr S., Stöckigt D., Weiske J., Barak N., Mowat J., Brumby T., Christ C. D., ter Laak A., Lang T., Fernandez-Montalvan A. E., Badock V., Weinmann H., Hartung I. V., Barsyte-Lovejoy D., Szewczyk M., Kennedy S., Li F., Vedadi M., Brown P. J., Santhakumar V., Arrowsmith C. H., Stellfeld T., Stresemann C., Discovery and characterization of a highly potent and selective aminopyrazoline-based in vivo probe (BAY-598) for the protein lysine methyltransferase SMYD2. J. Med. Chem. 59, 4578–4600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sweis R. F., Wang Z., Algire M., Arrowsmith C. H., Brown P. J., Chiang G. G., Guo J., Jakob C. G., Kennedy S., Li F., Maag D., Shaw B., Soni N. B., Vedadi M., Pappano W. N., Discovery of A-893, a new cell-active benzoxazinone inhibitor of lysine methyltransferase SMYD2. ACS Med. Chem. Lett. 6, 695–700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minn A. J., Interferons and the immunogenic effects of cancer therapy. Trends Immunol. 36, 725–737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kciuk M., Kołat D., Kałuzińska-Kołat Ż., Gawrysiak M., Drozda R., Celik I., Kontek R., PD-1/PD-L1 and DNA damage response in cancer. Cells 12, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato H., Niimi A., Yasuhara T., Permata T. B. M., Hagiwara Y., Isono M., Nuryadi E., Sekine R., Oike T., Kakoti S., Yoshimoto Y., Held K. D., Suzuki Y., Kono K., Miyagawa K., Nakano T., Shibata A., DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 8, 1751 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu X., Tang M., Zhu Q., Yang Q., Li Z., Bao Y., Liu G., Hou T., Lv Y., Zhao Y., Wang H., Yang Y., Cheng Z., Wen H., Liu B., Xu X., Gu L., Zhu W. G., GLP-catalyzed H4K16me1 promotes 53BP1 recruitment to permit DNA damage repair and cell survival. Nucleic Acids Res 47, 10977–10993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang M., Li Z., Zhang C., Lu X., Tu B., Cao Z., Li Y., Chen Y., Jiang L., Wang H., Wang L., Wang J., Liu B., Xu X., Wang H., Zhu W. G., SIRT7-mediated ATM deacetylation is essential for its deactivation and DNA damage repair. Sci. Adv. 5, eaav1118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang M., Lu X., Zhang C., du C., Cao L., Hou T., Li Z., Tu B., Cao Z., Li Y., Chen Y., Jiang L., Wang H., Wang L., Liu B., Xu X., Luo J., Wang J., Gu J., Wang H., Zhu W. G., Downregulation of SIRT7 by 5-fluorouracil induces radiosensitivity in human colorectal cancer. Theranostics 7, 1346–1359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma A., Tang M., Zhang L., Wang B., Yang Z., Liu Y., Xu G., Wu L., Jing T., Xu X., Yang S., Liu Y., USP1 inhibition destabilizes KPNA2 and suppresses breast cancer metastasis. Oncogene 38, 2405–2419 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Singh J. K., Smith R., Rother M. B., de Groot A. J. L., Wiegant W. W., Vreeken K., D’Augustin O., Kim R. Q., Qian H., Krawczyk P. M., González-Prieto R., Vertegaal A. C. O., Lamers M., Huet S., van Attikum H., Zinc finger protein ZNF384 is an adaptor of Ku to DNA during classical nonhomologous end-joining. Nat. Commun. 12, 6560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z., Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18, 248–262 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs S1 to S6